Abstract
The plastid transformation approach offers a number of unique advantages, including high-level transgene expression, multi-gene engineering, transgene containment, and a lack of gene silencing and position effects. The extension of plastid transformation technology to monocotyledonous cereal crops, including rice, bears great promise for the improvement of agronomic traits, and the efficient production of pharmaceutical or nutritional enhancement. Here, we report a promising step towards stable plastid transformation in rice. We produced fertile transplastomic rice plants and demonstrated transmission of the plastid-expressed green fluorescent protein (GFP) and aminoglycoside 3′-adenylyltransferase genes to the progeny of these plants. Transgenic chloroplasts were determined to have stably expressed the GFP, which was confirmed by both confocal microscopy and Western blot analyses. Although the produced rice plastid transformants were found to be heteroplastomic, and the transformation efficiency requires further improvement, this study has established a variety of parameters for the use of plastid transformation technology in cereal crops.
Keywords: Cereal Crop, Chloroplast Genome, Mono-cotyledonous Plant Species, Plastid Transformation, Rice
Introduction
The plastid transformation approach has been shown to have a number of advantages, most notably with regard to its high transgene expression levels (De Cosa et al., 2001; Devine and Daniell, 2004), capacity for multi-gene engineering in a single transformation event (Daniell and Dhingra, 2002; De Cosa et al., 2001; Lossl et al., 2003; Quesada-Vargas et al., 2005; Ruiz et al., 2003) and ability to accomplish transgene containment via maternal inheritance (Daniell, 2002; Daniell et al., 1998; Svab and Maliga, 1993). Moreover, chloroplast appears to be an ideal compartment for the accumulation of certain proteins, or their biosynthetic products, which would be harmful if they accumulated in the cytoplasm (Daniell et al., 2001; 2005a; Lee et al., 2003; Leelavathi and Reddy, 2003; Ruiz and Daniell, 2005). In addition, no gene silencing has been observed in association with this technique, whether at the transcriptional or translational level (DeCosa et al., 2001; Dhingra et al., 2004; Lee et al., 2003). Because of these advantages, the chloroplast genome has been engineered to confer several useful agronomic traits, including herbicide resistance (Daniell et al., 1998; Kang et al., 2003), insect resistance (Kota et al., 1999; McBride et al., 1995), disease resistance (DeGray et al., 2001), drought tolerance (Lee et al., 2003), salt tolerance (Kumar et al., 2004a), and phytoremediative ability (Ruiz et al., 2003). The chloroplast genome has also been utilized in the field of molecular farming, for the expression of biomaterials, human therapeutic proteins, and vaccines for use in humans or animals (Fernandez-San Milan et al., 2003; Guda et al., 2000; Koya et al., 2005; Leelavathi et al., 2003; Molina et al., 2004; 2005; Staub et al., 2000; Vitanen et al., 2004; Watson et al., 2004; for review, see Bock, 2001; Daniell et al., 2004a; 2004b; 2005a; Grevich and Daniell, 2005; Maliga, 2004).
Although plastid transformation has been accomplished via organogenesis in a number of dicotyledonous plant species, including tobacco (Svab and Maliga, 1993), tomato (Ruf et al., 2001), potato (Nguyen et al., 2005; Sidorov et al., 1999), Arabidopsis (Sikdar et al., 1998), Lesquerella (Skarjinskaia et al., 2003), oilseed rape (Hou et al., 2003), petunia (Zubko et al., 2004) and lettuce (Lelivelt et al., 2005), this technology has proven to be highly efficient only in tobacco. In the Arabidopsis, 1 transplastomic line per 40 or 151 bombarded plates was obtained, but these lines were sterile. In the potato, 1 transplastomic line was obtained per 18 or 25 bombarded plates, but no data was reported with regard to the fertility of these lines. In the tomato and petunia, 1 transplastomic line was obtained per 10 bombarded plates. However, in the tobacco, up to 14 transplastomic lines were obtained per bombarded leaf (Daniell et al., 2001a). In the case of Lesquella, transplastomic clones had to be grafted onto Brassica napus rootstock in order to obtain seeds. Plastid transformation of oilseed rape produced heteroplastomic plants, and no data was reported regarding the fertility of these lines. The aforementioned studies have all reported attempts to accomplish plastid transformation in dicotyledonous plant species, and have apparently met with varying degrees of success.
Major obstacles to the extension of plastid transformation technology to crop plants which regenerate via somatic embryogenesis include: (i) the difficulty of expressing transgenes in non-green plastids, in which gene expression and gene regulation systems are quite distinct from those of mature green chloroplasts, and (ii) our current inability to generate homoplastomic plants via subsequent rounds of regeneration, using leaves as explants. Despite these aforementioned limitations, plastid transformation has recently been accomplished in several primary dicotyledonous crops, including soybean, carrot, and cotton, via somatic embryogenesis (Dufourmantel et al., 2004; 2005; Kumar et al., 2004a; 2004b). Breakthroughs in the plastid transformation of recalcitrant crops, such as cotton and soybean, have raised the possibility of engineering plastid genomes of other major crops, via somatic embryogenesis. However, despite the great interest to extend plastid transformation to major cereal crops, there has not yet been even a single report of a fertile transplastomic plant which verifiably transmitted transgenes to the next generation in any monocotyledonous plant species. To date, only a couple of fragmentary data were reported in association with rice (Khan and Maliga, 1999). However, the purpose of the study was to test the feasibility of using a fluorescent antibiotic resistance marker for tracking of plastid transformation in higher plants, including rice suspension cells, not to test the feasibility of plastid transformation in rice itself.
Here, we report a promising step towards stable plastid transformation in rice. The transplastomic rice plants generated in this study exhibited stable integration and expression of the aadA and sgfp transgenes in their plastids. Moreover, the transplastomic rice plants generated viable seeds, which were confirmed to transmit the transgenes to the T1 progeny plants. However, it was impossible to convert the transplastomic rice plants to homoplasmy, even after two generations of continuous selection. Potential limiting factors in the routine application of plastid transformation to rice are discussed in this article, as are some possible solutions to these problems.
Materials and Methods
Construction of pLD-RCtV-sGFP
A rice-specific plastid transformation vector, pLD-RCtV-sGFP, targets insertions into the trnI-trnA inverted repeat regions of the rice plastid genome (at nucleotides 93072-94942 and 120176-122046; GenBank accession No. NC_001320). The trnI-trnA region was amplified from the rice plastid genome by PCR, using the primers, OS6P (anneals to nucleotides 92731-92755 and 122363-122387) and OS6M (anneals to 94919-94942 and 120176-120199). The 2,212 bp PCR product was then cloned into pCR2.1-TOPO (Invitrogen, USA). The StuI/EcoRV 1.89 kb fragment was inserted into the PvuII site of pUC19. The selection marker gene, aadA, which is driven by the 16S ribosomal RNA promoter (Prrn), chloroplast ribosome binding site (GGAGG), and the 3′ UTR of the plastid psbA gene were excised as a NotI-NotI fragment, from a basic chloroplast transformation vector which had been constructed in the Daniell lab. The ends were then filled in with Klenow enzyme in order to clone them into the unique PvuII site located between the trnI and trnA regions (94117-94122 or 120996-121001), to obtain pLD-RCtV. The sgfp gene sequence (Chiu et al., 1996) was then amplified by PCR, using the pSK-RG plasmid (Jang et al., 2002) as a template, as well as primers designed to introduce a ribosome binding site, which was to be located upstream of the start codon and the XbaI flanking sites. The PCR-amplified fragment was then cloned into the XbaI site, which was located between the aadA gene and the 3′ UTR of psbA of pLD-RCtV, resulting in the generation of pLD-RCtV-sGFP.
