Abstract
Purpose: Deformable registration of medical images often requires initial rigid alignment. Because of variations in the articulation of bony structures, rigid alignment can capture only limited regions of the image. We propose a method that allows us to compensate for misalignment of mobile parts, which leads to improved accuracy of deformable registration. The method is based on matching landmarks using radial basis functions (RBF) with adaptive radius.
Methods: Based on the assumption that the compactly positioned landmarks likely delineate an anatomic structure whose position needs to be corrected, the algorithm incorporates unsupervised clustering of landmarks based on their positions within the reference image. It calculates an appropriate RBF radius based on the set of pairwise distances between landmarks within the cluster. The algorithm distinguishes between clusters of different size and between clusters of spherical and elongated shape, and assigns the optimal RBF radius for each cluster in order to restrict the deformation field to the closest vicinity of the structure of interest.
Results: Experiments with synthetic images demonstrate sensitivity of registration results to the choice of the radius of RBF support. We have statistically validated the methods on a large set of pulmonary landmarks. We also tested the method on medical use cases that show that it is potentially advantageous for initial registration of images with large spatial dislocations.
Conclusions: The results of registration of CT images demonstrate that an automated selection of the RBF radius simplifies the registration routine and improves the registration quality. The selection is based on two criteria of preserving diffeomorphism of deformation and localization of the deformation within a desired area of the image.
Keywords: image registration, point landmark, manual, radial basis functions, adaptive algorithm
INTRODUCTION
Deformable registration of medical images is a necessary tool for addressing various therapeutic problems, and there is an increasing demand for fast and accurate registration methods. Modern registration algorithms, both parametric and nonparametric, produce high quality registration results in a reasonable time.1 To achieve better accuracy of deformable registration it is usually a requirement to have a prior rigid registration to minimize an initial distance between the images. However, rigid-body approximation usually fails when applied to images including body parts of substantial relative mobility. As a result, better alignment of some of them causes larger misalignment of the others. For example, in head and neck region, there are at least four bony structures that can move independently: the skull, mandible, upper and lower neck.2, 3 The relative position variation of these parts makes atlas-based registrations especially challenging. For intrasubject images, a variation of mutual position of the parts might increase during the course of radiotherapy because patient weight loss leads to a loosening of immobilization devices. In the pelvic region, rigid registration is usually done with respect to bony structures. The soft tissue organs, however, can be largely misaligned due to natural volume variation of the bladder and rectum. Therefore, rigid registration sometimes does not serve as the best initial alignment and an additional intermediate step is needed to proceed further with deformable registration.
In this paper, we propose a landmark-based method for initial alignment of images with large spatial dislocations. The method is based on well-established radial basis function (RBF) formalism.4 The choice of the RBF shape is dictated by the adopted model of the deformation linking the reference and target images. Physical models, such as minimization of elastic energy, lead to thin-plate splines (TPS),5 which have global support. Treating registration as an abstract interpolation problem removes the physical constraint, and various decreasing shapes of RBFs have been suggested, including multiquadrics,4 Gaussian,6, 7 Wendland,8, 9 and Wu10 functions. Kohlrausch et al.11 suggested a hybrid approach, by formally postulating a Gaussian distribution of the force around a point landmark in the elastic medium, and explicitly calculating the resulting displacement field, which can be expressed as a rather complicated decreasing, radially symmetric function. The kernel width of this function depends both on the width of the underlying force distribution, and the elastic moduli of the material. Overall, the choice of the RBF shape is largely left to the expert judgment of the algorithm designer.
Direct interpolation of the displacement field using RBF requires solving a system of linear equations to find the weights of individual functions. That leaves the algorithm designer with very few degrees of freedom, namely, the mathematical shape of the radial basis functions and the radius of RBF support. Given the RBF shape, however, explicit criteria for choosing the RBF radius can be established. First, the transformations must be smooth and preserve image topology, which requires a positive Jacobian of the transformation everywhere in the image. Obviously, RBF with small radius results in large field gradients, and can easily lead to topology violations and noninvertibility. Kohlrausch et al.11 found a lower bound for the kernel width of the Gaussian elastic force that guarantees the preservation of topology, by enforcing the strictly positive Jacobian for displacement field of a single landmark. However, applying this constraint to each point separately is not sufficient to guarantee the topology preservation of the whole transformation. To assure the invertibility of the landmark-based transformation field, constrained optimization for a width of the Gaussian force can be used.12, 13 Similarly, Rohde et al.10 proposed an iterative multiscale registration algorithm using Wu's polynomial RBFs, and derived a constraint on the norm of the RBF coefficients ensuring positiveness of the Jacobian. The RBF radius was selected automatically based on the size of the misregistered regions and was identical for all RBFs. As mentioned above, all of these methods establish the lower bound on the radius.
Second, a very large RBF radius, even though it satisfies all possible invertibility conditions, is not suitable for alignment of independently moving body parts. Indeed, if the RBF radius is comparable with image dimensions, all of the landmarks begin to influence all voxels. This is undesirable if only a certain region needs realignment.
