Abstract
We propose guidelines to evaluate the cumulative evidence of gene–environment (G × E) interactions in the causation of human cancer. Our approach has its roots in the HuGENet and IARC Monographs evaluation processes for genetic and environmental risk factors, respectively, and can be applied to common chronic diseases other than cancer. We first review issues of definitions of G × E interactions, discovery and modelling methods for G × E interactions, and issues in systematic reviews of evidence for G × E interactions, since these form the foundation for appraising the credibility of evidence in this contentious field. We then propose guidelines that include four steps: (i) score the strength of the evidence for main effects of the (a) environmental exposure and (b) genetic variant; (ii) establish a prior score category and decide on the pattern of interaction to be expected; (iii) score the strength of the evidence for interaction between the environmental exposure and the genetic variant; and (iv) examine the overall plausibility of interaction by combining the prior score and the strength of the evidence and interpret results. We finally apply the scheme to the interaction between NAT2 polymorphism and tobacco smoking in determining bladder cancer risk.
Keywords: Genetics, environment, interactions, evaluations
Introduction
With a few exceptions, cancer is a set of diseases with multifactorial aetiology. Causative factors operate at one or more steps of complex pathogenetic networks, which are only partially understood. Traditionally, the causes of cancer, which are operationalized as ‘risk factors’ in epidemiological studies, are distinguished as ‘environmental’ and ‘genetic’, and the interplay between these two categories is referred to as ‘gene–environment (G × E) interaction’. At first sight, ‘environmental risk factors’ include modifiable agents to which humans are exposed through a variety of routes, and ‘genetic risk factors’ include inherited variants that affect the probability of developing cancer. A closer look, however, reveals a more complex picture, including other important factors, such as reactive oxygen species and other endogenously formed molecules, which act through the same pathways as environmental factors and may play an important role in cancer development without being ‘environmental’ in a strict sense, and inherited epigenetic characteristics resulting from the influence of environmental factors on gene expression through methylation and other mechanisms.1
Because of the complexity of carcinogenesis, identifying the causal nature of associations observed between risk factors and cancer is not a straightforward process. Guidelines and recommendations have been developed to aid in the synthesis of our evolving knowledge on environmental causes of cancer. The best known are the rigorous IARC Monographs for assessing available evidence on environmental causes of cancer.2 The Human Genome Epidemiology Network3 has developed interim criteria for assessing the credibility of cumulative knowledge on gene–disease associations.4 Nevertheless, an evaluation framework has not been available so far to assess accumulating cumulative evidence on joint effects of genes and environments.
Evidence of a G × E interaction influencing cancer susceptibility is important because it should provide insights into cancer aetiology; help to identify human carcinogens and genetic variants; help explain distributions of disease in populations; clarify dose–response relationships between genetic or environmental factors and risk of disease; evaluate low levels of risk; dissect effects of complex mixtures with components differentially affected by various genes; identify population subgroups with greatest cancer susceptibility and potential to benefit from interventions; and provide clues to potentially effective cancer prevention, intervention and treatment strategies.
The purpose of this article is to propose criteria to evaluate the cumulative evidence of G × E interactions in the causation of human cancer. Our approach has its roots in the HuGENet and IARC evaluation processes for genetic and environmental risk factors, respectively, and can be applied to common chronic diseases other than cancer. Before proposing these criteria, we review issues of definitions of G × E interactions, discovery and modelling methods for G × E interactions, and issues in systematic reviews of evidence for G × E interactions, since these form the foundation for appraising the credibility of evidence in this contentious field.
Definitions
G × E interactions have been defined in many ways. In the Dictionary of Epidemiology,5 interaction is defined as: ‘1. The interdependent operation of two or more causes to produce, prevent or control an effect… 2. Differences in the effect measures for one factor at different levels of another factor. 3. The necessity for a product term in a linear model.’ These definitions clearly reflect multiple uses of the word interaction in epidemiology (see also refs6–8).
Ottman9 and Khoury et al.10 view G × E interactions as ‘a different effect of an environmental exposure on disease risk in persons with different genotypes’ or equivalently ‘a different effect of a genotype on disease risk in persons with different environmental exposures’.3
To describe G × E interactions, Haldane11 identified the three possible combinations of the four relative risks (RRs) in a 2 × 2 table based on a dichotomous exposure to an environmental factor (E) and a dichotomous status with respect to a genotype (G), in which the group of ‘non-susceptible’ and ‘unexposed’ (G–E−) is chosen as referent category, and both E and G either are neutral or increase the risk of disease. Plus or minus signs after ‘E’ or ‘G’ throughout this article denote the exposed (+) or unexposed (−) group and the group with the ‘risk’ allele(s), ‘+’, or those without, ‘−’, respectively. In combination A, those exposed to both risk factors are at greater risk than for either factor alone; in combination B, those exposed to both are at lower risk than for either alone (but still greater than the reference); and in combination C, the direction of the effect of one factor depends upon the other. Yang and Khoury12 and Ottman9 extended these classifications to include combinations D, in which there is an effect only when both E and G are present; and E, where there is no effect of one factor in the absence of the other, which however has an independent effect. These five patterns of interactions are illustrated in Figure 1 (RRs are on arbitrary scale). For patterns A, B, C and E, the data in Figure 1 illustrate examples in which the effect of E is greater than that of G; the same pattern of interaction would apply to a situation in which the RRs for E and G are reversed. A review of typology of interactions has been recently published.13
Figure 1.

Patterns of G × E interaction
Although some strong G × E interactions have been reported, it is considered that most cancers and other common diseases involve multiple genetic and environmental factors, each with relatively modest individual effects. Establishing the existence of and interpreting G × E interactions is difficult for many reasons, including, but not limited to, the selection of theoretical and statistical models and the ability to measure accurately both the G and E components. In the literature, G × E interactions are described and measured in highly variable ways.
Discovery and modelling methods for G × E interactions
Most reports of human G × E interactions in the literature have tended to be ‘empirical’ in nature, simply describing patterns of risk in various combinations of G and E factors and at most testing whether that pattern could be described in terms of some simple model involving only the main effects of the interacting factors or would require additional interaction terms. When main effects are considered, the presence of interaction is dependent on the scale used, e.g. additive or multiplicative on a scale of RRs, and issues regarding scale are complex.13,14 Such descriptive models have seldom gone beyond the consideration of pair-wise interactions between a single gene and a single environmental factor, although there have been some exceptions (e.g. a four-way interaction of smoking and well-done red meat with CYP1A2 and NAT2).15 Numerous exploratory data analysis and machine learning techniques, such as Classification and Regression Trees or Multivariate Adaptive Regression Splines,16 Multifactor Dimension Reduction,17,18 the Focused Interaction Testing Framework,19 logic regression,20 Neural Networks,21 Support Vector Machines,22 to name a few, are available for searching for higher-order interactions with no prior hypotheses.
Given the sample-size requirements for testing even a single G × E interaction specified a priori and the exponential growth in the number of possible comparisons as multiple factors, it is hardly surprising there are few, if any, replicated examples of higher-order interactions. Indeed, even the widely quoted four-way interaction mentioned above depended on only 12 cases and 2 controls in the highest risk stratum and did not cross-validate in multifactor dimension reduction.23 A more promising approach is to formally incorporate external information into the modelling process, using some form of Bayesian hierarchical modelling strategy.24,25 This generally entails treating coefficients from a conventional logistic regression model for the epidemiological study data (the logRRs for G and E main effects and their interactions) as random variables in a second-level regression model, using external information on characteristics of these variables as predictors. One of the first applications of such an approach was data on bladder cancer in relation to genes involved in metabolic activation and detoxification of carcinogens, oxidative stress, and DNA repair and their interactions with tobacco smoke and other exposures, using simple pathway indicator variables as the second-level ‘prior covariates’.26 Other applications to lung cancer,27 and melanoma,28 have relied on various other types of bioinformatic or genomic tools, functional assays, pathway ontologies or literature mining to provide prior covariate information.29,30
Stochastic Search Variable Selection,31 Bayes model averaging,32 Monte Carlo logic regression33 or Algorithm for Learning Pathway Structure34 can also be used to sift through alternative models containing subsets of the various main effects and interactions. Such methods can also be applied to genome-wide association studies (GWASs),35,36 including genome-wide interaction scans. Key to the future success of such approaches will be development of better ontologies to provide comprehensive integration of external knowledge, not only about genes, but also environmental factors, disease risks and toxicological information.37,38 The database being developed in the HuGE Navigator (http://www.hugenavigator.net) by the Human Genome Epidemiology Network39 is a valuable step in this direction.
In some areas, however, it has been possible to build physiologically based pharmacokinetic (PBPK) models and mechanistic models, notably in relation to the multistage theory of carcinogenesis. Population PBPK models40–42 typically involve mathematical modelling of a metabolic process through a system of differential equations with rates for each step that vary across individuals following some distribution whose parameters are to be estimated, so as to relate environmental exposures to some health outcomes. In principle, incorporating measurable determinants of inter-individual variation, such as genotypes at relevant loci or biomarker measurements of intermediate metabolite concentrations or enzyme activity levels, should be straightforward,43,44 but this has seldom been done. Among mechanistic models, the classic Armitage–Doll multistage model45 and the Moolgavkar–Knudson two-stage clonal-expansion model46 are best developed, with more recent work focusing on the role of genomic instability47 and bystander effects.48 These have generally been more focused on describing exposure-time–response relationships for environmental main effects than on G × E interactions, although in principle genetic modifiers could be incorporated into the modelling of the underlying event rates.