Transformation and regeneration
We have previously optimized the conditions for the efficient regeneration of plants from the japonica rice (Oryza sativa L.) variety, Hwa-Chung (Lee et al., 2002), and closely followed the protocols for the regeneration of transplastomic plants. Mature seeds were positioned on N6 medium (Chu et al., 1975), which had been supplemented with 2,4-dichlorophenoxy acetic acid (2,4-D) (2 mg/L), casein hydrolysate (1 g/L) and sucrose (30 g/L), and had been semi-solidified by the addition of 0.2% (w/v) gelrite (Duchefa, Haarlem, Netherlands), in order to elicit callus formation. After 15–20 days of seed-planting, the total calli, both embryogenic and non-embryogenic (Lee et al., 2002; Wang et al., 2004), were collected in the central region of the regeneration medium, which contained Murachige and Skoog (MS) salts and vitamins (Murashige and Skoog, 1962), kinetin (2 mg/L), 1-naphtaleneacetic acid (NAA) (2 mg/L), myo-inositol (100 mg/L), sucrose (30 g/L), and gelrite (2 g/L). The total calli were then bombarded with the pLD-RCtV-sGFP plasmid DNA-coated gold particles of 0.6 μm diameter, using a PDS-1000/He Biolistic gun device and 1,100 p.s.i. rupture disks (Bio-Rad, USA). After bombardment, the calli were incubated in darkness at 27°C for 1–2 d prior to transfer into a selective regeneration medium supplemented with 200 mg/L of streptomycin sulfate (Sigma, USA) and 14 g/L of phytagar (Gibco-BRL, USA). In an additional selection step, the streptomycin-resistant calli which produced shoots were transferred to fresh selective regeneration medium supplemented with 300 mg/L of streptomycin. The streptomycin-resistant shoots were then rooted on MS medium which contained MS salts, MS vitamins, sucrose (30 g/L), and gelrite (2 g/L), supplemented with 500 mg/L of streptomycin. The plantlets were maintained in the selective MS medium for 2–3 months, then transplanted into pots filled with soil.
In order to detect functional sGFP expression in the T1 progeny, the seeds from the self-pollinated T0 transplastomic plants were germinated, then allowed to grow for 8 weeks on MS medium supplemented with 100 mg/L of streptomycin. The progeny seedlings were then transferred to fresh MS selective medium supplemented with 300–500 mg/L of streptomycin, and maintained for 3 months before being transferred to a greenhouse.
PCR analyses
Total genomic DNA was extracted from the leaf tissues of both the transformed and untransformed plants, according to the methods described by Doyle and Doyle (1987). PCR analysis was conducted using Excel Taq DNA polymerase (Corebio, Korea), using 100 ng genomic DNA as a template. For the PCR analyses of the T0 transplastomic plants, we amplified the sgfp gene with the forward primer (5′-GACCCTGAAGTT-CATCTGAC-3′) and the reverse primer (5′-ACTTGTACAGCT CGTCCATG-3′). The left border fragment was amplified with the forward primer (5′-TCAGCCATACGGCGGTGAATCCG-3′) and the reverse primer (5′-CCGCG-TTGTTTCATCAAG-CCTTACG-3′). The right border fragment was amplified with the forward primer (5′-CGGGATCACTCACGGCATGGACG-AG-3′) and the reverse primer (5′-TACCATAGAGGCCAACG-ATAGACAATAA-3′). The left and right border PCR products were cloned into pGEM-T Easy vector (Promega, USA) for DNA sequencing. For the PCR analysis of the T1 progeny plants, a set of 3 primers, trnI-F (5′-TGATTCTCTCCCAATTGGTT-GGATCGTA-3′), aadA-R (5′-CGTCGTGCACAACAATGGTG-ACTTCTAC-3′) and trnA-R (5′-CAATTAGACAGCCAACCC-3′), at a ratio of 1:1:0.05 (trnI-F: aadA-R: trnA-R), was used to detect the level of transplastome relative to untransformed plastome. The primer locations are all shown in Fig. 1.
Fig. 1.
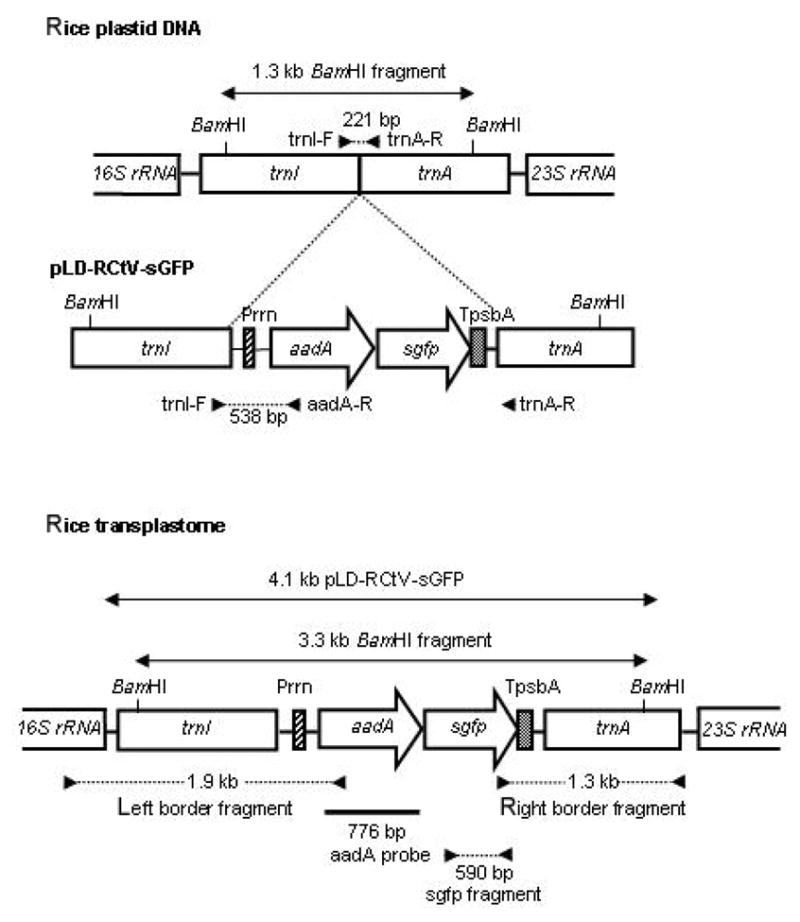
Rice transplastome resulting from the homologous recombination between the rice plastid transformation vector, pLD-RCtV-sGFP, and rice plastid DNA. The aadA and sgfp genes are driven by the rRNA operon promoter (Prrn). TpsbA indicates the 3′ UTR of the psbA gene. The filled triangles indicate the primer sites used for PCR analyses. The probe used for Southern analysis is marked with a thick line.
Southern analysis
Samples representing 18 μg of the BamHI-cut total genomic DNA were separated on 0.7% agarose gel, then transferred to Hybond-N+ nylon membranes (Amersham, Germany). These blots were hybridized with aadA-specific probe labeled with [α-32P]dCTP (Amersham, Germany). The probe was generated by PCR, using the following set of primers: aadAP1 (5′-GATCGCCGAAGTATCGACTC-3′) and aadAP2 (5′-ATTTGCCGACTACCTTGGTG-3′). The membrane was autoradiographed using X-ray film.
