Abstract
Pigeon ‘milk’ and mammalian milk have functional similarities in terms of nutritional benefit and delivery of immunoglobulins to the young. Mammalian milk has been clearly shown to aid in the development of the immune system and microbiota of the young, but similar effects have not yet been attributed to pigeon ‘milk’. Therefore, using a chicken model, we investigated the effect of pigeon ‘milk’ on immune gene expression in the Gut Associated Lymphoid Tissue (GALT) and on the composition of the caecal microbiota. Chickens fed pigeon ‘milk’ had a faster rate of growth and a better feed conversion ratio than control chickens. There was significantly enhanced expression of immune-related gene pathways and interferon-stimulated genes in the GALT of pigeon ‘milk’-fed chickens. These pathways include the innate immune response, regulation of cytokine production and regulation of B cell activation and proliferation. The caecal microbiota of pigeon ‘milk’-fed chickens was significantly more diverse than control chickens, and appears to be affected by prebiotics in pigeon ‘milk’, as well as being directly seeded by bacteria present in pigeon ‘milk’. Our results demonstrate that pigeon ‘milk’ has further modes of action which make it functionally similar to mammalian milk. We hypothesise that pigeon ‘lactation’ and mammalian lactation evolved independently but resulted in similarly functional products.
Introduction
Pigeon ‘milk’ is a substance produced in the crop of both male and female pigeons for the nourishment of their young. Similarly, male and female flamingos [1] and male emperor penguins [2] can produce crop ‘milk’, but there is a paucity of information available about these processes. Like mammalian lactation, pigeon ‘milk’ production is regulated by the lactogenic hormone prolactin [3]. The resulting pigeon crop ‘milk’ consists of lipid-filled, protein rich keratinocytes that have proliferated and separated from the germinal epithelium of the crop sac to form a curd-like substance that is regurgitated to the squab [4]. This cheesy substance also contains bacteria [5]. Like mammalian milk, pigeon ‘milk’ is highly nutritious, consisting of protein (60%), fat (32–36%), carbohydrate (1–3%) and minerals (calcium, potassium, sodium and phosphorus) [6]; it also contains IgA antibodies [7]. Interestingly, if squabs are fed a nutritional replacement of pigeon ‘milk’ they die or fail to thrive [8], which suggests that there are factors aside from nutrition in pigeon ‘milk’ that influence development of the young. Like mammalian milk components, these factors in pigeon ‘milk’ may play a role in immune development. Mammalian milk can modulate the development of the immune system directly, by delivering immune molecules such as immunoglobulins and cytokines [9], [10], and indirectly by influencing the microbiota through prebiotics [11].
The bacterial composition of the gut of breast fed infants is very different to formula fed infants, as it is influenced by prebiotics in the breast milk [12]. Similarly, the gut microbial composition of mother-fed piglets differs to formula-fed piglets [13]. These differences in microbiota are significant as it has been shown that the gut microflora of the developing infant can play a role in the developing immune system [14] and in energy and nutrient capture [15]. The first contact between the immune system and the gut microflora is by the Gut Associated Lymphoid Tissue (GALT), which comprises the largest lymphoid tissue mass in the human body [16]. The GALT is also the largest site of IgA production in the body, synthesising over 60% of all IgA produced [16]. Development of IgA B cells is dependent on microbial colonisation [17], and consequently, colostrum contains high levels of IgA [9], as the infant has not yet established a microbiome to facilitate production of IgA.
Not only does mammalian milk modulate the microbiota of the developing infant and provide copious amounts of IgA, it also contains a gamut of other immune modulators that contribute to the immune protection of the immunologically naive infant by either modulating development of the immune system or providing passive immunity [18]. At birth, the human infant is deficient in certain cytokines and cells of the myeloid lineage, and others have impaired function [19], which renders the infant reliant on maternal passive immunity and on milk components that aid in the development of the immune system. These components include cytokines, chemokines and colony stimulating factors [20], as well as maternally-derived immune cells [21], [22]. A breast fed human infant consumes an estimated 108 immune cells per day, which consist of 55–60% macrophages, 30–40% neutrophils and 5–10% lymphocytes [21], [22]. Other beneficial substances found in milk include hormones such as epidermal growth factor [23], [24], enzymes such as lysozyme (which also has antimicrobial activity) [25], and other antimicrobial proteins such as lactoferrin [26], [27].
Pigeon ‘milk’ has been shown to contain a number of bioactive proteins including IgA [7], a pigeon ‘milk’ growth factor with biological activity similar to epidermal growth factor [28], [29], and transferrin [30], a glycoprotein with a similar sequence and structure to lactoferrin [31]. In addition, it has been shown that chickens fed pigeon ‘milk’ had a higher rate of growth than chickens not receiving pigeon ‘milk’ [32], [33], which could be attributed to the increased caloric intake and/or the beneficial effect of bacteria and bioactive molecules in pigeon ‘milk’. However, there have been no studies explicitly examining whether pigeon ‘milk’ can modulate immune tissues. Previous studies in chickens have shown that bacteria is important for the development of the GALT [34]. Here we test the hypothesis that pigeon ‘milk’ will alter the intestinal microbiota and effect expression of genes in the GALT. We show that pigeon ‘milk’-fed chickens had a different microbial composition in their caeca to control chickens, and they also showed significant enrichment of immune-related genes among genes differentially expressed in GALT tissues.
Results
Chickens fed pigeon ‘milk’ had increased body mass
At the start of the experiment (day 0) and at day 4, there was no significant difference between the body mass of pigeon ‘milk’ (PM)-fed chickens and control chickens (Table 1). After 7 days, PM-fed chickens had grown on average 12.5% heavier than control chickens. A nutritional replacement of pigeon ‘milk’ had no effect on the growth of chickens compared to the control group (Figure S1). Interestingly, the breast muscle made up a significantly (p<0.05) higher proportion of total body mass in the PM-fed chickens and the wing span of PM-fed chickens was wider compared to normally fed chickens (Table 1). The leg span of PM-fed chickens tended to be wider (p = 0.0558; Table 1) as did the height (p = 0.0820; Table 1). This increase in size was also accompanied by a decrease in feed conversion ratio (FCR); PM-fed chickens had an average FCR of 1.34 compared to 1.47 for control chickens (Table 1).
Table 1. Comparison of chicken body measurements by group.
| Measurement | Control (n = 8) | PM-fed (n = 8) | p value |
| Day 0 body mass | 43.14 g±1.024 g | 41.90 g±1.647 g | 0.2672 |
| Day 4 body mass | 67.63 g±2.337 g | 72.25 g±3.807 g | 0.1590 |
| Day 7 body mass | 137.0 g±7.530 g | 154.2 g±5.467 g | 0.0426* |
| Breast muscle mass | 6.793 g±0.6869 g | 9.289 g±0.7624 g | 0.0145* |
| Proportion of breast muscle to body mass | 4.868±0.3180 | 5.973±0.3780 | 0.0210* |
| Height | 14.75 cm±0.2113 cm | 15.19 cm±0.2100 cm | 0.0820 |
| Wing span | 7.563 cm±0.1752 cm | 8.000 cm±0.1336 cm | 0.0335* |
| Leg span | 8.850 cm±0.1615 cm | 9.219 cm±0.1451 cm | 0.0558 |
Body measurements of control and PM-fed chickens were analysed statistically using an unpaired t-test and the results are presented as the mean ± standard deviation.
significantly different (p<0.05)
Pigeon ‘milk’ affected gene expression in the GALT
Differential gene expression in the GALT was analysed using tissue from ileum and caecal tonsil because they contain a high proportion of GALT. A comparison of gene expression in the ileum of PM-fed chickens to control chickens revealed 2202 differentially expressed genes (p<0.05); 1586 of these genes were up-regulated and 616 were down-regulated. In addition, a comparison of gene expression in the caecal tonsil of PM-fed chickens to control chickens revealed 1131 differentially expressed genes (p<0.05); 522 of these genes were up-regulated and 609 were down-regulated.
