Abstract
During embryogenesis the establishment of chromatin states permits the implementation of genetic programs that allow the faithful development of the organism. However, these states are not fixed and there is much evidence that stochastic or chronic deterioration of chromatin organization, as correlated by transcriptional alterations and the accumulation of DNA damage in cells, occurs during the lifespan of the individual. Whether causal or simply a by-product of macromolecular decay, these changes in chromatin states have emerged as potentially central conduits of mammalian aging. This review explores the current state of our understanding of the links between chromatin organization and aging.
Keywords: chromatin, epigenetics, histone, heterochromatin, senescence
Chromatin as a potential modulator of aging
One of the hallmarks of aging is the loss of homeostatic mechanisms that offset the macromolecular wear and tear that occurs during a lifetime of exposure to low doses of extrinsic (ultraviolet and g-irradiation, nutritional input) and intrinsic (reactive oxygen species, telomere shortening, protein synthesis errors and replication/transcription-coupled DNA damage) stimuli [1]. Like other macromolecules in the cell, chromatin is subjected to these stresses that can impinge on its structural integrity and function [Figure 1]. The nucleosome is the repeat unit of chromatin and consists of a dimer of each core histone (H2A, H2B, H3 and H4)[2]. The physical addition and removal of the linker histone H1 and additional non-histone chromatin proteins facilitates transitions between higher order chromatin states during the cell cycle. These transitions are important for rendering DNA accessible for transcription, replication, and packaging it into highly condensed metaphase chromosomes for transmission during mitosis. Chromatin is the template of epigenetic mechanisms such as DNA methylation and histone modifications including lysine methylation, lysine acetylation, lysine ubiquitylation and serine/threonine phosphorylation. By definition, epigenetic modifications do not involve changes in the DNA sequence and should be heritable but reversible.
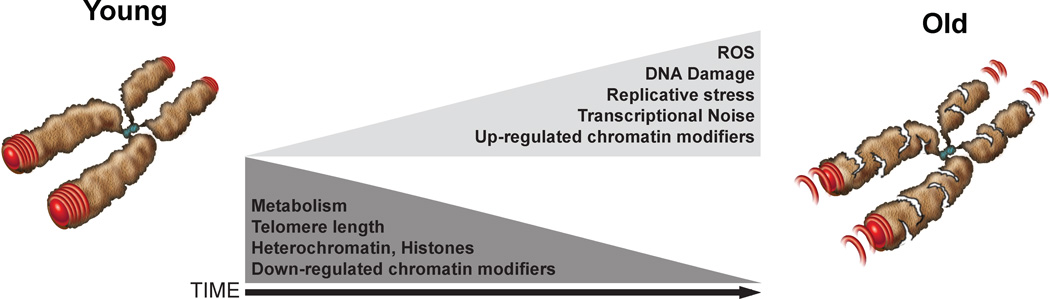
Figure 1. Aging is due to an increased disequilibrium in cellular homeostasis.
The decline in metabolic rates and telomere shortening over time can contribute to structural and gene expression changes that are associated with aging. By contrast, chronic exposure to reactive oxygen species (ROS), DNA damage and replicative stress can cooperatively cause elevated stochastic transcriptional noise. Structural changes in chromatin and the regulation of chromatin modifiers might be common denominators that underlie how these factors affect chromosomal stability and the cellular processes that drive the aging process.
It has been proposed that the aging phenotype is, at least in part, due to the progressive divergence from a youthful chromatin configuration to one that contributes to the molecular signatures of aging [3]. In humans, analyses of longevity data on Danish twins and other populations of related individuals indicate that 20–25% of the variation in adult lifespan can be attributed to genetic shifts between identical individuals [4]. This might be due to an increased impact of stochastic somatic mutations as survival extends into old age, including their influence on cognitive and physical ability. However, chromatin modification patterns increasingly diverged with age in monozygotic twins [5]. These changes in chromatin might underlie those subtle phenotypic variations that become more pronounced with extended survival of twins and other closely related individuals. This seminal observation lent additional support to the speculation that aging might be largely the remit of chromatin based epigenetic regulatory mechanisms. How chromatin is altered during aging has been extensively investigated, perhaps most notably with respect the reshaping of chromatin during the diverse contexts of cellular senescence. Cellular senescence is an irreversible state of cell-cycle arrest and is thought to reflect aging. In this review, we discuss recent developments that elaborate the functional links between chromatin, cellular senescence and longevity.
Age-associated deregulation of chromatin modifiers
Perhaps the most commonly invoked feature of aging is the increase in stochastic cell-to-cell variation in gene expression [6]. Additionally, a wide range of changes in the expression of chromatin modifiers has been identified in senescence and during aging. This can alter the levels and distribution of chromatin modifications throughout the nucleus and at aging-associated loci, causing the activation of physiological responses that promote aging [Table 1]. The following candidates might represent paradigms as to how chromatin and epigenetic mechanisms contribute to the wear and tear of tissues with age.
Table 1.
Chromatin changes correlated with aging.
| Histone Variant ¶ | Change with age | Factors Involved | Function | Organism | Aging Regimen | Ref. |
| H3 | ||||||
| - H3.1, H3.2 | down | SLBP, ASF1 | Replication-coupled chromatin assembly | Hs, Sc | Rep Sen, Chron | [71,73,93] |
| - H3.3 | up | HIRA | Transcription-coupled chromatin assembly | Hs | Rep Sen | [93] |
| H4 | down | SLBP, ASF1 | Replication-coupled chromatin assembly | Hs | Rep Sen, Chron | [71] |
| H2A | ||||||
| - H2A.1 | down | SWI, SNF | Chromatin assembly | Hs, Sc | Rep Sen | [73,93] |
| - H2A.2 | up | SWI, SNF | Chromatin assembly | Hs | Rep Sen | [93] |
| - γH2AX | up | ATM, ATR | DDR Signaling | Hs, Mus | Rep Sen, OIS | [71,94] |
| Modification § | Change with age | Factors Involved≠ | Function | Organism | Aging Regimen | Ref. |
| - H3K4me2/3 | down | ASH-2, MLL-1 (KMT2A), WDR-5 | Transcriptional activation | C.ele | Chron | [88] |
| - H3K9ac | up | TIP60 (KAT5), SIRT6? | Transcriptional activation | Hs, Mus | Rep Sen, Chron, HGPS | [41,53,60] |
| - H3K9me3 | down up |
SUV39H1/H2 (KMT1A/KMT1B), HP1α SUV39H1/H2 (KMT1A/KMT1B), HP1γ |
Heterochromatin formation Transcription regulation/DDR suppression |
Hs, Dm Hs |
Rep Sen, Chron, HGPS OIS |
[60,71,91] [66,69] |
| - H3K27me3 | down | EZH2 (KMT6), JMJD3 (KDM6B), UTX-1 (KDM6A) | Transcriptional Regulation | Hs, C.ele | OIS, Chron, CR | [31,95] |
| - H3K56ac | down | P300/CBP (KAT2A), HDAC1, SIRT2 | DNA replication/DNA damage/Transcription | Hs, Sc | Rep Sen, Chron | [71,73,76, 96] |
| - H4K5ac | down | P300/CBP (KAT2A), SATB-1 | Transcriptional activation | C.ele | Chron, CR | [97] |
| - H4K12ac | down | MYST4 (KAT6B), GCN5 (KAT2A), HDAC1 | Transcriptional activation | Mus | Chron | [98] |
| - H4K16ac | up down |
SAS2 (KAT8), SIR2 hMOF (KAT8), SIRT2 |
Telomere silencing/rDNA silencing Chromatin compaction (Mitosis/Transcription) |
Sc. Hs, Mus |
Chron Rep Sen, Chron, HGPS |
[76] [71] |
| - H4K20me2 | up | SUV420H1/H2 (KMT5A/KMT5B) | DNA damage/DNA replication | Hs | Rep Sen, Chron | [71,99] |
| - H4K20me3 | down | SUV420H1/H2 (KMT5A/KMT5B) | Heterochromatin formation | Hs | Rep Sen, Chron | [71] |
Abbreviations.
Organism Abbreviations: Hs (Homo sapiens), Sc (S. cerevisiae), Mus (Mus musculus), C.ele (C.elegans).
Aging Regimen Abbreviations: Res Sen; Replicative Senescence, OIS; Oncogene Induced Senescence, Chron; chronological aging, CR; Caloric Restriction, HGPS; Hutchison-Gilford Progeria Syndrome.