Western analysis
Total soluble proteins were extracted from the leaves of the transformed and untransformed plants in a greenhouse, using protein extraction buffer (0.1 M Tris-HCl pH 8.3, 0.5 M NaCl, 5 mM dithiothreitol, 5 mM EDTA, 2 mM phenyl-methylsulfonylfluoride). Samples which represented 2 μg of total soluble proteins were then electrophoresed on 12% SDS-PAGE gel, and transferred to nitrocellulose membranes (Amersham, Germany) for immunoblotting. Monoclonal anti-GFP (Sigma) was used as a primary antibody at a dilution of 1:500, and we used alkaline phosphatase-conjugated goat anti-mouse IgG (Sigma, USA) as a secondary antibody, at a dilution of 1:10,000. The BCIP/NBT Combo (Gibco-BRL, USA) was used for detection.
Detection of GFP fluorescence by confocal laser scanning microscope
Fluorescence was observed in the T1 progeny of the plastid transformants with a Carl Zeiss 510 CLSM (Jena, Germany). The laser was operated at an excitation wavelength of 488 nm and an emission wavelength of 510 nm. All images were then viewed and processed with Zeiss LSM Image Browser software. At 2 weeks after germination, leaf tissues from T1 progeny seedlings were used as samples for the confocal microscopic analysis.
Results
Construction of rice plastid transformation vector
The rice-specific plastid transformation vector, pLD-RCtV-sGFP (Fig. 1), contains the sgfp as a reporter gene, which encodes a synthetic GFP (Chiu et al., 1996; Hass et al., 1996), and the aadA gene, which confers streptomycin resistance for transformant selection (Goldschmidt-Clermont, 1991). Unlike the insoluble wild-type gfp gene product (Haseloff et al., 1997), the protein encoded by sgfp gene has been previously shown to exhibit no cytotoxicity in the rice plants (Jang et al., 2002). Both of the genes are driven by the strong plastid rRNA promoter, Prrn (Rapp et al., 1992), which was obtained from tobacco, and both employ the 3′ untranslated region (UTR) of the psbA gene for the stabilization of the mRNA (Stern and Gruissem, 1987). The capacity of the plastid for dicistronic expression of the aadA and gfp genes has been previously shown (Jeong et al., 2004). The suitability of the tobacco Prrn for use in transgenic expression in embryogenic cells and leaves of rice has also been demonstrated (Khan and Maliga, 1999; Silhavy and Maliga, 1998). The trnI-trnA intergenic region, located in the inverted repeat region of the plastid genomes, has been extensively exploited as the homologous recombination sequences for plastid transformation in tobacco, as well as in other crop species (Devine and Daniell, 2004; Dhingra et al., 2004). Hence, the trnI-trnA intergenic region was selected for plastid transformation in rice. The coding regions in the trnI-trnA genes are highly conserved among the higher plants (Hiratsuka et al., 1989; Koch et al., 1981; Michel et al., 1989), whereas introns in the trnI-trnA genes are not well conserved. For example, the introns in this region of rice and tobacco differ in 1 large and 5 small insertions and deletions and 25 point mutations. Therefore, the trnI-trnA flanking sequence in pLD-RCtV-sGFP was derived from the rice plastid genome. This flanking sequence also contains the oriA sequence (located within the trnI gene), and this is believed to enhance transgene integration (Kumar et al., 2004a; 2004b).
Evaluation of streptomycin selection conditions for rice plastid transformants
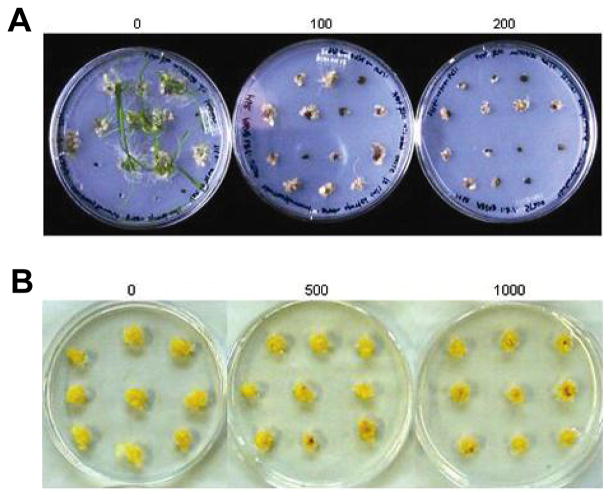
The aadA gene, which inactivates spectinomycin and streptomycin, is the most extensively used selection marker in the process of plastid transformation (Goldschmidt-Clermont, 1991; Svab and Maliga, 1993). The majority of homoplastomic plants produced thus far have been selected, under light conditions, and using spectinomycin (Ruf et al., 2001; Sidorov et al., 1999; Svab and Maliga, 1993). However, most cereal crops, including rice, exhibit natural spectinomycin resistance, due to the presence of point mutations in their 16S rRNAs sequence, which prevents spectinomycin binding (Fromm et al., 1987). Therefore, streptomycin resistance was employed in the selection of plastid transformants in rice. In order to determine the optimal streptomycin levels for use in the selection of rice plastid transformants, the total calli obtained 2–3 weeks after germination, containing both embryogenic and non-embryogenic ones, were positioned on shoot regeneration medium supplemented with a variety of streptomycin concentrations. Under light conditions, callus growth was partially inhibited at 100 mg/L of streptomycin, and no shoots developed, even after 2 months of culture. At streptomycin levels of between 200–500 mg/L, callus growth was inhibited to a greater extent (Fig. 2A). At 1,000 mg/L of streptomycin, all of the calli turned brown within 3 weeks of culture (data not shown), revealing that the selection had been too stringent. However, when selection progressed in darkness, callus growth did not appear to be significantly affected by the presence of streptomycin levels up to 1,000 mg/L, after 1 month of culture (Fig. 2B). We observed only a moderate degree of growth inhibition at streptomycin levels of more than 1,000 mg/L (data not shown). This suggests that the calli, when grown in darkness, are less sensitive to streptomycin. Therefore, the selection of transformants in darkness would be less efficient than transformant selection in the light. Therefore, in the following plastid transformation experiments, we conducted transformant selection in the light, using 200 mg/L of streptomycin as the optimal concentration for the initial selection phase. For the subsequent rounds of selection, we used 300–500 mg/L of streptomycin.
Fig. 2.
Determination of the optimal streptomycin concentration for inhibition of callus growth and shoot induction. The growth response of the total calli (2–3 weeks after germination) on selective regeneration medium containing (A) 0 (left), 100 (middle) and 200 (right) mg/L of streptomycin, after 2 months of culture in the light condition, and (B) 0 (left), 500 (middle) and 1,000 (right) mg/L of streptomycin, after 1 month of culture in the dark condition.
Plastid transformation and recovery of transplastomic plants
Plastid transformation was conducted by the bombardment of the total calli with pLD-RCtV-sGFP. After the bombardment, the total calli were incubated in the light, on shoot regeneration medium which contained 200 mg/L of streptomycin. The streptomycin-resistant calli exhibited green spots after one month of growth on the selective medium, and shoots emerged from the calli after another month (Fig. 3A). During the second round of selection, the shoot-producing streptomycin-resistant calli were transferred to a fresh selective medium, containing 300 mg/L of streptomycin, and were maintained for a month. The streptomycin-resistant shoots were then rooted in 500 mg/L of streptomycin, and were maintained for 2–3 months on the selection medium. In 10 independent transformation experiments, in which approximately 4,000 total calli were employed (on 100 to 120 bombarded plates), we selected two independent lines, and these were regenerated into plantlets (Fig. 3B). These two streptomycin-resistant lines were designated as sGFP-1 and sGFP-2, respectively.
Fig. 3.