The biological interpretation of measured interactions has been debated.7,14,49 It is not always clear whether or not sensible biological mechanistic understanding can come from a particular statistical formula of interaction.13
Issues in systematic reviews focusing on G × E interaction
One of the main motivations for meta-analysis of data on G × E interactions is the realization that very large sample sizes will be necessary to confirm evidence of G × E joint effects50 especially in light of exposure measurement error and genotyping error inherent in these types of studies.51,52 Mechanisms to combine studies appropriately will be crucial in meeting these sample size requirements. Assessing the evidence for G × E interactions in a systematic review requires consideration and reporting of potential sources of bias, study design and other issues in the individual studies and across studies.
Publication bias and selective reporting
Challenges to integrating evidence include publication bias and selective reporting of studies.53,54 The large number of potential comparisons implicit in the concept of multiple interacting variables increases the potential for selective reporting. ‘Positive’ results may be inappropriately favoured for publication.
The proportion of articles on human genome epidemiology reporting G × E interactions has been fairly small, ∼14%.55 So far, most of them examine pair-wise joint effects between a single candidate gene and a single environmental factor. Some investigators have argued that a pre-specified hypothesis is more credible, whereas others consider the prior probability of the hypothesis, irrespective of when it is specified, as more important, and that replication will determine its credibility.3,56 In a review of the reporting of interaction in general medicine, epidemiology and clinical specialty journals between 2001 and 2007, 12% of articles that addressed interaction explicitly stated that examination of interaction was an objective of the study or that interaction analyses were pre-specified.3,56 Investigators are now integrating analyses of genome-wide variation and environmental factors, extending the ‘agnostic’ approach used for genetic associations in GWASs to gene–environment-wide interaction studies.55
We propose investigators present web supplement tables of data on G × E joint effects (i.e. primary analyses if the analysis is based on a priori hypotheses, ‘top’ hits if based on an agnostic approach); this would not limit the freedom of investigators to present findings according to their preference in the main paper. Furthermore, it would be helpful if exposure categorization could be done in a comparable way across studies.
Study design
The use of different study designs has implications for integration of evidence. Most studies of G × E interaction use the classical case–control design. Increasingly, nested case–control or case–cohort designs within prospective cohorts are being reported, which may be less vulnerable to selection and information biases.3,57 Case-only designs can also be used to test for departure from a multiplicative model for the joint effects of genetic and environmental factors in disease aetiology, but cannot assess marginal effects of genotype or exposure.12,58–61 The validity of such an approach depends on the independence of genotype and exposure in the population at risk.62,63 Various hybrid case-only and case–control approaches have been proposed to overcome the need for assuming independence of gene and environment effects in the population,64,65 as well as an extension to a two-step approach to genome-wide scans that considering G × E interactions is more powerful than a standard single-step approach or pre-filtering on the basis of main effects.66
The case–parent trio design can also test for departure from a multiplicative model for joint effects of genotype and environment67–69 and assess effects of maternal vs infant genotype and imprinting, but not the marginal effects of exposure. This approach has greatest potential for investigations of cancer aetiology in children and young people, but is rarely used. A similar two-step approach for genome-wide scans for G × E interactions based on this design has also been proposed.70
Selection bias
Morimoto et al.71 have shown that estimates of G × E interaction, defined as departure from a multiplicative joint effect, will not be subject to selection bias when genotype itself does not influence participation in the study. This applies even when selection is influenced by exposure and disease status and genotype may be associated with either or both of these. Similarly, in a hospital-based case–control study, Wacholder et al.72 noted even if exposure, or genotype, or both are associated with the condition leading to hospitalization of control subjects, a departure from multiplicative effects can be estimated without bias. Although including controls with more than one type of disease might reduce bias resulting from one disease being associated with exposure, genotype or both, pooling controls with different diseases can lead to bias in assessing departure from multiplicative interaction, even if there is no such interaction in each individual disease-specific control set.72
Information bias
An important challenge is that the case–control design is susceptible to misclassification of exposure,73 and, although differential misclassification may not bias estimation of departure from multiplicative joint effects, a small level of differential misclassification can have a marked effect on the estimation of the marginal effects of exposure.74
Exposure misclassification can bias estimation of interaction effects, the magnitude of which depends on the prevalence of the misclassified exposure and on the interaction model.75,76 If interaction is defined as lack of fit to a multiplicative model, the test for interaction will be conservative.14 However, if misclassification of exposure does not vary by genotype, differential misclassification between cases and controls may not be a serious problem.14 Simulation analyses also suggest that many regular tests for the hypothesis of no interaction maintain correct type I error rates in the presence of differential misclassification when there is no or a weak marginal effect of genotype.77 The impact of misclassification on departures from additive effects is more difficult to predict.75 In theory, case–control studies are more vulnerable to differential misclassification than cohort studies, thus considerable investments have been, and continue to be, made in developing prospective cohort studies and associated biobanks.51,78–81
Confounding
Whereas main genetic effects are not generally confounded, except by population stratification,82 this is not necessarily true of G × E joint effects. Any factor associated with the environmental component of a particular G × E interaction will itself demonstrate some evidence of interaction with the genotype in question.83 Consider, e.g. the interaction reported between alcohol intake and alcohol dehydrogenase (ADH1B) variation in relation to head and neck cancer (Figure 2).84,85 When stratified by alcohol status, the variant does not affect cancer risk among never drinkers, whereas substantial protection of the rare variant is seen among alcohol drinkers, in a dose–response fashion with the amount of alcohol consumed. Smoking is associated with alcohol consumption. Thus, when stratified by smoking status, there is an apparent interaction between smoking status and ADH1B variation, which is entirely driven by the association between smoking and alcohol consumption.
Figure 2.
Risk of upper aerodigestive cancer by ADH1B genetic variation, stratified by drinking intensity and smoking status. OR and 95% CI of upper aerodigestive cancer by re1229984 variant in ADH1B. Rare allele (dominant model) carriers vs common allele homozygous genotype. ORs are derived from fixed effects models and are standardized by age, sex, centre, cumulative alcohol consumption and tobacco smoking. Source: ref.84
Large-scale measurement platforms and large-scale studies
One challenge for new generation biobank studies is to combine massive measurements of both genetic and environmental variables across studies.86 Large-scale GWAS designs typically use a case–control or nested case–control study design.87 When exposure information is available, they can also be used to investigate G × E interactions.55 GWAS studies often involve pooling individual-level data from multiple studies. Investigators typically need to harmonize the data and compare exposure variables across studies in order to ensure variables across studies are measuring the same thing and can be combined. An example of a tool to help perform the harmonization of metadata from large biobanks and other studies is the Public Population Project in Genomics (P3G: www.p3g.org), which has established the Data Schema and Harmonization Platform for Epidemiological Research (DATAShaper).88 PhenX (www.phenx.org), Consensus Measures for Phenotypes and Exposures for use in Genome-wide Association Studies, is another initiative that provides a recommended minimal set of high priority measures for many research domains related to complex diseases and environmental measures.89 Adoption of these standardized measures in GWAS and other large-scale genomic research efforts will obviate the need for harmonizing variables across studies and reduce any measurement bias derived from harmonizing data from studies in which exposure variables were not identical.
Combining studies
Apart from assessing risk for bias of single studies, an important issue in combining studies is the use of different methods of exposure assessment and, in the situation when individual participant data are not available, its categorization. For both cohort and case–control studies, assessment of measurement error and correction for it in analysis enhance the rigour of combined studies.90
Further issues in combining data are the comparability of definition of phenotype, and for combination of information at the level of studies, the nature of the genetic model considered and analytical strategy. With regard to the type of analysis done, in assessment of papers addressing interaction of any type between 2001 and 2007, the most frequent reporting approach was the presentation of stratum-specific effect estimates.56 P values or statements regarding statistical significance were reported in over half of the articles, but this statistical test was often unclear. Only one-tenth of the studies reported individual effects of both exposures and their joint effect, and few studies reported product terms or a synergy index. Therefore, increased transparency of analysis and reporting is needed, as called for in the STROBE91 and STREGA92 statements.
Proposed guidelines for assessing cumulative evidence of G × E interactions in cancer
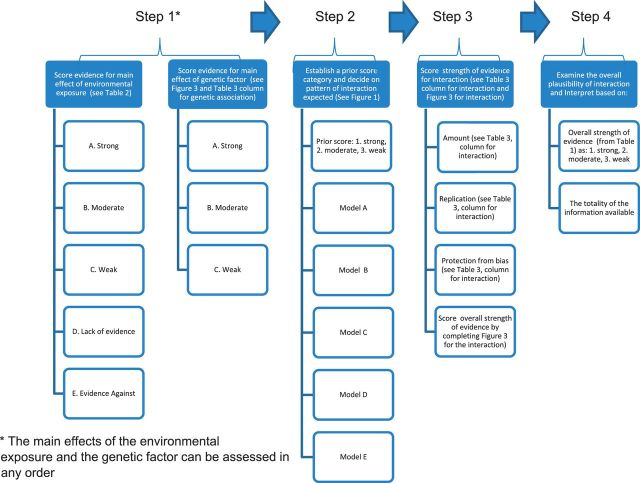
A schema for assessing cumulative evidence of G × E interactions should be based on explicit criteria and standardized procedures. The process of assessment should enable the reader to understand steps through which the final evaluation has been reached. The process should include the four steps shown in Figure 3:
Score the strength of the evidence for main effects of the (a) environmental exposure and (b) genetic variant;
Establish a prior score category and decide on the pattern of interaction to be expected;
Score the strength of the evidence for interaction between the environmental exposure and the genetic variant; and
Examine the overall plausibility of interaction by combining the prior score and the strength of the evidence and interpret results.
