SUMMARY
The plant hormone auxin controls numerous aspects of plant growth and development by regulating the expression of hundreds of genes. SMALL AUXIN UP RNA (SAUR) genes comprise the largest family of auxin-responsive genes, but their function is unknown. Although prior studies have correlated the expression of some SAUR genes with auxin-mediated cell expansion, genetic evidence implicating SAURs in cell expansion has not been reported. The Arabidopsis SAUR19, SAUR20, SAUR21, SAUR22, SAUR23, and SAUR24 (SAUR19– 24) genes encode a subgroup of closely related SAUR proteins. We demonstrate that these SAUR proteins are highly unstable in Arabidopsis. However, the addition of an N-terminal GFP or epitope tag dramatically increases the stability of SAUR proteins. Expression of these stabilized SAUR fusion proteins in Arabidopsis confers numerous auxin-related phenotypes indicative of increased and/or unregulated cell expansion, including increased hypocotyl and leaf size, defective apical hook maintenance, and altered tropic responses. Furthermore, seedlings expressing an artificial microRNA targeting multiple members of the SAUR19–24 subfamily exhibit short hypocotyls and reduced leaf size. Together, these findings demonstrate that SAUR19– 24 function as positive effectors of cell expansion. This regulation may be achieved through the modulation of auxin transport, as SAUR gain-of-function and loss-of-function seedlings exhibit increased and reduced basipetal indole-3-acetic acid transport, respectively. Consistent with this possibility, SAUR19–24 proteins predominantly localize to the plasma membrane.
Keywords: auxin, SAUR, Arabidopsis, cell expansion, protein stability, auxin transport
INTRODUCTION
The plant hormone indole-3-acetic acid (IAA or auxin) regulates many aspects of plant growth and development, including stem elongation, lateral branching of roots and shoots, establishment of embryonic polarity, vascular development, and tropic growth responses (Woodward and Bartel, 2005; Chapman and Estelle, 2009). These processes are controlled by auxin-mediated changes in cell division, expansion, and differentiation. Genomic expression studies have revealed that auxin regulates the expression of hundreds of genes (Nemhauser et al., 2006). Members of the Aux/IAA family of proteins repress auxin-inducible transcription by negatively regulating the transcriptional activity of AUXIN RESPONSE FACTOR (ARF) transcription factors. Upon an auxin stimulus, members of the TIR1/AFB family of F-box proteins bind the hormone, which stabilizes their interaction with Aux/IAA proteins, resulting in the ubiquitination of Aux/IAA proteins by the SCFTIR1/AFB ubiquitinligase (Dharmasiri et al., 2005; Kepinski and Leyser, 2005; Tan et al., 2007). Subsequent proteolysis of the ubiquitinated Aux/IAA proteins by the 26S proteasome results in de-repression of ARF transcriptional activity, leading to changes in the expression of auxin-regulated genes.
Among the genes most rapidly and strongly induced by auxin are several members of the SMALL AUXIN UP RNA (SAUR) gene family (Hagen and Guilfoyle, 2002). Originally identified as auxin-induced transcripts in soybean hypocotyls (McClure and Guilfoyle, 1987), SAUR genes have subsequently been identified in a wide range of plants, and genomic sequencing projects have revealed that SAURs are present as large gene families in monocots (58 in rice), dicots (78 in Arabidopsis), and moss (18 in Physcomitrella). These genes encode small proteins (86–189 amino acids in Arabidopsis) that are unique to the plant kingdom and contain no recognizable motifs suggestive of a possible biochemical function.
In the 25 years or so since their initial discovery, SAUR genes have been widely employed as auxin-inducible reporters. Notably, several SAUR genes have been found to be highly expressed in tissues undergoing differential cell expansion such as occurs during tropic growth (McClure and Guilfoyle, 1989; Knauss et al., 2003; Esmon et al., 2006). Molecular studies have also demonstrated that many SAUR genes contain a conserved downstream element (DST) in their 3′-untranslated region (UTR) that confers mRNA instability in an auxin-independent manner (Newman et al., 1993; Gil and Green, 1996). SAUR expression may also be regulated at the protein level, as one report found that the maize ZmSAUR2 protein is very short-lived (Knauss et al., 2003). Likewise, analysis of Arabidopsis plants expressing a SAUR15–luciferase fusion protein suggested that SAUR proteins are unstable (Zenser et al., 2003). Although limited in both scope and depth, these studies suggest that SAUR expression is closely associated with auxin-mediated cell expansion and is regulated at the transcriptional, posttranscriptional, and protein levels.
Surprisingly little information has been garnered regarding the functions of SAUR proteins. Most likely this is due to genetic redundancy within the large SAUR gene family, which greatly complicates forward genetic studies and may explain why no loss-of-function saur mutant has been described. ZmSAUR2 and a few additional maize and Arabidopsis SAUR proteins have been shown to bind calmodulin in vitro (Yang and Poovaiah, 2000; Knauss et al., 2003; Popescu et al., 2007), but the biological significance of these findings remains uncertain. Two recent reports have begun to examine SAUR functions. First, overexpression of SAUR32 in Arabidopsis interferes with apical hook maintenance during etiolated growth (Park et al., 2007). SAUR32, however, does not appear to be auxin responsive. Secondly, OsSAUR39 overexpression in rice confers several modest phenotypes including reductions in lateral root development, yield, and shoot and root lengths (Kant et al., 2009). These plants also exhibited reduced basipetal IAA transport and slightly lower IAA levels than controls.
Here, we present a reverse genetic analysis of a subgroup of closely related Arabidopsis SAUR genes, designated SAUR19, SAUR20, SAUR21, SAUR22, SAUR23, and SAUR24 (SAUR19–24). We find that these SAUR proteins are highly unstable. However, when expressed as N-terminal fusion proteins they are dramatically stabilized, resulting in increased activity. Arabidopsis plants expressing these stabilized SAUR fusion proteins exhibit several cell expansion- related growth defects, including root waving, increased hypocotyl elongation, larger leaf size, reduced phototropism, and impaired apical hook maintenance. Furthermore, plants expressing an artificial microRNA (ami- RNA) targeting three members of the SAUR19–24 subfamily exhibit reductions in hypocotyl elongation and leaf size, thus providing loss-of-function genetic support that SAUR proteins function as important regulators of plant cell growth.
RESULTS
Auxin induction of SAUR19–24 gene expression
The SAUR19–24 genes of Arabidopsis are found in a tandem array on chromosome V and encode highly related (93–96% identity) proteins. Prior phylogenetic analysis revealed that SAUR19–24 form a unique clade in Arabidopsis (Jain et al., 2006). At 88–91 amino acids, members of the SAUR19–24 clade are among the smallest SAUR proteins. In general, larger SAURs have N- or C-terminal extensions, with their middle regions being most closely related to SAUR19–24 (Figure S1). As such, SAUR19–24 may represent a ‘minimal’ or ‘core’ SAUR sequence.
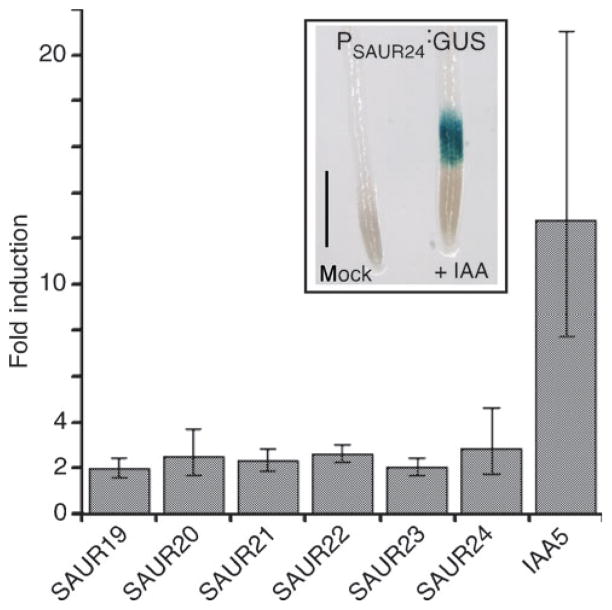
While the expression of several SAUR genes has been shown to be auxin-inducible, the SAUR19–24 family has not been examined in detail. SAUR23 is represented on the Affymetrix 22K gene chip and has been shown to be auxin responsive (Nemhauser et al., 2006). However, the oligo probe set (250012_x_at*) is not specific for SAUR23 and detects multiple members of this subfamily. Using gene-specific primers from SAUR19–24 UTR regions and RNA prepared from whole seedlings, our quantitative RT-PCR analysis detected a two- to three-fold up-regulation of all six SAUR19–24 family members following a 30-min auxin treatment (Figure 1). While auxin-inducible expression of the SAUR19–24 family members was relatively modest compared to the strongly auxin-inducible IAA5 gene, tissue- specific increases in expression were observed with SAUR19–24 promoter-GUS reporters in some instances. Most strikingly, PSAUR24-GUS expression was strongly auxin-induced in the root elongation zone (Figure 1).