Factors Involved Abbreviations: SLBP; Stem-loop binding protein, ASF1; Anti-silencing factor-1, HIRA; HIR cell cycle regulator defective homolog A, SWI/SNF; SWItch/Sucrose Non-Fermentable, ATM; Ataxia-telangiectasia mutated, ATR; Ataxia-telangiectasia and Rad3 related, ASH2; absent-small or homeotic-2, MLL1; mixed lineage leukemia-1, WDR5; WD domain repeat protein 5, TIP60; tat-interactive protein 60kDA, SIRT6; silent mating type information regulation 2 homolog 6, SUV39h1/2; suppressors of variegation homologs 1 and 2, HP1a and HP1g; heterochromatin protein 1- alpha and gamma isoforms, EZH2; enhancer of zeste-2. JMJD3, jumonji domain containing 3, p300/CBP; C-terminal binding protein, HDAC1; histone deacetylase 1, SIRT2; silent mating type information regulation 2 homolog 2 SATB1; special AT-rich sequence binding protein 1, MYST4; Monocytic leukemia zinc finger protein-related factor, GCN5, General control of amino acid synthesis protein 5, HDAC1; histone deacetylase 1, SAS2; Something about silencing protein 2, SIR2; silent mating type information regulation 2, hMOF; ortholog of Drosophila males absent on the first, SUV420H1/H2; suppressors of variegation 4–20 homologs 1 and 2.
histone variants are indicated beneath individual core histone. i.e. core histone (H3), histone H3 variants (H3.1, H3.2, H3.3)
Histone modifications are presented according to the Brno nomenclature which distinguishes individual histone methylation states; mono-methylation (me1), di-methylation (me2) and tri-methylation (me3).
the new name of chromatin modifiers is indicated between brackets following previous conventional name; KAT; lysine acetyltranferase, KMT; lysine methyltransferase, KDM; lysine demethylase.
Epigenetic regulation of p16INK4A expression
p16INK4A (also known as CDKN2a) is an important regulator in the Retinoblastoma, pRb, tumor suppressor pathway [7]. The primary function of p16INK4A is to stifle proliferation by attenuating cyclin dependent kinase CDK4/CDK6-mediated phosphorylation of pRb. Increased expression of p16INK4a maintains pRb in its active hypo-phosphorylated state, sequestering E2F transcription factors to maintain cell-cycle arrest. The expression of p16INK4A increases in mammalian cells with senescence and age [8,9] and correlates with increased levels of cellular stress. As a result, p16INK4A has become indelibly associated with cellular senescence and was suggested as a biomarker of aging [9]. The link was recently bolstered in a study where p16INK4A-positive cells were specifically eliminated by directed programmed cell death (apoptosis) in the murine BubR1 haplo-insufficiency model of premature aging [10]. Purging p16INK4A-expressing senescent cells profoundly affected aging and healthspan. Eliminating p16INK4A-positive cells throughout life delayed the onset of age-related tissue degeneration and led to lifespan extension, but doing so only in aged adult mice also delayed and improved the manifest traits of aging [10]. These experiments strongly support the idea that p16INK4A expressing cells reflect aging in vivo.
Epigenetic chromatin modification at the p16INK4A gene participates in the regulation of senescence and aging. Particularly, the polycomb (PcG) and trithorax (TrxG) proteins maintain chromatin in 'off' or 'on' states, thereby preventing or promoting expression, respectively [2]. In young cells, the polycomb repressive complexes PRC1 and PRC2 establish repressive chromatin throughout the p16INK4A locus [11]. The PRC2 complex consists of embryonic ectoderm development (EED), suppressor of zeste 12 homolog (SUZ12), and enhancer of zeste 2 (EZH2), catalyzing tri-methylation of H3 at lysine 27 (H3K27me3) [12] and tri-methylation of H1 at lysine 26 (H1K26me3) [13]. This in turn recruits the PRC1 complex, comprising polycomb group proteins BMI1, RING1A, RING1B, MEL18 and chromobox homolog CBX7 [14], which mono-ubiquitylates histone H2A at lysine 119 (K119) and silences gene expression [15] [Figure 2a]. In fact, ectopic over-expression of BMI1 and CBX7 extends the proliferative lifespan of cells in vitro by suppression of p16INK4A [16,17].
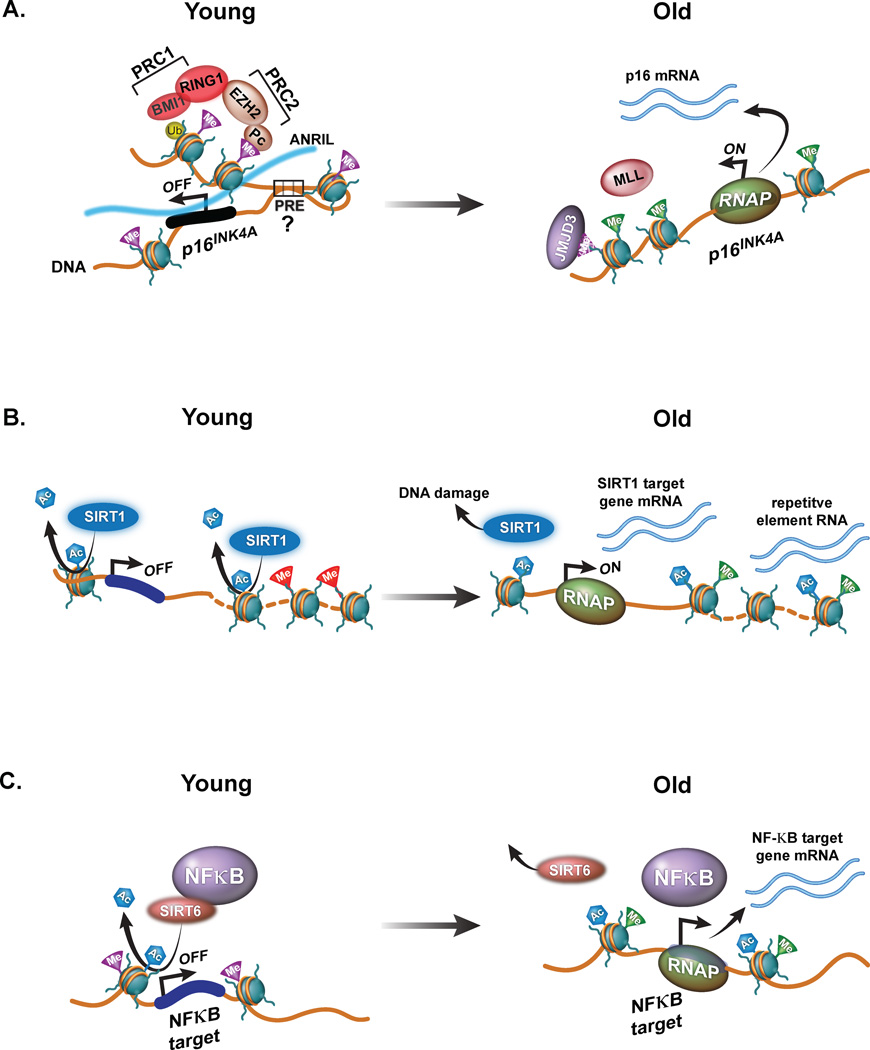
Figure 2. Chromatin modifiers linked with age-related transcriptional deregulation.
(a) Left: In young cells the repression of the p16INK4A gene is under the purview of the PRC1 and PRC2 polycomb repressive complexes. PRC2 consists of polycomb group proteins such as Pc and Enhancer of zeste-2, EZH2. EZH2 methylates chromatin at H3K27me3 (purple triangle) throughout the locus. PRC2, in turn, recruits the PRC1 complex, consisting of BMI1 and RING1 which ubiquitylate H2AK119 (yellow circle) and ‘lock in’ the inactivate state of the locus. The long non-coding RNA, ANRIL (shown as dark blue line) and looping between distal PREs might contribute in p16INK4A suppression. Right: In old cells this repressive domain switches from polycomb-mediated repression to trithorax (shown as mixed lineage leukemia 1, MLL1)-dependent transcriptional activation. First, H3K27me3 is removed by the H3K27me3 specific lysine demethylase, KDM Jumonji D3, JMJD3, and then MLL1 mediates H3K4me3 (green triangle) at the p16INK4A promoter to facilitate gene expression (RNA shown as blue lines).
(b) Left: in young cells SIRT1 localizes to repetitive elements (broken DNA line) and genes to deacetylate H1K26 (blue hexagon); this maintains transcriptional repression and promotes heterochromatic chromatin modifications such as H3K9me3 (red triangle). Right: following DNA damage, SIRT1 is mobilized to DNA damage sites to participate in DNA repair. This leads to activation of SIRT1-regulated genes and transcription of repetitive elements. As the expression of SIRT1 decreases, elevated transcriptional deregulation and the accumulation of unrepaired DNA damage lesions can promote aging.
(c) Left: in young cells SIRT6 binds to NFKB and blocks transcriptional activation by de-acetylating H3K9ac (blue hexagon) at NFKB target promoters. Transcriptional silencing of NFKB target promoters might be maintained by H3K27me3 (purple triangle)-dependent repression. Right: in old cells, SIRT6 might be titrated away from NFKB. As a result, the full transcriptional activation of NFKB and its target genes is unleashed. This is likely to involve the trithorax complex and MLL1-mediated H3K4me3 (green triangle).
