Abstract
The characterization of encapsulation efficiency and in vitro drug release from nanoparticle-based formulations often requires the separation of nanoparticles from unencapsulated drug. Inefficient separation of nanoparticles from the medium in which they are dispersed can lead to inaccurate estimates of encapsulation efficiency and drug release. This study establishes dynamic light scattering as a simple method for substantiation of the effectiveness of the separation process. Colistin-loaded liposomes, as an exemplar nano-sized delivery particle, were diluted to construct a calibration curve relating the amount of light scattering to liposome concentration. Dynamic light scattering revealed that, in the case of ultracentrifugation and centrifugal ultrafiltration, approximately 2.9% of the total liposomes remained in supernatants or filtrates, respectively. In comparison, filtrates obtained using pressure ultrafiltration contained less than 0.002% of the total liposomes from the formulation. Subsequent release studies using dialysis misleadingly implied a slow release of colistin over >48 h. In contrast, pressure ultrafiltration revealed immediate equilibration to the equilibrium distribution of colistin between the liposome and aqueous phases upon dilution. Pressure ultrafiltration is therefore recommended as the optimal method of choice for studying release kinetics of drug from nanomedicine carriers.
Keywords: Nanoparticle, dynamic light scattering, encapsulation efficiency, in vitro release, centrifuge ultrafiltration, ultracentrifugation, pressure ultrafiltration
Introduction
Drug loading, and the amount of drug that remains associated with the colloidal carrier over time following administration, are both important considerations in the formulation of a drug using a nanoparticle carrier [1]. Drug loading is frequently described by the encapsulation efficiency (EE), which is defined as the percentage or fraction of the drug added in the process of particle manufacture that is associated with the nanoparticle carrier. The time course of drug release from nanoparticles is also important, as it defines the amount of free drug available over time which is able to provide either the therapeutic effect or, in some cases, modify the toxicity profile of the product [2–3]. The in vitro drug release profile measured in a bio-relevant medium is therefore an important performance parameter to be considered in the development of new nanoparticle formulations. When performed under appropriately selected conditions, in vitro release profiles can allow prediction of the in vivo behaviour of the encapsulated drug [1, 4]. Drug release profiles may also provide information about the mechanism of release, which is essential information to enable optimization of the formulation to achieve the desired release rate properties.
The measurement of both EE and in vitro drug release from colloidal particles typically requires methods for the rapid physical separation of particles from their surrounding dispersion medium to enable real-time determination of the proportion of free drug. For large particles this may be achieved by a simple filtration approach. However, separation can be a challenge for nanoparticles due to their small size [5]. Most methods for measurement of encapsulation and in vitro release separate the particles from the medium in which they were dispersed, and rely on the quantification of the ‘free’ fraction of drug to indirectly measure the nanoparticle-bound fraction. Numerous methods for the separation of free and nanoparticle-associated drug are reported in the literature including dialysis-based methods, ultracentrifugation [6–7], centrifugal ultrafiltration [8–10] and pressure ultrafiltration [11–13].
A potential issue associated with the use of any of the aforementioned physical separation methods is that the separation may be incomplete or inefficient. It is impossible to visually detect the presence of a small amount of nanoparticles present in the filtrate or supernatant of a separated sample. However, their presence is likely to lead to significant measurement errors, particularly early in the release timescale when the concentration of drug in the carrier particles is high relative to that free in solution. The application of such separation methods is frequently reported in the drug delivery literature, however to our knowledge there has been no method proposed to validate the efficiency of separation of nanoparticles from the surrounding medium in which they are dispersed to produce a ‘clean’ sample of unbound drug. Consequently, in the first part of the current study, the application of dynamic light scattering (DLS) was investigated and applied to the different separation techniques described above, to demonstrate the presence of liposomes in apparently ‘separated’ free drug solution.
Dialysis techniques [14–16], including side-by-side dialysis, sac dialysis [17–20] and reverse dialysis [9, 17, 21], are almost exclusively used in the literature for the measurement of release kinetics. Dialysis methods achieve physical separation of particles from a ‘sink’ acceptor chamber by a semi-permeable membrane with an appropriately selected molecular weight cut-off (MWCO) [22]. In principle, unencapsulated drug molecules passage across the membrane, while nanoparticle-bound drug is unable to penetrate the membrane. Release of drug from nanoparticles is indicated by the appearance of drug in the acceptor compartment. Drug concentration in the acceptor compartment, however, may not always reflect the true free concentration of drug in the donor compartment and so the appropriateness of dialysis in estimating drug release from nanoparticles has come under question in the literature [1, 12, 23]. The passage of drug into the acceptor compartment can often be dictated by membrane transport effects which mask the true release rate of drug from nanoparticles [12].
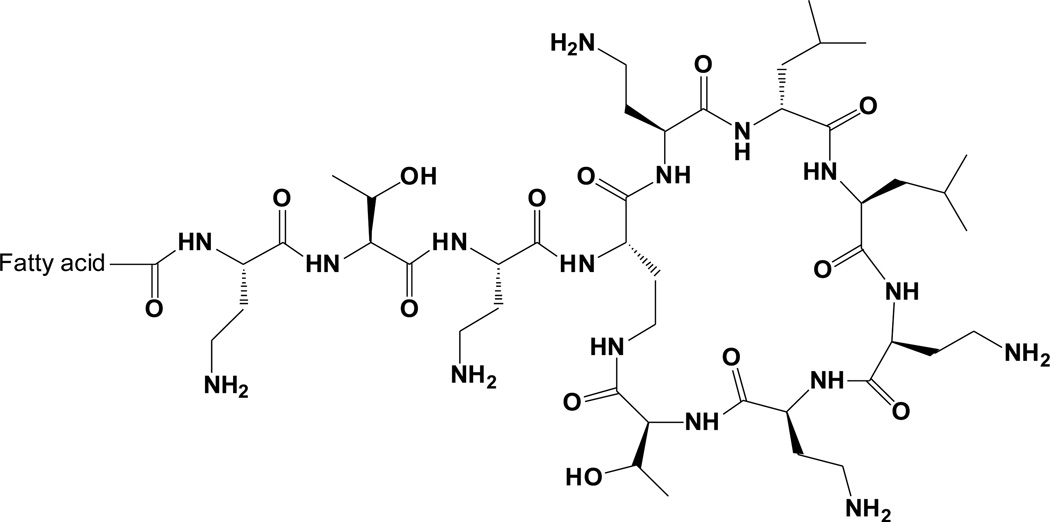
The interest in proposing validated drug release methodology has arisen from the desire to understand the mechanisms of association of an antimicrobial lipopeptide, colistin, with liposomes. The recent resurgence of colistin (Figure 1), an ‘old’ antibiotic, back into the clinic as a last-line therapy against Gram-negative ‘superbugs’, has highlighted the potential to optimize colistin therapy through formulation strategies [24–26]. Liposomal encapsulation has been suggested as an approach to enhance drug delivery and overcome colistin-associated toxicities [8]. As a result of the findings in the first part of the current work on validation of separation methods, in the second part of this paper, colistin-loaded dioleoylphosphatidylcholine (DOPC)/cholesterol liposomes were used as a model nanoparticle system for comparison of in vitro drug release profiles of colistin obtained by pressure ultrafiltration and equilibrium dialysis.
Figure 1.
