Summary
P-glycoprotein (P-gp) is an ATP-binding cassette (ABC) transporter that confers multidrug resistance in cancer cells1,2. It also affects the absorption, distribution, and clearance of cancer-unrelated drugs and xenobiotics. For these reasons, the structure and function of P-gp have been studied extensively for decades3. Here we present biochemical characterization of P-gp from C. elegans and its crystal structure at 3.4 Å resolution. This work provides the following new information towards a mechanistic understanding of P-gp: 1. The apparent affinities of P-gp for anticancer drugs actinomycin D and paclitaxel are approximately 4,000 and 100 times higher, respectively, in the membrane bilayer than in detergent. This affinity enhancement highlights the importance of membrane partitioning when drug accesses the transporter in the membrane4. 2. The transporter in the crystal structure opens its drug pathway at the level of the membrane’s inner leaflet. In the helices flanking the opening to the membrane we observe extended loops that may possibly mediate drug binding and/or function as hinges to gate the pathway. 3. The interface between the transmembrane and nucleotide-binding domains, which couples ATP hydrolysis to transport, contains a ball-and-socket joint and salt bridges similar to the ABC importers5, suggesting that ABC exporters and importers may share a similar mechanism to achieve alternating access for transport. 4. A carefully derived model of human P-gp, based on the C. elegans P-gp structure, not only is compatible with decades of biochemical analysis6–12, but also provides insights to explain perplexing functional data regarding the F335A mutant13,14.
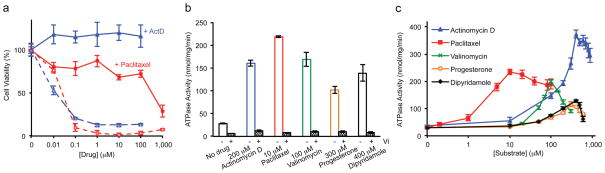
P-gp utilizes the energy from ATP hydrolysis to pump substrates across the membrane. Drug transport depends on ATP hydrolysis15,16, and the ATPase activity of P-gp is stimulated by the transported drugs17–19. C. elegans P-gp is 46% identical to human P-gp. To ensure that this level of sequence identity translates into functional similarity, we tested whether over-expression of C. elegans P-gp confers cellular resistance to cytotoxic drugs known to be transported by human P-gp. Two potent anti-cancer drugs, actinomycin D and paclitaxel (taxol) kill S. frugiperda (Sf9) cells at concentrations greater than 0.1 μM (Fig. 1a). In comparison, cells infected by recombinant baculoviruses carrying C. elegans P-gp gene are resistant to greater than 1,000 fold higher drug concentrations (Fig. 1a and Supplementary Fig. 1a). Using detergent-purified protein, we measured whether substrates of human P-gp also stimulate the ATPase activity of C. elegans P-gp. Among the 30 compounds we screened, actinomycin D, paclitaxel, progesterone, dipyridamole, and valinomycin increased the ATPase activity of C. elegans P-gp more than five times (Fig. 1b and Supplementary Fig. 1b). As expected for an ABC transporter, both the basal and drug-stimulated levels of ATPase activity are inhibited by vanadate (Fig. 1b). The ATPase activity plotted as a function of drug concentration shows the characteristic biphasic response; with increasing drug concentrations, ATPase activity increases to a maximum value and then decreases (Fig. 1c). This behavior has been studied extensively with human P-gp20,21. Like the P-glycoproteins identified from other species22, the substrate profile of C. elegans P-gp only partially overlaps with that of human P-gp. Nevertheless, their similarities in amino acid sequence and functional properties suggest that the structure of C. elegans P-gp would be a reasonable starting point for a mechanistic understanding of how human P-gp functions as a multidrug pump.
Figure 1. C. elegans P-gp is a multidrug transporter.
a, Cytotoxicity assay. Sf9 cells expressing P-gp were cultured with various concentrations of actinomycin D (blue line) or paclitaxel (red line). Uninfected cells were cultured in the presence of the same drugs as controls (dashed lines). b, ATPase activity in the presence and absence of 1 mM orthovanadate. c, ATPase activity as a function of substrate concentration. The protein concentration was kept at 0.15 μM for all measurements. Data points represent the means ± standard deviation (S.D.) of triplicate measurements from the same preparation.
