Abstract
Leptospirosis is considered a neglected infectious disease of human and veterinary concern. Although extensive investigations on host-pathogen interactions have been pursued by several research groups, mechanisms of infection, invasion and persistence of pathogenic Leptospira spp. remain to be elucidated. We have reported the ability of leptospires to bind human plasminogen (PLG) and to generate enzimatically active plasmin (PLA) on the bacteria surface. PLA-coated Leptospira can degrade immobilized ECM molecules, an activity with implications in host tissue penetration. Moreover, we have identified and characterized several proteins that may act as PLG-binding receptors, each of them competent to generate active plasmin. The PLA activity associated to the outer surface of Leptospira could hamper the host immune attack by conferring the bacteria some benefit during infection. The PLA-coated leptospires obstruct complement C3b and IgG depositions on the bacterial surface, most probably through degradation. The decrease of leptospiral opsonization might be an important aspect of the immune evasion strategy. We believe that the presence of PLA on the leptospiral surface may (i) facilitate host tissue penetration, (ii) help the bacteria to evade the immune system and, as a consequence, (iii) permit Leptospira to reach secondary sites of infection.
1. Introduction
The spirochete Leptospira interrogans is a highly invasive pathogen and the causal agent of leptospirosis, one of the most widespread zoonosis of human and veterinary concern. The disease represents a great economic burden because it has an effect on the public health system and the livestock [1–4]. The disease occurs mainly in urban regions lacking adequate sanitary conditions, associated with activities that involve direct contact with contaminated water, soil, or animals [3, 5, 6]. The leptospires chronically infect mammal species, which harbor the bacteria in their renal tubules, shedding them through the urine into the environment, thus constituting a source of reinfection to other animals. Humans are accidental and terminal hosts in the transmission process of leptospirosis [1, 7]. The leptospires enter the body via abrasions on skin or actively through mucosa, spreading to any tissue, but specially colonizing kidneys and liver [2].
The understanding of molecular aspects of the pathogenesis, the virulence, and invasion processes by which the leptospires infect the hosts and initiate tissue colonization remains to be elucidated, despite the availability of genomic sequencing of five strains of Leptospira that have identified more than 200 putative membrane proteins that might be involved in pathogenesis [8–12]. Yet, only few virulence factors related to the pathogenesis of the disease have been reported [13–16].
The ability of the leptospires to adhere to extracellular matrix (ECM) proteins is assumed to be crucial during the first steps of pathogenesis [17]. Indeed, this ability has been recently and firstly described by our group [18] and a number of adhesins, ECM-binding proteins, have been identified [15, 18–26]. After leptospiral adherence, the next fundamental step must be to overcome the barriers imposed by epithelial tissues and extracellular matrixes, to reach the bloodstream and secondary sites of infection within the hosts. Proteolytic activity has been demonstrated to be important during the penetration of several pathogenic microorganisms [27]. Because the presence of ECM degrading enzymes is limited to few bacterial pathogens, one of the strategies employed to surmount this deficiency is the interaction with protease-mediated systems of the hosts [27].
Plasmin is a broad-spectrum serine protease component of the fibrinolytic system, which has PLG as a main component. PLG is a circulating single-chain zymogen that is converted to plasmin by cleavage of the peptide bond between Arg-560 and Val-561, mediated by PLG activators-like uPA (urokinase-type PLG activator) and tPA (tissue-type PLG activator). Once activated to plasmin the bacteria are endowed with membrane-associated proteolytic activity, a characteristic that have been demonstrated to contribute to the degradation of ECM components, tissue penetration, and invasion. It has been reported that several pathogens, including the spirochetes Borrelia spp. and Treponema spp., bind PLG on the surface and converts it to plasmin by host activators [28–30]. The binding of PLG and plasmin generation promotes ECM molecules degradation and it is essential for dissemination of the bacteria through the host tissues, thus suggesting its importance in invasiveness process [27, 31–35].
In this paper, we highlight and compile the recent studies performed by our group describing and characterizing the leptospiral binding to PLG [36], the identification of possible PLG receptors [37–40], and the aspect of the leptospiral immune escape strategy associated to plasmin proteolytic activity [41]. The disclosure of this proteolytic system on the surface of Leptospira spp. and the implications associated to bacterial infectivity are discussed.
2. Materials and Methods
2.1. Bacteria Isolates and Culture Conditions
Virulent L. interrogans serovar Copenhageni strain Fiocruz L1-130 is routinely cultured at Faculdade de Medicina Veterinária da Universidade de Sao Paulo by iterative passages in Golden Syrian hamsters for maintenance of virulence [1]. The organs-derived leptospires were cultured at 28°C in semisolid modified Elinghausen-McCullough-Johnson-Harris (EMJH) medium supplemented with 10% rabbit sera. Nonvirulent culture-attenuated L. interrogans serovar Copenhageni strain M20 are equally routinely cultured by maintenance in liquid-modified EMJH medium supplemented with 10% rabbit sera [42]. For the experiments of PLG binding, to exclude the PLG interference from the rabbit serum supplementing the culture medium, serum-free leptospires were obtained by three passages in liquid modified EMJH medium supplemented with 10% Leptospira enrichment EMJH (BD, Difco), cultured at 28°C.
2.2. Labeling of Leptospires with Plasmin
The leptospires were treated with PLG and uPA based in the protocol described by Coleman et al. [43], with modifications. A total of 7.0 × 109 leptospires were centrifuged at 6,000 ×g for 10 min at 25°C, resuspended in 1.4 mL of EMJH culture medium supplemented with 10% Leptospira enrichment EMJH, divided into 7 aliquots of 0.2 mL each (1.0 × 109 leptospires) in 2 mL microcentrifuge tubes, and recentrifuged. The 7tubes containing the leptospires received different treatments: (a) 30% plasma and 3UuPA (urokinase, Sigma) in 100 μL low-salt PBS (lsPBS—with 50 mM NaCl), (b) 5 μg PLG (native PLG purified from human plasma was from Calbiochem) and 3 U uPA in 100 μL lsPBS, (c) 2 μg PLG and 3 U uPA in 100 μL lsPBS, (d) 0.5 μg PLG and 3 U uPA in 100 μL lsPBS, (e) 5 μg PLG in 100 μL lsPBS, (f) 3 U uPA in 100 μL lsPBS, and (g) 100 μL lsPBS. All the preparations were incubated for 1 h at 37°C, under gentle shaking, prior to the addition of the uPA, and followed by 1 h incubation at 37°C. Leptospires were then centrifuged and washed three times with 0.7 mL lsPBS.
2.3. Measurement of Enzymatic Activity of Plasmin-Coated Leptospires
The treated leptospires (1.0 × 109 per sample) were resuspended in 300 μL lsPBS and divided into 3aliquots. Each aliquot received 100 μL of 0.5 mg/mL of the chromogenic substrate D-Val-Leu-Lys 4-nitroanilide dihydrochloride (Sigma) in lsPBS, to a final substrate concentration of 0.25 mg/mL. The suspensions were incubated for 1.5 h at 37°C, under gentle shaking, and then centrifuged at 6,000 ×g for 10 min at 25°C. The supernatants (150 μL) were transferred to 96-well microplates and the cleavage of the specific plasmin substrate was quantified with a microplate reader set at a wavelength of 405 nm.
2.4. 6-Aminocaproic Acid (ACA) Binding-Inhibition Assay
Low-passage virulent L. interrogans serovar Copenhageni strain Fiocruz L1-130 (1.0 × 109 leptospires/sample) was treated with PLG and uPA, as described above, except for the addition of increase concentrations of ACA (Sigma) ranging from 0 to 1,000 mM, added together with the PLG. PLG quantity was set at 5 μg and uPA at 3 U. Aliquots treated with only PLG, uPA, or ACA, and just lsPBS treated, were used as controls. The enzymatic plasmin activity was measured as described before.
2.5. Liquid-Phase Immunofluorescence Assay (L-IFA)
Live bacteria suspensions (2.5 × 109) were harvested at 12,800 ×g for 15 min, washed twice with lsPBS, resuspended in 200 μL lsPBS containing 8 μg of human PLG, and incubated for 45 min at 37°C. After the incubation, 6 μg/mL of propidium iodide (Sigma) were added to stain the nuclei, and the suspensions were incubated for more 45 min at 37°C. After this time, the leptospires were gently washed three times with lsPBS and incubated for 45 min at 37°C with mouse-produced antiserum against human PLG (Sigma) at a 1 : 50 dilution. The leptospires were washed three times and incubated with goat anti-mouse IgG antibodies conjugated to fluorescein isothiocyanate (FITC; Sigma) at a dilution of 1 : 50 for 45 min at 37°C. After this incubation, the leptospires were washed twice and resuspended in lsPBS-antifading solution (ProLong Gold; Molecular Probes). The immunofluorescence-labeled leptospires were examined by the use of a confocal LSM 510 META immunofluorescence microscope (Zeiss, Germany). As control for cell integrity, we used antibodies against recombinant Leptospira proteins LipL32 or GroEL, produced in mice, following all the mentioned procedures for PLG.
2.6. SDS-PAGE and Affinity Blotting
Total leptospiral protein extracts for SDS-PAGE were prepared from 10 mL of ~109 bacteria in EMJH serum-free cultures. The cells were harvested by centrifugation, washed three times with 5 mM MgCl2 in lsPBS, and resuspended in 100 μL PBS. The proteins were loaded into 10% SDS-PAGE and transferred to nitrocellulose membranes (Hybond-ECL, GE Healthcare) in semidry equipment. The membranes were blocked for 2 h at 37°C with 5% BSA, washed three times (10 min for each wash) with PBS-0.5% Tween-20 solution (PBS-T), and incubated overnight with 3 μg/mL PLG or 3 μg/mL PLG + 100 mM ACA at 4°C, followed by a 2 h incubation at room temperature. Then, the membranes were washed three times and incubated with mouse anti-human PLG (1 : 750, Sigma) for 3 h at room temperature, following by more than three washings and 1 h incubation at room temperature with anti-mouse IgG-peroxidase conjugated (1 : 5,000, Sigma). The membranes were washed and the protein's reactivity was revealed by ECL reagent (GE Healthcare) with subsequent exposition to X-Ray films (Kodak).
2.7. Assay for the Degradation of Immobilized ECM Macromolecules
96-well plates were coated overnight at 37°C with 0.5 μg/well of cellular fibronectin (Sigma) or laminin (Sigma), washed four times with 200 μL per washing with PBS-T, and blocked with 2% BSA in PBS-T for 2 h at 37°C, followed by two washings. The spirochetes (1.0 × 108 leptospires/per sample) were treated as described above with 10 μg PLG and 3 U uPA or lsPBS (untreated). Bacteria were washed, resuspended in 100 μL lsPBS, and transferred to the plate previously coated wells. The plates were centrifuged at 180 ×g for 15 min to ensure the contact of the leptospires with the immobilized ECM components. The plates were incubated at 37°C for 20 h and washed 5 times to remove the bacteria. The degradation of the ECM components were detected by reduction in absorbance followed by incubation with antifibronectin or antilaminin IgG antibodies (1 : 5,000 dilution in 100 μL PBS-T for 45 min at 37°C, Sigma), anti-IgG peroxidase conjugated antibodies (1 : 5,000 dilution in 100 μL PBS-T for 45 min at 37°C, Sigma), and 100 μL/well of 1 mg/mL o-phenylenediamine—OPD—in citrate phosphate buffer, pH 5.0 plus 1 μL/mL H2O2. The reaction was stopped with 50 μL/well 4N H2SO4, and the absorbance was measured at 492 nm. Percentage degradation was calculated by de formula (A − B)/A(100), where A = the mean PBS samples absorbance (positive control group) and B = the mean experimental group absorbance (untreated or plasmin).