This repressive chromatin domain might be reinforced by chromatin looping [18] and by retention of the long intergenic non-coding RNA (lincRNA) ANRIL [19]. The initial recruitment of PRC2 can occur by various means. It could involve tethering of PRC2 to ANRIL [20], or alternatively, BMI1 could recruit PRC2 to the 5’ regulatory domain (RDINK/ARF) from where PRC2 could then spread downstream to create the repressive chromatin domain [21]. RDINK/ARF was identified as a master transcriptional regulator of the p16INK4A locus [22] and recruitment of BMI1 via cell cycle division regulator 6 homolog (CDC6) to RDINK/ARF also appears to regulate the replication timing of the locus, implying a dual role for PcG in DNA replication and transcription at the p16INK4A locus [21]. In senescent and aged cells PRC1 and PRC2 dissociate from chromatin. This might be due to pRb inactivation and the imposition of the G1 checkpoint that negatively regulates E2F driven expression of EZH2 and BMI1 [23]. As the expression of EZH2 and BMI1 subsides, the expression of the histone lysine demethylase (KDM) Jumonji D3 (JMJD3) rapidly increases and removes p16INK4A-associated H3K27me3 [24]. This enables the association of the TrxG protein mixed-lineage leukemia 1 (MLL1), which catalyzes tri-methyl lysine 4 of histone H3 (H3K4me3) to promote gene expression [25] [Figure 2a]. The switch from PcG-dependent H3K27me3 to TrxG-mediated H3K4me3 at the p16INK4A gene could be a central node of a pRb-dependent regulatory loop that drives constitutive p16INK4A expression and senescence.
It has emerged that deviations in the epigenetic regulation of p16INK4A expression might influence the aging process by constraining the self-renewal capacity of certain stem cell compartments. Elevated p16INK4A has been shown to limit the proliferation and self-renewal of pancreatic β-islet cells, hematopoietic stem cells (HSCs) and neuronal progenitor cells (NPCs) in mice [26–28]. Likewise, the absence of PcG-enforced gene repression though BMI1 deficiency severely compromises HSC and NPC self-renewal [29,29,30]. Reduced expression of EZH2 and BMI1 and diminished levels of H3K27me3 were evident in pancreatic b-islets [31], coinciding with elevated p16INK4A expression. The inhibitory effect of p16INK4A on stem cell function is further supported by the observation that senescent p16INK4A-expressing fibroblasts have proved incompatible for induced pluripotent stem (IPS) cell reprogramming, and that genetic ablation of p16INK4A significantly improves this process [32]. Furthermore, inhibition of the core components of the PRC1 and PRC2 including EZH2 markedly reduced reprogramming efficiency [33]. These findings not only illustrate the role of chromatin modifiers in aging but also suggest that gradual changes in their expression and activity could influence a given tissues tolerance of stress and capacity for renewal and regeneration that could diminish healthspan earlier in life.
SIRT1 and SIRT6 in DNA repair and aging
The sirtuin family of NAD+-dependent lysine deacetylases has a long and controversial relationship with aging [34]. Originally identified in yeast, Sir2 has been shown to participate in silencing of ribosomal RNA (rRNA) genes and telomeres [35]. This ability becomes compromised as yeast age due to the progressive loss of Sir2 expression. De-repression of rRNA expression due to loss of Sir2 resulted in the excision of rDNA arrays from the genome, creating circular episomal DNAs [36]. Sustained Sir2 expression led to the suppression of these arrays and extended the proliferative lifespan of yeast [36]. Although Sir2 homologs do not appear to directly influence longevity in Drosophila melanogaster [37] and Caenorhabditis elegans [38,39], a growing line of evidence indicates that the mammalian Sir2 homologues SIRT1 and SIRT6 have crucial roles in directing the response to DNA damage and cellular stress [40,41].
SIRT1 modifies chromatin through deacetylation of H1K26, H3K9 and H4K16 [42]. In mouse cells, chromatin immuno-precipitation (ChIP) experiments revealed that SIRT1 binds to repetitive elements, such as the satellite pericentromeric repeats [40], where it appears to be involved in the regulation of centromeric heterochromatin by the lysine methyl transferase (KMT) suppressor of variegation 3–9 homolog 1 (SUV39h1) [43]. Following oxidative DNA damage SIRT1 undergoes genome-wide redistribution to damage sites, where it deacetylates H1 to promote DNA repair [40] [Figure 2b]. Though nominally a histone deacetylase, SIRT1 also deeacetylates and regulates a growing number of non-histone substrates, including p53 [44]; the longevity modulator forkhead box transcription factor FOXO; [45] and Ku70, regulating its apoptotic function [46]. The recruitment of SIRT1 to damage sites requires activation of the ataxia telangiectasia-mutated (ATM)-dependent pathway, and once recruited it also binds to and deacetylates Nijmegan Breakage Syndrome protein (NBS1) [47], which modulates downstream steps of the DNA damage response. However, SIRT1 remobilization leads to damage-dependent transcriptional deregulation in cell culture and in vivo, particularly in brain tissue [40]. Transcriptional analyses revealed that ~2/3 of genes that are deregulated during normal aging depend on intact SIRT1 function for their repression. Thus, SIRT1 remobilization highlights a catch-22 between the immediate need to maintain genomic stability and accelerated tissue wear and tear during aging. The expression of SIRT1 declines during aging in mice [40]. Though SIRT1 over-expression does not markedly extend longevity in mice, it does improve metabolism, prevent diabetes and increase tumor resistance [48].
One of the primary functions of SIRT6 is the transcriptional regulation of the NFΚB-dependant inflammatory response [49]. SIRT6 directly binds to the NFΚB subunit RELA, to curtail its transcriptional activating potential by deacetylating H3K9 at NFΚB target gene promoters [Figure 2c]. SIRT6 depletion induces rampant expression of NFΚB targets, unleashing the inflammatory response. The link between SIRT6 and NFΚB has been substantiated by the observation that the premature aging phenotype of SIRT6−/− mice can be rescued by haplo-insufficiency of RELA, highlighting the proactive role of NFΚB in the SIRT6-dependent aging phenotype [49]. In addition, SIRT6 over-expressing mice exhibit lifespan extension [50]; this is most evident in male mice, whose lifespan is ~15% greater than females. This was associated with the prolonged activation of a youthful transcription program into later stages of adulthood [50].
SIRT6−/− mice also display an abrogated response to DNA damage, and SIRT6-deficient human cells have an increased sensitivity to DNA damaging agents [41]. SIRT6 participates in homologous recombination (HR) through deacetylation of C-terminal binding protein interacting protein (CtIP) [51]. SIRT6 also regulates the localization of one of the first responders to DNA damage, Poly [ADP-ribose] polymerase (PARP-1) [52]. SIRT6 might also contribute to aging by affecting telomere structure [53]. In normal cells, SIRT6 localizes to telomeres in S-phase, where it deacetylates H3K9, promoting the association of Werner helicase WRN, which unravels telomeric tracts during DNA replication [54]. Depletion of SIRT6 abrogates WRN function, promotes telomere dysfunction and might influence the replicative lifespan [53].
Given that DNA repair pathways and the stress response are linked with lifespan, the overlapping functions of SIRT1 and SIRT6 provide evidence that chromatin modifiers can integrate distinct cellular processes such as transcriptional regulation and the DNA damage response to influence longevity. Finally, though acting to affect localized changes in chromatin, the chromatin modifiers that we have discussed here illustrate the cascade of consequences that their deregulation can have beyond merely transcription but also fundamentally altering the structure and physiology of chromatin throughout the cell in ways that further contribute to aging.
Structural changes to chromatin during aging
Loss of heterochromatin during aging?
One of the earliest examinations of chromatin structure and aging revealed a significant reorganization of nucleosomes [55], most notably for condensed regions of constitutive heterochromatin. Constitutive heterochromatin is associated with permanently silent chromatin domains such as centromeres and other repetitive DNA regions and is characterized by enrichments of H3K9me3 and Heterochromatin Protein (HP1). Constitutive heterochromatin is also important in maintaining the integrity of repetitive DNA elements like rDNA arrays and suppressing the transcription of transposable elements. Such condensed heterochromatic regions were nearly absent from the nuclei of late passage diploid human and rodent fibroblasts [55]. The ensuing ‘heterochromatic island hypothesis’ postulated that loss of heterochromatin and nucleosomes instigates transcriptional deregulation, accelerating aging [56]. Despite the apparent framework for the role of chromatin in aging and our expanded knowledge of the components of heterochromatin, this hypothesis been only recently been interrogated. Recent comprehensive single cell and biochemical analysis of chromatin of cells derived from patients of Hutchison-Gilford Progeria Syndrome has considerably added to our understanding of this phenomenon.
