Abstract
Rationale
Cardiomyocytes differentiated from human pluripotent stem cells (PSCs) are increasingly being used for cardiovascular research including disease modeling and hold promise for clinical applications. Current cardiac differentiation protocols exhibit variable success across different PSC lines and are primarily based on the application of growth factors. However, extracellular matrix (ECM) is also fundamentally involved in cardiac development from the earliest morphogenetic events such as gastrulation.
Objective
We sought to develop a more effective protocol for cardiac differentiation of human PSCs by using ECM in combination with growth factors known to promote cardiogenesis.
Methods and Results
PSCs were cultured as monolayers on Matrigel, an ECM preparation, and subsequently overlayed with Matrigel. The matrix sandwich promoted an epithelial-to-mesenchymal transition as in gastrulation with the generation of N-cadherin+ mesenchymal cells. Combining the matrix sandwich with sequential application of growth factors (Activin A, BMP4, and bFGF) generated cardiomyocytes with high purity (up to 98%) and yield (up to 11 cardiomyocytes/input PSC) from multiple PSC lines. The resulting cardiomyocytes progressively mature over 30 days in culture based on myofilament expression pattern and mitotic activity. Action potentials typical of embryonic nodal, atrial and ventricular cardiomyocytes were observed, and monolayers of electrically coupled cardiomyocytes modeled cardiac tissue and basic arrhythmia mechanisms.
Conclusions
Dynamic ECM application promoted EMT of human PSCs and complemented growth factor signaling to enable robust cardiac differentiation.
Keywords: Cardiogenesis, cardiomyocytes, embryonic stem cells, induced pluripotent stem cells, extracellular matrix
INTRODUCTION
Cardiac differentiation protocols for human pluripotent stem cells (PSCs) have evolved from the inefficient and variable serum-based embryoid body (EB) methods to monolayer and EB protocols in defined conditions using the sequential application of growth factors that reflect the changes in Wnt, Activin/Nodal and BMP signaling during normal cardiac development.1–5 The optimization of cardiac differentiation protocols has focused primarily on the timing of application and concentration of growth factors. Variability in the endogenous signaling pathways among different human PSC lines during differentiation has required cell line-specific titration of growth factors and sometimes stage-specific inhibition of signaling pathways.3, 6 However, cell behavior and stem cell fate choices are also critically impacted by extracellular matrix (ECM).7, 8 Although studies have described ECM preparations, including synthetic surfaces, effective for attachment and culture of undifferentiated human embryonic stem cells (ESCs) and induced pluripotent stem cells (iPSCs),9–13 the ability of ECM to promote in vitro cardiac differentiation has not been described.
Human ESCs and iPSCs resemble epiblast cells present in the bilaminar embryo based on gene expression patterns and active cell signaling pathways.14, 15 During gastrulation, some epiblast cells undergo an epithelial-mesenchymal transition (EMT) and ingress in the primitive streak to generate the mesodermal cells fated to give rise to heart muscle.16 Primitive streak formation requires not only appropriate growth factor signaling, but ECM is critical for the process.8 The ECM provides an adhesive substrate for cell migration as well as allowing essential crosstalk between ECM receptors and growth factor receptors. In addition, ECM can sequester and store growth factors to enable spatio-temporally controlled release necessary for morphogenesis. The ECM can also sense and transduce mechanical signals which impact cell fate decisions.7, 17 ECM signaling is dynamic during development with constant remodeling by proteases and changes in expression of ECM proteins, such as increased fibronectin expression associated with mesoderm induction.18–20
Here we report that overlaying monolayer-cultured human PSCs with the Matrigel, a commercially available extracellular matrix preparation, promotes EMT required for generation of precardiac mesoderm. Combining ECM with growth factor signaling, we describe a robust method for differentiation of multiple human PSC lines to functional cardiomyocytes (CMs) in defined conditions.
METHODS
Human iPSC and ESC culture
Three iPSC lines derived from foreskin fibroblasts without integration of vector and transgene sequences (DF6-9-9T, DF19-9-7T and DF19-9-11T) and the lentiviral-generated iPS cell line IMR90 clone 4 (IMR90 C4) were used in this study.21, 22 Human ES cell lines of H1, H9 were used for comparison.23 The iPSCs and ESCs were either maintained on irradiated mouse embryonic fibroblasts (MEFs) in 80% DMEM/F12 basal medium, 20% KnockOut serum replacer, 0.1 mmol/L non-essential amino acids, 1 mmol/L L-glutamine (all from Invitrogen), and 0.1 mmol/L β-mercaptoethanol (Sigma), supplemented with 100 ng/ml zebrafish basic fibroblast growth factor (zbFGF) (purified from a bacterial expression system by us) for iPSCs, and 4 ng/ml human recombinant bFGF (Invitrogen) for ESCs as previously described. 21, 24, 25 Cell lines were used continuously between passages of 25–60.
Cardiac differentiation using the matrix sandwich protocol
Human iPSCs and ESCs maintained on MEFs were passaged on Matrigel (GFR, BD Biosciences) thin coated 6-well plates (8.7 ug/cm2) and cultured in mTeSR1 medium (WiCell Institute) for 5–6 days to deplete the feeder cells. Cells were washed with PBS and incubated with 1 ml/well Versene solution (Invitrogen) at 37°C for 5 minutes to singularize the cells and seeded on Matrigel coated plate at the density of 100,000 cells/cm2 in mTeSR1 medium supplemented with 10 μM ROCK inhibitor (Y-27632, CalBiochem). The medium was changed daily, and after 3–4 days when the monolayer of cells reached 80–90% confluence, a thin layer of Matrigel was overlaid by freshly mixing Matrigel (GFR), 0.5 mg (faster growing lines, i.e. DF19-9-11T, DF19-9-7T, IMR90 C4 or H9) or 1 mg (slower growing lines, i.e. DF6-9-9T, H1), in 15 ml ice cold mTeSR1 medium and replacing the medium in each well of a 6-well plate with 2.5 ml of Matrigel containing mTeSR1. Cells were cultured in mTeSR1 medium for another 1–2 days until the cells were 100% confluent, which is referred to as day 0 when the medium was replaced with 2.5 ml of RPMI 1640 basal medium (Invitrogen) plus B-27 without insulin supplement (Invitrogen) containing Activin A (100 ng/ml, R&D Systems) and Matrigel (0.5 mg Matrigel/6-well plate). After 24 hours, the medium was changed with the same medium as day 0 (3 ml/well) without Matrigel but supplemented with BMP4 (5–10 ng/ml, R&D Systems) and bFGF (5–10 ng/ml, Invitrogen) for another 4 days without medium change. At day 5, the medium was changed to RPMI plus B27 complete supplement (Invitrogen), and the medium was changed every 2–3 days. For H9 cells, 1% KnockOut serum replacer (Invitrogen) was added at day 0 in a subset of experiments.
Calculation of the yield of cardiomyocytes from matrix sandwich protocol
The input iPSCs or ESCs were initially seeded at 100,000 cells/cm2 at day -5. The output CMs were determined by flow cytometry of cTnT+ cells at 15 days differentiation. The yield of CMs was calculated by the number of cTnT+ cells divided by the number of input iPSCs or ESCs.
Electron microscopy
Human iPSCs and ESCs cultured in 6-well plate with or without matrix sandwich were fixed overnight at 4°C in a 2.5% gluteraldehyde, 2% paraformaldehyde, 0.1 M phosphate buffer solution and then post-fixed with 1% osmium tetroxide. Samples were dehydrated with increasing concentrations of ethanol up to 100% and kept in a 1:1 mixture of 100% EtOH: Durcupan embedding medium overnight. Samples were desiccated under vacuum for 3 hours, exchanged with fresh Durcupan and then kept at 60°C overnight for polymerization of embedding medium. A 1 cm2 sample of the culture plate was cut with a jeweler’s saw for microtome processing. Ultrathin 60 nm sections were cut perpendicular to the culture surface at the interface of the plate and embedded cells. The sections were placed on a copper grid and stained with uranyl acetate and lead citrate. Samples were visualized on a Phillips CM120 STEM. Photographs were taken with an attached digital camera.
RT-PCR and Quantitative RT-PCR
Cell samples were collected using 0.25% trypsin-EDTA (Invitrogen) to remove the cells from cell culture plates. Total RNA was purified using QIAGEN RNeasy® Mini kit. Possible genomic DNA contamination was removed by DNase I (Invitrogen) treatment for 15 minutes at room temperature. 500 ng of total RNA was used for Oligo(dT)20 – primed reverse transcription using SuperScript III™ First-Strand Synthesis System (Invitrogen). Standard RT-PCR was carried out using Platinum™ Taq DNA Polymerase (Invitrogen) or Gotaq Master Mix (Promega) and then subjected to 2% agarose gel electrophoresis. The primer sequences used in RT-PCR are listed in Online Table I. PCR conditions included denaturation at 94°C for 30 seconds, annealing at 60°C for 30 seconds, and extension at 72°C for 1 minute, for 35 cycles, with 72°C extension for 7 minutes at the end. ACTB (β-actin) was used as an endogenous control. Quantitative RT-PCR was performed using Taqman PCR Master Mix and Gene Expression Assays (Applied Biosystems) in triplicate for each sample and each gene. 0.5 μl from the total 20 μl of RT reaction was added as template for each Q-PCR reaction. The relative expression compared the expression of the gene of interest to the expression of the endogenous control β-actin. Mean Ct value was first calculated for technical replicates which is the average of triplicates or quadruplicates for each gene of each experiment, then ΔCt was calculated as each gene’s mean Ct value minus the mean Ct value of the endogenous control. Relative expression was expressed as the fold change calculated using the formula: fold change = 2 (−ΔCt).