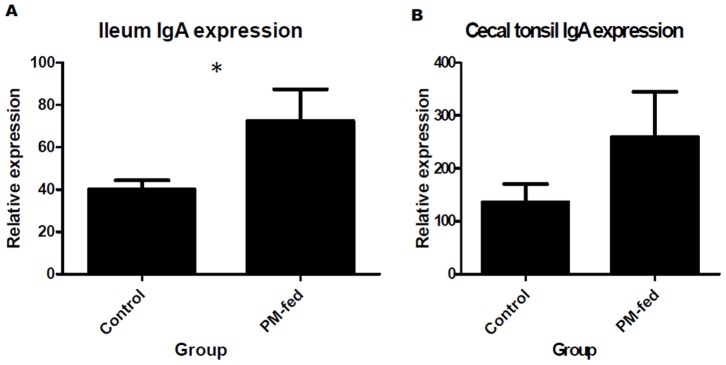
Functional analysis of the up-regulated genes by gene ontology in PM-fed chickens identified four immune-specific gene ontology biological processes in the ileum and 23 in the caecal tonsil (Table S1). Regulation of B cell activation was enriched in both ileum and caecal tonsil (Table S1) and analysis of the transcription of IgA heavy chain (transcribed in B cells) revealed that PM-fed chickens had a significantly higher level of IgA expression than control chickens in the ileum (p<0.05), and a trend toward higher expression in the caecal tonsil (p = 0.1265) (Figure 1). There were no immune-specific gene ontology biological processes down-regulated in either the ileum or caecal tonsil (Table S2). Down-regulated GO biological processes in the ileum related to cell cycle control and apoptosis, and lipid synthesis and metabolism in the caecal tonsil (Table S2). Three up-regulated immune-specific KEGG pathways were identified in the ileum and only one in the caecal tonsil. There were no down-regulated KEGG pathways in the ileum. In the caecal tonsil, there were two down-regulated KEGG pathways related to the splicesome and the actin cytoskeleton (Table S3).
Figure 1. IgA mRNA expression in the GALT.
Expression of IgA heavy chain mRNA was significantly higher in PM-fed chickens in the ileum (p = 0.033) and also tended to be higher in the caecal tonsil (p = 0.11), as compared to control chickens.
Six interferon-stimulated genes (ISGs) were up-regulated in the ileum and ten in the caecal tonsil (Table 2). The majority of these ISGs relate to host defence (five), antiviral (five), transcription factor or activator (four) or immune modulation (three) (Table 2).
Table 2. Interferon stimulated genes up-regulated in the gut of PM-fed chickens.
| Gene | Functional classification | Probe name | p value | Fold change |
| Ileum | ||||
| similar to complement component C2 | ComplementImmune modulation | RIGG20413 | 0.009 | 1.25 |
| Fibroblast growth factor 2 (basic) | AngiogenesisDevelopmentGrowth factor | RIGG16507 | 0.015 | 1.21 |
| CLIGg_41549 | 0.049 | 1.17 | ||
| Macrophage stimulating 1 (hepatocyte growth factor-like) | Growth factorSignaling | CLIGg_00552 | 0.009 | 2.15 |
| RIGG07902 | 0.003 | 2.14 | ||
| Interferon regulatory factor 7 | Host defenseTranscription factorTranscriptional activator | RIGG17886 | 0.019 | 1.53 |
| CLIGg_00887 | 0.023 | 1.38 | ||
| Interferon regulatory factor 1 | Host defenseImmune modulationSignalingTranscription factorTranscriptional activator | CLIGg_00658 | 0.035 | 1.34 |
| Interferon regulatory factor 4 | OncogeneTranscription factorTranscriptional activator | RIGG09155 | 0.032 | 1.24 |
| Caecal tonsil | ||||
| Interferon-induced protein with tetratricopeptide repeats 5 | Unknown | CLIGg_28648 | 0.006 | 2.76 |
| RIGG13336 | 0.010 | 2.52 | ||
| RIGG07326 | 0.006 | 2.47 | ||
| Myxovirus (influenza virus) resistance 1, interferon-inducible protein p78 (mouse) | AntiviralGTP-bindingHost defense | RIGG18960 | 0.005 | 2.20 |
| Misc_00001 | 0.005 | 1.97 | ||
| 2′-5′-oligoadenylate synthetase-like | AntiviralHost defense | CLIGg_00435 | 0.019 | 2.17 |
| RIGG01751 | 0.045 | 1.94 | ||
| Fibrinogen gamma chain | Blood clotting | RIGG14995 | 0.031 | 1.69 |
| Beta-2-microglobulin precursor | Antigen presentationHost defense | RIGG10931 | 0.009 | 1.35 |
| Interferon induced with helicase C domain 1 (MDA5) | Apoptosis [61]Antiviral [62], [63] | RIGG16089 | 0.033 | 1.30 |
| RIGG07546 | 0.029 | 1.24 | ||
| Misc_00005 | 0.042 | 1.23 | ||
| Zinc finger CCCH-type, antiviral 1 (ZAP) | Antiviral [64] | RIGG19894 | 0.010 | 1.22 |
| Similar to interferon-induced membrane protein 1 (IFITM1) | Antiviral [65], [66] | CLIGg_06123 | 0.003 | 1.21 |
| RIGG12134 | 0.005 | 1.19 | ||
| Complement component 1, q subcomponent, C chain | ComplementImmune modulation | CLIGg_08804 | 0.047 | 1.20 |
| V-ets erythroblastosis virus E26 oncogene homolog 2 (avian) | DevelopmentTranscription factorTranscriptional activator | CLIGg_04698 | 0.014 | 1.10 |
Genes up-regulated in PM-fed chicken (n = 6) gut which are known interferon-stimulated genes. No known interferon-stimulated genes were down-regulated.
Pigeon ‘milk’ influenced bacterial diversity and abundance
Statistical analysis of comparative abundance of bacteria between control and PM-fed chickens revealed that the PM had caused very significant changes in the population structure of the caecal microflora of PM-fed chickens. Many groups of bacteria were differentially abundant between control and PM-fed chickens at the levels of phylum, class, order, family and genus. Comparative analysis of bacterial abundance at a phylum level (Table 3) showed that bacterial 16S sequences were assigned to three bacterial phyla, and bacterial abundance was statistically different between control and PM-fed chickens in one of these phyla (Proteobacteria). The most abundant phyla in both groups was Firmicutes, constituting 99.622% of all control chicken bacteria and 96.630% of PM-fed bacteria, which had a more diverse range of bacteria (Table 3). The remainder of PM-fed bacteria belonged to Proteobacteria (0.318%) or were unassigned (3.052%). PM-fed chickens had no detected Bacteroidetes, whereas control chickens had 0.003% Bacteroidetes, and the remaining were Proteobacteria (0.021%) and unassigned (0.354%).