Lessons from Hutchison-Gilford Progeria Syndrome (HGPS)
Hutchison-Gilford Progeria Syndrome, HGPS, is a rare genetic disease where the symptoms of aging are very pre-maturely manifested. The symptoms of HGPS are first observed in infancy and include limited growth and alopecia with accelerated wrinkling of skin and accelerated tissue degeneration, particularly of the skeleton and heart. Individuals with HGPS rarely live more than 15 years, often succumbing to heart failure. HGPS is caused by the deterioration of the nuclear envelope, which is attributed to a point mutation (C1824T) within the LMNA gene [57]. This activates a cryptic donor splice site, leading to the production of a prelamin A mRNA that contains an internal deletion of 150 base pairs. This transcript is translated into a protein known as progerin, which lacks 50 amino acids in the C-terminal portion of the normal Lamin A protein [57]. Progerin abrogates the structural role of wild type Lamin A, leading to the characteristic blebbing of the nuclear envelope that is associated with HGPS. HGPS cells are prone to premature senescence in culture, exhibit chromosome segregation defects, bi-nucleation, accelerated telomere shortening and elevated levels of DNA damage [58]. HGPS cells also exhibit loss of heterochromatic structures, notably reduction in H3K9me3 and H3K27me3, elevated histone acetylation, downregulation of HP1 and increased transcription of pericentromeric satellite III repeats [59,60]. The loss of heterochromatin has been attributed to deregulation of the nucleosome remodeling and histone deacetylase (NuRD) complex [61]. In normal cells NuRD is physically tethered to lamin A at the nuclear envelope. Progerin abolishes this interaction and NuRD expression and activity is subsequently deregulated. As a chromatin remodeler, NuRD participates in the assembly of centromeric histone variant CENPA, and as a deacetylase it sets the stage for H3K9me3 by removing acetyl at the same lysine [61]. The targeted depletion of NuRD subunits RBBP4 and RBBP7 in non-HGPS cells recapitulates the defective heterochromatic phenotype of HGPS [61]. Therefore, HGPS is believed to reveal the hierarchical role of chromatin over age-dependent changes that are due to DNA damage and telomere shortening. NuRD also regulates the transcriptional activity of some genes involved in the NOTCH and WNT dependent developmental pathways [62]. The evident tissue degeneration and premature aging in HGPS patients is linked to the ablation of mesenchymal stem cell proliferation and differentiation that is controlled, in part, by both WNT and NOTCH signaling pathways [62]. Thus, HGPS provides a good example of how the deregulation of heterochromatin promotes aging by perturbing nuclear structure with consequences for stem cell differentiation and renewal.
Interestingly, progerin has also been detected in tissues of normal elderly individuals that exhibit reduced heterochromatic H3K9me3 and HP1 [60,63]. However, the cause-and-effect relationship between normal physiological aging and progerin production is relatively unclear. Recent findings suggest that telomere dysfunction could instigate progerin production through an as yet unidentified mechanism [64]. Morpholino-based gene therapy reverses the C1824T mutation to permit splicing of the full length LMNA transcript and establishes the normal pattern of heterochromatin in the cell [65]. A recent study showed that treating HGPS cells with rapamycin, the inhibitor of mTOR (mammalian target of rapamycin), eliminated progerin from HGPS cells and enhanced proliferation, likely by activating a process termed autophagy, which removes excess protein aggregates. Given that inhibition of the mTOR pathway by rapamycin is strongly associated with promoting longevity, the intersection of this pathway and HGPS supports the link between pre-mature aging of HGPS with normal physiological aging.
The case of senescence associated heterochromatic foci (SAHF)
One of the problems in delineating the role of heterochromatin with aging has been that reports have suggested that heterochromatin can be either reduced or increased during senescence. This may be due to the equivocation of different forms of senescence, like replicative senescence and oncogene-induced senescence (OIS), where cell-cycle arrest is observed but the physiological context and molecular details of each are quite distinct. Another problem is the dual role for senescence in both promoting aging and suppressing tumorigenesis. The latter might involve the phenomenon of senescence associated heterochromatic foci (SAHF). Senescent cells often exhibit the accumulation and compaction of chromatin, frequently of entire chromosomes, into large sub-nuclear heterochromatic domains, termed SAHF [66]. These structures contain heterochromatic features, such as H3K9me3 and HP1γ, the histone variant macroH2A and HMGA (high- mobility group A) [66,67]. The histone chaperone anti-silencing factor 1a (ASF1a), histone cell cycle regulator HIRA, and pro-myelocytic leukemia protein (PML), are involved in the early stages of SAHF formation [68] [Figure 3]. SAHF was initially suggested to participate in the pRb/p16INK4A tumor suppressive pathway by orchestrating heterochromatinization and repression of pro-proliferative genes to impose senescence [66]. However, this is contradicted by the observation that SAHF formation is not a universal feature of cell cycle arrest. Rather, SAHF appears to be induced primarily following expression of oncogenic RAS, a potent suppressor of both p16INK4A and p53 tumor suppression pathways [69]. SAHF formation is less prevalent in replicative senescence or quiescence and is highly variable between cell types. Therefore, SAHF might not be absolutely required for p16INK4A cell cycle arrest and could be influenced by the origin of the primary stress.
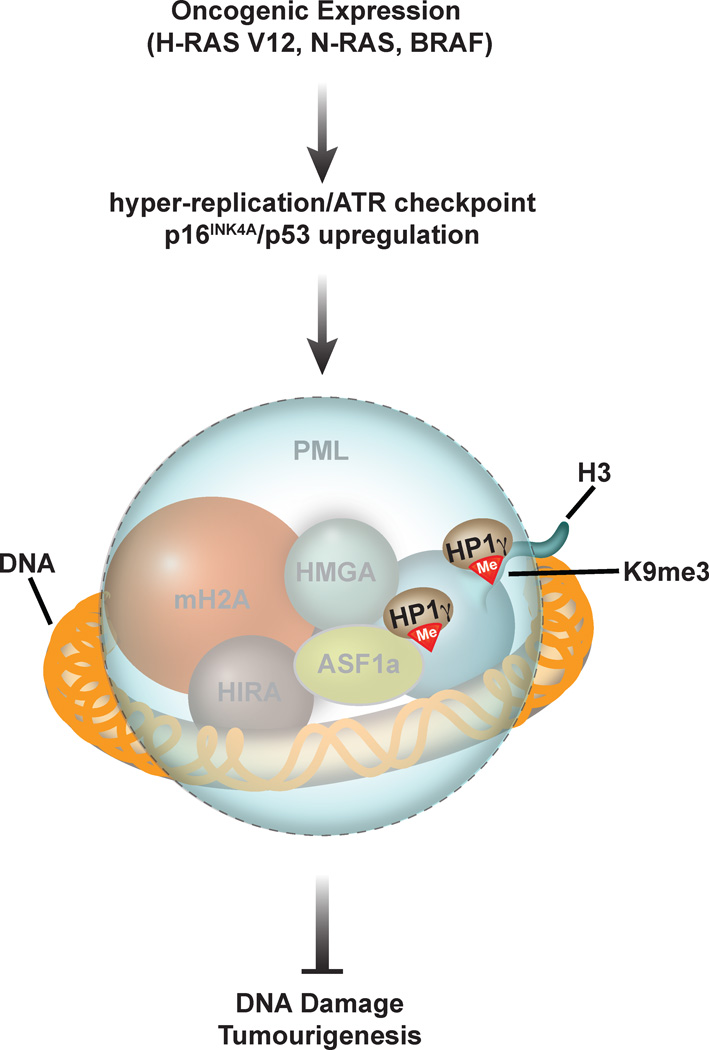
Figure 3. Senescence associated heterochromatic foci (SAHF).
Senescence associated heterochromatic foci are sub-nuclear structures that consist of DNA, chromatin and proteins such as histone macroH2A, high-mobility group protein A, HMGA, Anti-silencing factor 1, ASF1, HIR histone cell cycle regulation defective homolog A, HIRA, heterochromatin protein 1-γ isoform, HP1γ and H3K9me3 (red triangle), that are encapsulated in large Pro-Myelocytic leukemia, PML, bodies (shown as large blue circle with dashed border line). SAHF are formed in response to oncogene-induced hyper-replication, which and leads to activation of the ATR checkpoint and the up-regulation of p16INK4A/p53 tumour suppressors. SAHF are postulated to subdue the potency of the DNA damage response and prevent tumorigenesis.
The origin of genomic stress in replicative senescence, where SAHF is less prevalent, is the chronic attrition of telomeres. Though increased p16INK4A expression is observed, the induction of senescence is largely p53-dependent [70]. In addition, quantitative mass spectroscopic analysis of chromatin modification revealed a reduction of total H3K9me3 levels [71]. By contrast, oncogene induced senescence (OIS), which coincides with SAHF formation, arises from an acute stress at stalled replication forks, leading to irreparable DNA damage [72]. A recent study has teased apart the relationship between SAHF and OIS [69]. Though the abrogation of the ATM/p53-G1/S-dependent checkpoint in human cells maintained the expression of pro-proliferative genes and alleviated the oncogene-enforced cell cycle block, many cells retained SAHF. This indicates that localized repression of pro-proliferative genes does not require SAHF or its associated H3K9me3. By contrast, cells lacking ataxia-telangiectasia and rad3 related kinase (ATR), which regulates the intra-S phase replicative checkpoint, bore no evidence of SAHF. ATR is a key regulator of DNA replication and the DNA damage response (DDR), therefore these data strongly suggest that SAHF is a response to oncogenic stimuli [Figure 3]. But what is SAHF responding to? Treatment of OIS cells with drugs that inhibit the H3K9me3 KMT SUV39H1 and therefore disrupt heterochromatin, including SAHF, had the unexpected result of provoking an enhanced DDR.
