Abstract
Antennal olfaction is extremely important for insect survival, mediating key behaviors such as host preference, mate choice, and oviposition site selection. Multiple antennal proteins are involved in olfactory signal transduction pathways. Of these, odorant receptors (ORs) and ionotropic receptors (IRs) confer specificity on olfactory sensory neuron responses. In this study, we identified the olfactory gene repertoire of the economically important agricultural pest moth, Helicoverpa armigera, by assembling the adult male and female antennal transcriptomes. Within the male and female antennal transcriptomes we identified a total of 47 OR candidate genes containing 6 pheromone receptor candidates. Additionally, 12 IR genes as well as 26 odorant-binding proteins and 12 chemosensory proteins were annotated. Our results allow a systematic functional analysis across much of conventional ORs repertoire and newly reported IRs mediating the key olfaction-mediated behaviors of H. armigera.
Introduction
Olfaction, the sense of smell, plays a predominant role in mediating insect behavior including food source identification, oviposition site selection, mate choice, kin recognition and predator avoidance. Of insect olfactory events, sexual communication in moths is an excellent model system for understanding the mechanism of animal sensory perception at the molecular level because of the complexity, specificity and extreme sensitivity of males to specific pheromone molecules emitted from conspecific females [1], [2].
The surface of insect antennae is covered with several different types of sensilla that are small sensory hair structures in which olfactory receptor neurons extend dendrites into the antennal lymph where peripheral olfactory signal transduction events occur. Previous studies have shown diverse olfactory genes including at least odorant-binding proteins (OBPs), chemosensory proteins (CSPs), Sensory neuron membrane proteins (SNMPs), odorant-degrading enzymes (ODEs), ionotropic receptors (IRs) and odorant receptors (ORs) involved in different steps in signal transduction pathway [3], [4], [5]. All of these, ORs play a central role in chemosensory signal transduction processes that occur in olfactory receptor neurons. ORs located on the surface of olfactory sensory neuronal dendrites in the antennae possess seven transmembrane domains (TMDs). In insects, it is generally thought that odor recognition is mediated by a single set of odor receptor heterodimers composed of a conserved, nonconventional ORCO protein acting as an ion channel and a variable, conventional OR that apparently mediates odorant-binding specificity [6], [7], [8], [9], [10], [11]. In addition, a novel family of candidate chemosensory ionotropic receptors (IRs) involved in odorant recognition was recently characterized by a genome-based bioinformatics screen in Drosophila melanogaster [12]. IRs are not closely related to ionotropic glutamate receptors in the phylogenetic analysis. However, they possess an obviously similar modular organization to ionotropic glutamate receptors [12], [13]. Like ORs, IRs are extremely divergent and expresssed in sensory dendrites. Misexpression of several Drosophila IRs suggested that they might be tuned to a small odor panel such as small amine-like volatile compounds [12], [14], [15].
In addition, the aforementioned multiple proteins may interact with ORs/IRs and are required for olfactory signal transduction. The soluble OBPs are thought to facilitate the transport of odorant molecules through the sensillum lymph and are sometimes thought to be directly involved in ligand binding [16], [17], [18]. The CSPs are another class of soluble proteins in the sensillum lymph with abundant expression, however their function in olfactory transduction remains largely unknown [16], [19]. SNMPs locate in the dendritic membrane of peripherally olfactory receptor neurons just adjoining ORs and are presumed to trigger ligand delivery to the receptor [20], [21], [22]. Within the olfactory sensilla, ODEs are reputedly involved in the signal inactivation step serving to rapidly remove stimulatory odorant molecules [23], [24].
Genome sequencing and molecular studies have together characterized the complete OR repertoires and other olfactory genes in several insect species such as D. melanogaster, Anopheles gambiae, Bombyx mori and others, promoting understanding of olfactory signal pathways in these insects. However, a systematic analysis of the olfactory genes especially ORs/IRs is still largely absent in from most insect species, including important crop pest insects where a genome sequence is not yet unavailable. Traditional homology-based cloning strategies could identify some conserved genes such as ORCO and OBPs, but would not readily identify divergent genes, especially the divergent ORs and IRs families. Therefore, recent studies of the molecular mechanisms of olfactory signal transduction in moths have largely been limited to B. mori due to the availability of genome sequence. In other Lepidoptera species, only a few ORs and PRs have been identified [25], [26], [27], [28], [29], [30], [31]. Expressed Sequence Tag (EST) sequencing strategies have been successfully used to identify olfactory genes in Lepidoptera without genomic data such as Manduca sexta [32], Epiphyas postvittana [33] and Spodoptera littoralis [34], [35]. But most identified genes are OBPs; relatively few are divergent genes such as ORs and IRs. The identification of moth ORs and IRs is still a challenging exercise. Recently, RNA-seq approaches have been used to identify olfactory genes in species where a genome sequence is not yet available. To date, next-generation sequencing of antennal transcriptome has been successfully used to identify substantial numbers of candidate ORs and IRs in M. sexta [36], Cydia pomonella [37], and Bactrocera dorsalis [38].
Moths are a diverse group of insects that include many economically important crop pests. The subfamily heliothinae contains approximately 365 species of noctuid moths including the world’s most injurious crop pests such as the Old World bollworm and relatives, and the New World tobacco budworm, which cause huge economic loss annually [39], [40], [41]. Better understanding on the molecular mechanisms of olfactory recognition in these moths could ultimately lead to identify new targets for developing environment-friendly control strategies.
To explore the olfaction-related transcriptome of the worldwide agricultural pest, H. armigera, we conducted 454 pyrosequencing of RNA extracted from adult male and female antennae. Our goals were to identify olfaction-related genes and olfactory signal transduction mechanisms in H. armigera antennae. Here we report the identification of 47 candidate OR genes and 12 IR genes as well as 26 OBP genes and 12 CSP genes in the male and female antennal transcriptomes.
Methods
Insect Rearing
Helicoverpa armigera used in all experiments were obtained from a colony maintained at the Institute of Plant Protection, Chinese Academy of Agricultural Sciences, Beijing, China. Larvae were reared on an artificial diet at 27±1°C under a photoperiod of 14∶10 (L: D). Male and female pupae were placed in separate glass tubes. Adults were fed 10% honey solution. Antennae of female or male individuals were dissected 1–3 days after eclosion and immediately frozen in liquid nitrogen, and stored at –70°C until extraction.
Extraction of Total RNA
Frozen antennae were transferred to a liquid nitrogen cooled mortar and ground with a pestle. The homogenate was covered with 1 mL of TriZol reagent (Invitrogen, Carlsbad, CA, USA) and total RNA extractions were performed following the manufacturer’s instructions. Total RNA was dissolved in H2O and RNA integrity was verified by gel electrophoresis. RNA quantity was determined on a Nanodrop ND-1000 spectrophotometer (NanoDrop products, Wilmington, DE, USA).
Sequencing
PolyA+mRNA was separated from 10 µg of total RNA extracted from approximately 300 antennae of 1–3 day old adult male or female moths using the PolyA+ Ttract mRNA Isolation System (Promega, Madison, WI, USA) and further purified by using the RNeasy MinElute Clean up Kit (Qiagen, Hilden, Germany) following the manufacturer’s protocol. mRNA integrity and quantity were verified on an Agilent 2100 Bioanalyzer. mRNA was fragmented in the RNA fragment reagent for 1 min according to the NEBNext® Magnesium RNA Fragmentation Module Protocol (NEB, Ipswich, MA, USA). After recovered by RNeasy MinElute Clean up Kit, the fragmented mRNA larger than 250 bp was used to synthesize double-stranded–cDNA (ds-cDNA). The first strand cDNA was synthesized using random primers and MMLV reverse transcriptase (Promega, Madison, WI, USA). The second strand was then synthesized using E. coli DNA Polymerase I and RNase H. The resulting ds-cDNAs were used as a template for NextGen sequencing on a Roche 454 FLX by using standard chemistry according to the manufacturer's instructions (Roche, Branford, CT, USA). One full picotiterplate (PTP) with two regions was used for sequencing the male and female samples. 1/2 run data of each sample was generated separately for next analysis.
Unigene Generation
The raw 454 sequences in SFF files were extracted using the Python script sff_extract.py developed by COMAV (http://bioinf.comav.upv.es). All the raw sequences were then processed to remove low quality and adaptor sequences using programs TAGDUST [42], LUCY [43] and SeqClean [44]. The resulting sequences were then screened against the NCBI UniVec database and bacterial genome sequences to remove possible contaminants. Sequences shorter than 50 bp were discarded. The cleaned 454 read sequences from both males and females were assembled into a single assembly by Newbler version 2.5.3 using default parameters under the cDNA option (Roche, Branford, CT, USA).After assembly, the clean reads of both male and females samples were mapped to the assembled unigenes using GS Reference Mapper (Roche, Branford, CT, USA). These reads in male and female samples mapped to each unigene were derived and counted separately. If all the reads mapped certain unigene were from male or female samples, this unigene was determined as male or female special.