Chemical structure of colistin
Materials and methods
Materials
Water was purified using a Milli-Q® water system (Millipore, Milford, MA). Soy dioleylphosphatidylcholine (DOPC) (stored at −20°C) was obtained from Phospholipid GmbH (Cologne, Germany). Cholesterol, sodium chloride, potassium chloride, sodium phosphate dibasic, sodium hydroxide and potassium dihydrogen orthophosphate were purchased from Sigma (St Louis, MO). Colistin sulfate was from Zhejiang Shenghua Biok Biology Co. Ltd (Grade EP5, Hangzhou, China). Chloroform and methanol were from Mallincrodt Baker. Nitrogen gas was from BOC Gases (Sydney, Australia). All reagents and chemicals used for HPLC analysis were of HPLC grade.
Methods
Preparation of liposomes
Liposomes were produced by the dry film method [27]. Briefly, 500 mg DOPC was weighed into a 100-mL round-bottomed flask with or without cholesterol (mole ratio DOPC:cholesterol 2:1) and was dissolved in 1.5 mL of chloroform/methanol (2:1). The solvent was evaporated under vacuum at 40°C using a rotary evaporator (Buchi Labortechnik, Switzerland). The resulting dry lipid film was flushed with a continuous stream of nitrogen gas to remove trace solvent and was stored at −20°C until required. After equilibration to room temperature, the dry lipid film was dispersed in 10 mL of colistin sulfate aqueous solution in Milli-Q® water (for encapsulation efficiency measurements) or phosphate buffered saline, PBS (for in vitro release experiments) to achieve the final DOPC concentration (64.5 mM). On an iced water bath, liposomes were ultrasonicated using a probe ultrasonicator with a 3-mm titanium probe (Misonix Incorporated, Farmingdale, NY, USA) for 30 min on pulse mode (1 sec pulses interrupted by 1 sec break, 15 min total sonication time).
Dynamic light scattering (DLS)
Detection of nanoparticles in separated samples and particle size measurements were both conducted using a Zetasizer Nano ZS DLS instrument (Malvern Instruments, Worcestershire, UK). The instrument uses a 4 mW He-Ne laser (λ = 632.8 nm) with backscatter detection at 173° and a thermostatted sample chamber set to 25°C. The viscosity and refractive index of water at 25°C (0.8937 and 1.333 cP respectively [28, 29]) were used for all measurements. The derived count rate, in kilo counts per second (kcps) was recorded during particle size measurements.
In the absence of nanoparticles, separated samples of ultrafiltrate or supernatant completely free of nanoparticles are expected to scatter light comparable to the scattering of pure dispersant. The light scattered by Milli-Q® water according to DLS measurements ranged from 45 – 50 kcps. In contrast, samples obtained after incomplete separation of nanoparticles from the dispersion medium (e.g. penetration of particles through ultrafilter, or incomplete sedimentation during ultracentrifugation) that still contain nanoparticles, scatter a significant amount of light which can be quantified by DLS. Because light scattered is directly proportional to the size and number of particles present in a sample, the absolute light scattering (derived count rate) can be used to indicate the presence of nanoparticles in the sample. Comparison of the efficiency of separation methods was obtained by measurement of the light scattered by the ‘free solution’ (ultrafiltrate or supernatant) obtained using the different separation methods.
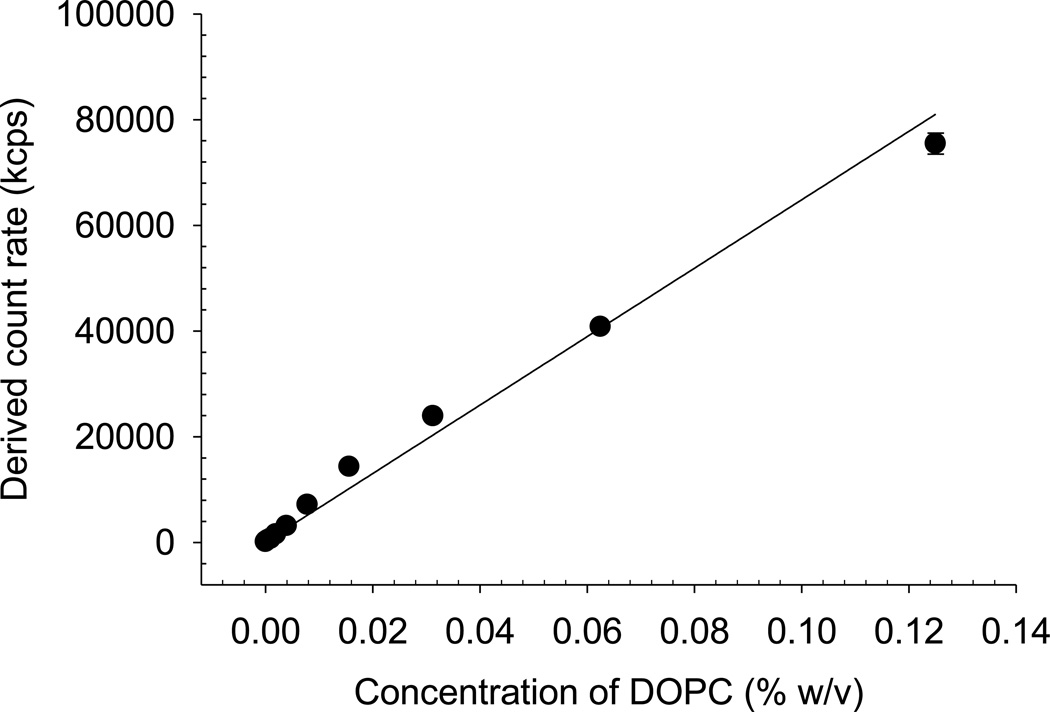
To establish a calibration curve relating counts to particle concentration, DOPC liposomes were diluted and the amount of light scattering was measured. A suspension of 1% (w/v) DOPC in water was prepared by the dry film method described above. A series of standards at concentrations up to 0.125% (w/v) DOPC in water were prepared by serial dilution of the 1% (w/v) DOPC preparation in water. The amount of light scattered by each standard was measured during particle size measurements in a low volume polystyrene cuvette. The Zetasizer Nano ZS automatically adjusted the attenuator setting to optimize the amount of light scattered by a sample. Consequently, the derived count rate (as opposed to the reported attenuated count rate), was used to indicate the absolute light scattered by the liposome dispersions.
Liposome separation methods
Ultracentrifugation
The ultracentrifugation of DOPC liposome suspensions containing 1 mg/mL colistin (sulfate) prepared by the dry film method, was carried out using a Beckman Optima L-90 K preparative ultracentrifuge with a SW 60 Ti rotor. Formulations were ultracentrifuged at 300,000 g for up to 8 h at 25°C. The efficiency of the separation of liposomes from the dispersion medium after ultracentrifugation was assessed by visual inspection and also by DLS measurements of the supernatant.
Centrifugal ultrafiltration
Centrifugal ultrafiltration was performed using Nanosep 300K (MWCO 300 kDa) and 100K (MWCO 100 kDa) modified polyethersulphone ultrafiltration devices (Pall Life Sciences, NY, USA) and Microcon YM50 (MWCO 50 kDa) and YM30 (MWCO 30 kDa) regenerated cellulose ultrafiltration devices (Millipore Corp., MS, USA). A 0.5-mL aliquot of DOPC liposomes containing 1 mg/mL colistin was loaded into each of the centrifuge ultrafiltration units and centrifuged at 25°C for 5 min at 9,300 g using a microcentrifuge. The filtrate was collected and the scattering by the filtrate was measured using DLS.
