Abstract
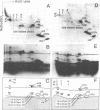
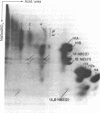
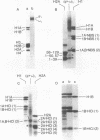
The topography of the interaction between histone H1 and the histone octamer has been investigated. Bovine thymus nuclei or enzymatically fragmented chromatin were treated 1-ethyl-3(3-dimethylaminopropyl)carbodiimide, which catalyzes the formation of covalent bonds between residues of proteins in electrostatic contact. Histone H1-core histone dimers were identified and the segments of molecules participating in crosslinking were elucidated. The results demonstrate that the major histone H1-core histone dimer generated upon carbodiimide crosslinking of intact nuclei, chromatin, or mononucleosomes consists of the segment of histone H1 containing amino acids 74-106 crosslinked to the segment of histone H2A containing amino acids 58-129. Thus, the central globular region of histone H1 intimately contacts the histone octamer. Besides histone H1-H2 dimers, two other histone H1-containing crosslinked products were detected. In these instances, the segments of histone H1 molecules containing amino acids 1-72 were shown to participate in crosslinking. The histone H1 contact points defined here all occur within mononucleosomes and not between nucleosomes. These results permit the formulation of a testable model for the arrangement of histone H1 along polynucleosome chains.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Albright S. C., Nelson P. P., Garrard W. T. Histone molar ratios among different electrophoretic forms of mono- and dinucleosomes. J Biol Chem. 1979 Feb 25;254(4):1065–1073. [PubMed] [Google Scholar]
- Bina-Stein M., Singer M. F. The effect of H1 histone on the action of DNA-relaxing enzyme. Nucleic Acids Res. 1977 Jan;4(1):117–127. doi: 10.1093/nar/4.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonner W. M. Proximity and accessibility studies of histones in nuclei and free nucleosomes. Nucleic Acids Res. 1978 Jan;5(1):71–85. doi: 10.1093/nar/5.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonner W. M., Stedman J. D. Histone 1 is proximal to histone 2A and to A24. Proc Natl Acad Sci U S A. 1979 May;76(5):2190–2194. doi: 10.1073/pnas.76.5.2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradbury E. M., Carpenter B. G., Rattle H. W. Magnetic resonance studies of deoxyribonucleoprotein. Nature. 1973 Jan 12;241(5385):123–126. doi: 10.1038/241123a0. [DOI] [PubMed] [Google Scholar]
- Bradbury E. M., Chapman G. E., Danby S. E., Hartman P. G., Riches P. L. Studies on the role and mode of operation of the very-lysine-rich histone H1 (F1) in eukaryote chromatin. The properties of the N-terminal and C-terminal halves of histone H1. Eur J Biochem. 1975 Sep 15;57(2):521–528. doi: 10.1111/j.1432-1033.1975.tb02327.x. [DOI] [PubMed] [Google Scholar]
- Cleveland D. W., Fischer S. G., Kirschner M. W., Laemmli U. K. Peptide mapping by limited proteolysis in sodium dodecyl sulfate and analysis by gel electrophoresis. J Biol Chem. 1977 Feb 10;252(3):1102–1106. [PubMed] [Google Scholar]
- Elgin S. C., Weintraub H. Chromosomal proteins and chromatin structure. Annu Rev Biochem. 1975;44:725–774. doi: 10.1146/annurev.bi.44.070175.003453. [DOI] [PubMed] [Google Scholar]
- Finch J. T., Lutter L. C., Rhodes D., Brown R. S., Rushton B., Levitt M., Klug A. Structure of nucleosome core particles of chromatin. Nature. 1977 Sep 1;269(5623):29–36. doi: 10.1038/269029a0. [DOI] [PubMed] [Google Scholar]
- Franklin S. G., Zweidler A. Non-allelic variants of histones 2a, 2b and 3 in mammals. Nature. 1977 Mar 17;266(5599):273–275. doi: 10.1038/266273a0. [DOI] [PubMed] [Google Scholar]