Table 3. Proportions of bacterial phyla present in control and PM-fed chickens.
| Classification | Control (%) (n = 8) | PM-fed (%) (n = 8) | p value |
| Phylum | |||
| Bacteroidetes | 0.003±0.003 | 0.000±0.000 | 1 |
| Firmicutes | 99.622±0.234 | 96.630±1.705 | 0.082 |
| Proteobacteria | 0.021±0.009 | 0.318±0.126 | 0.004* |
| Unassigned | 0.354±0.238 | 3.052±1.634 | 0.120 |
| Class | |||
| Bacilli | 77.117±5.666 | 57.917±6.345 | 0.022* |
| Bacteroidia | 0.003±0.003 | 0.000±0.000 | 1 |
| Betaproteobacteria | 0.000±0.000 | 0.315±0.127 | 0.013* |
| Clostridia | 22.026±5.323 | 37.378±5.452 | 0.045* |
| Erysipelotrichi | 0.059±0.031 | 0.088±0.051 | 0.551 |
| Gammaproteobacteria | 0.021±0.009 | 0.003±0.003 | 0.068 |
| Unclassified | 0.775±0.483 | 4.298±1.564 | 0.030* |
| Order | |||
| Bacillales | 0.269±0.216 | 0.096±0.055 | 0.441 |
| Bacteroidales | 0.003±0.003 | 0.000±0.000 | 1 |
| Burkholderiales | 0.000±0.000 | 0.315±0.127 | 0.013* |
| Clostridiales | 22.026±5.323 | 37.312±5.426 | 0.040* |
| Enterobacteriales | 0.021±0.0090 | 0.003±0.003 | 0.059 |
| Erysipelotrichales | 0.059±0.031 | 0.088±0.051 | 0.596 |
| Lactobacillales | 76.848±5.791 | 57.821±6.373 | 0.023* |
| Unclassified | 0.775±0.483 | 4.364±1.556 | 0.024* |
| Family | |||
| Alcaligenaceae | 0.000±0.000 | 0.315±0.127 | 0.013* |
| Bacillaceae | 0.269±0.216 | 0.096±0.055 | 0.472 |
| Bacteroidaceae | 0.003±0.003 | 0.000±0.000 | 1 |
| Enterobacteriaceae | 0.021±0.009 | 0.003±0.003 | 0.068 |
| Enterococcaceae | 0.464±0.32 | 1.802±0.381 | 0.007* |
| Erysipelotrichaceae | 0.059±0.031 | 0.088±0.051 | 0.644 |
| Eubacteriaceae | 0.065±0.034 | 0.043±0.026 | 0.625 |
| Incertae Sedis XIII | 0.012±0.009 | 0.008±0.006 | 0.710 |
| Incertae Sedis XIV | 0.017±0.007 | 0.007±0.007 | 0.329 |
| Lachnospiraceae | 7.643±2.45 | 9.208±1.704 | 0.622 |
| Lactobacillaceae | 76.179±5.923 | 55.262±6.423 | 0.017* |
| Peptostreptococcaceae | 5.21±1.894 | 1.14±0.273 | 0.039* |
| Ruminococcaceae | 7.367±1.2 | 9.3±1.656 | 0.370 |
| Streptococcaceae | 0.144±0.069 | 0.527±0.489 | 0.471 |
| Unclassified | 2.546±0.733 | 20.59±4.746 | 0.001* |
| Veillonellaceae | 0.000±0.000 | 1.611±0.999 | 0.111 |
| Genus | |||
| Anaerotruncus | 1.725±0.583 | 1.555±0.251 | 0.781 |
| Bacteroides | 0.003±0.003 | 0.000±0.000 | 1 |
| Blautia | 0.017±0.007 | 0.007±0.007 | 0.344 |
| Butyricicoccus | 0.323±0.162 | 0.419±0.138 | 0.660 |
| Enterococcus | 0.464±0.32 | 1.802±0.381 | 0.008* |
| Escherichia/Shigella | 0.021±0.009 | 0.003±0.003 | 0.058 |
| Eubacterium | 0.065±0.034 | 0.043±0.026 | 0.613 |
| Faecalibacterium | 0.113±0.103 | 0.657±0.439 | 0.264 |
| Lactobacillus | 70.772±6.107 | 52.351±6.442 | 0.037* |
| Oscillibacter | 0.194±0.107 | 0.371±0.083 | 0.219 |
| Roseburia | 0.119±0.048 | 0.23±0.107 | 0.378 |
| Sporacetigenium | 5.194±1.897 | 1.14±0.273 | 0.034* |
| Streptococcus | 0.144±0.069 | 0.527±0.489 | 0.470 |
| Subdoligranulum | 0.000±0.000 | 0.037±0.023 | 0.120 |
| Sutterella | 0.000±0.000 | 0.315±0.127 | 0.015* |
| Unclassified | 20.846±4.02 | 38.93±6.31 | 0.017* |
| Veillonella | 0.000±0.000 | 1.611±0.999 | 0.113 |
The proportion of bacteria present in each phylum, by chicken group. Proportional abundance of bacteria in each phylum was calculated using Metastats and the results are presented as the mean ± the standard error.
p<0.05
At the class level bacteria from the two groups of chickens were classified into 6 classes (Table 3); three of which were significantly differentially abundant between PM-fed and control chickens (Bacilli, Betaproteobacteria, and Clostridia) (Table 3). Bacilli was the most abundant class of bacteria in both groups of chickens (77.117% in control chickens and 57.917% in PM-fed chickens) followed by Clostridia (22.026% in control chickens and 37.378% in PM-fed chickens) (Table 3). At the order level, there were three bacterial orders significantly differentially abundant between PM-fed and control chickens out of seven orders classified (Table 3). These were Burkholdierales (not present in control chickens and 0.315% in PM-fed), Clostridiales (22.026% in controls and 37.312% in PM-fed) and Lactobacillales (76.848% in controls and 57.821% in PM-fed) (Table 3).
16S sequences from both chicken groups were assigned to 15 families, four of which were significantly differentially abundant (Table 3). These were Alcaligenaceae (not present in control and 0.315% of PM-fed), Enterococcaceae (0.464% of control and 1.802% of PM-fed), Lactobacillaceae (76.179% of control and 55.262% PM-fed) and Peptostreptococcaceae (5.21% of control and 1.14% of PM-fed) (Table 3). In addition to Alcaligenaceae, control chickens had no Veillonellaceae (PM-fed 1.611%) (Table 3). Conversely, PM-fed chickens had no Bacteroidaceae (control 0.003%) (Table 3).
At the genus level, sequences were classified into 16 genera, four of which were significantly differentially abundant (Table 3). These were Enterococcus (control 0.464%, PM-fed 1.802%), Lactobacillus (control 70.772%, PM-fed 52.351%), Sporacetigenium (control 5.194%, PM-fed 1.14%), and Sutterella (control not present, PM-fed 0.315%) (Table 3). In addition to Suterella, control chickens had no Veillonella (PM-fed 1.611%) or Subdoligranulum (PM-fed 0.037%) (Table 3). PM-fed chickens had no Bacteroides (control 0.003%) (Table 3).
PM-fed chickens shared a number of bacteria present in PM
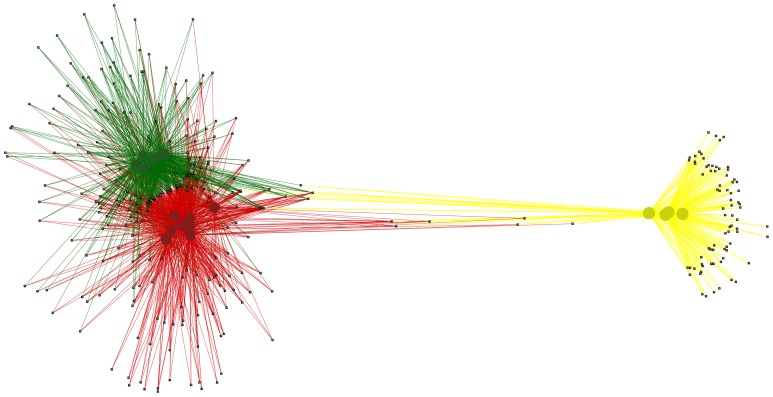
Network analysis of Operational Taxonomic Units (OTUs) shared between groups (Figure 2) revealed that PM-fed chickens share several OTUs with PM that are not present in control chickens, and control chickens share only one OTU with PM that is not present in PM-fed chickens. Additionally, control chickens and PM-fed chickens share many OTUs that are not present in PM, but they cluster as distinct groups (Figure 2).