This has led to the new hypothesis that the role of SAHF is to curtail the strength of the DDR and force the cell into senescence rather than apoptosis and suggests that the heterochromatinization associated with SAHF might be a barrier to tumorigenesis. Indeed, several cancer cell lines display elevated levels of H3K9me3 and HP1 and suggests that the inactivation of tumor suppressors, like p53, in cancer cells might allow for the hijacking of SAHF to maintain a low DDR in cancer cells and maintain their proliferation [69]. Therefore, opposing senescence-associated changes in heterochromatin, here H3K9 and HP1, occur but represent distinct physiological contexts. As age is a significant risk factor for cancer a greater understanding of the context that drives changes in chromatin is crucial to better interpret their relationship with aging and might aid in the effort to stifle both cancer and other aging associated pathologies.
Histone loss during aging
As mentioned, early EM studies of aged chromatin revealed extended patches of nucleosome-free regions [55], suggesting extensive remobilization of nucleosomes throughout the genome and attenuated nucleosome assembly during aging. Restraining the supply of histones could affect every facet of chromatin structure and chromatin-templated processes. Recently, age-related histone loss has been observed during aging of budding yeast and in human cells nearing replicative senescence [71,73] [Figure 4].
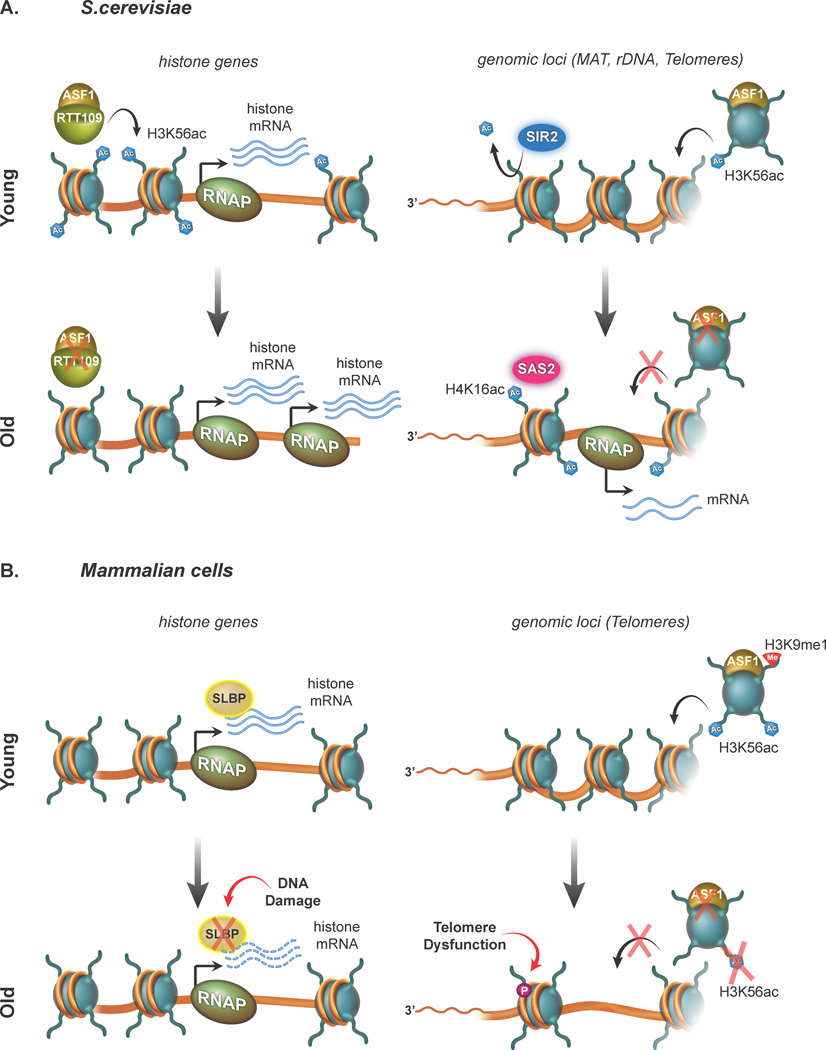
Figure 4. Histone loss during aging in S. cerevisiae and human cells.
(a) Yeast cells. Left panels: regulation of histone genes. Top: In young yeast cells the functional cooperation between Regulator of Ty1 transposition protein 109, RTT109 and Anti-silencing factor 1, ASF1 (green and yellow ovals, respectively) promotes H3K56ac (blue hexagons) at histone gene promoters to drive histone gene expression. Bottom: in old yeast cells there are less histone proteins available. This leads to nucleosome loss at histone genes, permitting increased accessibility for RNA Polymerase II (RNAP) and leading to increased histone transcription and greater histone mRNA yields. Though the precise mechanism of histone loss is unclear, it might be due to changes in the expression of RTT109 and ASF1 and reduced levels of H3K56ac. Right panels: certain heterochromatic genomic loci. Top: in young cells, the integrity of genomic loci such as the MAT locus, rDNA loci and telomeres is maintained by the histone deacetylase SIR2 (blue oval), which removes H4K16ac and potentially other acetylation chromatin marks (blue hexagons). Bottom: in the absence of histones, ASF1, and H3K56ac, the KAT SAS2 acetylates H4K16ac at target loci. This might promote transcription of these loci and inhibit nucleosome assembly.
(b) Mammalian cells. Left panels: histone genes. Top: In young human cells, histone mRNA is stabilized by the association of SLBP (yellow oval) and translated to new histone proteins. Bottom: in old cells, DNA damage might constrain expression and activity of SLBP (red arrow) leading to the disruption of histone mRNA maturation and translation. Right panels: genomic loci (telomeres). Top: In young cells, ASF1 might regulate the supply of histones at genomic loci including telomeres. ASF1 promotes the establishment of chromatin modifications, H3K56ac (blue hexagon) and H3K9me1 (red triangle) at chromatin. Bottom: In old cells, in conjunction with diminished levels of histone mRNA, the absence of ASF1 might diminish chromatin assembly dynamics and lead to loss of nucleosomes at telomeres and potentially other regions. Reduced nucleosome occupancy might promote telomere dysfunction (red arrow) and accumulation of γH2AX (purple circle).
Aged wildtype yeast exhibited a significant protein level decline of histones H3 and H2A. ChIP analysis revealed a ~50–75% reduction in H3 occupancy at the mating type locus, rDNA and telomere proximal loci [73] [Figure 4a]. This was suggested to be due to age-related changes in the activity of the histone chaperone ASF1, which regulates histone metabolism by coordinating histone expression and subsequent exchange into chromatin [74]. ASF1 and the histone KAT Repressor of Ty 1 transposition 109 (RTT109) cooperatively regulate H3K56ac in budding yeast[75], which promotes nucleosome assembly and facilitates histone gene expression. In comparison with wildtype strains, ASF1 and RTT109 single mutants exhibit shortened lifespan, which is exacerbated in ASF1/RTT109 double mutants[73]. These mutants have reduced levels of H3K56ac, and yeast in which H3K56ac is blocked by a K-to-R substitution display a similar reduction in lifespan [76]. This suggested that ASF1/RTT109-dependent H3K56ac might have a role in yeast aging by altering histone metabolism. However, unlike ASF1 and RTT109 mutants, aged wildtype yeast show dramatically more histone mRNA levels [73] [Figure 4a]. It has been proposed that the scarcity of histones causes discrepant nucleosome assembly, which leads to aberrant ASF1/RTT109/H3K56ac-independent transcription at several genes, including histone genes themselves [73]. This is corroborated by the observation that nucleosome depletion by deletion of histone H4 caused the deregulated expression of ~25% of yeast genes [77]. Deleting the histone regulatory (HIR) complex, which represses histone gene expression outside of S-phase [78], increased the proportion of chromatin associated histones and prolonged lifespan. Similarly, in a proof-of-principle, the overexpression of histones H3 and H4 prolonged lifespan [73].
In cultured human cells, comparative quantitative analysis of global histone protein expression between early passage and late passage (hereafter termed young and old) revealed a ~30% reduction in total histones H3 and H4, and a ~40% decrease in turnover of H3 and H4 in old cells. In addition, reduced nucleosome occupancy was observed at telomeres but not at other repetitive elements, such as Alu repeats, indicting non-uniform changes in nucleosome loss[71]. In humans, the reduced levels of histones H3 and H4 in old cells might also be linked with the observed reduced expression of stem loop binding protein (SLBP), the crucial regulator of histone H3.1, H3.2 and H4 mRNA maturation. The association of SLBP with the 3’loop of histone mRNAs is exquisitely regulated and cellular stress, particularly during S-phase, significantly reduces the half-life of both SLBP protein and mRNA as well as histone mRNAs by activating the nonsense mediated decay pathway in an ATR-dependent manner [79]. Therefore, the DNA damage-induced dissociation of SLBP from histone mRNAs could have the dual effects of promoting histone mRNA degradation and blocking their translation, yielding a cumulative reduction in histone H3 and H4 [Figure 4b].