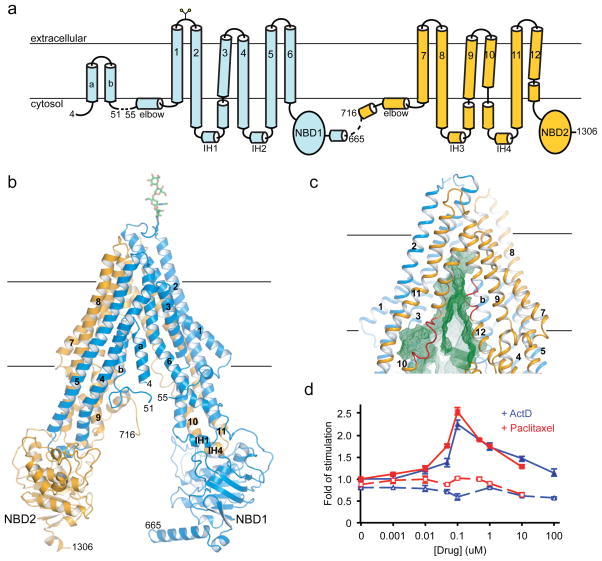
P-gp is a single polypeptide with two homologous halves, each containing a transmembrane domain (TMD) and a cytoplasmic nucleotide-binding domain (NBD) (Fig. 2a). Crystals of C. elegans P-gp were obtained in the absence of nucleotides and transport substrates. The structure was determined to 3.4 Å resolution and the register was confirmed by 34 selenium-labeled methionines and three mercury-labeled cysteines (Supplementary Fig. 2 and Table I).
Figure 2. The molecular architecture of P-gp.
a, The secondary structure. b, Ribbon presentation. c, The TM cavity (green mesh) open to the cytosol and continuous with the membrane inner leaflet. The loops in TM10 and TM12 are colored in red. d, Drug-stimulated ATPase activities in isolated membranes. The differences measured in the absence and presence of vanadate (1 mM) are plotted. Membranes from untransfected cells were used as controls (dashed lines). The data points show the means and S.D. of three determinations from the same preparation. These results have been reproduced from different protein preparations.
Compared with the crystal structure of mouse P-gp determined at 3.8 Å resolution23, the C. elegans P-gp shows a similar inward-facing conformation (i.e. open to the cytoplasm), with a larger degree of separation between the two NBDs (Fig. 2b and Supplementary Fig. 3). Superposition of the TM helices individually shows that sequence assignments of six comparable TM helices (TM1, 2, 6, 7, 8, and 11) agree well. The two structures have different conformations in TM9, 10 and 12 and thus, these helices cannot easily be compared. TM helices 3, 4, and 5, which can be compared, show important differences due to register shifts in model building (Supplementary Fig. 4, 5, 6). As we will discuss later, these corrections in the structure are relevant to the identification of drug interacting amino acids and an accurate definition of the NBD/TMD interface.
In the crystal structure the drug transport pathway is open to the cytoplasmic surface and also continuous with the membrane inner leaflet (Fig. 2c). Thus, in principle, drugs could gain access to the transport pathway from the aqueous phase as well as the membrane. Previously Higgins and Gottesman proposed that P-gp interacts with drugs from the inner leaflet of the membrane instead of the cytoplasm4 (the “hydrophobic vacuum” model). Based on this hypothesis, one would expect P-gp to be more sensitive to drug stimulation in membranes than in detergents, because most P-gp substrates are highly concentrated in the membrane with partition coefficients ranging between 100 and 10,00024. In detergents, 400 μM of actinomycin D or 10 μM of paclitaxel were required to obtain the maximum ATPase activity of purified P-gp (Fig. 1c). Addition of lipid molecules does not change the apparent drug affinities. Using membranes from yeast cells over-expressing C. elegans P-gp, we show that both drugs stimulated the ATPase activities at 0.1 μM concentration (Fig. 2d), a 4,000 fold increase in sensitivity for actinomycin D and 100 fold increase for paclitaxel. Although the magnitude of stimulation was lower using the membrane sample compared with purified P-gp in detergent, this is likely due to baseline activity of other cellular ATPases in the yeast cell membranes. We note that the drug concentration-dependence of ATPase stimulation in membranes matches well the concentration dependence observed in the cell protection assay (Fig. 1a and Fig. 2d). The shift in drug sensitivity in membranes supports the hypothesis that drugs enter the transporter through the membrane’s inner leaflet. Such a mechanism whereby transport depends on an elevated local concentration of drug due to membrane partitioning also implies that the intrinsic affinity of drugs for the transporter itself is low, consistent with the ability of P-gp to transport many different compounds.