Figure 3.
Categories for the credibility of cumulative epidemiological evidence for genetic associations
For assessments (i)–(iii), a prerequisite is the systematic review of the respective evidence with caveats discussed above. The rationale here is that it is more likely to observe an interaction when the evidence is stronger for both the environmental exposure and the genetic variant, and conversely an interaction between an agent and a variant with only weak evidence of any effect on cancer risk. The criteria for combining the two lines of evidence are outlined in Table 1.
Table 1.
Score categories for an interaction between an environmental agent and a genetic variant based on the strength of evidence for a main effect of each of them (1 = strong, 2 = moderate, 3 = weak)
| Evidence for environmental main effect |
||||
|---|---|---|---|---|
| Evidence for genetic main effect | A (strong) | B (moderate) | C (weak) | D (lack); E (evidence against) |
| Strong | 1 | 2 | 2 | 3 |
| Moderate | 2 | 2 | 3 | 3 |
| Weak | 2 | 3 | 3 | 3 |
| Lack; evidence against | 3 | 3 | 3 | 3 |
The prior score (from Table 1) is defined as the likelihood on a scale of 1–3 (with 1 being the most likely) that a G × E interaction can be expected. When the evidence is strong for both E and G, and such evidence that both operate through the same causal pathway, there would be a strong rationale for expecting some interaction (prior score category 1). A moderate probability for an interaction (prior score category 2) can result from interactions of (i) established environmental carcinogens and genotypes with less than strong evidence, (ii) genotypes with strong evidence and environmental agents with moderate or weak evidence and (iii) moderate evidence for the effect of both environmental agents and genotype. Other combinations of the evidence for main effects support only a weak probability for an interaction (prior score category 3), although one cannot fully exclude the possibility that interactions may also exist in the absence of any main effect. Finally, the default prior scores can be modified based on mechanistic or biological information, when such evidence is available, e.g. for tobacco-related cancer.93
Details on each step are described further.
Step 1. Score the strength of the evidence for main effect of the environmental exposure
The assessment of evidence of the main effect of the environmental exposure is based on a systematic review of human studies and other relevant data available in the scientific literature.
In the case of carcinogenic effects, comprehensive systematic reviews and evaluations such as those of the IARC Monographs2 and those of the World Cancer Research Fund (WCRF) (specifically for nutritional factors)94 should be used whenever appropriate. The criteria used in the two systems are described in Box 1. It is important however to specify whether the evaluation comes from a systematic programme such as the IARC Monographs or the WCRF Reports, or from an ad hoc exercise.
Box 1 Criteria for assessing the evidence of carcinogenicity in the IARC Monograph Program and the WCRF Report on Nutrition and Cancer.
| IARC Monographs |
WRCF Report |
||
|---|---|---|---|
| Category | Criteria | Category | Criteria |
| Sufficient | A causal relationship has been established between exposure to the agent and human cancer. That is, a positive relationship has been observed between the exposure and cancer in studies in which chance, bias and confounding could be ruled out with reasonable confidence. | Convincing | All of the following criteria are generally required:
|
| Limited | A positive association has been observed between exposure to the agent and cancer for which a causal interpretation is considered to be credible, but chance, bias or confounding could not be ruled out with reasonable confidence. | Probable | All the following criteria are generally required:
|
| Suggestive | All the following were generally required:
|
||
| Inadequate | The available studies are of insufficient quality, consistency or statistical power to permit a conclusion regarding the presence or absence of a causal association between exposure and cancer. | No conclusion | The evidence might be limited by the amount of evidence in terms of the number of studies available, by inconsistency of direction of effect, by poor quality of studies (e.g. lack of adjustment for known confounders), or by any combination of these factors. |
| Evidence suggesting lack of carcinogenicity | There are several adequate studies covering the full range of levels of exposure that humans are known to encounter, which are mutually consistent in not showing a positive association between exposure to the agent and any studied cancer at any observed level of exposure. The results from these studies alone or combined should have narrow confidence intervals with an upper limit close to the null value (e.g. an RR of 1.0). Bias and confounding should be ruled out with reasonable confidence, and the studies should have an adequate length of follow-up. | Substantial effect unlikely | All of the following criteria are generally required:
|
In Box 1, the criteria used in the IARC Monographs evaluations of carcinogenic risks to humans2 and the WCRF Report on Nutrition and Cancer94 are summarized. Although the categories of evidence do not match perfectly and the criteria vary in several important details, the two approaches are broadly in agreement. The WCRF criteria are more explicit than the IARC ones.
It should be noted that some exposures might be potentially carcinogenic, whereas others could be putatively protective and the same exposure, e.g. oestrogen plus progestin hormone replacement therapy may be protective for some health outcomes but a risk factor for others,95 suggesting the need to consider potential harms as well as benefits for potential preventive exposures.
In all cases, evidence for main effects of the environmental factor is classified according to a qualitative scale comprising five categories, such as those proposed in Table 2.
Table 2.
Proposal of a common scheme for the evaluation of main effects of environmental agents
| Common scheme | IARC Monographsa | WCRF Reportb |
|---|---|---|
| A (strong evidence) | Sufficient; limited with supporting datac | Convincing |
| B (moderate evidence) | Limited | Probable |
| C (weak evidence) | Inadequate with supporting datac | Suggestive; limited |
| D (lack of evidence) | Inadequate | No conclusions |
| E (evidence against an effect) | Substantial effect unlikely |
Score the strength of the evidence for main effect of the genetic variant
Scoring the main effect of the environmental factor and the genetic variant can occur in any order. The evidence of main effects of the genetic variant is assessed according to the HuGENet criteria for assessing the epidemiological credibility of cumulative evidence,4 also called the Venice criteria (Figure 4). On the basis of combination of three criteria (amount of evidence, degree of replication and protection from bias) (each of which can be scored A, B and C), the epidemiological evidence for an effect of the genotype is classified as strong, moderate or weak. Tables 3 and 4 show the main considerations for the three criteria for genetic associations, and address the special issues regarding protection from bias. Evidence is considered to be strong if the genetic variant scores A on all three criteria; moderate if any B is present but no C; and weak, if any C is present for any of the three criteria.
Figure 4.
Steps in assessing G × E interactions
Table 3.
Considerations for epidemiological credibility in the assessment of cumulative evidence on genetic associations and G × E interactions (adapted from ref.4)
| Proposed operationalization |
|||
|---|---|---|---|
| Criteria | Categories | Genetic association | G × E interaction |
| Amount of evidence | A: large-scale evidence | Thresholds may be defined based on sample size, power or false-discovery rate considerations. The frequency of the genetic variant of interest should be accounted for | Same as for genetic association plus: The frequency of the environmental variant of interest should also be accounted for |
| B: moderate amount of evidence | |||
| C: little evidence | |||
| Replication | A: extensive replication including at least one well-conducted meta-analysis with little between-study inconsistency | Between-study inconsistency entails statistical considerations and also epidemiological considerations for the similarity/standardization or at least harmonization of phenotyping, genotyping and analytical models across studies | Between-study consistency in a variation in risk estimates by genetic and environmental standards that entail considerations for replication of genetic associations listed at left |
| B: well-conducted meta-analysis with some methodological limitations or moderate between-study inconsistency | |||
| C: no association; no independent replication; failed replication; scattered studies; flawed meta-analysis or large inconsistency | |||
| Protection from bias | A: bias, if at all present, could affect the magnitude but probably not the presence of the association | A prerequisite for A is that the bias due to phenotype measurement, genotype measurement, confounding and selective reporting can be appraised as not being high plus there is no other demonstrable bias in any other aspect of the design, analysis or accumulation of the evidence that could invalidate the presence of the proposed association | Same as for genetic associations except that only certain biases listed for genetic associations apply |
| B: no obvious bias that may affect the presence of the association but there is considerable missing information on the generation of evidence | In category B, although no strong biases are visible, there is no such assurance that major sources of bias have been minimized or accounted for because information is missing on how phenotyping, genotyping and confounding have been handled | ||
| C: considerable potential for or demonstrable bias that can affect even the presence or absence of the association | |||
Table 4.