2.8. Bioinformatics Characterization of the Proteins
Predicted coding sequences (CDSs) were analyzed as their cellular localization predictions by PSORT program, http://psort.hgc.jp/ [44, 45]. The SMART http://smart.embl-heidelberg.de/ [46, 47], PFAM http://www.sanger.ac.uk/resources/software/ [48], and LipoP, http://www.cbs.dtu.dk/services/LipoP/ [49] web servers were used to search for predicted functional and structural domains within the amino acid sequences of the CDSs.
2.9. Cloning, Expression, and Purification of Recombinant Proteins
Amplification of the CDSs was performed by PCR from L. interrogans serovar Copenhageni genomic DNA using complementary primer pairs. The gene sequences were amplified without the signal peptide tag and predicted by SignalP (http://www.cbs.dtu.dk/services/SignalP/). The final constructs were verified by DNA sequencing on an ABI Prism 3730_L sequencer (Seq-Wright, Houston, TX, USA) with appropriate vector-specific T7 (F: TAATACGACTCACTATAGGG) and pAE (R: CAGCAGCCAACTCAGTTCCT) primers. Cloning, expression, and purification of the recombinant proteins have been previously described as summarized in Table 1.
Table 1.
Leptospiral selected proteins for characterization of the binding to PLG. Gene locus, protein name, NCBI reference sequence, features, gene conservation, sequence of the primers employed for DNA amplification, and molecular mass of expressed recombinant proteins.
| Gene locu1 | Recombinant protein given name | NCBI reference sequence number2 | Description/Function | Conservation (identity)3 |
Sequence of primers for PCR amplification | Recombinant protein molecular mass (kDa) |
|---|---|---|---|---|---|---|
| LIC11352 | LipL32a, h | YP_001316 | Major outer membrane protein (MOMP), LipL32 lipoprotein | Lai (100%); LBH (98%) |
F: 5′CACCGGTGCTTTCGGTGGTCTG 3′ R: 5′ATTACTTAGTCGCGTCAGAAGC 3′ |
30.2 |
| LIC10314 | Lsa63b, h | YP_000304 | Conserved hypothetical protein with Borrelia_P83 domain | Lai (98%); LBH (87%); LBP(39%) |
F: 5′GGATCCTTATTTTCTCAGGAAAG 3′ (BamH I) R: 5′GGTACCCTAAGGTTTAATTTTTTT 3′ (Kpn I) |
63.0 |
| LIC10509 | rLIC10509c, h | YP_000493 | Putative lipoprotein | Lai (98%) | F: 5′CCGGGATCCAAAAAGAGCAAAGAAG 3′ (BamH I) R: 5′GGTACCCTACTCGAGACAGCCAGGACCTTC 3′ (Kpn I) |
22.0 |
| LIC12892 | Lp29d, h | YP_002808 | Putative lipoprotein | F: 5′CTCGAGGCAGTACATTACAATCTTGCT 3′ (Xho I) R: 5′CCATGGCTCTTAGGAGCCTGGAAA 3′ (Nco I) |
29.6 | |
| LIC10793 | Lp49d, h | YP_000772 | Putative lipoprotein, Surface antigen | Lai (99%); LBH (86%) |
F: 5′CTCGAGAGCGGAGACTTTTCTTTACTT 3′ (Xho I) R: 5′CCATGGTTAAAAACCATCTCTACGATAAAC 3′ (Nco I) |
49.1 |
| LIC12895 | Lsa27e, h | YP_002811 | Putative lipoprotein | Lai (79%) | F: 5′GGATCCCTGAAATATACGAA 3′ (EcoRI) R: 5′GAATTCTTACTGTTCTCCTTC 3′ (BamHI) |
27.0 |
| LIC13131 | MPL21a, f, h | YP_003039 | Hypothetical protein with Ycel domain | Lai (98%); LBP (45%) |
F: 5′CACCACGTCTCAAAGTTACGCTTCAG 3′ R: 5′ TTCTCACCATCCAGCTCGG 3′ |
21.9 |
| LIC10765 | MPL17a, f, h | YP_000745 | Conserved hypothetical protein | Lai (100%); LBH (80%); LBP (41%) |
F: 5′ CACCGAAAGTCCCGTAAGGTTCAAA 3′ R: 5′TGCAGGAGTTCCCACATTTTA 3′ |
15.4 |
| LIC10091 | LipL40a, h | YP_000088 | Putative lipoprotein | Lai (100%); LBH (84%) |
F: 5′CCATGGGACTCGAGACGCCTCCTCCTAAAGATCC 3′ R: 5′CTCCATGGTCATTTCAAAACTTCTACGGGGC 3′ |
39.0 |
| LIC10054 | MPL36a, h | YP_000054 | Putative lipoprotein with Rare lipoprotein A (RplA) like domain | Lai (100%); LBH (88%); LBP (50%) |
F: 5′CACCACGTCTTGTGCGTCGGTAGAG 3′ R: 5′CCAAGTATTCTATTTATACGTCCGAG 3′ |
35.1 |
| LIC10494 | rLIC10494g, h | YP_000478 | Putative lipoprotein | Lai (99%) | F: 5′CACCACTGCTAGGGCTGCAGAAA 3′ R: 5′ACTTTGAGAGCTTCGTCTCGT 3′ |
25.8 |
| LIC12730 | rLIC12730g, h | YP_002650 | Hypothetical protein with TPR domain and 4 NHL repetition | Lai (100%); LBH (90%); LBP(37%) |
F: 5′CACCAGTTCTGACGGACTTCCCAA 3′ R: 5′TCTTGCGAATGAGTTGATCC 3′ |
77.4 |
| LIC12922 | rLIC12922h | YP_002837 | Conserved hypothetical protein | Lai (100%); LBH (89%); LBP (48%) |
F: 5′CACCGAATCACTCAACAGAGTCATTGC 3′ R: 5′ATCAATCTAAATGAAACGTCTCTTC 3′ |
40.0 |
| LIC12238 | rLIC12238h, i | YP_002173 | Hypothetical protein | Lai (99%); LBH (77%); LBP (39%) |
F: 5′CTCGAGTGTTTTAAACCTACCGGAG 3′ (Xho I) R: 5′AAGCTTCTACTTCATCGCTTTTTCTATATC 3′ (Hind III) |
17.6 |
| LIC10258 | Lsa66i | YP_000249 | Hypothetical protein with ompA domain | Lai (99%); LBH (79%) |
F: 5′GGATCCGAAGCCTTCTCACCCAATTG 3′ (BamH I) R: 5′CCATGGTTAAAGTGAAAGATAAAAATCGATTC 3′ (Nco I) |
65.7 |
| LIC12880 | Lp30i | YP_002796 | Putative lipoprotein | Lai (99%); LBH (76%) |
F: 5′CTCGAGGAAGTTGTCCGAGTCTAT 3′ (Xho I) R: 5′CCATGGTTATTGATTGTTTAATTCAG 3′ (Nco I) |
30.7 |
| LIC11469 | Lsa20j | YP_001430 | Hypothetical protein | Lai (100%); LBH (82%); LBP (29%) |
F: 5′CTCGAGCCAATTTCTTTCGATCCAAATC 3′ (Xho I) R: 5′AAGCTTTCAATCCTCTACTGCAGCCC 3′ (Hind III) |
20.0 |
| LIC11030 | rLIC11030g, j | YP_001000 | Putative lipoprotein with unknown domain (DUF1565) | Lai (100%) | F: 5′CTCGAGTGTACAAACGAAAAAGAAGGT 3′ (Xho I) R: 5′AAGCTTTTAGTTGCAAGGATTTGGA 3′ (Hind III) |
37.0 |
| LIC11834 | rLIC11834k (Lsa33) | YP_001783 | Putative lipoprotein | Lai (99%); LBH (87%); LBP (31%) |
F: 5′CTCGAGGATCTACAAGGTGGGGTTTTTAC3′ (Xho I ) R:5′CCATGGTTACTGAGGTTTTACTTGGTCC3′ (Nco I) |
33.1 |
| LIC12253 | rLIC12253k (Lsa25) | YP_002188 | Conserved hypothetical protein | Lai (100%); LBH (77%); LBP (39%) |
F: 5′CTCGAGGAGGAGAAACCGGACGATAC 3′ (Xho I) R: 5′CCATGGTTAGGGAAGACTTCTAACACATC 3′ (Nco I) |
24.0 |
3Protein BLAST—http://www.ncbi.nlm.nih.gov/blast/Blast.cgi.
aPreviously published by Gamberini et al. [78];
bPreviously published by Vieira et al. [37];
cPreviously published by Gómez et al. [79];
dPreviously published by Neves et al. [80];
ePreviously published by Longhi et al. [19];
fPreviously published by Oliveira et al. [81];
gPreviously published by Barbosa et al. [18];
hPreviously published by Vieira et al. [37];
iPreviously published by Oliveira et al. [38];
jPreviously published by Mendes et al. [39];
kPreviously published by Domingos et al. [40];
2.10. Plasminogen Binding to Recombinant Proteins
The binding of the recombinant proteins to PLG was evaluated by a modified ELISA, based in the protocol described by Brissette et al. [50], as follows: 96-well plates (Costar High Binding, Corning) were coated overnight in PBS at 4°C with 100 μL of 10 μg/mL of the recombinant proteins or bovine serum albumin (BSA, Sigma) as negative control. Plates were washed once with PBS-T and blocked for 2 h at 37°C with PBS with 10% (wt/vol) nonfat dry milk. The blocking solution was discarded and 100 μL of 10 μg/mL human PLG (Calbiochem) in PBS was incubated for 2 h at 37°C. Wells were washed four times with PBS-T and incubated for 1 h at 37°C with mouse anti-human PLG (1 : 4,000 in PBS, Sigma). Plates were washed again and incubated with horseradish peroxidase-conjugated anti-mouse immunoglobulin G (IgG, Sigma), diluted 1 : 5,000 in PBS. After three washings, 100 μL/well of 1 mg/mL OPD plus 1 μL/mL H2O2 in citrate phosphate buffer (pH 5.0) were added. The reactions were carried out for 5 min and stopped by the addition of 50 μL/well of H2SO4 (4 N). Readings were taken at 492 nm.
2.11. Enzymatic Activity Assay of Plasmin Bound to Recombinant Proteins
ELISA plates were coated overnight with 10 μg/mL recombinant proteins or BSA in PBS at 4°C. Plates were then washed once with PBS-T and blocked with PBS with 10% (wt/vol) nonfat dry milk for 2 h at 37°C. The blocking solution was discarded and 100 μL/well of 10 μg/mL human PLG (Calbiochem) was added, followed by incubation for 2 h at 37°C. Wells were washed three times with PBS-T, and then 4 ng/well of human uPA (Sigma) were added. Subsequently, 100 μL/well of the plasmin-specific substrate D-valyl-leucyl-lysine 4-p-nitroanilide dihydrochloride (Sigma) was added at a final concentration of 0.4 mM in PBS. Plates were incubated overnight at 37°C and substrate was measured by taking readings at 405 nm.
2.12. Dose-Response Curves and Dissociation Constant of the Proteins-PLG Interactions
ELISA plates were coated overnight in PBS at 4°C with 100 μL of 10 μg/mL PLG. Plates were then blocked and increasing concentrations of the purified recombinant proteins (0-1 μM) were added (100 μL/well in PBS). The assessment of bound proteins was performed by incubation for 1 h at 37°C with the antiserum raised against each protein at appropriate dilutions, followed by horseradish peroxidase-conjugated anti-mouse IgG (1 : 10,000 in PBS, Sigma). The binding was evaluated by the peroxidase substrate OPD and readings were taken at 492 nm. The ELISA data were used to calculate the dissociation constant (KD) according to the method described by Pathirana et al. [51] and Lin et al. [52], based on the equation: A = Amax [protein]/(KD + [protein]), where A is the absorbance at a given protein concentration, Amax is the maximum absorbance for the ELISA plate reader (equilibrium), [protein] is the protein concentration, and KD is the dissociation equilibrium constant for a given absorbance at a given protein concentration (ELISA data point).