Flow cytometry
Cells were detached from cell culture plates by incubation with 0.25% trypsin-EDTA (Invitrogen) plus 2% chick serum (Sigma) for 5 minutes at 37°C. The chick serum is added for a more gentle dissociation to single cells without clumping. Cells were vortexed to disrupt the aggregates followed by neutralization by adding equal volume of EB20 medium.26 About one million cells were used for each flow sample. Cells were fixed in 1% paraformaldehyde in a 37°C water bath for 10 minutes in the dark, permeabilized in ice-cold 90% methanol for 30 minutes on ice. Cells were washed once in FACS buffer (PBS without Ca/Mg2+, 0.5% BSA, 0.1% NaN3) plus 0.1% Triton, centrifuged, and the supernatant was discarded leaving about 50 μl. Primary antibody was diluted in 50 μl/sample FACS buffer plus 0.1% Triton and aliquoted to each sample for a total sample volume of 100 μl. Samples were incubated with the primary antibodies overnight at 4°C. Please refer to Online Supplemental Material for detail of the primary antibodies. Cells were washed once in 3 ml FACS buffer plus 0.1% Triton, centrifuged, and supernatant discarded leaving ~ 50 μl. Secondary antibody specific to the primary IgG isotype was diluted in FACS buffer plus Triton in a final sample volume of 100 μl at 1:1000 dilution. Samples were incubated for 30 minutes in the dark at room temperature, washed in FACS buffer plus Triton and resuspended in 300 – 500 μl FACS buffer plus Triton for analysis. Data were collected on a FACS Caliber flow cytometer (Beckton Dickinson) and analyzed using FlowJo v8.5.2.
Immunolabeling
Single CMs were isolated from the matrix sandwich culture using 0.25% trypsin-EDTA (Invitrogen) plus 2% chick serum (Sigma) for 5–10 minutes at 37°C. Cells were washed and plated on glass coverslips coated with 0.1% gelatin solution in EB20 medium for 2 days to allow attachment. Monolayer (control) and matrix sandwich cell culture were prepared by directly seeding the PSCs on Matrigel-coated coverslips in 12-well plates and differentiated using the matrix sandwich protocol. Cells were fixed in 4% paraformaldehyde for 15 minutes at room temperature, permeabilized in 0.2% Triton X-100 (Sigma) for 1 hour at room temperature. Samples were blocked with 5% non-fat dry milk (Bio-Rad) in 0.2% Triton X-100 solution and incubated for 2 hours at room temperature on a rotator followed by two washes with PBS. Primary antibodies (please refer to Online Supplemental Material for details of the primary antibodies) were added in 0.1% Triton X-100, 1% BSA in PBS solution and incubated overnight at 4°C. Samples were washed with 0.2% Tween 20 in PBS twice and 1X PBS twice. Secondary antibodiesy specific to the primary IgG isotype were diluted (1:1000) in the same solution as the primary antibodies and incubated at room temperature for 1.5 hours in dark on a rotator. Samples were washed with 0.2% Tween 20 in PBS twice and 1X PBS twice. Glycoproteins on plasma membrane were labeled with wheat germ agglutinin (WGA) fluorescein conjugates (Invitrogen) for fixed cells by incubation with WGA conjugate in PBS for 30 minutes at room temperature, followed by 2 washes with PBS. Coverslips were sealed with Gold Anti-fade Reagent with DAPI (Invitrogen) on glass slides. Slides were examined with an epifluorescence microscope (Leica DM IRB) with QImaging® Retiga 4000R camera for imaging analysis or a confocal microscope (Nikon, A1R) with the image analysis by the software of NIS-Elements BR3.0.
Electrophysiology
Microdissected beating areas were maintained in EB2 medium for 1 – 10 days prior to recording. CM electrical activity was measured using sharp microelectrodes (50 – 100 MΩ; 3 M KCl) in a 37°C bath continuously perfused with Tyrode’s solution (mmol/L): 140 NaCl, 5.4 KCl, 1.8 CaCl2, 1 MgCl2, 10 Hepes, 10 glucose, pH 7.4 NaOH.26 Junction potentials and capacitance were nulled and data were acquired at 10 kHz using an AxoClamp2A amplifier and pClamp 9.2 software (Molecular Devices, Sunnyvale CA). Electrical field stimulation was performed using two platinum electrodes coupled to a Grass SD-9 stimulator (Quincy, MA). For analysis, data were filtered off-line using a low pass Gaussian filter with a cut-off frequency of 2 kHz.
Optical mapping of monolayer cardiomyocytes derived from human ESCs/iPSCs
For high resolution optical mapping of action potential propagation, iPSC or ESC-CMs were isolated from day 30 matrix sandwich differentiated cells using 0.25% trypsin-EDTA (Invitrogen) plus 2% chick serum (Sigma) for 5–10 minutes at 37°C. CMs were then plated on fibronectin coated 18×18mm coverslips (20μg/coverslip) at a density of 250,000 cells per monolayer in EB20 supplemented with 10 μM blebbistatin. After 24 hours in EB20, the medium was switched to RPMI supplemented with B27 and 10 μM blebbistatin and cultured for an additional 48 hours. On the day of optical mapping recordings the monolayers were switched to HBSS (Sigma) and stained with the voltage sensitive dye, Di-8ANEPPS (40 μM, Invitrogen). Optical action potential recordings were made using an ultra-sensitive CCD camera (80×80 pixels, SciMeasure) as described before.27 First spontaneously occurring APs were recorded, followed by overdrive electrical pacing. The basic cycle length (BCL) was gradually decreased (1000–333ms) until loss of 1:1 capture or reentry was induced. Conduction velocity (CV) for spontaneous activity and for electrically paced propagation were calculated as described previously.27 Immunohistochemistry of the monolayer CMs with connexin 40 (Millipore), connexin 43 (Abcam) and α-actinin (Sigma) antibodies was performed using the similar procedure as described previously.28
Statistics
Data are presented as mean ± standard error of the mean (SEM) or mean ± standard deviation as indicated. Data were first analyzed using the Shapiro-Wilk normality test. For datasets with normal distributions, statistical significance was determined by Student’s t-test (two-tailed) for two groups or one-way ANOVA for multiple groups with post-hoc test using Tukey method. For the datasets failing the normality test, statistical significance was determined by non-parametric test using the Mann-Whitney test for two groups or Kruskal-Wallis one-way analysis of variance. Statistical analysis was performed using Microcal Origin, v7.5, P < 0.05 was considered statistically significant.
RESULTS
Extracellular matrix promotes epithelial-mesenchymal transition of human PSCs
Human ESCs and iPSCs, when seeded as single cells and grown as monolayer in defined mTeSR1 medium on Growth Factor Reduced (GFR) Matrigel, exhibit a polarized epithelial cell phenotype with the basal surface attached to ECM and the apical surface demonstrating microvilli and surface glycoproteins (Figures 1A and 1B). The epithelial adhesion junction protein E-cadherin is present on the lateral surfaces of the cells (Figure 1C and Online Figure IA). Robust expression of OCT4, NANOG and SSEA4 (Online Figure IB) in a polarized epithelium is consistent with these cells exhibiting an epiblast-like phenotype. ECM contributes to major morphogenetic events such as gastrulation,8 so we tested the effect of adding ECM in the form of GFR Matrigel to monolayer cultured human PSCs, creating a sandwich of extracellular matrix between which the cells grow. The matrix sandwich resulted in rapid changes in cellular organization within 24 hours with focal areas becoming multilayered (Figure 1A, right), composed of mesenchymal cells below a layer of epithelial cells (Figure 1B, right). The overlaying matrix was quickly internalized by the epithelium as evidenced by spaces between cells present in electron micrographs after 24 hours (Figure 1B, right) and by immunolabeling for laminin, a major ECM component of Matrigel, detected between cells rather than apically (Figure 1A, right). A hallmark change of EMT is the transition in cadherin expression from the epithelial E-cadherin to mesenchymal N-cadherin. Co-labeling the cells with E-cadherin and N-cadherin antibodies showed prominent N-cadherin+ foci present in the multilayered areas (Figure 1C) with 6-fold more N-cadherin+ foci in the matrix sandwich compared to the control culture (Figure 1D). These N-cadherin positive cells were present as clusters of lower layer cells which lacked regular cell-cell contacts and apicobasal polarity and no longer expressed OCT4 (Figure 1B, right and Figure 1E), suggesting these cells represent migratory mesendoderm cells. Gene expression analysis demonstrated that the matrix sandwich resulted in a significant increase in expression of multiple genes associated with EMT during gastrulation including the transcription factors SNAIL1, SNAIL2, GSC (goosecoid), the cytoskeletal protein VIM (vimentin), and the ECM protein FN1 (fibronectin) (Figure 2). Gene expression also demonstrated evidence of the EMT cadherin switch with an increase of CDH2 (N-cadherin) expression, but a decrease in CDH1 (E-cadherin) expression (Figure 2). Together these findings demonstrate that the matrix sandwich promoted formation of multilayered areas of cells undergoing EMT.