Gene Identification and Functional Annotation
The unigenes were searched against the NCBI non-redundant protein database on a local server using the National Center for Biotechnology Information (NCBI) BLASTALL program [45]. GO Annotation was performed by using Blast2GO (GO association done by a BLASTX against the NCBI NR database) [46], [47]. The ORFs of the unigenes were predicted by using ORF finder (http://www.ncbi.nlm.nih.gov/gorf/gorf.html). The signal peptide of the protein sequences were predicted using SignalP 4.0 [48]. The TMDs of annotated genes were predicted using TMHMM Server Version2.0 (http://www.cbs.dtu.dk/services/TMHMM). H. armigera antennal unigenes were searched with B. mori ORs [49], OBPs [50] and IRs [15] as queries using TBLASTN in the free software BioEdit program. Read numbers of each unigene in male and female samples were derived and counted separately.
Phylogenetic Analyses
The phylogenetic reconstruction implemented for the analysis of OR, IR, OBP and CSP was performed based on the amino sequences of the candidate olfaction genes and the collected data sets. The OR data set contained OR sequences identified in Lepidoptera (12 from H. armigera, 21 from H. virescens and 64 from B. mori) [29], [30], [49], [51], [52]. The IR data set contained 12, 18 and 66 IR sequences from S. littoralis, B. mori and D. melanogaster, respectively [15], [35]. The OBP data set contained 15 sequences from H. armigera, 17 sequences from H. virescens and 35 sequences from B. mori [50], [53]. The CSP data set contained the 7 sequences from H. armigera [53], 9 sequences from H. virescens [54], two sequences from Spodoptera exigua and the 16 sequences from B. mori [19]. The protein name and accession number of the genes used for phylogenetic tree building are listed in supplementary material S1. Amino acid sequences were aligned using ClustalW2 [55]. Unrooted trees were constructed by the neighbor-joining method, with Poisson correction of distances, as implemented in MEGA5 software [56]. Node support was assessed using a bootstrap procedure base on 1000 replicates.
Expression Analysis of the Candidate Receptors by Semi-quantitative Reverse Transcription PCR
To illustrate and compare the expression of candidate receptors in male and female antennae, semi-quantitative reverse transcription PCR was performed using cDNAs prepared from male antennae, female antennae and legs (male and female mixture). Legs were used as a control to verify that the candidate receptors were antennae enriched. Total RNAs were extracted as described above. Prior to cDNA synthesis, RNA were treated with DNase I (Fermentas, Vilnius, Lithuania) to remove trace amounts of genomic DNA. The cDNA was synthesized by First Strand cDNA Synthesis Kit (Fermentas, Vilnius, Lithuania) and was used as a template in PCR reactions with gene-specific primers. An actin gene fragment was used as control. Primers were designed using the Primer Premier 5 software (PREMIER Biosoft International). The primer sequences are available in supplementary material S2. PCR was performed with Veriti Thermal Cycler (Applied Biosystems, Carlsbad, CA, USA) under the following conditions: 94°C for 2 min, 33 cycles of 94°C for 30 s, 55–60°C for 30 s; 72°C for 30 s, and 72°C for 8 min. The cycle number was reduced to 26 and 29 for Actin and OR2 amplifications because of their high expression level. The experiment was repeated three times using three independently isolated RNA samples. PCR amplification products were run on a 2% agarose gel and verified by DNA sequencing.
Results
Sequencing and Unigene Assembly
Using GS/FLX 454 pyrosequencing technology, a total of 753,643 raw reads were obtained from the male sample and 518,746 raw reads from female sample. After removing low quality, adaptor, and contaminating sequence reads, male and female antennae yielded 731,001 (average read length 522 bp) and 463,908 (average read length 431 bp) clean reads, respectively. The total bases of sequence data were approximately 382 million and 200 million from male and female samples, respectively. All clean reads from male and female samples were combined into an assembly that generated 37,920 unigenes larger than 100 bp with 991 bp average length. Of these, 8,706 (23.0%) unigenes were from the male antennae and 3,698 (9.8%) unigenes were from female antennae. A flow chart of sequencing and unigene assembly are shown in supplementary material S3. The gene length, ORF length, read number in male or female samples of each unigene were integrated in supplement material S4.
Gene Identification and Functional Annotation
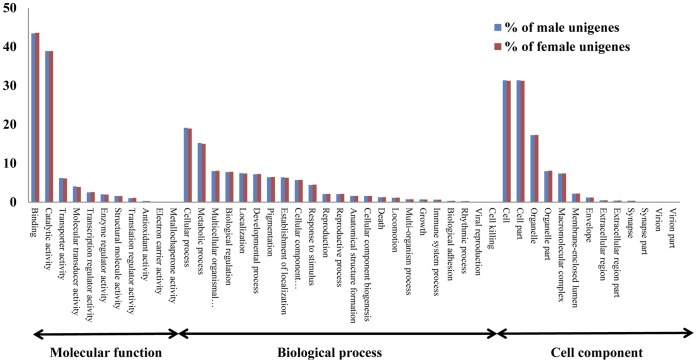
The gene functional annotation was first performed by GO annotation using Blast2GO. Figure 1 illustrates the distribution of the unigene set in GO terms. Among the 37,920 unigenes, 11,233 (29.6%) corresponded to at least one GO term (Figure 1), 9,164 were assigned to a molecular function (24.2%), 8,329 to a biological processes (22%), and 6,588 to a cellular component (17.4%). There was no difference between the GO terms of male and female sets. In the molecular function category, binding and catalytic activities were the most abundant and enriched GOs terms in both male and female sets. In the biological process terms, cellular and metabolic processes were the most represented. In the cellular component terms, cell, cell part and organelle were the most abundant (Figure 1). GO annotations of the unigenes are presented in supplementary material S4.
Figure 1. Distribution and comparison of male and female H. armigera unigenes annotated at GO level 2.
The Y-axis shows the percentage of the sequences. The X-axis shows three areas of annotation, and in each area the sequences are further divided into subgroups at GO level 2.
The unigenes were then searched against the NCBI non-redundant nucleotide and protein database using BLASTN and BLASTX. Of the 37,920 unigenes, 24,675 (65.1%) showed similarity to known proteins supplementary material S1). The ORs, IRs, OBPs and CSPs were annotated according to the BLAST result. The BLASTN and BLASTX best hit result is listed in supplementary material S4.
Identification of Candidate Chemosensory Receptors
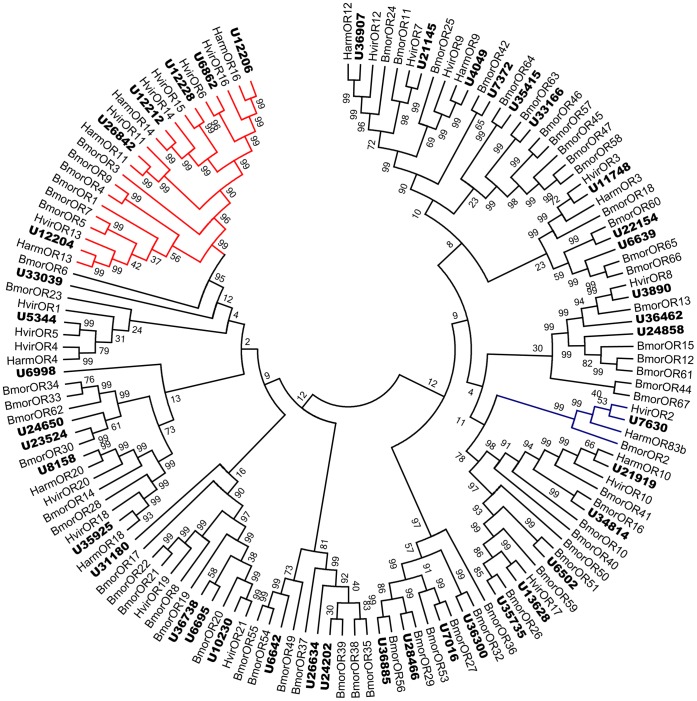
All the contigs were searched by BLASTX and further by TBLASTN using 63 and 21 known ORs from B. mori and H. virescens, respectively, leading to identification of 47 different contigs that were putative OR genes. All 47 sequences possessed overlapping regions with low identity, and therefore, likely represent unigenes. Of these, 13 HarmOR sequences had full-length open reading frames (ORF) with 5–6 transmembrane domains characteristic of typical insect ORs. Not surprisingly, a HarmOR sequence that shared very high identity (∼up to 90%) with the conserved insect co-receptor was identified. As other insect ORs, most HarmORs were highly divergent and shared low similarity with other insect ORs except for closely related species such as H. virescens. However, six HarmORs shared considerable similarity and were classified into a subgroup in the phylogenetic tree with previously characterized lepidopteran pheromone receptors (Figure 2). Almost all odorant receptor candidates were clustered with at least one lepidopteran orthologous gene in the phylogenetic tree except for two ORs (unigene6998 and 31180) appear to be more distant from their closest homolog because of their short length. We named the candidate OR unigenes according to their similarity to known ORs. The unigenes which had high similarity to H. virescens ORs were named following their orthologous ORs. Unigene6695 was named as HarmOR21.2 because of its relatively low similarity to HvirOR21. The remaining 27 ORs were named HarmOR22 through HarmOR48. The information including unigene reference, length, BLASTx best hit of all 47 ORs and 1 GR are listed in Table 1. The nucleotide sequences of all 47 ORs and 1 GR are listed in supplementary material S5.