2.13. Two-Dimensional Gel Electrophoresis (2-DE Gels)
Virulent low passage leptospires culture was harvested by centrifugation at 12,800 ×g at 4°C for 10 min. Pellet was washed (×5) by resuspension in PBS containing 5 mM MgCl2. The pellet was resuspended in DeStreak rehydration solution(GE Healthcare, USA) and lysed by vigorous vortexing. The cellular debris was separated by centrifugation at 20,800 ×g for 10 min, and the supernatant was collected. Total protein content was determined according to the Bradford method (Pierce Biotechnology, USA). Samples containing 200 μg of protein were adjusted to 125 μL with DeStreak rehydration solution (GE Healthcare, USA), along with 0.8% (v/v) IPG buffer, with a pH range of 3–10 (GE Healthcare).
First-dimension isoelectric focusing was performed using the IPGphor-System (GE Healthcare, USA), and the second dimension was conducted on the Ettan DALTsix system (GE Healthcare). The IPG gel strips (7 cm) with a linear separation of immobilized pH ranging from 3 to 10 were rehydrated directly with the solubilized samples. The focusing protocol was 30 V for 180 Vh, 150 V for 300 Vh, 350 V for 350 Vh, 500 V for 500 Vh, 1000 V for 1,000 Vh, 3,000 V for 3,000 Vh, and 5,000 V for 40,000 Vh, with a 50 μA/strip maximum-setting at 20°C. The strips were equilibrated twice (reduced and alkylated) for 15 min in 15 mL equilibration solution (0.05 M Tris-HCl, pH 8.8, 6.0 M urea, 30% [v/v] glycerol, and 2% [w/v] SDS), first with the addition of 1% DTT, and finally with 2.5% iodoacetamide. After equilibration, the strips were attached to the 12% SDS-PAGE. The gels were stained by Coomassie Blue R350 (PhastGel BlueR-GE Healthcare). The gels were analyzed with Image Master-2D Platinum version 6.0 software (GE Healthcare, USA).
2.14. Mass Spectrometry and Protein Identification
The samples were analyzed by MALDI-TOF (Matrix Assisted Laser Desorption Ionization-Time of Flight) mass spectrometry, using α-cyano-4-hydroxycinnamic acid as the matrix on an Ettan MALDI-TOF/Pro instrument (Amersham Biosciences, USA). The spots preparation, mass spectrometry, and database search for protein identification were performed according to the protocol already described in Vieira et al. [53]. Prediction for protein localization was performed by LipoP program [49].
2.15. Human Sera and Microscopic Agglutination Test
Confirmed-leptospirosis serum samples were obtained from Instituto Adolfo Lutz collection, Sao Paulo, Brazil, as previously described [41]. In brief, a laboratory-confirmed case of leptospirosis was defined by the demonstration of a four-fold microagglutination titer rise between paired serum samples.The probable predominant serovar was considered the titer that was the highest sample dilution with 50% of agglutination. MAT (microscopic agglutination test) was considered negative when the titer was below 100. In addition to the MAT-negative and MAT-positive paired samples of the same individuals with laboratory and clinic leptospirosis confirmations, we also employed sera from normal healthy donors (normal human sera—NHS), without a known history of leptospirosis, with confirmed negative MAT.
2.16. Human IgG and C3b Deposition on Leptospires
L. interrogans serovar Pomona strain LPF (2.5 × 107 cells/well) were diluted in 50 μL lsPBS, coated onto microplates, and allowed to stand at 37°C for 1 h, followed by overnight at 4°C. The plates were washed twice with 200 μL lsPBS and blocked with 5% nonfat dry milk and 2.5% BSA for 2 h at 37°C. After addition of 50 μL of pooled sera/well (NHS, MAT-negative or MAT-positive) as a source of IgG, the plates were allowed to incubate for 30 min on ice, and then washed three times. The coated leptospires were treated with 50 μL/well of 5 μg/mL uPA (urokinase, Sigma) and 40 μg/mL PLG (Calbiochem) for 2 h at 37°C, or only with uPA, only with PLG, or no additions, as negative controls. After three washings, the plates were incubated with anti-human IgG Fc region specific antibodies (Calbiochem) at a dilution of 1 : 5,000 for 1 h at 37°C, and then with secondary antibodies conjugated with peroxidase at a dilution of 1 : 10,000 for 1 h at 37°C. The IgG depositions on leptospires were evaluated by the peroxidase substrate OPD and readings were taken at 492 nm. When dilutions of NHS were used, they were performed with lsPBS.
The protocol of C3b deposition on leptospires was similar to the IgG deposition described before, with exception of the incubation with NHS, placed at 37°C for 50 min, and use of anti-human C3b primary antibodies (1 : 5,000) (Calbiochem), followed by secondary antibodies conjugated with peroxidase (1 : 5,000).
2.17. Human C3b Degradation by Leptospires
L. interrogans serovar Pomona strain LPF (108 leptospires/sample) were treated in 200 μL lsPBS with the addition of 10 μg PLG and 3 U uPA (plasmin), 10 μg PLG, or no additions (untreated). The cells were incubated for 1 h at 37°C with the PLG, and for one more hour after the addition of uPA. The cells were washed three times with lsPBS, and then resuspended in 100 μL lsPBS containing 15 μg/mL human purified C3b (Calbiochem), being incubated for 20 h at 37°C. The leptospires were removed by centrifugation, 20 μL of the supernatants were separated by 10% SDS-PAGE and then transferred to nitrocellulose membranes in semi-dry equipment. The membranes were blocked by incubating overnight at 4°C with 5% nonfat dry milk and 1% BSA. The C3b detection was made by incubations with anti-human C3b antibodies and secondary antibodies conjugated with peroxidase, followed by ECL (GE Healthcare) development.
2.18. Serum Susceptibility Testing for Treated Leptospires
Complement-mediated killing of leptospires was evaluated after incubation with nonimmune NHS, as described by Meri et al. [54], with some modifications. Approximately 5 × 108 L. interrogans serovar Pomona strain LPF per sample were treated with 40 μg PLG and 4 U uPA in 200 μL lsPBS for 3 h at 37°C. As a control, one aliquot of L. interrogans serovar Pomona strain LPF was incubated only with PBS. Then, the samples were divided into aliquots containing 1.0 × 108 cells, which were added by 80 μL NHS and 80 μL EMJH medium supplemented with 10% Leptospira enrichment. The bacteria suspensions were incubated at 37°C for one hour, followed by a incubation of 5 min on ice for stopping complement activation. After, 20 μL of each sample (four replicates) were transferred to microplates filled with 180 μL of EMJH medium/well. The plates were sealed with a sterile adhesive film and incubated at 30°C for 4 days. Bacterial growth was determined by counting leptospires in a Petroff-Hausser chamber under dark-field microscopy.
2.19. Statistics
For the data that were tested for statistical significance, the student's two-tailed test was applied, considering the minimum of P < 0.05.
3. Results and Discussion
3.1. Binding of Human Plasminogen by L. interrogans Cells
Live-immunofluorescence microscopy (L-IFA) was employed in order to evaluate the ability of L. interrogans cells to bind human PLG. L-IFA revealed that leptospires bind PLG in their outer surface along the entire cell, as seen when the bacteria were probed with antibodies anti-PLG (Figure 1). PLG bound to leptospires does not seem to damage the bacterial membrane because no fluorescence of GroEL, a cytoplasmatic heat-shock protein [55], was achieved when serum anti-GroEL was employed (Figure 1). The leptospiral recombinant proteins LipL32, an abundant outer membrane protein of Leptospira [56], used as positive control, showed an intense surface fluorescence. We also observed that the spirochete regular movement was conserved, suggesting that the flagella remained undamaged. Taken together, these data suggest that the PLG-binding does not interfere with the membrane structure or dynamics [36].
Figure 1.
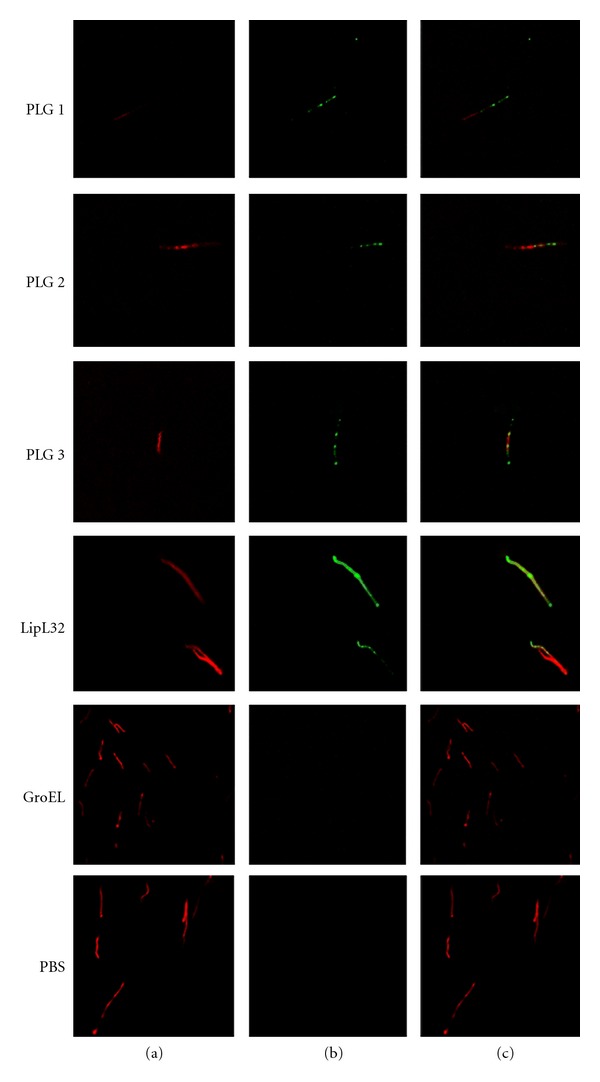
Recognition of PLG binding to Leptospira by L-IFA. Live virulent L. interrogans serovar Copenhageni isolates were treated with PLG, and the recognition was assessed through polyclonal anti-PLG antibodies under a confocal immunofluorescence microscope (PLG 1 to 3). PLG-treated leptospires stained for the proteins LipL32 (outer surface protein marker) or GroEL (a protoplasmic-cylinder marker) and untreated leptospires (PBS), are shown as controls (a) DNA propidium iodide-stained, (b) FITC-stained and (c) A+B composite images.
3.2. Activation and Enzymatic Activity of Plasminogen-Bound Leptospira
The binding of PLG to leptospiral surface was further analyzed by plasmin enzymatic activity. Leptospires were incubated with purified PLG, followed by plasmin activation after addition of uPA, conferring the bacteria a surface-associated plasmin activity (Figure 2), as measured by the degradation of plasmin's chromogenic specific substrate [36].
Figure 2.
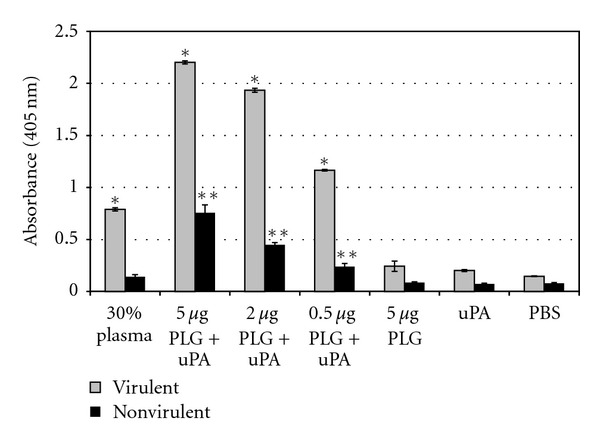
Cleavage of the plasmin-specific chromogenic substrate by plasmin-bound leptospires. Live low-passage virulent and high-passage nonvirulent L. interrogans serovar Copenhageni cells received the following treatments: PBS only (PBS), uPA alone (uPA), 5 μg PLG alone (PLG), PLG in crescent quantities (0.5, 2 and 5 μg) together with uPA, and 30% human plasma together with uPA (30% plasma). Bars represent mean absorbance as a measure of relative substrate degradation ± the standard deviation of three replicates for each experimental group and are representative of three independent experiments. *Virulent leptospires experiments: statistically significant (P < 0.0001) in comparison to the PBS control; **nonvirulent leptospires experiments: statistically significant (P < 0.01) in comparison to the PBS control. The 30% plasma, as PLG source, and all the PLG + uPA samples of the virulent leptospires were statistically significant (P < 0.001) in comparison to the same nonvirulent bacteria tested samples.