The structure of C. elegans P-gp presents an interesting variation on the theme of membrane access in that only one lateral opening is patent (Fig. 2c). An N-terminal helical hairpin is inserted into the other lateral opening observed in the bacterial exporter MsbA25 and mouse P-gp23 between TM4 and TM6 (Fig. 2b). A truncation mutant devoid of the helical hairpin functions similarly to the full-length protein in a cytotoxic assay (Supplementary Fig. 7a) but has a reduced maximum level of stimulation in an ATPase activity assay (Supplementary Fig. 7b). The drug concentration dependence, however, is unaltered for the truncation mutant (Supplementary Fig. 7b).
Another notable feature of C. elegans P-gp is a discontinuity of helices TM10 and TM12 lining the lateral opening (Fig. 2c). We imagine two possible reasons why the helical secondary structure gives rise to more disordered loop structures in this region. The first is that a break down of secondary structure creates a greater number of possible interactions between protein atoms and drugs entering the pathway. In other transporters where discontinuous TM helices have been observed, the loops inside the membrane have been shown to bind substrates26. Thus, loops flanking the lateral opening could assist drug recognition and entry. The second possible reason is that these loop regions could function as flexible hinges to gate the pathway and/or mediate conformational changes associated with drug transport.
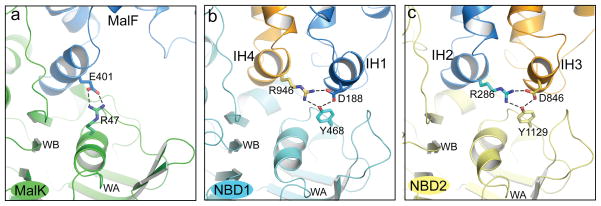
The NBD/TMD interface is important in ABC transporters because it transmits conformational changes associated with ATP hydrolysis to substrate translocation. In all ABC importers for which structures have been determined the TMD is connected to the NBD through a “ball-and-socket” joint5. This joint consists of a cytoplasmic helix from the TMD known as the coupling helix or EAA loop (the ball), which docks into a cleft on the NBD surface (the socket) (Fig. 3a). A similar structural feature is observed in P-gp, in which helices IH2 and IH4 on the TMDs resemble the coupling helices and engage the same surface of the NBDs (Fig. 3b,c). In addition to the ball-and-socket joint, ABC exporters, including P-gp, have an additional set of interactions in which IH1 and IH3 also interact with the NBDs, creating a more extensive interaction surface between the TMDs and NBDs (Fig. 3b,c). The importance of this interface in P-gp to both structure and function is underscored by the high degree of amino acid conservation, which approaches 80% identity between C. elegans and human P-gp (Supplementary Fig. 8, 9).
Figure 3. Interactions between the TMDs and NBDs.
a, the NBD/TMD interface of the maltose importer. MalF is the TM subunit and MalK is the NBD. E401 is the conserved glutamate residue in the “EAA” loop. b, and c, the NBD1/TMD (b) and NBD2/TMD (c) interfaces in P-gp. Dash lines indicate the conserved salt-bridges and hydrogen bonds. The Walker A and Walker B motifs are labeled as WA and WB, respectively.
Multiple crystal structures of the maltose importer show that transition between different structures is accompanied by rotations of the EAA loop inside the cleft27. Salt bridge interactions appear to play a central role in tethering a pivot point in these conformational changes; elimination of the salt-bridges by double mutations results in a defective transporter and dissociation of the MalK subunit28. For the NBD1/TMD interface in P-gp, three highly conserved residues, R946 (IH4), D188 (IH1), and Y468 (NBD1) are engaged in a network of salt-bridge interactions (Fig. 3b). The same atomic interactions are also observed in the NBD2/TMD interface involving residues R286 (IH2), D846 (IH3), and Y1129 (NBD2) (Fig. 3c). It is likely that these interactions in P-gp play a role similar to the salt bridge interactions in importers such as the maltose transporter.