Figure 2. Phylogenetic tree of candidate ORs from lepidopterans including PR (red) and ORCO (Blue) clades.
Harm: H. armigera, Hvir: H. virescens , Bmor: B. mori. The H. armigera unigenes are shown in bold and the letter-unigene in the unigene reference was abbreviated as U.
Table 1. Unigenes of candidate olfactory receptors and gustatory receptor.
| Unigene reference | Gene name | Length (bp) | ORF (aa) | BLASTx best hit (Reference/Name/Species) | E value | Identity | Full length | TMD (No) | Sexual specificity |
| Co-receptor | |||||||||
| unigene7630 | HarmOR2 | 2588 | 473 | gb|ADQ13177.1| olfactory receptor OR83b [Helicoverpa armigera] | 0.0 | 99% | Yes | 7 | No |
| Pheromone receptors | |||||||||
| unigene26842 | HarmOR11 | 1736 | 430 | gb|ACS45305.1| candidate odorant receptor 2 [Helicoverpa armigera] | 0.0 | 99% | Yes | 6 | No |
| unigene12204 | HarmOR13 | 1596 | 425 | gb|ACS45304.1| candidate odorant receptor 1 [Helicoverpa armigera] | 0.0 | 99% | Yes | 7 | Male |
| unigene12212 | HarmOR14 | 1378 | 313 | gb|ACF32964.1| olfactory receptor 14 [Helicoverpa armigera] | 0.0 | 99% | No | 4 | Male |
| unigene12228 | HarmOR15 | 1231 | 387 | emb|CAG38116.1| putative chemosensory receptor 15 [Heliothis virescens] | 0.0 | 86% | No | 4 | Male |
| unigene12206 | HarmOR16 | 1616 | 422 | gb|ACS45306.1| candidate odorant receptor 3 [Helicoverpa armigera] | 0.0 | 97% | Yes | 3 | Male |
| unigene6862 | HarmOR6 | 719 | 191 | emb|CAD31948.1| putative chemosensory receptor 6 [Heliothis virescens] | 1e−103 | 85% | No | 0 | No |
| Olfactory receptors | |||||||||
| unigene11748 | HarmOR3 | 699 | 200 | emb|CAD31852.1| putative chemosensory receptor 3 [Heliothis virescens] | 6e−129 | 94% | No | 2 | No |
| unigene5344 | HarmOR5 | 430 | 142 | emb|CAD31947.1| putative chemosensory receptor 5 [Heliothis virescens] | 3e−93 | 96% | No | 2 | No |
| unigene21145 | HarmOR7 | 814 | 271 | emb|CAD31853.1| putative chemosensory receptor 7 [Heliothis virescens] | 1e−172 | 96% | No | 4 | No |
| unigene3890 | HarmOR8 | 1331 | 351 | emb|CAD31949.1| putative chemosensory receptor 8 [Heliothis virescens] | 0.0 | 73% | No | 6 | No |
| unigene4049 | HarmOR9 | 764 | 243 | emb|CAD31950.1| putative chemosensory receptor 9 [Heliothis virescens] | 2e−140 | 86% | No | 4 | No |
| unigene21919 | HarmOR10 | 666 | 221 | gb|ACC63238.1| olfactory receptor 10 [Helicoverpa armigera] | 0.0 | 100% | No | 3 | No |
| unigene36907 | HarmOR12 | 1368 | 408 | gb|ACF32963.1| olfactory receptor 12 [Helicoverpa armigera] | 0.0 | 97% | Yes | 6 | No |
| unigene13628 | HarmOR17 | 1339 | 396 | emb|CAG38118.1| putative chemosensory receptor 17 [Heliothis virescens] | 0.0 | 93% | Yes | 6 | Male |
| Unigene35925 | HarmOR18 | 1356 | 398 | gb|ACL81187.1| putative olfactory receptor 18 [Helicoverpa armigera] | 0 | 100% | Yes | 5 | No |
| unigene186 | HarmOR19 | 429 | 142 | emb|CAG38120.1| putative chemosensory receptor 19 [Heliothis virescens] | 5e−49 | 61% | No | 1 | Female |
| unigene8158 | HarmOR20 | 1269 | 387 | gb|ACC63240.1| olfactory receptor 20, partial [Helicoverpa armigera] | 0 | 99% | Yes | 7 | No |
| unigene10230 | HarmOR21 | 939 | 278 | emb|CAG38122.1| putative chemosensory receptor 21 [Heliothis virescens] | 2e−160 | 85% | No | 5 | No |
| unigene6695 | HarmOR21.2 | 1237 | 396 | emb|CAG38122.1| putative chemosensory receptor 21 [Heliothis virescens] | 2e−86 | 37% | No | 6 | No |
| unigene36738 | HarmOR22 | 1040 | 346 | gb|ADM32898.1| odorant receptor OR-5 [Manduca sexta] | 6e−54 | 36% | No | 4 | No |
| unigene6998 | HarmOR23 | 411 | 135 | gb|AEF32141.1| odorant receptor [Spodoptera exigua] | 2e−63 | 73% | No | 2 | No |
| unigene35735 | HarmOR24 | 1197 | 358 | dbj|BAF31195.1| candidate olfactory receptor [Bombyx mori] | 7e−151 | 67% | No | 4 | No |
| unigene24858 | HarmOR25 | 1167 | 389 | tpg|DAA05974.1| TPA_exp: odorant receptor 15 [Bombyx mori] | 4e−115 | 48% | No | 4 | No |
| unigene6502 | HarmOR26 | 693 | 212 | dbj|BAH66346.1| olfactory receptor [Bombyx mori] | 5e−95 | 67% | No | 2 | No |
| unigene36300 | HarmOR27 | 2136 | 413 | dbj|BAH66328.1| olfactory receptor [Bombyx mori] | 3e−108 | 51% | Yes | 6 | No |
| unigene24202 | HarmOR28 | 671 | 185 | dbj|BAH66335.1| olfactory receptor [Bombyx mori] | 3e−54 | 57% | No | 2 | No |
| unigene36462 | HarmOR29 | 1619 | 395 | ref|NP_001166603.1| olfactory receptor 13 [Bombyx mori] | 7e−127 | 48% | Yes | 7 | No |
| unigene34814 | HarmOR30 | 703 | 229 | ref|NP_001104832.2| olfactory receptor 16 [Bombyx mori] | 2e−111 | 70% | No | 4 | No |
| unigene28466 | HarmOR31 | 975 | 325 | ref|NP_001166894.1| olfactory receptor 29 [Bombyx mori] | 2e−160 | 71% | No | 6 | No |
| unigene24650 | HarmOR32 | 1069 | 349 | ref|NP_001103623.1| olfactory receptor 33 [Bombyx mori] | 2e−99 | 42% | No | 3 | No |
| unigene26634 | HarmOR33 | 1195 | 398 | ref|NP_001103476.1| olfactory receptor 35 [Bombyx mori] | 4e−138 | 51% | No | 4 | No |
| unigene30070 | HarmOR34 | 550 | 183 | tpg|DAA05992.1| TPA_exp: odorant receptor 36 [Bombyx mori] | 8e−69 | 53% | No | 3 | No |
| unigene7372 | HarmOR35 | 867 | 241 | ref|NP_001091818.1| olfactory receptor 42 [Bombyx mori] | 3e−27 | 31% | No | 3 | No |
| unigene31113 | HarmOR36 | 321 | 106 | ref|NP_001091818.1| olfactory receptor 42 [Bombyx mori] | 6e−38 | 58% | No | 2 | No |
| unigene14199 | HarmOR37 | 455 | 136 | ref|NP_001166607.1| olfactory receptor 44 [Bombyx mori] | 7e−78 | 86% | No | 2 | Male |