The labeling with PLG alone did not trigger proteolytic activity suggesting that L. interrogans serovar Copenhageni is dependent on the host PLG activation system, probably not possessing an endogenous mechanism, as already reported for other pathogens [57–64]. In contrast, the ability of PLG activation is shown for a number of pathogenic microorganisms [57–64].
Both the virulent and nonvirulent strains of L. interrogans serovar Copenhageni tested were capable of capturing PLG, although the virulent strain seems to be more efficient in the PLG-binding than the nonvirulent bacteria (~2.5 fold) (Figure 2) [36]. PLG is present in plasma at a concentration of approximately 20.8 ± 1.9 mg/100mL [65]. Our data shows that virulent L. interrogans, but not the attenuated strain, have the ability to sequester PLG from human plasma in the conditions assayed. At any rate, once in the host, the infectious leptospires can acquire PLG when reach the blood circulation.
3.3. Inhibition by 6-Aminocaproic Acid (ACA) of Proteolytic Activity Acquired by L. Interrogans
It is well known that the lysine-binding sites of PLG kringle domains frequently mediate interactions with lysine residues of the cellular receptors [27]. ACA, a lysine analogue, decreased the proteolytic activity of virulent L. interrogans serovar Copenhageni incubated with PLG, in a dose-dependent manner (Figure 3). The total inhibition (100%) of plasmin activity achieved with the higher ACA concentrations employed shows that the PLG interaction with these spirochetes occurs trough the lysine-binding sites of the kringle domains [36].
Figure 3.
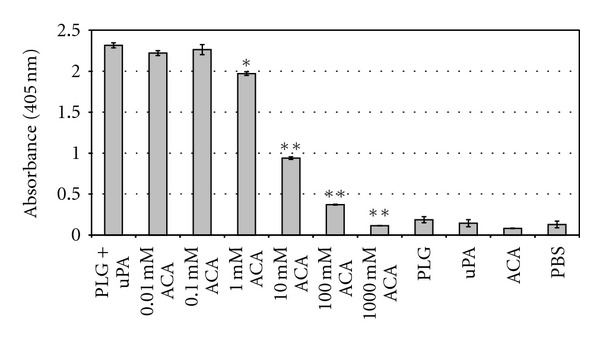
Inhibition of PLG binding to L. interrogans by ACA. Low-passage virulent L. interrogans serovar Copenhageni cells were treated with the following: PLG together with uPA (PLG + uPA), PLG together with uPA with the addition of crescent concentrations of ACA (0.01 to 1,000 mM ACA), PLG alone (PLG), uPA alone (uPA), ACA alone (ACA), and no additions (PBS). The cleavage of the plasmin-specific substrate D-Val-Leu-Lys 4-nitroanilide dihydrochloridein by the treated spirochetes was measured by absorbance readings at 405 nm. Bars represent mean absorbance ± the standard deviation of three replicates for each experimental group and are representative of three independent experiments. Statistically significant substrate degradation inhibition results in comparison to the positive control (PLG + uPA) are shown: P < 0.01 (*) and P < 0.0001 (**).
3.4. Analysis of Plasminogen-Binding L. Interrogans Proteins on Cell Lysates
Assessment of leptospiral proteins binding to PLG was first visualized by affinity blotting using total whole-cell lysates (Figure 4(a)). Our results are indicative that multiple leptospiral proteins are capable of binding PLG, although these affinity blotting experiments do not simulate the protein-PLG interaction in vivo. As expected, the data also demonstrate that there are similarities between the PLG-binding proteins of the virulent and nonvirulent strains, but also revealed the presence of some reactive bands on the low-passage virulent strain that are absent in the nonvirulent one, indicating that virulent strains have more PLG binding proteins. Thus, based on the data from enzymatic activity and affinity blotting presented here we cannot discriminate whether the increased bound activity of the virulent strain is due to only differential expression of a different set of membrane receptors, an increased expression of the same receptors, or both. Comparable to the results obtained with the intact leptospires, the affinity blotting was inhibited by the presence of the lysine analog, ACA, supporting the central role of lysine residues of the proteins in the PLG interactions (Figure 4(b)) [36].
Figure 4.
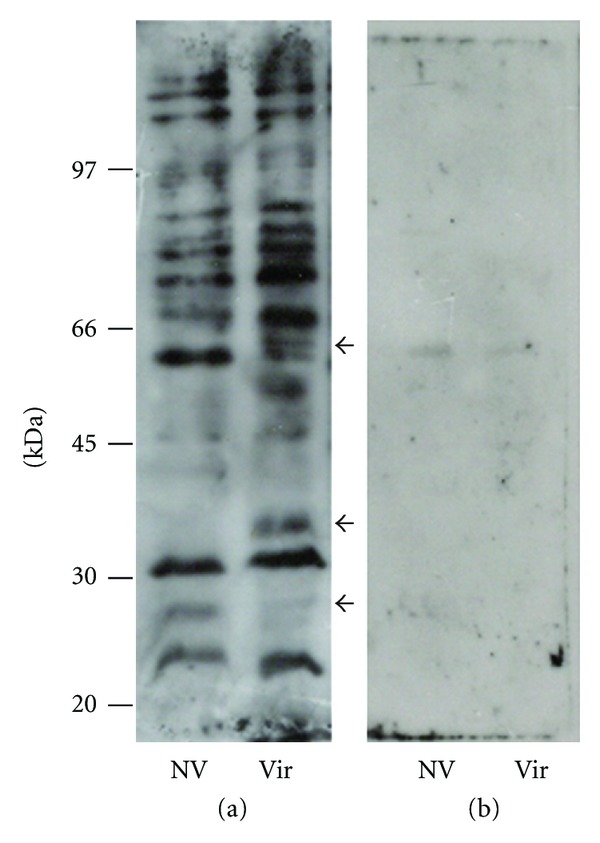
Binding of human PLG to L. interrogans proteins by affinity blotting. Total protein extracts were resolved by 10% SDS-PAGE and electroblotted into nitrocellulose membranes. The membranes were incubated with PLG (a) or PLG + ACA (b), and the binding was detected by anti-PLG antibodies and peroxidase-conjugated antibodies. The reactivity was revealed by ECL (GE Healthcare) and exposure to X-Ray films. NV: high passage, nonvirulent L. interrogans serovar Copenhageni strain M 20; Vir: low-passage, virulent L. interrogans serovar Copenhageni strain Fiocruz L1-130. The arrows indicate reactivity protein regions seen in the virulent but absent in the nonvirulent bacterial extract. Positions of protein molecular mass markers are shown on the left.
3.5. Plasmin-Coated Leptospira Degrades Immobilized ECM Molecules
Plasmin is a broad-spectrum serine protease that is capable to degrade directly laminin and fibronectin. To evaluate whether the plasmin associated to the leptospires surface can directly break these ECM components, purified soluble human fibronectin and laminin were adhered into 96-well plates, and the reaction was evaluated by exposing the ECM proteins with low-passage virulent L. interrogans, pretreated with PLG/uPA. As shown in Figure 5, the plasmin-coated leptospires were able to significantly cleave down the immobilized ECM macromolecules, when compared to the ones exposed to untreated bacteria [36].
Figure 5.
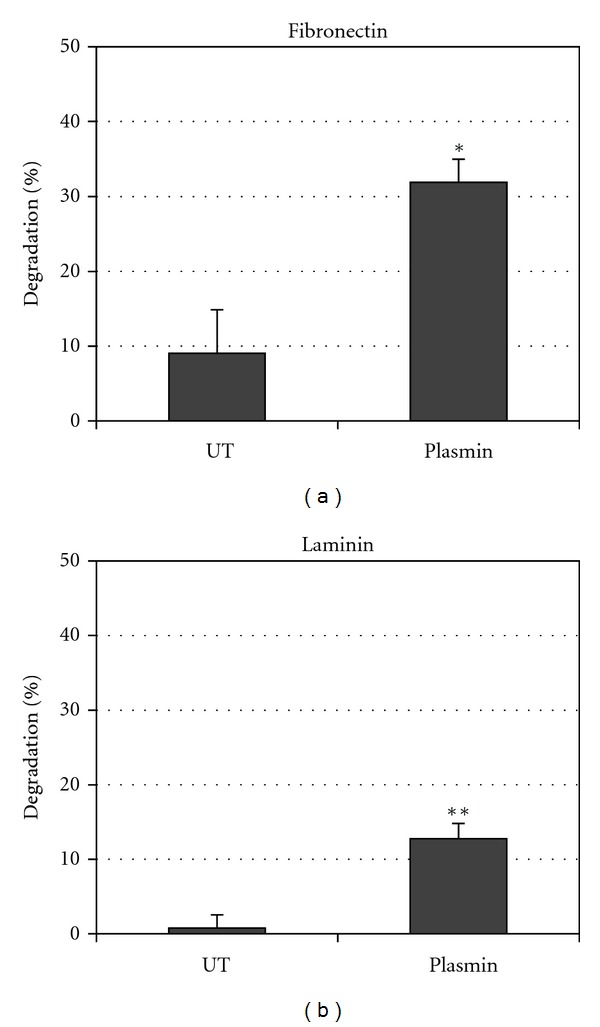
Fibronectin and laminin degradation by plasmin-coated leptospires. Spirochetes were incubated in lsPBS with no additions (UT) and with addition of PLG and uPA (plasmin). ELISA plate wells coated with 0.5 μg/well fibronectin (a) or laminin (b) were incubated for 20 h with the spirochetes from each experimental group to allow substrate digestion. Plates were then incubated with antibodies antilaminin or antifibronectin. A reduction in absorbance was interpreted as substrate digestion, relative to controls lacking bacteria (0.5 μg/well purified laminin or cellular fibronectin, corresponding to 0% degradation). Percent degradation (a reduction in absorbance value relative to control lacking bacteria) was calculated as described in Section 2. Bars represent mean percent substrate degradation relative to the positive control (0% degradation) ± the standard deviation of three replicates. The experiment was independently performed three times with similar results. P < 0.001 (*) and P < 0.01(**).
Given that leptospires are highly invasive pathogens that are thought to penetrate the skin or break in the skin to initiate infection, the ability of digesting ECM macromolecules via plasmin activity might be an important step for leptospiral pathogenesis. To date, this is the first proteolytic activity detected in Leptospira that can promote ECM degradation and it is a plausible mechanism that can contribute to leptospiral invasiveness [1, 66].
3.6. Recombinant Leptospiral Proteins Bind Human Plasminogen
In order to investigate PLG binding proteins in Leptospira we benefitted from the availability of several recombinant proteins of L. interrogans available in our laboratory. We evaluated the ability of these proteins to interact with PLG in vitro. Table 1 summarizes the features of these proteins and gene conservation within the sequenced genomes [8–12]. The binding of these proteins to PLG was quantified by ELISA and the results are shown in Figure 6. Proteins LipL32, Lp29, Lp49, LipL40, MLP36, rLIC10494, rLIC12730, rLIC12238, Lsa66, Lp30, Lsa20, and Lsa33 (rLIC11834) presented significant binding to PLG while proteins Lsa63, Lsa27, rLIC10509, MPL21, MPL17, rLIC12922, rLIC11030, Lsa25 (rLIC12253), BSA, and gelatin did not show significant binding activity (Figure 6) [37–40].