Pressure ultrafiltration
The pressure ultrafiltration method used an Amicon 8010 10-mL pressure ultrafiltration cell fitted with a 25-mm diameter regenerated cellulose YM10 ultrafiltration membrane (MWCO 10 kDa, Millipore Corp., Bedford, MA). The membranes were stored in 10% ethanol and were soaked in Milli-Q® water overnight prior to use. A minimum volume of 5 mL of DOPC liposomes containing 1 mg/mL colistin was loaded into the pressure ultrafiltration cell. Pressure was applied to the surface using nitrogen gas (< 400 kPa) while the cell was stirred using the inbuilt magnetic stirrer. The filtrate was collected and the back scattering of the filtrate was measured using DLS.
Equilibrium dialysis
Equilibrium dialysis, conducted in a side-by-side diffusion cell (Austin Health, Melbourne, Australia) was employed to measure the release of colistin from DOPC/cholesterol (2:1) liposomes. A 3-mL aliquot of a 1 mg/mL colistin liposome suspension, or a 1 mg/mL colistin solution (both dispersed in (PBS) pH 7.4), was filled into one side of the dialysis cell and blank PBS was filled into the other side. Separating the two sides of the dialysis cell was a circular piece of Spectra-Por®, 12,000 - 14,000 MWCO dialysis membrane (Spectrum Medical Industries, CA, USA) with a surface area of ~ 4.9 cm2. Membranes were soaked overnight in PBS prior to use. The dialysis cells were maintained at 37°C in a shaking water bath.
In vitro release studies
Dialysis
For in vitro release studies, samples (150 µL) were carefully removed from each compartment via the sampling port at appropriate time points and were retained for HPLC analysis. The experiment was conducted in three replicates.
Pressure ultrafiltration
The adsorption of colistin on the ultrafiltration equipment was first quantified to ensure that the concentrations of drug obtained in the filtrate reflected the free concentration of drug in the dispersion medium. Solutions containing colistin (sulfate, 100 µg/mL) were filtered through the ultrafiltration cell and the recovery of colistin was determined by HPLC. To ensure that the concentration of drug in the ultrafiltrate was within 95% of that inside the cell, the volume of solution required to saturate the membrane was determined.
The colistin-containing DOPC/cholesterol liposomes (10 mg/mL) were diluted into PBS at a 1:5 ratio (v/v) in a 50-mL polypropylene tube in order to disturb the equilibrium distribution of drug between particles and the medium to stimulate release. The diluted liposome suspension was incubated at 37°C in a shaking water bath. At predetermined time points, a 5-mL sample was removed and filtered through the pressure ultrafiltration cell. The filtrate was collected in two separate 500-µL fractions. The first 500-µL fraction of ultrafiltrate was retained for the measurement of separation efficiency. Samples exhibiting light scattering > 500 kcps were discarded due to penetration of the liposomes through the ultrafiltration membrane. In that case, the separation was repeated with a new ultrafiltration membrane. The second 500-µL fraction of ultrafiltrate was retained for determination of colistin concentration in the ultrafiltrate (free drug concentration) by HPLC as described below. All experiments were repeated in triplicate.
HPLC analysis of colistin in filtrate samples
The HPLC system was similar to that used in an earlier study, except that a SPD-10A UV detector was used [30]. A C18 Phenosphere Next column (250 mm×4.60 mm, 5µm, Phenomonex, Torrance, CA, USA) was used to separate the two major components of colistin, colistin A and B. All analyses were carried out at 25°C. The mobile phase consisting of 0.1% trifluoroacetic acid (phase A) and acetonitrile (ACN, phase B) was eluted at 1 mL/min under a gradient program: 0 – 2 min: 74% A, 26% B, 2 – 2.5 min 67% A, 33% B, 2.5 – 6 mins: 74% A, 26% B. Peaks for colistin A and B were detected at 210 nm, at 5.3 and 5.0 min, respectively. The peak areas for colistin A and B peaks were summed to obtain the total colistin concentration. The assay was linear in the range between 0.05 and 2 mg/mL with reproducibility and accuracy within 10%. The lower limit of quantification was 0.05 mg/mL. For measurement of concentrations in ultrafiltrate, samples were diluted 1:1 with ACN prior to injection. The sample pretreatment step involved the addition of 200 µL of ACN to 200 µL of sample. The sample was then vortex mixed before 10 µL was injected directly onto the HPLC column.
Results and discussion
Calibration curves for liposome concentration obtained using DLS
The calibration curves using highly-diluted DOPC liposome suspensions showed an approximately linear relationship (R2 = 0.969) between liposome concentration and amount of light scattering up to approximately 0.6% w/v DOPC (Figure 2 shows a section of this curve). The detection of liposomes using DLS was highly sensitive, detecting as low as 0.002% w/v DOPC. Readers should recognize that alternative colloidal systems are likely to produce different calibration curves due to differences in absolute scattering power between systems, hence preparation of a calibration curve by dilution of the particular dispersion under study is necessary in appropriate application of this method.
Figure 2.
Light scattering (derived count rate, kcps) of DOPC liposomes in water with increasing concentrations of DOPC. Data are presented as mean ± SD (n = 3).
Comparison of particle separation effectiveness
Ultracentrifugation
Separation is achieved by forcing particles to sediment into a pellet under high centrifugal force often over very long centrifugation times when the densities of the particle and surrounding medium are closely matched [31]. For example, small unilamellar liposomes have been reported to be sedimented by ultracentrifugation at 200,000 g for 10 to 20 h in a preparative ultracentrifuge [32]. After sedimentation, the concentration of the drug in the supernatant and/or pellet is measured to estimate the encapsulated and unencapsulated proportions of drug.
In this study, ultracentrifugation of colistin liposomes at 300,000 g, a relatively high centrifugal force, yielded a supernatant that scattered light at ~37,000 kcps. According to the calibration plot in Figure 2 this corresponded to approximately 0.06% DOPC, or 1.13% of the total liposomes in the formulation, remaining in the supernatant.