Typical biases and their typical impact on genetic associations depending on the status of the evidence (adapted from ref.4)
| Likelihood of bias to invalidate an observed association |
|||||
|---|---|---|---|---|---|
| Biases | Status of the evidence | Small OR <1.15 | Typical OR 1.15–1.8 | Large OR >1.8 | Application to evidence for G × E interaction assessment |
| Bias in phenotype definition | Not reported what was done | Unknown | Unknown | Unknown | Not applicable unless differential by environmental exposure |
| Unclear phenotype definitions | Possible/high | Possible/high | Possible/high | ||
| Clear widely agreed definitions of phenotypes | Low/none | Low/none | Low/none | ||
| Efforts for retrospective harmonization | Possible/high | Low | Low/none | ||
| Prospective standardization of phenotypes | Low/none | Low/none | Low/none | ||
| Bias in genotyping | Not reported what was done | Unknown | Unknown | Unknown | Not applicable unless differential by environmental exposure |
| No quality control checks | Possible/high | Low | Low | ||
| Appropriate quality control checks | Low | Low | Low/none | ||
| Population stratification | Not reported what was done | Unknown | Unknown | Unknown | Not applicable |
| Nothing donea | Possible/high | Possible/high | Possible/high | ||
| Same descent groupb | Possible/high | Low | Low/none | ||
| Adjustment for reported descent | Possible/high | Low | Low/none | ||
| Family-based design | Low/none | Low/none | Low/none | ||
| Genomic control, principal component analysis or similar method | Low/none | Low/none | Low/none | ||
| Selective reporting biases | Meta-analysis of published data | Possible/high | Possible | Possible | Possible |
| Retrospective efforts to include unpublished data | Possible/high | Possible | Possible | Possible | |
| Meta-analysis within consortium | Low/none | Low/none | Low/none | Possible | |
Category decreases from A to B, if the ‘Unknown’ are considered to be a major issue for the appraisal of the evidence. Any ‘Possible/high’ item confers category C status. ‘Possible’ (selective reporting biases for non-consortium/prospective meta-analysis) does not necessarily decrease the category grade (from A to C); this may need to be appraised separately in each field and may be facilitated by using tests for selective reporting biases (tests for small-study effects and excess of significant studies), although probably no test has high sensitivity and specificity for such biases. Clear demonstrable biases in other aspects of the design, conduct and analysis of the evidence (besides the four aspects considered in this table) also result in shift to category C for protection from bias.
The majority of nominally significant genetic associations that emerged in the candidate gene era have weak credibility under this scheme. For example, out of 31 nominally significant associations in associations between DNA polymorphisms and diverse cancers, only 1 has strong credibility based on the Venice criteria.96 Conversely, there were 92 associations of genetic variants with diverse cancers from GWASs with P < 10−7 97 and, with three exceptions, all had sufficient amount of evidence to be characterized as A for the first Venice criterion, whereas the attainment of genome-wide significance is sufficient for getting an A in replication. Protection from bias in prospectively conducted and fully reported GWASs is also considered sufficient for an A on this criterion. Thus, the vast majority of these associations seem to have strong credibility.
Step 2. Establish a prior score category and decide on the pattern of interaction to be expected
The second step consists of establishing a prior score, rated as 1 for strong, 2 for moderate and 3 for weak, and considering different possible patterns of interaction and identifying those more likely to occur.
If there is strong evidence that the genetic variant and the environmental agent act in ways directly relevant to each other (e.g. the variant affects the metabolism of the agent), the default prior score can be upgraded (e.g. from score 2 to 1). Due to limitations in the current understanding of carcinogenesis, lack of biological or mechanistic evidence should not be used to downgrade these prior scores.
The decision on expected patterns of interaction should be based on a priori knowledge about the biological interplay of the environmental factor and the genotype. When such knowledge cannot predict any pattern of interaction, the default pattern of interaction should be the one requiring fewer assumptions (pattern A in Figure 1). Similarly, a priori decisions should be made on whether the expected scale of interaction should be additive or multiplicative. Usually, it is very difficult to make a strong a priori choice for additive vs multiplicative effects. It should be stressed that pattern A may be consistent with a pure main effects model, but depending upon the choice of scale (additive vs multiplicative), an interaction term might be needed.
If more than one model of interaction is plausible, the subsequent steps of the process can be repeated under each model.
Step 3. Score the strength of the evidence for the interaction between the environmental exposure and the genetic variant
Assessment of the evidence for the presence of an interaction according to the pre-defined scale and pattern should be done based on an extension of the HuGENet Venice criteria used for assessing cumulative evidence for genetic associations.4,96 As discussed above, the amount of evidence, consistency of replication and protection from bias of proposed G × E interactions should be critically assessed. With respect to amount of evidence, one may use either power considerations, or Bayesian or false-discovery approaches or the simplified operational approach of sample size,4,96 focusing on the available sample size of the smallest compared subgroup: in this case this group would be the smallest group defined by the combination of G and E. Usually, G × E interactions are evaluated in small studies, and thus it is expected that evidence would get a low grade in most cases. Alternatively, one could grade using a rough power estimate based on the smallest cell in the 2 × 2 × 2 table for the gene by environment by disease interaction. For example, a grade of B could correspond to 80% power to detect an interaction RR of 1.5 (corresponding to a cell size >100) or a C for an interaction RR >2 (and a cell size of 50–99). Consistency of replication is an even greater challenge for G × E interactions, since results are often generated as secondary aims in studies focused on main effects, and there is no tradition of immediately replicating these results in multiple datasets. Finally, to protect from bias, one needs to consider three aspects: protection from bias for the epidemiological exposure, for the genotype and for the interaction effect.
Even if the evidence in Step 1 for either G or E is lacking or there is evidence against an effect (i.e. there is good evidence of an absence of marginal effects of E or G), Step 3 should be undertaken because a G × E interaction may exist even in the absence of main effects. However, this situation would probably only merit a low prior score (e.g. 3 in the schema of Table 2).
There are situations in which caution in interpretation is important. For example, case–parent trio studies cannot discriminate between exposures that are neutral in the absence of a variant allele and deleterious in its presence, and exposures beneficial in the absence of a variant allele and neutral in its presence.98
This difficulty also applies to the case-only design. Furthermore, in the case-only design, a submultiplicative interaction can occur even when there is no mechanistic interaction between genetic and environmental factors.99
Step 4. Examine the overall plausibility of interaction by combining a prior score and strength of the evidence and interpreting results.
In this step all of the information from the previous steps is put together. There are nine possibilities for overall plausibility of the proposed interaction from the combination of Table 1 and Step 3. A proposed interaction with a high prior score and strong evidence has the best plausibility. A proposed interaction with a low prior score and weak evidence has the worst possible plausibility. The other seven combinations are between these extremes. Of note, a different model for the interaction between the same genetic and environmental factors may have a high prior score but moderate or weak evidence for the presence of the interaction. Moreover, even in case of a low prior score the presence of interaction should not be excluded (e.g. in case of a genotype with no overall effect and opposite effects depending on the presence of the environmental factor). On the other hand, some apparent interactions with effects going in opposite directions, may be simply statistical artefacts or the complete absence of effect in one stratum of the exposure or genetic factor. Overall, given such required stringency, few G × E interactions previously proposed would be graded as having strong credibility, if such criteria were used. Finally, additional support for interaction may come from knowledge about the gene, the environment and the interaction from pharmacokinetics, animal models and other sources.
Data should be interpreted by examining the overall strength of the evidence from the previous steps and taking into account any additional information to put the findings into context, in other words, the interpretation of the evidence for a G × E interaction should take into account the totality of information available. For example, the extent to which assumptions are made in studies and meta-analyses should be reviewed and the validity of those assumptions should be assessed in interpreting the overall plausibility of a G × E interaction. For example, if the data are primarily from case-only studies, has the assumption needed under Mendelian randomization for independence of the genotype on the exposure been adequately addressed?100 A final stage of interpretation of G × E interactions relates to the evidence regarding the causal nature of the association of the environmental factor with disease risk.101,102 When there is clear evidence that an effect of an environmental risk factor can be modified by genetic variation, particularly when the genotype has a clear relation to components of the exposure, then this increases the strength of evidence regarding the causal nature of the environmental factor itself, independent of genotype.
Examples of evaluation of G × E interactions
Some examples of G × E interactions for which a relatively extensive database is available are shown in Table 5: it is noteworthy that these examples concern either carcinogens characterized by strong associations, i.e. factors whose exposure results in ≥3-fold increase in cancer, or high-penetrance genotypes, whose carriers have a very high cumulative risk of one or more cancers. It is plausible that many G × E interactions act at the interface of environmental carcinogens and genotypes entailing small RRs, but such interactions have not yet been robustly identified and replicated, despite their potential cumulative role in determining the burden of cancer in a population. Khoury et al. have shown that ‘weak’ genetic or environmental marginal effect size can hide considerable underlying interactions10 leading to a loss of statistical power in measuring both genetic and environmental effects separately. The problem, however, is to disentangle these true interactions from a sea of false-positive findings when multiple genetic and environmental factors are analysed, especially in the era of GWASs.55
Table 5.
Examples of suggested G × E interactions
| Evidence for main effect of genetic factor | Evidence for main effect of environmental factor | Target cancer | Possible mechanism | Evidence for G × E interaction |
|---|---|---|---|---|
| NAT2 slow polymorphism103 | Tobacco smoking102 | Bladder cancer | Reduced detoxification of aromatic amines104 | Ref.103 |
| ALDH fast polymorphism105,106 | Alcohol drinking101 | Head and neck and oesophageal cancer | Increased exposure to acetaldehyde107 | Refs84,85 |
| ATM mutation108 | Ionizing radiation101 | Breast cancer | Reduced repair of radiation-induced DNA damage110 | Ref.111 |
In order to test the performance of these interim guidelines, we applied them to an example from Table 5. The other examples are listed as suggestions for future reviews and evaluations.
