Summary
Background
In multicellular organisms, cell-cell junctions are involved in many aspects of tissue morphogenesis. α-catenin links the cadherin-catenin complex (CCC) to the actin cytoskeleton, stabilizing cadherin-dependent adhesions.
Results
To identify modulators of cadherin-based cell adhesion, we conducted a genome-wide RNAi screen in C. elegans and uncovered MAGI-1, a highly conserved protein scaffold. Loss of magi-1 function in wild-type embryos results in disorganized epithelial migration and occasional morphogenetic failure. MAGI-1 physically interacts with the putative actin regulator AFD-1/afadin; knocking down magi-1 or afd-1 function in a hypomorphic α-catenin background leads to complete morphogenetic failure and actin disorganization in the embryonic epidermis. MAGI-1 and AFD-1 localize to a unique domain in the apical junction and normal accumulation of MAGI-1 at junctions requires SAX-7/L1CAM, which can bind MAGI-1 via its C-terminus. Depletion of MAGI-1 leads to loss of spatial segregation and expansion of apical junctional domains and greater mobility of junctional proteins.
Conclusions
Our screen is the first genome-wide approach to identify proteins that function synergistically with the CCC during epidermal morphogenesis in a living embryo. We demonstrate novel physical interactions between MAGI-1, AFD-1/afadin and SAX-7/L1CAM, which are part of a functional interactome that includes components of the core CCC. Our results further suggest MAGI-1 helps to partition and maintain a stable, spatially ordered apical junction during morphogenesis.
Introduction
Adhesion between epithelial cells is essential for the dramatic changes in cell shape, cell-cell contacts and cellular rearrangements that occur during morphogenesis [1]. Caenorhabditis elegans provide a powerful genetic model in which to study modulation of cell-cell junctions during development due to the simplicity of the C. elegans apical junction (CeAJ). The CeAJ contains three domains [2]: an apical membrane domain, a classical cadherin-catenin complex (CCC) [3], and a more basal DLG-1/AJM-1 complex [4, 5]. While important for morphogenesis, the C. elegans CCC is not essential for general cell-cell adhesion in embryos [3], unlike other systems [6–8], suggesting that other proteins act redundantly with the CCC during development.
We used a genome-wide RNAi screen to identify proteins that exhibit functional redundancy or synergy with the CCC during embryonic development and uncovered MAGI-1, an inverted MAGUK (Membrane Associated GUanylate Kinase) family protein. MAGUKs can assemble multiprotein complexes at sites of cell-cell adhesion, and are key organizers of cell-cell junctions [9]. The C. elegans magi-1 locus encodes a short isoform, MAGI-1S, containing two WW and five PDZ domains, and a long isoform, MAGI-1L, which has an additional N-terminal GUK domain [10]. Reduction of MAGI-1L results in defective mechanosensory memory acquisition and olfactory and gustatory associative learning [10, 11]. Furthermore, memory consolidation and nose-touch response depend on the ability of MAGI-1 to interact with β-catenin at synapses [11].
Here we show MAGI-1 physically interacts with AFD-1/afadin, which itself synergizes with the CCC during morphogenesis. In contrast to a previous report [12], we use immunolocalization of endogenous proteins to show that MAGI-1 localizes between the CCC and DLG-1/AJM-1 complex and that localization is partially dependent on the MAGI-1 binding partner SAX-7/L1CAM. Finally, we show that MAGI-1 acts to maintain the spatial segregation of apical junctional complexes during embryonic morphogenesis. Our screen is the first genome-wide approach to identifying proteins that function synergistically with the CCC in a living organism and identifies in vivo roles for a MAGI protein during epithelial morphogenesis.