| unigene31180 | HarmOR38 | 428 | 142 | ref|NP_001166607.1| olfactory receptor 44 [Bombyx mori] | 1e−23 | 75% | No | 1 | No |
| unigene6642 | HarmOR39 | 1272 | 377 | ref|NP_001166616.1| olfactory receptor 54 [Bombyx mori] | 9e−73 | 38% | No | 6 | No |
| unigene7016 | HarmOR40 | 910 | 303 | ref|NP_001166893.1| olfactory receptor 27 [Bombyx mori] | 8e−113 | 63% | No | 5 | No |
| unigene36885 | HarmOR41 | 1803 | 179 | ref|NP_001166617.1| olfactory receptor 56 [Bombyx mori] | 1e−87 | 78% | No | 3 | No |
| unigene22154 | HarmOR42 | 1237 | 392 | ref|NP_001155301.1| olfactory receptor 60 [Bombyx mori] | 0 | 70% | No | 5 | No |
| unigene33166 | HarmOR43 | 1247 | 393 | ref|NP_001166620.1| olfactory receptor 63 [Bombyx mori] | 6e−132 | 52% | Yes | 7 | No |
| unigene35415 | HarmOR44 | 1245 | 375 | ref|NP_001166621.1| olfactory receptor 64 [Bombyx mori] | 7e−91 | 53% | Yes | 7 | No |
| unigene6639 | HarmOR45 | 1513 | 434 | ref|NP_001166622.1| olfactory receptor 65 [Bombyx mori] | 2e−79 | 63% | Yes | 7 | Female |
| unigene33039 | HarmOR46 | 1041 | 346 | ref|NP_001116817.1| olfactory receptor-like [Bombyx mori] | 3e−147 | 69% | No | 5 | No |
| unigene5523 | HarmOR47 | 320 | 106 | tpg|DAA05974.1| TPA_exp: odorant receptor 15 [Bombyx mori] | 8e−22 | 40% | No | 1 | No |
| unigene23524 | HarmOR48 | 639 | 197 | tpg|DAA05986.1| TPA_exp: odorant receptor 30 [Bombyx mori] | 1e−73 | 54% | No | 1 | No |
| Gustatory receptors | |||||||||
| unigene14001 | HarmGR1 | 632 | 210 | tpg|DAA06395.1| TPA_inf: gustatory receptor 63 [Bombyx mori] | 1e−49 | 47% | No | No | |
Identification of Candidate Ionotropic Receptors
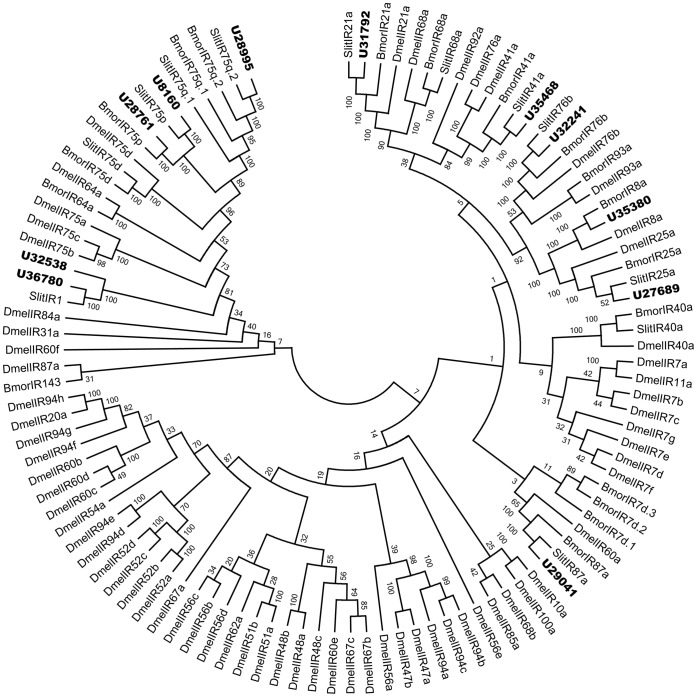
The IRs sequences in the H. armigera antennal transcriptome assembly were represented according to the similarity with known insect IRs. Bioinformatic analysis led to the identification of 12 candidates IRs. Sequence analysis identified 9 unigenes with a full length ORF. The insect IRs contained three transmembrane domains. TMHMM2.0 predicted 11 candidate IRs with three transmembrane domains (Table 2). Eight of the 12 putative IRs had at least 68% identity with the corresponding IRs of S. littoralis. These may be the orthologous genes in H. armigera. One candidate IR was represented to be IR8a due to its high identity to BmorIR8a. The remaining three putative IRs had relatively low similarity to other insect IRs. Unigene27689 had 65% identity with IR25a of D. melanogaster. Unigene32538 had 34% identity with IR1 of S. littoralis. Unigene28761 had 61% identity with IR75p of S. littoralis. The orthologs of these genes probably haven’t been identified in S. littoralis. The phylogenetic analyses validated accurate prediction of the IRs. In the neighbor-joining tree of IRs, all candidate H. armigera IRs clustered with their orthologs of S. littoralis and B. mori into a separate clade (Figure 3). Eleven of the 12 candidate IR unigenes were named according to their similarity to known IRs. The new IR unigene32538 was named HarmIR1.2. The information including unigene reference, length, and BLASTx best hit of all the12 IRs are listed in Table 2. The nucleotide sequences of all 12 IRs were listed in supplementary material S5.
Table 2. Unigenes of candidate ionotropic receptors.
| Unigene reference | Gene name | Length (bp) | ORF (aa) | Blastx best hit (Reference/Name/Species) | E value | Identity | Full length | TMD (No) |
| unigene36780 | HarmIR1 | 2070 | 617 | gb|ADR64688.1| putative chemosensory ionotropic receptor IR1 [Spodoptera littoralis] | 0.0 | 68% | Yes | 3 |
| unigene32538 | HarmIR1.2 | 1862 | 620 | gb|ADR64688.1| putative chemosensory ionotropic receptor IR1 [Spodoptera littoralis] | 4e−42 | 34% | Yes | 2 |
| unigene35380 | HarmIR8a | 3456 | 895 | gb|AFC91764.1| putative ionotropic receptor IR8a, partial [Cydia pomonella] | 0.0 | 81% | Yes | 3 |
| unigene31792 | HarmIR21a | 2927 | 857 | gb|ADR64678.1| putative chemosensory ionotropic receptor IR21a [Spodoptera littoralis] | 0.0 | 82% | Yes | 3 |
| unigene27689 | HarmIR25a | 2966 | 918 | gb|AFC91757.1| putative ionotropic receptor IR25a [Cydia pomonella] | 0.0 | 87% | Yes | 3 |
| unigene35468 | HarmIR41a | 2014 | 608 | gb|ADR64681.1| putative chemosensory ionotropic receptor IR41a [Spodoptera littoralis] | 0.0 | 81% | Yes | 3 |
| unigene21502 | HarmIR75d | 840 | 231 | gb|ADR64683.1| putative chemosensory ionotropic receptor IR75d [Spodoptera littoralis] | 1e−71 | 81% | No | 3 |
| unigene8160 | HarmIR75p | 1681 | 547 | gb|ADR64684.1| putative chemosensory ionotropic receptor IR75p [Spodoptera littoralis] | 0.0 | 90% | No | 3 |
| unigene28761 | HarmIR75p.2 | 2368 | 654 | gb|ADR64684.1| putative chemosensory ionotropic receptor IR75p [Spodoptera littoralis] | 0.0 | 61% | Yes | 3 |
| unigene28995 | HarmIR75q.2 | 4238 | 628 | gb|ADR64685.1| putative chemosensory ionotropic receptor IR75q.2 [Spodoptera littoralis] | 0.0 | 83% | No | 3 |
| unigene32241 | HarmIR76b | 1879 | 558 | gb|ADR64687.1| putative chemosensory ionotropic receptor IR76b [Spodoptera littoralis] | 0.0 | 84% | Yes | 3 |
| unigene29041 | HarmIR87a | 1919 | 583 | gb|ADR64689.1| putative chemosensory ionotropic receptor IR87a [Spodoptera littoralis] | 0.0 | 94% | Yes | 3 |
Figure 3. Phylogenetic tree of candidate IRs from insects.
Slit: S. littoralis, Bmor: B. mori, Dmel: D. melanogaster, the H. armigera unigenes are shown in bold and the letter-unigene in the unigene reference was abbreviated as U.