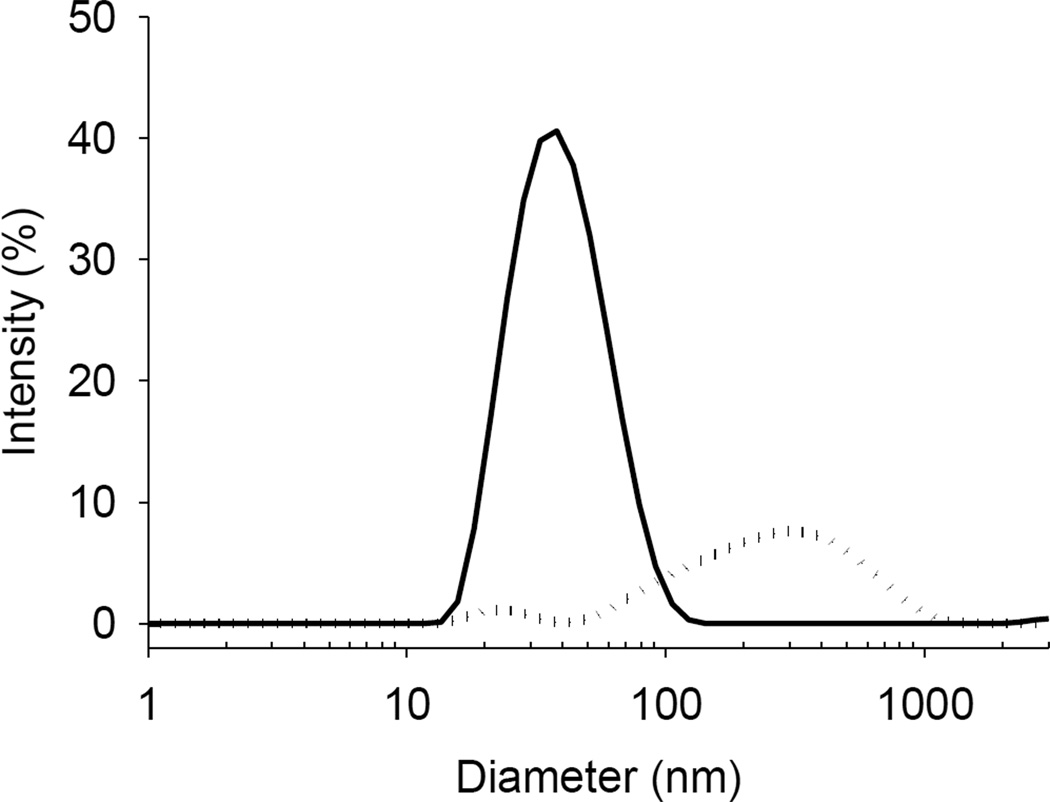
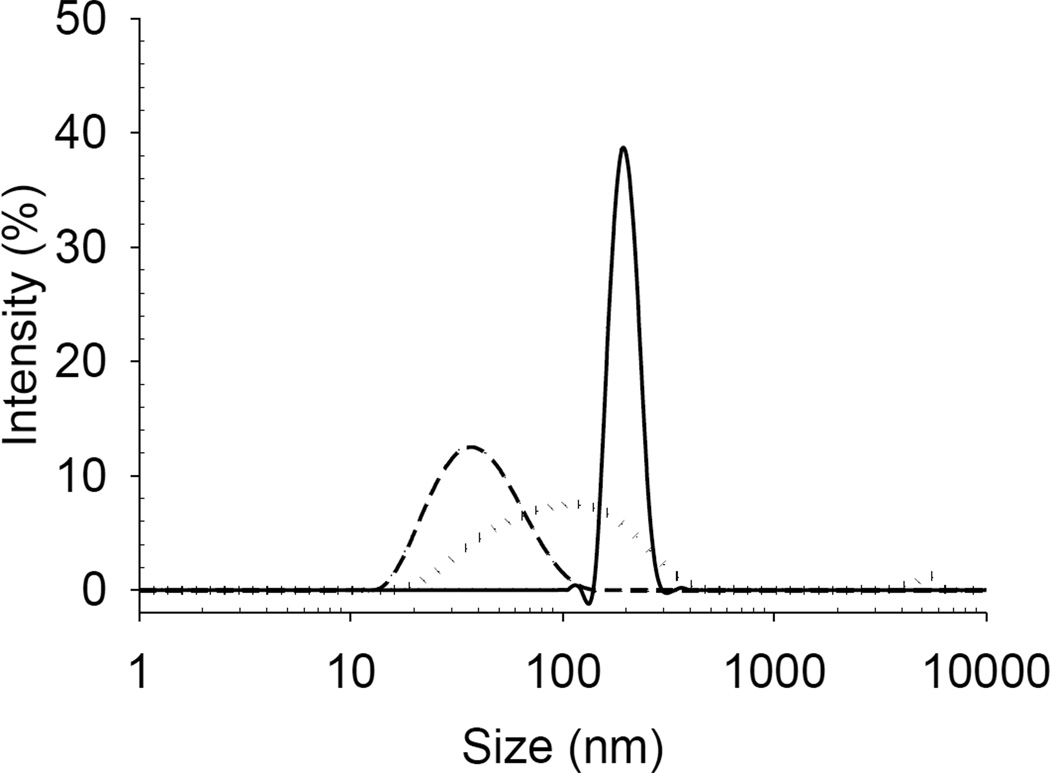
The particle size distributions shown in Figure 3 indicate that the ultracentrifugation technique was unable to sediment liposomes smaller than 100 nm under these experimental conditions. The z-average particle size of liposomes prior to separation was ~123.87 ± 1.13 nm with polydispersity index 0.492 ± 0.031. The difference between the densities of the liposomes and the dispersing solution was insufficient to enable quantitative sedimentation, even at extremely high gravitational force. Although only ~1% of the liposomes remained in the supernatant, for a highly lipophilic drug this may incorrectly imply a much greater amount of drug ‘free’ in solution than is actually the case.
Figure 3.
Particle size distribution of the supernatant before (dotted line) and after (solid line) ultracentrifugation of liposomes containing colistin (1 mg/mL).
In addition to the lack of complete separation of liposomes from free drug, the long centrifugation times required to sediment particles prohibited obtaining a ‘snapshot’ of drug distribution between nanoparticles and the dispersing medium, with the resolution between time points being defined by the long centrifugation time. Furthermore, the high centrifugal forces involved in ultracentrifugation are of concern due to the potential for disturbance of the drug-nanoparticle interactions and potential to induce artifactual release [33].
Centrifugal ultrafiltration
The separation of samples using centrifugal ultrafiltration can be problematic for dispersions with high particle content due to impaction and clogging of the filter. On the other hand, this method has several advantages over ultracentrifugation: separation is rapid in the order of minutes and low centrifugal forces (e.g. 2,000 – 4,000 g) can be used compared to ultracentrifugation. Due to the relatively low centrifugal force used, there may be less potential for particle deformation and therefore less compromise to particle integrity.