Therefore, the lower limit on the RBF radii is determined by the invertibility condition, while the upper limit is set by the requirement to keep the image alignment local and independent. Both constraints are therefore amenable to automated treatment, thus eliminating the user input for the RBF radius. Moreover, if the RBF radius is determined automatically, there is no need to keep it constant across all landmarks, as different radii do not add any computational complexity.
Recently, Yang et al.14 suggested that the RBF radius can be determined from the distance between the landmarks in the reference image. In 2D, they considered the deformation field needed to register two landmarks separated by a distance L, and found the RBF radius ρ from the condition that the inflexion, or saddle point of the deformation field is unique and lies exactly halfway between the landmarks. For a specific shape of truncated TPS function, they found ρ = kL(2e−3/2), where k ≈ 3 is an empirically determined constant to ensure invertibility. For the case of multiple landmarks, they defined Li as Li = minj ≠ idij, where dij is the Euclidean distance between landmarks i and j on the reference image, and calculated ρi accordingly. Using artificially deformed brain images, Yang et al. showed that RBF with adaptive radii result in a better registration than RBF of equal radii, either large or small.
Here, we propose a method for initial image alignment that takes into account independent movement of body parts, and is accelerated by an adaptive, automatic determination of RBF radii. Landmark positions are grouped together, reflecting the user's desire to align specific, localized areas within the image, but allows a different radius for each group. This method provides a smooth crossover from RBF-based registration with a constant radius for all landmarks to the one with a unique radius for each landmark. Its advantage is an independent alignment of large parts of images considered as separate entities.
Considering all-to-all pairwise distances between the landmarks, we automatically split the landmarks into distinct clusters and second, we determine the RBF radii (constant for all landmarks within a cluster) from within-cluster distances. Furthermore, to align linear anatomic structures such as the spine, we analyze cluster shape (anisotropy) and treat spherical and elongated clusters differently. Therefore, we are able to substantially simplify landmark-based registration procedure by assigning the RBF radius automatically for each manually identified misaligned region.
METHOD
Let us consider two 3D images, reference R and target T with a position of each voxel given by a vector with coordinates (x, y, z).
Radial basis functions
Spatially limited ψ31-functions of Wendland have been applied to registration of medical images that differ only locally9
| (1) |
where the support is determined by the radius ρ. This polynomial function is smooth, differentiable and strongly resembles the Gaussian function.
The landmark displacement field u at position is a linear combination of RBF, each RBF being parameterized by the distances from the point to the RBF origin. The origins of each RBF is the position of the landmark in the reference image, , so we can write
| (2) |
where is a unit vector along p-axis, M is the total number of landmarks in the reference image.
The coefficients αip in Eq. 2 are found from the interpolation condition that requires an exact matching of the corresponding landmarks on reference () and target () images
| (3) |
This condition determines the following set of 3 × M linear equations
| (4) |
where p = x, y, z and 1 < j < M, which we solve numerically for coefficients αip by matrix inversion using singular value decomposition (SVD).
Adaptive radius from landmark clustering
In this section, we present an algorithm for automatic choice of the RBF radius. The algorithm involves analyzing pairwise distances between landmarks in the reference image,14 and choosing the minimum radius so that RBF from neighboring landmarks would overlap.10, 14 It also distinguishes among the regions of the image that require different radii.
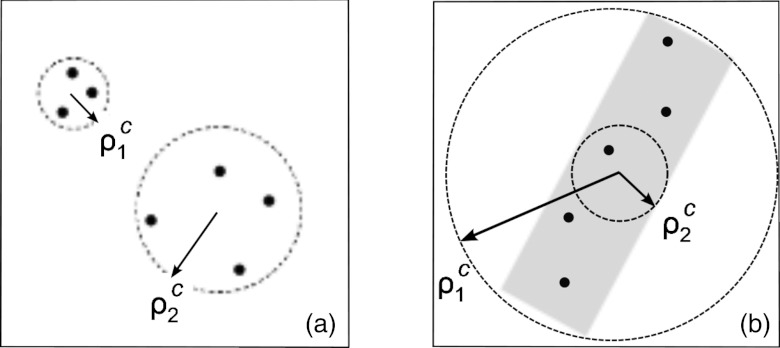
The later problem is somewhat harder, as the algorithm must identify separate regions requiring correction, and determine radii accordingly. As shown in Fig. 1a, our expectation for the choice of radii would be to use a smaller radius, ρ1, for a closely located group of landmarks at the top left corner, and a larger radius, ρ2, for the sparse group at the bottom right corner. Such a choice would ensure a well-localized correction. Automatic partitioning of landmarks into separate groups is a nontrivial task, but can be achieved by unsupervised clustering. Here, we assume that the expert user has manually specified the number of clusters based on the anatomic structures that need correction, but in principle this number can be determined automatically as well.15 Once the number of groups of landmarks is known, grouping of landmarks can be achieved by k-means++ clustering algorithm.16, 17 After the clusters of Mc landmarks are identified, the average pairwise distance within each cluster is used as the appropriate RBF radius, ρc,
| (5) |
where dij is the Euclidean distance between landmarks i and j on the reference image.