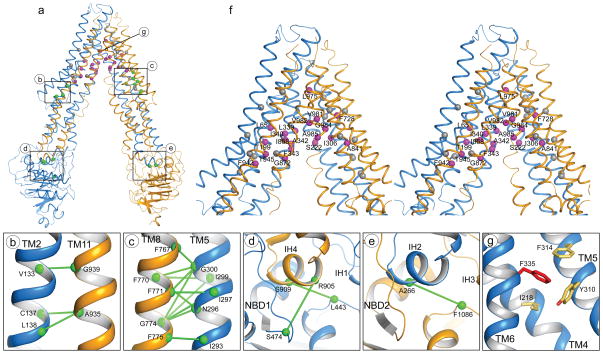
To assist structural and functional analysis of human P-gp we generated a homology model of human P-gp based on the structure of the C. elegans P-gp determined in this study. The modeled structure is consistent with a large body of biochemical and biophysical data on human P-gp (Fig. 4 and Supplementary Fig. 10). For example, pairs of residues in the TMDs shown by cysteine mutagenesis to form disulfide bonds6,7 are located in close proximity to each other (Fig. 4b,c). The NBD/TMD interfaces are consistent with data showing that A266 in TMD1 is in close proximity to F1086 in NBD229 and that cysteines introduced at positions 443 and 474 in NBD1 cross-link with residues 909 and 905 in TMD2, respectively12 (Fig. 4d,e). Residues protected by drug substrates from inhibition by thiol-reactive analogues are suggested to form the drug binding region8–11. In the modeled structure, 17 of the 19 residues are distributed on the surface of the drug-translocation pathway (Fig. 4f). Most of these residues are non-polar, consistent with the hydrophobic nature of the substrates. In contrast, the 17 residues that were not protected by substrate, and therefore are presumed to be not involved in drug binding11, are scattered throughout the TMDs (Fig. 4f). Previously the orientations of TM3, 4 and 5 with regards to the drug-translocation pathway predicted by arginine-scanning mutagenesis were incompatible with the crystal structure of the mouse P-gp30. Upon correction of amino acid registry (Supplementary Fig. 5,6), these biochemical data are now consistent with the homology modeled human P-gp (Supplementary Fig. 10).
Figure 4. A model of human P-gp.
a, The overall structure. b,c, Pairs of residues in TMDs that formed disulfide bonds (green line) when mutated to cysteines6,7. d,e, Pairs of residues at the NBD/TMD interfaces that were crosslinked12,29. f, Stereo view of the drug transport pathway. Drug-interacting residues8–11 are labeled and shown as magenta balls. The non-protected residues, Y118, V125, V133, C137, Q195, N296, G300, Y310, F314, A729, F759, S766, G774, N842, A871, S943 and F957, are shown in grey11. g, F335 in TM6 (red) interacts with residues in TM4 and TM5 (yellow sticks), thereby stabilizing the inward-facing conformation.
The atomic structure of P-gp offers insights into functional data that otherwise are difficult to explain. For example, mutants that enhance drug-stimulated ATPase activity often show stronger resistance to the same drug, likely due to increased affinity for the drug. However, this correlation does not hold for the F335A mutant, which has a higher ATPase activity but confers lower or similar drug resistance13,14. The structure shows that F335 is located at the apex of the drug transport pathway, making van der Waals interactions with Y310 and F314 in TM5, and I218 in TM4 (Fig. 4a,g). These contacts will be broken in the outward-facing conformation, in which the transport pathway has to be continuous with the extracellular side of the membrane25. Mutating the phenylalanine to alanine, if it destabilized the inward-facing conformation, would facilitate the transition to the outward-facing conformation and thus give rise to higher basal and drug-stimulated ATPase activity. In this description, the F335A mutant would be reminiscent of mutants identified in the maltose transporter that allow constitutive ATP hydrolysis in the absence of the binding protein27.
In summary, the structure of the C. elegans P-gp provides an accurate model to interpret decades of functional and biochemical data on P-gp. The functional data complement the crystal structure to support a model in which P-gp uses the energy from ATP hydrolysis to expel lipophilic molecules from the inner leaflet of the membrane. Many details of this molecule, such as how substrate binding activates the ATPase activity, how ATP hydrolysis is coupled to substrate flipping, and how multiple substrates are recognized by a single transporter, remain to be elucidated.