- Fulmer A. W., Fasman G. D. Analysis of chromatin reconstitutiion. Biochemistry. 1979 Feb 20;18(4):659–668. doi: 10.1021/bi00571a017. [DOI] [PubMed] [Google Scholar]
- Hardison R. C., Zeitler D. P., Murphy J. M., Chalkley R. Histone neighbors in nuclei and extended chromatin. Cell. 1977 Oct;12(2):417–427. doi: 10.1016/0092-8674(77)90118-0. [DOI] [PubMed] [Google Scholar]
- Hartman P. G., Chapman G. E., Moss T., Bradbury E. M. Studies on the role and mode of operation of the very-lysine-rich histone H1 in eukaryote chromatin. The three structural regions of the histone H1 molecule. Eur J Biochem. 1977 Jul 1;77(1):45–51. doi: 10.1111/j.1432-1033.1977.tb11639.x. [DOI] [PubMed] [Google Scholar]
- Henderson L. E., Oroszlan S., Konigsberg W. A micromethod for complete removal of dodecyl sulfate from proteins by ion-pair extraction. Anal Biochem. 1979 Feb;93(1):153–157. [PubMed] [Google Scholar]
- Kornberg R. D. Structure of chromatin. Annu Rev Biochem. 1977;46:931–954. doi: 10.1146/annurev.bi.46.070177.004435. [DOI] [PubMed] [Google Scholar]
- Langmore J. P., Wooley J. C. Chromatin architecture: investigation of a subunit of chromatin by dark field electron microscopy. Proc Natl Acad Sci U S A. 1975 Jul;72(7):2691–2695. doi: 10.1073/pnas.72.7.2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardian J. K., Isenberg I. Preparative gel electrophoresis: detection, excision, and elution of protein bands from unstained gels. Anal Biochem. 1978 Nov;91(1):1–12. doi: 10.1016/0003-2697(78)90811-4. [DOI] [PubMed] [Google Scholar]
- Müller U., Zentgraf H., Eicken I., Keller W. Higher order structure of simian virus 40 chromatin. Science. 1978 Aug 4;201(4354):406–415. doi: 10.1126/science.208155. [DOI] [PubMed] [Google Scholar]
- Nelson P. P., Albright S. C., Wiseman J. M., Garrard W. T. Reassociation of histone H1 with nucleosomes. J Biol Chem. 1979 Nov 25;254(22):11751–11760. [PubMed] [Google Scholar]
- Pardon J. F., Worcester D. L., Wooley J. C., Cotter R. I., Lilley D. M., Richards R. M. The structure of the chromatin core particle in solution. Nucleic Acids Res. 1977 Sep;4(9):3199–3214. doi: 10.1093/nar/4.9.3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer D. S., Singer M. F. Studies on the interaction of H1 histone with superhelical DNA: characterization of the recognition and binding regions of H1 histones. Nucleic Acids Res. 1976 Oct;3(10):2531–2547. doi: 10.1093/nar/3.10.2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smart J. E., Bonner J. Studies on the role of histones in relation to the template activity and precipitability of chromatin at physiological ionic strengths. J Mol Biol. 1971 Jun 28;58(3):675–684. doi: 10.1016/0022-2836(71)90032-5. [DOI] [PubMed] [Google Scholar]
- Thoma F., Koller T. Influence of histone H1 on chromatin structure. Cell. 1977 Sep;12(1):101–107. doi: 10.1016/0092-8674(77)90188-x. [DOI] [PubMed] [Google Scholar]
- Thomas J. O., Kornberg R. D. An octamer of histones in chromatin and free in solution. Proc Natl Acad Sci U S A. 1975 Jul;72(7):2626–2630. doi: 10.1073/pnas.72.7.2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd R. D., Garrard W. T. Overall pathway of mononucleosome production. J Biol Chem. 1979 Apr 25;254(8):3074–3083. [PubMed] [Google Scholar]
- Todd R. D., Garrard W. T. Two-dimensional electrophoretic analysis of polynucleosomes. J Biol Chem. 1977 Jul 10;252(13):4729–4738. [PubMed] [Google Scholar]
- Worcel A., Benyajati C. Higher order coiling of DNA in chromatin. Cell. 1977 Sep;12(1):83–100. doi: 10.1016/0092-8674(77)90187-8. [DOI] [PubMed] [Google Scholar]
- Zweidler A. Resolution of histones by polyacrylamide gel electrophoresis in presence of nonionic detergents. Methods Cell Biol. 1978;17:223–233. [PubMed] [Google Scholar]