Figure 1. Extracellular matrix overlay promotes epithelial-mesenchymal transition of human PSCs.
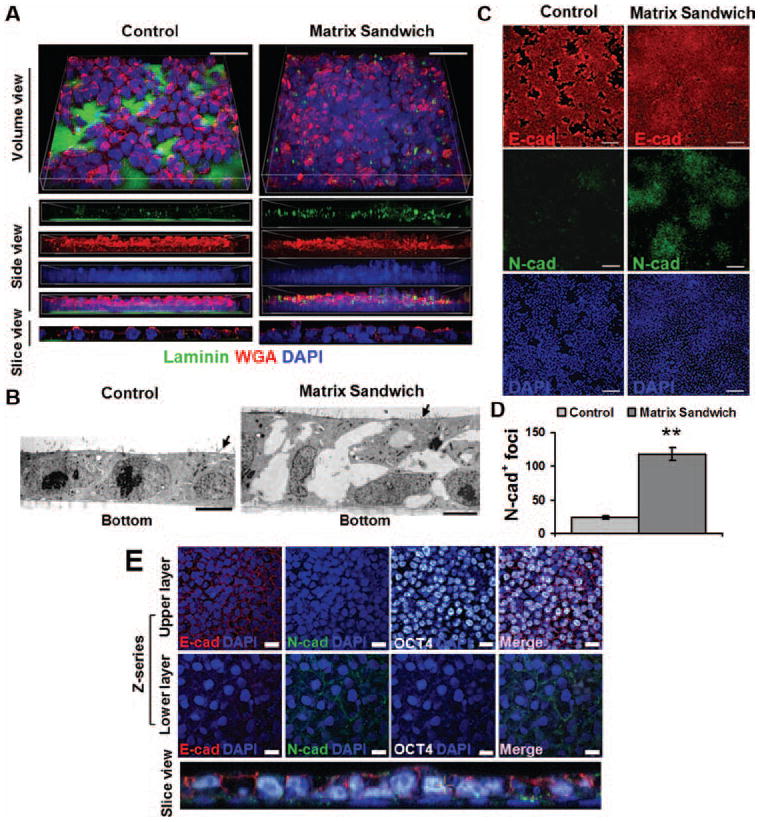
(A) Confocal imaging of iPSCs (DF19-9-11T) seeded as single cells on Matrigel coated surface and propagated in mTeSR1 for 4 days without Matrigel overlay (Control) or with a Matrigel overlay (Matrix Sandwich) for the last 24 hours. 3D reconstructed z-series of images labeled with an antibody to laminin, fluorescent-conjugated WGA for glycoproteins, and DAPI for nuclei. Scale bars are 20 μm. (B) Electron micrographs of control cultures demonstrate confluent monolayers compared to matrix sandwich cultures which are multilayered with an upper epithelial layer and mesenchymal cells below. Arrows indicate microvilli. Scale bars are 5 μm. (C) Epifluorescence images of control and matrix sandwich cell culture 24 hours after Matrigel overlay immunolabeled with E-cadherin and N-cadherin antibodies. Scale bars are 100 μm. (D) Comparison of average number of N-cadherin+ foci (per 1.8 cm2 surface area) in control and matrix sandwich cell cultures (DF19-9-11T). Error bars represent SEM, N=3. Data were compared using Student’s t-test with ** indicating significantly different, P < 0.01. (E) Confocal images of the multilayered areas in the matrix sandwich culture as described in (A) immunolabeled for E-cadherin, N-cadherin, and Oct4. DAPI identifies nuclei. The bottom panel shows a slice view reconstructed from the z-series highlighting the different cell layers. Scale bars are 20 μm.
Figure 2. Quantitative RT-PCR of EMT-related gene expression in control and matrix sandwich culture (DF19-9-11T).
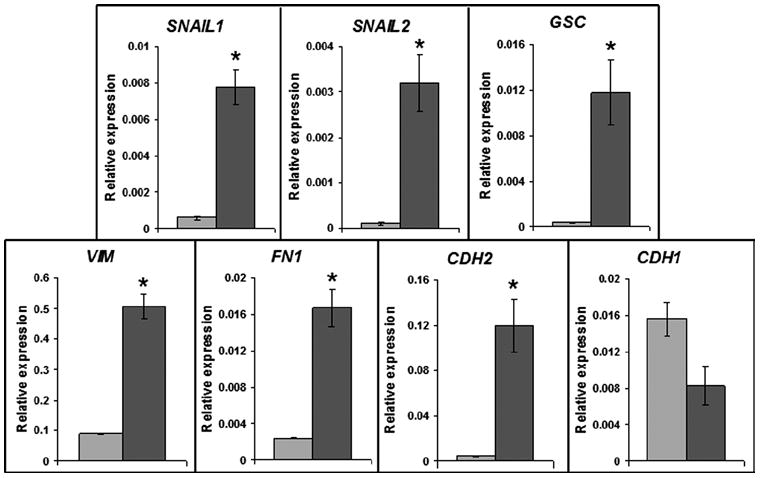
Light grey bars are the control and dark grey bars are the matrix sandwich culture. Total RNA was isolated from monolayer and matrix sandwich culture 48 hours after Matrigel overlay. The relative gene expression was analyzed from three replicates and normalized to the endogenous β-actin. Error bars represent SEM, N=3. Data were compared using Student’s t-test with * indicating significantly different, P < 0.05.
The matrix sandwich protocol for robust cardiac differentiation of human PSC lines
Effective cardiac differentiation of monolayer cultured H7 human ESCs was previously demonstrated using a protocol employing the sequential application of Activin A and BMP4;1 however, this protocol was not broadly successful on other human PSC lines such as the vector-free iPSC line DF19-9-11T (Figure 4A, control) or H1 ESCs.6 Building on the results that Matrigel promotes EMT of the monolayer cultured human PSCs, we combined ECM application with Activin A and BMP4 treatment to develop a three-stage protocol for cardiac differentiation using DF19-9-11T iPSCs (Figure 3A). With Matrigel overlays at day -2 and day 0, we observed a marked reduction in cell death that occurs 24 hours after the addition of Activin A and the change in medium (Figure 3B). Furthermore, the cell number increased markedly from day 2 to day 5 in the matrix sandwich culture in contrast to the control (Figure 3B). To investigate for mesodermal committed cells at this stage, we examined the expression of Brachyury. Immunolabeling revealed that on day 2 cells were primarily growing as clusters of Brachyury+ cells, and the number and size of Brachyury+ clusters were markedly increased by the Matrigel overlays compared to the control (Figure 3C). Flow cytometry showed that the protocol produced a highly enriched population of Brachyury+ cells with 98% of the cells being Brachyury+ on day 2 (Figure 3D). Insulin was found to inhibit early cardiogenesis, so it was not included in the RPMI/B27 medium at stage 2 in agreement with the results of others (Online Figure II).29, 30 On day 5, the medium was changed to basal RPMI/B27 with insulin. Contracting cells were observed as early as day 7, and the developing CMs formed contracting sheets of cells by day 15 (Online Movies I and II). Gene expression analysis during the differentiation protocol showed the sequential up-regulation of early mesoderm genes T and MESP1, followed by the cardiac transcription factors of GATA4, ISL1 and NKX2-5, and finally the cardiac myofilament proteins TNNT2, TNNI3, MYL7, and MYL2 (Figure 3E). OCT4 and NANOG were concurrently down-regulated. Lack of expression of SOX1 and PAX6 suggests the absence of neuroectoderm, and only transient expression of SOX17 and no FOXA2 expression argues that definitive endoderm derivatives are likely not present (Figure 3E). Immunolabeling of the matrix sandwich cultures at 15 days showed dense networks of cTnT labeled CMs (Figure 3F). Up to 98% of the cells in the preparations at day 30 were cTnT+ CMs demonstrated by flow cytometry (Figure 3G). Single CMs exhibited sarcomeric organization demonstrated by the immunolabeling of myofilament proteins α-actinin and MLC2a (Figure 3H).
Figure 4. ECM requirements for matrix sandwich protocol-based cardiac differentiation and robust application to multiple hPSC lines.