Figure 6.
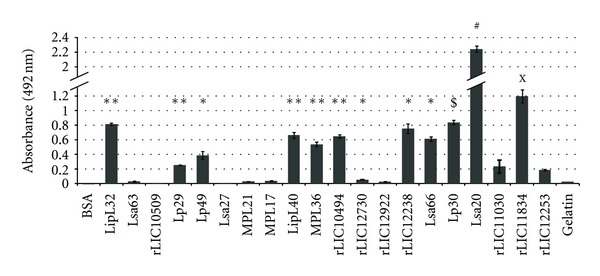
Binding of leptospiral recombinant proteins to human plasminogen. The recombinant proteins LipL32, Lsa63, rLIC10509, Lp29, Lp49, Lsa27, MPL21, MPL17, LipL40, MPL36, rLIC10494, rLIC12730, rLIC12922, rLIC12238, Lsa66, Lp30, Lsa20, rLIC11030, rLIC11834, and rLIC12253 were coated onto 96-well ELISA plates (10 μg/mL) and allowed to interact with purified human PLG (10 μg/mL). BSA and gelatin were used as a negative control for nonspecific binding. Binding was detected and quantified by specific antibodies. Bars represent the mean absorbance values at 492 nm ± the standard deviation of four replicates for each protein and are representative of three independent experiments. Statistically significant binding in comparison to negative control BSA are shown by *(P < 0.001); **(P < 0.0001); $(P < 0.02); #(P < 0.0005). The binding of rLIC11834 and rLIC12253 was compared to its binding to gelatin X(P < 0.005).
3.7. Activation of Plasminogen-Bound Proteins
To evaluate whether purified PLG-binding proteins were independently capable to generate active plasmin in the presence of uPA PLG activator, we measured the degradation of plasmin specific chromogenic substrate in a modified ELISA. The PLG captured by the proteins could be converted into plasmin, as demonstrated indirectly by specific proteolytic activity (Figure 7). The negative control BSA, that showed no PLG binding, produced no significant proteolytic activity, as well as the controls lacking PLG, uPA, or the chromogenic substrate [37–40].
Figure 7.
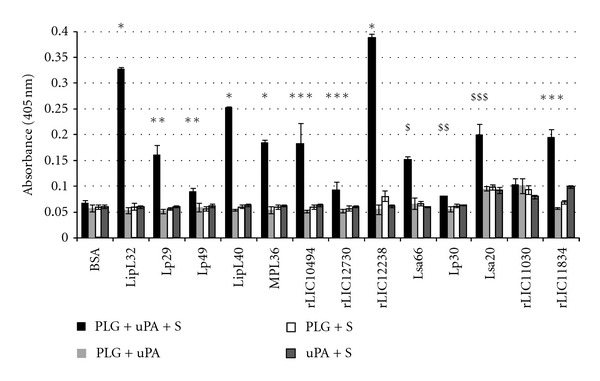
Activation of recombinant protein-PLG to enzymatically active plasmin. Cleavage of specific plasmin substrate by PLG bound to recombinant proteins was assayed by modified ELISA as immobilized proteins received the following treatment: PLG + uPA + specific plasmin substrate (PLG + uPA + S) or controls lacking one of the three components (PLG + uPA; PLG + S; uPA + S). BSA was employed as a negative control. Bars represent the mean absorbance values at 405 nm, as a measure of relative substrate cleavage, ± the standard deviation of four replicates for each experimental group and are representative of two independent experiments. Statistically significant differences are shown by *(P < 0.00001), **(P < 0.005), ***(P < 0.05), $(P < 0.001), $$(P < 0.02), and $$$(P < 0.003).
3.8. Characterization of the Binding of Recombinant Proteins to PLG
The binding between each recombinant protein and PLG was assessed on a quantitative basis as illustrated in Figures 8(a), 8(b), and 8(c). Dose-dependent and saturable binding was observed when increasing concentrations (0 to 1,000 nM) of the recombinant proteins MPL36, LipL40, LipL32, rLIC10494, rLIC12238, Lsa66, Lp30, Lsa33 (rLIC11834), and Lsa20 was allowed to individually adhere to a fixed amount of PLG (1 μg), indicating the specificity of the binding. For the proteins Lp29, Lp49, rLIC12730, Lsa25 (rLIC12253), and rLIC11030, the saturation level was not reached, even at the highest concentration tested (1,000 nM). Based on the ELISA data, the calculated dissociation equilibrium constants (KD) for the recombinant proteins with PLG are depicted in Figure 8(d); for the ones that reached equilibrium, the highest and the lowest KD values were for Lsa20 (509.13 ± 77.47 nM) and MPL36 (3.52 ± 3.95 nM), respectively, [37–40].
Figure 8.
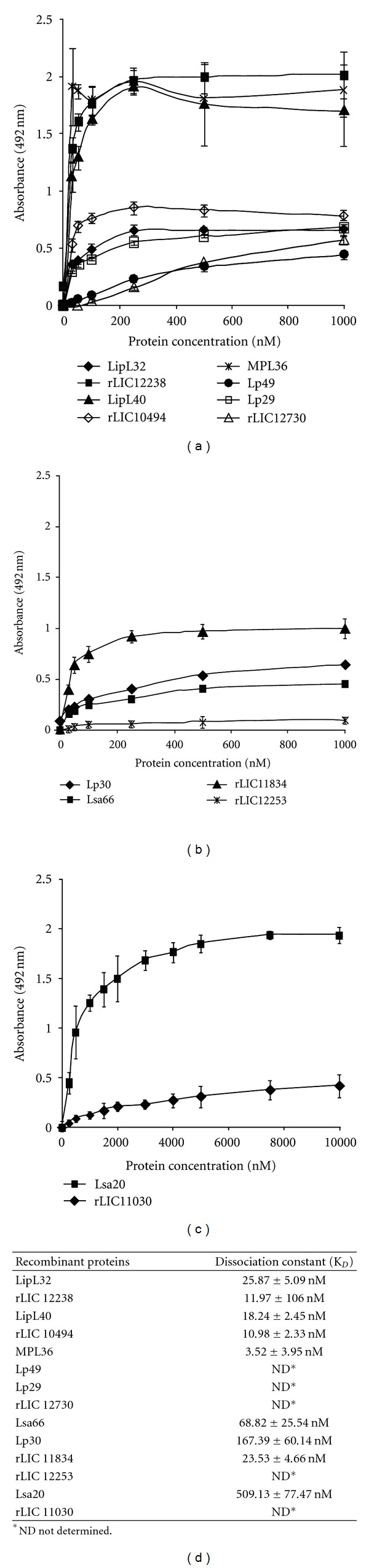
Characterization of recombinant proteins binding to PLG. In (a),(b), and (c) PLG (10 μg/mL) was immobilized in 96-well ELISA plates, and each recombinant protein at 0 to 1,000 nM was added for interaction. The binding was detected using antiserum raised in mice against each protein at appropriate dilutions (1 : 4,000 for LipL32; 1 : 5,000 for rLIC12238, LipL40, and MPL36; 1 : 1,000 for Lp29, Lp49, Lsa20, and rLIC11030; 1 : 500 for rLIC12730, Lsa66, and Lp30; 1 : 750 for rLIC11834 and rLIC12253), followed byhorseradish peroxidase-conjugated anti-mouse IgG. Data represent the mean absorbance values ± the standard deviation of six replicates for each experimental group. The results are representative of two independent experiments. In (d) The dissociation constant (KD) was calculated based on ELISA data for the recombinant proteins that reached equilibrium up to a concentration of 1,000 nM.
3.9. Proteomic Approach to Leptospiral PLG Binding Protein Identification
In order expand our knowledge on PLG binding proteins, we decided to employ two-dimensional gel electrophoresis followed by affinity immunoblotting, as an alternative strategy to identify novel leptospiral PLG-binding proteins. Total protein extracts of low passage L. interrogans were separated by isoelectric point and molecular mass (Figure 9(a)), transferred to membranes, and incubated with PLG and anti-PLG antibodies for detection of reactive spots (Figure 9(b)). The reactive spots were excised and prepared for MALDI-TOF mass spectrometry and protein identification. Under the conditions assayed, we could identify seven proteins capable of binding PLG, as depicted in Table 2. Two were annotated as membrane and five as cytoplasmatic proteins (Table 2). Along with the membrane proteins, one is hypothetical and the other is a putative flagellar filament sheath protein (FlaA-1); the cytoplasmic proteins are citrate lyase, Tuf (elongation factor), acetyl-CoA acetyltransferase, glutamine synthetase, and the heat shock protein DnaK. While the role of membrane proteins on the PLG sequestering within the host is expected, the meaning of cytoplasmatic proteins remains to be investigated. Intriguingly, none of the PLG-binding recombinant proteins was detected in these experiments. One possible explanation is that the identified proteins, except LIC12816, are expressed in high amounts per cell, while the proteins assayed in our laboratory were below the detection limit, as reported by quantitative proteomics [67]. However, LipL32 that was shown to have the highest expression level per leptospiral cell, was not detected in our proteomics studies. Although the reason for this is unknown, we might speculate that because LipL32 is always presented as isoforms in 2D gels [53, 68, 69], it is possible that these isoforms do not bind PLG. Several cytoplasmatic proteins of other pathogenic microrganisms have been identified as PLG binding proteins, such as Bifidobacterium animalis DnaK [70], Paracoccidioides brasiliensis, Schistosoma bovis and Streptcoccus pneumoniae enolases [71] and Streptococcus pneumoniae GAPDH [72]. For some of these proteins, there is the possibility that at some moment they could be exposed in the bacterial outer membrane. Indeed, it has been recently reported that B. burgdorferi enolase, which is both a cytoplasmic and membrane-associated protein, is also a PLG-binding protein [73]. It is also possible that the PLG binding to these cytoplasmic proteins are not relevant for the bacteria.
Figure 9.
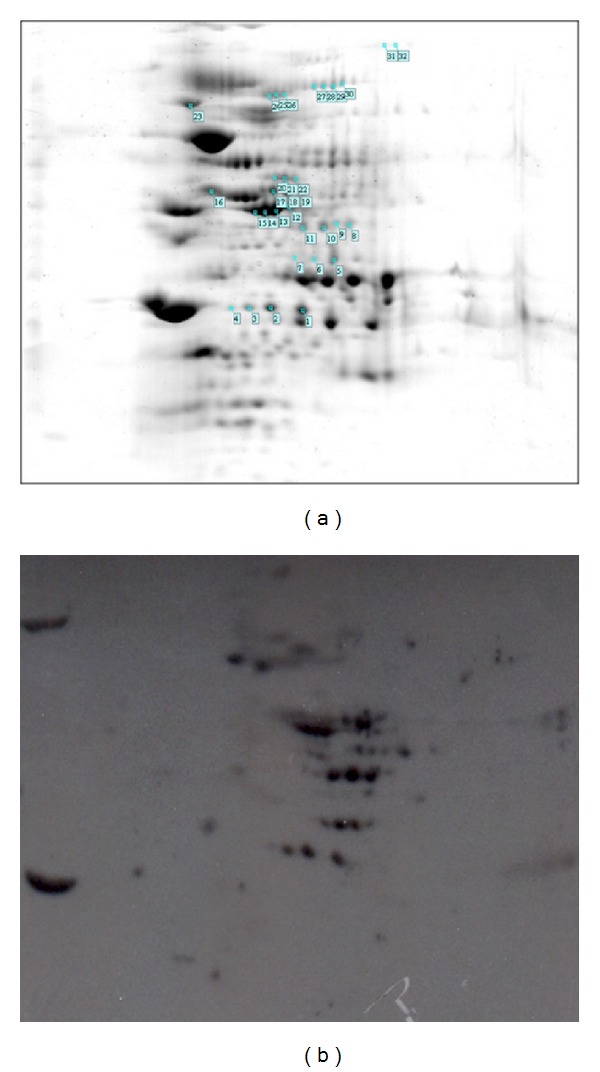
Two-dimensional gel analysis of L. interrogans PLG binding proteins. Leptospiral total cell lysates were separated by isoelectric point (pH 3–10) and molecular mass on 12% SDS-PAGE. Replicate gels were Coomassie Blue stained (a) or transferred to membranes and submitted to PLG affinity immunoblotting (b). The reactive spots are enumerated on (a). The results are representative of three independent experiments performed in duplicates.