Figure 2. Network analysis of OTUs present in PM, PM-fed chickens and control chickens.
PM-fed chickens (large red circles) and control chickens (large green circles) form distinct groups based on OTU (small black squares) abundance, although they still share many OTUs. PM (large yellow circles) was distinct from both groups of chickens. PM-fed chickens and PM shared six OTUs that were not present in control chickens. There were eight OTUs shared by all three groups. PM and control chickens shared only one OTU that was not present in PM-fed chickens.
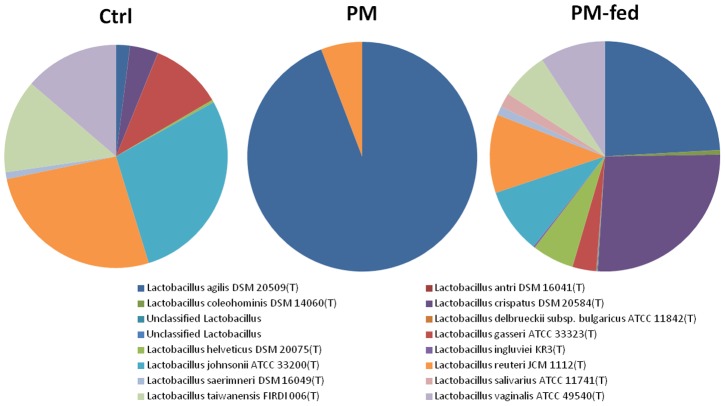
Analysis of the six OTUs present only in PM and PM-fed chickens revealed that four of the six OTUs are most closely related to Veillonella species (V. criceti, V. caviae, V. magna and V. ratti), one was identified as Enterococcus columbae, and one was most closely related to Sutterella stercoricanis (Table 4). The one OTU that was shared by PM and control chickens was most closely related to Bacteroides paurosaccharolyticus, and was present in very low abundance in control chickens (Table 4). The eight OTUs that are shared between all three groups were all identified as two Lactobacillus species; L. reuteri and L. agilis (Table 4). Analysis of the total Lactobacillus population in all groups (Figure 3) revealed that L. agilis and L. reuteri made up the entire Lactobacillus population of PM (94.19% and 5.82% respectively). L. agilis constituted 24.11% of PM-fed chicken Lactobacillus, whereas it constituted only 2.01% of control chicken Lactobacillus (Figure 3). L. reuteri constituted a higher percentage of control chicken Lactobacillus (26.47%) than PM-fed chicken Lactobacillus (11.01%), whereas PM-fed chicken total Lactobacillus had a higher proportion of L. crispatus and L. helveticus (26.23% and 5.89% respectively) than control chickens (4.10% and 0.38% respectively) (Figure 3). The PM-fed chicken total Lactobacillus population was more diverse than in control chickens, with 16 Lactobacillus species present compared to 12 in control chickens (Figure 3). The four species not present in control chickens make up a small percentage of the total PM-fed Lactobacillus population (L. coleohominis 0.62%, L. delbruckii subsp. Bulgaricus 0.09%, L. ingluvei 0.24% and L. salivarius 2.02%) (Figure 3).
Table 4. OTUs shared with PM.
| OTU | Closest cultured isolate | Similarity(%) | Rarefied abundance | ||
| PM (n = 4) | PM-fed (n = 8) | Ctrl (n = 8) | |||
| Present in PM and PM-fed chickens only | |||||
| 17 | Veillonella criceti ATCC 17747(T) | 94.41 | 33.53 | 35.95 | 0.00 |
| 86 | Sutterella stercoricanis CCUG 47620(T) | 95.42 | 2.27 | 7.64 | 0.00 |
| 88 | Veillonella caviae DSM 20738(T) | 94.82 | 18.44 | 0.91 | 0.00 |
| 183 | Enterococcus columbae LMG 11740(T) | 98.954 | 3.52 | 0.19 | 0.00 |
| 203 | Veillonella magna lac18(T) | 94.207 | 0.60 | 0.54 | 0.00 |
| 311 | Veillonella ratti DSM 20736(T) | 93.017 | 0.58 | 1.41 | 0.00 |
| Present in PM, PM-fed chickens and control chickens | |||||
| 3 | Lactobacillus reuteri JCM 1112(T) | 98.34 | 0.59 | 35.20 | 137.15 |
| 4 | Lactobacillus agilis DSM 20509(T) | 100 | 7.21 | 236.30 | 27.29 |
| 53 | Lactobacillus reuteri JCM 1112(T) | 99.349 | 0.10 | 51.00 | 241.19 |
| 97 | Lactobacillus agilis DSM 20509(T) | 99.554 | 0.51 | 11.06 | 1.14 |
| 107 | Lactobacillus agilis DSM 20509(T) | 98.718 | 0.11 | 6.58 | 0.25 |
| 217 | Lactobacillus agilis DSM 20509(T) | 99.111 | 0.10 | 4.27 | 0.26 |
| 334 | Lactobacillus agilis DSM 20509(T) | 98.95 | 1.02 | 39.41 | 2.31 |
| 393 | Lactobacillus agilis DSM 20509(T) | 97.976 | 0.25 | 17.46 | 6.60 |
| Present in PM and control chickens only | |||||
| 42 | Bacteroides paurosaccharolyticus WK042 | 90.798 | 54.22 | 0.00 | 0.06 |
OTUs (bacterial identifiers) present in PM and another group were classified to their closest cultured isolate using EZTaxon. The rarefied abundance is mean number of times a bacteria was present in a random sampling of 1000.
Figure 3. Proportion of Lactobacillus species present in PM, PM-fed chickens and control chickens.
The genus Lactobacillus was represented by only 2 species of bacteria in PM, whereas control and PM-fed chickens had a greater number of species that constitute the total population of Lactobacillus. PM-fed chickens had a more diverse Lactobacillus population than control chickens (16 species and 12 species, respectively), and the species abundance as a proportion of the total Lactobacillus population was also very different between the two groups.
Discussion
This is the first study to investigate the effects of pigeon ‘milk’ on intestinal microbiota and gut gene expression. Our results demonstrate that, like mammalian milk, PM modulates the development of both the gut immune system and the gut microbiota. Pigeon ‘lactation’ and mammalian lactation, although produced by very different biological processes (one being a secretive process and the other a cellular exudate), have resulted in similarly functional products. Mammalian milk fulfils the needs of the developing young both nutritionally and immunologically. Here, we have shown that PM also appears to fulfil both these roles, as immune-related genes are significantly enriched in the gut of PM-fed chickens and there are significant differences between the microbiota of PM-fed chickens and control chickens.
A previous study found that pigeons fed a nutritional replacement of PM died or failed to thrive [8], so in order to make a comparison between newly hatched young that were fed PM and those that received a control diet, we used chickens, which are precocial and do not require any parental care. Previous studies have investigated the rate of growth of PM-fed chickens, reporting large increases in growth without any ill effects [32], [33]. Despite the great advances of the past decades in chicken breeding, which have provided massive gains in growth performance, the modern broiler chickens in our study still showed a significant improvement in growth when fed PM. A nutritional replacement of PM had no significant affect on chicken growth (Figure S1). PM-fed chickens had a 12.5% higher body mass than control chickens, but they were not significantly taller or with longer leg span (Table 1). Interestingly, there was an altered body composition, with the proportion of breast muscle to body mass significantly greater (23%) in PM-fed chickens (Table 1) which could suggest that the increased rate of growth is not only attributable to the slightly higher caloric intake of the PM-fed chickens. It could also be influenced by growth hormones such as Pigeon Milk Growth Factor (PMGF) [28] and/or bioactive molecules and bacteria in the PM.