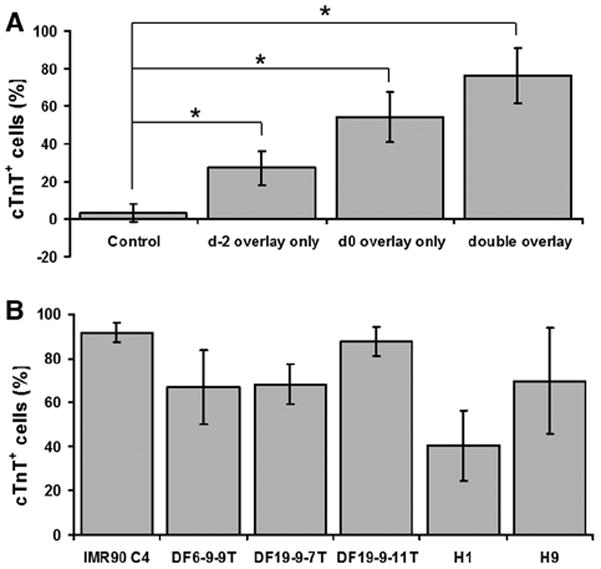
(A) Effect of single or double Matrigel overlays on cardiac differentiation using the protocol defined in Figure 3(A) with Activin A (100 ng/ml), BMP4 (10 ng/ml) and bFGF (5 ng/ml). Cardiogenesis efficiency was measured by flow cytometry for cTnT+ CMs at 15 days differentiation of the iPSC line DF19-9-11T. Control is the cell culture without Matrigel overlay. Error bars represent SD, N=9. (B) Cardiac differentiation of multiple hPSC lines assessed by flow cytometry for cTnT at 15 days differentiaton using the matrix sandwich protocol. Error bars represent SD, N=15 for IMR90 C4; N=15 for DF6-9-9T; N=13 for DF19-9-7T; N=21 for DF19-9-11T; N=24 for H1; N=16 for H9. Data were compared using one-way ANOVA with * indicating significantly different, P < 0.05.
Figure 3. Matrix sandwich method for efficient cardiogenesis of iPSC line DF19-9-11T.
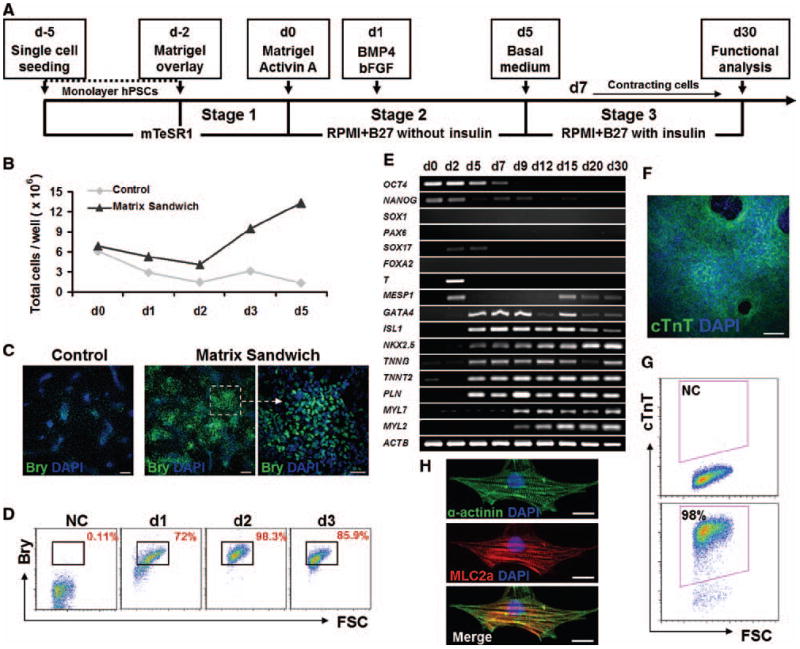
(A) Schematic of the matrix sandwich protocol. (B) Total cells present per 35 mm well of the control (without Matrigel overlay) or the matrix sandwich culture for days 0 to 5 with Activin A (100 ng/ml) added on d0 followed by BMP4 (10 ng/ml) and bFGF (5 ng/ml) added on d1–d5. Data from one experiment are shown which are representative of 4 experiments. (C) Fluorescence images of day 2 cells differentiated using the protocol defined in (A) of the Control (without Matrigel overlay) or the Matrix sandwich culture immunolabeled with Brachyury (Bry) antibody. Scale bars are 200 μm for the left and middle panel, 50 μm for the right panel. (D) Quantification of Brachyury+ cells at day 1, 2 and 3 by flow cytometry. NC indicates negative control of the cell sample without primary antibody. (E) RT-PCR analysis of gene expression of matrix sandwich differentiated cells over 30 days of differentiation. (F) Immunofluorescence image of day 15 matrix sandwich culture identifying cTnT with DAPI for nuclei. Scale bar is 100 μm. (G) Flow cytometry of cells from matrix sandwich culture for cTnT at 30 days differentiation, NC shows the negative control with secondary antibody only. (H) Epifluorescence images of single CM isolated from the matrix sandwich culture shows sarcomeric organization. Re-plated CMs from the matrix sandwich culture were co-labeled with anti-α-actinin antibody which marks the Z-lines and anti-MLC2a antibody that shows the A-bands in the sarcomere. Scale bars are 20 μm.
The specific ECM requirements for the successful induction of cardiogenesis using the protocol were investigated. Comparing a single Matrigel overlay at day-2 or day 0 to two sequential overlays revealed that the combination of two overlays was optimal for generation of CMs (Figure 4A). Phase contrast photomicrographs of cultures at various time points during the protocol with and without the double matrigel overlay demonstrate the typical appearance of cultures during the early differentiation process and highlight the impact of the Matrigel overlay (Online Figure III).
Because the Matrigel application may impact the growth factor requirements originally described for monolayer-based cardiac differentiation,1 we also examined a range of different growth factor combinations and concentrations during stage 2 for the iPSC line DF19-9-11T. We tested the application of a single growth factor, Activin A, for 24 hours of treatment. Activin A alone led to a concentration-dependent increase in purity and yield of cTnT+ cells peaking at 50 ng/ml which produced 20% cTnT+ cells (Online Figure IV, A); however, this was inferior to the combination of 100 ng/ml Activin A with 10 ng/ml BMP4 and 5 ng/ml bFGF initially tested. Therefore, we tested varying concentrations of Activin A and bFGF on day 0 followed by the addition of BMP4 (10 ng/ml) and bFGF (5 ng/ml) on days 1–5 (Online Figure IV, B). The addition of bFGF during the exposure to Activin A had little effect, but Activin A was required for efficient cardiogenesis and displayed a concentration-dependent effect on the purity and yield of cTnT+ cells. At the optimal concentration of 100 ng/ml Activin A, the matrix sandwich protocol generated 80% cTnT+ cells with yield of 11 cTnT+ cells per input iPSC (Online Figure IV, B). We also tested various concentrations of BMP4 and bFGF on day 1–5 after Activin A treatment and found that concentrations of BMP4 of 10 ng/ml or less resulted in a similar purity of CMs (Online Figure IV, C). Increasing the concentration of BMP4 to 25 ng/ml inhibited cardiogenesis (Online Figure IV, C). Addition of bFGF (5–25 ng/ml) during this stage generally increased the yield of CMs (Online Figure IV, C). In conclusion, we found that 100 ng/ml Activin A added on day 0 followed by 5–10 ng/ml BMP4 with 5–25 ng/ml bFGF on days 1–5 resulted in the highest purity and yield of cTnT+ CMs of the iPSC line DF19-9-11T.