Telomere dysfunction might be a major contributor to some of the chromatin changes that occur during aging. Though speculative, it is notable that short-lived telomerase-deficient yeast show significantly reduced histone transcript levels [80]. Nucleosome assembly at telomeres has been shown to be particularly prone to age-dependant defects in yeast [76]. Telomeres in aged yeast have reduced levels of histone H3 and H3K56ac levels, but demonstrably higher levels of SAS2-dependent H4K16ac, which renders chromatin into an accessible configuration that abolishes telomeric silencing [76] [Figure 4a]. In contrast to ASF1 and RTT109 mutants, SAS2 mutants exhibit prolonged lifespan [76], suggesting that the switch in the balance from H3K56ac to H4K16ac might converge with telomere dysfunction to regulate lifespan in yeast.
In humans there are two ASF1 isoforms, ASF1a and ASF1b, with both involved in regulating the supply and transfer of nucleosomes for chromatin assembly during DNA replication[81]. ASF1b also regulates a cytosolic pool of histones that are kept in reserve and utilized during recovery from replication stress and DNA damage [82]. It is not yet known whether the ASF1b-associated free histone reservoir or the chromatin associated ASF1-dependent nucleosomes are drained during aging. Like in yeast, ASF1 is absolutely required for H3K56ac [83]. H3K56ac is present at telomeres but it is not known whether the levels are altered in old cells. Then again, the localization of SIRT6 to human telomeres, which can deacetylate H3K56, allows this speculation[84]. Reduced histone occupancy was observed at telomeres and not at chromatin of internal repetitive elements (Alu repeats) suggestive of regional changes in nucleosome occupancy during aging [71]. However, genome wide analysis of nucleosome occupancy and chromatin modification must be performed to resolve exactly where age-dependent changes in nucleosome assembly occur. Only then can the consequences of nucleosome loss be directly assessed. The restoration of telomere length by ectopic expression of the catalytic telomerase subunit hTERT in old cells increased histone levels and turnover to near that of young cells and somewhat restored ASF1 and SLBP expression, likely by eliminating DNA damage signaling from short telomeres [71].
In theory, the restoration of a ‘youthful’ histone supply could provide a powerful mechanism to restore balance to the epigenome. Despite the regulatory stringency of mammalian histone biosynthesis, there are hints that modulating ASF1 and SLBP could extend the replicative lifespan of cells. Depletion of ASF1a from primary cells yields a brief delay in senescence induction [68]. Similarly, the expression of SLBP restored chromosomal and cell cycle defects in S-phase, brought about by cessation of histone biosynthesis [85]. Interestingly, elevated histone transcript have been detected in studies investigating the effects of caloric restriction on the transcriptome in C. elegans and mice [86,87]. Finally, histone loss, as we have discussed here, has not only been observed in yeast and human models of aging, but also been suggested to occur in mitotic mouse tissues [73]. Therefore, though these observations might be merely scratching the surface, future studies into histone metabolism will yield a more complete picture of how perturbations in chromatin metabolism could affect aging.
Concluding remarks
In this review we have discussed some of the paradigms of chromatin and epigenetic regulation that have been linked with aging. Their relationship with aging is substantiated by a certain degree of evolutionary conservation, at least between humans and various model organisms. For instance, the epigenetic switch between polycomb and trithorax that we discussed for the mammalian p16INK4A locus has been shown to heritably regulate lifespan through successive generations of C. elegans. The inactivation of the H3K4 methylase (set-2/ash-2/wdr-5) or demethylase (rbr-2) in nematodes led to lifespan extension or shortening of the parental strains, respectively [88,89]. This lifespan extension could be reverted upon re-establishment of H3K4me3. Inactivating the H3K27me3 demethylase, ubiquitously transcribed tetratricopeptide on X-chromosome, utx-1, thereby maintaining elevated H3K27me3, similarly promotes longevity [90]. The H3K4 methylase dependant lifespan extension was inherited over several generations, providing an elegant example of epigenetic trans-generational inheritance of longevity [89]. Though suspected, it is still unclear whether this phenomenon is due to the activation or repression of groups of genes that are uniquely linked with longevity, and these studies raise important questions pertaining to the mechanism of trans-generational epigenetic inheritance. Similarly, the loss of heterochromatic features (H3K9me3 and HP1) that has been observed in HGPS and during replicative senescence has now been observed in the fruitfly, Drosophila melanogaster. Reversing the loss of heterochromatin during aging of drosophila promotes genome stability, improved fitness and longevity of aged flies [91]. Future investigations of other chromatin modifiers in these genetically tractable model systems will undoubtedly provide insights as to whether the divergence from ‘youthful’ chromatin occurs as either a consequence or is a direct cause of aging.
The combination of genetics with regimens that control nutritional intake, like dietary or caloric restriction, has proven to be one of the most effective means to identify factors that modulate aging in nematodes, flies, primates and mice [92]. There has been a growing interest in the direct relationship between metabolism and chromatin. For instance, high glucose levels have been shown to affect the KMT activity of MLL5, and lowering cellular levels of NAD+ inhibits global SIRT-mediated deacetylation. Metabolites such as nicotinamide adenine dinucleotide (NAD+), S-adenosyl methionine (SAM), and acetyl-coA serve as co-factors in the enzymatic reactions that establish most chromatin modifications. Fluctuations in the availability of these metabolites due to extrinsic (nutritional input) or intrinsic constraints (ROS-induced mitochrondrial DNA damage, circadian rhythms) could compromise the ability of chromatin modifiers to fulfil their functions. This raises the intriguing question of whether the levels of these metabolites change during aging and whether this can cause systemic changes in cellular physiology that drive aging. It will be exciting and technically challenging to address this question, but having fleshed out the parts-list of factors involved in chromatin regulation and metabolism, it could be the next great step forward if one identifies where and how these pathways intersect to regulate human health and lifespan.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Campisi J, Vijg J. Does damage to DNA and other macromolecules play a role in aging? If so, how? J Gerontol A Biol Sci Med Sci. 2009;64:175–178. doi: 10.1093/gerona/gln065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Margueron R, Reinberg D. Chromatin structure and the inheritance of epigenetic information. Nat Rev Genet. 2010;11:285–296. doi: 10.1038/nrg2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sedivy JM, et al. Aging by epigenetics--a consequence of chromatin damage? Exp. Cell Res. 2008;314:1909–1917. doi: 10.1016/j.yexcr.2008.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Herskind AM, et al. The heritability of human longevity: a population-based study of 2872 Danish twin pairs born 1870–1900. Hum. Genet. 1996;97:319–323. doi: 10.1007/BF02185763. [DOI] [PubMed] [Google Scholar]
- 5.Fraga MF, et al. Epigenetic differences arise during the lifetime of monozygotic twins. Proc Natl Acad Sci USA. 2005;102:10604–10609. doi: 10.1073/pnas.0500398102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bahar R, et al. Increased cell-to-cell variation in gene expression in ageing mouse heart. Nature. 2006;441:1011–1014. doi: 10.1038/nature04844. [DOI] [PubMed] [Google Scholar]
- 7.Serrano M, et al. A new regulatory motif in cell-cycle control causing specific inhibition of cyclin D/CDK4. Nature. 1993;366:704–707. doi: 10.1038/366704a0. [DOI] [PubMed] [Google Scholar]
- 8.Zindy FF, et al. Expression of the p16INK4a tumor suppressor versus other INK4 family members during mouse development and aging. Oncogene. 1997;15:203–211. doi: 10.1038/sj.onc.1201178. [DOI] [PubMed] [Google Scholar]
- 9.Krishnamurthy J, et al. Ink4a/Arf expression is a biomarker of aging. J Clin Invest. 2004;114:1299–1307. doi: 10.1172/JCI22475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baker DJ, et al. Clearance of p16Ink4a-positive senescent cells delays ageing-associated disorders. Nature. 2011;479:232–236. doi: 10.1038/nature10600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bracken AP, et al. The Polycomb group proteins bind throughout the INK4A-ARF locus and are disassociated in senescent cells. Genes & Development. 2007;21:525–530. doi: 10.1101/gad.415507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cao R, et al. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 13.Kuzmichev A, et al. Different Ezh2-containing complexes target methylation of histone H1 or nucleosomal histone H3. Mol Cell. 2004;14:183–193. doi: 10.1016/s1097-2765(04)00185-6. [DOI] [PubMed] [Google Scholar]