This study has shown that, like mammalian milk, PM clearly influences the composition of the caecal microbiota. PM-fed chickens had a more diverse microbiota than control chickens at the level of phylum, class, order, family and genus (Table 3). Pigeon ‘milk’ could be a source of both probiotics and prebiotics. Three genera of bacteria were present in PM-fed chickens but not controls; Subdoligranulum, Sutterella and Veillonella (Table 3). Of these three genera, Veillonella and Sutterella were also present in PM but not control chickens (Table 4). Only one OTU, closest to the culturable isolate Bacteroides paurosaccharolyticus, was shared between control chickens and PM, but it was present in very low abundance in control chickens (0.06 as compared to 54.22 in PM)(Table 4), suggesting that the apparent absence in PM-fed chickens could simply be a depth of sampling issue.
Species of Veillonella, one of the two genera shared by PM and PM-fed chickens, has been characterised as having inhibitory activity against the enteropathogenic bacterial species Listeria monocytogenes [35], Salmonella Typhimurium [36], and Salmonella Enteritidis [37]. It is to that end that Veillonella is included in a probiotic product designed for poultry [38], which suggests that Veillonella species could be important probiotics in pigeon ‘milk’. All four of the Veillonella species shared by PM and PM-fed chickens have a 16S rRNA sequence divergence of more than 3% from the closest cultured isolate (Table 4), which suggests that the Veillonella species present in PM and PM-fed chickens could be novel species [39]. In addition, the Sutterella species shared by PM and PM-fed chickens is more than 3% divergent from the closest culturable isolate (Table 4), so it is also likely to be a novel species.
The variation in microbiota between PM-fed and control chickens and the relatively modest overlap in shared species between the PM and PM-fed chickens indicates that the PM is likely to be exerting its influence more by prebiotic effects rather than by the direct seeding of new microbiota. The presence of oligosaccharides in pigeon ‘milk’ [40] is indicative of one class of potential prebiotic. Composition of Lactobacillus populations varied greatly between groups, with PM-fed chickens having a more diverse Lactobacillus population than control chickens (Figure 3). This could be due to putative PM prebiotics, as there are many species of Lactobacillus that are amenable to the addition of prebiotics [41], [42]. In addition, there were more bacteria that were unclassified at the phylum level in PM-fed chickens (3.052%) than control chickens (0.354%) that could be potentially novel bacteria, some of which could be important in the functional modulation of the gut by PM.
Changes in gut microbiota can modulate the immune capabilities of the GALT, particularly by modulating IgA B cell development [17]. Consequently, the up-regulation of IgA heavy chain mRNA in the GALT of PM-fed chickens (Figure 1) and the up-regulation of various other genes implicated in immune processes (Table S1, Table S3) suggests that there could be modulation of the PM-fed chicken GALT by the microbiota. Gene ontology processes that were significantly enriched in GALT tissues of PM-fed chickens included the innate immune response, regulation of cytokine production and regulation of B cell activation and proliferation (Table S1), which are all suggestive of an immune effect of PM. Aside from the effect of microbiota, this could also be due to the effects of other as yet unidentified PM components such as cytokines and other bioactive peptides. In a study where chickens were given different bacterial inocula from chicken caeca, there was no up-regulation of any immune pathways or groups in the chicken GALT [43], which, aside from the differences in PM bacteria and chicken caecal bacteria, could suggest that PM modulates GALT development with immunomodulatory components that are in addition to the microbiota. Six ISGs are up-regulated in the ileum of PM-fed chickens, and ten in the caecal tonsil (Table 2). Four of these ISGs are also differentially expressed in breast-fed versus formula-fed infants [44]. In the chicken, these ISGs could have multiple interferon inducers from PM, including hormones. Two of the ISGs up-regulated in PM-fed chickens have been identified as targets of prolactin (interferon regulatory factor 1)[45] and the prolactin receptor (2′-5′-oligoadenylate synthetase)[46] which could suggest that, like mammalian milk [47], [48], PM production is not only induced by prolactin, but prolactin could be delivered to the young through the milk. Interestingly, four of the ISGs up-regulated in the caecal tonsil have antiviral activity (Table 2), which indicates PM may confer antiviral activity, which is again, functionally similar to mammalian milk [49], [50]. It is possible that the up-regulation of some of these immune genes is a response by the chicken to foreign antigens in the PM. However, the increase in body mass and bacterial diversity indicates PM is having a more beneficial effect on the chicken.
PM and mammalian milk both have nutritional and immune modulatory components, and the ability to modulate the microbiota of the gut. This is fascinating from an evolutionary point of view when one considers that mammals and birds evolved these processes independently. To this end, it would be interesting to investigate other bird species that have altricial young, as it may reveal additional ‘lactating’ bird species that were previously thought to be regurgitating seeds or insects to their young. This would allow comparative studies that could elucidate the evolutionary pressures that resulted in birds producing crop ‘milk’. Additionally, this would make for an interesting comparison with the evolutionary history of mammalian lactation.
Conclusions
This study is the first to investigate the effects of pigeon ‘milk’ on the GALT and gut microbiota. Gene expression in the GALT of PM-fed chickens was significantly enriched with immune-related pathways, in particular ISGs, other components of the innate immune response, regulation of cytokine production and regulation of B cell activation and proliferation. The microbiota of PM-fed chickens was significantly more diverse than control chickens, and appears to be effected by prebiotics in pigeon ‘milk’, as well as being directly seeded by bacteria present in PM. Taken together, these results suggest that PM is more functionally similar to mammalian milk than was previously thought. PM and mammalian milk both have nutritional and immune modulatory components, and the ability to modulate the microbiota of the gut. This is fascinating from an evolutionary point of view when one considers that mammals and birds evolved these processes independently.
Methods
Ethics statement
All work using animals was conducted in accordance with the Australian Code of Practise for the Care and Use of Animals for Scientific Purposes (7th edition), and in accordance with institutional animal ethics guidelines (Commonwealth Scientific and Industrial Research Organisation (CSIRO) Australian Animal Health Laboratory (AAHL) Animal Ethics Committee approval numbers 1289,1357 and 1446; and Deakin University Animal Ethics Committee approval numbers AEX56/2008 and AEX57/2008).
Collection of pigeon ‘milk’
Breeding pairs of King pigeons were purchased from Kooyong Squab Producers (Moama, New South Wales, Australia) and housed in temperature controlled cabinets (between 21°C to 24°C) with a 12 hour light cycle (lights on 6 am). They were supplied with nest bowls and materials and had ad libitum access to pigeon mix (pro-vit-min, Ivorsons, Geelong, Australia) and water. Pigeons were allowed to breed, and were culled, along with their squabs, at either the time the squab hatched, or 2 days after the squab hatched. Pigeon ‘milk’ was collected from the crop of the parents and the squabs into sterile 2 mL tubes and frozen at −80°C until use. Samples were thawed at 4°C and pooled before use.
Chicken husbandry
Sixteen newly hatched male Ross308 chickens were purchased from a commercial supplier (Bartter Enterprises, Bannockburn, Victoria, Australia). They were randomly assigned into 2 groups, wing-tagged for identification and weighed. The chicks were housed in separate cages within the same cabinet, to prevent access to the other group’s feed. Heat lamps were provided at one side of each cage to establish a temperature gradient. To keep the pigeon ‘milk’ fresh, the chicks were fed three times a day by mixing the pigeon ‘milk’ into a pre-weighed amount of antibiotic-free chicken feed (Country Heritage Feeds OPO05, Queensland, Australia), which was placed on a tray in the cage. Before each feed the amount of feed consumed by each group was calculated. Each chicken received on average 5 grams of pigeon ‘milk’ per day for 7 days.