Methods
Cloning, expression and purification
A synthetic gene encoding the full-length C. elegans P-gp (pgp-1, Genebank accession code AB01232.1) was subcloned into the pPICZ (Invitrogen) and pVL1393 vectors (BD Biosciences). The C-terminus of P-gp was fused to the enhanced green fluorescence protein (eGFP) plus a deca-histidine tag or the Protein A tag derived from pEZZ18 (GE Healthcare). A PreScission protease cleavage site was engineered between P-gp and the C-terminal tag.
Native protein was expressed in P. pastoris strain SMD1163 (Invitrogen). Cells were grown at 28°C in minimal methanol medium and induced with 0.5% methanol for 24 hours. After harvesting by centrifugation (4,000 g for 15 minutes), the cells were frozen in liquid nitrogen and broken by cryomilling (Retsch model MM400). Broken cells were resuspended in the lysis buffer containing 50 mM Tris-HCl pH 8.0, 150 mM NaCl, 20% glycerol, 5 mM MgCl2, 1 mM PMSF, 1 mM Benzamidine, 0.1 mg/mL trypsin inhibitor, 3 ug/mL DNAse, 1 ug/mL pepstatinA, leupeptin and aprotinin. Membranes were solubilized by adding 1% (w/v) n-dodecyl-β-D-maltopyranoside (DDM; Affymetrix) to the lysis buffer and incubating at 4°C for 2 hours. Insoluble membrane was removed by centrifugation at 80,000 g for 40 minutes and supernatant was loaded onto cobalt affinity resin (Clontech). After on-column cleavage by PreScission protease, protein was eluted and reloaded onto a GST column to remove the GST-tagged PreScission protease. Protein was concentrated using an Amicon Ultra (MWCO 50K, Millipore) centrifugal device and further purified by Superdex 200 size-exclusion chromatography (GE Healthcare) in a buffer containing 20 mM MES pH 6.5, 200 mM NaCl, 10% glycerol, 5 mM DTT and 0.02% (w/v) n-undecyl-β-D-thiomaltopyranoside (UDTM; Affymetrix).
SeMet-labeled protein was expressed in Hi5 cells (Invitrogen) adapted to methionine-free medium (Expression Systems). Baculovirus-infected Hi5 cells were cultured for 16–20 hours at 28°C, supplemented with 200 mg/L L-(+)-selenomethione (Fisher Scientific), and then incubated for an additional 48 hours. Cells were harvested and broken by a high pressure homogenizer (Emulsiflex-C5; Avestin) in the lysis buffer plus 5 mM DTT. Cell debris was removed by a low-speed centrifugation (6,000g, 20 minutes) and membranes were collected by a high-speed centrifugation (40,000rpm, 1 hour). The membranes were homogenized and solubilized with 1% (w/v) DDM for 2 hours. Extracted protein was purified using IgG sepharose affinity resin (GE Healthcare) followed by PreScission cleavage to remove the Protein A tag. The protein sample was further purified by the GST affinity resin and gel-filtration chromatography. All purification steps were carried out at 4°C.
Cytotoxicity assay
Sf9 cells infected with recombinant baculovirus carrying the P-gp+eGFP gene were incubated at 28°C for 24 hours before actinomycin D or paclitaxel (Sigma-Aldrich) were added to the media. Cells were monitored by counting the cell densities every 6 hours for 1 day in the presence of actinomycin D or every 24 hours for 3 days in the presence of paclitaxel. The percent cell viability was calculated by dividing the number of live cells cultured with drug by that of the infected cells cultured without drug. In the presence of actinomycin D, cytotoxic effects were observed after 6 hours of drug exposure and reached maximum at 12 hours. Paclitaxel-induced cell death was observed after 24 hours of drug exposure and reached maximum at 48 hours. P-gp expression was monitored by fluorescence microscopy. Each assay was repeated in triplicate.
Preparation of microsomes
Cryomilled yeast cells were resuspended in the lysis buffer and the suspension was centrifuged at 6,000 rpm for 15 minutes to remove the cell debris. The supernatant was ultracentrifuged at 40,000 rpm for 1 hour and the pellet containing the microsome was stored at −80°C.