It was recently reported that precise optimization of Activin A and BMP4 signaling for each individual human ESC and iPSC line is needed for efficient cardiac differentiation in EBs, 3 so we tested whether the optimal concentrations of Activin A (100 ng/ml) and BMP4 (5 ng/ml) and bFGF (10 ng/ml) determined using the matrix sandwich protocol for the iPSC line DF19-9-11T (Online Figure IV) can result in efficient cardiac differentiation of other cell lines. Lentiviral generated iPSC line (IMR90 C4),21 and non-integrating iPSC lines (DF6-9-9T and DF19-9-7T),22 and 2 hESC lines (H1 and H9)23 underwent differentiation by the matrix sandwich protocol yielding an average 40 to 92% cTnT+ cells (Figure 4B). Furthermore, the matrix sandwich protocol resulted in high yields of CMs ranged from 4–11 CMs per iPSC/ESC initially seeded. Differentiation of these same cell lines in EBs using a serum-based approach demonstrated much lower efficiencies and much greater variability from line to line (Online Figure V). Six disease iPSC lines were also differentiated to CMs efficiently using the matrix sandwich protocol without further growth factor optimization (data not shown). In addition, although all presented data are from human PSC lines maintained on MEFs, in other experiments we observed similar results for human PSCs maintained on Matrigel in mTeSR as previously described.9
Characterization of human PSC-derived cardiomyocytes
The expression pattern of cardiac myofilament proteins can provide information regarding diversity and maturity of the CMs, so we examined the expression of the two major myosin light chain 2 isoforms expressed in heart, MLC2a and MLC2v. In developing mouse and human hearts, MLC2a expression is present in all chambers. In the postnatal heart, MLC2a expression becomes restricted to the atria in mouse, but persists in both atria and to some extent in ventricles in humans.24, 31, 32 In both mouse and human cardiac development, MLC2v expression is restricted to the ventricular chambers, and a ventricular-specific distribution persists into adulthood.24, 31, 32 In both mouse and human embryoid bodies, MLC2a mRNA is detectable before MLC2v mRNA suggesting that 2v is a marker not only of myocyte cell type but also maturity.31, 33 Immunolabeling for MLC2a and MLC2v in 30 day differentiated CMs revealed three distinct patterns: MLC2a only, MLC2a and MLC2v, and MLC2v only (Figure 5A). To provide a quantitative assessment of the expression pattern of MLC2a and MLC2v, we performed flow cytometry on cells after 15 and 30 days differentiation. Co-labeling with cTnT/MLC2a demonstrated that almost all of the cTnT+ cells express MLC2a at both day 15 and day 30 of differentiation. Co-labeling with cTnT/MLC2v and MLC2a/MLC2v revealed little or no expression of MLC2v at 15 days differentiation. However, a substantial population of cells expressing MLC2v was present after 30 days of differentiation, and the majority of these cells co-expressed MLC2a (Figures 5B and 5C).
Figure 5. Expression pattern of myosin light chain 2 isoforms, MLC2a and MLC2v, during cardiac differentiation of hPSCs using matrix sandwich protocol.
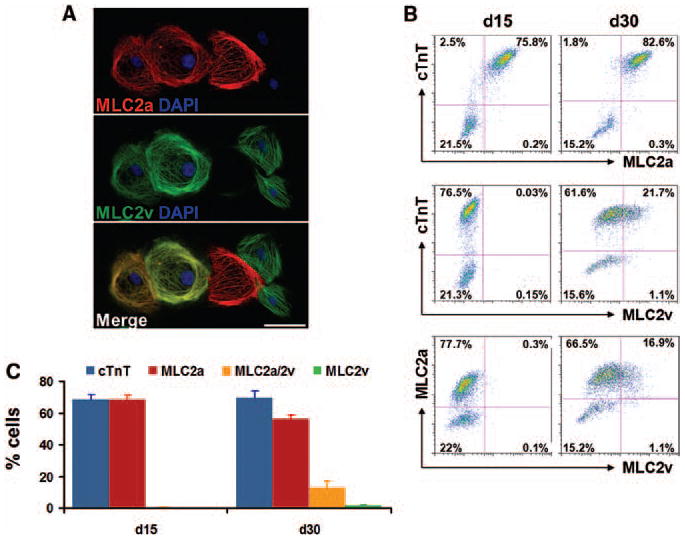
(A) Co-labeling of the re-plated CMs from matrix sandwich culture after 30 days differentiation of iPSCs (DF19-9-11T) with MLC2a and MLC2v antibodies. Scale bar is 50 μm. (B) Dot plots of flow cytometry analysis of cells after 15 and 30 days differentiation of the iPSCs (DF19-9-11T) for expression of cardiac myofilament proteins cTnT, MLC2a and MLC2v. (C) Average of the percentage of cells expressing cTnT; MLC2a (without MLC2v); MLC2a and MLC2v; and MLC2v (without MLC2a) measured by flow cytometry at 15 and 30 days differentiation of the iPSCs (DF19-9-11T). Error bars represent SEM, N=3. Isotype controls were performed for each antibody combination used in the flow cytometry, data not shown.
As another assessment of the maturity of the differentiating CMs, we examined the expression of smooth muscle actin (SMA), which is expressed in the earliest embryonic CMs but not in later-stage CMs.34 In addition, this protein is expressed in smooth muscle cells and fibroblasts. Flow cytometry of cTnT/SMA co-labeling revealed that 53% cells were cTnT+/SMA+ double positive at 15 days differentiation; however, by 30 days differentiation, the cTnT+/SMA+ population decreased to 22% with a concurrent increase in the cTnT+/SMA− population (Online Figure VI, A). Immunolabeling re-plated cells from the matrix sandwich culture with antibodies to SMA and cTnT confirmed distinct populations of cells expressing these proteins (Online Figure VI, B). These results are consistent with the CMs undergoing maturation during this time in culture.
The presence of a small (7%) cTnT−/SMA+ population suggests that either fibroblasts and/or smooth muscle cells are also present. To examine what cell types are present in the small cTnT− population in the matrix sandwich culture after 30 days differentiation, we characterized the cells by flow cytometry using antibodies for fibroblasts (clone TE-7), smooth muscle myocytes (SM-MHC), endothelial cells (CD31) and undifferentiated stem cells (OCT4). In the cell preparations that contained on average 74% CMs (cTnT+), there were 8% fibroblasts and 2% smooth muscle cells (SM-MHC+) (Online Figure VII). Endothelial cells (CD31+) and undifferentiated stem cells (Oct4+) were not detected in the culture. Thus, besides the majority population of CMs resulting from matrix sandwich protocol, relatively small fractions of smooth muscle and fibroblast cells were present in culture. Approximately 16% of the cells were not recognized by any of these antibodies.
As CMs mature during normal cardiac development, they become post-mitotic; therefore, we examined the proliferative activity of CMs by co-labeling cells for sarcomeric myosin (MF20) and the proliferation marker Ki-67. After 15 days of differentiation for two iPSC lines (DF6-9-9T and DF19-9-11T) and one ESC line (H1) approximately 60% of the MF20+ CMs were also Ki67+ (Figure 6). After 30 days, approximately 30% of the MF20+ CMs were Ki-67+. Thus, there is a continued loss of mitotic activity in CMs during the 30 days of observation consistent with ongoing maturation.
Figure 6. Proliferation of human PSC-derived cardiomyocytes in the matrix sandwich culture.
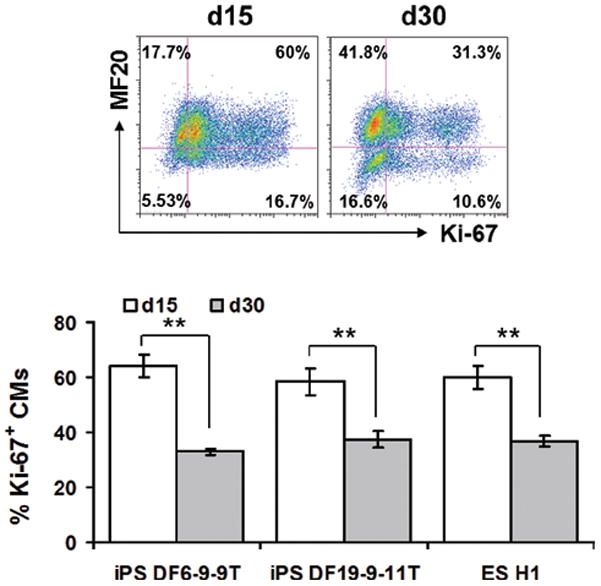
Top panel shows the dot plots of flow cytometry of cells after 15 and 30 days differentiation of iPSCs (DF19-9-11T) labeled with MF20 and Ki-67 antibodies. Bottom panel shows the average of the percentage of Ki-67+ CMs measured by flow cytometry at 15 and 30 days differentiation in multiple cell lines. Error bars represent SEM, N=10 for DF6-9-9T; N=10 for DF19-9-11T; N=8 for H1. Data were compared using Student’s t-test with ** indicating significantly different, P < 0.01.
The electrophysiological phenotype of matrix sandwich protocol differentiated CMs was also investigated to assess the heterogeneity and maturity of CMs. Sharp microelectrode recordings of spontaneous action potentials (APs) from iPSC- and ESC-CMs at 30 days differentiation revealed three distinct AP morphologies comparable to prior studies using serum-based EB protocols (Figure 7A).26, 35 The ventricular-like AP morphology was the predominant form (64/76, 84%) with atrial-like APs (9/76, 12%) and nodal-like APs (3/76, 4%) being less commonly observed. AP properties showed variability among the ventricular-like CMs studied within each cell line, and generally the ventricular AP properties were not significantly different comparing the cell lines with the exception of H9 CMs (Figure 7B). The AP properties are more comparable to human embryonic CMs than adult CMs based on the lower upstroke maximum rate (dV/dtmax) and more depolarized maximum diastolic potential (MDP).36, 37 However, the observed AP properties are similar to those from EBs differentiated for 60–90 days,26 suggesting the matrix sandwich protocol enables more rapid maturation of functional CMs than serum-based EB protocols.26, 35
Figure 7. Electrophysiology properties of human PSC-derived cardiomyocytes from matrix sandwich culture.