A subsequent trial investigating the effect of the protein and fat components of pigeon ‘milk’ was set up as described above, where the replacement pigeon ‘milk’ consisted of peptone proteose (Becton Dickson, Australia) equivalent to 45% and pig lard (Fonterra, Australia) equivalent to 11%. These were chosen as they had the most similar amino acid and fatty acid compositions to pigeon ‘milk’.
Chicken measurements and sample collection
Body mass of each chicken was determined on day 4. The chickens were culled after 7 days and their final weight was recorded. The following measurements were taken: from the top of the cranium to the cloaca (height), from the end of the furthermost wing digit on the left to the furthermost digit on the right (wing span), and from the patella to the posterior end of the tarsometatarsus (leg span). The breast muscle was removed from the breast bone with a scalpel and weighed. The caecal tonsils and ileum (adjacent to the caecal tonsils) were removed and collected in RNALater (Invitrogen) and frozen at −20°C until RNA extraction. The contents of the cecum was collected in sterile 5 mL containers and frozen at −20°C until DNA extraction.
Statistical analysis of chicken body measurements
A statistical comparison of control and PM-fed chicken body measurements was performed with an unpaired t-test. Average percent body mass gain of PM-fed and PM replacement-fed chickens was calculated by normalising the weight gain of each experimental group chicken to the median weight gain of the corresponding control group chickens. A Kruskal-Wallis test with Dunns post-hoc test was used to identify any statistically significant difference in body mass gain between control, PM-fed and PM replacement-fed chickens.
RNA isolation, labelling and microarray hybridisation
RNA was extracted from the caecal tonsil and ileum tissue of 6 control and 6 PM-fed chickens (mean weights) using a Cartagen RNA extraction kit (Inbio, Eltham, Australia) according to the manufacturer's instructions. cDNA was synthesised from 5 µg RNA using SuperScript III (Invitrogen) with oligodt primer. This was purified with a Qiagen PCR Purification Kit and labelled with Cy3 using a Roche One-Color DNA Labelling Kit according to the manufacturer's instructions. The labelled microarray probes were resuspended with a sample tracking control and hybridisation buffer and loaded on 12-plex 135 k custom chicken microarrays (NimbleGen design #10309). The array contains 65,850 probes printed in duplicate, of which there are 32,357 probes with unique UniGene IDs. Most unique genes have 2 or more probes. Information on the custom array is available from ArrayExpress using the accession number A-MEXP-2133. These were hybridised for 20 hours in a NimbleGen Hybridisation Station (Roche) at 42°C and then washed using the NimbleGen wash buffer kit (Roche) according to the manufacturer's instructions. Each subarray was scanned at 2 µm on autogain with a NimbleGen MS200 microarray scanner (Roche).
Microarray quality control and statistical analysis
Sample tracking controls and control spots were used to autoalign a grid over each subarray using NimbleGen MS200 software (Roche), and Robust Multichip Average (RMA) analysis [51] was used to background correct and normalise the spot signal intensity. The datasets, along with probe annotation information, were exported into GeneSpring (Agilent) and differentially expressed genes were identified using Student's t-test, assuming unequal variances, with a false discovery rate of p = 0.05. Control ileum was compared to PM-fed ileum, and control caecal tonsil was compared to PM-fed caecal tonsil. All results have been deposited into the ArrayExpress database with accession number E-MTAB-1127.
IgA expression analysis
The relative expression level of the IgA heavy chain (probe CLIGG_34917) was calculated from the RMA normalised spot signal intensity by dividing each probe by the total probe intensity and multiplying by 10 million. The relative signal intensity in the ileum and caecal tonsil for PM-fed chickens and control chickens was subjected to an unpaired t-test, and the mean and standard error of the mean was calculated and graphed using GraphPad5.
Gene functional analysis
The DAVID functional annotation tool [52] was used to identify pathways and biological functions up-regulated in the caecal tonsil and ileum in association with pigeon ‘milk’. An ease score of 0.05 was used to determine enriched Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways and Gene Ontology (GO) FAT biological functions.
Interferon-stimulated genes were functionally annotated using the Interferon Stimulated Gene Database [53] and/or a literature search.
Caecal DNA extraction and 16S amplification
Total DNA was extracted from caecal contents as per the method of Yu and Morrison [54]. DNA quality and quantity was measured on a NanoDrop ND-1000 spectrophotometer. The V1-V3 region of bacterial 16S rRNA was amplified from caecal DNA following the method of Stanley et al using the primers and conditions previously detailed [55].
High throughput 16S amplicon sequencing and data pre-processing
The amplified 16S rRNA gene samples from each bird were pooled using approximately equal amounts of each PCR product. The pooled sample was sequenced using the Roche/454 FLX Genome Sequencer and Titanium chemistry according to the manufacturer's instructions. Sff files were split into fasta and qual files using PyroBayes [56], and data was analysed with Qiime v1.3.0 software [57], except for OTU picking, denoising and chimera detection which was done using Otupipe [58]. Two samples (C1 and C3) were removed from analysis due to low sequence numbers per sample. Additional filtering of samples was performed to remove OTUs present in less than 3 samples or with less than 5 sequences. The default Qiime analysis parameters were used except as follows: sequence length 300–600 bases, no ambiguous sequences allowed, maximum of 6 homopolymers and classification by RDP. OTU sequences have been deposited in the European Molecular Biology Laboratory EMBL-Bank with accession numbers HE814242-HE814562.
Network analysis of OTUs
Filtered, multiple rarefied OTU abundance data was used to generate a network of shared OTUs in Cytoscape v2.8.
Analysis of bacteria that are differentially abundant in the cecum of PM-fed chickens and control chickens
Raw filtered OTU reads for each control chicken and PM-fed chicken sample were imported into Metastats [59] for statistical analysis, using 1000 permutations, to identify OTUs that were differentially abundant between control chickens and PM-fed chickens. OTUs were considered differentially abundant if the p value was less than 0.05.
Identification of shared OTUs in PM, ctrl and PM-fed chickens
OTUs were called as present if the filtered, multiple rarefied count was greater than zero. For shared OTUs, the representative OTU sequence was uploaded to EZTaxon [60] and the closest cultured isolate was identified.
Supporting Information
Body mass gain of PM and PM replacement-fed chickens. PM-fed chickens (n = 8) gained significantly more body mass than control chickens over 7 days. There was no difference between body mass gain of control chickens (n = 16) and PM-replacement-fed chickens (n = 8).
(JPG)
UniFrac analysis of bacteria present in PM, PM-fed and control chickens. Principal Coordinate Analysis plot based on unweighted UniFrac. Rarefied samples of PM are represented by yellow circles, PM-fed chickens by red triangles and control chickens by green squares.
(JPG)
Biological processes up-regulated in the gut of PM-fed chickens. Gene ontology biological processes that were identified as enriched amongst genes up-regulated in ileum or caecal tonsil of PM-fed chickens (n = 6).
(DOCX)
Biological processes down-regulated in the gut of PM-fed chickens. Gene ontology biological processes that were identified as enriched amongst genes down-regulated in ileum or caecal tonsil of PM-fed chickens (n = 6).
(DOCX)
Enriched KEGG pathways in the gut of PM-fed chickens. KEGG pathways that were identified as enriched amongst differentially expressed genes in ileum or caecal tonsil of PM-fed chickens (n = 6).
(DOCX)
Acknowledgments
The authors would like to thank Kate Goossens and Sarah Fardy for their assistance with sample collection, and Nic Kieselbach and Adam Stein for pigeon and chicken husbandry. Thank you to Susanne Wilson for animal husbandry and assistance with chicken feeding, sample measurement and collection. Thank you to Leona McLaren from Kooyong Squab for supplying the pigeons.