Figure 1. SAUR19–24 are rapidly induced by auxin.
Quantitative PCR analysis of SAUR gene expression in wild-type seedlings following a 30-min treatment with 0.35 μM indole-3-acetic acid (IAA). Fold induction over mock-treated control seedlings is presented ± standard deviation.
Inset: Col seedlings harboring a PSAUR24:GUS transgene were treated with 1.0 μM IAA for 2 h and stained for GUS activity. The increase in GUS expression was primarily confined to the root elongation zone. Scale bar = 0.5 mm.
Overexpression of SAUR fusion proteins confers auxin-related phenotypes
To explore potential roles for SAUR19–24 in auxin-regulated growth and development, we employed an overexpression approach by generating transgenic plants expressing a GFP-SAUR19 fusion protein from the cauliflower mosaic virus 35S promoter. While the results described below focus primarily on GFP-SAUR19 seedlings, similar findings were obtained with seedlings expressing GFP fusions to SAUR21, SAUR23, and SAUR24, which is expected given the high degree of sequence identity among SAUR19–24 family members.
Seedlings expressing the GFP–SAUR19 fusion protein displayed several auxin-related phenotypes. Light-grown GFP–SAUR19 seedlings exhibited longer hypocotyls than wild-type controls (Figure 2a). This increase in hypocotyl length was due to increased cell expansion, as the change in hypocotyl epidermal cell length (two-fold) was very similar to the change in total organ length (1.96-fold) (Figure 2b). When grown in the dark, hypocotyl elongation of GFP– SAUR19 seedlings was comparable to wild-type controls (Figure 2a). However, as previously reported for SAUR32 overexpression (Park et al., 2007), GFP–SAUR19 seedlings were defective in apical hook maintenance (Figure 2c). Surprisingly, unlike 35S:GFP–SAUR19–24 transgenic lines, plants expressing untagged 35S:SAUR19 or 35S:SAUR24 constructs appeared indistinguishable from wild-type controls (Figure 2a). These findings led us to consider the possibility that the GFP–SAUR19 phenotypes may be an artifact of the GFP tag. We therefore generated additional N-terminal SAUR19 and SAUR24 fusion constructs that contained the much smaller StrepII epitope tag. Similar to the GFP–SAUR19–24 transgenic lines, seedlings expressing 35S:StrepII–SAUR19 or 35S:StrepII–SAUR24 constructs exhibited an approximately two-fold increase in hypocotyl elongation, as well as the other phenotypes seen with the GFP-tagged lines (Figure 2a). Together, these findings suggest that the addition of a protein or epitope tag to the N-termini of SAUR19–24 family members uniquely alters the activity of these SAUR proteins.
Figure 2. Phenotypes resulting from SAUR19–24 fusion protein overexpression.
(a) Hypocotyl length of 10-day-old (d.o.) light-grown seedlings and 7 d.o. dark-grown seedlings. All transgenes were expressed from the 35S promoter. S19 = SAUR19, S21 = SAUR21, etc. Specific transgenic lines are indicated by the alphanumeric designations following the semicolons.
(b) Mean hypocotyl epidermal cell lengths were calculated by measuring 12 epidermal cells from the mid-hypocotyl region of 10, 10 d.o. light-grown seedlings.
(c) Seven-day-old etiolated seedlings.
(d) Root waving phenotype of GFP-SAUR19 seedlings grown on vertically oriented plates.
(e) Lateral roots/cm primary root of 10 d.o. seedlings.
(f) Enhanced root waving when grown on a 45° incline.
(g) Reduced phototropic growth of GFP-SAUR19 seedlings. Five-day-old etiolated seedlings were photo-stimulated with lateral blue light and the angle of hypocotyl bending measured at the indicated time points. All error bars indicate standard deviation (n ≥ 10). P-values (*P ≤ 0.05; **P ≤ 0.01) as determined using an unpaired Student’s t-test of each transgenic line with Col-0.
In addition to the long hypocotyl phenotype, GFP– and StrepII–SAUR19–24 seedlings exhibited increased root waving on vertically oriented agar plates (Figure 2d). These seedlings also displayed a modest increase in lateral root development (Figure 2e). However, this is probably a consequence of the exaggerated wavy root growth habit, as root waving is known to elicit lateral root development on the convex side of a curved root (De Smet et al., 2007; Richter et al., 2009). Wild-type Arabidopsis seedlings exhibit root waving when grown on inclined agar plates. This growth response has been proposed to result from the combined influences of gravitropism and thigmotropism, as well as root circumnutation (Migliaccio and Piconese, 2001). Growth on inclined agar plates further exacerbated the root waving phenotype of GFP-SAUR19 seedlings (Figure 2f). Auxin transport is required to propagate the differential cell growth underlying root waving, as mutants in the auxin efflux carrier PIN2/WAV6/EIR1/AGR1 do not exhibit wavy root growth on inclined agar plates (Okada and Shimura, 1990; Luschnig et al., 1998). We therefore examined whether PIN2 was required for GFP–SAUR19-mediated root waving. Loss of PIN2 noticeably diminished the enhanced root waving phenotype of GFP–SAUR19 seedlings both on vertical and inclined agar plates (Figure 2f).
The wavy root phenotype of GFP–SAUR19 seedlings suggested that tropic growth may be altered. While root gravitropism assays with GFP–SAUR19 seedlings were complicated by the root waving phenotype, we detected a clear reduction in hypocotyl phototropism when etiolated seedlings were photo-stimulated with unilateral blue light (Figure 2g). Similar findings were obtained in bending assays with post-photomorphogenic seedlings (Figure S2). This reduction in phototropic growth was not due to diminished blue light signaling, as GFP–SAUR19 hypocotyl elongation was inhibited by blue light comparably to Col-0 controls (Figure S2). Together, these findings demonstrate that GFP–SAUR19 expression interferes with normal tropic growth.
GFP–SAUR19 expression confers increased leaf size
The increased hypocotyl length and cell size of N-terminally tagged SAUR19–24 transgenic seedlings prompted us to examine whether other plant organs might be similarly affected. Two GFP–SAUR19 transgenic lines were grown alongside Col-0 control plants and several vegetative parameters were measured. After 20 days of growth, both GFP–SAUR19 lines displayed increased vegetative biomass, conferring approximately 30 and 20% increases in fresh and dry weights, respectively (Figure 3a,b). Furthermore, GFP–SAUR19 expression resulted in increased leaf areas at almost all positions of the rosette (Figure 3c). Final leaf size is determined by both cell division and cell expansion. To analyze the contributions of cell proliferation and cell expansion to the enlargement in leaf area of GFP–SAUR19 plants, epidermal cell number and size were analyzed with leaf #3 harvested from 21-day-old plants. While the total number of leaf cells was comparable to wild-type controls, the average leaf cell area was 26 and 40% greater in the two GFP–SAUR19 lines (Figures 3d,e). Taken together, these results demonstrate that overexpression of GFP–SAUR19 promotes leaf cell expansion, leading to increased leaf size and vegetative biomass.
Figure 3. GFP-SAUR19 promotes increased leaf size.
Fresh (a) and dry weights (b) of vegetative tissue from 20-day-old plants grown on MS plates (n = 8–12).
(c) Leaf area measurements from 20-day-old plants. Numbers on the x-axis indicate leaf position.
(d) Mean leaf epidermal cell area and (e) cell number obtained from leaf #3 of 21-day-old plants. All error bars represent standard deviation. *P ≤ 0.01; **P ≤ 0.01 for one of the two GFP–SAUR19 lines shown in panel (c).
Induction of SAUR19–24 expression in elongating tissues
The phenotypes displayed by plants expressing N-terminally tagged SAUR19–24 fusion proteins described above can be explained by increased or improper regulation of cell expansion. Based on prior expression studies, SAUR genes have been proposed to function in some aspect of auxin-mediated cell elongation (McClure and Guilfoyle, 1989; Knauss et al., 2003; Esmon et al., 2006). We therefore examined the expression of PSAUR-GUS reporters under conditions that promote cell expansion. We first compared expression between 5-day-old light- or dark-grown seedlings. Compared with light-grown seedlings, GUS activity was dramatically higher in the elongating hypocotyls of etiolated seedlings for all four SAUR promoter-GUS reporters examined (Figure 4a). We next examined expression in seedlings grown under a low red:far-red (R:FR) light ratio to simulate shade conditions and promote the shade avoidance response. This growth response is known to involve a transient increase in IAA levels and the induction of auxin responsive gene expression (Tao et al., 2008). Once again, we observed strong induction of PSAUR-GUS expression in the elongating hypocotyls of seedlings grown under simulated shade conditions (Figure 4b). Consistent with these reporter findings, Tao et al. (2008) observed a 7.7-fold increase in SAUR23* (250012_x_at*) expression after 1 h of transfer to simulated shade.