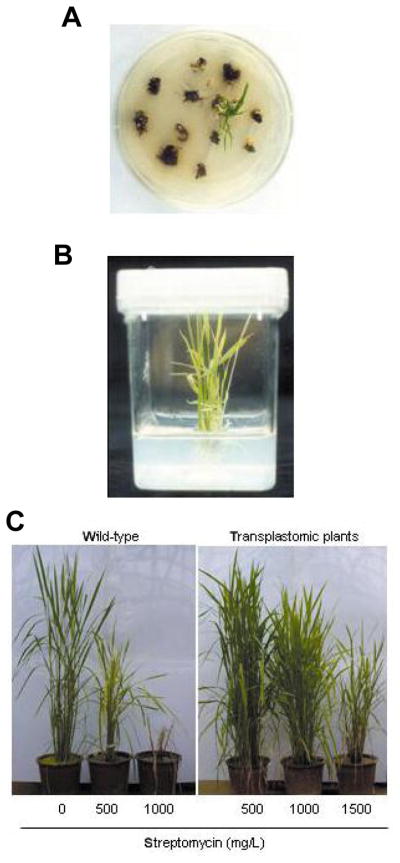
Recovery of transplastomic rice plants. (A) shoots induced from the bombarded calli after 2 months’ incubation on regeneration medium containing 200 mg/L of streptomycin, (B) streptomycin-resistant shoots regenerated into a plantlet, approximately 1 month after transferring to rooting medium containing 500 mg/L of streptomycin and (C) phenotype of the mature transplastomic plants in the greenhouse, indicating streptomycin resistance at different concentrations.
The putative transformants were then transferred into pots filled with soil, and grown to maturity in a greenhouse. In order to ensure continuous selection pressure, we added 500 mg of streptomycin into one liter of the soil per week. When sufficient numbers of tillers had appeared from sGFP-1 and sGFP-2, the tillers of sGFP-1 and sGFP-2 were divided into 3 separate parts, and transplanted into 3 different soil-containing pots. These tillers were then treated weekly with 500, 1,000, and 1,500 mg/L of streptomycin, respectively. In the presence of 500 mg/L of streptomycin, untransformed plants exhibited moderately inhibited growth, whereas the growth of the sGFP-1 and sGFP-2 plants appeared normal, and both of these lines produced viable seeds for T1 progeny analyses (Fig. 3C). At 1,000 mg/L of streptomycin, the vegetative growth of sGFP-1 and sGFP-2 was not affected significantly, but both lines were found to have produced either no, or few viable seeds. With a selection pressure exerted by 1,500 mg/L of streptomycin, both sGFP-1 and sGFP-2 exhibited significantly inhibited growth, and neither lines produced viable seeds.
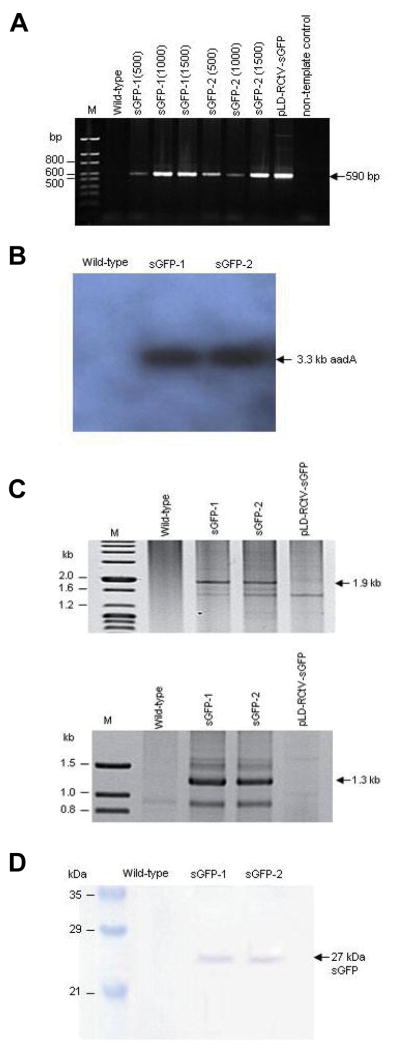
The putative transformants were initially tested by PCR screening for the presence of the sgfp gene, using total genomic DNA. All six of the plants derived from sGFP-1 and sGFP-2, which had been selected with 500–1,500 mg/L of streptomycin in the greenhouse, were found to harbor 590 bp sgfp fragments (Fig. 4A). This indicates that streptomycin-resistant phenotype constitutes a reliable indicator for the determination of transformants. Further molecular analyses were conducted with sGFP-1 and sGFP-2 lines grown on 500 mg/L of streptomycin, which generated viable seeds for T1 progeny analyses. Southern analysis using the aadA probe detected a 3.3 kb fragment in the Bam HI-digested genomic DNA (Fig. 4B), further verifying the integration of the aadA gene in the transformants. Then, in order to confirm the site-specific integration of the sgfp and aadA genes into the plastid genome, sGFP-1 and sGFP-2 were subjected to PCR analysis, using one internal primer which binds to the sgfp or aadA gene, and a second primer, which binds to the native plastid genome adjacent to the flanking sequences utilized in homologous recombination (Fig. 1). Both of the lines generated both the left and right border DNA fragments of 1.9 and 1.3 kb in size, respectively, as was expected with the transplastomes (Fig. 4C). The identity of these amplified PCR products was confirmed via DNA sequencing, thus eliminating the possible attribution of the amplified DNA fragments to a PCR artifact. We also tested sGFP-1 and sGFP-2 plants for the expression of sGFP, via immunoblotting using total soluble leaf proteins and GFP antibody. Both of the transplastomic plants produced the expected band of 27 kDa, indicating the stable expression and accumulation of sGFP in the transplastomic plants (Fig. 4D). These transplastomic plants were grown under continuous streptomycin selection conditions, and the seeds were harvested for subsequent T1 progeny analyses.
Fig. 4.
Characterization of T0 transplastomic plants. (A) PCR analysis indicates the presence of the 590 bp sgfp gene fragment in all six of the streptomycin-resistant plants derived from sGFP-1 and sGFP-2; streptomycin concentrations used in these selections are shown in parentheses, (B) Southern analysis using the aadA probe detects the 3.3 kb BamHI fragment in both sGFP-1 and sGFP-2, (C) PCR amplification of 1.9 and 1.3 kb left and right border DNA fragments, respectively and (D) the detection of sGFP accumulation via Western analysis.
T1 progeny analyses
For dicotyledonous plant species that regenerate via organogenesis, homoplasmy can be accomplished by multiple cycles of regeneration from transformed leaf tissues under the selective pressures. However, this process of shoot subculturing is not applicable for the elimination of untransformed plastomes from rice plants. For this reason, we expected our primary transplastomic lines might be heteroplastomic. Therefore, we applied further selection pressure to the sGFP-1 and sGFP-2 progeny. All of the seeds from the self-pollinated sGFP-1 and sGFP-2 were germinated on a selective medium that contained 100 mg/L of streptomycin. A small portion of the progeny seedlings bleached, after being transferred to a selective medium that contained 300–500 mg/L of streptomycin. In contrast, only some of the seeds from the untransformed control plants were germinated in the presence of 100 mg/L of streptomycin, but these green seedlings were ultimately bleached on 300–500 mg/L of streptomycin (data not shown). After another 3 months of culturing, the streptomycin-resistant progeny plants were transplanted into soil that contained 500 mg/L of streptomycin, and grown to maturity under continuous selection pressure.