Results
A genome-wide RNAi screen identifies genetic interactors with the CCC
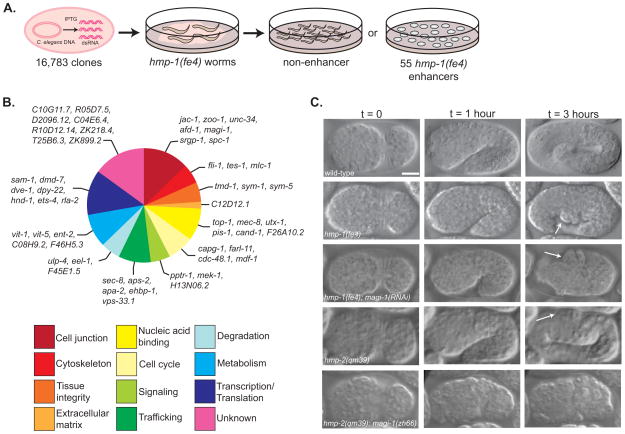
To identify functional interactors with the C. elegans CCC, we screened an RNAi bacterial expression library that targets ~86% of C. elegans open reading frames [13] in worms homozygous for a hypomorphic allele of α-catenin, hmp-1(fe4). hmp-1(fe4) homozygotes exhibit ~70% lethality and defects in actin organization and body morphology [14]. Induced bacteria from the library were fed to hmp-1(fe4) worms and clones that increased lethality in hmp-1(fe4) worms were identified (Figure 1A). Clones that had not been previously reported to cause lethality were rescreened in hmp-1(fe4) and wild-type worms; we eliminated clones that caused significant lethality in wild-type worms in our hands, even though they had not been previously reported to do so. The remaining clones were screened a final time in hmp-1(fe4) and wild-type worms to count the percentage of dead embryos. Our screen identified 55 genes that when knocked down by feeding RNAi enhanced the hmp-1(fe4) phenotype to >83% lethality but did not produce severe phenotypes in wild-type worms (Figure 1B, Table S1).
Figure 1. An enhancer screen identifies putative adhesion modulators during C. elegans morphogenesis.
(A) We screened a library of 16,783 RNAi feeding clones to identify enhancers of a hypomorphic allele of α-catenin, hmp-1(fe4). Each bacterial clone was fed to wild-type and hmp-1(fe4) worms and phenotypes were assessed in the next generation. Enhancers increased lethality in hmp-1(fe4) embryos to >83% while causing <10% lethality in wild-type embryos. (B) Putative functions for enhancers were determined using literature searches, BLAST and ClustalW comparisons of protein sequence, identification of domains using Pfam, and NCBI KOG description. Functional groups were based on previously published categories [37, 38]. (C) Knocking down magi-1 enhances the penetrance and severity of hmp-1(fe4) and hmp-2(qm39) phenotypes. Wild-type embryos enclose (t = 0), turn (t = 1 hour), and elongate ~4-fold their original length before hatching (t = 3 hours). hmp-1(fe4) embryos enclose (t = 0), but fail to fully elongate, developing body shape defects (arrow, t = 3 hours). The majority of hmp-1(fe4); magi-1(RNAi) embryos enclose (t = 0), but almost immediately retract dorsally and arrest, with a characteristic humpback phenotype (arrows, t = 1 and 3 hours). hmp-2(qm39) embryos enclose (t = 0) and elongate, occasionally developing mild body shape defects (arrow, t = 3 hours). A significant fraction of hmp-2(qm39); magi-1(zh66) embryos fail to enclose and rupture (t = 0). Scale bar is 10μm.
Knocking down magi-1 enhances the penetrance and severity of hmp-1(fe4) phenotypes
Our screen identified several putative junctional proteins including magi-1. magi-1(RNAi) enhances the lethality of hmp-1(fe4) embryos to 99% (n=399) (Figure 1C; Movie 1). 88% of embryos fail to elongate (n=41); the remaining 12% of embryos rupture before elongation starts, a more severe phenotype than normally seen in hmp-1(fe4) embryos, which only rupture 4% of the time (n=35). Targeting only the long isoform of MAGI-1 using dsRNA against the GUK domain only moderately enhances hmp-1(fe4) (data not shown). In contrast, targeting both the long and the short isoforms via dsRNA against the 3rd and 4th PDZ motifs significantly enhances hmp-1(fe4). Thus it seems likely that the long isoform alone plays a relatively minor role in synergizing with the CCC during morphogenesis.