Figure 1.
Schematic representation of (a) two landmark clusters requiring different RBF radii, ; (b) a cluster of anisotropic shape with two possible definitions of RBF radius, .
All RBF centered on landmarks belonging to the same cluster are assigned the same radius. For clusters containing just one landmark, a default constant radius value ρc = 50 mm is used. Since in practice the total number of landmarks is at most several dozen, the time required to run the clustering algorithm is negligible with respect to the time it takes to interpolate the vector field and warp the target image.
Shape-dependent adaptive radius
The algorithm outlined above works well for quasispherical clusters of landmarks, where the dimensions of the clusters in every direction are roughly the same. However, as shown in Fig. 1b, elongated clusters occurring in practice by marking bones or vessels require a different approach. In this case, the average pairwise distance between landmarks is dominated by the pairs of landmarks situated at the opposite ends of the cluster, , and results in an overestimate of the optimum RBF radius. Using such a large radius would strongly deform the image in directions perpendicular to the cluster axis and is clearly undesirable.
Therefore, to optimize the RBF radius we distinguish between clusters of round (isotropic) and elongated (anisotropic) shape. For elongated anisotropic clusters, we define the RBF radius ρc as the maximum distance across all pairs of nearest neighboring landmarks,
| (6) |
so the deformation is concentrated along the cluster rather than in the perpendicular direction [ in Fig. 1b].
To distinguish between spherical-like and elongated clusters, we first calculate the principal moments of inertia Ii, i = 1, 2, 3 of each cluster, assuming all landmark have an equal mass of 1. From the moments of inertia, we calculate the principal radii of inertia , and select the minimum and maximum ones Lmin and Lmax. Anisotropy of the cluster is then defined by the shape factor s,
| (7) |
The shape factor s equals zero for spherical clusters, when all of the principal moments of inertia are equal, and equals one for very long, slender clusters, having very different moments of inertia along the different axes.
Therefore, to ensure a continuous change of the adaptive RBF radius as cluster shapes change from spherical to strongly elongated, we define the adaptive radius by
| (8) |
Equation 8 has two free parameters, the exponent a of the asymmetry parameter, and the prefactor b. Statistical testing on pulmonary images using a grid of small integer values for a and b showed that the best registration accuracy was achieved at b = 2, a = 4. For a = 2, variation of b between 1 and 4 reduced registration accuracy within 20% off its optimum value at b = 2. Changing a for b = 2 showed that the registration accuracy was not very sensitive to a, although a very shallow minimum at a = 4 was found. The optimality of a = 4 was further confirmed by visual analysis of the registration of the clinical images with strongly anisotropic clusters.
RESULTS
Registration of synthetic images
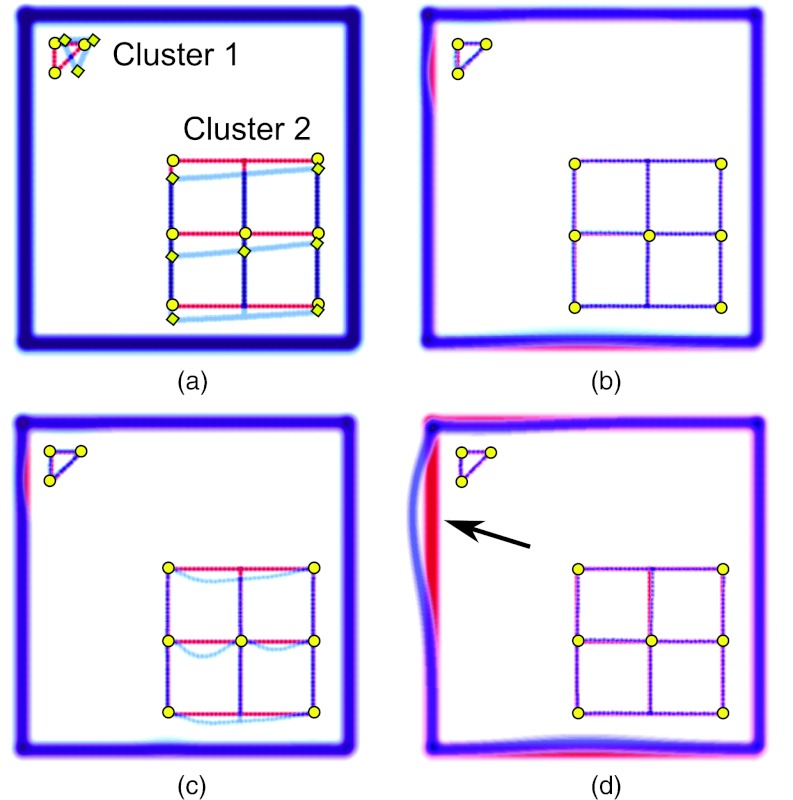
To test our algorithm for adaptive choice of the RBF radius, we generated synthetic images mimicking misalignment of two sites of substantially different sizes, as seen in Fig. 2a, and used our algorithm to register them. The only information supplied to the algorithm was the coordinates of landmarks, and the number of misregistered regions. The k-means++ clustering algorithm correctly partitioned the landmarks between the small and large clusters. The calculated RBF radius for the small cluster is much smaller than that of the large cluster [see Fig. 2b].