At 2 weeks after germination, the progeny seedlings were subjected to confocal laser scanning microscopic analysis, in order to examine the transgene transmission and the proportion of the transplastome in the progeny. Confocal microscopic analysis confirmed that the expression of sGFP was limited to only a small fraction of the chloroplasts (marked with arrows in Fig. 5A), in some of the T1 progeny plants. Due to the heteroplastomic state of the T0 plastid transformants, 100% transmission of the transgenes was not expected (Skarjinskaia et al., 2003). Nevertheless, the localization of sGFP fluorescence in the transgenic chloroplasts constituted convincing evidence that both sGFP-1 and sGFP-2 were, in fact, plastid transformants, and that fully functional transgenes had been transmitted to the T1 progeny.
Fig. 5.
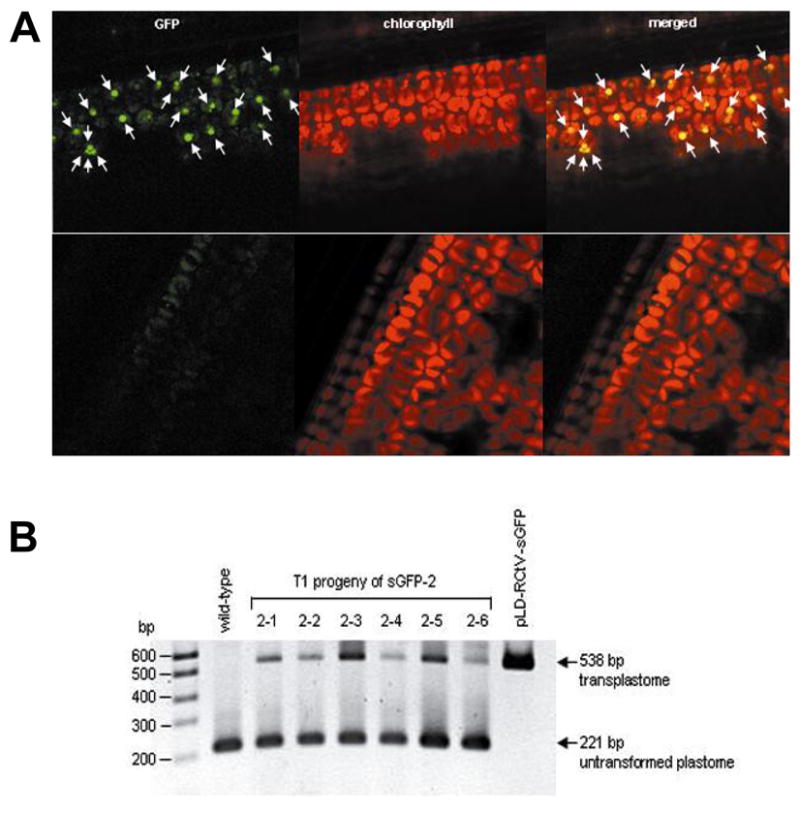
T1 progeny analyses. (A) Plastid-localized sGFP expression (marked with arrows) is observed from transplastomic line (top), whereas only a background level of autofluorescence is detectable from non-transformed plant (bottom) and (B) PCR analysis showing the presence of the 538 and 221 bp fragments, representing the transplastome and untransplastome, respectively.
PCR analysis was also conducted with approximately 6 months-old transplastomic plants, in order to determine the levels of heteroplasmy inherent to the mature progeny. To enhance the detection limit of the transplastome and to allow approximate comparisons of the levels between the transplastomes and untransformed plastomes, PCR analysis was performed using a set of 3 primers, trnI-F, trnA-R and aadA-R, hybridizing to sequences of the trnI-trnA regions and the aadA gene (Fig. 1). Both fragments of 538 and 221 bp, corresponding to the transplastome and untransformed plastome, respectively, were detectable at different levels in the T1 progeny plants (Fig. 5B), when approximately 20-fold higher amount of the transgene-specific primer was used, than that for the rice plastid gene, trnA (the primer ratio of trnI-F: aadA-R: trnA-R = 1:1:0.05). This primer ratio was necessary due to the low level of the transplastomes relative to the untransformed plastomes and the bias of the DNA polymerase towards shorter amplification products in PCR reactions. Although this PCR analysis could not reveal the exact levels of heteroplasmy, differences in the intensities of the transplastomic DNA bands are likely to reflect differences in the levels of transplastomes among the individual T1 progeny plants. This result, along with the data from the microscopic analysis, indicated that the proportion of transplastomes relative to the untransformed plastomes was low, even after two generations of continuous selection.
Discussion
Rice is the primary staple crop for more than one-third of the global population. Khan and Maliga previously reported plastid transformation of rice (1999), demonstrating stable transgene integration in the rice plastome. However, the transmission of this transgene to subsequent generations could not be achieved. In the present study, we generated fertile transplastomic rice plants, in which the aadA and sgfp genes were stably integrated and expressed. Furthermore, for the first time, we have demonstrated inheritance of plastid-expressed transgenes to the progeny of a transplastomic cereal crop, although low transformation efficiency and heteroplasmy remain to be solved.
Rice plastid transformation is quite inefficient, compared to dicotyledonous plant species. There are several possible reasons for this relatively low transformation efficiency. First, efficient subculture systems with sustained plant regeneration capability for transplastomic plants are lacking in monocotyledonous cereal crops, including rice. Second, undeveloped plastids (proplastids) are used as the transformation target, rather than chloroplasts. Our transmission electron microscopic observations showed that plastids in the dark-grown calli of rice were undeveloped and approximately 1 μm in size (data not shown). This is approximately 5-fold smaller than well-developed chloroplasts in the green leaf tissue. Hence, the amount of cells exhibiting irreversible physical damage due to biolistic bombardment with 0.6 μm gold particles might be greater in the proplastids. In fact, recent work by Langbecker et al. (2004) indicated that the use of smaller particle sizes (0.4 μm) resulted in 3- to 4-fold increase in plastid transformation frequency during the transformation of proplastids in dark-grown tobacco suspension cells.
In addition, transcription and translation levels are lower in proplastids than in chloroplasts (Mullet, 1993; Silhavy and Maliga, 1998). This results in lower levels of selection marker gene expression to confer resistance to selective chemicals. Therefore, the recovery of the newly transformed cells is attenuated further during the initial selection phase. The Prrn plastid-encoded RNA polymerase (PEP) promoter was used to drive transgene expression in the rice plants. Although the Prrn promoter has been shown to be active in both the proplastids and chloroplasts of rice, its activity has been shown to be 7-fold lower in the proplastids than in the chloroplasts (Silhavy and Maliga, 1998). In fact, the proplastids are a plastid type in which most PEP promoters are either weak, or completely inactive (Silhavy and Maliga, 1998; Vera and Sugiura, 1995). While the Prrn nuclear-encoded RNA polymerase (NEP) promoter has been demonstrated to be active in tobacco cell suspensions and carrot cultures, it was transcriptionally silent in rice cell suspension cultures (Silhavy and Maliga, 1998). Therefore, it is unlikely that the use of full-length 16S rRNA promoter, encompassing both the PEP and NEP promoters, would increase the transgene transcription levels in the proplastids of rice. Thus far, no other promoters that accumulate high transcript levels in both the proplastids and chloroplasts of rice are currently available. As the choice of promoter appears to be restricted to Prrn, current efforts are likely to focus on the augmentation of protein accumulation levels in the proplastids by employing a variety of 5′ UTRs or 5′ translational control regions, which are free from light and developmental regulation (Dhingra et al., 2004; Eibl et al., 1999; Kumar et al., 2004a; Maliga, 2003).