Figure 4. Expression of SAUR19 family members in elongating tissues.
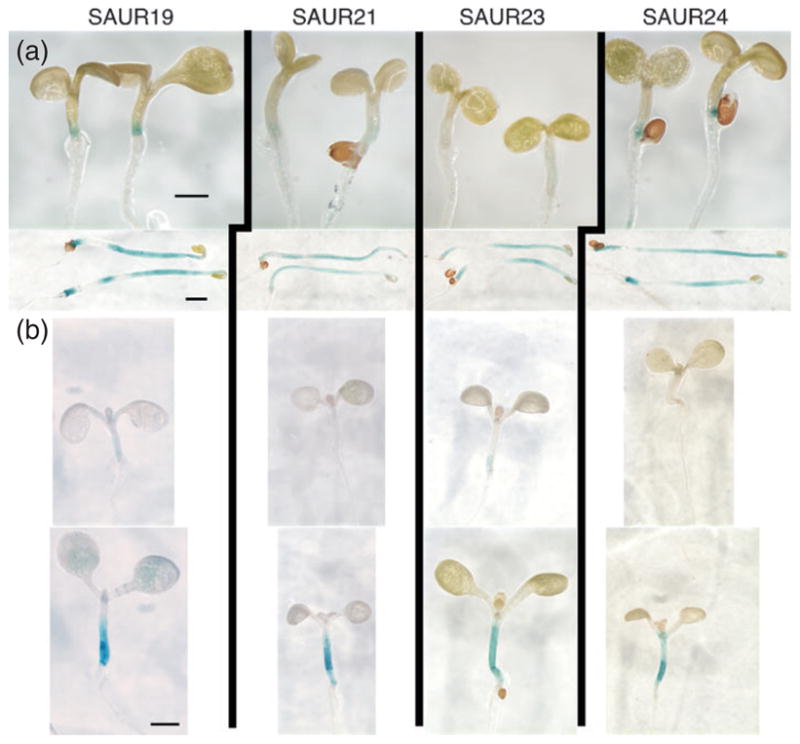
Promoter–GUS reporter constructs for SAUR19, -21, -23, and -24 were introduced into Col-0 plants.
(a) Five-day-old light- (top) or dark-grown (bottom) seedlings were stained for β-glucuronidase activity. Seedlings were stained for the same duration (1.5 h) to compare expression levels between the two growth conditions.
(b) Five-day-old light-grown seedlings were shifted to Wc (top) or low red:far-red (bottom) lighting for 24 h and stained for β-glucuronidase activity. Size bars = 1 mm.
Auxin-mediated cell expansion is also required for tropic growth. We examined the expression of different SAUR promoter–GUS reporter lines at various times following phototropic or gravitropic stimulation. However, we could not convincingly detect differential expression between the elongating and non-elongating sides of seedling hypocotyls or roots.
N-terminal tags stabilize SAUR19, facilitating overexpression
Our finding that plants expressing N-terminally tagged SAUR fusion proteins displayed several striking phenotypes, but plants overexpressing untagged SAUR proteins exhibited no obvious phenotypes, prompted us to examine the basis for this difference. We first confirmed that these two types of SAUR19 transgenes were expressed at comparable levels in our transgenic lines. Northern blots hybridized with a SAUR19 cDNA probe detected very high transcript levels for both SAUR19 transgenes (Figure 5a). We next generated polyclonal antisera against recombinant SAUR19 protein to assess protein levels (Figure 5b). While our α-SAUR19 antisera detected a strong signal for the GFP– and StrepII– SAUR19 fusion proteins, untagged SAUR19 protein was only detected following prolonged film exposures (Figure 5c). The endogenous SAUR19 protein was undetectable. This finding demonstrates that the GFP- and Strep-tagged fusion proteins accumulate to a much higher level than untagged SAUR19. To better quantify the steady-state levels of the GFP–SAUR19 and untagged SAUR19 transgenic proteins, we probed blots containing a dilution series of 35S:GFP–SAUR19 extract and 30 μg 35S:SAUR19 extract (Figure S3). This analysis demonstrated that the GFP– SAUR19 fusion protein is more than 30 times more abundant than untagged SAUR19. To determine whether the level of SAUR19 fusion protein correlated with the severity of the resulting phenotypes, we compared GFP– SAUR19 levels in two transgenic lines that consistently exhibited differences in hypocotyl length and root waving severity. The α-GFP western blots of seedling extracts revealed that GFP–SAUR19 abundance and phenotypic severity were indeed correlated (Figure 5d).
Figure 5. The RNA and protein expression of SAUR19 transgenic lines.

(a) Northern blot of seedling RNA probed with SAUR19 cDNA. The endogenous SAUR19 transcript is not seen in this exposure. Extraneous lanes between lanes 2 and 3 were spliced out of the image.
(b) α-GFP and α-SAUR19 western blots of 7-day-old seedling extracts.
(c) α-SAUR19 western blot. Panels I and II are equivalent exposures (15 sec). Panel III is a longer exposure (20 min) of the same blot shown in panel II. Panel IV shows a non-specific cross-reacting band used as a loading control.
(d) Hypocotyl length and GFP–SAUR19 abundance in two 35S:GFP–SAUR19 transgenic lines. Error bars indicate standard deviation (n = 12). †Indicates cross-reacting upper band used as a loading control.
These findings demonstrate a direct relationship between the accumulation of the transgenic SAUR19 protein and the resulting phenotypes. We therefore hypothesized that SAUR19–24 family members might be highly unstable proteins, and that the N-terminal tags have a stabilizing effect, leading to accumulation of the respective fusion protein and gain-of-function phenotypes. Consistent with this idea, the addition of N-terminal epitope or protein tags has previously been found to confer increased stability to several proteins (Breitschopf et al., 1998; Reinstein et al., 2000; Bloom et al., 2003; Coulombe et al., 2004). We first examined the stability of SAUR19 and N-terminally tagged derivatives in an in vitro protein degradation assay. Glutathione S-transferase (GST)–SAUR19 was expressed and purified from Escherichia coli, and the resulting fusion protein was subjected to a partial thrombin cleavage to release SAUR19 from the N-terminal GST tag. The resulting mixture of GST–SAUR19 and untagged SAUR19 proteins was then incubated with wild-type seedling extracts and degradation over a 30-min time course assessed by western blotting. While the GST–SAUR19 substrate persisted throughout the course of this assay, levels of the untagged SAUR19 substrate rapidly diminished (Figure 6a). Inclusion of the 26S proteasome inhibitor MG132 in these assays significantly stabilized SAUR19, suggesting that SAUR stability is regulated by the ubiquitin/26S proteasome pathway.
Figure 6. N-terminal tags stabilize SAUR19.

(a) In vitro degradation assay comparing GST–SAUR19 (GST-S19) and SAUR19 (S19) stability. Time (t) is in minutes. The right panel shows a shorter time course.
(b) α-SAUR19 western blot of 6-day-old etiolated seedlings expressing both 35S:GFP-SAUR19 and 35S:SAUR19 transgenes. Seedlings were treated with 40 μg ml−1 cycloheximide (CHX) or solvent control and extracts prepared at 10-min intervals.
To directly compare the stabilities of SAUR19 and a SAUR19 fusion protein in vivo, we transformed 35S:GFP– SAUR19 (line 1-1) plants with the 35S:SAUR19 transgene. Etiolated seedlings homozygous for both transgenes were then used for cycloheximide-chase assays. While GFP– SAUR19 was stable over the time-course examined, untagged SAUR19 was highly labile, with levels showing a rapid decline within 10 min of cycloheximide addition (Figure 6b). Together, our in vitro and in vivo studies clearly demonstrate that N-terminal tags have a dramatic stabilizing effect on the SAUR19 protein.