Figure 2.
Example of landmark-based image registration with two landmark clusters of essentially different sizes. (a) Overlaid reference and target images; the size of Cluster 1 is smaller than the size of Cluster 2; (b) registration using clustering algorithm and Wendland RBF with two calculated radii; (c) registration using Wendland RBF with small cluster radius for all landmarks; (d) registration using Wendland RBF with large cluster radius for all landmarks. The voxel grid is 500 × 500.
If one chooses either of these radii for all of the RBF, registration fails as shown in Figs. 2c, 2d. For the radius ρ = 87 [Fig. 2c], the small site is accurately aligned; the large site remains misaligned because matching of landmarks with strongly localized RBF of small radius leads to undesirable distortion of the lattice grid between the landmarks.
For the larger radius ρ = 343 [Fig. 2d], both sites are registered correctly. The outer thick frame represents the image neighborhood around the clusters that is initially aligned and; therefore, any distortion of the alignment will cause a degradation of overall registrations. As shown in Fig. 2d, the deformation centered on the smaller cluster extends over a large distance causing misalignment of the frame (shown by an arrow). These results are in qualitative agreement with the previously obtained results.14
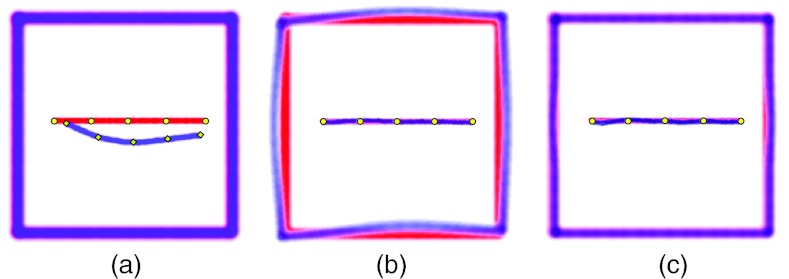
In Fig. 3, we present the results of application of our shape-dependent algorithm to a synthetic image. The images mimic misalignment of a detail with anisotropic shape. The algorithm recognized the anisotropy of the cluster and set the RBF radius to the maximum distance between nearest neighboring landmarks, 140 voxels in this case. The result of registration shown in Fig. 3c demonstrates a perfect alignment of the detail.
Figure 3.
Example of landmark-based image registration with one landmark cluster of anisotropic shape. (a) Overlaid reference and target images; (b) registration using shape-independent algorithm and Wendland RBF with calculated ; the domain of deformation expands too far leading to a misalignment of the frame; (c) registration using clustering shape-dependent algorithm and Wendland RBF with calculated ρc = 140. The voxel grid is 500 × 500.
For comparison, in Fig. 3b we show the result of registration using the algorithm that does not take into account cluster anisotropy. Since the average pairwise distance is large due to a contribution from the distance between the ends of the long cluster, the calculated RBF radius is very large, ρ = 416. The large radius, in turn, translates into a long-range deformation causing unwanted distortions of the image far from the detail being registered.
Statistical analysis using pulmonary images
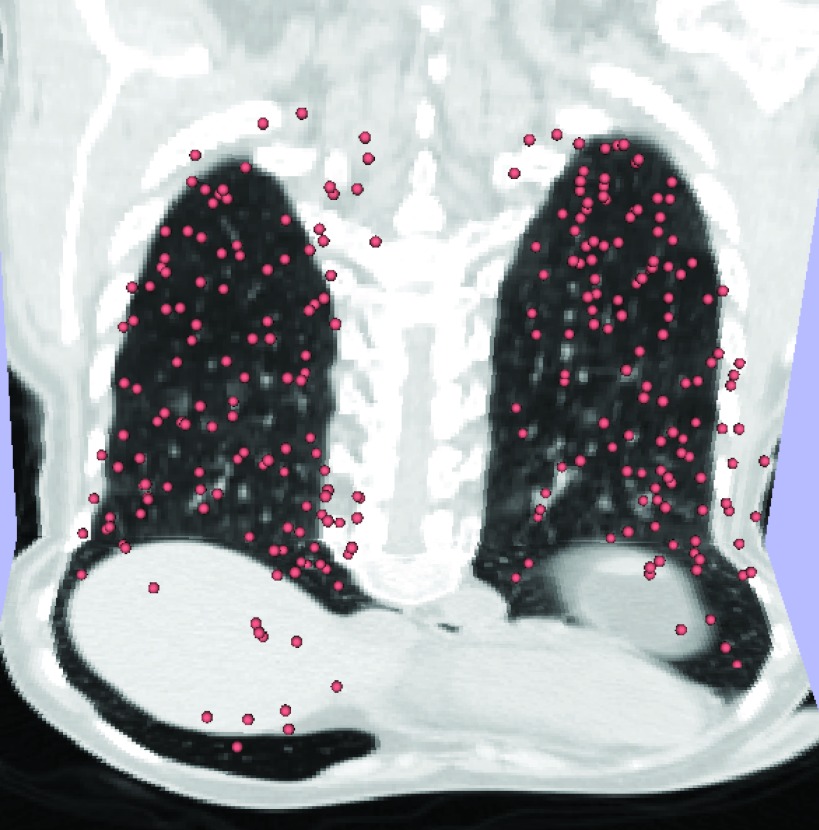
To assess the performance of our radius selection method, and compare it with fixed radius RBF registration and the adaptive method by Yang14 we did an analysis of a large number of registrations, using the publicly available sets of pulmonary landmarks from the DIR-Lab web site.18 The data set consists of 4DCT images, with 300 landmarks manually identified on each extreme inhalation and exhalation phases of the breathing cycle19 (see Fig. 4).