Identification of Putative Odorant-binding Proteins
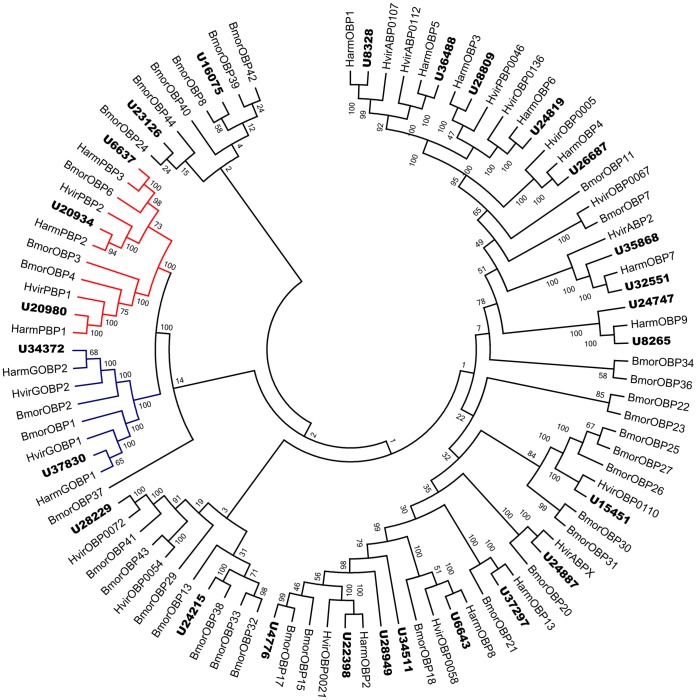
Within the H. armigera antennal transcriptome 26 different sequences encoding odorant binding proteins were identified, including three PBPs and two GOBPs. Sequence analysis identified 19 unigenes with a full length ORF with a predicted signal peptide sequence. Signal peptide sequence was not detected in the remainder of putative OBPs due to incomplete N-termini. All 26 putative OBPs had high similarity to known lepidopteran OBPs. Unigene35868 and unigene24747 had very high similarity with HarmOBP7 and HarmOBP9 (94% and 95%, respectively). They may be isoforms of the respective H. armigera OBPs or potentially new OBP genes. As expected, the PBP and GOBP sequences were clustered in separate clade in the OBP neighbor-joining tree. All the candidate OBP sequences were clustered with at least one lepidopteran ortholog, in congruence with the BLAST results (Figure 4). HarmOBPs were named according to their similarities with previously annotated H. armigera OBPs. Two of the putative OBP isoforms, unigene35868 and unigene24747, were named as HarmOBP7.2 and HarmOBP9.2, respectively. The information including unigene reference, length, and BLASTx best hit and so on of all the 26 OBPs was listed in table 3. The nucleotide sequences of all the 26 OBPs were listed in supplement material S5.
Figure 4. Phylogenetic tree of candidate odorant binding protein from lepidopterans including PBP (red), GOBP (Blue) and other OBP.
Harm: H. armigera, Hvir: H. virescens, Bmor: B. mori, Dple: D. plexippus, the H. armigera unigenes are shown in bold and the letter-unigene in the unigene reference was abbreviated as U.
Table 3. Unigenes of candidate odorant binding proteins.
| Unigene reference | Gene name | Length (bp) | ORF (aa) | Blastx best hit (Reference/Name/Species) | E value | Identity | Full length | Signal peptide |
| Pheromone binding protein | ||||||||
| unigene20980 | HarmPBP1 | 1253 | 170 | gb|AEB54585.1| PBP1 [Helicoverpa armigera] | 2e−108 | 99% | Yes | Yes |
| unigene20934 | HarmPBP2 | 989 | 165 | gb|AEB54583.1| PBP2 [Helicoverpa armigera] | 2e−114 | 98% | Yes | Yes |
| unigene6637 | HarmPBP3 | 704 | 164 | gb|AAO16091.1| pheromone binding protein 3 [Helicoverpa armigera] | 3e−106 | 99% | Yes | Yes |
| General odorant binding protein | ||||||||
| unigene37830 | HarmGOBP1 | 1097 | 172 | emb|CAA65605.1| general odorant binding protein 1 [Heliothis virescens] | 4e−101 | 96% | Yes | Yes |
| unigene34372 | HarmGOBP2 | 847 | 162 | gb|AAG54078.1| general odorant binding protein 2 [Helicoverpa zea] | 3e−113 | 100% | Yes | Yes |
| Other odorant binding protein | ||||||||
| unigene8328 | HarmOBP1 | 789 | 147 | gb|AEB54580.1| OBP1 [Helicoverpa armigera] | 0 | 100% | Yes | Yes |
| unigene22398 | HarmOBP2 | 831 | 143 | gb|AEB54586.1| OBP2 [Helicoverpa armigera] | 3e−99 | 99% | Yes | Yes |
| unigene28809 | HarmOBP3 | 580 | 147 | gb|AEB54582.1| OBP3 [Helicoverpa armigera] | 6e−100 | 99% | Yes | Yes |
| unigene26687 | HarmOBP4 | 507 | 137 | gb|AEB54584.1| OBP4 [Helicoverpa armigera] | 3e−90 | 98% | No | No |
| unigene36488 | HarmOBP5 | 924 | 147 | gb|AEB54581.1| OBP5 [Helicoverpa armigera] | 1e−99 | 99% | Yes | Yes |
| unigene24819 | HarmOBP6 | 572 | 147 | gb|AEB54587.1| OBP6 [Helicoverpa armigera] | 9e−90 | 100% | Yes | Yes |
| unigene32551 | HarmOBP7 | 570 | 148 | gb|AEB54591.1| OBP7 [Helicoverpa armigera] | 2e−80 | 100% | Yes | Yes |
| unigene35868 | HarmOBP7.2 | 737 | 148 | gb|AEB54591.1| OBP7 [Helicoverpa armigera] | 9e−85 | 94% | Yes | Yes |
| unigene6643 | HarmOBP8 | 495 | 139 | gb|AEB54589.1| OBP8 [Helicoverpa armigera] | 6e−98 | 100% | Yes | Yes |
| unigene8265 | HarmOBP9 | 1655 | 148 | gb|AEB54592.1| OBP9 [Helicoverpa armigera] | 1e−101 | 100% | Yes | Yes |
| unigene24747 | HarmOBP9.2 | 631 | 147 | gb|AEB54592.1| OBP9 [Helicoverpa armigera] | 2e−101 | 95% | Yes | Yes |
| unigene37297 | HarmOBP13 | 610 | 141 | gb|AEB54588.1| OBP13 [Helicoverpa armigera] | 9e−92 | 100% | Yes | Yes |
| unigene15451 | HarmOBP14 | 339 | 111 | gb|ACX53795.1| odorant binding protein [Heliothis virescens] | 1e−70 | 93% | No | No |
| unigene24215 | HarmOBP15 | 1374 | 217 | gb|ADY17882.1| odorant binding protein [Spodoptera exigua] | 4e−75 | 77% | No | No |
| unigene28229 | HarmOBP16 | 702 | 186 | gb|ACX53761.1| odorant binding protein [Heliothis virescens] | 3e−71 | 58% | Yes | Yes |
| unigene24887 | HarmOBP17 | 1184 | 158 | antennal binding protein X [Heliothis virescens] | 5e−59 | 100% | Yes | Yes |
| unigene4776 | HarmOBP18 | 929 | 121 | gb|AFG72998.1| odorant-binding protein 1 [Cnaphalocrocis medinalis] | 8e−70 | 83% | No | No |
| unigene16075 | HarmOBP19 | 423 | 122 | ref|NP_001140188.1| odorant-binding protein 4 [Bombyx mori] | 2e−29 | 47% | No | No |
| unigene23126 | HarmOBP20 | 1100 | 251 | gb|ADD71058.1| odorant-binding protein [Chilo suppressalis] | 3e−118 | 65% | Yes | Yes |
| unigene34511 | HarmOBP21 | 2207 | 121 | gb|EHJ65654.1| antennal binding protein 4 [Danaus plexippus] | 8e−50 | 66% | No | No |
| unigene28949 | HarmOBP22 | 1417 | 121 | gb|AFG72998.1| odorant-binding protein 1 [Cnaphalocrocis medinalis] | 1e−49 | 58% | No | No |
Identification of Candidate Chemosensory Proteins
Bioinformatic analysis led to the identification of 12 different sequences encoding candidate CSPs. Ten sequences were predicted to have full length and all of them had a signal peptide. Neighbor-joining tree showed all the 12 sequences were clustered with one lepidopterans orthologous gene and the candidate CSPs could be well identified (Figure 5). The unigenes corresponding to CSP genes were named following the identified CSPs. The rest 6 CSPs named from HarmCSP8 to HarmCSP13 following the known HarmCSP7. The information of all the 12 CSPs was listed in table 4. The nucleotide sequences of all the 12 CSPs were listed in supplement material S5.
Figure 5. Phylogenetic tree of candidate o chemosensory protein from lepidopterans.
Harm: H. armigera, Hvir: H. virescens , Bmor: B. mori, Se: S. exigua, the H. armigera unigenes are shown in bold and the letter-unigene in the unigene reference was abbreviated as U.