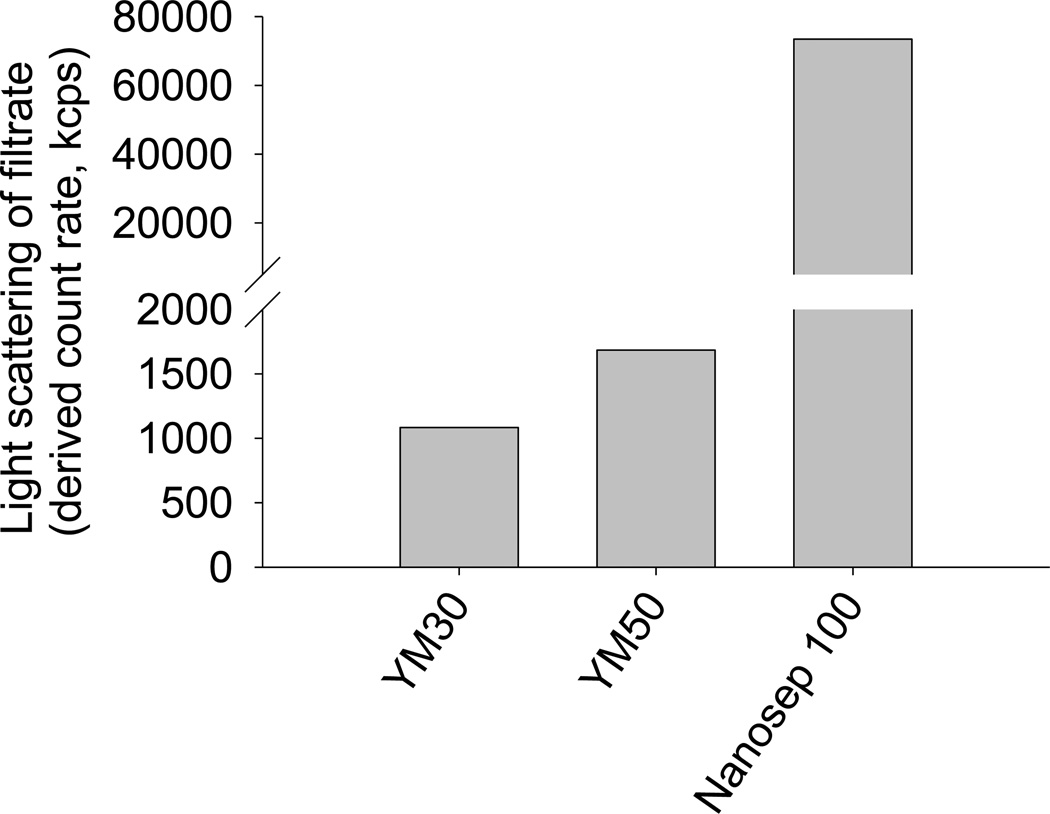
The amount of light scattered by filtrates obtained by the separation of liposomes using different centrifuge ultrafiltration devices is shown in Figure 4. The scattered light increased as the MWCO of the membranes in the devices increased, indicating that an increased proportion of liposomes penetrated the membrane. Based on the light scattering data, the YM30 device achieved the best separation, however, only an extremely small volume of filtrate (< 20 µL) could be recovered from each separation due to the clogging of the filter by the larger liposome particles under the centrifugal force. A slightly larger filtrate yield (~ 50 µL) could be obtained using the YM50 device, however light scattering measurements revealed that the separation efficiency of the YM50 device was lower than that of the YM30 device. As determined from the calibration plot (Figure 2), the amount of DOPC present was 0.002%, 0.003% and 0.113% for the YM30, YM50 and Nanosep 100 filtrate samples, respectively, indicating that 0.03%, 0.05% and 2.26% of the total liposomes in the formulation had penetrated the filter membranes, respectively.
Figure 4.
Light scattered by filtrates obtained with centrifugal ultrafiltration after separation of DOPC liposomes containing colistin (1 mg/mL) using membranes with 30, 50 and 100 kDa molecular weight cut-offs.
The particle size distributions in Figure 5 clearly show that liposomes with size approximating 100 nm in diameter were able to penetrate the 100kDa MWCO centrifuge ultrafiltration membranes. The lower MWCO membranes also showed peaks in the particle size distributions, however the counts obtained were very low, meaning that the particle size data should be interpreted with caution. The broad peak obtained with the 50kDa MWCO membranes may be reflective of the size of the liposomes passing the membrane, however the multimodal distribution evident for the filtrate from the YM30 membrane is typical of samples with insufficient concentration to obtain a reliable measurement. Although the lower MWCO devices produced relatively ‘clean’ filtrates, the yield of filtrate in volume terms was too low for these methods to be practically useful. Low volume yield is a particular problem during the separation of formulations containing drugs that adsorb to the filtration membrane, due to the requirement to saturate binding to the membrane prior to obtaining a representative sample.
Figure 5.
Particle size distribution of filtrates obtained after centrifugal ultrafiltration of liposome samples through YM30 ••••••, YM50 --- and Nanosep 100 —— filtration devices.
Pressure ultrafiltration
Pressure ultrafiltration was first described for the measurement of drug release by Magenheim et al. [5]. In contrast to centrifugal ultrafiltration, pressure ultrafiltration uses pressure applied to the top surface of the dispersion to exert a gentle hydrostatic force upon the particles. The contents of a pressure ultrafiltration cell are stirred continuously to prevent particles from clogging the filter pores. Separation is rapid and a representative sample may be obtained in less than 5 minutes, depending on the concentration of particles in the formulation [5].
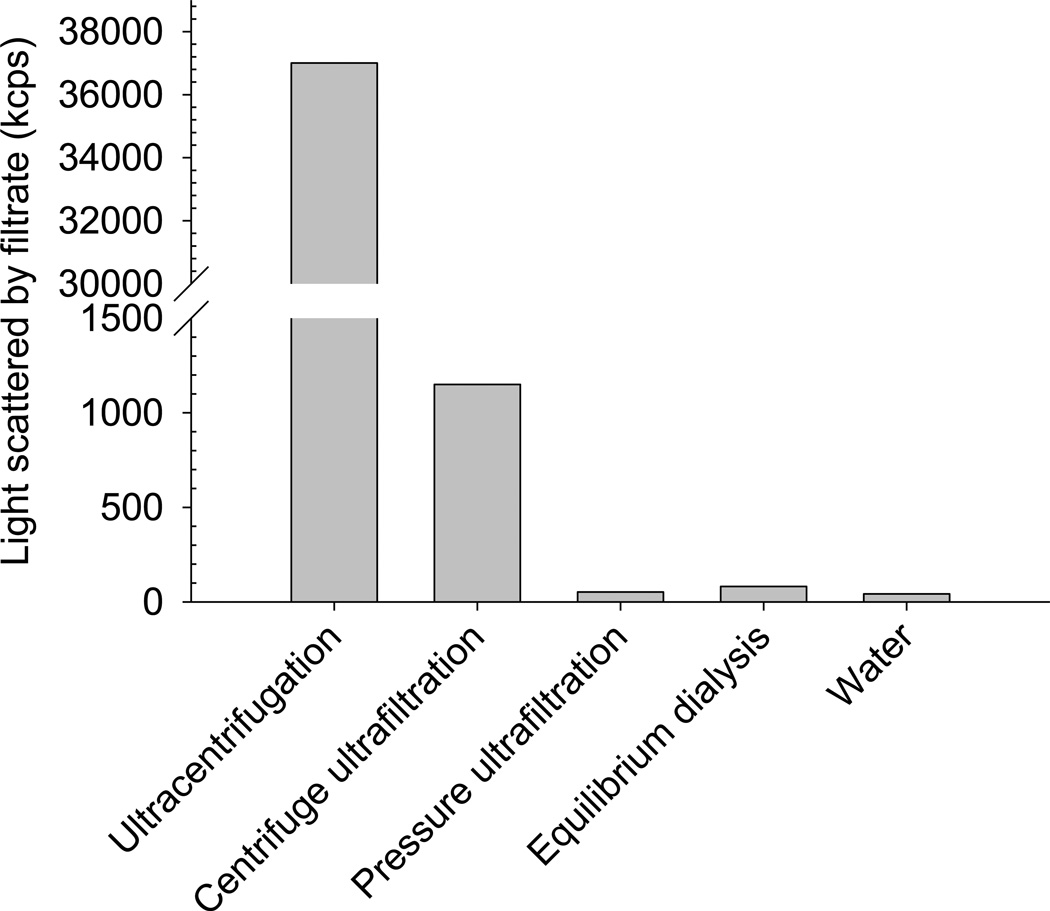
Separation of liposomes from free solution using the pressure ultrafiltration method in this study consistently produced filtrates with light scattering comparable to that of Milli-Q® grade water (Figure 6). No size data could be obtained due to quantitative exclusion of particles. This clearly demonstrates that pressure ultrafiltration produces filtrates that are free from liposomes, and is by far the most efficient method for the separation of liposomes from their surrounding dispersion medium. The method also yields a useful volume of filtrate in a sufficiently short time to enable informative time resolution for dynamic release studies. The devices are available at smaller nominal membrane cut-offs than the 10 kDa MWCO used in this study, but would be expected to pass a lower volume of ultrafiltrate under the same condition as the present study. Pressure ultrafiltration was subsequently used to measure the encapsulation efficiency of colistin in DOPC/cholesterol liposomes. Measurement of the light scattered by the ultrafiltrate was routinely conducted to ensure consistent separation in subsequent release measurements.