Figure 4.
Visualization of 300 manually placed landmarks on 3D image of extreme exhalation phase of 4DCT (Ref. 18).
We first present the results of our algorithm applied to five pairs of images acquired from five different patients. We randomly selected M = 10 and 50 landmarks out of 300 to be used for registration. For each M, 100 different random choices for the set of landmarks were considered. We performed registration for each landmark set, and calculated the average distance between the remaining 290 and 250 landmarks (error of registration). The smooth solid curves in Fig. 5 show that the registration accuracy initially improves as the radius increases, and reaches a plateau at the radius of about 200 mm and 150 mm, respectively.
Figure 5.
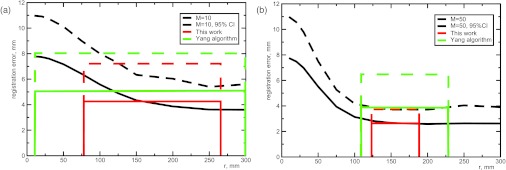
(a) Registration error of pulmonary images as a function of the RBF radius averaged among 5 cases registered using 10 landmarks. Root mean square average registration error (solid) and 95% confidence interval (dashed) are shown for the fixed RBF radius, our algorithm and the Yang's algorithm (Ref. 14). Our adaptive algorithm clearly outperforms the Yang's algorithm in registration accuracy. The horizontal ranges show the minimum and maximum radii selected by the two algorithms. (b) Same data, but using 50 landmarks for registration.
For comparison, we calculated the error of registration using our clustering algorithm. Setting the number of clusters to two, corresponding to the left and right lungs, we first determined the RBF radius for each cluster, and then performed the registration. For each set of images, the procedure was repeated for 100 different random choices of M = 10 or 50 landmarks, and the error of registration of the remaining 290 or 250 landmarks has been determined. We chose the root mean square error of registration, and the 95% confidence interval RCI as the quality metrics. The meaning of the confidence interval is that a landmark will be registered to within RCI of its correct position (or better) with a probability P = 0.95; unlike the average landmark error, RCI provides a measure of the worst-case, rather than average registration.
The rectangles in Fig. 5 show the minimum and maximum radii, and mean and 95% confidence intervals of the registration error observed over the five sets of images. First, we note that our fully automated procedure made a reasonable choice of the RBF radius, between 77 and 260 mm for M = 10 and between 122 and 188 mm for M = 50. The narrower range of radii for M = 50 is fully expected on statistical grounds, as clusters made of a larger number of landmarks become more similar. Remarkably, the radii selected by our algorithm lie near the transition to the plateau of the registration error, where a further increase in radius does not lead to an increase of registration accuracy. The average and confidence interval values provided by our algorithm are also consistent with constant radius ones.
We also compared the results of our algorithm to that of Yang et al.,14 setting k = 3. The results of the Yang algorithm applied to 100 random choices of landmarks on each of the 5 sets of images are shown in Fig. 5 in green. As the Yang algorithm selects the distance between the nearest landmarks as the radius, the minimum radius is consistently smaller and the maximum is larger than the adaptive radius based on clustering. As a consequence, the Yang radius might be either too small to match corresponding landmarks without excessive gradients or so large that the registration between the landmarks becomes ruined.
Table 1 compares the performance metrics of the two adaptive radius RBF algorithms. A good registration would have lower average error and a small confidence interval. As seen from Table 1, the adaptive-radius algorithm outperforms the Yang algorithm in every metric, for both 10 and 50 landmarks.
Table 1.
Comparison of average registration error and 95% confidence interval for registration error (in mm) of two adaptive radii algorithms.