Table 2.
L. interrogans PLG-binding proteins identified by MALDI-TOF mass spectrometry. The proteins are identified as the genomic nomenclature (LIC: Leptospira interrogans serovar Copenhageni), and are listed and respective NCBI reference sequence, predicted name/function and predicted cellular localization, in theoric molecular mass and isoelectric point (pI).
| Gene locus1 | NCBI reference | Gene/protein function | Localization3 | Theoric molecular mass (kDa) | pI |
|---|---|---|---|---|---|
| sequence number2 | |||||
| LIC12816 | YP_002733 | Hypothetical protein | Membrane | 14.7 | 9.5 |
| LIC10788 | YP_000767 | flaA-1 (Flagellar filament sheath protein) | Membrane | 35.0 | 7.6 |
| LIC11194 | YP_001164 | Putative citrate lyase | Cytoplasm | 37.2 | 6.1 |
| LIC12875 | YP_002791 | tuf(Elongation factor Tu) | Cytoplasm | 43.6 | 5.7 |
| LIC12795 | YP_002712 | Acetyl-CoA acetyltransferase | Cytoplasm | 47.4 | 6.0 |
| LIC12407 | YP_002339 | Glutamine synthetase protein | Cytoplasm | 55.4 | 5.9 |
| LIC10524 | YP_000508 | dnaK (Heat shock protein) | Cytoplasm | 69.1 | 5.1 |
3Lipo P: Juncker et al., [49].
3.10. Human IgG and C3b Deposition on Leptospires
Plasmin-generation on surface of L. interrogans may facilitate the bacteria to degrade opsonizing IgG and C3b molecules, and therefore contribute to immune evasion. To test this hypothesis, microtiter plates coated with virulent L. interrogans serovar Pomona were incubated with NHS, as a source of C3b and nonspecific IgG molecules. Bacteria were then treated with PLG and uPA, and the resulting surface-associated opsonins were quantified by specific antibodies. The results show that the incubation of bacteria with PLG and uPA resulted in a decrease in IgG Fc region (Figure 10(a)) and C3b depositions (Figure 10(b)) at the leptospiral surface. No effect on IgG or C3b binding to L. interrogans was observed with only PLG or uPA [41].
Figure 10.
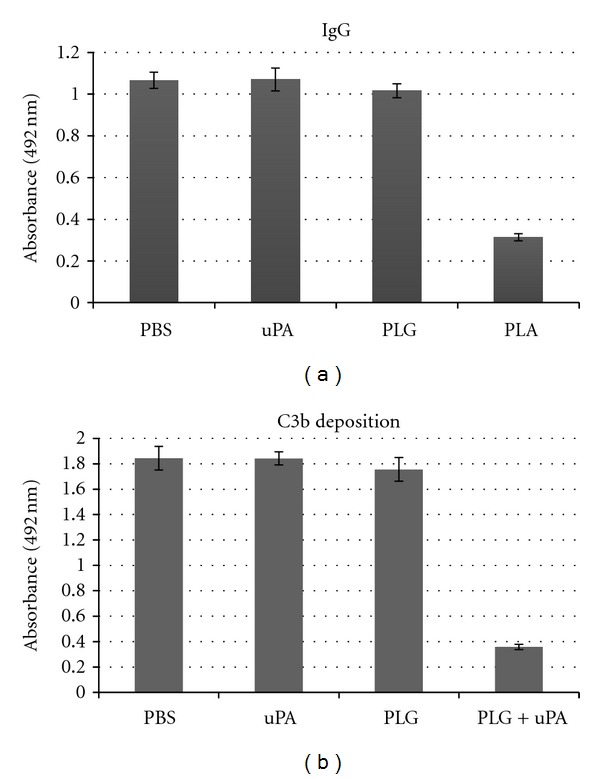
Human IgG and C3b binding to L. interrogans. Bacteria were coated onto microtiter plates (25 × 106 leptospires/well) and IgG (a) or C3b (b)were bound by incubating bacteria with 100% NHS. Bacteria were treated with PBS, 40 μg/mL PLG (PLG), 5 μg/mL uPA (uPA), or PLG + uPA (Pla). Presence of human IgG or C3b deposited on leptospires was determined by ELISA. Bars represent the mean absorbance values at 492 nm ± the standard deviation of three replicates for each experimental group and are representative of two independent experiments.
3.11. IgG Binding from Human Leptospirosis Immune Serum to L. Interrogans
To evaluate whether PLA activity observed against nonspecific human IgG bound to leptospires could also be functional to immune sera from patients diagnosed with leptospirosis, we performed the same experiment using MAT negative (preimmune phase) and MAT positive (convalescent immune phase) pools of human sera. As expected, MAT positive sera resulted in significant higher IgG depositions on bacterial surface when compared to NHS and MAT negative sera (Figure 11). When PLA was generated on leptospires, a noticeable decrease in the amount of IgG deposition from both negative and positive MAT sera was detected [41].
Figure 11.
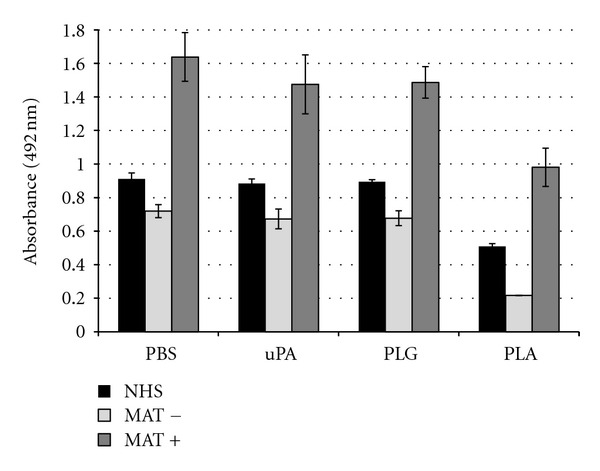
IgG binding from human leptospirosis immune serum to L. interrogans. Bacteria were coated onto microtiter plates (25 × 106 leptospires/well) and IgG was bound by incubating bacteria 50% nonimmune human sera (NHS) or confirmed paired leptospirosis patients sera: MAT negative (MAT−) or MAT positive (MAT+). Bacteria were treated with PBS, 40 μg/mL PLG (PLG), 5 μg/mL uPA (uPA), or PLG + uPA (Pla). Presence of human IgG deposited on leptospires was determined by ELISA using Fc-specific anti-human IgG antibodies. Bars represent the mean absorbance values at 492 nm ± the standard deviation of three replicates for each experimental group and are representative of two independent experiments.
Opsonization obstruction by antibodies or complement system appears to be an interesting approach for the bacteria to evade the first line of human immune defense upon infection. In fact, the expression of proteins on the bacterial surface that sequester host complement regulators FH, FHL-1, or C4BP and therefore the inhibition of the complement cascade by the alternative or classical pathways has been employed by several pathogens [74–77]. Moreover, the proteolytic cleavage of antibodies that prevents the activation of complement classical pathway or Fc receptor-mediated phagocytosis, is another well-known strategy employed by pathogenic bacteria [74–77].
3.12. Human C3b Degradation by Leptospires
Meri and colleagues [54] reported the capability of Leptospira-bound factor H (FH) to act as a cofactor for the serum protease factor I in cleaving C3b. Thus, we decided to assess whether the decrease in C3b deposition by Leptospira-generated PLA was due to C3b cleavage. L. interrogans serovar Pomona were coated with PLA by treatment with PLG and uPA, and then incubated with human purified C3b in fluid-phase. The resulting supernatant together with anti-C3b-specific antibodies was analyzed by Western blotting (Figure 12). The data revealed the appearance of two-lower molecular mass bands of estimated 60 and 20 kDa, concurrently with the decrease in the masses of the alpha (105 kDa) and beta (75 kDa) C3b chains that took place only with PLA-coated leptospires. Untreated or leptospires only treated with PLG showed no gradation. The results unmistakably show that the human C3b is cleaved by leptospires-coated PLA, indicating that the reduction in deposition is most probably due to degradation [41].
Figure 12.
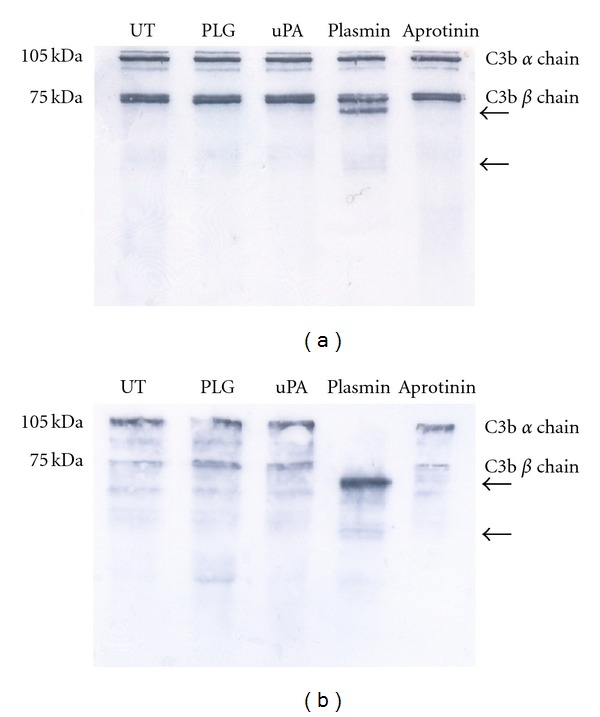
Detection of human C3b. Leptospires (108/sample) were treated with 10 μg PLG and 3 U uPA (Plasmin), only with PLG (PLG), or left untreated (UT). The cells were incubated with 15 μg/mL human purified C3b for 20 h at 37°C, and the supernatants were resolved by SDS-PAGE and transferred to membranes. The C3b was detected by specific antibodies followed by ECL reagent development and exposition to X-Ray films. There are indicated the native human C3b alpha chain (105 KDa) and beta-chain (75 KDa), as well as the degradation products (arrows).
3.13. Serum Susceptibility Testing for Treated Leptospires
It has been described that L. interrogans serovar Pomona is partially susceptible to complement killing upon incubation with normal human sera [54]. To evaluate whether the PLG/PLA binding to leptospires confers protection on exposure to NHS, leptospires were treated with PLG+uPA (Pla) or with PBS, as negative control, and incubated for one hour with pooled NHS. The bacteria were harvested and counted in a dark-field microscopy using Petroff-Hausser chamber, at the fourth day of incubation. The number of untreated leptospires was almost four times less when compared to the plasmin-coated leptospires (0.35 × 108 ± 0.1 × 108 versus 1.28 × 108 ± 0.3 × 108 leptospires/mL), suggesting that the leptospires endowed with active plasmin are less vulnerable to components present in NHS [41].