To confirm the RNAi phenotype, we attempted to generate a strain carrying hmp-1(fe4) and a protein null allele of magi-1, magi-1(zh66) [11] however, we were unable to recover viable progeny. magi-1(RNAi) also enhances the penetrance and severity of phenotypes caused by a hypomorphic allele of hmp-2, hmp-2(qm39) [15], and we were able to generate a strain carrying hmp-2(qm39) and magi-1(zh66). Embryos from qm39/qm39; zh66/+ worms exhibited 29.3% lethality (n=868) and embryos from qm39/+; zh66/zh66 worms exhibited 39.3% lethality (n=916). 25% of the (presumably) qm39/qm39; zh66/zh66 embryos ruptured (Figure 1C; Movie 1) and the remaining 75% embryos died at the onset of elongation. That homozygosity for one mutation renders the other haploinsufficient further supports a functional interaction between MAGI-1 and the CCC. Additionally, worms homozygous for either hmp-2(qm39) or magi-1(zh66) and heterozygous for the other allele had body shape defects, including blunted tails, shortened body length, and midsection bulges.
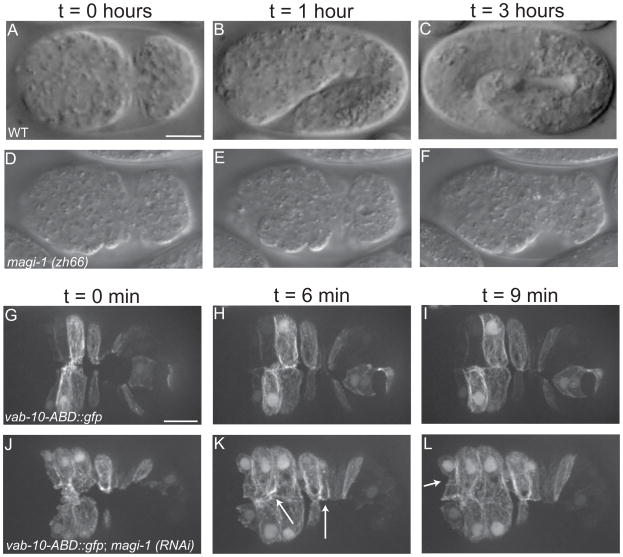
Loss of MAGI-1 disrupts cell migration events during enclosure
Our screen was designed to identify genetic modifiers of CCC-dependent processes during morphogenesis, so we examined ventral enclosure in magi-1 mutant embryos in more detail [16] (Figure 2). A small number of magi-1(zh66) homozygotes (1.3%, n=719; compared to 0.3% lethality, n=1130, in wild-type embryos) arrest during enclosure (Figure 2D–F; Movie 1). To better understand this phenotype, we performed live imaging of magi-1(RNAi) embryos expressing an F-actin reporter [15, 17]. In wild-type embryos, two pairs of anterior leading cells are the first to extend protrusions ventrally. The cells then meet at the ventral midline, forming nascent junctions (Figure 2G; Movie 2). This behavior continues in more posterior cells (pocket cells), which become wedge shaped as they elongate toward the ventral midline (Figure 2G–H). Finally, anterior epidermal cells encase the anterior-most region of the embryo in epidermis prior to elongation [16] (Figure 2I). In magi-1(RNAi) embryos, leading cells extend protrusions, but they often lack directionality and show prolonged protrusive activity (Figure 2J, Movie 2). While leading cells eventually meet and form junctions, these junctions are not straight along the midline; misalignment also occurs in more posterior ventral cells (Figure 2K, arrows). Finally, in contrast to the rectangular shape seen in wild-type embryos, anterior ventral cells in magi-1(RNAi) embryos often have a more oblong shape (Figure 2L). We observed these defects in 70% of magi-1 depleted embryos filmed (n=20) compared to the 1 wild-type embryo in which we observed minor defects in (n=15). These non-uniform cell movements and misalignments of cells at the ventral midline are likely the cause of the morphogenetic failures seen in dead magi-1(zh66) embryos.
Figure 2. Loss of MAGI-1 perturbs ventral enclosure of the epidermis.