Figure 6.
Light scattering of separated samples obtained using ultracentrifugation, centrifugal ultrafiltration (30 kDa cut-off) and pressure ultrafiltration (10 kDa cut-off). The light scattering of pure water is shown for the purposes of comparison.
Equilibrium dialysis
Equilibrium dialysis was also shown to efficiently separate liposomes between the donor and receptor compartments, with dialysate samples scattering light at count rates similar to that of pure water indicating that drug-loaded liposomes were not able to penetrate the dialysis membrane (Figure 6). However, the dialysis methods have inherent limitations in the measurement of encapsulation efficiency and in vitro drug release and these limitations are discussed in subsequent sections.
In vitro release studies for colistin from liposomes by equilibrium dialysis and pressure ultrafiltration
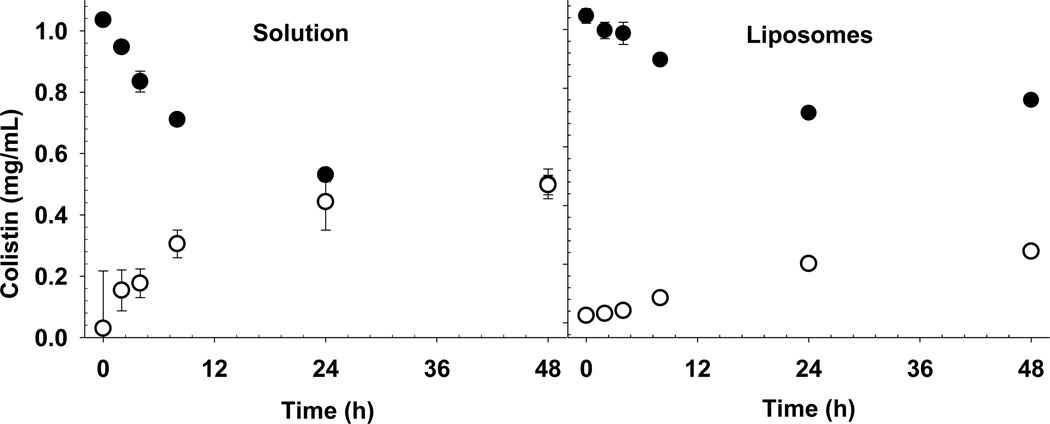
It is apparent that the release of colistin from liposomes, measured using equilibrium dialysis, was dictated by the slow diffusion of free colistin across the dialysis membrane (Figure 7). The time required for the free colistin to equilibrate across the membrane was >24 h in the presence and absence of liposomes. Therefore, release profiles obtained by dialysis of liposomes must be interpreted with care. The long time required to reach equilibrium (>24 h), demonstrates that the concentration of colistin in the acceptor compartment at any time point did not necessarily reflect the true ‘free’ concentration of drug in the donor chamber. The limited membrane surface area available for passage of colistin into the acceptor chamber may have contributed to the long equilibration time.
Figure 7.
Appearance of colistin in the equilibrium dialysis acceptor chamber (○), and disappearance from the donor chamber (●) where a colistin solution (left panel) or colistin DOPC/chol 2:1 liposomes (right panel) were dialyzed against PBS. Data are presented as mean ± SD (n = 3).
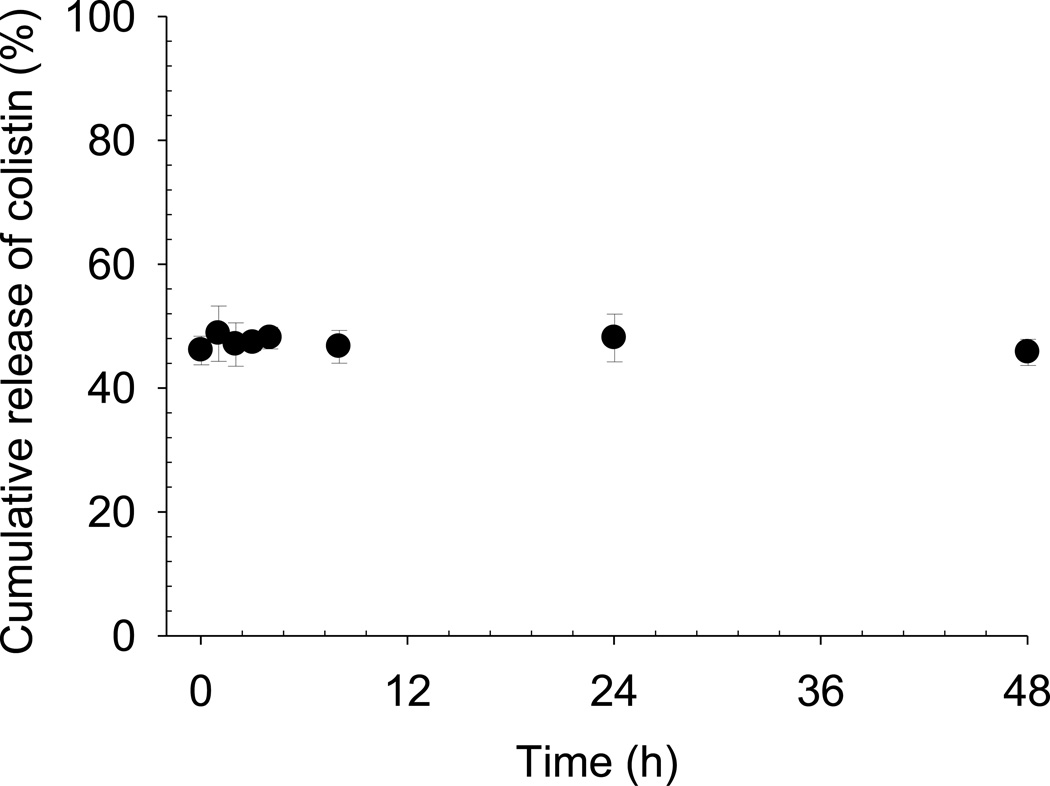
One must also consider that the concentration gradient driving the diffusion of colistin across the dialysis membrane is lower in the case of dialysis of the liposome formulation than for the ‘free’ solution. The encapsulated proportion of colistin does not contribute to the concentration gradient driving the diffusion of colistin across the dialysis membrane. As such, the slow rate of colistin appearance in the acceptor chamber may then be wrongly attributed to slow release of colistin from the liposomes, when in fact that may not be the case. The true release rate must be deconvoluted from the kinetics of drug diffusion across the membrane, which requires a priori knowledge of the appropriate ‘free’ concentration to enable an appropriate control experiment to be conducted [1]. In the present study, the in vitro release profile obtained by pressure ultrafiltration of a diluted colistin-containing liposome dispersion (Figure 8) was markedly different from that obtained by equilibrium dialysis of the same formulation (Figure 7). Following dilution into the in vitro release media, the concentration of free colistin was between 45 - 50% of the total and was independent of time. In other words, a typical ‘release’ phase was not observed. The encapsulation efficiency of colistin in undiluted liposomes when measured by pressure ultrafiltration was 46%. Taken together these results indicate that spontaneous re-establishment of the liposome-bound fraction of colistin occurs upon dilution, due to the rapid partitioning of colistin across the liposomal bilayer.