ATP hydrolysis assay
The ATPase activity of C. elegans P-gp was analyzed by a coupled assay in which the regeneration of hydrolyzed ATP is coupled to the oxidation of NADH31. Purified protein was added to the ATPase reaction buffer (50 mM potassium HEPES pH 8.0, 10 mM MgCl2, 60 μg/mL pyruvate kinase, 32 μg/mL lactate dehydrogenase, 4 mM phosphoenolpyruvate, 0.3 mM NADH, and 5 mM ATP) at 25°C in the presence or absence of drug substrates. For vanadate inhibition, the protein samples were mixed with 1 mM vanadate in the presence of ATP-Mg for 10 minutes at room temperature before measuring the ATPase activity. To determine the drug-stimulated ATPase activity of P-gp in the membrane, microsomes (5ug) were preincubated in the reaction buffer with actinomycin D or paclitaxel for 5 minutes at room temperature. The fold of drug stimulation was calculated by deducing the rate of the ATP hydrolysis in the presence of vanadate (1 mM) from that in the absence of vanadate.
Crystallization and data collection
Native crystals were obtained by mixing 1 μL of protein sample (10 mg/mL) and 1 μL of reservoir solution containing 100 mM HEPES pH 6.6–7.2, 200 mM sodium malonate and 19–22% PEG2000MME at 4°C by the sitting-drop vapor diffusion method. Mercury-labeled crystals were prepared by adding 1 mM methyl mercury chloride (II) into crystal-containing drops for 4 hours at 4°C. Crystals of SeMet-substituted protein were obtained with 100 mM HEPES pH 6.6–7.2, 200 mM ammonium phosphate monobasic and 19–22% PEG1500 at 4°C. All crystals appeared within two days and continued to grow to full size in one week. Crystals were dehydrated and cryoprotected by a serial increase of PEG concentrations in 5% steps in the reservoir (final 30%) for 48 hours followed by 2.5% steps in the drop (final 35%) for 2 hours. Crystals were flash-frozen in liquid nitrogen. Diffraction data were collected at the 23-ID beamline at the Advanced Photon System (APS). Diffraction images were indexed, integrated and scaled with the HKL2000 package (HKL Research Inc.).
Structure Determination and homology modeling
The structure was determined by molecular replacement (Phaser, CCP4)32 using separate domains of bacterial Sav1866 and mouse Pgp structures as search models. The molecular replacement phase was combined with SAD phasing (PhaserEP, CCP4) to identify methionine and cysteine sites. The model was built by iterative modeling in COOT33 and refined by CNS34 and REFMAC535. The crystallographic statistics are summarized in Supplementary Table 1. Residues of the N-terminus (1–3, 52–54), the linker region (666–715) and the C-terminus (1307–1321) were not visible in the electron density map and were not included in the final structure. No residues lie in the disallowed region of the Ramachandran plot. The homology model of human P-gp was generated by the program MODELLER based on the structure of C. elegans P-gp in which the N-terminal and the linker region were excluded due to low sequence conservation (Supplementary Fig 9). All figures were generated using the program PyMOL (www.pymol.org).
Supplementary Material
Acknowledgments
We thank the beamline staff of the GM/CA CAT at APS for assistance with data collection, Yun-Kyung Cho for assistance in protein purification, and Dr. Amy Davidson for comments on this manuscript. We also thank the MacKinnon laboratory for reagents and advice on the P. pastoris expression system. This work was supported by Howard Hughes Medical Institute (J.C.), Purdue Center for Cancer Research (NCI CCSG CA23168), and postdoctoral fellowships (M.S.J.) from the National Research Foundation of Korea and the International Human Frontier Science Program.
Footnotes
Author Information
All authors designed the study and analyzed the data. M.S.J. and M.L.O. determined the crystal structure. M.S.J. and Q.Z. performed the biochemical experiments. M.S.J., M.L.O. and J.C. wrote the manuscript. Coordinates and structure factors have been deposited in the Protein Data Bank under accession number 4F4C. Reprints and permissions information are available at www.nature.com/reprints. The authors declare no competing financial interests.