(A–C) Wild-type embryos enclose (A), and elongate (B–C), eventually reaching ~4-fold their original length. (D–F) A small percentage of embryos homozygous for the null allele magi-1(zh66) initiate enclosure (D), but never finish (E–F) and die. Scale bar is 10 μm. (G–I) Wild-type embryos expressing an actin reporter during ventral enclosure. During enclosure, cells migrate ventrally, meet their opposing neighbors (G) and form junctions (H) along the ventral midline. (J–L) In magi-1(RNAi) treated embryos, cells still migrate ventrally, but the migration is not as neatly ordered (J). Cells do eventually meet and form junctions, although junctions are often oblique to the ventral midline (arrows, K) and anterior ventral cells often have an irregular shape (arrow, L). Scale bar is 10 μm.
MAGI-1 localizes to a unique domain in the C. elegans apical junction
To address the function of MAGI-1, we examined MAGI-1 localization in comma-stage embryos using an antibody that recognizes both isoforms of MAGI-1 [10]. MAGI-1 localizes to cell junctions in all epithelial tissues in C. elegans. Triple immunostaining of wild-type embryos for MAGI-1, HMP-2/β-catenin and AJM-1 revealed MAGI-1 localizes basal to the CCC and apical to the DLG-1/AJM-1 complex (n=7). There was some overlap between the CCC and MAGI-1; however, HMP-2 signal extended more apically than MAGI-1. We also performed pair-wise colocalization analysis of MAGI-1 and JAC-1::GFP or AJM-1 separately, which confirmed the results obtained via triple immunolabeling, including the slight overlap between the CCC and MAGI-1. (Figure S1).
SAX-7 is important for localizing MAGI-1 at junctions
We next assessed effects on junctional localization of MAGI-1 in living embryos when components of the CCC and DLG-1/AJM-1 complex are depleted. MAGI-1::GFP localizes to junctions during epithelial morphogenesis where it persists through development (Figure 3A–C). We found MAGI-1::GFP is initially targeted to junctions following RNAi-mediated depletion of both HMR-1/cadherin and DLG-1/Discs large, as has been reported in fixed embryos [12]. However, as embryos age, MAGI-1::GFP begins to lose its tight association with junctions (data not shown), indicating the CCC and DLG-1/AJM-1 complex are not crucial for MAGI-1’s initial recruitment, but can affect its maintenance at junctions.
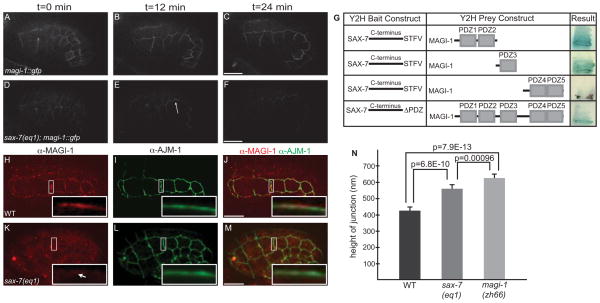
Figure 3. MAGI-1 localization partially depends on SAX-7/L1CAM.
(A–C) Wild-type embryos expressing MAGI-1::GFP. (D–F) The total amount of MAGI-1::GFP at junctions in sax-7(eq1) embryos is decreased, but it is not completely absent. (G) In a directed yeast-two hybrid test, PDZ1-2 and PDZ3 of MAGI-1 can interact with the C-terminus of SAX-7 while PDZ4-5 of MAGI-1 does not. The ability of MAGI-1 to interact with SAX-7 is reduced when the last four amino acids of SAX-7, a PDZ binding motif, are deleted. (H–J) In wild-type embryos, MAGI-1 localizes to junctions (H), apical to AJM-1 (Inset, J). (K–M) In sax-7(eq1) embryos, MAGI-1 accumulation is greatly reduced at junctions (white arrow, inset, K) and the region of AJM-1 expression appears expanded (inset, K). (N) Quantification of the area of AJM-1 expression sax-7(eq1) (556±52 nm, mean ± SEM, n=17) and magi-1(zh66) (623±51 nm, n=15) embryos compared to wild-type (421±46 nm, n=23). Significance was calculated using a two-tailed Student’s t-test. Scale bars are 10 μm.