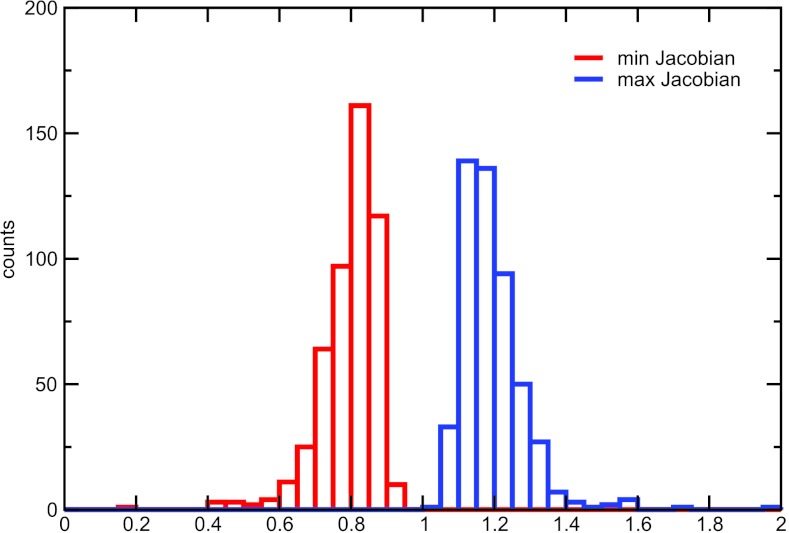
To check whether our adaptive algorithm provides a smooth and invertible registration, we have calculated the minimum and maximum values of the Jacobian of the deformation field. The histograms of the Jacobian (Fig. 6) demonstrate that the minimum value of the Jacobian remained positive in this test.
Figure 6.
Histograms of the minimum and maximum values of the Jacobian of the deformation field for our adaptive algorithm, M = 10. Five hundred independent registrations with random choices of landmarks were performed (100 registrations for each of the 5 images).
By performing a rigorous statistical evaluation of registration accuracy, we established that our adaptive-radius algorithm results in significantly better registrations than a previously reported algorithm,14 and is similar conventional fixed-radius RBF in this test. The performance gains were observed for the case of pulmonary landmarks, with a rather uniform spatial distribution of landmarks, Fig. 4. We expect that our algorithm will perform significantly better in case of registrations of elongated or asymmetrical anatomical structures. Unfortunately, we are not aware of large-scale public datasets that would allow thorough statistical testing of the clustering algorithm in a more realistic setting.
Registration of CT images
Having validated our method statistically, we will now present results of registration of clinical case images. We use our clustering algorithm to register CT images of the upper body which are common in radiotherapy of head and neck cancer. The registration of these images ought to compensate for position error. The images include the head and chest with different head and neck position resulting in a pitch and yaw of the head and bending of the spine. Since angular rotation of the head is independent of the position of the chest and shoulders, rigid-body registration cannot be applied to the whole image. At the same time, deformable registration fails because of a large spatial shift of the head position. To solve this registration problem, we used a combination of two methods. Initially, a coarse registration was performed using our proposed clustering algorithm with Wendland RBF, and a fine registration was then performed by deformable B-spline registration.20
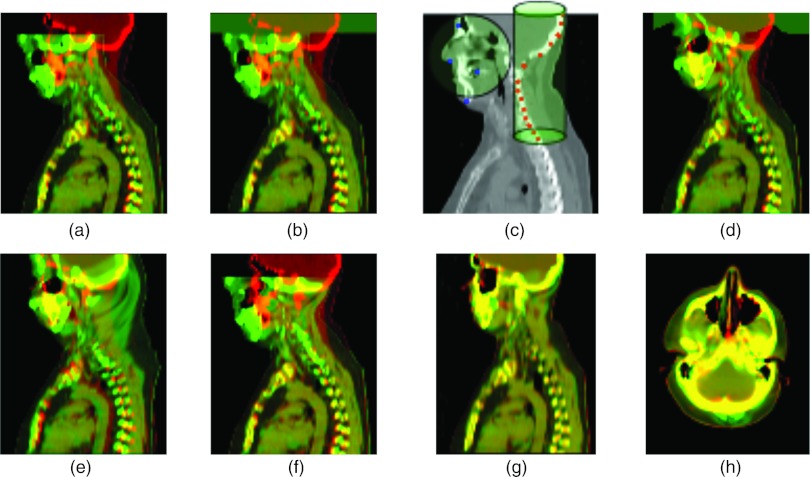
In Fig. 7a, we show two overlaid images, reference (red) and target (green). B-spline registration of downsampled images with a very course grid fails to register a large spacial shift of the head [Fig. 7b]. To apply RBF method, we placed two groups of landmarks to roughly align bony structures of the face and the spine [see Fig. 7c]. The facial landmarks are distributed among several slices and the spine landmarks are placed on two sagittal slices. The algorithm with a given number of two clusters successfully divides the landmarks between them and automatically assigns the RBF radii for both clusters. The cluster that includes landmarks uniformly distributed in 3D around the face is defined as spherical and is assigned a large radius in order to include the whole front head area. The other cluster includes landmarks placed along the spine and skull region on the sagittal view and is assigned a small radius to include an elongated area around the upper spine. The parameters of the clusters are given in Table 2.