References
- 1.Juliano RL, Ling V. A surface glycoprotein modulating drug permeability in Chinese hamster ovary cell mutants. Biochimica et biophysica acta. 1976;455:152–162. doi: 10.1016/0005-2736(76)90160-7. [DOI] [PubMed] [Google Scholar]
- 2.Ueda K, et al. The mdr1 gene, responsible for multidrug-resistance, codes for P-glycoprotein. Biochemical and biophysical research communications. 1986;141:956–962. doi: 10.1016/s0006-291x(86)80136-x. [DOI] [PubMed] [Google Scholar]
- 3.Sharom FJ. The P-glycoprotein multidrug transporter. Essays in biochemistry. 2011;50:161–178. doi: 10.1042/bse0500161. [DOI] [PubMed] [Google Scholar]
- 4.Higgins CF, Gottesman MM. Is the multidrug transporter a flippase? Trends in biochemical sciences. 1992;17:18–21. doi: 10.1016/0968-0004(92)90419-a. [DOI] [PubMed] [Google Scholar]
- 5.Oldham ML, Davidson AL, Chen J. Structural insights into ABC transporter mechanism. Current opinion in structural biology. 2008;18:726–733. doi: 10.1016/j.sbi.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Loo TW, Bartlett MC, Clarke DM. Val133 and Cys137 in transmembrane segment 2 are close to Arg935 and Gly939 in transmembrane segment 11 of human P-glycoprotein. The Journal of biological chemistry. 2004;279:18232–18238. doi: 10.1074/jbc.M400229200. [DOI] [PubMed] [Google Scholar]
- 7.Loo TW, Bartlett MC, Clarke DM. Disulfide cross-linking analysis shows that transmembrane segments 5 and 8 of human P-glycoprotein are close together on the cytoplasmic side of the membrane. The Journal of biological chemistry. 2004;279:7692–7697. doi: 10.1074/jbc.M311825200. [DOI] [PubMed] [Google Scholar]
- 8.Loo TW, Bartlett MC, Clarke DM. Transmembrane segment 1 of human P-glycoprotein contributes to the drug-binding pocket. The Biochemical journal. 2006;396:537–545. doi: 10.1042/BJ20060012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Loo TW, Bartlett MC, Clarke DM. Transmembrane segment 7 of human P-glycoprotein forms part of the drug-binding pocket. The Biochemical journal. 2006;399:351–359. doi: 10.1042/BJ20060715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Loo TW, Bartlett MC, Clarke DM. Suppressor mutations in the transmembrane segments of P-glycoprotein promote maturation of processing mutants and disrupt a subset of drug-binding sites. The Journal of biological chemistry. 2007;282:32043–32052. doi: 10.1074/jbc.M706175200. [DOI] [PubMed] [Google Scholar]
- 11.Loo TW, Clarke DM. Do drug substrates enter the common drug-binding pocket of P-glycoprotein through “gates”? Biochemical and biophysical research communications. 2005;329:419–422. doi: 10.1016/j.bbrc.2005.01.134. [DOI] [PubMed] [Google Scholar]
- 12.Zolnerciks JK, Wooding C, Linton KJ. Evidence for a Sav1866-like architecture for the human multidrug transporter P-glycoprotein. Faseb J. 2007;21:3937–3948. doi: 10.1096/fj.07-8610com. [DOI] [PubMed] [Google Scholar]
- 13.Loo TW, Clarke DM. Functional consequences of phenylalanine mutations in the predicted transmembrane domain of P-glycoprotein. The Journal of biological chemistry. 1993;268:19965–19972. [PubMed] [Google Scholar]
- 14.Loo TW, Clarke DM. Rapid purification of human P-glycoprotein mutants expressed transiently in HEK 293 cells by nickel-chelate chromatography and characterization of their drug-stimulated ATPase activities. The Journal of biological chemistry. 1995;270:21449–21452. doi: 10.1074/jbc.270.37.21449. [DOI] [PubMed] [Google Scholar]
- 15.Azzaria M, Schurr E, Gros P. Discrete mutations introduced in the predicted nucleotide-binding sites of the mdr1 gene abolish its ability to confer multidrug resistance. Molecular and cellular biology. 1989;9:5289–5297. doi: 10.1128/mcb.9.12.5289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hamada H, Tsuruo T. Purification of the 170- to 180-kilodalton membrane glycoprotein associated with multidrug resistance. 170- to 180-kilodalton membrane glycoprotein is an ATPase. The Journal of biological chemistry. 1988;263:1454–1458. [PubMed] [Google Scholar]
- 17.Al-Shawi MK, Senior AE. Characterization of the adenosine triphosphatase activity of Chinese hamster P-glycoprotein. The Journal of biological chemistry. 1993;268:4197–4206. [PubMed] [Google Scholar]