- 14.Fischle W, et al. Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 bv Polvcomb and HP1 chromodomains. Genes & Development. 2003;17:1870–1881. doi: 10.1101/gad.1110503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cao R, et al. Role of Bmi-1 and Ring1A in H2A ubiquitylation and Hox gene silencing. Mol Cell. 2005;20:845–854. doi: 10.1016/j.molcel.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 16.Jacobs JJ, et al. The oncogene and Polycomb-group gene bmi-1 regulates cell proliferation and senescence through the ink4a locus. Nature. 1999;397:164–168. doi: 10.1038/16476. [DOI] [PubMed] [Google Scholar]
- 17.Gil J, et al. Polycomb CBX7 has a unifying role in cellular lifespan. Nat Cell Biol. 2004;6:67–72. doi: 10.1038/ncb1077. [DOI] [PubMed] [Google Scholar]
- 18.Kheradmand Kia S, et al. EZH2-dependent chromatin looping controls INK4a and INK4b, but not ARF, during human progenitor cell differentiation and cellular senescence. Epigenetics Chromatin. 2009;2:16. doi: 10.1186/1756-8935-2-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yap KL, et al. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol Cell. 2010;38:662–674. doi: 10.1016/j.molcel.2010.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kotake Y, et al. Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15INK4B tumor suppressor gene. Oncogene. 2010 doi: 10.1038/onc.2010.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Agherbi H, et al. Polycomb Mediated Epigenetic Silencing and Replication Timing at the INK4a/ARF Locus during Senescence. PLoS ONE. 2009;4:e5622–e5622. doi: 10.1371/journal.pone.0005622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gonzalez S, et al. Oncogenic activity of Cdc6 through repression of the INK4/ARF locus. Nature. 2006;440:702–706. doi: 10.1038/nature04585. [DOI] [PubMed] [Google Scholar]
- 23.Kotake Y, et al. pRB family proteins are required for H3K27 trimethylation and Polycomb repression complexes binding to and silencing p16INK4alpha tumor suppressor gene. Genes & Development. 2007;21:49–54. doi: 10.1101/gad.1499407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Agger K, et al. The H3K27me3 demethylase JMJD3 contributes to the activation of the INK4A-ARF locus in response to oncogene- and stress-induced senescence. Genes & Development. 2009;23:1171–1176. doi: 10.1101/gad.510809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kotake Y, et al. DDB1-CUL4 and MLL1 mediate oncogene-induced p16INK4a activation. Cancer Res. 2009;69:1809–1814. doi: 10.1158/0008-5472.CAN-08-2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krishnamurthy J, et al. p16INK4a induces an age-dependent decline in islet regenerative potential. Nature. 2006;443:453–457. doi: 10.1038/nature05092. [DOI] [PubMed] [Google Scholar]
- 27.Janzen V, et al. Stem-cell ageing modified by the cyclin-dependent kinase inhibitor p16INK4a. Nature. 2006;443:421–426. doi: 10.1038/nature05159. [DOI] [PubMed] [Google Scholar]
- 28.Molofsky AV, et al. Increasing p16INK4a expression decreases forebrain progenitors and neurogenesis during ageing. Nature. 2006;443:448–452. doi: 10.1038/nature05091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Molofsky AV, et al. Bmi-1 dependence distinguishes neural stem cell self-renewal from progenitor proliferation. Nature. 2003;425:962–967. doi: 10.1038/nature02060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Molofsky AV, et al. Bmi-1 promotes neural stem cell self-renewal and neural development but not mouse growth and survival by repressing the p16Ink4a and p19Arf senescence pathways. Genes & Development. 2005;19:1432–1437. doi: 10.1101/gad.1299505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dhawan S, et al. Bmi-1 regulates the Ink4a/Arf locus to control pancreatic beta-cell proliferation. Genes & Development. 2009;23:906–911. doi: 10.1101/gad.1742609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li H, et al. The Ink4/Arf locus is a barrier for iPS cell reprogramming. Nature. 2009;460:1136–U101. doi: 10.1038/nature08290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Onder TT, et al. Chromatin-modifying enzymes as modulators of reprogramming. Nature. 2012 doi: 10.1038/nature10953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Imai S-I, Guarente L. Ten years of NAD-dependent SIR2 family deacetylases: implications for metabolic diseases. Trends Pharmacol Sci. 2010;31:212–220. doi: 10.1016/j.tips.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kennedy BK, et al. Redistribution of silencing proteins from telomeres to the nucleolus is associated with extension of life span in S. cerevisiae. Cell. 1997;89:381–391. doi: 10.1016/s0092-8674(00)80219-6. [DOI] [PubMed] [Google Scholar]
- 36.Sinclair DA, Guarente L. Extrachromosomal rDNA circles--a cause of aging in yeast. Cell. 1997;91:1033–1042. doi: 10.1016/s0092-8674(00)80493-6. [DOI] [PubMed] [Google Scholar]
- 37.Rogina B, et al. Sir2 Mediates Longevity in the Fly through a Pathway Related to Calorie Restriction. Proc Natl Acad Sci USA. 2004;101:15998–16003. doi: 10.1073/pnas.0404184101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tissenbaum H, Guarente L. Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature. 2001;410:227–230. doi: 10.1038/35065638. [DOI] [PubMed] [Google Scholar]
- 39.Burnett C, et al. Absence of effects of Sir2 overexpression on lifespan in C. elegans and Drosophila. Nature. 2011;477:482–485. doi: 10.1038/nature10296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oberdoerffer P, et al. SIRT1 Redistribution on Chromatin Promotes Genomic Stability but Alters Gene Expression during Aging. Cell. 2008;135:907–918. doi: 10.1016/j.cell.2008.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mostoslavsky R, et al. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell. 2006;124:315–329. doi: 10.1016/j.cell.2005.11.044. [DOI] [PubMed] [Google Scholar]
- 42.Vaquero A, et al. Human SirT1 interacts with histone H1 and promotes formation of facultative heterochromatin. Mol Cell. 2004;16:93–105. doi: 10.1016/j.molcel.2004.08.031. [DOI] [PubMed] [Google Scholar]
- 43.Vaquero A, et al. SIRT1 regulates the histone methyl-transferase SUV39H1 during heterochromatin formation. Nature. 2007;450:440–444. doi: 10.1038/nature06268. [DOI] [PubMed] [Google Scholar]
- 44.Vaziri HH, et al. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 2001;107:149–159. doi: 10.1016/s0092-8674(01)00527-x. [DOI] [PubMed] [Google Scholar]
- 45.Brunet A, et al. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303:2011–2015. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- 46.Cohen HY, et al. Calorie Restriction Promotes Mammalian Cell Survival by Inducing the SIRT1 Deacetylase. Science, New Series. 2004;305:390–392. doi: 10.1126/science.1099196. [DOI] [PubMed] [Google Scholar]
- 47.Yuan ZZ, et al. SIRT1 regulates the function of the Nijmegen breakage syndrome protein. Mol Cell. 2007;27:149–162. doi: 10.1016/j.molcel.2007.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Herranz D, Serrano M. SIRT1: recent lessons from mouse models. Nat. Rev. Cancer. 2010;10:819–823. doi: 10.1038/nrc2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kawahara TLA, et al. SIRT6 Links Histone H3 Lysine 9 Deacetylation to NF-kappa B-Dependent Gene Expression and Organismal Life Span. Cell. 2009;136:62–74. doi: 10.1016/j.cell.2008.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kanfi Y, et al. The sirtuin SIRT6 regulates lifespan in male mice. Nature. 2012 doi: 10.1038/nature10815. [DOI] [PubMed] [Google Scholar]
- 51.Kaidi A, et al. Human SIRT6 Promotes DNA End Resection Through CtIP Deacetylation. Science. 2010;329:1348–1353. doi: 10.1126/science.1192049. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 52.Mao Z, et al. SIRT6 promotes DNA repair under stress by activating PARP1. Science. 2011;332:1443–1446. doi: 10.1126/science.1202723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Michishita E, et al. SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature. 2008;452:492–U16. doi: 10.1038/nature06736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Crabbe L, et al. Defective telomere lagging strand synthesis in cells lacking WRN helicase activity. Science. 2004;306:1951–1953. doi: 10.1126/science.1103619. [DOI] [PubMed] [Google Scholar]
- 55.Ishimi Y, et al. Changes in chromatin structure during aging of human skin fibroblasts. Exp. Cell Res. 1987;169:458–467. doi: 10.1016/0014-4827(87)90206-0. [DOI] [PubMed] [Google Scholar]
- 56.Villeponteau B. The heterochromatin loss model of aging. Exp. Gerontol. 1997;32:383–394. doi: 10.1016/s0531-5565(96)00155-6. [DOI] [PubMed] [Google Scholar]