NAT2 polymorphism, tobacco smoking and bladder cancer
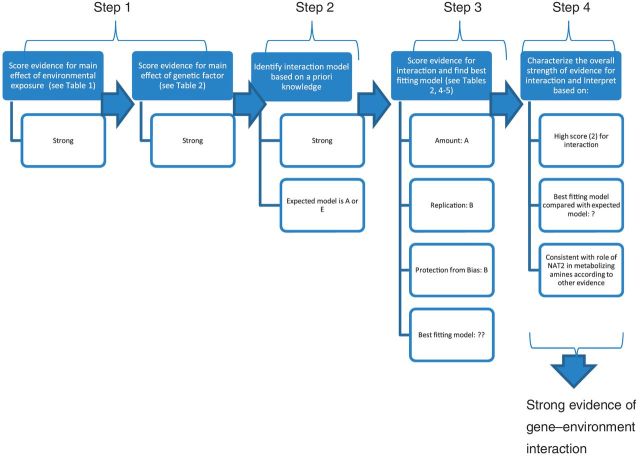
The application of the interim guidelines to the example of NAT2 polymorphism, tobacco smoking and bladder cancer risk is summarized in Figure 5.
Figure 5.
Assessment of NAT2, tobacco smoking and bladder cancer G × E interactions
Step 1. There is abundant and consistent evidence for carcinogenicity of tobacco smoking on the urinary bladder. The IARC Monographs evaluated evidence in 1986, 2002 and 2009.102,112,113 In all occasions, the evaluation has been of sufficient evidence for tobacco smoking being a cause of human cancer, so the assessment of the evidence for the main effect of the exposure, tobacco smoking, on bladder cancer can be classified as ‘strong’ based on language in Table 2.
Several meta-analyses of the association between NAT2 slow acetylator genotype and bladder cancer have shown a moderate but consistent increase in risk.103,114 Using the Venice criteria, there is strong evidence of a genetic association in persons of European ancestry and moderate credibility when all ethnic groups are considered. Of note, the region containing NAT2 has not emerged in GWASs of bladder cancer.115,116 However, this does not rule out a role for NAT2.
Step 2. Given the evidence for main effects of smoking and NAT2 on bladder cancer, the prior score category is 2 when all ethnic groups are considered (strong E, moderate G), and prior score category is 1 when only European Caucasians are considered (strong E, strong G). This corresponds to a moderate or strong, respectively, a priori likelihood of a G × E interaction. Given the role of NAT2 in metabolizing aromatic amines, which are among the likely bladder carcinogens in tobacco smoke, one can expect the interaction of the two agents to follow model A in Figure 1 (or model E in case tobacco smoke is the only source of exposure to aromatic amines).
Step 3. Next we examine the evidence for interaction for pattern A. A case-only meta-analysis was conducted117 that found an interaction odds ratio (OR) of 1.21 [95% confidence interval (CI) 1.04–1.42]. The association was observed especially among Europeans based on 13 studies (OR 1.38; 95% CI 1.13–1.68) and not among US Caucasians or Asians. The estimated I2 value was 0% for Europeans (P for heterogeneity 0.89), suggesting considerable consistency in this population. The same case-only meta-analysis117 found a borderline significant interaction when all ethnicities were considered (OR 1.21, 95% CI 1.04–1.42) and no significant between-study heterogeneity (I2 = 0%, P = 0.54). A prior meta-analysis103 concluded the OR for the acetylators who were current or ever cigarette smokers as 2.73 (85% CI 1.70–4.31).
According to Table 3, the score for replication is ‘A’, extensive replication. The numbers of cases in the meta-analysis vary, but some are quite small and the CIs for the interaction ORs were quite large, suggesting a ‘B’ on the amount of evidence for the interaction would be an appropriate score. Examining the interaction ORs by date of publication suggests most recent studies tend to have tighter CIs and generally have ORs above 1. However, these meta-analyses are retrospective and there are some potential biases that are not possible to address and exclude. This suggests a score of B for protection from bias. Applying scores of A for replication, B for amount, and B for protection from bias suggests overall moderate strength of the evidence for the interaction.
Step 4. This proposed interaction has a strong prior score category (score 1) and moderate evidence among Europeans and a moderate prior score category (score 2) and moderate evidence when all ethnicities are considered. Overall, this is the second best possibility for the plausibility of an interaction among Europeans and a moderate scenario for the plausibility of an interaction across all ethnicities. Moreover, the evidence of a G × E interaction is well supported by animal and pharmacokinetic studies. Overall, the conclusion for some G × E interaction seems quite plausible, although not fully documented yet. This observed interaction can be taken as providing evidence regarding the causal nature of cigarette smoking on bladder cancer risk given pattern of a strong genotypic effect amongst cigarette smokers, but little evidence of any effect amongst non-smokers would be unlikely to be seen unless tobacco smoke increases the risk of bladder cancer.
Conclusions
Any formal assessment of evidence of interaction may fall short of being conclusive. More G × E interactions have been suggested from cancer genetic association studies in the past decade. Most of these were not confirmed in replication studies, when these were performed; and, in general, the amount of available evidence is weak. This also applies to G × E interactions for diseases other than cancer: a well-known example is the interaction between variants in the serotonin transporter gene and stressful life events in determining the risk of depression, which was recently shown not to be robust.118 This stresses the need for guidelines to assess the strength of evidence for G × E interactions. We have developed an interim set of recommendations and guidelines; further research is needed to test their performances and apply them to diseases other than cancer.
Acknowledgements
This manuscript is based on a workshop on assessment of Gene-Environment Interaction which was organized in 2009 by the National Cancer Institute (NCI) of the United States, the HuGENet project, and the International Agency for Research on Cancer (IARC). The following individuals either attended the workshop or contributed a presentation: Christine Ambrosone (USA), Paolo Boffetta (France, USA), Paul Brennan (France), Graham Byrnes (France), Harry Campbell (United Kingdom), Vincent J. Cogliano (France), George Davey Smith (United Kingdom), Stephen S. Hecht (USA), John P.A. Ioannidis (Greece, USA), Muin Khoury (USA), Julian Little (Canada), Paul McKeigue (United Kingdom), Nathaniel Rothman (USA), Daniela Seminara (USA), Kurt Straif (France), Sean Tavtigian (France, USA), Duncan C. Thomas (USA), Paolo Vineis (United Kingdom), Christopher P. Wild (France), Sholom Wacholder (USA), Deborah M. Winn (USA). J.L. holds a Canada Research Chair in Human Genome Epidemiology.
Conflict of interest: None declared.
KEY MESSAGES.
Joint effects of genetic and environmental factors are presumed to be important for determining cancer risk, but the evaluation of G × E interactions poses several challenges.
We propose criteria to evaluate the cumulative evidence of G × E interactions in the causation of human cancer.
The criteria appraise systematically the strength of the evidence on genetic main effects and on environmental main effects to generate a prior score and then incorporate the strength of the evidence of interaction effects for specific patterns of postulated interaction.