Figure 7.
Registration of intra-subject CT images of upper body. (a) Reference image overlaid with target image, showing pinched position of the head; (b) overlaying warped image obtained by B-spline registration with 200 × 200 × 200 mm grid size and (16 × 16 × 8) resolution; (c) a slice of the reference image with landmarks grouped in two clusters, spherical cluster includes landmarks placed around the face and cylindrical cluster with landmarks placed along the skull and the spine; (d) overlaying warped image obtained by landmark-based registration with adaptive radius; (e) overlaying warped image obtained by landmark-based registration with a large constant radius; (f) overlaying warped image obtained by Yang's method (Ref. 14) with RBF radius individual for each landmark; (g)–(h) result of B-spline registration performed after initial alignment of two landmark clusters with adaptive radii, sagittal and axial views.
Table 2.
Characteristics of landmark clusters for the case shown in Fig. 7.
| Cluster | Num. landmarks | Dmax (mm) | Dmean (mm) | ρ (mm) |
|---|---|---|---|---|
| Spherical | 25 | 152 | 78 | 179 |
| Cylindrical | 29 | 254 | 100 | 57 |
The result of landmark-based registration with adaptive radius is shown in Fig. 7d. Sagittal view demonstrates that the position of the head is recovered without any change in the chest position. Using RBF registration with a large constant radius, we are able to recover the head position but previous accurate registration of the chest become corrupted [Fig. 7e]. With the Yang algorithm we were not able to register a pitched head [Fig. 7f].
After initial alignment of two landmark clusters [Fig. 7d], we performed B-spline deformable registration and the final result is shown in Figs. 7g, 7h where both sagittal and axial views demonstrate high-quality registration.
DISCUSSION
Our results clearly demonstrate the feasibility of an automatic choice of RBF radius in landmark-based registration of medical images. In principle, the radius could be determined using (i) information about image intensities, (ii) relative positions of landmarks within reference (or target) image, or (iii) the distance Δ between the corresponding landmarks on the reference and target images. As follows from Rohde et al.,10 the first option is more computationally expensive than distance-based methods. To rationalize our choice of intra-, rather than interimage landmark distances, we note that information about the shape of misaligned anatomic features is implicitly contained in the relative positions of the landmarks within the reference image, rather than in the distances between the corresponding landmarks on reference and target images. Given a RBF radius, large distances between the corresponding landmarks are automatically addressed by the larger values of the RBF coefficients αip, and do not always indicate the need of a large RBF radius.
Invertibility and diffeomorphism of the resulting displacement field impose significant constraints on the RBF radius.10, 11, 14 Since in our case the RBF radius is determined automatically, these constraints are at least as important as in the case of manual selection of the radius. Generally, the Jacobian of a RBF-based transformation depends on ρ, as well as on the distance(s) Δ between the landmarks being matched. For an isolated landmark, an exact lower bound of ρ/Δ ensuring invertibility can be established.9 Contrary, no exact result is known for an arbitrary spatial distribution of multiple landmarks, where the extreme values of the Jacobian depend on ρ, all possible distances Δi, i = 1…M, and the spatial arrangement of the landmarks. Simulations9 reveal that in specific geometries the (Wendland) RBF radius must be three to four times larger than landmark displacement Δ. This condition was satisfied in our experiments, and registration of clinical images did not reveal visible folds or other artifacts.
Further studies of the mutual influence of RBF radius ρ, landmark displacement D, and landmark geometry on the Jacobian of the image transformation will establish specific lower limits on RBF radii, which should be incorporated into future versions of this algorithm.
CONCLUSIONS
Successful image registration using radial basis functions crucially depends on the proper choice of their radii. Here, we presented an algorithm for automated, adaptive choice of the RBF radii. Using landmark locations, and the user-supplied number of clusters the landmarks must be partitioned into, our algorithm assigns landmarks to the clusters, and calculates the appropriate RBF radius for each cluster. This procedure is especially advantageous when the images contain misaligned structures of very different sizes, and the traditional one-radius-fits-all approach leads to noticeable artifacts. In addition, our algorithm is able to distinguish between isotropic and elongated clusters, and appropriately decrease the RBF radius for the elongated clusters. Together, our results prove that an automatic choice of RBF radius can be achieved in a simple and computationally efficient way, and provide a proof of concept demonstration of semiautomated image registration using RBF, with minimum user input. Clinical examples on upper body images registration demonstrate the potential of our method.
ACKNOWLEDGMENTS
Funding for this work was provided by an Ira J. Spiro translational research award, by the National Institutes of Health (NIH) NCI 6-PO1CA21239, and by the Federal share of program income earned by MGH on NIH NCI-C06CA059267. This work is part of the National Alliance for Medical Image Computing (NA-MIC), funded by the National Institutes of Health through the NIH Roadmap for Medical Research, Grant No. 2-U54EB005149-06.