Table 4. Unigenes of candidate chemosensory protein.
| Unigene reference | Gene name | Length (bp) | ORF (aa) | Blastx best hit (Reference/Name/Species) | E value | Identity | Full length | Signal peptide |
| Chemosensory protein | ||||||||
| unigene23136 | HarmCSP | 706 | 127 | gb|AAK53762.1|AF368375_1 chemosensory protein [Helicoverpa armigera] | 5e−77 | 100% | Yes | Yes |
| unigene27156 | HarmCSP2 | 515 | 120 | gb|AEX07265.1| CSP2 [Helicoverpa armigera] | 7e−83 | 100% | Yes | Yes |
| unigene24935 | HarmCSP4 | 856 | 128 | gb|AEX07269.1| CSP4 [Helicoverpa armigera] | 7e−74 | 100% | Yes | Yes |
| unigene34049 | HarmCSP5 | 686 | 127 | gb|AEB54579.1| CSP5 [Helicoverpa armigera] | 1e−86 | 100% | Yes | Yes |
| unigene21552 | HarmCSP6 | 1196 | 122 | gb|AEX07267.1| CSP6 [Helicoverpa armigera] | 1e−79 | 98% | Yes | Yes |
| unigene35951 | HarmCSP7 | 923 | 111 | gb|AEX07268.1| CSP7 [Helicoverpa armigera] | 2e−69 | 100% | Yes | Yes |
| unigene21097 | HarmCSP8 | 570 | 128 | gb|ACX53700.1| chemosensory protein [Heliothis virescens] | 4e−77 | 86% | Yes | Yes |
| unigene12229 | HarmCSP9 | 1084 | 126 | gb|ACX53745.1| chemosensory protein [Heliothis virescens] | 8e−72 | 95% | Yes | Yes |
| unigene35427 | HarmCSP10 | 926 | 107 | dbj|BAF91720.1| chemosensory protein [Papilio xuthus] | 2e−52 | 94% | Yes | Yes |
| unigene33025 | HarmCSP11 | 1333 | 158 | gb|EHJ76401.1| chemosensory protein CSP1 [Danaus plexippus] | 6e−53 | 57% | Yes | Yes |
| unigene15064 | HarmCSP12 | 473 | 116 | gb|ACX53817.1| chemosensory protein [Heliothis virescens] | 7e−56 | 75% | No | No |
| unigene34785 | HarmCSP13 | 801 | 122 | gb|ACX53719.1| chemosensory protein [Heliothis virescens] | 5e−81 | 98% | Yes | Yes |
Identification of Candidate Sensory Neuron Membrane Proteins
SNMPs were first identified in pheromone-sensitive neurons of Lepidoptera [20] and are thought to play a role in pheromone detection [21]. Two kinds of SNMPs (SNMP1 and SNMP2) have been identified in insects and both kinds of SNMPs were discovered in H. armigera transcriptome. The nucleotide sequence of contig5289 was Identical to the HarmSNMP published in Genbank. Contig5355 had 61% identity with SNMP2 of H. virescens and was annotation to be SNMP2 of H. armigera (Table 5). The nucleotide sequences of the 2 SNMPs were listed in supplement material S5.
Table 5. Unigenes of candidate sensory neuron membrane protein.
| Unigene reference | Gene name | Length (bp) | ORF (aa) | BLASTx best hit (Reference/Name/Species) | E value | Identity | Full length |
| Sensory neuron membrane protein | |||||||
| unigene28770 | HarmSNMP1 | 2175 | 523 | gb|AAO15604.1|AF462067_1 sensory neuron membrane protein [Helicoverpa armigera] | 0.0 | 99% | Yes |
| unigene32002 | HarmSNMP2 | 1727 | 520 | emb|CAP19028.1| sensory neuron membrane protein-2 [Heliothis virescens] | 0.0 | 95% | Yes |
Tissue- and Sex- specific Expression of Candidate H. armigera OR, GR and IR Genes
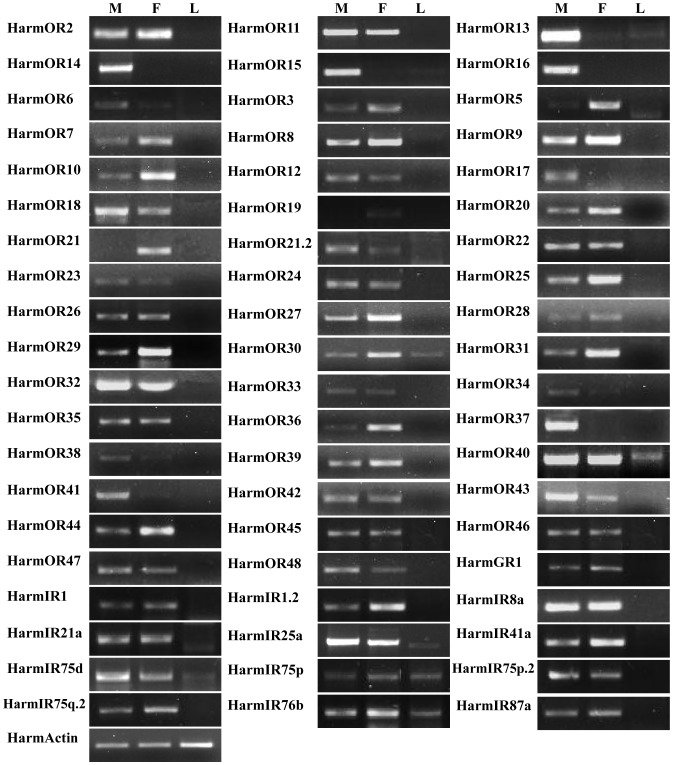
The expression patterns of the candidate 47 ORs, 1 GR and 12 IRs in male antennae, female antennae and legs were analyzed by semi-quantitative reverse transcription PCR. Figure 6 shows the detection of all the 60 candidate receptors in antennae of H. armigera.
Figure 6. Tissue- and sex- specific expressions of candidates H. armigera ORs, GRs and IRs genes.
M: male antennae, F: female antennae, L: legs.
The expressions of all the 47 ORs were detected in antennae. Of the six candidate PRs, four PRs were found to be exclusive to the male antennae and this result was congruent with the expression profile calculated during unigene assembly. The antennal expression level of HarmOR6 was very low. RT-PCR results demonstrated that the ORs HarmOR17, HarmOR37, and HarmOR41 are male-specific; and the ORs HarmOR5and HarmOR21 are female-specific. The remaining 36 ORs were expressed in both sexes, with some of them differentially expressed in male or female antennae. The single GR identified in this study, HarmGR1, was found to be highly expressed at equal levels in antennae of both sexes. Compared to ORs, the expressions of all IRs had no significant difference between males and females.
Discussion
We used transcriptomic sequencing to identify putative olfactory system genes consisting of 47 ORs, 12 IRs, 26 OBPs, 12 CSPs, and 2 SNMPs in the antennae of H. armigera. The olfactory system genes identified in this study are comparable to recently reported insect antennal transcriptome sequence of M. sexta with 47 ORs, 6 IRs, 18 OBPs, and 21 CSPs, and C. pomonella with 43 ORs and 15 IRs [36], [37]. All the previously annotated or characterized ORs [51], [52], OBPs [53], CSPs [53], and SNMPs of H. armigera were included in the candidate olfaction genes identified in this work. Many nonreceptor olfaction genes including OBPs, CSPs and SNMPs were also identified in our antennal transcriptome. The number identified was slightly less than B. mori. Probably the remaining OR genes are exclusively expressed in other olfaction organ such as maxillary palp and proboscis or developmental period.
Activated ORs are the first critical step mediating odorant recognition in the peripheral olfactory signal transduction pathway. Only a few OR genes could be identified according to homolog-based strategy on the condition of ORs of closely related species. Previous studies have suggested that single olfactory receptor neuron (ORN) class generally expresses a single OR (except ORCO) [57], and ORN innervates a corresponding glomerulus in the insect olfactory system [57]. While the relationship is not exactly 1∶1:1, the number of glomeruli could form the basis of a rough estimate of the number of ORs in a species [3], [36]. In H. armigera, 65 distinct glomeruli have been found in each sex [58] and therefore, the total number of ORs should correspond with the number of glomeruli. Despite much effort searching for ORs in H. armigera, only the coreceptor (HaOR2) and ten OR-coding genes or fragments have been identified and deposited in GenBank [51], [52]. In this work, we identified 47 ORs in H. armigera antennae and largely extend the number of ORs. Obviously, the number of ORs identified in this study is still less than expected based on the number of glomeruli. There are several possibilities to address the phenomena. Firstly, we only sequenced the antennal transcriptomes of male and female adult. Some OR genes might specifically expressed in different developmental stage such as larval stage or other olfactory organs of adults such as maxillary palp and proboscis [49]. Previous reports indicated that at least 6 ORs and 1o ORs specifically expressed in Bombyx mori and Drosophila larvae antennae, respectively, which supported our hypothesis (Current Biology 2008; Neuron, 2005 Kreher). Secondly, some glomeruli should be innervated by OSNs expressing other classes of chemoreceptors such as ionotropic receptos and gustatory receptors identified in this study. Thirdly, some OR paralogs with highly sequence similarity could be missing in current analysis since they are difficult to be separated with polymorphism without genome sequence(2012 PLOS one Bengtsson). Finally, we couldn’t exclude the possibility that 454 pyrosequencing is not powerful enough to exhaustedly obtain all ORs, especially those ORs with extremely low expression level in the antennae.