Figure 8.
Cumulative release of colistin from liposomes measured using pressure ultrafiltration. Data are presented as mean ± SD (n = 3).
Given that colistin is amphiphilic and has the capacity to interact with biological membranes [34], the prospect of colistin spontaneously partitioning into and out of liposomes by passage across the lipid bilayer is reasonable. Considering the high aqueous solubility of colistin, it is unlikely that the incomplete ‘release’ obtained in this study can be attributed to the presence of non-sink conditions. Therefore we conclude that release of colistin from liposomes was instantaneous on dilution, and that the apparent slow release of colistin in the equilibrium dialysis measurement was an artifact of the methodology. Because the separation of liposomes is rapid and efficient using pressure ultrafiltration, a true picture of the distribution of drug between the free and nanoparticle-bound fraction was obtained by this method.
An example of a likely misinterpretation of in vitro release data when using dialysis to measure release of colistin from liposomes, is in the study published by Wang et al.[8] In that study, the in vitro release of colistin from undiluted liposomes was measured using sac dialysis. The authors claimed an initial “burst” release of colistin, followed by what was reported to be a “slow” release over 24 h. Based on the differences between the release profiles of free colistin in solution and liposome-encapsulated colistin, the authors that claimed an initial ‘burst release’ of 40% was attributed to the passage of unencapsulated colistin across the dialysis membrane, and that the passage of the remaining colistin was due to the slow release of the drug from the liposome interior. We agree that the initial ‘burst release’ was a result of the passage of unencapsulated colistin across the dialysis membrane. However, we disagree with the conclusion that the passage of the remaining colistin was due to a ‘slow’ or ‘controlled release’ effect. Our ultrafiltration experiments have shown the spontaneous, rapid redistribution of colistin between the aqueous and liposomal phases upon dilution. This behavior, under dialysis conditions, was wrongly interpreted by Wang et al. as a sustained release.
The misrepresentation of drug release from nanoparticles by equilibrium dialysis is not limited to the example of drug release from liposomes, but also applies to other nanoparticle systems. Benita et al. demonstrated the advantage of pressure ultrafiltration in measuring the release rate of diazepam from the commercial emulsion formulation, Diazemuls® [35]. The burst release phase, reaching 100% release within the first 5 min after dilution, could be identified using pressure ultrafiltration [35]. The dialysis method, on the other hand, indicated a slow release of diazepam from Diazemuls®, with only 20% release achieved after 3 h [36]. Similarly, vastly different drug release profiles were also obtained using equilibrium dialysis compared to pressure ultrafiltration when used to measure drug release from cubosomes (dispersed particles of lipid cubic phase, analogous to liposomes) [12]. Equilibrium dialysis measurements indicated a slow release of diazepam from cubosomes over 24 h, however pressure ultrafiltration revealed much more rapid release, with a plateau due to non-sink conditions occurring after only 20 min [12]. Further, Rosenblatt et al. Independently measured drug release from cubosomes using a non-separation in situ electrochemical method and concluded that burst release of small molecules occurs from cubosome particles on dilution [37]. Hence, the validity of equilibrium dialysis in measuring the release rate from particles is clearly limited by the rate of drug diffusion across the dialysis membrane, which itself is highly dependent upon the concentration gradient across the membrane, amongst other variables.
Conclusion
The DLS approach presented in this study provides a simple, new method to establish the effectiveness of separation techniques for drug encapsulation and release measurements from nanoparticulate drug carriers such as liposomes. The approach will find application as an in-use method for confirming the adequate separation of the nanoparticles from a sample of the dispersion medium. Although the equilibrium dialysis method can achieve separation of nanoparticles from the surrounding solution, this method can produce misleading in vitro release data. To date, no standardized technique for the assessment of drug release from nanomedicines has been issued by regulatory authorities. In view of the shortcomings of dialysis methods, pressure ultrafiltration is proposed to be more likely to produce a release profile that is representative of the true distribution of drug between nanoparticle and dispersing medium at any point in time. The example of instant release and re-equilibration of colistin from liposomes on dilution is consistent with previous findings for other drugs in cubosomes and emulsions using pressure ultrafiltration. Therefore, the potential for pressure ultrafiltration to be implemented in the routine quality analysis of nanomedicines is worthy of further investigation.
Acknowledgements
JL is an Australian National Health and Medical Research Council Senior Research Fellow. This study was supported by Award Number R01AI079330 and Award Number R01AI070896 from the National Institute of Allergy and Infectious Diseases. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Allergy and Infectious Diseases or the National Institutes of Health.
References
- 1.Washington C. Drug release from microdisperse systems: a critical review. Int J Pharm. 1990;58:1–12. [Google Scholar]
- 2.Herman EH, Rahman A, Ferrans VJ, Vick JA, Schein PS. Prevention of chronic doxorubicin cardiotoxicity in beagles by liposomal encapsulation. Cancer Res. 1983;43:5427–5432. [PubMed] [Google Scholar]
- 3.Barenholz Y. Relevancy of drug loading to liposomal formulation therapeutic efficacy. J Liposome Res. 2003;13:1–8. doi: 10.1081/lpr-120017482. [DOI] [PubMed] [Google Scholar]
- 4.Washington C, Koosha F. Drug release from microparticles: denconvolution of measurement errors. Int J Pharm. 1990;59:79–82. [Google Scholar]
- 5.Magenheim B, Levy MY, Benita S. A new in vitro rechnique for the evaluation of drug release profile from colloidal carriers - ultrafiltration technique at low pressure. Int J Pharm. 1993;94:115–123. [Google Scholar]
- 6.Ricci M, Giovagnoli S, Blasi P, Schoubben A, Perioli L, Rossi C. Development of liposomal capreomycin sulfate formulations: effects of formulation variables on peptide encapsulation. Int J Pharm. 2006;311:172–181. doi: 10.1016/j.ijpharm.2005.12.031. [DOI] [PubMed] [Google Scholar]
- 7.Zheng Y, Wu Y, Yang W, Wang C, Fu S, Shen X. Preparation, characterization, and drug release in vitro of chitosan-glycyrrhetic acid nanoparticles. J Pharm Sci. 2006;95:181–191. doi: 10.1002/jps.20399. [DOI] [PubMed] [Google Scholar]