The other major difficulty in rice plastid transformation involves the acquisition of homoplasmy, which is of critical importance with regard to genetic stability. The T1 progeny plants of the transplastomic lines were shown to remain heteroplastomic, even after the continuous selection throughout the regeneration and vegetative/reproductive growth cycles on streptomycin-containing medium and soil. Our results indicate that the application of selection pressure after the transformed callus has been regenerated into a plantlet may not be an effective method for the achievement of homoplasmy. All of the homoplastomic plants produced thus far had been selected under light conditions, through a repeated process of subculturing of transformed leaf tissues (Ruf et al., 2001; Sidorov et al., 1999; Sikdar et al., 1998; Skarjinskaia et al., 2003; Svab and Maliga, 1993) or transformed calli/cells (Dufourmantel et al., 2004; Kumar et al., 2004a; 2004b). However, this subculturing process can not be applied to cereal crops for prolonged periods under light conditions, as it results in the rapid reduction of regenerability in the transformed tissues. Therefore, further selection pressure must be applied at the undifferentiated callus stage until plastid segregation is complete to achieve homoplastomic status, as it was achieved in the carrot plastid transformation (Kumar et al., 2004a).
Alternatively, in order to achieve homoplasmy, it may be necessary to develop new selection markers for a monocot-specific selection scheme. The use of suitable selection markers has been shown to be critical for success in the transformation of plastids in tobacco and cotton (Carrer et al., 1993; Kumar et al., 2004b; Svab and Maliga, 1993). Another strategy might be to use transgenic rice plants with a reduced number of plastids per cell as the transformation material, for the reduction of the selection period to achieve homoplasmy. In the Arabidopsis and tobacco plants, antisense suppression or over-expression of the ftsZ gene, which is involved in chloroplast division, has been shown to generate transgenic plants with fewer and larger chloroplasts in the mesophyll cells (Jeong et al., 2002; Osteryoung et al., 1998; Stokes et al., 2000). However, it remains to be determined whether the same approach can generate transgenic rice plants with fewer chloroplasts per cell.
Acknowledgments
We would like to thank Dr. J.-K. Kim, at Myong Ji University, for generously providing the pSK-RG plasmid, and Dr. H. J. Koh, at Seoul National University, for the seeds of the japonica rice var. Hwa-Chung, and for looking after the plastid transformants in the greenhouse. Special thanks also go to Dr. Yang Do Choi, at Seoul National University, for his critical scientific communication throughout this work. This study was supported by research grants from the Crop Functional Genomics Center of the 21st Century Frontier Research Program funded by the Ministry of Science and Technology and from the Korea Science and Engineering Foundation through the Center for Plant Molecular Genetics and Breeding Research to Minkyun Kim as well as USDA grant (3611-2100-017-001) to Henry Daniell. Kyungsu Kang and Hyunsup Chung were supported by the Korean Ministry of Education, through the Brain Korea 21 Project.
References
- Bock R. Transgenic plastids in basic research and plant biotechnology. J Mol Biol. 2001;312:425–438. doi: 10.1006/jmbi.2001.4960. [DOI] [PubMed] [Google Scholar]
- Bogorad L. Engineering chloroplasts: an alternative site for foreign genes, proteins, reactions and products. Trends Biotechnol. 2000;18:257–263. doi: 10.1016/s0167-7799(00)01444-x. [DOI] [PubMed] [Google Scholar]
- Carrer H, Hockenberry T, Svab Z, Maliga P. Kanamycin resistance as a selectable marker for plastid trnasformation in tobacco. Mol Gen Genet. 1993;232:33–39. doi: 10.1007/BF00280200. [DOI] [PubMed] [Google Scholar]
- Chiu W-L, Niwa Y, Zeng W, Hirano T, Kobayashi H, et al. Engineered GFP as a vital reporter in plants. Curr Biol. 1996;6:325–330. doi: 10.1016/s0960-9822(02)00483-9. [DOI] [PubMed] [Google Scholar]
- Chu C, Wang C, Sun CC, Yin K, Chu C. Establishment of an efficient medium for anther culture of rice through comparative experiments on the nitrogen sources. Scienta Sinic. 1975;18:659–668. [Google Scholar]
- Daniell H. Molecular strategies for gene containment in transgenic crops. Nat Biotechnol. 2002;20:581–586. doi: 10.1038/nbt0602-581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniell H, Dhingra A. Multigene engineering: dawn of an exciting new era in biotechnology. Curr Opin Biotechnol. 2002;13:136–141. doi: 10.1016/s0958-1669(02)00297-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniell H, Datta R, Varma S, Gray S, Lee SB. Containment of herbicide resistance through genetic engineering of the chloroplast genome. Nat Biotechnol. 1998;16:345–348. doi: 10.1038/nbt0498-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniell H, Lee SB, Pahchal T, Wiebe P. Expression of the native cholera toxin B subunit gene and assembly as functional oligomers in transgenic tobacco chloroplasts. J Mol Biol. 2001a;311:1001–1009. doi: 10.1006/jmbi.2001.4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniell H, Muthukumar B, Lee SB. Marker free transgenic plants: the chloroplast genome without the use of antibiotic selection. Curr Genet. 2001b;39:109–116. doi: 10.1007/s002940100185. [DOI] [PubMed] [Google Scholar]
- Daniell H, Muhammad S, Allison L. Milestones in chloroplast genetic engineering: an environmentally friendly era in biotechnology. Trends Plant Sci. 2002;7:84–91. doi: 10.1016/s1360-1385(01)02193-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniell H, Camrmona-Sanchez O, Burns B. Chloroplast derived antibodies, biopharmaceuticals and edible vaccines. In: Rischer R, Schill-berg S, editors. Molecular Farming. Wiley-VCH Verlag; Weinheim: 2004a. pp. 113–133. [Google Scholar]
- Daniell H, Cohill P, Kumar S, Dufourmantel N, Dubald M. Chloroplast genetic engineering. In: Daniell H, Chase C, editors. Molecular Biology and Biotechnology of Plant Organelles. Kluwer Academic Publishers; Dordrecht: 2004b. pp. 423–468. [Google Scholar]
- Daniell H, Chebolu S, Kumar S, Singleton M, Falconer R. Chloroplast-derived vaccine antigens and other therapeutic proteins. Vaccine. 2005a;23:1779–1783. doi: 10.1016/j.vaccine.2004.11.004. [DOI] [PubMed] [Google Scholar]
- Daniell H, Kumar S, Dufourmantel N. Breakthrough in chloroplast genetic engineering of agronomically important crops. Trends Biotechnol. 2005b;23:238–245. doi: 10.1016/j.tibtech.2005.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Cosa B, Moar W, Lee SB, Miller M, Daniell H. Overexpression of the Bt cry2Aa2 operon in chloroplasts leads to formation of insecticidal crystals. Nat Biotechnol. 2001;19:71–74. doi: 10.1038/83559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeGray G, Rajasekaran K, Smith F, Saford J, Daniell H. Expression of an antimicrobial peptide via the chloroplast genome to control phytopathogenic bacteria and fungi. Plant Physiol. 2001;127:852–862. [PMC free article] [PubMed] [Google Scholar]