SAUR19 localization
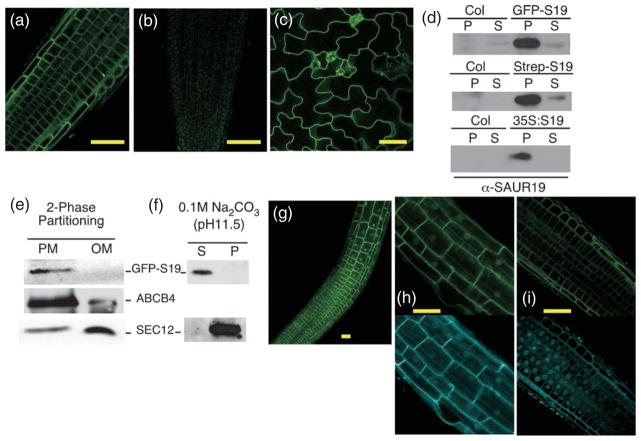
Prior studies examining SAUR protein localization utilizing fusion proteins with GFP or GUS have yielded mixed results. Whereas some studies have detected SAUR fusion proteins primarily in nuclei (Knauss et al., 2003; Park et al., 2007), one recent study examining GFP-tagged OsSAUR39 found exclusive localization in the cytoplasm (Kant et al., 2009). Upon examining light-grown GFP–SAUR seedlings by confocal microscopy, we were surprised to find GFP fusions to SAUR19, -21, -23, and -24 all localized primarily to the cell surface, suggesting association with the plasma membrane (Figures 7a–c). Microsomal fractionation studies confirmed that the GFP–SAUR19 protein is membrane associated (Figure 7d). To confirm that the membrane association observed with the GFP–SAUR19 fusion protein was not an artifact of the GFP tag, we examined the fractionation of untagged SAUR19 as well as the StrepII–SAUR19 fusion protein. Consistent with our GFP–SAUR19 findings, the untagged and StrepII-tagged SAUR19 derivatives were also highly enriched in microsomal pellets (Figure 7d). To confirm that membrane association was with plasma membranes as suggested by our microscopy studies, microsomal fractions were subjected to two-phase partitioning assays. The GFP–SAUR19 protein was highly enriched in plasma membrane-enriched fractions, whereas the endoplasmic reticulum-associated SEC12 protein was found predominantly in the ‘other membrane’ fraction (Figure 7e). Consistent with the lack of a predicted transmembrane spanning domain in the SAUR19 sequence, GFP–SAUR19 could be extracted from membrane fractions with high-pH buffer, indicating that SAUR19 is a peripheral rather than an integral membrane protein (Figure 7f).
Figure 7. SAUR19 localizes primarily to the plasma membrane.
Confocal images of 35S:GFP–SAUR19 fluorescence in (a) root basal meristem, (b) Col control root meristem, and (c) leaf epidermal cells.
(d) α-SAUR19 western blots of seedling extracts fractionated into soluble (S) and microsomal pellet (P) fractions.
(e) α-GFP western blot of 35S:GFP–SAUR19 seedling extracts subjected to two-phase membrane partitioning. ABCB4 and SEC12 are shown as plasma membrane (PM) and other membrane (OM) controls.
(f) Microsomal fractions of 35S:GFP–SAUR19 extracts were treated with 0.1 M Na2CO3 to extract peripheral membrane proteins. GFP–SAUR19 is present in the extraction supernatant (S), whereas the integral membrane protein SEC12 remains in the extracted pellet (P).
(g) GFP–SAUR19 fluorescence in 7-day-old root meristem and elongation zones. Higher magnification of GFP–SAUR19 fluorescence in cells of the root elongation zone (h) and root meristem region (i). The bottom panels in (h) and (i) show nuclei stained with Hoechst dye. Size bars = 50 μm.
While GFP–SAUR19 localized to the plasma membrane in the vast majority of cells, nuclear fluorescence was also observed in cells comprising the root elongation zone (Figure 7g,h). Counter-staining nuclei with Hoechst dye confirmed that GFP–SAUR19 was present within the nuclei of cells comprising the root elongation zone, but largely absent from nuclei of cells in the root meristem (Figure 7i).
GFP–SAUR19 seedlings exhibit altered IAA response and transport
To begin to elucidate the mechanisms responsible for the phenotypes conferred by tagged SAUR19–24 transgenes, we examined the effects of increased SAUR19–24 activity on auxin response and transport. We first examined auxin responsiveness by monitoring root growth inhibition by the synthetic auxin 2,4-dichlorophenoxy acetic acid (2,4-D). The GFP–SAUR19 and StrepII–SAUR19 seedlings both displayed significant auxin resistance compared with Col-0 control seedlings (Figure 8a). However, this resistance was considerably weaker than that conferred by the tir1-1 mutation, which itself is a weak auxin response mutant. To examine auxin-induced gene expression, we crossed the DR5-GUS reporter into plants harboring the GFP–SAUR19 transgene. While the overall pattern of GUS expression was not noticeably altered by the expression of the SAUR transgene, we consistently detected a slight reduction in the intensity of DR5-GUS staining following short inductions with auxin (Figure 8b). Together, these findings indicate that these stabilized SAUR19 derivatives confer a slight reduction in auxin response.
Figure 8. Auxin response and transport are altered in 35S:GFP–SAUR19 seedlings.
(a) Inhibition of root elongation by increasing concentrations of the synthetic auxin 2,4-dichlorophenoxy acetic acid (2,4-D). Five-day-old seedlings were transferred to medium containing 2,4-D and grown for an additional 3 days. Error bars depict standard deviation from the mean (n ≥ 12).
(b) Ten-day-old seedlings were treated with 1 μM indole-3-acetic acid (IAA) for 0, 2, or 4 h and stained for β-glucuronidase activity for 1 h.
(c) Five-day-old Col-0 and 35S::GFP–SAUR19 seedlings were transferred to media containing 10 μM 1- naphthylphthalamic acid (NPA) and grown for an additional 4 days.
(d) Ten nanoliters of radiolabeled IAA was deposited at the shoot apex of light-grown seedlings (10 per genotype) and transport down the hypocotyls was measured as described by Christie et al. (2011). Error bars indicate standard deviation from three experiments.
Our finding that GFP–SAUR19 localized primarily to plasma membranes led us to consider the possibility that these SAUR proteins play a role in IAA transport. To assess potential differences in IAA transport, we examined root growth of GFP–SAUR19 seedlings in the presence of the polar auxin transport inhibitor, 1-naphthylphthalamic acid (NPA). The GFP–SAUR19 seedlings exhibited hypersensitivity to NPA, as indicated by strongly agravitropic root growth on concentrations of NPA that had comparatively minor effects on wild-type controls (Figures 8c, S4). When basipetal IAA transport was measured directly, we detected an approximately 75% increase in basipetal IAA transport in the hypocotyls of light-grown seedlings (Figure 8d).
Reduced expression of SAUR19–24 confers diminished cell expansion
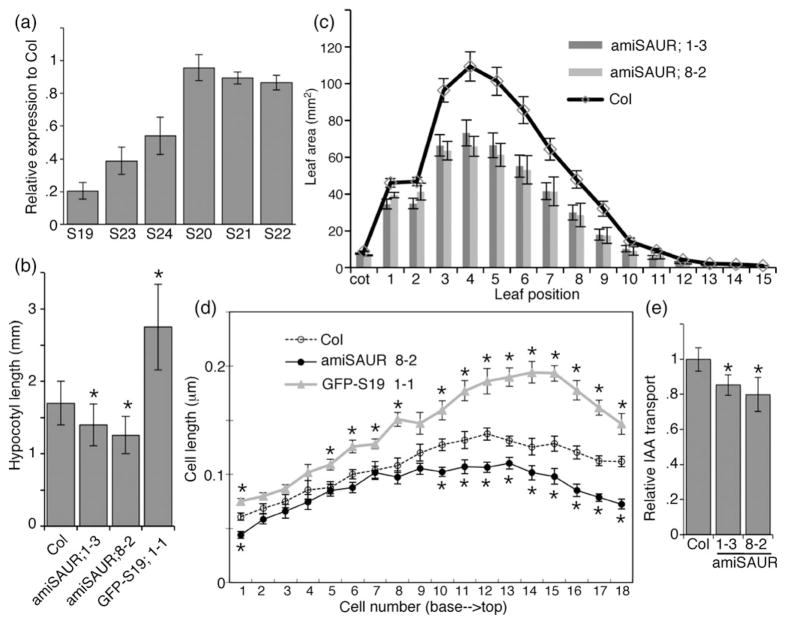
To obtain loss-of-function genetic support for our hypothesis that SAUR19–24 promote cell expansion, we constructed several amiRNA constructs designed to target multiple members of this gene family. While most of these constructs failed to effectively knock down SAUR19–24 transcript levels, the amiSAUR19/23/24 construct diminished transcript levels of these three target genes (Figures 9a, S5). We therefore examined amiSAUR19/23/24 lines for phenotypes related to those conferred by overexpression of the N-terminally tagged SAUR19–24 constructs. We detected slight but consistent reductions in both hypocotyl length and leaf size in multiple lines expressing the amiSAUR19/23/24 transgene (Figures 9b,c). Mean epidermal cell length of amiSAUR19/23/24 hypocotyls was significantly shorter than that of wild-type controls (Figure 9d). Since all tested members of the SAUR19–24 family are strongly induced by growth under shade avoidance conditions (Figure 4b), we examined the shade avoidance response in amiSAUR19/23/ 24 seedlings. However, while amiSAUR19/23/24 seedlings exhibited shorter hypocotyls than Col-0 controls under both low white and low R:FR lighting, the magnitude of increase in hypocotyl length in response to shade avoidance conditions was comparable (Figure S6).