Funding Statement
MG was supported by Deakin University and CSIRO PhD scholarships. This study was funded by CSIRO. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Studer-Thiersch A (1967) Beitrage zur brutbiologie der flamingos (Gattung phoenicopterus). Der Zoologische garten 34: 159–229. [Google Scholar]
- 2. Prevost J, Vilter V (1962) Histologie de la secretion oesophagienne du manchot empereur. Proceedings of the 13th international ornithological congress 2: 1085–1094. [Google Scholar]
- 3. Riddle O, Bates RW, Dykshorn S (1933) The preparation, identification and and assay of prolactin - a hormone of the anterior pituitary. Am J Physiol 105: 191–216. [Google Scholar]
- 4. Gillespie MJ, Haring VR, McColl KA, Monaghan P, Donald JA, et al. (2011) Histological and global gene expression analysis of the 'lactating' pigeon crop. BMC Genomics 12: 452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Shetty S, Sridhar KR, Shenoy KB, Hegde SN (1990) Observations on bacteria associated with pigeon crop. Folia Microbiol (Praha) 35: 240–244. [DOI] [PubMed] [Google Scholar]
- 6. Davies WL (1939) The composition of the crop milk of pigeons. Biochem J 33: 898–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Goudswaard J, van der Donk JA, van der Gaag I, Noordzij A (1979) Peculiar IgA transfer in the pigeon from mother to squab. Dev Comp Immunol 3: 307–319. [DOI] [PubMed] [Google Scholar]
- 8. Guareschi C (1936) Necessita di fattori alimentari materni per l'accrescimento del giovanissimi colombi. Boll Soc Ital Biol Sper 11: 411–412. [Google Scholar]
- 9. Stelwagen K, Carpenter E, Haigh B, Hodgkinson A, Wheeler TT (2009) Immune components of bovine colostrum and milk. J Anim Sci 87: 3–9. [DOI] [PubMed] [Google Scholar]
- 10. Wagstrom EA, Yoon KJ, Zimmerman JJ (2000) Immune components in porcine mammary secretions. Viral Immunol 13: 383–397. [DOI] [PubMed] [Google Scholar]
- 11. Chichlowski M, German JB, Lebrilla CB, Mills DA (2011) The influence of milk oligosaccharides on microbiota of infants: opportunities for formulas. Annu Rev Food Sci Technol 2: 331–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zuppa AA, Alighieri G, Scorrano A (2010) Chapter 27 - Prebiotics and Probiotics in Infant Nutrition. In: Ronald Ross W, Victor RP, editors. Bioactive Foods in Promoting Health. Boston: Academic Press. pp. 441–477.
- 13. Poroyko V, White JR, Wang M, Donovan S, Alverdy J, et al. (2010) Gut microbial gene expression in mother-fed and formula-fed piglets. PLoS One 5: e12459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cebra JJ (1999) Influences of microbiota on intestinal immune system development. Am J Clin Nutr 69: 1046S–1051S. [DOI] [PubMed] [Google Scholar]
- 15. Musso G, Gambino R, Cassader M (2011) Interactions between gut microbiota and host metabolism predisposing to obesity and diabetes. Annu Rev Med 62: 361–380. [DOI] [PubMed] [Google Scholar]
- 16. Brandtzaeg P, Halstensen TS, Kett K, Krajci P, Kvale D, et al. (1989) Immunobiology and immunopathology of human gut mucosa: humoral immunity and intraepithelial lymphocytes. Gastroenterology 97: 1562–1584. [DOI] [PubMed] [Google Scholar]
- 17.Honjo T, Melchers F (2006) Gut-associated lymphoid tissues. Berlin: Springer-Verlag. 284 p.
- 18. Field CJ (2005) The immunological components of human milk and their effect on immune development in infants. J Nutr 135: 1–4. [DOI] [PubMed] [Google Scholar]
- 19. West LJ (2002) Defining critical windows in the development of the human immune system. Hum Exp Toxicol 21: 499–505. [DOI] [PubMed] [Google Scholar]
- 20. Garofalo R (2010) Cytokines in human milk. J Pediatr 156: S36–40. [DOI] [PubMed] [Google Scholar]
- 21. Michie CA, Tantscher E, Schall T, Rot A (1998) Physiological secretion of chemokines in human breast milk. Eur Cytokine Netw 9: 123–129. [PubMed] [Google Scholar]
- 22. Goldman AS (1993) The immune system of human milk: antimicrobial, antiinflammatory and immunomodulating properties. Pediatr Infect Dis J 12: 664–671. [DOI] [PubMed] [Google Scholar]
- 23. Malo C, Menard D (1982) Influence of epidermal growth factor on the development of suckling mouse intestinal mucosa. Gastroenterology 83: 28–35. [PubMed] [Google Scholar]
- 24. Opleta-Madsen K, Hardin J, Gall DG (1991) Epidermal growth factor upregulates intestinal electrolyte and nutrient transport. Am J Physiol 260: G807–814. [DOI] [PubMed] [Google Scholar]
- 25. Cooper CA, Brundige DR, Reh WA, Maga EA, Murray JD (2011) Lysozyme transgenic goats' milk positively impacts intestinal cytokine expression and morphology. Transgenic Res 20: 1235–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jenssen H, Hancock RE (2009) Antimicrobial properties of lactoferrin. Biochimie 91: 19–29. [DOI] [PubMed] [Google Scholar]
- 27. Benkerroum N (2010) Antimicrobial peptides generated from milk proteins: a survey and prospects for application in the food industry. A review. International Journal of Dairy Technology 63: 320–338. [Google Scholar]
- 28. Shetty S, Hegde SN (1993) Pigeon milk: a new source of growth factor. Experientia 49: 925–928. [DOI] [PubMed] [Google Scholar]
- 29. Shetty S, Hegde SN, Bharathi L (1992) Purification of a growth factor from pigeon milk. Biochim Biophys Acta 1117: 193–198. [DOI] [PubMed] [Google Scholar]
- 30. Frelinger JA (1971) Maternally derived transferrin in pigeon squabs. Science 171: 1260–1261. [DOI] [PubMed] [Google Scholar]
- 31. Wally J, Buchanan SK (2007) A structural comparison of human serum transferrin and human lactoferrin. Biometals 20: 249–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pace DM, Landolt PA, Mussehl FE (1952) The effect of pigeon crop-milk on growth in chickens. Growth 16: 279–285. [PubMed] [Google Scholar]
- 33. Hegde SN (1973) Composition of pigeon milk and its effect on growth in chicks. Indian J Exp Biol 11: 238–239. [PubMed] [Google Scholar]
- 34. Brisbin JT, Gong J, Sharif S (2008) Interactions between commensal bacteria and the gut-associated immune system of the chicken. Anim Health Res Rev 9: 101–110. [DOI] [PubMed] [Google Scholar]
- 35. Hinton A Jr, Hume ME (1997) Research note: in vitro inhibition of Listeria monocytogenes growth by veillonellae cultures grown on tartrate media. J Appl Microbiol 82: 780–782. [DOI] [PubMed] [Google Scholar]
- 36. Hinton A Jr, Hume ME (1996) Inhibition of Salmonella typhimurium by the products of tartrate metabolism by a Veillonella species. J Appl Bacteriol 81: 188–190. [DOI] [PubMed] [Google Scholar]
- 37. Hinton A Jr, Hume ME (1995) Synergism of lactate and succinate as metabolites utilized by Veillonella to inhibit the growth of Salmonella typhimurium and Salmonella enteritidis in vitro. Avian Dis 39: 309–316. [PubMed] [Google Scholar]
- 38.LeLoach J.R CDE, Hinton jr A. (1992) Probiotic for control of Salmonella. United States of America.