- Devine A, Daniell H. Chloroplast genetic engineering for enhanced agronomic traits and expression of proteins for medical/industrial applications. In: Moller S, editor. Plastids. Blackwell publishing; Oxford: 2004. pp. 283–323. [Google Scholar]
- Dhingra A, Portis A, Jr, Daniell H. Enhanced translation of a chloroplast-expressed RbcS gene restores small subunit levels and photosynthesis in nuclear RbcS antisense plants. Proc Natl Acad Sci USA. 2004;101:6315–6320. doi: 10.1073/pnas.0400981101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle J, Doyle J. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytomchem Bull. 1987;19:11–15. [Google Scholar]
- Dufourmantel N, Pelissier B, Garcon F, Peltier G, Ferullo J-M, et al. Generation of fertile transplastomic soybean. Plant Mol Biol. 2004;55:479–489. doi: 10.1007/s11103-004-0192-4. [DOI] [PubMed] [Google Scholar]
- Dufourmantel N, Tissot G, Goutorbe F, Garcon F, Muhr C, et al. Generation and analysis of soybean plastid transformants expressing Bacillus thuringiensis Cry1Ab protoxin. Plant Mol Biol. 2005;58:659–668. doi: 10.1007/s11103-005-7405-3. [DOI] [PubMed] [Google Scholar]
- Eibl C, Zou Z, Beck A, Kim M, Mullet J, et al. In vivo analysis of plastid psbA, rbcL and rpl32 UTR elements by chloroplast trasformation: tobacco plastid gene expression is controlled by modulation of transcript levels and translation efficiency. Plant J. 1999;19:333–345. doi: 10.1046/j.1365-313x.1999.00543.x. [DOI] [PubMed] [Google Scholar]
- Fernandez-San MA, Mingeo-Castel AM, Miller M, Daniell H. A chloroplast transgenic approach to hyper-express and purify human serum albumin, a protein highly susceptible to proteolytic degradation. Plant Biotechnol J. 2003;1:71–79. doi: 10.1046/j.1467-7652.2003.00008.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromm H, Edelman M, Aviv D, Galun E. The molecular basis of rRNA-dependent spectinomycin resistance in Nicotiana chloroplasts. EMBO J. 1987;11:3233–3237. doi: 10.1002/j.1460-2075.1987.tb02640.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldschmidt-Clermont M. Transgenic expression of aminoglycoside adenine transferase in the chloroplast: a selectable marker for site-directed transformation of chlamydomonas. Nucleic Acids Res. 1991;19:4083–4089. doi: 10.1093/nar/19.15.4083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grevich JJ, Daniell H. Chloroplast genetic engineering: Recent advances and future perspectives. Crit Rev Plant Sci. 2005;24:83–108. [Google Scholar]
- Guda C, Lee SB, Daniell H. Stable expression of biodegradable protein based polymer in tobacco chloroplasts. Plant Cell Rep. 2000;19:257–262. doi: 10.1007/s002990050008. [DOI] [PubMed] [Google Scholar]
- Haseloff J, Siemering K, Prasher D, Hodge S. Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proc Natl Acad Sci USA. 1997;94:2122–2127. doi: 10.1073/pnas.94.6.2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hass J, Park E-C, Seed B. Codon usage limitation in the expression of HIV-1 envelope glycoprotein. Curr Biol. 1996;6:315–324. doi: 10.1016/s0960-9822(02)00482-7. [DOI] [PubMed] [Google Scholar]
- Hiratsuka J, Shimada H, Whittier S, Ishibashi T, Sakamoto M, et al. The complete sequence of the rice (Oryza sativa) chloroplast genome: intermolecular recombination between distinct tRNA genes accounts for a major plastid DNA inversion during the evolution of the cereals. Mol Gen Genet. 1989;217:185–194. doi: 10.1007/BF02464880. [DOI] [PubMed] [Google Scholar]
- Hou B, Zhou Y, Wan L, Zhang Z, Shen G, et al. Chloroplast transformation in oil seed rape. Transgenic Res. 2003;12:111–114. doi: 10.1023/a:1022180315462. [DOI] [PubMed] [Google Scholar]
- Jang EC, Choi WB, Lee KH, Song S, Nahm B, et al. High-level and ubiquitous expression of the rice cytochrome c gene OsCc1 and its promoter activity in transgenic plants provides a useful promoter for transgenesis of monocots. Plant Physiol. 2002;129:1–9. doi: 10.1104/pp.002261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong WJ, Park Y-I, Suh K, Raven JA, Yoo OJ, et al. A large population of small chloroplasts in tobacco leaf cells allows more effective chloroplast movement than a few enlarged chloroplasts. Plant Physiol. 2002;129:112–121. doi: 10.1104/pp.000588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong S-W, Jeong W-J, Woo J-W, Choi D-W, Park YI, et al. Dicistronic expression of the green fluorescent protein and antibiotic resistance genes in the plastid for selection and tracking of plastid-transformed cells in tobacco. Plant Cell Rep. 2004;22:747–751. doi: 10.1007/s00299-003-0740-4. [DOI] [PubMed] [Google Scholar]
- Kang TJ, Seo JE, Loc NH, Yang MS. Herbicide resistance of tobacco chloroplasts expressing the bar gene. Mol Cells. 2003;16:60–66. [PubMed] [Google Scholar]
- Khan M, Maliga P. Fluorescent antibiotic resistance marker for tracking plastid transformation in higher plants. Nat Biotechnol. 1999;17:910–915. doi: 10.1038/12907. [DOI] [PubMed] [Google Scholar]
- Koch W, Edwards K, Kossel H. Sequencing of the 16S–23S spacer in a ribosomal RNA operon of Zea mays chloroplast DNA reveals two split genes. Cell. 1981;25:203–213. doi: 10.1016/0092-8674(81)90245-2. [DOI] [PubMed] [Google Scholar]
- Kota M, Daniell H, Varma S, Garczynski S, Gould F, et al. Overexpression of the Bacillus thuringiensis (Bt) Cry2Aa2 protein in chloroplasts confers resistance to plants against susceptible and Bt-resistant insects. Proc Natl Acad Sci USA. 1999;96:1840–1845. doi: 10.1073/pnas.96.5.1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koya V, Moayeri M, Leppla SH, Daniell H. Plant based vaccine: mice immunized with chloroplast-derived anthrax protective antigen survive anthrax lethal toxin challenge. Infect Immun. 2005;73:8266–8274. doi: 10.1128/IAI.73.12.8266-8274.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Dhingra A, Daniell H. Plastid-expressed betaine aldehyde dehydrogenase gene in carrot cultured cells, roots and leaves confers enhanced salt tolerance. Plant Physiol. 2004a;136:2843–2854. doi: 10.1104/pp.104.045187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Dhingra A, Daniell H. Stable transformation of the cotton plastid genome and maternal inheritance of transgenes. Plant Mol Biol. 2004b;56:203–216. doi: 10.1007/s11103-004-2907-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langbecker C, Ye GN, Broyles D, Duggan L, Xu C, et al. High-frequency transformation of undeveloped plastids in tobacco suspension cells. Plant Physiol. 2004;135:39–46. doi: 10.1104/pp.103.035410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K, Jeon H, Kim M. Optimization of a mature embryo-based in vitro culture system for high-frequency somatic embryogenic callus induction and plant regeneration from japonica rice cultivars. Plant Cell Tiss Org Cult. 2002;71:237–244. [Google Scholar]
- Lee SB, Kwon H, Kwon S, Park S, Jeong M, et al. Accumulation of trehalose within transgenic chloroplasts confers drought tolerance. Mol Breed. 2003;11:1–13. [Google Scholar]