- 8.Wang D, Kong L, Wang J, He X, Li X, Xiao Y. Polymyxin E sulfate-loaded liposome for intravenous use: preparation, lyophilization, and toxicity assessment in vivo. PDA J Pharm Sci Technol. 2009;63:159–167. [PubMed] [Google Scholar]
- 9.Santos Magalhaes NS, Fessi H, Puisieux F, Benita S, Seiller M. An in vitro release kinetic examination and comparative evaluation between submicron emulsion and polylactic acid nanocapsules of clofibride. J Microencapsul. 1995;12:195–205. doi: 10.3109/02652049509015290. [DOI] [PubMed] [Google Scholar]
- 10.Ozer AY, Talsma H. Preparation and stability of liposomes containing 5-fluorouracil. Int J Pharm. 1989;55:185–191. [Google Scholar]
- 11.Cui J, Li C, Deng Y, Wang Y, Wang W. Freeze-drying of liposomes using tertiary butyl alcohol/water cosolvent systems. Int J Pharm. 2006;312:131–136. doi: 10.1016/j.ijpharm.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 12.Boyd BJ. Characterisation of drug release from cubosomes using the pressure ultrafiltration method. Int J Pharm. 2003;260:239–247. doi: 10.1016/s0378-5173(03)00262-x. [DOI] [PubMed] [Google Scholar]
- 13.Magenheim B, Levy MY, Benita S. A new in vitro technique for the evaluation of drug release profile from colloidal carriers - ultrafiltration technique at low pressure. Int J Pharm. 1993;94:115–123. [Google Scholar]
- 14.Ammoury N, Fessi H, Devissaguet JP, Puisieux F, Benita S. In vitro release kinetic pattern of indomethacin from poly(D,L-lactide) nanocapsules. J Pharm Sci. 1990;79:763–767. doi: 10.1002/jps.2600790902. [DOI] [PubMed] [Google Scholar]
- 15.Henriksen I, Sverre AS, Gro S, Karlsen J. In vitro evaluation of drug release kinetics from liposomes by fractional dialysis. Int J Pharm. 2005;119:231–238. [Google Scholar]
- 16.Johnston MJ, Edwards K, Karlsson G, Cullis PR. Influence of drug-to-lipid ratio on drug release properties and liposome integrity in liposomal doxorubicin formulations. J Liposome Res. 2008;18:145–157. doi: 10.1080/08982100802129372. [DOI] [PubMed] [Google Scholar]
- 17.Chidambaram N, Burgess DJ. A novel in vitro release method for submicron sized dispersed systems. AAPS PharmSci. 1999;1:E11. doi: 10.1208/ps010311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Avgoustakis K, Beletsi A, Panagi Z, Klepetsanis P, Karydas AG, Ithakissios DS. PLGA-mPEG nanoparticles of cisplatin: in vitro nanoparticle degradation, in vitro drug release and in vivo drug residence in blood properties. J Control Release. 2002;79:123–135. doi: 10.1016/s0168-3659(01)00530-2. [DOI] [PubMed] [Google Scholar]
- 19.Saarinen-Savolainen P, Jarvinen T, Taipale H, Urtti A. Methods for evaluating drug release from liposomes in sink conditions. Int J Pharm. 1997;159:27–33. [Google Scholar]
- 20.Sezer AD, Akbuga J, Bas AL. In vitro evaluation of enrofloxacin-loaded MLV liposomes. Drug Deliv. 2007;14:47–53. doi: 10.1080/10717540600640146. [DOI] [PubMed] [Google Scholar]
- 21.Muthu MS, Singh S. Poly (D, L-lactide) nanosuspensions of risperidone for parenteral delivery: formulation and in-vitro evaluation. Curr Drug Deliv. 2009;6:62–68. doi: 10.2174/156720109787048302. [DOI] [PubMed] [Google Scholar]
- 22.D'Souza SS, DeLuca PP. Methods to assess in vitro drug release from injectable polymeric particulate systems. Pharm Res. 2006;23:460–474. doi: 10.1007/s11095-005-9397-8. [DOI] [PubMed] [Google Scholar]
- 23.Rosenblatt KM, Douroumis D, Bunjes H. Drug release from differently structured monoolein/poloxamer nanodispersions studied with differential pulse polarography and ultrafiltration at low pressure. J Pharm Sci. 2007;96:1564–1575. doi: 10.1002/jps.20808. [DOI] [PubMed] [Google Scholar]
- 24.Nation RL, Li J. Colistin in the 21st century. Curr Opin Infect Dis. 2009;22:535–543. doi: 10.1097/QCO.0b013e328332e672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Falagas ME, Kasiakou SK. Colistin: the revival of polymyxins for the management of multidrug-resistant gram-negative bacterial infections. Clin Infect Dis. 2005;40:1333–1341. doi: 10.1086/429323. [DOI] [PubMed] [Google Scholar]
- 26.Wallace SJ, Li J, Nation RL, Prankerd RJ, Velkov T, Boyd BJ. Self-assembly behavior of colistin and its prodrug colistin methanesulfonate: implications for solution stability and solubilization. J Phys Chem B. 2010;114:4836–4840. doi: 10.1021/jp100458x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bangham AD. Membrane models with phospholipids. Prog Biophys Mol Biol. 1968;18:29–95. doi: 10.1016/0079-6107(68)90019-9. [DOI] [PubMed] [Google Scholar]
- 28.Gales AC, Reis AO, Jones RN. Contemporary assessment of antimicrobial susceptibility testing methods for polymyxin B and colistin: review of available interpretative criteria and quality control guidelines. J Clin Microbiol. 2001;39:183–190. doi: 10.1128/JCM.39.1.183-190.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lide DR. Handbook of Chemistry and Physics. 76th ed. New York: CRC Press/Lewis Publishers; 1995. [Google Scholar]
- 30.Wallace SJ, Li J, Rayner CR, Coulthard K, Nation RL. Stability of colistin methanesulfonate in pharmaceutical products and solutions for administration to patients. Antimicrob Agents Chemother. 2008;52:3047–3051. doi: 10.1128/AAC.00103-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lasch J. Isothermic microcalorimetry. In: Torchillin VP, Weissig V, editors. Liposomes: a practical approach. 2nd ed. Oxford, UK: Oxford University Press; 2003. [Google Scholar]
- 32.New RRC. Liposomes: A practical approach. Oxford: IRL Press; 1990. [Google Scholar]
- 33.Heeremans JL, Gerritsen HR, Meusen SP, Mijnheer FW, Gangaram Panday RS, Prevost R, et al. The preparation of tissue-type Plasminogen Activator (t-PA) containing liposomes: entrapment efficiency and ultracentrifugation damage. J Drug Target. 1995;3:301–310. doi: 10.3109/10611869509015959. [DOI] [PubMed] [Google Scholar]
- 34.Clausell A, Garcia-Subirats M, Pujol M, Busquets MA, Rabanal F, Cajal Y. Gram-negative outer and inner membrane models: insertion of cyclic cationic lipopeptides. J Phys Chem B. 2007;111:551–563. doi: 10.1021/jp064757+. [DOI] [PubMed] [Google Scholar]
- 35.Benita S, Levy MY. Submicron emulsions as colloidal drug carriers for intravenous administration: comprehensive physicochemical characterization. J Pharm Sci. 1993;82:1069–1079. doi: 10.1002/jps.2600821102. [DOI] [PubMed] [Google Scholar]
- 36.Levy MY, Benita S. Drug release from submicronised o/w emulsion: a new in vitro kinetic evaluation model. Int J Pharm. 1990;66:29–37. [Google Scholar]
- 37.Rosenblatt KM, Douroumis D, Bunjes H. Drug release from differently structured monoolein/poloxamer nanodispersions studied with differential pulse polarography and ultrafiltration at low pressure. J Pharm Sci. 2007;96:1564–1575. doi: 10.1002/jps.20808. [DOI] [PubMed] [Google Scholar]