Figure 9. Phenotypes of seedlings expressing an artificial microRNA (ami-RNA) targeting SAUR19–24 family members.
(a) Quantitative PCR analysis of SAUR19–24 expression in amiSAUR19/23/24 line 8-2. Expression levels are shown relative to wild-type Col-0 seedlings.
(b) Mean hypocotyl length of 7-day-old (d.o.) seedlings. Error bars indicate standard deviation (n = 50). Asterisks indicate significant difference from Col control (P < 0.05).
(c) Mean leaf size of 25 d.o. plants. Error bars indicate standard deviation.
(d) Mean hypocotyl epidermal cell length of 9 d.o. seedlings. Error bars indicate standard error (n = 10).
(e) Relative basipetal indole-3-acetic acid (IAA) transport through seedling hypocotyls. Values depict the mean of three replicate assays (10 seedlings each). Asterisks indicate significant difference from the Col control (P < 0.05).
Given that we detected a significant increase in basipetal IAA transport in hypocotyls of GFP–SAUR19 seedlings (Figure 8d), we conducted similar assays with amiSAUR19/ 23/24 seedlings. In contrast to the increased IAA transport displayed by GFP–SAUR19 seedlings, amiSAUR seedlings exhibited a 15–20% reduction in polar auxin transport (Figure 9e). Thus, in both phenotypic assays and auxin transport assays, plants expressing the amiSAUR19/23/24 transgene behave in an opposite way to plants overexpressing N-terminally tagged SAUR19–24 fusion proteins.
DISCUSSION
SAUR, Aux/IAA, and GH3 genes comprise three large gene families that are rapidly induced in response to auxin. While the molecular functions of Aux/IAA and GH3 genes have begun to be elucidated (Woodward and Bartel, 2005), the function of SAUR genes has remained elusive. Loss-of-function studies investigating SAUR activities are severely hampered by functional redundancy amongst the many members of this gene family in Arabidopsis and other plants. Furthermore, since the most closely related SAUR genes are typically found in tandem arrays on the chromosome, construction of higher-order mutants is exceedingly difficult. Thus, as was the case for the Aux/IAA gene family, gain-of-function genetic studies may be needed to gain insight into SAUR functions.
In this study, we find that expression of N-terminally tagged SAUR19–24 proteins confers numerous cell expansion phenotypes. In marked contrast, overexpression of untagged SAUR19 family members does not noticeably alter plant growth. Our findings indicate that the phenotypes conferred by the N-terminally tagged SAUR19–24 fusion proteins are the result of gain-of-function activity conferred by the tag. Molecular studies demonstrate that the addition of an N-terminal tag to SAUR19 family members confers a stabilizing effect on the resulting fusion protein. We demonstrate that SAUR19 is highly unstable in both in vitro and in vivo assays. In contrast, N-terminally tagged SAUR19 fusion proteins are stable, and consequently accumulate when expressed in plants, thus facilitating overexpression and increased SAUR activity. Since SAUR19 degradation is inhibited by the 26S proteasome inhibitor, MG132, the presence of an N-terminal tag may stabilize SAUR proteins by interfering with recognition by a ubiquitin protein ligase. Fusions to the N-termini of SAUR19–24 could potentially disrupt N-end rule regulation of SAUR protein stability. All six of these SAUR proteins contain an alanine at the second position, raising the possibility that these proteins are subject to the recently discovered N-acetylation branch of the N-end rule pathway (Hwang et al., 2010). Alternatively, the tags could interfere with SAUR ubiquitination directly. While ubiquitin is typically conjugated to an ε-NH2 internal lysine residue, it can also be conjugated to the α-NH2 group of an N-terminal residue (Ciechanover and Ben-Saadon, 2004; Coulombe et al., 2004). Regardless of the mechanism, our discovery of the stabilizing effect of N-terminal tags provides an additional approach for investigating the functions of SAUR proteins and points toward potential regulatory mechanisms involving the ubiquitin/26S proteasome pathway for modulating SAUR activity. Interestingly, compared to the SAUR19–24 clade, most other Arabidopsis SAUR proteins contain N-terminal extensions of 12–50 amino acids (Figure S1). It is tempting to speculate that these N-terminal differences may confer important differences in protein stability or other regulatory aspects of SAUR protein function.
Analysis of amiSAUR19/23/24 lines further supports our conclusion that the N-terminally tagged SAUR19–24 fusion proteins confer increased SAUR activity. In sharp contrast to GFP–SAUR19 plants, amiSAUR seedlings consistently exhibit short hypocotyl and small leaf phenotypes, and diminished basipetal IAA transport. While all of these effects are relatively modest, this is probably because the amiSAUR construct only efficiently knocks down expression of a subset of the SAUR19–24 family. In addition to SAUR20– 22, several additional SAUR genes, including SAUR26–29, share a high degree of sequence similarity with the SAUR19–24 clade and thus may provide overlapping functions. We are currently generating amiRNA constructs targeting additional SAUR family members to test this possibility.
The strong induction of SAUR19–24 expression in elongating tissues is consistent with our hypothesis that the encoded proteins positively regulate cell expansion as indicated by our gain- and loss-of-function genetic studies. All four of the SAUR19–24 family members included in our promoter–GUS studies were strongly up-regulated in the elongating hypocotyls of seedlings grown in the dark or under shade avoidance conditions. Furthermore, in a separate study (Franklin et al., 2011), we found that these SAUR genes are also up-regulated in hypocotyls in response to high temperature, a third environmental condition that promotes hypocotyl elongation (Gray et al., 1998).
The precise mechanism by which SAUR19–24 promote expansion is unclear. Overexpression of N-terminally tagged SAUR19 fusion proteins conferred reduced auxin response in both root growth and DR5-GUS assays. This reduction was slight, however, as the weak tir1-1 mutant was considerably more resistant to exogenous auxin than the tagged SAUR19 lines. No significant difference in auxin sensitivity was apparent in root growth assays with ami- SAUR19/23/24 seedlings (Figure S7). In contrast, both gain-and loss-of-function SAUR19–24 lines displayed altered auxin transport. Thus, the primary role of SAUR19–24 proteins in the auxin pathway may involve IAA transport. Numerous studies have shown that perturbation of IAA transport confers altered cell expansion phenotypes similar to those we observe with our SAUR transgenic lines. For example, NPA-mediated inhibition of polar auxin transport inhibits hypocotyl elongation (Jensen et al., 1998), as do mutations in some members of the ABCB family of auxin transporters (Lin and Wang, 2005). Reciprocally, overexpression of ABCB1 results in long hypocotyls as a result of increased cell expansion (Sidler et al., 1998). Likewise, genetic studies of several IAA influx and efflux carriers have demonstrated important roles for IAA transport during apical hook development and opening (Vandenbussche et al., 2010; Wu et al., 2010; Zádníková et al., 2010). Additionally, PIN3 was recently found to play a critical role in the cell elongation associated with both phototropic bending and shade avoidance (Keuskamp et al., 2010; Ding et al., 2011). Finally, mutations in the PINOID-like kinases WAG1 and WAG2 confer an exaggerated root waving phenotype (Santner and Watson, 2006). While there is less evidence implicating auxin transport in regulating cell expansion associated with leaf growth, one recent report did find that ABCB19-mediated IAA transport promotes cotyledon expansion (Lewis et al., 2009). Given that SAUR19–24 gain-of-function plants exhibit defects in all of the above processes, and amiSAUR19/23/24 loss-of-function plants display reciprocal defects in a subset, it seems likely that altered auxin transport may be the basis for these phenotypes.
Our confocal microscopy and cell fractionation studies revealed association with the plasma membrane. Thus, SAUR19 is properly positioned to participate in some aspect of auxin transport. While it seems unlikely that SAUR proteins transport auxin directly, the possibility remains that SAUR19 family members modulate the activities of established auxin transporters such as members of the PIN, ABCB, or AUX1/LAX families. Prior studies examining the localization of other SAUR proteins from a variety of plants utilizing GFP or GUS fusions have reported localization to the nucleus or cytoplasm (Knauss et al., 2003; Park et al., 2007; Kant et al., 2009), but not the plasma membrane. However, none of these prior studies have included SAUR proteins closely related to the SAUR19–24 family. Together, these findings suggest that different SAUR proteins may have distinct intracellular localizations and functions. Alternatively, SAUR localization may vary depending on the tissue examined, the developmental state, or in response to endogenous or exogenous signals. Indeed, in addition to the plasma membrane localization, we also detected nuclear GFP–SAUR19 in certain cells, such as within the root elongation zone. We considered the possibility that SAUR protein localization might be regulated by auxin, but we observed no detectable effect of IAA treatments on GFP– SAUR19 localization.