References
- 1.Fletcher O, Houlston RS. Architecture of inherited susceptibility to common cancer. Nat Rev Cancer. 2010;10:353–61. doi: 10.1038/nrc2840. [DOI] [PubMed] [Google Scholar]
- 2.International Agency for Research on Cancer. Preamble to the IARC Monographs (amended January 2006) Lyon, France: International Agency for Research on Cancer; 2009. [Google Scholar]
- 3.Little J. Reporting and review of human genome epidemiology studies (Chapter 10) In: Khoury MJ, Little J, Burke W, editors. Human Genome Epidemiology: A Scientific Foundation for Using Genetic Information to Improve Health and Prevent Disease. New York: Oxford University Press; 2003. pp. 168–92. [Google Scholar]
- 4.Ioannidis JP, Boffetta P, Little J, et al. Assessment of cumulative evidence on genetic associations: interim guidelines. Int J Epidemiol. 2008;37:120–32. doi: 10.1093/ije/dym159. [DOI] [PubMed] [Google Scholar]
- 5.Porta M. A Dictionary of Epidemiology. 5th. New York: Oxford University Press; 2008. pp. 1–289. [Google Scholar]
- 6.Weinberg CR. Less is more, except when less is less: studying joint effects. Genomics. 2009;93:10–12. doi: 10.1016/j.ygeno.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thomas D. Gene-environment-wide association studies: emerging approaches. Nat Rev Genet. 2010;11:259–72. doi: 10.1038/nrg2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clayton DG. Prediction and interaction in complex disease genetics: experience in type 1 diabetes. PLoS Genet. 2009;5:e1000540. doi: 10.1371/journal.pgen.1000540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ottman R. Gene-environment interaction: definitions and study designs. Prev Med. 1996;25:764–70. doi: 10.1006/pmed.1996.0117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khoury MJ, Adams MJ, Jr, Flanders WD. An epidemiologic approach to ecogenetics. Am J Hum Genet. 1988;42:89–95. [PMC free article] [PubMed] [Google Scholar]
- 11.Haldane JB. The interaction of nature and nurture. Ann Eugenics. 1946;13:197–205. doi: 10.1111/j.1469-1809.1946.tb02358.x. [DOI] [PubMed] [Google Scholar]
- 12.Yang Q, Khoury MJ. Evolving methods in genetic epidemiology. III. Gene-environment interaction in epidemiologic research. Epidemiol Rev. 1997;19:33–43. doi: 10.1093/oxfordjournals.epirev.a017944. [DOI] [PubMed] [Google Scholar]
- 13.Berrington de Gonzalez A, Cox DR. Interpretation of interaction: a review. Ann Appl Stat. 2007;1:371–85. [Google Scholar]
- 14.Clayton D, McKeigue PM. Epidemiological methods for studying genes and environmental factors in complex diseases. Lancet. 2001;358:1356–60. doi: 10.1016/S0140-6736(01)06418-2. [DOI] [PubMed] [Google Scholar]
- 15.Le Marchand L, Hankin JH, Wilkens LR, et al. Combined effects of well-done red meat, smoking, and rapid N-acetyltransferase 2 and CYP1A2 phenotypes in increasing colorectal cancer risk. Cancer Epidemiol Biomarkers Prev. 2001;10:1259–66. [PubMed] [Google Scholar]
- 16.Cook NR, Zee RY, Ridker PM. Tree and spline based association analysis of gene-gene interaction models for ischemic stroke. Stat Med. 2004;23:1439–53. doi: 10.1002/sim.1749. [DOI] [PubMed] [Google Scholar]
- 17.Hahn LW, Ritchie MD, Moore JH. Multifactor dimensionality reduction software for detecting gene-gene and gene-environment interactions. Bioinformatics. 2003;19:376–82. doi: 10.1093/bioinformatics/btf869. [DOI] [PubMed] [Google Scholar]
- 18.Ritchie MD, Motsinger AA. Multifactor dimensionality reduction for detecting gene-gene and gene-environment interactions in pharmacogenomics studies. Pharmacogenomics. 2005;6:823–34. doi: 10.2217/14622416.6.8.823. [DOI] [PubMed] [Google Scholar]
- 19.Millstein J, Conti DV, Gilliland FD, Gauderman WJ. A testing framework for identifying susceptibility genes in the presence of epistasis. Am J Hum Genet. 2006;78:15–27. doi: 10.1086/498850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schwender H, Ickstadt K. Identification of SNP interactions using logic regression. Biostatistics. 2008;9:187–98. doi: 10.1093/biostatistics/kxm024. [DOI] [PubMed] [Google Scholar]
- 21.Motsinger AA, Lee SL, Mellick G, Ritchie MD. GPNN: power studies and applications of a neural network method for detecting gene-gene interactions in studies of human disease. BMC Bioinformatics. 2006;7:39. doi: 10.1186/1471-2105-7-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen SH, Sun J, Dimitrov L, et al. A support vector machine approach for detecting gene-gene interaction. Genet Epidemiol. 2008;32:152–67. doi: 10.1002/gepi.20272. [DOI] [PubMed] [Google Scholar]
- 23.Thomas D. Methods for investigating gene-environment interactions in candidate pathway and genome-wide association studies. Annu Rev Public Health. 2010;31:21–36. doi: 10.1146/annurev.publhealth.012809.103619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Greenland S. Hierarchical regression for epidemiologic analyses of multiple exposures. Environ Health Perspect. 1994;102(Suppl 8):33–39. doi: 10.1289/ehp.94102s833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Witte JS. Genetic analysis with hierarchical models. Genet Epidemiol. 1997;14:1137–42. doi: 10.1002/(SICI)1098-2272(1997)14:6<1137::AID-GEPI96>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 26.Hung RJ, Brennan P, Malaveille C, et al. Using hierarchical modeling in genetic association studies with multiple markers: application to a case-control study of bladder cancer. Cancer Epidemiol Biomarkers Prev. 2004;13:1013–21. [PubMed] [Google Scholar]
- 27.Hung RJ, McKay JD, Gaborieau V, et al. A susceptibility locus for lung cancer maps to nicotinic acetylcholine receptor subunit genes on 15q25. Nature. 2008;452:633–37. doi: 10.1038/nature06885. [DOI] [PubMed] [Google Scholar]
- 28.Capanu M, Orlow I, Berwick M, Hummer AJ, Thomas DC, Begg CB. The use of hierarchical models for estimating relative risks of individual genetic variants: an application to a study of melanoma. Stat Med. 2008;27:1973–92. doi: 10.1002/sim.3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rebbeck TR, Spitz M, Wu X. Assessing the function of genetic variants in candidate gene association studies. Nat Rev Genet. 2004;5:589–97. doi: 10.1038/nrg1403. [DOI] [PubMed] [Google Scholar]
- 30.Thomas PD, Mi H, Lewis S. Ontology annotation: mapping genomic regions to biological function. Curr Opin Chem Biol. 2007;11:4–11. doi: 10.1016/j.cbpa.2006.11.039. [DOI] [PubMed] [Google Scholar]
- 31.Wakefield J, De VF, Hung RJ. Bayesian mixture modeling of gene-environment and gene-gene interactions. Genet Epidemiol. 2010;34:16–25. doi: 10.1002/gepi.20429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Conti DV, Cortessis V, Molitor J, Thomas DC. Bayesian modeling of complex metabolic pathways. Hum Hered. 2003;56:83–93. doi: 10.1159/000073736. [DOI] [PubMed] [Google Scholar]
- 33.Kooperberg C, Ruczinski I. Identifying interacting SNPs using Monte Carlo logic regression. Genet Epidemiol. 2005;28:157–70. doi: 10.1002/gepi.20042. [DOI] [PubMed] [Google Scholar]
- 34.Baurley JW, Conti DV, Gauderman WJ, Thomas DC. Discovery of complex pathways from observational data. Stat Med. 2010;29:1998–2011. doi: 10.1002/sim.3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen GK, Witte JS. Enriching the analysis of genomewide association studies with hierarchical modeling. Am J Hum Genet. 2007;81:397–404. doi: 10.1086/519794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lewinger JP, Conti DV, Baurley JW, Triche TJ, Thomas DC. Hierarchical Bayes prioritization of marker associations from a genome-wide association scan for further investigation. Genet Epidemiol. 2007;31:871–82. doi: 10.1002/gepi.20248. [DOI] [PubMed] [Google Scholar]
- 37.Conti DV, Lewinger JP, Tyndale RF, Benowitz NL, Swan GE, Thomas PD. Using ontologies in hierarchical modeling of genes and exposure in biological pathways. In: Swan GE, Baker TB, Chassin L, Conti DV, Lerman C, Perkins KA, editors. Monograph 20: Phenotypes and Endophenotypes Foundations for Genetic Studies of Nicotine Use and Dependence. NIH Publication No. 09-6366.Bethesda, MD: National Cancer Institute; 2009. pp. 539–84. [Google Scholar]
- 38.De Roos AJ, Smith MT, Chanock S, Rothman N. Toxicological considerations in the application and interpretation of susceptibility biomarkers in epidemiological studies. IARC Sci Publ. 2004;157:105–25. [PubMed] [Google Scholar]
- 39.Lin BK, Clyne M, Walsh M, et al. Tracking the epidemiology of human genes in the literature: the HuGE Published Literature database. Am J Epidemiol. 2006;164:1–4. doi: 10.1093/aje/kwj175. [DOI] [PubMed] [Google Scholar]
- 40.Lunn DJ, Best N, Thomas A, Wakefield J, Spiegelhalter D. Bayesian analysis of population PK/PD models: general concepts and software. J Pharmacokinet Pharmacodyn. 2002;29:271–307. doi: 10.1023/a:1020206907668. [DOI] [PubMed] [Google Scholar]
- 41.Racine-Poon A, Wakefield J. Statistical methods for population pharmacokinetic modelling. Stat Methods Med Res. 1998;7:63–84. doi: 10.1177/096228029800700106. [DOI] [PubMed] [Google Scholar]
- 42.Wakefield J. The Bayesian analysis of population pharmacokinetic models. J Am Stat Assoc. 1996;91:62–75. [Google Scholar]