References
- Hill D. L., Batchelor P. G., Holden M., and Hawkes D. J., “Medical image registration,” Phys. Med. Biol. 46, R1–R45 (2001). 10.1088/0031-9155/46/3/201 [DOI] [PubMed] [Google Scholar]
- Polat B., Wilbert J., Baier K., Flentje M., and Guckenberger M., “Nonrigid patient setup errors in the head-and-neck region,” Strahlenther. Onkol. 183, 506–517 (2007). 10.1007/s00066-007-1747-5 [DOI] [PubMed] [Google Scholar]
- Ove R., Cavalieri R., Noble D., and Russo S., “Variation of neck position with image-guided radiotherapy for head and neck cancer,” Am. J. Clin. Oncol. 35, 1–5 (2012). 10.1097/COC.0b013e3181fe46bb [DOI] [PubMed] [Google Scholar]
- Buhmann M. D., “Radial basis functions,” Acta Numerica 9, 1–38 (2000). 10.1017/S0962492900000015 [DOI] [Google Scholar]
- Bookstein F. L., “Principal warps: Thin-plate splines and the decomposition of deformations,” IEEE Trans. Pattern Anal. Mach. Intell. 11, 567–585 (1989). 10.1109/34.24792 [DOI] [Google Scholar]
- Shusharina N. and Sharp G., “Analytic regularization for landmark-based image registration,” Phys. Med. Biol. 57, 1477–1498 (2012). 10.1088/0031-9155/57/6/1477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arad N., Dyn N., Reisfeld D., and Yeshurun Y., “Image warping by radial basis functions: Application to facial expression,” CVGIP: Graph. Models Image Process. 56, 161–172 (1994). 10.1006/cgip.1994.1015 [DOI] [Google Scholar]
- Arad N. and Reisfeld D., “Image warping using few anchor points and radial functions,” Comput. Graph. Forum 14, 35–46 (1995). 10.1111/1467-8659.1410035 [DOI] [Google Scholar]
- Fornefett M., Rohr K., and Stiehl H. S., “Radial basis functions with compact support for elastic registration of medical images,” Image Vis. Comput. 19, 87–96 (2001). 10.1016/S0262-8856(00)00057-3 [DOI] [Google Scholar]
- Rohde G. K., Aldroubi A., and Dawant B. M., “The adaptive bases algorithm for intensity-based nonrigid image registration,” IEEE Trans. Med. Imag. 22, 1470–1479 (2003). 10.1109/TMI.2003.819299 [DOI] [PubMed] [Google Scholar]
- Kohlrausch J., Rohr R., and Stiehl H. S., “A new class of elastic body splines for nonrigid registration of medical images,” J. Math. Imaging Vision 23, 253–280 (2005). 10.1007/s10851-005-0483-7 [DOI] [Google Scholar]
- Pekar V. and Gladilin E., “Deformable image registration by adaptive Gaussian forces,” Lect. Notes Comput. Sci. 3117, 317–328 (2004). 10.1007/b98995 [DOI] [Google Scholar]
- Pekar V., Gladilin E., and Rohr R., “An adaptive irregular grid approach for 3D deformable image registration,” Phys. Med. Biol. 51, 361–377 (2006). 10.1088/0031-9155/51/2/012 [DOI] [PubMed] [Google Scholar]
- Yang X., Zhang Z., and Pei J., “Elastic image deformation using adaptive support radial basis function,” in Proceedings of the International Conference on Wavelet Analysis and Pattern Recognition, Hong Kong, 2008 (IEEE, 2008), pp. 158–162.
- Bischof H., Leonardis A., and Selb A., “MDL principle for robust vector quantization,” Pattern Anal. Appl. 2, 59–72 (1999). 10.1007/s100440050015 [DOI] [Google Scholar]
- MacQueen J. B., “Some methods for classification and analysis of multivariate observations,” in Proceedings of the Fifth Berkeley Symposium on Mathematical statistics and Probability, Berkeley, CA, 1967 (University of California Press, 1967), pp. 281–297.
- Arthur D. and Vassilvitskii S., “k-means++: The advantages of careful seeding,” in Proceedings of the Eighteen Annual ACM-SIAM Symposium on Discrete Algorithms, New Orleans, LA, 2007 (Society for Industrial and Applied Mathematics, 2007), pp. 1027–1735.
- http://www.dir-lab.com.
- Castillo R., Castillo E., Guerra R., Johnson V. E., McPhail T., Garg A. K., and Guerrero T., “A Framework for evaluation of deformable image registration spatial accuracy using large landmark point sets,” Phys. Med. Biol. 54, 1849–1870 (2009). 10.1088/0031-9155/54/7/001 [DOI] [PubMed] [Google Scholar]
- Rueckert D., Sonoda L. I., Hayes C., Hill D. L. G., Leach M. O., and Hawkes D. J., “Nonrigid registration using free-form deformations: Application to breast MR images,” IEEE Trans. Med. Imag. 18, 712–721 (1999). 10.1109/42.796284 [DOI] [PubMed] [Google Scholar]