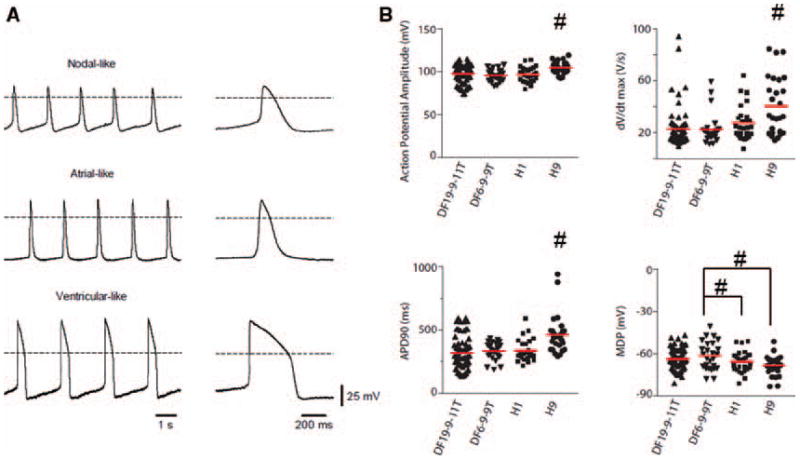
(A) Representative sharp microelectrode recordings of nodal-, atrial-, and ventricular-like action potentials from CMs differentiated from the iPSCs (DF19-9-11T). Dotted line indicates 0 mV. Right, Single action potentials at an expanded timescale taken from traces on the left. (B) Comparison of action potential properties measured from CMs differentiated using the matrix sandwich protocol. Solid lines through distributions indicate population means. Data were compared using one-way ANOVA with # indicating significantly different from all other cell lines or from cell lines indicated, P < 0.05.
Human PSC- derived cardiomyocytes form functionally coupled monolayers
Heart muscle acts as a functional syncytium with individual CMs tightly coupled by adhesion junctions which provide mechanical and electrical connections. Cultured neonatal rat ventricular myocytes have been used to generate electrically coupled monolayers of CMs to study impulse conduction and mechanisms of arrhythmias,38–40 and so we tested if the matrix sandwich-differentiated CMs exhibited the purity and functional characteristics which would enable formation of 2-dimensional monolayers of coupled CMs. The enriched CMs preparations from the matrix sandwich culture were re-plated on fibronectin coated coverslips at a high density (250,000 cells/coverslip) to form monolayers. After two days of culture, the monolayers were labeled with the voltage sensitive dye, Di-8-ANEPPS and optical mapping was performed. Optical signals demonstrated that the monolayers were spontaneously active, and activation maps showed a relatively uniform spread of the electrical impulse with average conduction velocities ranging from 13–18 cm/s in iPSC- and ESC-derived CMs monolayers (Figures 8A–D). Overdrive pacing the monolayers resulted in basic cycle length (BCL) dependent conduction velocity (BCL) typical of cardiac muscle (Figure 8C). Single pixel APs demonstrated predominantly ventricular type AP morphology (Figure 8E). Immunolabeling revealed that the coupling between CMs was provided by Cx43 not Cx40 gap junctions (Figure 8F). Rapid electrical stimulation could induce periodic reentrant waves of conduction called rotors (Figure 8E and Online Movie III), demonstrating that this culture system provides a powerful model for studying human arrhythmia mechanisms.
Figure 8. Optical mapping of transmembrane voltage from monolayer cultures of PSC-derived CMs.
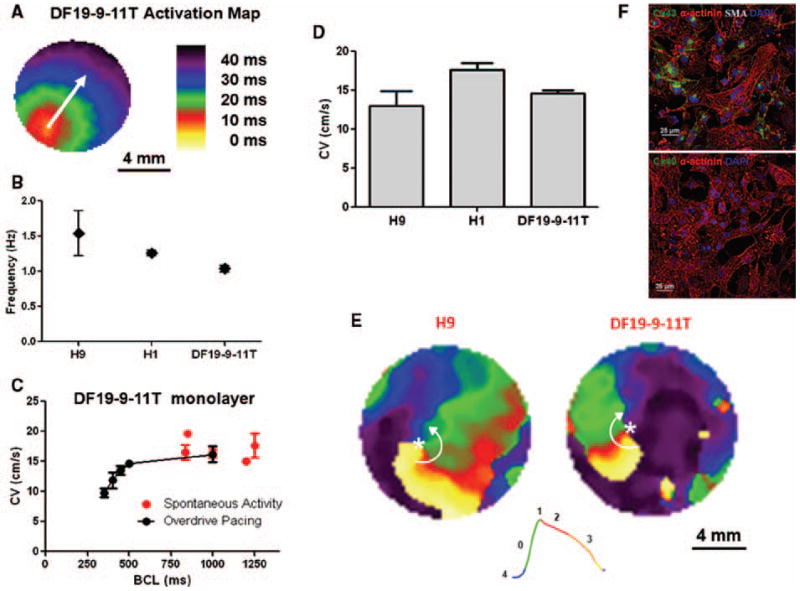
(A) Activation map showing uniform action potential propagation across the monolayer. (B) Spontaneous activation rate for H9, H1, and the DF19-9-11T monolayers. The spontaneous activation rates were 1.54 ± 0.32 (H9, N=8), 1.26 ± 0.04 (H1, N=6), and 1.04 ±0.17 Hz (DF19-9-11T, N=13). (C) Spontaneous and average conduction velocities of the DF19-9-11T cell line measured as a function of pacing frequency (basic cycle length, BCL): Spontaneous (N=13); BCL 1000 (N=5); BCL 500 (N=5); BCL 450 (N =5); BCL 400 (N=5); BCL 350 (N=5). (D) Average conduction velocities for the H9 (N=3), H1 (N=6), and DF19-9-11T (N=5) monolayers measured at 2 Hz pacing. (E) Representative snapshots from phase movies showing electrical rotors in H9 and DF 19-9-11T CM monolayers. Green represents the depolarization phase of the propagating action potential, phase zero; red represents phase 2 or the plateau phase of the action potential; finally orange and yellow represent phase 3 repolarization of the action potential. The white asterisk indicates the phase singularity, the point where all phases of the action potential converge which is the organizing center of the arrhythmic reentry. The white arrows denote the direction of the rotation of the reentrant waves. (F) Immunolabeling of iPSC-CM monolayer (DF19-9-11T) for sacromeric protein α-actinin in combination with α-smooth muscle actin (SMA) and gap junction protein Cx43 and Cx40, respectively. Note abundant staining for α-actinin and Cx43 but no staining for SMA or Cx40. Scale bars are 25 μm. Error bars where shown represent SEM.
DISCUSSION
Here we demonstrate that appropriately timed application of extracellular matrix and key growth factors results in the robust cardiac differentiation of human PSCs. The overlay of monolayer-cultured hPSCs with Matrigel along with Activin A application promoted the initial EMT process leading to the generation of a highly enriched population of mesodermal progenitors (Brachyury+). Subsequent application of BMP4 and bFGF resulted in the efficient generation of CMs from multiple hPSCs lines with high purity (up to 98% cTnT+) and high yield (up to 11 CMs/seeded ESC or iPSC). Furthermore, the matrix sandwich protocol does not require any subsequent steps such as Percoll density gradient or other methods to select CMs. The resulting CMs exhibit progressive maturation in culture, but after 30 days in culture the CMs display significant heterogeneity in their maturity based on myofilament expression pattern, mitotic activity, and electrophysiological properties. As observed with other hPSC differentiation protocols, a mixture of CM cell types arise, including nodal-like, atrial-like and ventricular-like CMs based on their electrophysiological properties. However, most of the CMs (>80%) are ventricular-like and can functionally couple in monolayers to exhibit classic cardiac electrophysiological properties and basic arrhythmia mechanisms.
Prior cardiac differentiation protocols have exhibited significant variability across different cell lines and at different passages when examined, and cell line-specific titration of growth factors is required for efficient differentiation in an EB-based protocol.3 In some cases, inhibiting endogenous signaling in particular lines was necessary to enable cardiogenesis3 and in another case activating an additional signaling pathway (Wnt) was needed for differentiation of a given cell line.6 Although the matrix sandwich protocol shows a steep concentration dependence primarily of Activin A, all of the cell lines tested differentiated relatively well under the same conditions without individualization of growth factor concentrations, which suggests that addition of ECM provides a favorable microenvironment complementing the added growth factors to promote a more robust differentiation process. The ECM effect likely reflects the behavior of cells during normal development in which dynamic changes in both ECM and growth factors are critical. However, the detailed of mechanisms by which the Matrigel promotes EMT and cardiogenesis will require future investigations focused on the individual components of this complex ECM mixture and potential cellular receptors and signaling pathways.
Although the matrix sandwich protocol was broadly successful across the human PSC lines studied, this required optimal culture conditions. Several factors were noted to be critical to the success of the protocol and may be considered for troubleshooting purposes:
The starting hPSC cultures must robustly express markers of pluripotency and exhibit minimal or no background differentiation which can dramatically hinder success of the protocol.