4. Conclusions
The interaction of the human PLG system has been suggested to be a feature that significantly contributes to the virulence of many bacterial pathogens by facilitating the initial anchoring to endothelium and penetration [27]. Our group has been studying the interaction of Leptospira with PLG/plasmin generation system and the possible implications for pathogenesis. We demonstrated that leptospires interact with PLG and can acquire plasmin activity associated to the surface with no apparent damage to the surface, what occurs through multiple proteins. Although nonvirulent leptospires can interact with PLG, there seems to be a correlation between the efficiency of PLG capturing and virulence, suggesting a role in virulence and infection. Moreover, the generation of enzymatically active PLA on the leptospiral surface decreased C3b and IgG depositions that may constitute a novel mechanism by which leptospires could evade the immune system until reach immunologically safe environments. This latter assertion will also have implication to the bacterial persistence within the host. We believe that the disclosure of this enzymatically system will shed light on the molecular mechanism of leptospiral pathogenesis.
Acknowledgments
The authors are deeply indebted to Alexsander Seixas de Souza (Departamento de Parasitologia, Instituto Butantan, Sao Paulo, Brazil) for the use of the confocalmicroscope facilities and the helpful discussion. This work was supported by FAPESP, CNPq, and Fundação Butantan, Brazil; M.L.Vieira; M.V.Atzingen; R.Oliveira; R.S.Mendes; R.F.Domingos have scholarships from FAPESP (Brazil).
References
- 1.Faine S, Adler B, Bolin C, Perolat P. Leptospira and Leptospirosis. 2nd edition. Melbourne, Australia: MediSci; 1999. [Google Scholar]
- 2.Levett PN. Leptospirosis. Clinical Microbiology Reviews. 2001;14(2):296–326. doi: 10.1128/CMR.14.2.296-326.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vinetz JM. Leptospirosis. Current Opinion in Infectious Diseases. 2001;14(5):527–538. doi: 10.1097/00001432-200110000-00005. [DOI] [PubMed] [Google Scholar]
- 4.Bharti AR, Nally JE, Ricaldi JN, et al. Leptospirosis: a zoonotic disease of global importance. Lancet Infectious Diseases. 2003;3(12):757–771. doi: 10.1016/s1473-3099(03)00830-2. [DOI] [PubMed] [Google Scholar]
- 5.Ko AI, Galvão Reis M, Ribeiro Dourado CM, Johnson WD, Riley LW. Urban epidemic of severe leptospirosis in Brazil. Lancet. 1999;354(9181):820–825. doi: 10.1016/s0140-6736(99)80012-9. [DOI] [PubMed] [Google Scholar]
- 6.Haake DA, Dundoo M, Cader R, et al. Leptospirosis, water sports, and chemoprophylaxis. Clinical Infectious Diseases. 2002;34(9):e40–43. doi: 10.1086/339942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Plank R, Dean D. Overview of the epidemiology, microbiology, and pathogenesis of Leptospira spp. in humans. Microbes and Infection. 2000;2(10):1265–1276. doi: 10.1016/s1286-4579(00)01280-6. [DOI] [PubMed] [Google Scholar]
- 8.Ren SX, Fu G, Jiang XG, et al. Unique physiological and pathogenic features of Leptospira interrogans revealed by whole-genome sequencing. Nature. 2003;422(6934):888–893. doi: 10.1038/nature01597. [DOI] [PubMed] [Google Scholar]
- 9.Nascimento ALTO, Verjovski-Almeida S, Van Sluys MA, et al. Genome features of Leptospira interrogans serovar Copenhageni. Brazilian Journal of Medical and Biological Research. 2004;37(4):459–478. doi: 10.1590/s0100-879x2004000400003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nascimento ALTO, Ko AI, Martins EAL, et al. Comparative Genomics of two Leptospira interrogans serovars reveals novel insights into physiology and pathogenesis. Journal of Bacteriology. 2004;186(7):2164–2172. doi: 10.1128/JB.186.7.2164-2172.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bulach DM, Zuerner RL, Wilson P, et al. Genome reduction in Leptospira borgpetersenii reflects limited transmission potential. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(39):14560–14565. doi: 10.1073/pnas.0603979103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Picardeau M, Bulach DM, Bouchier C, et al. Genome sequence of the saprophyte Leptospira biflexa provides insights into the evolution of Leptospira and the pathogenesis of leptospirosis. PLoS One. 2008;3(2) doi: 10.1371/journal.pone.0001607.e1607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matsunaga J, Barocchi MA, Croda J, et al. Pathogenic Leptospira species express surface-exposed proteins belonging to the bacterial immunoglobulin superfamily. Molecular Microbiology. 2003;49(4):929–945. doi: 10.1046/j.1365-2958.2003.03619.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ristow P, Bourhy P, da Cruz McBride FW, et al. The OmpA-like protein Loa22 is essential for Leptospiral virulence. PLoS pathogens. 2007;3(7, article e97) doi: 10.1371/journal.ppat.0030097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Atzingen MV, Barbosa AS, De Brito T, et al. Lsa21, a novel leptospiral protein binding adhesive matrix molecules and present during human infection. BMC Microbiology. 2008;8, article no. 70 doi: 10.1186/1471-2180-8-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luo Y, Liu Y, Sun D, et al. InvA protein is a Nudix hydrolase required for infection by pathogenic Leptospira in cell lines and animals. The Journal of Biological Chemistry. 2011;286(42):36852–36863. doi: 10.1074/jbc.M111.219931. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 17.Boyle EC, finlay BB. Bacterial pathogenesis: exploiting cellular adherence. Current Opinion in Cell Biology. 2003;15(5):633–639. doi: 10.1016/s0955-0674(03)00099-1. [DOI] [PubMed] [Google Scholar]
- 18.Barbosa AS, Abreu PAE, Neves FO, et al. A newly identified leptospiral adhesin mediates attachment to laminin. Infection and Immunity. 2006;74(11):6356–6364. doi: 10.1128/IAI.00460-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Longhi MT, Oliveira TR, Romero EC, et al. A newly identified protein of Leptospira interrogans mediates binding to laminin. Journal of Medical Microbiology. 2009;58(10):1275–1282. doi: 10.1099/jmm.0.011916-0. [DOI] [PubMed] [Google Scholar]
- 20.Choy HA, Kelley MM, Chen TL, Møller AK, Matsunaga J, Haake DA. Physiological osmotic induction of Leptospira interrogans adhesion: LigA and LigB bind extracellular matrix proteins and fibrinogen. Infection and Immunity. 2007;75(5):2441–2450. doi: 10.1128/IAI.01635-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stevenson B, Choy HA, Pinne M, et al. Leptospira interrogans endostatin-like outer membrane proteins bind host fibronectin, laminin and regulators of complement. PLoS One. 2007;2(11) doi: 10.1371/journal.pone.0001188.e1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hauk P, Macedo F, Romero EC, et al. In LipL32, the major leptospiral lipoprotein, the C terminus is the primary immunogenic domain and mediates interaction with collagen IV and plasma fibronectin. Infection and Immunity. 2008;76(6):2642–2650. doi: 10.1128/IAI.01639-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoke DE, Egan S, Cullen PA, Adler B. LipL32 is an extracellular matrix-interacting protein of Leptospira spp. and Pseudoalteromonas tunicata. Infection and Immunity. 2008;76(5):2063–2069. doi: 10.1128/IAI.01643-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Atzingen MV, Gómez RM, Schattner M, et al. Lp95, a novel leptospiral protein that binds extracellular matrix components and activates e-selectin on endothelial cells. Journal of Infection. 2009;59(4):264–276. doi: 10.1016/j.jinf.2009.07.010. [DOI] [PubMed] [Google Scholar]
- 25.Carvalho E, Barbosa AS, Gómez RM, et al. Leptospiral TlyC is an extracellular matrix-binding protein and does not present hemolysin activity. FEBS Letters. 2009;583(8):1381–1385. doi: 10.1016/j.febslet.2009.03.050. [DOI] [PubMed] [Google Scholar]
- 26.Vieira ML, de Morais ZM, Gonçales AP, Romero EC, Vasconcellos SA, Nascimento ALTO. Lsa63, a newly identified surface protein of Leptospira interrogans binds laminin and collagen IV. Journal of Infection. 2010;60(1):52–64. doi: 10.1016/j.jinf.2009.10.047. [DOI] [PubMed] [Google Scholar]
- 27.Lähteenmäki K, Kuusela P, Korhonen TK. Bacterial plasminogen activators and receptors. FEMS Microbiology Reviews. 2001;25(5):531–552. doi: 10.1111/j.1574-6976.2001.tb00590.x. [DOI] [PubMed] [Google Scholar]
- 28.Coleman JL, Sellati TJ, Testa JE, Kew RR, Furie MB, Benach JL. Borrelia burgdorferi binds plasminogen, resulting in enhanced penetration of endothelial monolayers. Infection and Immunity. 1995;63(7):2478–2484. doi: 10.1128/iai.63.7.2478-2484.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klempner MS, Noring R, Epstein MP, et al. Binding of human plasminogen and urokinase-type plasminogen activator to the Lyme disease spirochete, Borrelia burgdorferi. Journal of Infectious Diseases. 1995;171(5):1258–1265. doi: 10.1093/infdis/171.5.1258. [DOI] [PubMed] [Google Scholar]
- 30.Hu LT, Perides G, Noring R, Klempner MS. Binding of human plasminogen to Borrelia burgdorferi. Infection and Immunity. 1995;63(9):3491–3496. doi: 10.1128/iai.63.9.3491-3496.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Coleman JL, Gebbia JA, Piesman J, Degen JL, Bugge TH, Benach JL. Plasminogen is required for efficient dissemination of B. burgdorferi in ticks and for enhancement of spirochetemia in mice. Cell. 1997;89(7):1111–1119. doi: 10.1016/s0092-8674(00)80298-6. [DOI] [PubMed] [Google Scholar]
- 32.Coleman JL, Benach JL. Use of the plasminogen activation system by microorganisms. Journal of Laboratory and Clinical Medicine. 1999;134(6):567–576. doi: 10.1016/s0022-2143(99)90095-1. [DOI] [PubMed] [Google Scholar]
- 33.Coleman JL, Benach JL. The generation of enzymatically active plasmin on the surface of spirochetes. Methods. 2000;21(2):133–141. doi: 10.1006/meth.2000.0984. [DOI] [PubMed] [Google Scholar]
- 34.Nordstrand A, Shamaei-Tousi A, Ny A, Bergström S. Delayed invasion of the kidney and brain by Borrelia crocidurae in plasminogen-deficient mice. Infection and Immunity. 2001;69(9):5832–5839. doi: 10.1128/IAI.69.9.5832-5839.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Haile WB, Coleman JL, Benach JL. Reciprocal upregulation of urokinase plasminogen activator and its inhibitor, PAI-2, by Borrelia burgdorferi affects bacterial penetration and host-inflammatory response. Cellular Microbiology. 2006;8(8):1349–1360. doi: 10.1111/j.1462-5822.2006.00717.x. [DOI] [PubMed] [Google Scholar]
- 36.Vieira ML, Vasconcellos SA, Gonçales AP, De Morais ZM, Nascimento ALTO. Plasminogen acquisition and activation at the surface of Leptospira species lead to fibronectin degradation. Infection and Immunity. 2009;77(9):4092–4101. doi: 10.1128/IAI.00353-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vieira ML, Atzingen MV, Oliveira TR, et al. In vitro identification of novel plasminogen-binding receptors of the pathogen Leptospira interrogans. PLoS One. 2010;5(6):p. e11259. doi: 10.1371/journal.pone.0011259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oliveira R, de Morais ZM, Gonçales AP, Romero EC, Vasconcellos SA, Nascimento ALTO. Characterization of novel OmpA-like protein of Leptospira interrogans that binds extracellular matrix molecules and plasminogen. PLoS One. 2011;6(7) doi: 10.1371/journal.pone.0021962.e21962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mendes RS, Von Atzingen M, de Morais ZM, et al. The novel leptospiral surface adhesin Lsa20 binds laminin and human plasminogen and is probably expressed during infection. Infection and Immunity. 2011;79(11):4657–4667. doi: 10.1128/IAI.05583-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Domingos RF, Vieira ML, Romero EC, et al. Features of two proteins of Leptospira interrogans with potential role in host-pathogen interactions. BMC Microbiology. 2012;12(article 50) doi: 10.1186/1471-2180-12-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vieira ML, de Morais ZM, Vasconcellos SA, Romero EC, Nascimento ALTO. In vitro evidence for immune evasion activity by human plasmin associated to pathogenic Leptospira interrogans. Microbial Pathogenesis. 2011;51(5):360–365. doi: 10.1016/j.micpath.2011.06.008. [DOI] [PubMed] [Google Scholar]