Six PRs were identified which were named OR6, OR11, OR13, OR14, OR15, OR16 according H. virescens [29]. Only four PRs (OR11, OR13, OR14, and OR16) had been identified in H. armigera before. Of the 47 ORs identified in this work, six belong to the PR group. They were orthologous genes of six PRs identified in H. virescens. The male-specific or male-enriched expression profiles of these six ORs (Figure 6) were consistent with the PR expression in closely related species, H. virescens [29].
All the 41 normal ORs were also specifically expressed in antennae. Experiments were conducted to identify ORs with differential expression patterns between male and female because they might perform specific functions in each gender. The RT-PCR showed that three ORs (HarmOR17, HarmOR37, and HarmOR41) were male specific and two other ORs (HarmOR5 and HarmOR21) were female specific (Figure 6). However, the expression profile calculated according to number of copies found in transcriptomes gave a different result and inconsistencies were also found in other 454 sequences [37]. This may have occurred because 454 reads are suboptimal for expression profiling because the number of reads acquired are relatively low (less than million) compared to Illumina RNA-Seq. In B. mori, BmorOR19 and BmorOR30 were found to be specifically expressed in female [49], [59]. We identified one homolog of BmorOR30 (HarmOR48) and two homologs of BmorOR19 (HarmOR21.2 and HarmOR22). But expression of OR48, OR21.2, and OR22 was detected in both male and female H. armigera antennae. This may be occurred because the sequences and expression patterns of normal ORs in different insects showed greater specificity than PRs. The sex-specific ORs need further study in H. armigera.
Recently, a new family of candidate chemosensory ionotropic receptors was discovered, first in D. melanogaster [12] and then in several other species through genome analyses [15]. D. melanogaster antennal IRs have been reported to detect a variety of molecules [14]. In D. melanogaster, 66 IRs were identified 15 of which proved to be antennae-specific [12], [15]. Twelve IRs were identified in the antennae of S. littoralis [35]. We also found 12 IRs in H. armigera antennae including two co-receptors, IR8a and IR25a [14]. This is the first report of IRs in H. armigera. Sequences alignments showed that the putative H. armigera IRs have higher similarity with known IRs than ORs. Unlike ORs, the expression of the IRs appeared to be similar between male and female. Similar results were also observed in the expression study of S. littoralis IRs [35]. The relatively high sequence conservation and expression of IRs implies a probable functional conservation. The antennal IRs are a novel group of chemosensory receptors.
Conclusions
The main objective of antennal transcriptome sequencing was to identify genes potentially involved in olfactory signal detection in H. armigera. The numbers of ORs, IRs, OBPS, CSPs and SNMPs identified in this species are close to the complete repertoire of olfactory genes identified from the antennae of other Lepidopteran species. This study demonstrates that high-throughput 454 pyrosequencing enables the recovery of rare or low copy number expressed genes, especially receptor genes with low expression levels in a species without an available genome sequence. Our findings lay the foundation for future research on the molecular basis of olfactory system of H. armigera and provide information for comparative and functional genomic analyses of related species. In addition, further studies of olfactory systems, exploring the potential for olfaction-based management of pest moth populations will be feasible with the findings reported in this antennal transcriptome sequencing study.
Supporting Information
Accession numbers for amino acid sequences of ORs, IRs, OBPs and CSPs used in phylogenetic analyses.
(DOCX)
Primers for RT-PCR expression analyses of H. armigera ORs, GR and IRs.
(XLSX)
Flow chart of RNA sequencing, unigene assemble and functional annotation. The experiment performed in male or female sets was marked in red or green separately. The experiment performed in the blended set of male and female was marked in blue. The numbers without unit in the parentheses were the sequence numbers. The numbers with bp unit in the parentheses were the average base numbers of sequences.
(TIF)
The gene length, ORF length, expression, GO annotation, and BLAST best hit of each unigene.
(XLSX)
Unigenes of candidate olfactory genes identified in this study, FASTA formatted file.
(TXT)
Acknowledgments
We thank Drs C. Liang and O.P. Perera for discussions and comments on the manuscript and to Dr. K.S. Shelby for editorial assistance.
Funding Statement
This work was supported by National Natural Science Foundation of China (31071752) and the China National “973” Basic Research Program (2012CB114104). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Hildebrand JG (1995) Analysis of chemical signals by nervous systems. Proc Natl Acad Sci U S A 92: 67–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mustaparta H (1990) Chemical information processing in the olfactory system of insects. Physiol Rev 70: 199–245. [DOI] [PubMed] [Google Scholar]
- 3. Bruyne M, Baker TC (2008) Odor Detection in Insects: Volatile Codes. Journal of Chemical Ecology 34: 882–897. [DOI] [PubMed] [Google Scholar]
- 4. Rützler M, Zwiebel LJ (2005) Molecular biology of insect olfaction:recent progress and conceptual models. Journal of Comparative Physiology A 191: 777–790. [DOI] [PubMed] [Google Scholar]
- 5. Sato K, Touhara K (2009) Insect olfaction: receptors, signal transduction, and behavior. Results Probl Cell Differ 47: 121–138. [DOI] [PubMed] [Google Scholar]
- 6. Benton R, Sachse S, Michnick SW, Vosshall LB (2006) Atypical Membrane Topology and Heteromeric Function of Drosophila Odorant Receptors In Vivo. PLoS Biology 4: e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Neuhaus EM, Gisselmann G, Zhang W, Dooley R, Stortkuhl K, et al. (2005) Odorant receptor heterodimerization in the olfactory system of Drosophila melanogaster. Nat Neurosci 8: 15–17. [DOI] [PubMed] [Google Scholar]
- 8. Sato K, Pellegrino M, Nakagawa T, Nakagawa T, Vosshall LB, et al. (2008) Insect olfactory receptors are heteromeric ligand-gated ion channels. Nature 452: 1002–1006. [DOI] [PubMed] [Google Scholar]
- 9. Wicher D, Schäfer R, Bauernfeind R, Stensmyr MC, Heller R, et al. (2008) Drosophila odorant receptors are both ligand-gated and cyclic-nucleotide-activated cation channels. Nature 452: 1007–1011. [DOI] [PubMed] [Google Scholar]
- 10. Larsson MC, Domingos AI, Jones WD, Chiappe ME, Amrein H, et al. (2004) Or83b Encodes a Broadly Expressed Odorant Receptor Essential for Drosophila Olfaction. Neuron 43: 703–714. [DOI] [PubMed] [Google Scholar]
- 11. Jones PL, Pask GM, Rinker DC, Zwiebel LJ (2011) Functional agonism of insect odorant receptor ion channels. Proc Natl Acad Sci U S A 108: 8821–8825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Benton R, Vannice KS, Gomez-Diaz C, Vosshall LB (2009) Variant Ionotropic Glutamate Receptors as Chemosensory Receptors in Drosophila. Cell 136: 149–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Stern DL, Croset V, Rytz R, Cummins SF, Budd A, et al. (2010) Ancient Protostome Origin of Chemosensory Ionotropic Glutamate Receptors and the Evolution of Insect Taste and Olfaction. PLoS Genetics 6: e1001064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Abuin L, Bargeton B, Ulbrich MH, Isacoff EY, Kellenberger S, et al. (2011) Functional architecture of olfactory ionotropic glutamate receptors. Neuron 69: 44–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Croset V, Rytz R, Cummins SF, Budd A, Brawand D, et al. (2010) Ancient protostome origin of chemosensory ionotropic glutamate receptors and the evolution of insect taste and olfaction. PLoS Genet 6: e1001064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pelosi P, Maida R (1995) Odorant-binding proteins in insects. Comp Biochem Physiol B Biochem Mol Biol 111: 503–514. [DOI] [PubMed] [Google Scholar]
- 17. Zhou JJ (2010) Odorant-binding proteins in insects. Vitam Horm 83: 241–272. [DOI] [PubMed] [Google Scholar]
- 18. Laughlin JD, Ha TS, Jones DNM, Smith DP (2008) Activation of pheromone-sensitive neurons is mediated by conformational activation of pheromone-binding protein. Cell 133: 1255–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Foret S, Wanner KW, Maleszka R (2007) Chemosensory proteins in the honey bee: Insights from the annotated genome, comparative analyses and expressional profiling. Insect Biochem Mol Biol 37: 19–28. [DOI] [PubMed] [Google Scholar]
- 20. Rogers ME, Krieger J, Vogt RG (2001) Antennal SNMPs (sensory neuron membrane proteins) of Lepidoptera define a unique family of invertebrate CD36-like proteins. J Neurobiol 49: 47–61. [DOI] [PubMed] [Google Scholar]
- 21. Benton R, Vannice KS, Vosshall LB (2007) An essential role for a CD36-related receptor in pheromone detection in Drosophila. Nature 450: 289–293. [DOI] [PubMed] [Google Scholar]
- 22. Vogt RG, Miller NE, Litvack R, Fandino RA, Sparks J, et al. (2009) The insect SNMP gene family. Insect Biochem Mol Biol 39: 448–456. [DOI] [PubMed] [Google Scholar]
- 23.Vogt RG, editor (2003) Biochemical diversity of odor detection OBPs, ODEs and SNMPs. London: Elsevier Academic Press. 391–445 p.