- 57.Eriksson M, et al. Recurrent de novo point mutations in lamin A cause Hutchinson-Gilford progeria syndrome. Nature. 2003;423:293–298. doi: 10.1038/nature01629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Goldman RD, et al. Accumulation of mutant lamin A causes progressive changes in nuclear architecture in Hutchinson-Gilford progeria syndrome. Proc Natl Acad Sci USA. 2004;101:8963–8968. doi: 10.1073/pnas.0402943101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shumaker DK, et al. Mutant Nuclear Lamin a Leads to Progressive Alterations of Epigenetic Control in Premature Aging. Proc Natl Acad Sci USA. 2006;103:8703–8708. doi: 10.1073/pnas.0602569103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Scaffidi P, Misteli T. Lamin A-Dependent Nuclear Defects in Human Aging. Science. 2006;312:1059–1063. doi: 10.1126/science.1127168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pegoraro G, et al. Ageing-related chromatin defects through loss of the NURD complex. Nat Cell Biol. 2009;11:1261–U251. doi: 10.1038/ncb1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Scaffidi P, Misteli T. Lamin A-dependent misregulation of adult stem cells associated with accelerated ageing. Nat Cell Biol. 2008;10:452–459. doi: 10.1038/ncb1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cao K, et al. A lamin A protein isoform overexpressed in Hutchinson-Gilford progeria syndrome interferes with mitosis in progeria and normal cells. Proc Natl Acad Sci USA. 2007;104:4949–4954. doi: 10.1073/pnas.0611640104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cao K, et al. Progerin and telomere dysfunction collaborate to trigger cellular senescence in normal human fibroblasts. J Clin Invest. 2011;121:2833–2844. doi: 10.1172/JCI43578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Scaffidi P, Misteli T. Reversal of the cellular phenotype in the premature aging disease Hutchinson-Gilford progeria syndrome. Nat Med. 2005;11:440–445. doi: 10.1038/nm1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Narita M, et al. Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell. 2003;113:703–716. doi: 10.1016/s0092-8674(03)00401-x. [DOI] [PubMed] [Google Scholar]
- 67.Narita M, et al. A novel role for high-mobility group a proteins in cellular senescence and heterochromatin formation. Cell. 2006;126:503–514. doi: 10.1016/j.cell.2006.05.052. [DOI] [PubMed] [Google Scholar]
- 68.Zhang R, et al. Formation of MacroH2A-containing senescence-associated heterochromatin foci and senescence driven by ASF1a and HIRA. Dev Cell. 2005;8:19–30. doi: 10.1016/j.devcel.2004.10.019. [DOI] [PubMed] [Google Scholar]
- 69.Di Micco R, et al. Interplay between oncogene-induced DNA damage response and heterochromatin in senescence and cancer. Nat Cell Biol. 2011;13:292–U244. doi: 10.1038/ncb2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Herbig U, et al. Telomere shortening triggers senescence of human cells through a pathway involving ATM, p53, and p21(CIP1), but not p16(INK4a) Mol Cell. 2004;14:501–513. doi: 10.1016/s1097-2765(04)00256-4. [DOI] [PubMed] [Google Scholar]
- 71.O'Sullivan RJ, et al. Reduced histone biosynthesis and chromatin changes arising from a damage signal at telomeres. Nature Structural & Molecular Biology. 2010;17:1218–1225. doi: 10.1038/nsmb.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bartkova J, et al. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature. 2006;444:633–637. doi: 10.1038/nature05268. [DOI] [PubMed] [Google Scholar]
- 73.Feser J, et al. Elevated histone expression promotes life span extension. Mol Cell. 2010;39:724–735. doi: 10.1016/j.molcel.2010.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sutton A, et al. Yeast ASF1 protein is required for cell cycle regulation of histone gene transcription. Genetics. 2001;158:587–596. doi: 10.1093/genetics/158.2.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kaplan T, et al. Cell cycle- and chaperone-mediated regulation of H3K56ac incorporation in yeast. PLoS Genet. 2008;4 doi: 10.1371/journal.pgen.1000270. e1000270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dang W, et al. Histone H4 lysine 16 acetylation regulates cellular lifespan. Nature. 2009;459:802–807. doi: 10.1038/nature08085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wyrick JJ, et al. Chromosomal landscape of nucleosome-dependent gene expression and silencing in yeast. Nature. 1999;402:418–421. doi: 10.1038/46567. [DOI] [PubMed] [Google Scholar]
- 78.Xu H, et al. Identification of a new set of cell cycle-regulatory genes that regulate S-phase transcription of histone genes in Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:5249–5259. doi: 10.1128/mcb.12.11.5249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kaygun H, Marzluff W. Regulated degradation of replication-dependent histone mRNAs requires both ATR and Upf1. Nature Structural & Molecular Biology. 2005;12:794–800. doi: 10.1038/nsmb972. [DOI] [PubMed] [Google Scholar]
- 80.Lesur I, Campbell JL. The transcriptome of prematurely aging yeast cells is similar to that of telomerase-deficient cells. Molecular Biology of the Cell. 2004;15:1297–1312. doi: 10.1091/mbc.E03-10-0742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Groth A, et al. Regulation of Replication Fork Progression Through Histone Supply and Demand. Science. 2007;318:1928–1931. doi: 10.1126/science.1148992. [DOI] [PubMed] [Google Scholar]
- 82.Jasencakova Z, et al. Replication stress interferes with histone recycling and predeposition marking of new histones. Mol Cell. 2010;37:736–743. doi: 10.1016/j.molcel.2010.01.033. [DOI] [PubMed] [Google Scholar]
- 83.Recht J, et al. Histone chaperone Asf1 is required for histone H3 lysine 56 acetylation, a modification associated with S phase in mitosis and meiosis. Proc Natl Acad Sci USA. 2006;103:6988–6993. doi: 10.1073/pnas.0601676103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Michishita E, et al. Cell cycle-dependent deacetylation of telomeric histone H3 lysine K56 by human SIRT6. Cell Cycle. 2009;8:2664–2666. doi: 10.4161/cc.8.16.9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wagner E, et al. Expression of an RNAi-resistant SLBP restores proper S-phase progression. Biochemical Society Transactions. 2005;33:471–473. doi: 10.1042/BST0330471. [DOI] [PubMed] [Google Scholar]
- 86.McColl G, et al. Pharmacogenetic analysis of lithium-induced delayed aging in Caenorhabditis elegans. J Biol Chem. 2008;283:350–357. doi: 10.1074/jbc.M705028200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Barger JL, et al. Short-term consumption of a resveratrol-containing nutraceutical mixture mimics gene expression of long-term caloric restriction in mouse heart. Exp. Gerontol. 2008;43:859–866. doi: 10.1016/j.exger.2008.06.013. [DOI] [PubMed] [Google Scholar]
- 88.Greer EL, et al. Members of the H3K4 trimethylation complex regulate lifespan in a germline-dependent manner in C. elegans. Nature. 2010;466:383–U137. doi: 10.1038/nature09195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Greer EL, et al. Transgenerational epigenetic inheritance of longevity in Caenorhabditis elegans. Nature. 2011;479:365–U204. doi: 10.1038/nature10572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Jin C, et al. Histone demethylase UTX-1 regulates C. elegans life span by targeting the insulin/IGF-1 signaling pathway. Cell Metab. 2011;14:161–172. doi: 10.1016/j.cmet.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 91.Larson K, et al. Heterochromatin Formation Promotes Longevity and Represses Ribosomal RNA Synthesis. PLoS Genet. 2012;8 doi: 10.1371/journal.pgen.1002473. e1002473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Fontana L, et al. Extending Healthy Life Span--From Yeast to Humans. Science. 2010;328:321–326. doi: 10.1126/science.1172539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Rogakou EP, Sekeri-Pataryas KE. Histone variants of H2A and H3 families are regulated during in vitro aging in the same manner as during differentiation. Exp. Gerontol. 1999;34:741–754. doi: 10.1016/s0531-5565(99)00046-7. [DOI] [PubMed] [Google Scholar]
- 94.di Fagagna FD, et al. A DNA damage checkpoint response in telomere-initiated senescence. Nature. 2003;426:194–198. doi: 10.1038/nature02118. [DOI] [PubMed] [Google Scholar]
- 95.Maures TJ, et al. The H3K27 demethylase UTX-1 regulates C. elegans lifespan in a germline-independent, insulin-dependent manner. Aging Cell. 2011;10:980–990. doi: 10.1111/j.1474-9726.2011.00738.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Miller KM, et al. Human HDAC1 and HDAC2 function in the DNA-damage response to promote DNA nonhomologous end-joining. Nature Structural & Molecular Biology. 2010;17:1144–1151. doi: 10.1038/nsmb.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zhang M, et al. Role of CBP and SATB-1 in aging, dietary restriction, and insulin-like signaling. PLoS Biol. 2009;7 doi: 10.1371/journal.pbio.1000245. e1000245–. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Peleg S, et al. Altered histone acetylation is associated with age-dependent memory impairment in mice. Science. 2010;328:753–756. doi: 10.1126/science.1186088. [DOI] [PubMed] [Google Scholar]
- 99.Sarg B, et al. Postsynthetic trimethylation of histone H4 at lysine 20 in mammalian tissues is associated with aging. J Biol Chem. 2002;277:39195–39201. doi: 10.1074/jbc.M205166200. [DOI] [PubMed] [Google Scholar]