- 42.Turner LH. Leptospirosis III. Maintenance, isolation and demonstration of leptospires. Transactions of the Royal Society of Tropical Medicine and Hygiene. 1970;64(4):623–646. doi: 10.1016/0035-9203(70)90087-8. [DOI] [PubMed] [Google Scholar]
- 43.Coleman JL, Roemer EJ, Benach JL. Plasmin-coated Borrelia burgdorferi degrades soluble and insoluble components of the mammalian extracellular matrix. Infection and Immunity. 1999;67(8):3929–3936. doi: 10.1128/iai.67.8.3929-3936.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nakai K, Horton P. PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends in Biochemical Sciences. 1999;24(1):34–35. doi: 10.1016/s0968-0004(98)01336-x. [DOI] [PubMed] [Google Scholar]
- 45.Nakai K, Kanehisa M. Expert system for predicting protein localization sites in gram-negative bacteria. Proteins: Structure, Function and Genetics. 1991;11(2):95–110. doi: 10.1002/prot.340110203. [DOI] [PubMed] [Google Scholar]
- 46.Letunic I, Copley RR, Pils B, Pinkert S, Schultz J, Bork P. SMART 5: domains in the context of genomes and networks. Nucleic Acids Research. 2006;34:D257–260. doi: 10.1093/nar/gkj079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schultz J, Milpetz F, Bork P, Ponting CP. SMART, a simple modular architecture research tool: identification of signaling domains. Proceedings of the National Academy of Sciences of the United States of America. 1998;95(11):5857–5864. doi: 10.1073/pnas.95.11.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Finn RD, Mistry J, Schuster-Böckler B, et al. Pfam: clans, web tools and services. Nucleic Acids Research. 2006;34:D247–251. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Juncker AS, Willenbrock H, Von Heijne G, Brunak S, Nielsen H, Krogh A. Prediction of lipoprotein signal peptides in Gram-negative bacteria. Protein Science. 2003;12(8):1652–1662. doi: 10.1110/ps.0303703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brissette CA, Haupt K, Barthel D, et al. Borrelia burgdorferi infection-associated surface proteins ErpP, ErpA, and ErpC bind human plasminogen. Infection and Immunity. 2009;77(1):300–306. doi: 10.1128/IAI.01133-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pathirana RD, O’Brien-Simpson NM, Veith PD, Riley PF, Reynolds EC. Characterization of proteinase-adhesin complexes of Porphyromonas gingivalis. Microbiology. 2006;152(8):2381–2394. doi: 10.1099/mic.0.28787-0. [DOI] [PubMed] [Google Scholar]
- 52.Lin YP, Lee DW, McDonough SP, Nicholson LK, Sharma Y, Chang YF. Repeated domains of Leptospira immunoglobulin-like proteins interact with elastin and tropoelastin. Journal of Biological Chemistry. 2009;284(29):19380–19391. doi: 10.1074/jbc.M109.004531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vieira ML, Pimenta DC, de Morais ZM, Vasconcellos SA, Nascimento AL. Proteome analysis of Leptospira interrogans virulent strain. The Open Microbiology Journal. 2009;3: 69–74. doi: 10.2174/1874285800903010069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Meri T, Murgia R, Stefanel P, Meri S, Cinco M. Regulation of complement activation at the C3-level by serum resistant leptospires. Microbial Pathogenesis. 2005;39(4):139–147. doi: 10.1016/j.micpath.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 55.Haake DA, Matsunaga J. Characterization of the leptospiral outer membrane and description of three novel leptospiral membrane proteins. Infection and Immunity. 2002;70(9):4936–4945. doi: 10.1128/IAI.70.9.4936-4945.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Haake DA, Chao G, Zuerner RL, et al. The leptospiral major outer membrane protein LipL32 is a lipoprotein expressed during mammalian infection. Infection and Immunity. 2000;68(4):2276–2285. doi: 10.1128/iai.68.4.2276-2285.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Young KC, Shi GY, Wu DH, et al. Plasminogen activation by streptokinase via a unique mechanism. Journal of Biological Chemistry. 1998;273(5):3110–3116. doi: 10.1074/jbc.273.5.3110. [DOI] [PubMed] [Google Scholar]
- 58.McCoy HE, Broder CC, Lottenberg R. Streptokinases produced by pathogenic group C streptococci demonstrate species-specific plasminogen activation. Journal of Infectious Diseases. 1991;164(3):515–521. doi: 10.1093/infdis/164.3.515. [DOI] [PubMed] [Google Scholar]
- 59.Lijnen HR, De Cock F, Van Hoef B, Schlott B, Collen D. Characterization of the interaction between plasminogen and staphylokinase. European Journal of Biochemistry. 1994;224(1):143–149. doi: 10.1111/j.1432-1033.1994.tb20005.x. [DOI] [PubMed] [Google Scholar]
- 60.Schlott B, Gührs KH, Hartmann M, Röcker A, Collen D. NH2-terminal structural motifs in staphylokinase required for plasminogen activation. Journal of Biological Chemistry. 1998;273(35):22346–22350. doi: 10.1074/jbc.273.35.22346. [DOI] [PubMed] [Google Scholar]
- 61.Okada K, Ueshima S, Takaishi T, Yuasa H, Fukao H, Matsuo O. Effects of fibrin and α2-Antiplasmin on plasminogen activation by staphylokinase. American Journal of Hematology. 1996;53(3):151–157. doi: 10.1002/(SICI)1096-8652(199611)53:3<151::AID-AJH1>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 62.Sodeinde OA, Subrahmanyam YVBK, Stark K, Quan T, Bao Y, Goguen JD. A surface protease and the invasive character of plague. Science. 1992;258(5084):1004–1007. doi: 10.1126/science.1439793. [DOI] [PubMed] [Google Scholar]
- 63.Leytus SP, Bowles LK, Konisky J, Mangel WF. Activation of plasminogen to plasmin by a protease associated with the outer membrane of Escherichia coli. Proceedings of the National Academy of Sciences of the United States of America. 1981;78(3 I):1485–1489. doi: 10.1073/pnas.78.3.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li Z, Ploplis VA, French EL, Boyle MDP. Interaction between group A streptococci and the plasmin(ogen) system promotes virulence in a mouse skin infection model. Journal of Infectious Diseases. 1999;179(4):907–914. doi: 10.1086/314654. [DOI] [PubMed] [Google Scholar]
- 65.Collen D, Tytgat G, Claeys H, Verstraete M, Wallén P. Metabolism of plasminogen in healthy subjects: effect of tranexamic acid. Journal of Clinical Investigation. 1972;51(6):1310–1318. doi: 10.1172/JCI106927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Barocchi MA, Ko AI, Galvão Reis M, McDonald KL, Riley LW. Rapid translocation of polarized MDCK cell monolayers by Leptospira interrogans, an invasive but nonintracellular pathogen. Infection and Immunity. 2002;70(12):6926–6932. doi: 10.1128/IAI.70.12.6926-6932.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Malmström J, Beck M, Schmidt A, Lange V, Deutsch EW, Aebersold R. Proteome-wide cellular protein concentrations of the human pathogen Leptospira interrogans. Nature. 2009;460(7256):762–765. doi: 10.1038/nature08184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cullen PA, Cordwell SJ, Bulach DM, Haake DA, Adler B. Global analysis of outer membrane proteins from Leptospira interrogans serovar Lai. Infection and Immunity. 2002;70(5):2311–2318. doi: 10.1128/IAI.70.5.2311-2318.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nally JE, Whitelegge JP, Bassilian S, Blanco DR, Lovett MA. Characterization of the outer membrane proteome of Leptospira interrogans expressed during acute lethal infection. Infection and Immunity. 2007;75(2):766–773. doi: 10.1128/IAI.00741-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Candela M, Centanni M, Fiori J, et al. DnaK from Bifidobacterium animalis subsp. lactis is a surface-exposed human plasminogen receptor upregulated in response to bile salts. Microbiology. 2010;156(6):1609–1618. doi: 10.1099/mic.0.038307-0. [DOI] [PubMed] [Google Scholar]
- 71.Nogueira SV, Fonseca FL, Rodrigues ML, et al. Paracoccidioides brasiliensis enolase is a surface protein that binds plasminogen and mediates interaction of yeast forms with host cells. Infection and Immunity. 2010;78(9):4040–4050. doi: 10.1128/IAI.00221-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lottenberg R, Broder CC, Boyle MD, Kain SJ, Schroeder BL, Curtiss 3rd R. Cloning, sequence analysis, and expression in Escherichia coli of a streptococcal plasmin receptor. Journal of Bacteriology. 1992;174(16):5204–5210. doi: 10.1128/jb.174.16.5204-5210.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Floden AM, Watt JA, Brissette CA. Borrelia burgdorferi enolase is a surface-exposed plasminogen binding protein. PLoS One. 2011;6(11) doi: 10.1371/journal.pone.0027502.e27502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Blom AM, Hallström T, Riesbeck K. Complement evasion strategies of pathogens-Acquisition of inhibitors and beyond. Molecular Immunology. 2009;46(14):2808–2817. doi: 10.1016/j.molimm.2009.04.025. [DOI] [PubMed] [Google Scholar]
- 75.Lambris JD, Ricklin D, Geisbrecht BV. Complement evasion by human pathogens. Nature Reviews Microbiology. 2008;6(2):132–142. doi: 10.1038/nrmicro1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zipfel PF, Würzner R, Skerka C. Complement evasion of pathogens: common strategies are shared by diverse organisms. Molecular Immunology. 2007;44(16):3850–3857. doi: 10.1016/j.molimm.2007.06.149. [DOI] [PubMed] [Google Scholar]
- 77.Rooijakkers SHM, van Strijp JAG. Bacterial complement evasion. Molecular Immunology. 2007;44(1–3):23–32. doi: 10.1016/j.molimm.2006.06.011. [DOI] [PubMed] [Google Scholar]
- 78.Gamberini M, Gómez RM, Atzingen MV, et al. Whole-genome analysis of Leptospira interrogans to identify potential vaccine candidates against leptospirosis. FEMS Microbiology Letters. 2005;244(2):305–313. doi: 10.1016/j.femsle.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 79.Gómez RM, Vieira ML, Schattner M, et al. Putative outer membrane proteins of Leptospira interrogans stimulate human umbilical vein endothelial cells (HUVECS) and express during infection. Microbial Pathogenesis. 2008;45(5-6):315–322. doi: 10.1016/j.micpath.2008.08.004. [DOI] [PubMed] [Google Scholar]
- 80.Neves FO, Abreu PAE, Vasconcellos SA, De Morais ZM, Romero EC, Nascimento ALTO. Identification of a novel potential antigen for early-phase serodiagnosis of leptospirosis. Archives of Microbiology. 2007;188(5):523–532. doi: 10.1007/s00203-007-0273-2. [DOI] [PubMed] [Google Scholar]
- 81.Oliveira TR, Longhi MT, De Morais ZM, et al. Evaluation of leptospiral recombinant antigens MPL17 and MPL21 for serological diagnosis of leptospirosis by enzyme-linked immunosorbent assays. Clinical and Vaccine Immunology. 2008;15(11):1715–1722. doi: 10.1128/CVI.00214-08. [DOI] [PMC free article] [PubMed] [Google Scholar]