To identify transmembrane proteins that initially target MAGI-1 to cell-cell junctions, we targeted 93 previously identified cell adhesion receptors [18] and orthologs of vertebrate transmembrane proteins that bind MAGIs using RNAi and mutants [19–23] (Table S2). We found loss of sax-7/L1CAM affected MAGI-1 localization. We crossed magi-1::gfp into sax-7(eq1) animals and found a reduced amount of MAGI-1::GFP at junctions (Figure 3D–F; Movie 3). Immunostaining sax-7(eq1) embryos for MAGI-1 and AJM-1 showed significantly reduced accumulation of MAGI-1 at junctions (cf. Figure 3H–J with K–M). We also quantified the apicobasal expression of AJM-1 in sax-7(eq1) and magi-1(zh66) embryos (556±52 nm, n=17; 623±51 nm, n=15, respectively) and found significant expansion compared to wild-type embryos (421±46 nm, n=23) (Figure 3N). Although sax-7 was not found in our feeding screen, injecting dsRNA against sax-7 into hmp-1(fe4) worms did enhance the penetrance and severity of fe4 phenotypes in a manner similar to depletion of magi-1 (Figure S2).
We used yeast two-hybrid analysis to test for a physical interaction between MAGI-1’s PDZ motifs and the C-terminus of SAX-7, which contains a type I consensus PDZ-binding motif [24]. We found that the SAX-7 C-terminus was able to bind MAGI-1 PDZ1-2, and PDZ3 and that this interaction was abrogated, although not completely eliminated, by deletion of the SAX-7 PDZ binding motif (Figure 3G). The decrease in junctional accumulation of MAGI-1 in sax-7 null mutants and the ability of SAX-7 and MAGI-1 to physically interact suggest SAX-7 is important for recruitment of MAGI-1 to apical junctions.
MAGI-1 influences the localization of AFD-1/afadin at junctions
Because MAGUKs are scaffold proteins [9], we examined the list of screen enhancers to identify potential interacting partners. A promising candidate was AFD-1/afadin/Canoe, an adaptor protein that localizes to cell junctions and can facilitate linkages to the cytoskeleton [25]. Vertebrate afadin can be spliced to produce a shorter variant, AF-6, whose domains–two Ras association (RA1-2) domains, a forkhead-associated (FHA) domain, a Dilute (DIL) domain, a PDZ domain, and a long C-terminus with no significant homology–are conserved in afd-1. Moreover, hmp-1(fe4); afd-1(RNAi) and hmp-1(fe4); magi-1(RNAi) treated embryos exhibit similar phenotypes (Figure S2). We used yeast-two hybrid assays to test for an interaction between MAGI-1 and AFD-1. The PDZ domains of MAGI-1 interact with the RA domains and the C terminus of AFD-1, but not a fragment encoding only the DIL-PDZ domains (Figure 4Q).
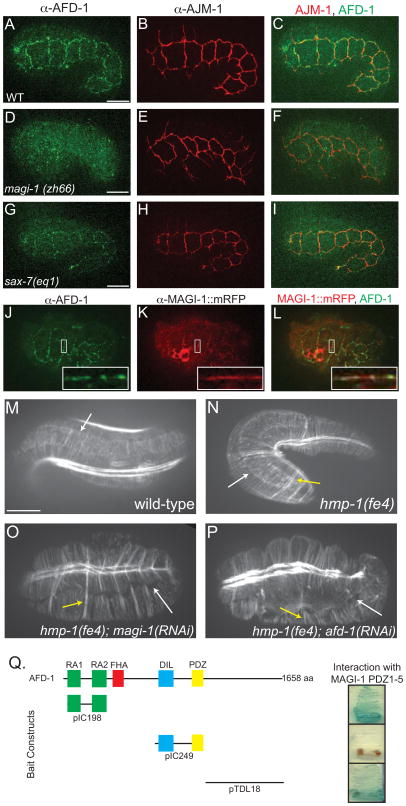
Figure 4. MAGI-1 influences localization of AFD-1.