In recent years, dramatic advances have been made in our understanding of auxin signaling, transport, and metabolism. The function of SAUR genes, however, has remained enigmatic. Our analysis of the closely related SAUR19–24 subgroup of Arabidopsis SAURs demonstrates that these proteins are important regulators of plant cell expansion. Furthermore, the expression, stability, and possibly the localization of these proteins are highly regulated, suggesting that plants tightly control SAUR protein activity. Future studies focused on these different regulatory mechanisms, as well as the biochemical functions of these proteins, will further our understanding of auxin-regulated growth.
EXPERIMENTAL PROCEDURES
Plant materials and growth conditions
All Arabidopsis lines employed in this study are in the Col-0 ecotype. Seedlings were grown under sterile conditions on ATS nutrient medium (Lincoln et al., 1990) under long-day lighting (60–80 μE m−2 sec−1) at 20°C unless specified otherwise. For shade avoidance experiments, seedlings were grown under 80 μE m−2 sec−1 fluorescent light (Wc) for 5 days, and then transferred to continuous 14 μE m−2 sec−1 Wc light plus light-emitting diode (LED) FR light (R:FR ratio = 0.9) or continuous Wc only and grown for an additional 3 days.
Plant growth measurements
Seedling hypocotyls were photographed using a SPOT Insight camera (http://www.spotimaging.com/) mounted on an Olympus SZX12 stereomicroscope (http://www.olympus.com/), and lengths measured using the accompanying software package. Hypocotyl epidermal cell length measurements were performed on a Nikon Diaphot 200 phase contrast microscope fitted with an ocular micrometer. Abaxial epidermal cells (40–100 cells) were drawn for four or five leaves with a DM LB microscope (Leica, http://www.leica.com/) fitted with a drawing tubus and a differential interference contrast objective. Photographs of leaves and drawings were used to measure the leaf area and calculate the average cell area, respectively, using IMAGEJ software (http://rsb.info.nih.gov/ij/). Leaf and cell areas were subsequently used to calculate cell numbers. For fresh and dry weight measurements, and the rosette leaf area measurements, 8–15 seedlings were grown on half-strength Murashige and Skoog medium (Murashige and Skoog, 1962), supplemented with 1% sucrose at 21°C under a 16-h day/8-h night regime or on soil for 20 days. Individual leaves (cotyledons and rosette leaves) were dissected and their area was measured using IMAGEJ software.
Confocal microscopy
Confocal images were obtained on a Nikon C1si Confocal system (Nikon USA, http://www.nikon.com/) using the 10 mW 405-nm line, the 488-nm line of the 30 mW argon laser, or the 18 mW 561-nm diode laser. For nuclear counter-staining, 5-day-old light grown seedlings were incubated in 1 μg ml−1 Hoechst 33258 (Invitrogen, http://www.invitrogen.com.), and processed with a Pelco model 3450 microwave oven equipped with a temperature controlled ColdSpot load cooler (Ted Pella Inc., http://www.tedpella.com/). Processing was done at 12°C for a total of 12 min, with two cycles of 2 min at 175 W, 2 min with no power, and 2 min at 175 W.
RT-PCR and northern blots
The RNA samples for RT-PCR studies were obtained from 7-day-old seedlings using the RNeasy Plant Kit (Qiagen, http://www.qiagen.com/ ). Complementary DNA synthesis from DNase-treated RNA samples was performed as previously described (Quint et al., 2005). Real-time PCR reactions were performed on the LightCycler System (Roche Applied Sciences, http://www.roche.com/) using the SYBR Green JumpStart kit (Sigma, http://www.sigmaaldrich.com/) and the primer pairs listed in Table S1. The RT-PCR results for each primer pair were normalized to those obtained with the ACT7 gene. All RT-PCR experiments include three or more replicate assays of at least three biologically independent samples. The RNA samples for northern blotting were prepared as above and blotted to Hybond N membrane (GE Healthcare, http://www3.gehealthcare.com/). Blots were probed with a full-length 32P-labeled SAUR19 cDNA.
SAUR expression constructs
The SAUR19–24 coding sequences were amplified (Table S1) from A. thaliana Col-0 genomic DNA and the resulting PCR products purified and recombined into pDONR207 using Gateway BP Clonase (Invitrogen, http://www.invitrogen.com/) according to the manufacturer’s protocol. Resulting plasmids were sequenced and then recombined into pMDC43 (N-term GFP), pMDC32 (no tag) (Curtis and Grossniklaus, 2003), pJ2B-StrepII-GW (Noël et al., 2007), or pEarleygate100 (no tag) (Earley et al., 2006) using Gateway LR Clonase to produce vectors containing the 35S promoter driving expression of SAUR fusion proteins.
For SAUR promoter–GUS constructs, 0.7–1.5 kb fragments containing the upstream genomic sequence through SAURx amino acid 7 were PCR amplified from Col-0 DNA and cloned into the BamHI or BamHI–SalI sites of pBI101.2 (Clontech, http://www.clontech.com/) to create in-frame fusions with β-glucuronidase. Primers SAUR19 (P) F, SAUR21 (P) F, SAUR23 (P) F, and SAUR 24 (P) F were used in combination with SAUR (P) R for amplification (Table S1).
The amiRNAs targeting SAUR19–24 family members were designed using the amiRNA designer within the on-line WMD interface (Schwab et al., 2006; Ossowski et al., 2008). The SAUR19/ 23/24 amiRNA was generated in the mi319a backbone of pRS300 (Ossowski et al., 2008) and subsequently cloned behind the 35S promoter in pEarleygate100 (Earley et al., 2006) for expression in plants. All of the above constructs were sequenced, introduced into Agrobacterium tumefaciens strain GV3101, and transformed into Col-0 plants using the floral dip method. Three to five independent transgenic lines were examined for each construct.
For bacterial expression, the SAUR19 coding sequence was amplified and cloned into the BamHI–EcoRI site of pGEX-2T and the BamHI–XhoI sites of pET28A (Table S1).
Antibodies and western blots
The α-GFP monoclonal antibody was purchased from Covance (http://www.covance.com/). To generate antibodies against SAUR19, a 6× His–SAUR19 fusion protein was purified from E. coli and used to immunize a New Zealand white rabbit (Cocalico Biologicals, http://www.cocalicobiologicals.com/). The SAUR19 antibodies were immunopurified against nitrocellulose-bound antigen (Pringle et al., 1989). For western blotting, 30–40 μg crude extract, soluble, or microsomal fractions were separated on 4–12% polyacrylamide gels and blotted to nitrocellulose. Blots were incubated with affinity-purified α-SAUR19 (1:800) antibody followed by α-rabbit IgG-HRP (Sigma), and detected by enhanced chemiluminescence using SuperSignal West Fempto Maximum Sensitivity Substrate (Thermo Scientific, http://www.thermofisher.com/).
In vitro degradation assays
In vitro degradation assays were conducted as per Wang et al. (2009) with minor modifications. Briefly, 10-day-old Col-0 seedlings were homogenized under liquid nitrogen and immediately placed in lysis buffer [50 mM HEPES-KOH, pH 7.0, 10 mM MgCl2, 10 mM NaCl, 10 mM potassium acetate (KOAc), 400 mM sucrose, and 5 mM DTT]. Homogenates were cleared by two successive 15-min spins at 18 000 g at 4°C. The GST-SAUR19 protein was purified from E. coli using glutathione-agarose beads, eluted with reduced glutathione, and dialyzed against thrombin digestion buffer [50 mM 2-amino-2-(hydroxymethyl)-1,3-propanediol (TRIS)– HCl, 150 mM NaCl, 2.5 mM CaCl2]. Approximately 2 μg of GST-SAUR19 was partially digested with thrombin (2.5 U ml−1) for 10 min and one-fifth of the digest was immediately added to 200 μg plant lysate and 2 mM ATP with or without 100 μM MG132 (Peptides International, http://pepnet.com/). Twenty-microliter aliquots were removed at given time points, placed in SDS sample buffer, boiled, and run on polyacrylamide gels for western blotting. Blots were probed with affinity purified α-SAUR19 antibody (1:500 dilution).
Indole-3-acetic acid transport assays
Basipetal transport assays in dark-acclimated light-grown seedlings were performed as previously described (Christie et al., 2011). Assays utilized [3H]-IAA (24 Ci mmol−1) deposited as a 10-nl droplet at the shoot apical meristem, with a 2 mm segment of tissue sampled at the root–shoot transition zone after 4.5 h. Triplicate assays were performed with 10 seedlings per assay.