- 43.Cortessis V, Thomas DC. Toxicokinetic genetics: an approach to gene-environment and gene-gene interactions in complex metabolic pathways. IARC Sci Publ. 2004;157:127–50. [PubMed] [Google Scholar]
- 44.Jonsson F, Johanson G. A Bayesian analysis of the influence of GSTT1 polymorphism on the cancer risk estimate for dichloromethane. Toxicol Appl Pharmacol. 2001;174:99–112. doi: 10.1006/taap.2001.9206. [DOI] [PubMed] [Google Scholar]
- 45.Armitage P, Doll R. The age distribution of cancer and a multi-stage theory of carcinogenesis. Br J Cancer. 1954;8:1–12. doi: 10.1038/bjc.1954.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moolgavkar SH, Knudson AG., Jr Mutation and cancer: a model for human carcinogenesis. J Natl Cancer Inst. 1981;66:1037–52. doi: 10.1093/jnci/66.6.1037. [DOI] [PubMed] [Google Scholar]
- 47.Little MP, Li G. Stochastic modelling of colon cancer: is there a role for genomic instability? Carcinogenesis. 2007;28:479–87. doi: 10.1093/carcin/bgl173. [DOI] [PubMed] [Google Scholar]
- 48.Mothersill C, Seymour C. Genomic instability, bystander effects and radiation risks: implications for development of protection strategies for man and the environment. Radiats Biol Radioecol. 2000;40:615–20. [PubMed] [Google Scholar]
- 49.Thompson WD. Effect modification and the limits of biological inference from epidemiologic data. J Clin Epidemiol. 1991;44:221–32. doi: 10.1016/0895-4356(91)90033-6. [DOI] [PubMed] [Google Scholar]
- 50.Hunter DJ. Gene-environment interactions in human diseases. Nat Rev Genet. 2005;6:287–98. doi: 10.1038/nrg1578. [DOI] [PubMed] [Google Scholar]
- 51.Burton PR, Hansell AL, Fortier I, et al. Size matters: just how big is BIG?: quantifying realistic sample size requirements for human genome epidemiology. Int J Epidemiol. 2009;38:263–73. doi: 10.1093/ije/dyn147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wong MY, Day NE, Luan JA, Chan KP, Wareham NJ. The detection of gene-environment interaction for continuous traits: should we deal with measurement error by bigger studies or better measurement? Int J Epidemiol. 2003;32:51–57. doi: 10.1093/ije/dyg002. [DOI] [PubMed] [Google Scholar]
- 53.Boffetta P, McLaughlin JK, La VC, Tarone RE, Lipworth L, Blot WJ. False-positive results in cancer epidemiology: a plea for epistemological modesty. J Natl Cancer Inst. 2008;100:988–95. doi: 10.1093/jnci/djn191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dwan K, Altman DG, Arnaiz JA, et al. Systematic review of the empirical evidence of study publication bias and outcome reporting bias. PLoS One. 2008;3:e3081. doi: 10.1371/journal.pone.0003081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Khoury MJ, Wacholder S. From genome-wide association studies to gene-environment-wide interaction studies–challenges and opportunities. Am J Epidemiol. 2009;169:227–30. doi: 10.1093/aje/kwn351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Knol MJ, Egger M, Scott P, Geerlings MI, Vandenbroucke JP. When one depends on the other: reporting of interaction in case-control and cohort studies. Epidemiology. 2009;20:161–66. doi: 10.1097/EDE.0b013e31818f6651. [DOI] [PubMed] [Google Scholar]
- 57.Vandenbroucke JP, Von EE, Altman DG, et al. Strengthening the Reporting of Observational Studies in Epidemiology (STROBE): explanation and elaboration. Ann Intern Med. 2007;147:W163–94. doi: 10.7326/0003-4819-147-8-200710160-00010-w1. [DOI] [PubMed] [Google Scholar]
- 58.Begg CB, Zhang ZF. Statistical analysis of molecular epidemiology studies employing case-series. Cancer Epidemiol Biomarkers Prev. 1994;3:173–75. [PubMed] [Google Scholar]
- 59.Khoury MJ, Flanders WD. Nontraditional epidemiologic approaches in the analysis of gene-environment interaction: case-control studies with no controls! Am J Epidemiol. 1996;144:207–13. doi: 10.1093/oxfordjournals.aje.a008915. [DOI] [PubMed] [Google Scholar]
- 60.Piegorsch WW, Weinberg CR, Taylor JA. Non-hierarchical logistic models and case-only designs for assessing susceptibility in population-based case-control studies. Stat Med. 1994;13:153–62. doi: 10.1002/sim.4780130206. [DOI] [PubMed] [Google Scholar]
- 61.Schaid DJ. Case-parents design for gene-environment interaction. Genet Epidemiol. 1999;16:261–73. doi: 10.1002/(SICI)1098-2272(1999)16:3<261::AID-GEPI3>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 62.Albert PS, Ratnasinghe D, Tangrea J, Wacholder S. Limitations of the case-only design for identifying gene-environment interactions. Am J Epidemiol. 2001;154:687–93. doi: 10.1093/aje/154.8.687. [DOI] [PubMed] [Google Scholar]
- 63.Gatto NM, Campbell UB, Rundle AG, Ahsan H. Further development of the case-only design for assessing gene-environment interaction: evaluation of and adjustment for bias. Int J Epidemiol. 2004;33:1014–24. doi: 10.1093/ije/dyh306. [DOI] [PubMed] [Google Scholar]
- 64.Li D, Conti DV. Detecting gene-environment interactions using a combined case-only and case-control approach. Am J Epidemiol. 2009;169:497–504. doi: 10.1093/aje/kwn339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mukherjee B, Ahn J, Gruber SB, Rennert G, Moreno V, Chatterjee N. Tests for gene-environment interaction from case-control data: a novel study of type I error, power and designs. Genet Epidemiol. 2008;32:615–26. doi: 10.1002/gepi.20337. [DOI] [PubMed] [Google Scholar]
- 66.Murcray CE, Lewinger JP, Gauderman WJ. Gene-environment interaction in genome-wide association studies. Am J Epidemiol. 2009;169:219–26. doi: 10.1093/aje/kwn353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lake SL, Laird NM. Tests of gene-environment interaction for case-parent triads with general environmental exposures. Ann Hum Genet. 2004;68(Pt 1):55–64. doi: 10.1046/j.1529-8817.2003.00073.x. [DOI] [PubMed] [Google Scholar]
- 68.Starr JR, Hsu L, Schwartz SM. Performance of the log-linear approach to case-parent triad data for assessing maternal genetic associations with offspring disease: type I error, power, and bias. Am J Epidemiol. 2005;161:196–204. doi: 10.1093/aje/kwi021. [DOI] [PubMed] [Google Scholar]
- 69.Umbach DM, Weinberg CR. The use of case-parent triads to study joint effects of genotype and exposure. Am J Hum Genet. 2000;66:251–61. doi: 10.1086/302707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gauderman WJ, Thomas DC, Murcray CE, Conti D, Li D, Lewinger JP. Efficient genome-wide association testing of gene-environment interaction in case-parent trios. Am J Epidemiol. 2010;172:116–22. doi: 10.1093/aje/kwq097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Morimoto LM, White E, Newcomb PA. Selection bias in the assessment of gene-environment interaction in case-control studies. Am J Epidemiol. 2003;158:259–63. doi: 10.1093/aje/kwg147. [DOI] [PubMed] [Google Scholar]
- 72.Wacholder S, Chatterjee N, Hartge P. Joint effect of genes and environment distorted by selection biases: implications for hospital-based case-control studies. Cancer Epidemiol Biomarkers Prev. 2002;11:885–89. [PubMed] [Google Scholar]
- 73.Thurigen D, Spiegelman D, Blettner M, Heuer C, Brenner H. Measurement error correction using validation data: a review of methods and their applicability in case-control studies. Stat Methods Med Res. 2000;9:447–74. doi: 10.1177/096228020000900504. [DOI] [PubMed] [Google Scholar]
- 74.Jurek AM, Greenland S, Maldonado G. How far from non-differential does exposure or disease misclassification have to be to bias measures of association away from the null? Int J Epidemiol. 2008;37:382–85. doi: 10.1093/ije/dym291. [DOI] [PubMed] [Google Scholar]
- 75.Garcia-Closas M, Rothman N, Lubin J. Misclassification in case-control studies of gene-environment interactions: assessment of bias and sample size. Cancer Epidemiol Biomarkers Prev. 1999;8:1043–50. [PubMed] [Google Scholar]
- 76.Garcia-Closas M, Wacholder S, Caporaso N, Rothman N. Inference issues in cohort and case-control studies of genetic effects and gene-environment interactions. In: Khoury MJ, Little J, Burke W, editors. Human Genome Epidemiology: A Scientific Foundation for Using Genetic Information to Improve Health and Prevent Disease. New York: Oxford University Press; 2003. pp. 127–44. [Google Scholar]
- 77.Cheng KF, Lin WJ. The effects of misclassification in studies of gene-environment interactions. Hum Hered. 2009;67:77–87. doi: 10.1159/000179556. [DOI] [PubMed] [Google Scholar]
- 78.Guttmacher AE, Collins FS. Genomic medicine–a primer. N Engl J Med. 2002;347:1512–20. doi: 10.1056/NEJMra012240. [DOI] [PubMed] [Google Scholar]
- 79.Knoppers BM. Biobanking: international norms. J Law Med Ethics. 2005;33:7–14. doi: 10.1111/j.1748-720x.2005.tb00205.x. [DOI] [PubMed] [Google Scholar]
- 80.Riboli E, Hunt KJ, Slimani N, et al. European Prospective Investigation into Cancer and Nutrition (EPIC): study populations and data collection. Public Health Nutr. 2002;5:1113–24. doi: 10.1079/PHN2002394. [DOI] [PubMed] [Google Scholar]
- 81.Swede H, Stone CL, Norwood AR. National population-based biobanks for genetic research. Genet Med. 2007;9:141–49. doi: 10.1097/gim.0b013e3180330039. [DOI] [PubMed] [Google Scholar]