- Leelavathi S, Reddy V. Chloroplast expression of His-tagged GUS fusions: a general strategy to overproduce and purify foreign proteins using transplastomic plants as bioreactors. Mol Breed. 2003;11:49–58. [Google Scholar]
- Leelavathi S, Gupta N, Maiti S, Ghosh A, Reddy VS. Overproduction of an alkali- and thermo-stable xylanase in tobacco chloroplasts and efficient recovery of the enzyme. Mol Breed. 2003;11:59–67. [Google Scholar]
- Lelivelt CLC, McCabe MS, Newell CA, de Snoo CB, van Dun KMP, et al. Stable plastid transformation in lettuce (Lactuca sativa L.) Plant Mol Biol. 2005;58:763–774. doi: 10.1007/s11103-005-7704-8. [DOI] [PubMed] [Google Scholar]
- Lossl A, Eibl C, Harloff HJ, Jung C, Koop H-U. Polyester synthesis in transplastomic tobacco (Nicotiana tabacum L.): significant contents of polyhydroxybutyrate are associated with growth reduction. Plant Cell Rep. 2003;21:891–899. doi: 10.1007/s00299-003-0610-0. [DOI] [PubMed] [Google Scholar]
- Maliga P. Progress towards commercialization of plastid transformation technology. Trends Biotechnol. 2003;21:20–28. doi: 10.1016/s0167-7799(02)00007-0. [DOI] [PubMed] [Google Scholar]
- Maliga P. Plastid transformation in higher plants. Annu Rev Plant Biol. 2004;55:289–313. doi: 10.1146/annurev.arplant.55.031903.141633. [DOI] [PubMed] [Google Scholar]
- McBride K, Svab Z, Schaaf D, Hogan P, Stalker D, et al. Amplification of a chimeric Bacillus gene in chloroplasts leads to an extraordinary level of an insecticidal protein in tobacco. Biotechnology. 1995;13:362–365. doi: 10.1038/nbt0495-362. [DOI] [PubMed] [Google Scholar]
- Michel F, Umesono K, Ozeki H. Comparative and functional anatomy of group II catalytic introns- a review. Gene. 1989;82:5–30. doi: 10.1016/0378-1119(89)90026-7. [DOI] [PubMed] [Google Scholar]
- Molina A, Herva-Stubbs S, Daniell H, Mingo-Castel AM, Veramendi J. High yield expression of a viral peptide animal vaccine in transgenic tobacco chloroplasts. Plant Biotechnol J. 2004;2:141–153. doi: 10.1046/j.1467-7652.2004.00057.x. [DOI] [PubMed] [Google Scholar]
- Molina A, Veramendi J, Hervas-Stubbs S. Induction of neutralizing antibodies by a tobacco chloroplast derived vaccine based on a B cell epitope from canine parvo virus. Virology. 2005;342:266–275. doi: 10.1016/j.virol.2005.08.009. [DOI] [PubMed] [Google Scholar]
- Mullet J. Dynamic regulation of chloroplast transcription. Plant Physiol. 1993;103:309–313. doi: 10.1104/pp.103.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bio-assays with tobacco tissue cultures. Physiol Plant. 1962;15:473–497. [Google Scholar]
- Nguyen TT, Nugent G, Cardi T, Dix PJ. Generation of homoplasmic plastid transformants of a commercial cultivar of potato (Solanum tuberosum L.) Plant Sci. 2005;168:1495–1500. [Google Scholar]
- Osteryoung KW, Stokes KD, Rutherford SM, Percival AL, Lee WY. Chloroplast division in higher plants requires members of two functionally divergent gene families with homology to bacterial FtsZ. Plant Cell. 1998;10:1991–2004. doi: 10.1105/tpc.10.12.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quesada-Vargas T, Ruiz ON, Daniell H. Characterization of heterologous multigene operons in transgenic chloroplasts: transcription, processing, translation. Plant Physiol. 2005;128:1746–1762. doi: 10.1104/pp.105.063040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapp J, Baumgartner B, Mullet J. Quantitative analysis of transcription and RNA levels of 15 barley chloroplast genes. J Biol Chem. 1992;267:21404–21411. [PubMed] [Google Scholar]
- Ruf S, Hermann M, Berger I, Carrer H, Bock R. Stable genetic transformation of tomato plastids and expression of a foreign protein in fruit. Nat Biotechnol. 2001;19:870–875. doi: 10.1038/nbt0901-870. [DOI] [PubMed] [Google Scholar]
- Ruiz ON, Daniell H. Engineering cytoplasmic male sterility via the chloroplast genome by expression of β-ketothiolase. Plant Physiol. 2005;138:1232–1246. doi: 10.1104/pp.104.057729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz O, Hussein S, Terry N, Daniell H. Phytoremediation of organomercurial compounds via chloroplast genetic engineering. Plant Physiol. 2003;132:1344–1352. doi: 10.1104/pp.103.020958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidorov V, Kasten D, Pang S-Z, Hajdukiewicz P, Staub J, et al. Stable chlroplast transformatio in potato: use of green fluorescent protein as a plastid marker. Plant J. 1999;19:209–216. doi: 10.1046/j.1365-313x.1999.00508.x. [DOI] [PubMed] [Google Scholar]
- Sikdar S, Seriono G, Chaudhuri S, Maliga P. Plastid transformation in Arabidopsis thaliana. Plant Cell Rep. 1998;18:20–25. [Google Scholar]
- Silhavy D, Maliga P. Plastid promoter utilization in a rice embryogenic cell culture. Curr Genet. 1998;34:67–70. doi: 10.1007/s002940050367. [DOI] [PubMed] [Google Scholar]
- Skarjinskaia M, Svab Z, Maliga P. Plastid transformation in Lesquerella fendleri, an oilseed Brassicacea. Transgenic Res. 2003;12:115–122. doi: 10.1023/a:1022110402302. [DOI] [PubMed] [Google Scholar]
- Staub JM, Garcia B, Graves J, Hajdukiewicz PTJ, Hunter P, et al. High yield production of a human therapeutic protein in tobacco chloroplasts. Nat Biotechnol. 2000;18:333–338. doi: 10.1038/73796. [DOI] [PubMed] [Google Scholar]
- Stern D, Gruissem W. Control of plastid gene expression: 3′ inverted repeats act as mRNA processing and stabilizing elements, but do not terminate transcription. Cell. 1987;51:1145–1157. doi: 10.1016/0092-8674(87)90600-3. [DOI] [PubMed] [Google Scholar]
- Stokes KD, McAndrew RS, Figueroa R, Vita S, Osteryoung KW. Chloroplast division and morphology are differentially affected by overexpression of ftsZ1 and ftsZ2 genes in Arabidopsis. Plant Physiol. 2000;124:1668–1677. doi: 10.1104/pp.124.4.1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svab Z, Maliga P. High-frequency plastid transformation in tobacco by selection for a chimeric aadA gene. Proc Natl Acad Sci USA. 1993;90:913–917. doi: 10.1073/pnas.90.3.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vera A, Sugiura M. Chloroplast rRNA transcription from structurally different tandem promoters: an additional novel-type promoter. Curr Genet. 1995;17:280–284. doi: 10.1007/BF00326161. [DOI] [PubMed] [Google Scholar]
- Viitanen PV, Devine AL, Khan MS, Deuel DL, Van-Dyk DE, et al. Metabolic engineering of the chloroplast genome using the E. coli ubiC gene reveals that chorismate is a readily abundant plant precursor for phydroxybenzoic acid biosynthesis. Plant Physiol. 2004;136:4048–4060. doi: 10.1104/pp.104.050054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Seliskar DM, Gallagher JL. Plant regeneration via somatic embryogenesis in the brackish wetland monocot Scirpus robustus. Aquat Bot. 2004;79:163–174. [Google Scholar]
- Watson J, Koya V, Leppla SH, Daniell H. Expression of Bacillus anthrax protective antigen in transgenic tobacco chloroplasts: development of an improved anthrax vaccine in a non-food/feed crop. Vaccine. 2004;22:4374–4384. doi: 10.1016/j.vaccine.2004.01.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubko MK, Zubko EI, van Zuilen K, Meyer P, Day A. Stable transformation of petunia plastids. Transgenic Res. 2004;13:23–530. doi: 10.1007/s11248-004-2374-x. [DOI] [PubMed] [Google Scholar]