Membrane fractionations
Microsomal fractionations and two-phase membrane partitioning experiments were conducted as previously described (Ito and Gray, 2006). Antibody against SEC12 was kindly provided by Dr A. Sanderfoot (University of Wisconsin-La Crosse, La Crosse, WI, USA). Peripheral membrane proteins were extracted from microsomes with 0.1 M Na2CO3 as per Boonsirichai et al. (2003).
Cycloheximide-chase assays
Six-day-old etiolated Col[35S:GFP-SAUR19/35S:SAUR19] seedlings were transferred to 5 ml of liquid ATS medium containing 40 μg ml−1 cycloheximide or solvent control. Seedlings were incubated under ambient lighting with gentle agitation for 0–30 min, and subsequently subjected to extraction and microsome fractionation as previously described (Stukkens et al., 2005). Western blots were performed as above.
Supplementary Material
Acknowledgments
We thank Klaus Palme and Edgar Spalding for seed stocks, Min Ni, Gary Gardner, and Doug Brinkman for assistance with light quality assays, and Mark Sanders and the University of Minnesota, College of Biological Sciences Imaging Center, St Paul, MN for assistance with confocal microscopy. We would also like to acknowledge Starchild Weivoda, Chen Gu, Liesbeth De Milde, Amber Chase, and Candace Pritchard for their technical assistance. This work was supported the National Science Foundation (MCB-0817205, WMG and PJO; IOS-0820648, ASM and WAP), the National Institutes of Health (GM067203, WMG), and the McKnight Land-Grant professorship (WMG). SHL was supported in part by the Korea Research Foundation (KRF-2007-357-C00108). DI and NG were supported by Ghent University (‘Bijzonder Onderzoeksfonds Methusalem project’ grant no. BOF08/ 01M00408).
Footnotes
ACCESSION NUMBERS
The Arabidopsis Genome Initiative accession numbers for genes referred to in this work are as follows: SAUR19 (At5g18010), SAUR20 (At5g18020), SAUR21 (At5g18030), SAUR22 (At5g18050), SAUR23 (At5g18060), SAUR24 (At5g18080), SAUR26 (At3g03850), SAUR27 (At3g03840), SAUR28 (At3g03830), SAUR29 (At3g03820), IAA5 (At1g15580), PIN2 (At5g57090), and TIR1 (At3g62980).
Additional Supporting Information may be found in the online version of this article:
TAIR annotation spreadsheet.
Figure S1. Clustal alignment of Arabidopsis SAUR proteins.
Figure S2. GFP-SAUR19 phototropism and blue light sensitivity.
Figure S3. Quantification of GFP-SAUR19 and SAUR19 steady-state levels.
Figure S4. 1-Naphthylphthalamic acid (NPA) hypersensitivity of GFP-SAUR19 seedlings.
Figure S5. The amiSAUR suppression of SAUR19 expression and GFP-SAUR19 phenotypes.
Figure S6. Shade avoidance response of amiSAUR19/23/24 seedlings.
Figure S7. Auxin response of amiSAUR19/23/24 seedlings.
Table S1. The DNA primers used in this study. Please note: As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer-reviewed and may be re-organized for online delivery, but are not copy-edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
References
- Bloom J, Amador V, Bartolini F, DeMartino G, Pagano M. Proteasome-mediated degradation of p21 via N-terminal ubiquitinylation. Cell. 2003;115:71–82. doi: 10.1016/s0092-8674(03)00755-4. [DOI] [PubMed] [Google Scholar]
- Boonsirichai K, Sedbrook JC, Chen R, Gilroy S, Masson PH. ALTERED RESPONSE TO GRAVITY is a peripheral membrane protein that modulates gravity-induced cytoplasmic alkalinization and lateral auxin transport in plant statocytes. Plant Cell. 2003;15:2612–2625. doi: 10.1105/tpc.015560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitschopf K, Bengal E, Ziv T, Admon A, Ciechanover A. A novel site for ubiquitination: the N-terminal residue, and not internal lysines of MyoD, is essential for conjugation and degradation of the protein. EMBO J. 1998;17:5964–5973. doi: 10.1093/emboj/17.20.5964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman EJ, Estelle M. Mechanism of auxin-regulated gene expression in plants. Annu Rev Genet. 2009;43:265–285. doi: 10.1146/annurev-genet-102108-134148. [DOI] [PubMed] [Google Scholar]
- Christie JM, Yang H, Richter GL, et al. phot1 inhibition of ABCB19 primes lateral auxin fluxes in the shoot apex required for phototropism. PLoS Biol. 2011;9:e1001076. doi: 10.1371/journal.pbio.1001076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciechanover A, Ben-Saadon R. N-terminal ubiquitination: more protein substrates join in. Trends Cell Biol. 2004;14:103–106. doi: 10.1016/j.tcb.2004.01.004. [DOI] [PubMed] [Google Scholar]
- Coulombe P, Rodier G, Bonneil E, Thibault P, Meloche S. N-Terminal ubiquitination of extracellular signal-regulated kinase 3 and p21 directs their degradation by the proteasome. Mol Cell Biol. 2004;24:6140–6150. doi: 10.1128/MCB.24.14.6140-6150.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U. A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 2003;133:462–469. doi: 10.1104/pp.103.027979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Smet I, Tetsumura T, De Rybel B, et al. Auxin-dependent regulation of lateral root positioning in the basal meristem of Arabidopsis. Development. 2007;134:681–690. doi: 10.1242/dev.02753. [DOI] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Estelle M. The F-box protein TIR1 is an auxin receptor. Nature. 2005;435:441–445. doi: 10.1038/nature03543. [DOI] [PubMed] [Google Scholar]
- Ding Z, Galván-Ampudia CS, Demarsy E, et al. Light-mediated polarization of the PIN3 auxin transporter for the phototropic response in Arabidopsis. Nat Cell Biol. 2011;13:447–452. doi: 10.1038/ncb2208. [DOI] [PubMed] [Google Scholar]
- Earley KW, Haag JR, Pontes O, Opper K, Juehne T, Song K, Pikaard CS. Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 2006;45:616–629. doi: 10.1111/j.1365-313X.2005.02617.x. [DOI] [PubMed] [Google Scholar]
- Esmon CA, Tinsley AG, Ljung K, Sandberg G, Hearne LB, Liscum E. A gradient of auxin and auxin-dependent transcription precedes tropic growth responses. Proc Natl Acad Sci USA. 2006;103:236–241. doi: 10.1073/pnas.0507127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin KA, Lee SH, Patel D, et al. PHYTOCHROME-INTERACTING FACTOR 4 (PIF4) regulates auxin biosynthesis at high temperature. Proc Natl Acad Sci USA. 2011;108:20231–20235. doi: 10.1073/pnas.1110682108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil P, Green PJ. Multiple regions of the Arabidopsis SAUR-AC1 gene control transcript abundance: the 3′ untranslated region functions as an mRNA instability determinant. EMBO J. 1996;15:1678–1686. [PMC free article] [PubMed] [Google Scholar]
- Gray WM, Ostin A, Sandberg G, Romano CP, Estelle M. High temperature promotes auxin-mediated hypocotyl elongation in Arabidopsis. Proc Natl Acad Sci USA. 1998;95:7197–7202. doi: 10.1073/pnas.95.12.7197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen G, Guilfoyle T. Auxin-responsive gene expression: genes, promoters and regulatory factors. Plant Mol Biol. 2002;49:373–385. [PubMed] [Google Scholar]
- Hwang CS, Shemorry A, Varshavsky A. N-terminal acetylation of cellular proteins creates specific degradation signals. Science. 2010;327:973–977. doi: 10.1126/science.1183147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H, Gray WM. A gain-of-function mutation in the Arabidopsis pleiotropic drug resistance transporter PDR9 confers resistance to auxinic herbicides. Plant Physiol. 2006;142:63–74. doi: 10.1104/pp.106.084533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain M, Tyagi AK, Khurana JP. Genome-wide analysis, evolutionary expansion, and expression of early auxin-responsive SAUR gene family in rice (Oryza sativa) Genomics. 2006;88:360–371. doi: 10.1016/j.ygeno.2006.04.008. [DOI] [PubMed] [Google Scholar]
- Jensen PJ, Hangarter RP, Estelle M. Auxin transport is required for hypocotyl elongation in light-grown but not dark-grown Arabidopsis. Plant Physiol. 1998;116:455–462. doi: 10.1104/pp.116.2.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kant S, Bi YM, Zhu T, Rothstein SJ. SAUR39, a small auxin-up RNA gene, acts as a negative regulator of auxin synthesis and transport in rice. Plant Physiol. 2009;151:691–701. doi: 10.1104/pp.109.143875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kepinski S, Leyser O. The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature. 2005;435:446–451. doi: 10.1038/nature03542. [DOI] [PubMed] [Google Scholar]