- 24.Durand N, Carot-Sans G, Chertemps T, Bozzolan F, Party V, et al.. (2010) Characterization of an Antennal Carboxylesterase from the Pest Moth Spodoptera littoralis Degrading a Host Plant Odorant. PLoS One 5. [DOI] [PMC free article] [PubMed]
- 25. Mitsuno H, Sakurai T, Murai M, Yasuda T, Kugimiya S, et al. (2008) Identification of receptors of main sex-pheromone components of three Lepidopteran species. European Journal of Neuroscience 28: 893–902. [DOI] [PubMed] [Google Scholar]
- 26. Miura N, Nakagawa T, Tatsuki S, Touhara K, Ishikawa Y (2009) A male-specific odorant receptor conserved through the evolution of sex pheromones in Ostrinia moth species. Int J Biol Sci 5: 319–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Patch HM, Velarde RA, Walden KKO, Robertson HM (2009) A Candidate Pheromone Receptor and Two Odorant Receptors of the Hawkmoth Manduca sexta. Chemical Senses 34: 305–316. [DOI] [PubMed] [Google Scholar]
- 28. Brigaud I, Montagné N, Monsempes C, François M-C, Jacquin-Joly E (2009) Identification of an atypical insect olfactory receptor subtype highly conserved within noctuids. FEBS Journal 276: 6537–6547. [DOI] [PubMed] [Google Scholar]
- 29. Krieger J (2004) Genes encoding candidate pheromone receptors in a moth (Heliothis virescens). Proceedings of the National Academy of Sciences 101: 11845–11850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Krieger J, Raming K, Dewer YM, Bette S, Conzelmann S, et al. (2002) A divergent gene family encoding candidate olfactory receptors of the moth Heliothis virescens. Eur J Neurosci 16: 619–628. [DOI] [PubMed] [Google Scholar]
- 31. Wanner KW, Nichols AS, Allen JE, Bunger PL, Garczynski SF, et al. (2010) Sex pheromone receptor specificity in the European corn borer moth, Ostrinia nubilalis. PLoS One 5: e8685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Robertson HM, Martos R, Sears CR, Todres EZ, Walden KK, et al. (1999) Diversity of odourant binding proteins revealed by an expressed sequence tag project on male Manduca sexta moth antennae. Insect Mol Biol 8: 501–518. [DOI] [PubMed] [Google Scholar]
- 33. Jordan MD, Stanley D, Marshall SD, De Silva D, Crowhurst RN, et al. (2008) Expressed sequence tags and proteomics of antennae from the tortricid moth, Epiphyas postvittana. Insect Mol Biol 17: 361–373. [DOI] [PubMed] [Google Scholar]
- 34. Legeai F, Malpel S, Montagné N, Monsempes C, Cousserans F, et al. (2011) An Expressed Sequence Tag collection from the male antennae of the Noctuid moth Spodoptera littoralis: a resource for olfactory and pheromone detection research. BMC Genomics 12: 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Olivier V, Monsempes C, Francois MC, Poivet E, Jacquin-Joly E (2011) Candidate chemosensory ionotropic receptors in a Lepidoptera. Insect Mol Biol 20: 189–199. [DOI] [PubMed] [Google Scholar]
- 36. Grosse-Wilde E, Kuebler LS, Bucks S, Vogel H, Wicher D, et al. (2011) Antennal transcriptome of Manduca sexta. Proceedings of the National Academy of Sciences 108: 7449–7454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bengtsson JM, Trona F, Montagne N, Anfora G, Ignell R, et al. (2012) Putative chemosensory receptors of the codling moth, Cydia pomonella, identified by antennal transcriptome analysis. PLoS One 7: e31620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Shen GM, Dou W, Niu JZ, Jiang HB, Yang WJ, et al. (2011) Transcriptome analysis of the oriental fruit fly (Bactrocera dorsalis). PLoS One 6: e29127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matthews A (1999) Heliothine Moths of Australia: A Reference Guide to Pest Bollworms & Related Noctuid Groups. Collingwood: CSIRO Publishing.
- 40. Fitt GP (1989) The ecology of Heliothis species in relation to agroecosystems. Annual Review of Entomology 34: 17–53. [Google Scholar]
- 41. Cho S, Mitchell A, Mitter C, Regier J, Matthews M, et al. (2008) Molecular phylogenetics of heliothine moths (Lepidoptera: Noctuidae: Heliothinae), with comments on the evolution of host range and pest status. Systematic Entomology 33: 581–594. [Google Scholar]
- 42. Lassmann T, Hayashizaki Y, Daub CO (2009) TagDust–a program to eliminate artifacts from next generation sequencing data. Bioinformatics 25: 2839–2840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chou HH, Holmes MH (2001) DNA sequence quality trimming and vector removal. Bioinformatics 17: 1093–1104. [DOI] [PubMed] [Google Scholar]
- 44. Chen YA, Lin CC, Wang CD, Wu HB, Hwang PI (2007) An optimized procedure greatly improves EST vector contamination removal. BMC Genomics 8: 416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, et al. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M, et al. (2005) Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21: 3674–3676. [DOI] [PubMed] [Google Scholar]
- 47. Gotz S, Garcia-Gomez JM, Terol J, Williams TD, Nagaraj SH, et al. (2008) High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res 36: 3420–3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Petersen TN, Brunak S, von Heijne G, Nielsen H (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8: 785–786. [DOI] [PubMed] [Google Scholar]
- 49. Tanaka K, Uda Y, Ono Y, Nakagawa T, Suwa M, et al. (2009) Highly Selective Tuning of a Silkworm Olfactory Receptor to a Key Mulberry Leaf Volatile. Current Biology 19: 881–890. [DOI] [PubMed] [Google Scholar]
- 50. Gong DP, Zhang HJ, Zhao P, Xia QY, Xiang ZH (2009) The odorant binding protein gene family from the genome of silkworm, Bombyx mori. BMC Genomics 10: 332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhang S, Zhang YJ, Su HH, Gao XW, Guo YY (2009) Cloning and tissue specific expression of olfactory receptors in Helicoverpa armigera (Hübner). Acta Entomologica Sinica 52: 728–735. [Google Scholar]
- 52. Zhang D-D, Zhu KY, Wang C-Z (2010) Sequencing and characterization of six cDNAs putatively encoding three pairs of pheromone receptors in two sibling species, Helicoverpa armigera and Helicoverpa assulta. Journal of Insect Physiology 56: 586–593. [DOI] [PubMed] [Google Scholar]
- 53. Zhang T, Gu S, Wu K, Zhang Y, Guo Y (2011) Construction and analysis of cDNA libraries from the antennae of male and female cotton bollworms Helicoverpa armigera (Hubner) and expression analysis of putative odorant-binding protein genes. Biochem Biophys Res Commun 407: 393–399. [DOI] [PubMed] [Google Scholar]
- 54. Picimbon JF, Dietrich K, Krieger J, Breer H (2001) Identity and expression pattern of chemosensory proteins in Heliothis virescens (Lepidoptera, Noctuidae). Insect Biochem Mol Biol 31: 1173–1181. [DOI] [PubMed] [Google Scholar]
- 55. Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, et al. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948. [DOI] [PubMed] [Google Scholar]
- 56. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, et al. (2011) MEGA5: Molecular Evolutionary Genetics Analysis Using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Molecular Biology and Evolution 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Vosshall LB, Wong AM, Axel R (2000) An olfactory sensory map in the fly brain. Cell 102: 147–159. [DOI] [PubMed] [Google Scholar]
- 58. Skiri HT, Ro H, Berg BG, Mustaparta H (2005) Consistent organization of glomeruli in the antennal lobes of related species of heliothine moths. J Comp Neurol 491: 367–380. [DOI] [PubMed] [Google Scholar]
- 59. Wanner KW, Anderson AR, Trowell SC, Theilmann DA, Robertson HM, et al. (2007) Female-biased expression of odourant receptor genes in the adult antennae of the silkworm, Bombyx mori. Insect Mol Biol 16: 107–119. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Accession numbers for amino acid sequences of ORs, IRs, OBPs and CSPs used in phylogenetic analyses.
(DOCX)
Primers for RT-PCR expression analyses of H. armigera ORs, GR and IRs.
(XLSX)
Flow chart of RNA sequencing, unigene assemble and functional annotation. The experiment performed in male or female sets was marked in red or green separately. The experiment performed in the blended set of male and female was marked in blue. The numbers without unit in the parentheses were the sequence numbers. The numbers with bp unit in the parentheses were the average base numbers of sequences.
(TIF)
The gene length, ORF length, expression, GO annotation, and BLAST best hit of each unigene.
(XLSX)
Unigenes of candidate olfactory genes identified in this study, FASTA formatted file.
(TXT)