- 18.Ambudkar SV. Drug-stimulatable ATPase activity in crude membranes of human MDR1-transfected mammalian cells. Methods in enzymology. 1998;292:504–514. doi: 10.1016/s0076-6879(98)92039-0. [DOI] [PubMed] [Google Scholar]
- 19.Sarkadi B, Price EM, Boucher RC, Germann UA, Scarborough GA. Expression of the human multidrug resistance cDNA in insect cells generates a high activity drug-stimulated membrane ATPase. The Journal of biological chemistry. 1992;267:4854–4858. [PubMed] [Google Scholar]
- 20.Al-Shawi MK, Polar MK, Omote H, Figler RA. Transition state analysis of the coupling of drug transport to ATP hydrolysis by P-glycoprotein. The Journal of biological chemistry. 2003;278:52629–52640. doi: 10.1074/jbc.M308175200. [DOI] [PubMed] [Google Scholar]
- 21.Litman T, Zeuthen T, Skovsgaard T, Stein WD. Structure-activity relationships of P-glycoprotein interacting drugs: kinetic characterization of their effects on ATPase activity. Biochimica et biophysica acta. 1997;1361:159–168. doi: 10.1016/s0925-4439(97)00026-4. [DOI] [PubMed] [Google Scholar]
- 22.Kim IW, Booth-Genthe C, Ambudkar SV. Relationship between drugs and functional activity of various mammalian P-glycoproteins (ABCB1) Mini reviews in medicinal chemistry. 2008;8:193–200. doi: 10.2174/138955708783744100. [DOI] [PubMed] [Google Scholar]
- 23.Aller SG, et al. Structure of P-glycoprotein reveals a molecular basis for poly-specific drug binding. Science (New York, N Y. 2009;323:1718–1722. doi: 10.1126/science.1168750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gatlik-Landwojtowicz E, Aanismaa P, Seelig A. Quantification and characterization of P-glycoprotein-substrate interactions. Biochemistry. 2006;45:3020–3032. doi: 10.1021/bi051380+. [DOI] [PubMed] [Google Scholar]
- 25.Ward A, Reyes CL, Yu J, Roth CB, Chang G. Flexibility in the ABC transporter MsbA: Alternating access with a twist. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:19005–19010. doi: 10.1073/pnas.0709388104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Screpanti E, Hunte C. Discontinuous membrane helices in transport proteins and their correlation with function. Journal of structural biology. 2007;159:261–267. doi: 10.1016/j.jsb.2007.01.011. [DOI] [PubMed] [Google Scholar]
- 27.Khare D, Oldham ML, Orelle C, Davidson AL, Chen J. Alternating access in maltose transporter mediated by rigid-body rotations. Molecular cell. 2009;33:528–536. doi: 10.1016/j.molcel.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mourez M, Hofnung M, Dassa E. Subunit interactions in ABC transporters: a conserved sequence in hydrophobic membrane proteins of periplasmic permeases defines an important site of interaction with the ATPase subunits. The EMBO journal. 1997;16:3066–3077. doi: 10.1093/emboj/16.11.3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Loo TW, Bartlett MC, Clarke DM. Processing mutations disrupt interactions between the nucleotide binding and transmembrane domains of P-glycoprotein and the cystic fibrosis transmembrane conductance regulator (CFTR) The Journal of biological chemistry. 2008;283:28190–28197. doi: 10.1074/jbc.M805834200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Loo TW, Bartlett MC, Clarke DM. Identification of residues in the drug translocation pathway of the human multidrug resistance P-glycoprotein by arginine mutagenesis. The Journal of biological chemistry. 2009;284:24074–24087. doi: 10.1074/jbc.M109.023267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Garrigos M, Belehradek J, Jr, Mir LM, Orlowski S. Absence of cooperativity for MgATP and verapamil effects on the ATPase activity of P-glycoprotein containing membrane vesicles. Biochemical and biophysical research communications. 1993;196:1034–1041. doi: 10.1006/bbrc.1993.2355. [DOI] [PubMed] [Google Scholar]
- 32.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 34.Brunger AT. Version 1.2 of the Crystallography and NMR system. Nat Protoc. 2007;2:2728–2733. doi: 10.1038/nprot.2007.406. [DOI] [PubMed] [Google Scholar]
- 35.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.