- Keuskamp DH, Pollmann S, Voesenek LA, Peeters AJ, Pierik R. Auxin transport through PIN-FORMED 3 (PIN3) controls shade avoidance and fitness during competition. Proc Natl Acad Sci USA. 2010;107:22740–22744. doi: 10.1073/pnas.1013457108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knauss S, Rohrmeier T, Lehle L. The auxin-induced maize gene ZmSAUR2 encodes a short-lived nuclear protein expressed in elongating tissues. J Biol Chem. 2003;278:23936–23943. doi: 10.1074/jbc.M212585200. [DOI] [PubMed] [Google Scholar]
- Lewis DR, Wu G, Ljung K, Spalding EP. Auxin transport into cotyledons and cotyledon growth depend similarly on the ABCB19 Multidrug Resistance-like transporter. Plant J. 2009;60:91–101. doi: 10.1111/j.1365-313X.2009.03941.x. [DOI] [PubMed] [Google Scholar]
- Lin R, Wang H. Two homologous ATP-binding cassette transporter proteins, AtMDR1 and AtPGP1, regulate Arabidopsis photomorphogenesis and root development by mediating polar auxin transport. Plant Physiol. 2005;138:949–964. doi: 10.1104/pp.105.061572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lincoln C, Britton JH, Estelle M. Growth and development of the axr1 mutants of Arabidopsis. Plant Cell. 1990;2:1071–1080. doi: 10.1105/tpc.2.11.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luschnig C, Gaxiola RA, Grisafi P, Fink GR. EIR1, a root-specific protein involved in auxin transport, is required for gravitropism in Arabidopsis thaliana. Genes Dev. 1998;12:2175–2187. doi: 10.1101/gad.12.14.2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure BA, Guilfoyle T. Characterization of a class of small auxin-inducible soybean polyadenylated RNAs. Plant Mol Biol. 1987;9:611–623. doi: 10.1007/BF00020537. [DOI] [PubMed] [Google Scholar]
- McClure BA, Guilfoyle T. Rapid redistribution of auxin-regulated RNAs during gravitropism. Science. 1989;243:91–93. doi: 10.1126/science.11540631. [DOI] [PubMed] [Google Scholar]
- Migliaccio F, Piconese S. Spiralizations and tropisms in Arabidopsis roots. Trends Plant Sci. 2001;6:561–565. doi: 10.1016/s1360-1385(01)02152-5. [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant. 1962;15:473–497. [Google Scholar]
- Nemhauser JL, Hong F, Chory J. Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell. 2006;126:467–475. doi: 10.1016/j.cell.2006.05.050. [DOI] [PubMed] [Google Scholar]
- Newman TC, Ohme-Takagi M, Taylor CB, Green PJ. DST sequences, highly conserved among plant SAUR genes, target reporter transcripts for rapid decay in tobacco. Plant Cell. 1993;5:701–714. doi: 10.1105/tpc.5.6.701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noël LD, Cagna G, Stuttmann J, Wirthmüller L, Betsuyaku S, Witte CP, Bhat R, Pochon N, Colby T, Parker JE. Interaction between SGT1 and cytosolic/nuclear HSC70 chaperones regulates Arabidopsis immune responses. Plant Cell. 2007;19:4061–4076. doi: 10.1105/tpc.107.051896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada K, Shimura Y. Reversible root tip rotation in Arabidopsis seedlings induced by obstacle-touching stimulus. Science. 1990;250:274–276. doi: 10.1126/science.250.4978.274. [DOI] [PubMed] [Google Scholar]
- Ossowski S, Schwab R, Weigel D. Gene silencing in plants using artificial microRNAs and other small RNAs. Plant J. 2008;53:674–690. doi: 10.1111/j.1365-313X.2007.03328.x. [DOI] [PubMed] [Google Scholar]
- Park JE, Kim YS, Yoon HK, Park CM. Functional characterization of a small auxin-up RNA gene in apical hook development in Arabidopsis. Plant Sci. 2007;172:150–157. [Google Scholar]
- Popescu SC, Popescu GV, Bachan S, Zhang Z, Seay M, Gerstein M, Snyder M, Dinesh-Kumar SP. Differential binding of calmodulin- related proteins to their targets revealed through high-density Arabidopsis protein microarrays. Proc Natl Acad Sci USA. 2007;104:4730–4735. doi: 10.1073/pnas.0611615104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pringle JR, Preston RA, Adams AE, Stearns T, Drubin DG, Haarer BK, Jones EW. Fluorescence microscopy methods for yeast. Methods Cell Biol. 1989;31:357–435. doi: 10.1016/s0091-679x(08)61620-9. [DOI] [PubMed] [Google Scholar]
- Quint M, Ito H, Zhang W, Gray WM. Characterization of a novel temperature-sensitive allele of the CUL1/AXR6 subunit of SCF ubiquitinligases. Plant J. 2005;43:371–383. doi: 10.1111/j.1365-313X.2005.02449.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinstein E, Scheffner M, Oren M, Ciechanover A, Schwartz A. Degradation of the E7 human papillomavirus oncoprotein by the ubiquitin-proteasome system: targeting via ubiquitination of the N-terminal residue. Oncogene. 2000;19:5944–5950. doi: 10.1038/sj.onc.1203989. [DOI] [PubMed] [Google Scholar]
- Richter GL, Monshausen GB, Krol A, Gilroy S. Mechanical stimuli modulate lateral root organogenesis. Plant Physiol. 2009;151:1855–1866. doi: 10.1104/pp.109.142448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santner AA, Watson JC. The WAG1 and WAG2 protein kinases negatively regulate root waving in Arabidopsis. Plant J. 2006;45:752–764. doi: 10.1111/j.1365-313X.2005.02641.x. [DOI] [PubMed] [Google Scholar]
- Schwab R, Ossowski S, Riester M, Warthmann N, Weigel D. Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell. 2006;18:1121–1133. doi: 10.1105/tpc.105.039834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidler M, Hassa P, Hasan S, Ringli C, Dudler R. Involvement of an ABC transporter in a developmental pathway regulating hypocotyl cell elongation in the light. Plant Cell. 1998;10:1623–1636. doi: 10.1105/tpc.10.10.1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stukkens Y, Bultreys A, Grec S, Trombik T, Vanham D, Boutry M. NpPDR1, a pleiotropic drug resistance-type ATP-binding cassette transporter from Nicotiana plumbaginifolia, plays a major role in plant pathogen defense. Plant Physiol. 2005;139:341–352. doi: 10.1104/pp.105.062372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan X, Calderon-Villalobos LI, Sharon M, Zheng C, Robinson CV, Estelle M, Zheng N. Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature. 2007;446:640–645. doi: 10.1038/nature05731. [DOI] [PubMed] [Google Scholar]
- Tao Y, Ferrer JL, Ljung K, et al. Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell. 2008;133:164–176. doi: 10.1016/j.cell.2008.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenbussche F, Petrásek J, Zádníková P, et al. The auxin influx carriers AUX1 and LAX3 are involved in auxin-ethylene interactions during apical hook development in Arabidopsis thaliana seedlings. Development. 2010;137:597–606. doi: 10.1242/dev.040790. [DOI] [PubMed] [Google Scholar]
- Wang F, Zhu D, Huang X, Li S, Gong Y, Yao Q, Fu X, Fan LM, Deng XW. Biochemical insights on degradation of Arabidopsis DELLA proteins gained from a cell-free assay system. Plant Cell. 2009;21:2378–2390. doi: 10.1105/tpc.108.065433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward AW, Bartel B. Auxin: regulation, action, and interaction. Ann Bot. 2005;95:707–735. doi: 10.1093/aob/mci083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Cameron JN, Ljung K, Spalding EP. A role for ABCB 19-mediated polar auxin transport in seedling photomorphogenesis mediated by cryptochrome 1 and phytochrome B. Plant J. 2010;62:179–191. doi: 10.1111/j.1365-313X.2010.04137.x. [DOI] [PubMed] [Google Scholar]
- Yang T, Poovaiah BW. Molecular and biochemical evidence for the involvement of calcium/calmodulin in auxin action. J Biol Chem. 2000;275:3137–3143. doi: 10.1074/jbc.275.5.3137. [DOI] [PubMed] [Google Scholar]
- Zádníková P, Petrásek J, Marhavy P, et al. Role of PIN-mediated auxin efflux in apical hook development of Arabidopsis thaliana. Development. 2010;137:607–617. doi: 10.1242/dev.041277. [DOI] [PubMed] [Google Scholar]
- Zenser N, Dreher KA, Edwards SR, Callis J. Acceleration of Aux/IAA proteolysis is specific for auxin and independent of AXR1. Plant J. 2003;35:285–294. doi: 10.1046/j.1365-313x.2003.01801.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.