- 39. Stackebrandt E, Goebel BM (1994) A Place for DNA-DNA Reassociation and 16s Ribosomal-Rna Sequence-Analysis in the Present Species Definition in Bacteriology. International Journal of Systematic Bacteriology 44: 846–849. [Google Scholar]
- 40. Shetty S, Salimath PV, Hegde SN (1994) Carbohydrates of pigeon milk and their changes in the first week of secretion. Arch Int Physiol Biochim Biophys 102: 277–280. [DOI] [PubMed] [Google Scholar]
- 41. Hernandez-Hernandez O, Muthaiyan A, Moreno FJ, Montilla A, Sanz ML, et al. (2012) Effect of prebiotic carbohydrates on the growth and tolerance of Lactobacillus. Food Microbiol 30: 355–361. [DOI] [PubMed] [Google Scholar]
- 42. Nagpal R, Kaur A (2011) Synbiotic effect of various prebiotics on in vitro activities of probiotic lactobacilli. Ecol Food Nutr 50: 63–68. [DOI] [PubMed] [Google Scholar]
- 43. Yin Y, Lei F, Zhu L, Li S, Wu Z, et al. (2010) Exposure of different bacterial inocula to newborn chicken affects gut microbiota development and ileum gene expression. ISME J 4: 367–376. [DOI] [PubMed] [Google Scholar]
- 44. Schwartz S, Friedberg I, Ivanov IV, Davidson LA, Goldsby JS, et al. (2012) A metagenomic study of diet-dependent interaction between gut microbiota and host in infants reveals differences in immune response. Genome Biol 13: r32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yu-Lee L (2001) Stimulation of interferon regulatory factor-1 by prolactin. Lupus 10: 691–699. [DOI] [PubMed] [Google Scholar]
- 46. McAveney KM, Book ML, Ling P, Chebath J, Yu-Lee L (2000) Association of 2′,5′-oligoadenylate synthetase with the prolactin (PRL) receptor: alteration in PRL-inducible stat1 (signal transducer and activator of transcription 1) signaling to the IRF-1 (interferon-regulatory factor 1) promoter. Mol Endocrinol 14: 295–306. [DOI] [PubMed] [Google Scholar]
- 47. Cregan MD, Mitoulas LR, Hartmann PE (2002) Milk prolactin, feed volume and duration between feeds in women breastfeeding their full-term infants over a 24 h period. Exp Physiol 87: 207–214. [DOI] [PubMed] [Google Scholar]
- 48. Lkhider M, Delpal S, Bousquet MO (1996) Rat prolactin in serum, milk, and mammary tissue: characterization and intracellular localization. Endocrinology 137: 4969–4979. [DOI] [PubMed] [Google Scholar]
- 49. van Hooijdonk AC, Kussendrager KD, Steijns JM (2000) In vivo antimicrobial and antiviral activity of components in bovine milk and colostrum involved in non-specific defence. Br J Nutr 84 (Suppl 1) S127–134. [DOI] [PubMed] [Google Scholar]
- 50. Matthews TH, Nair CD, Lawrence MK, Tyrrell DA (1976) Antiviral activity in milk of possible clinical importance. Lancet 2: 1387–1389. [DOI] [PubMed] [Google Scholar]
- 51. Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, et al. (2003) Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4: 249–264. [DOI] [PubMed] [Google Scholar]
- 52. Huang da W, Sherman BT, Lempicki RA (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57. [DOI] [PubMed] [Google Scholar]
- 53. de Veer MJ, Holko M, Frevel M, Walker E, Der S, et al. (2001) Functional classification of interferon–stimulated genes identified using microarrays. J Leukoc Biol 69: 912–920. [PubMed] [Google Scholar]
- 54. Yu Z, Morrison M (2004) Improved extraction of PCR-quality community DNA from digesta and fecal samples. Biotechniques 36: 808–812. [DOI] [PubMed] [Google Scholar]
- 55.Stanley D, Keyburn AL, Denman SE, Moore RJ (2012) Changes in the caecal microflora of chickens following Clostridium perfringens challenge to induce necrotic enteritis. Vet Microbiol. [DOI] [PubMed]
- 56. Quinlan AR, Stewart DA, Stromberg MP, Marth GT (2008) Pyrobayes: an improved base caller for SNP discovery in pyrosequences. Nat Methods 5: 179–181. [DOI] [PubMed] [Google Scholar]
- 57. Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, et al. (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7: 335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R (2011) UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27: 2194–2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. White JR, Nagarajan N, Pop M (2009) Statistical methods for detecting differentially abundant features in clinical metagenomic samples. PLoS Comput Biol 5: e1000352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Chun J, Lee JH, Jung Y, Kim M, Kim S, et al. (2007) EzTaxon: a web-based tool for the identification of prokaryotes based on 16S ribosomal RNA gene sequences. Int J Syst Evol Microbiol 57: 2259–2261. [DOI] [PubMed] [Google Scholar]
- 61. Kang DC, Gopalkrishnan RV, Lin L, Randolph A, Valerie K, et al. (2004) Expression analysis and genomic characterization of human melanoma differentiation associated gene-5, mda-5: a novel type I interferon-responsive apoptosis-inducing gene. Oncogene 23: 1789–1800. [DOI] [PubMed] [Google Scholar]
- 62. Berghall H, Siren J, Sarkar D, Julkunen I, Fisher PB, et al. (2006) The interferon-inducible RNA helicase, mda-5, is involved in measles virus-induced expression of antiviral cytokines. Microbes Infect 8: 2138–2144. [DOI] [PubMed] [Google Scholar]
- 63. Siren J, Imaizumi T, Sarkar D, Pietila T, Noah DL, et al. (2006) Retinoic acid inducible gene-I and mda-5 are involved in influenza A virus-induced expression of antiviral cytokines. Microbes Infect 8: 2013–2020. [DOI] [PubMed] [Google Scholar]
- 64. Gao G, Guo X, Goff SP (2002) Inhibition of retroviral RNA production by ZAP, a CCCH-type zinc finger protein. Science 297: 1703–1706. [DOI] [PubMed] [Google Scholar]
- 65. Lu J, Pan Q, Rong L, He W, Liu SL, et al. (2011) The IFITM proteins inhibit HIV-1 infection. J Virol 85: 2126–2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Brass AL, Huang IC, Benita Y, John SP, Krishnan MN, et al. (2009) The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell 139: 1243–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Body mass gain of PM and PM replacement-fed chickens. PM-fed chickens (n = 8) gained significantly more body mass than control chickens over 7 days. There was no difference between body mass gain of control chickens (n = 16) and PM-replacement-fed chickens (n = 8).
(JPG)
UniFrac analysis of bacteria present in PM, PM-fed and control chickens. Principal Coordinate Analysis plot based on unweighted UniFrac. Rarefied samples of PM are represented by yellow circles, PM-fed chickens by red triangles and control chickens by green squares.
(JPG)
Biological processes up-regulated in the gut of PM-fed chickens. Gene ontology biological processes that were identified as enriched amongst genes up-regulated in ileum or caecal tonsil of PM-fed chickens (n = 6).
(DOCX)
Biological processes down-regulated in the gut of PM-fed chickens. Gene ontology biological processes that were identified as enriched amongst genes down-regulated in ileum or caecal tonsil of PM-fed chickens (n = 6).
(DOCX)
Enriched KEGG pathways in the gut of PM-fed chickens. KEGG pathways that were identified as enriched amongst differentially expressed genes in ileum or caecal tonsil of PM-fed chickens (n = 6).
(DOCX)