Growth factor lots are variable and may require testing of new lots for optimization. We found that exceeding a threshold of BMP4>10 ng/ml can potently inhibit cardiogenesis. Furthermore, optimal storage of the labile growth factors is essential.
Matrigel lots are also variable, and it may be necessary to test more than one lot.
The initial Matrigel overlay can be optimized from 0.5 mg/6-well plate for rapidly growing lines to 1.0 mg/6-wellplate for slower growing lines as described in the Methods.
High passage cells (~>p65) or cells that begin to demonstrate a change in growth patterns have an increased risk of genetic or karotypic abnormalities impairing the differentiation.
For human iPSCs, lack of expression of exogenous transgenes and a stable reprogrammed phenotype after 20 or more passages promotes more reproducible differentiation.
A control human PSC line that works robustly in the protocol can be used in parallel with new cell lines to validate that conditions are optimal.
Robust differentiation protocols for human PSC lines in fully defined conditions to generate cell lineages of interest are essential to advance biomedical research and clinical applications. For example, research using the growing number of cardiac disease-specific and patient-specific iPSC lines will be greatly facilitated by the availability of robust cardiac differentiation protocols that do not require cell line-specific optimization. The matrix sandwich protocol is also amenable to scale-up using large parallel plate bioreactors technology, since the protocol starts with single cell seeding and does not require further cell processing until harvest of the CMs. Future advances are needed to generate homogenous populations of specific CM cell types and to enhance the maturation of CMs. Matrix-enhanced protocols could also prove valuable for generating a variety of other cell lineages in the future.
Supplementary Material
Novelty and Significance.
What Is Known?
Human pluripotent stem cells (PSCs), including human embryonic stem cells and induced pluripotent cells, are a promising source of human cardiomyocytes that can be used for basic research such as disease modeling, drug development and potentially in clinical applications.
Current protocols to generate cardiomyocytes from human PSCs are primarily based on the sequential application of key growth factors, but these protocols have had variable success and often require optimization for different human PSC lines.
Extracellular matrix (ECM) signaling plays critical roles in development, but the efficacy of ECM has not been tested in protocols for generation of cardiomyocytes from human PSCs.
What New Information Does This Article Contribute?
Sandwiching human PSC cultures between layers of Matrigel, a commercially available ECM preparation, promotes an epithelial-to-mesenchymal transition in cell phenotype and thus initiates the differentiation process needed to form cardiomyocytes.
The matrix sandwich protocol combines dynamic application of Matrigel with sequential applications of growth factors (Activin A, bFGF, BMP4) and results in efficient generation of functional cardiomyocytes with high purity (up to 98%) and high yield (up to 11 cardiomyocytes per input PSC) from multiple human PSCs lines.
Advances in stem cell technology including the ability to generate disease- and patient-specific induced pluripotent stem cells have led to an increasing need to obtain cardiomyocytes from various human PSC lines. However, existing protocols for cardiac differentiation are either inefficient or highly cell line-dependent. Most cardiac differentiation protocols employ either serum or sequential application of growth factors. In the present study, we tested whether application of ECM can promote cardiac differentiation. We find that sandwiching human PSCs by overlaying the monolayer cells with Matrigel promotes an epithelial-to-mesenchymal transition which is an essential first step in generating cardiomyocytes. Combining the Matrigel overlay with application of growth factors (Activin A, bFGF, and BMP4) results in the generation of cardiomyocytes in high yield and purity from multiple human PSC lines. The resulting cardiomyocytes are functionally competent, based on spontaneous contractions and action potentials. The matrix sandwich protocol could advance the use of human PSCs for cardiovascular applications by providing investigators access to a robust method for cardiac differentiation and enable research requiring large numbers of cardiomyocytes as well as future clinical applications.
Acknowledgments
The authors gratefully acknowledge the technical support for connexin immunostaining by Alexandra Bizyand Guadalupe Guerrero-Serna at the University of Michigan.
SOURCES OF FUNDING
The following funding sources supported this work NIH R01 HL08416 (TJK, GEL), NIH U01 HL099773 (JAT, TJK), NIH R01 EB007534 (SPP, TJK), NSF EFRI-0735903 (SPP, TJK), NIH P01-HL039707 (JJ); P01-HL087226 (JJ) and the Leducq Foundation (JJ).
Non-standard Abbreviations
- AP
action potential
- bFGF
basic fibroblast growth factor
- BMP4
bone morphogenetic protein 4
- CM
cardiomyocyte
- DAPI
4′,6′-Diamidino-2-Phenylindole
- EB
embryoid body
- ECM
extracellular matrix
- EMT
epithelial-mesenchymal transition
- ESC
embryonic stem cells
- GFR
growth factor reduced
- iPSC
induced pluripotent stem cell
- MEFs
mouse embryonic fibroblasts
- PSC
pluripotent stem cell
- WGA
wheat germ agglutinin
Footnotes
DISCLOSURES
We acknowledge that two authors (JAT and TJK) hold founding shares in Cellular Dynamics International, Inc, a company that uses cardiomyocytes derived from human stem cells for drug discovery and toxicity testing. JY is a full-time employee of Cellular Dynamics International, Inc.
References
- 1.Laflamme MA, Chen KY, Naumova AV, Muskheli V, Fugate JA, Dupras SK, Reinecke H, Xu C, Hassanipour M, Police S, O’Sullivan C, Collins L, Chen Y, Minami E, Gill EA, Ueno S, Yuan C, Gold J, Murry CE. Cardiomyocytes derived from human embryonic stem cells in pro-survival factors enhance function of infarcted rat hearts. Nat Biotechnol. 2007;25:1015–1024. doi: 10.1038/nbt1327. [DOI] [PubMed] [Google Scholar]
- 2.Yang L, Soonpaa MH, Adler ED, Roepke TK, Kattman SJ, Kennedy M, Henckaerts E, Bonham K, Abbott GW, Linden RM, Field LJ, Keller GM. Human cardiovascular progenitor cells develop from a kdr+ embryonic-stem-cell-derived population. Nature. 2008;453:524–528. doi: 10.1038/nature06894. [DOI] [PubMed] [Google Scholar]
- 3.Kattman SJ, Witty AD, Gagliardi M, Dubois NC, Niapour M, Hotta A, Ellis J, Keller G. Stage-specific optimization of activin/nodal and bmp signaling promotes cardiac differentiation of mouse and human pluripotent stem cell lines. Cell Stem Cell. 2011;8:228–240. doi: 10.1016/j.stem.2010.12.008. [DOI] [PubMed] [Google Scholar]
- 4.Elliott DA, Braam SR, Koutsis K, Ng ES, Jenny R, Lagerqvist EL, Biben C, Hatzistavrou T, Hirst CE, Yu QC, Skelton RJ, Ward-van Oostwaard D, Lim SM, Khammy O, Li X, Hawes SM, Davis RP, Goulburn AL, Passier R, Prall OW, Haynes JM, Pouton CW, Kaye DM, Mummery CL, Elefanty AG, Stanley EG. Nkx2-5(egfp/w) hescs for isolation of human cardiac progenitors and cardiomyocytes. Nat Methods. 2011;8:1037–1040. doi: 10.1038/nmeth.1740. [DOI] [PubMed] [Google Scholar]
- 5.Dubois NC, Craft AM, Sharma P, Elliott DA, Stanley EG, Elefanty AG, Gramolini A, Keller G. Sirpa is a specific cell-surface marker for isolating cardiomyocytes derived from human pluripotent stem cells. Nat Biotechnol. 2011;29:1011–1018. doi: 10.1038/nbt.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paige SL, Osugi T, Afanasiev OK, Pabon L, Reinecke H, Murry CE. Endogenous wnt/beta-catenin signaling is required for cardiac differentiation in human embryonic stem cells. PLoS One. 2010;5:e11134. doi: 10.1371/journal.pone.0011134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guilak F, Cohen DM, Estes BT, Gimble JM, Liedtke W, Chen CS. Control of stem cell fate by physical interactions with the extracellular matrix. Cell Stem Cell. 2009;5:17–26. doi: 10.1016/j.stem.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rozario T, DeSimone DW. The extracellular matrix in development and morphogenesis: A dynamic view. Dev Biol. 2010;341:126–140. doi: 10.1016/j.ydbio.2009.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ludwig TE, Bergendahl V, Levenstein ME, Yu J, Probasco MD, Thomson JA. Feeder-independent culture of human embryonic stem cells. Nat Methods. 2006;3:637–646. doi: 10.1038/nmeth902. [DOI] [PubMed] [Google Scholar]