- 82.Davey Smith G, Lawlor DA, Harbord R, et al. Clustered environments and randomized genes: a fundamental distinction between conventional and genetic epidemiology. PLoS Med. 2007;12:e352. doi: 10.1371/journal.pmed.0040352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Davey Smith G. Use of genetic markers and gene-diet-interactions for interrogating population-level-causal influences of diet on health. Genes Nutrition. 2011;6:27–43. doi: 10.1007/s12263-010-0181-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Brennan P, Lewis S, Hashibe M, et al. Pooled analysis of alcohol dehydrogenase genotypes and head and neck cancer: a HuGE review. Am J Epidemiol. 2004;159:1–16. doi: 10.1093/aje/kwh003. [DOI] [PubMed] [Google Scholar]
- 85.Cui R, Kamatani Y, Takahashi A, et al. Functional variants in ADH1B and ALDH2 coupled with alcohol and smoking synergistically enhance esophageal cancer risk. Gastroenterology. 2009;137:1768–75. doi: 10.1053/j.gastro.2009.07.070. [DOI] [PubMed] [Google Scholar]
- 86.Ioannidis JP, Loy EY, Poulton R, Chia KS. Researching genetic versus nongenetic determinants of disease: a comparison and proposed unification. Sci Transl Med. 2009;1:7ps8. doi: 10.1126/scitranslmed.3000247. [DOI] [PubMed] [Google Scholar]
- 87.Pearson TA, Manolio TA. How to interpret a genome-wide association study. JAMA. 2008;299:1335–44. doi: 10.1001/jama.299.11.1335. [DOI] [PubMed] [Google Scholar]
- 88.Fortier I, Burton P, Robson P, et al. Quality, quantity and harmony: the DataSHaPER approach to integrating data across bioclinical studies. Int J Epidemiol. 2010;39:1383–93. doi: 10.1093/ije/dyq139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.PhenX: Consensus measures for Phenotypes and eXposures. www.phenx.org (27 March 2012, date last accessed) [Google Scholar]
- 90.Kipnis V, Midthune D, Freedman L, et al. Bias in dietary-report instruments and its implications for nutritional epidemiology. Public Health Nutr. 2002;5:915–23. doi: 10.1079/PHN2002383. [DOI] [PubMed] [Google Scholar]
- 91.Von EE, Altman DG, Egger M, Pocock SJ, Gotzsche PC, Vandenbroucke JP. The Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) statement: guidelines for reporting observational studies. Lancet. 2007;370:1453–57. doi: 10.1016/S0140-6736(07)61602-X. [DOI] [PubMed] [Google Scholar]
- 92.Little J, Higgins JP, Ioannidis JP, et al. Strengthening the reporting of genetic association studies (STREGA): an extension of the strengthening the reporting of observational studies in epidemiology (STROBE) statement. J Clin Epidemiol. 2009;62:597–608. doi: 10.1016/j.jclinepi.2008.12.004. [DOI] [PubMed] [Google Scholar]
- 93.U.S. Department of Health and Human Services. How Tobacco Causes Disease: The Biology and Behavioral Basis for Tobacco-Attributable Disease. A Report of the Surgeon General. Rockville, MD: U.S. Dept. of Health and Human Services, Public Health Service, Office of Surgeon General; 2010. [Google Scholar]
- 94.World Cancer Research Fund and American Institute for Cancer Research. Food, Nutrition, Physical Activity and the Prevention of cancer: A global perspective. Washington DC: AICR; 2007. [Google Scholar]
- 95.Rossouw JE, Anderson GL, Prentice RL, et al. Risks and benefits of estrogen plus progestin in healthy postmenopausal women: principal results from the Women's Health Initiative randomized controlled trial. JAMA. 2002;288:321–33. doi: 10.1001/jama.288.3.321. [DOI] [PubMed] [Google Scholar]
- 96.Vineis P, Manuguerra M, Kavvoura FK, et al. A field synopsis on low-penetrance variants in DNA repair genes and cancer susceptibility. J Natl Cancer Inst. 2009;101:24–36. doi: 10.1093/jnci/djn437. [DOI] [PubMed] [Google Scholar]
- 97.Davey Smith G, Ebrahim S. ‘Mendelian randomization’: can genetic epidemiology contribute to understanding environmental determinants of disease? Int J Epidemiol. 2003;32:1–22. doi: 10.1093/ije/dyg070. [DOI] [PubMed] [Google Scholar]
- 98.Umbach DM, Weinberg CR. The use of case-parent triads to study joint effects of genotype and exposure. Am J Hum Genet. 2000;66:251–61. doi: 10.1086/302707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.VanderWeele TJ, Hernández-Díaz S, Hernán MA. Case-only gene-environment interaction studies: when does association imply mechanistic interaction? Genet Epidemiol. 2010;34:327–34. doi: 10.1002/gepi.20484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Davey Smith G, Ebrahim S. ‘Mendelian randomization’: can genetic epidemiology contribute to understanding environmental determinants of disease? Int J Epidemiol. 2003;32:1–22. doi: 10.1093/ije/dyg070. [DOI] [PubMed] [Google Scholar]
- 101.IARC Working Group on the Evaluation of Carcinogenic Risks to Humans. Aclohol consumption and ethyl carbamate (urethane) IARC Mongr Eval Carcinog Risks Hum. 2010;96:41–1284. [PMC free article] [PubMed] [Google Scholar]
- 102.IARC Working Group on the Evaluation of Carcinogenic Risks to Humans. Tobacco smoke and involuntary smoking. IARC Monogr Eval Carcinog Risks Hum. 2004;83:1–1452. [PMC free article] [PubMed] [Google Scholar]
- 103.Rothman N, Garcia-Closas M, Hein DW. Commentary: reflections on G.M. Lower and colleagues' 1979 study associating slow acetylator phenotype with urinary bladder cancer: meta-analysis, historical refinements of the hypothesis, and lessons learned. Int J Epidemiol. 2007;36:23–28. doi: 10.1093/ije/dym026. [DOI] [PubMed] [Google Scholar]
- 104.Rodrigues-Lima F, Dairou J, Dupret JM. Effect of environmental substances on the activity of arylamine N-acetyltransferases. Curr Drug Metab. 2008;9:505–9. doi: 10.2174/138920008784892092. [DOI] [PubMed] [Google Scholar]
- 105.Edenberg HJ. The genetics of alcohol metabolism: role of alcohol dehydrogenase and aldehyde dehydrogenase variants. Alcohol Res Health. 2007;30:5–13. [PMC free article] [PubMed] [Google Scholar]
- 106.Lewis SJ, Davey Smith G. Alcohol, ALDH2, and esophageal cancer: a meta-analysis which illustrates the potentials and limitations of a Mendelian randomization approach. Cancer Epidemiol Biomarkers Prev. 2005;14:1967–71. doi: 10.1158/1055-9965.EPI-05-0196. [DOI] [PubMed] [Google Scholar]
- 107.Boccia S, Hashibe M, Galli P, et al. Aldehyde dehydrogenase 2 and head and neck cancer: a meta-analysis implementing a Mendelian randomization approach. Cancer Epidemiol Biomarkers Prev. 2009;18:248–54. doi: 10.1158/1055-9965.EPI-08-0462. [DOI] [PubMed] [Google Scholar]
- 108.Concannon P, Haile RW, Borresen-Dale AL, et al. Variants in the ATM gene associated with a reduced risk of contralateral breast cancer. Cancer Res. 2008;68:6486–91. doi: 10.1158/0008-5472.CAN-08-0134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.El GF, Baan R, Straif K, et al. A review of human carcinogens–part D: radiation. Lancet Oncol. 2009;10:751–52. doi: 10.1016/s1470-2045(09)70213-x. [DOI] [PubMed] [Google Scholar]
- 110.Khanna KK, Chenevix-Trench G. ATM and genome maintenance: defining its role in breast cancer susceptibility. J Mammary Gland Biol Neoplasia. 2004;9:247–62. doi: 10.1023/B:JOMG.0000048772.92326.a1. [DOI] [PubMed] [Google Scholar]
- 111.Bernstein JL, Haile RW, Stovall M, et al. Radiation exposure, the ATM Gene, and contralateral breast cancer in the women's environmental cancer and radiation epidemiology study. J Natl Cancer Inst. 2010;102:475–83. doi: 10.1093/jnci/djq055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.IARC Working Group on the Evaluation of Carcinogenic Risks to Humans. Tobacco smoking. IARC Monogr Eval Carcinog Risks Hum. 1986;38:1–592. [PMC free article] [PubMed] [Google Scholar]
- 113.Secretan B, Straif K, Baan R, et al. A review of human carcinogens – Part E: tobacco, areca nut, alcohol, coal smoke, and salted fish. Lancet Oncol. 2009;10:1033–34. doi: 10.1016/s1470-2045(09)70326-2. [DOI] [PubMed] [Google Scholar]
- 114.Sanderson S, Salanti G, Higgins J. Joint effects of the N-acetyltransferase 1 and 2 (NAT1 and NAT2) genes and smoking on bladder carcinogenesis: a literature-based systematic HuGE review and evidence synthesis. Am J Epidemiol. 2007;166:741–51. doi: 10.1093/aje/kwm167. [DOI] [PubMed] [Google Scholar]
- 115.Kiemeney LA, Thorlacius S, Sulem P, et al. Sequence variant on 8q24 confers susceptibility to urinary bladder cancer. Nat Genet. 2008;40:1307–12. doi: 10.1038/ng.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wu X, Ye Y, Kiemeney LA, et al. Genetic variation in the prostate stem cell antigen gene PSCA confers susceptibility to urinary bladder cancer. Nat Genet. 2009;41:991–95. doi: 10.1038/ng.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Figueroa JD, Garcia-Closas M, Rothman N. Bladder cancer. In: Khoury MJ, Bedrosian SR, Gwinn M, Higgins JPT, Ioannidis JPA, Little J, editors. Human Genome Epidemiology. 2nd. New York: Oxford University Press; 2010. pp. 299–315. [Google Scholar]
- 118.Risch N, Herrell R, Lehner T, et al. Interaction between the serotonin transporter gene (5-HTTLPR), stressful life events, and risk of depression. JAMA. 2009;301:2462–71. doi: 10.1001/jama.2009.878. [DOI] [PMC free article] [PubMed] [Google Scholar]