- 10.Villa-Diaz LG, Nandivada H, Ding J, Nogueira-de-Souza NC, Krebsbach PH, O’Shea KS, Lahann J, Smith GD. Synthetic polymer coatings for long-term growth of human embryonic stem cells. Nat Biotechnol. 2010;28:581–583. doi: 10.1038/nbt.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Melkoumian Z, Weber JL, Weber DM, Fadeev AG, Zhou Y, Dolley-Sonneville P, Yang J, Qiu L, Priest CA, Shogbon C, Martin AW, Nelson J, West P, Beltzer JP, Pal S, Brandenberger R. Synthetic peptide-acrylate surfaces for long-term self-renewal and cardiomyocyte differentiation of human embryonic stem cells. Nat Biotechnol. 2010;28:606–610. doi: 10.1038/nbt.1629. [DOI] [PubMed] [Google Scholar]
- 12.Rodin S, Domogatskaya A, Strom S, Hansson EM, Chien KR, Inzunza J, Hovatta O, Tryggvason K. Long-term self-renewal of human pluripotent stem cells on human recombinant laminin-511. Nat Biotechnol. 2010;28:611–615. doi: 10.1038/nbt.1620. [DOI] [PubMed] [Google Scholar]
- 13.Klim JR, Li L, Wrighton PJ, Piekarczyk MS, Kiessling LL. A defined glycosaminoglycan-binding substratum for human pluripotent stem cells. Nat Methods. 2010;7:989–994. doi: 10.1038/nmeth.1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tesar PJ, Chenoweth JG, Brook FA, Davies TJ, Evans EP, Mack DL, Gardner RL, McKay RD. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature. 2007;448:196–199. doi: 10.1038/nature05972. [DOI] [PubMed] [Google Scholar]
- 15.Brons IG, Smithers LE, Trotter MW, Rugg-Gunn P, Sun B, Chuva de Sousa Lopes SM, Howlett SK, Clarkson A, Ahrlund-Richter L, Pedersen RA, Vallier L. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature. 2007;448:191–195. doi: 10.1038/nature05950. [DOI] [PubMed] [Google Scholar]
- 16.Evans SM, Yelon D, Conlon FL, Kirby ML. Myocardial lineage development. Circ Res. 2010;107:1428–1444. doi: 10.1161/CIRCRESAHA.110.227405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 18.George EL, Georges-Labouesse EN, Patel-King RS, Rayburn H, Hynes RO. Defects in mesoderm, neural tube and vascular development in mouse embryos lacking fibronectin. Development. 1993;119:1079–1091. doi: 10.1242/dev.119.4.1079. [DOI] [PubMed] [Google Scholar]
- 19.Duband JL, Thiery JP. Appearance and distribution of fibronectin during chick embryo gastrulation and neurulation. Dev Biol. 1982;94:337–350. doi: 10.1016/0012-1606(82)90352-9. [DOI] [PubMed] [Google Scholar]
- 20.Page-McCaw A, Ewald AJ, Werb Z. Matrix metalloproteinases and the regulation of tissue remodelling. Nat Rev Mol Cell Biol. 2007;8:221–233. doi: 10.1038/nrm2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, Slukvin, Thomson JA. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 22.Yu J, Hu K, Smuga-Otto K, Tian S, Stewart R, Slukvin, Thomson JA. Human induced pluripotent stem cells free of vector and transgene sequences. Science. 2009;324:797–801. doi: 10.1126/science.1172482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 24.Chuva de Sousa Lopes SM, Hassink RJ, Feijen A, van Rooijen MA, Doevendans PA, Tertoolen L, Brutel de la Riviere A, Mummery CL. Patterning the heart, a template for human cardiomyocyte development. Dev Dyn. 2006;235:1994–2002. doi: 10.1002/dvdy.20830. [DOI] [PubMed] [Google Scholar]
- 25.Amit M, Carpenter MK, Inokuma MS, Chiu CP, Harris CP, Waknitz MA, Itskovitz-Eldor J, Thomson JA. Clonally derived human embryonic stem cell lines maintain pluripotency and proliferative potential for prolonged periods of culture. Dev Biol. 2000;227:271–278. doi: 10.1006/dbio.2000.9912. [DOI] [PubMed] [Google Scholar]
- 26.Zhang J, Wilson GF, Soerens AG, Koonce CH, Yu J, Palecek SP, Thomson JA, Kamp TJ. Functional cardiomyocytes derived from human induced pluripotent stem cells. Circ Res. 2009;104:e30–41. doi: 10.1161/CIRCRESAHA.108.192237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hou L, Deo M, Furspan P, Pandit SV, Mironov S, Auerbach DS, Gong Q, Zhou Z, Berenfeld O, Jalife J. A major role for herg in determining frequency of reentry in neonatal rat ventricular myocyte monolayer. Circ Res. 2010;107:1503–1511. doi: 10.1161/CIRCRESAHA.110.232470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Herron TJ, Devaney E, Mundada L, Arden E, Day S, Guerrero-Serna G, Turner I, Westfall M, Metzger JM. Ca2+-independent positive molecular inotropy for failing rabbit and human cardiac muscle by alpha-myosin motor gene transfer. Faseb J. 2010;24:415–424. doi: 10.1096/fj.09-140566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Freund C, Ward-van Oostwaard D, Monshouwer-Kloots J, van den Brink S, van Rooijen M, Xu X, Zweigerdt R, Mummery C, Passier R. Insulin redirects differentiation from cardiogenic mesoderm and endoderm to neuroectoderm in differentiating human embryonic stem cells. Stem Cells. 2008;26:724–733. doi: 10.1634/stemcells.2007-0617. [DOI] [PubMed] [Google Scholar]
- 30.McLean AB, D’Amour KA, Jones KL, Krishnamoorthy M, Kulik MJ, Reynolds DM, Sheppard AM, Liu H, Xu Y, Baetge EE, Dalton S. Activin a efficiently specifies definitive endoderm from human embryonic stem cells only when phosphatidylinositol 3-kinase signaling is suppressed. Stem Cells. 2007;25:29–38. doi: 10.1634/stemcells.2006-0219. [DOI] [PubMed] [Google Scholar]
- 31.Kubalak SW, Miller-Hance WC, O’Brien TX, Dyson E, Chien KR. Chamber specification of atrial myosin light chain-2 expression precedes septation during murine cardiogenesis. J Biol Chem. 1994;269:16961–16970. [PubMed] [Google Scholar]
- 32.O’Brien TX, Lee KJ, Chien KR. Positional specification of ventricular myosin light chain 2 expression in the primitive murine heart tube. Proc Natl Acad Sci US A. 1993;90:5157–5161. doi: 10.1073/pnas.90.11.5157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Segev H, Kenyagin-Karsenti D, Fishman B, Gerecht-Nir S, Ziskind A, Amit M, Coleman R, Itskovitz-Eldor J. Molecular analysis of cardiomyocytes derived from human embryonic stem cells. Development, growth & differentiation. 2005;47:295–306. doi: 10.1111/j.1440-169X.2005.00803.x. [DOI] [PubMed] [Google Scholar]
- 34.Sugi Y, Lough J. Onset of expression and regional deposition of alpha-smooth and sarcomeric actin during avian heart development. Dev Dyn. 1992;193:116–124. doi: 10.1002/aja.1001930203. [DOI] [PubMed] [Google Scholar]
- 35.He JQ, Ma Y, Lee Y, Thomson JA, Kamp TJ. Human embryonic stem cells develop into multiple types of cardiac myocytes: Action potential characterization. Circ Res. 2003;93:32–39. doi: 10.1161/01.RES.0000080317.92718.99. [DOI] [PubMed] [Google Scholar]
- 36.Jezek K, Pucelik P, Sauer J, Bartak F. Basic electrophysiological parameters and frequency sensitivity of the ventricular myocardium of human embryos. Physiol Bohemoslov. 1982;31:11–19. [PubMed] [Google Scholar]
- 37.Mummery C, Ward-van Oostwaard D, Doevendans P, Spijker R, van den BS, Hassink R, van der HM, Opthof T, Pera M, de la Riviere AB, Passier R, Tertoolen L. Differentiation of human embryonic stem cells to cardiomyocytes: Role of coculture with visceral endoderm-like cells. Circulation. 2003;107:2733–2740. doi: 10.1161/01.CIR.0000068356.38592.68. [DOI] [PubMed] [Google Scholar]
- 38.Fast VG, Kleber AG. Microscopic conduction in cultured strands of neonatal rat heart cells measured with voltage-sensitive dyes. Circ Res. 1993;73:914–925. doi: 10.1161/01.res.73.5.914. [DOI] [PubMed] [Google Scholar]
- 39.Rohr S, Kucera JP, Kleber AG. Slow conduction in cardiac tissue, i: Effects of a reduction of excitability versus a reduction of electrical coupling on microconduction. Circ Res. 1998;83:781–794. doi: 10.1161/01.res.83.8.781. [DOI] [PubMed] [Google Scholar]
- 40.Munoz V, Grzeda KR, Desplantez T, Pandit SV, Mironov S, Taffet SM, Rohr S, Kleber AG, Jalife J. Adenoviral expression of iks contributes to wavebreak and fibrillatory conduction in neonatal rat ventricular cardiomyocyte monolayers. Circ Res. 2007;101:475–483. doi: 10.1161/CIRCRESAHA.107.149617. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