(A–C) Immunostaining of AFD-1 in wild-type embryos shows AFD-1 accumulates at cell junctions (A), but there is little overlap with AJM-1 (C). (D–F) AFD-1 does not accumulate at cell junctions in magi-1 null embryos (D), even though AJM-1 still localizes to junctions (E). (G–I) Consistent with the interaction between SAX-7 and MAGI-1, AFD-1 accumulation at junctions in sax-7(eq1) embryos is reduced (G). (J–L) Immunostaining against MAGI-1::mRFP (K) and AFD-1 (J) reveals a high degree of colocalization between the two proteins (L, inset). Scale bar is 10 μm. (M–P) Phalloidin staining of wild-type embryos shows accumulation of actin and ordered CFBs anchored at the junction (white arrow, M). (N) CFBs in hmp-1(fe4) embryos are also anchored at the junction (white arrow); however, some of the CFBs are clumped together (yellow arrow). Similar staining in hmp-1(fe4); magi-1(RNAi)(O) and hmp-1(fe4); afd-1(RNAi) (P) embryos reveals a loss of junctional proximal actin and CFBs are completely detached from the junction (white arrows). There are also several clumps of CFBs (yellow arrows). (Q) In a directed yeast two-hybrid test, the PDZ domains in MAGI-1 interacted with the RA domains and the C-terminus, but not the DIL-PDZ domains of AFD-1. Scale bars are 10 μm.
Based on the yeast two-hybrid interaction, we immunostained wild-type (Figure 4A–C), magi-1(zh66) (Figure 4D–F), and sax-7(eq1) (Figure 4G–I) embryos for AFD-1 using affinity purified polyclonal antibodies. AFD-1 accumulates at cell-cell junctions in wild-type embryos in the epidermis, pharynx, and intestine (Figure 4A). We saw dramatically reduced accumulation of AFD-1 at epithelial junctions in both magi-1(zh66) and sax-7(eq1) embryos (Figure 4D, G), indicating MAGI-1 is important for junctional localization of AFD-1. We also examined the apicobasal position of AFD-1 with respect to AJM-1 and found that, like MAGI-1, AFD-1 was predominantly apical to AJM-1 (data not shown). Reslicing through cell-cell junctions of wild-type worms expressing MAGI-1::mRFP immunostained for mRFP and AFD-1 revealed a high degree of overlap between the expression patterns of MAGI-1::mRFP and AFD-1 (Figure 4J–L; Figure 4L, inset).
hmp-1(fe4); magi-1(RNAi) and hmp-1(fe4); afd-1(RNAi) embryos have a disorganized actin cytoskeleton
afd-1 and magi-1 both synergize with hmp-1(fe4) to predominantly yield elongation failure. Elongation of C. elegans embryos depends on a highly organized array of circumferential actin filament bundles, CFBs, which terminate near junctional-proximal accumulations of actin at cell-cell borders in epidermal cells [16]. We used phalloidin staining to determine if AFD-1 or MAGI-1 impacted actin organization during morphogenesis. In wild-type embryos, CFBs are arranged in parallel arrays anchored at junctions, where there is an accumulation of junctional-proximal actin (Figure 4M). In hmp-1(fe4) embryos, junctional-proximal actin is still present and CFBs are still anchored at the junction, but there is some clumping of CFBs (Figure 4N). In hmp-1(fe4); magi-1(RNAi) and hmp-1(fe4); afd-1(RNAi) embryos, there is also clumping of CFBs; however, CFBs present in these embryos are detached from the junction, and the accumulation of junctional-proximal actin is severely disrupted (Figure 4O–P). Since these phenotypes are found in hmp-1 strong loss of function mutants [3], these results suggest MAGI-1 and AFD-1 may modulate the actin cytoskeleton in ways that normally act redundantly with the CCC. To test whether afd-1 and magi-1 act in the same or different genetic pathways, we examined magi-1(zh66); afd-1(RNAi) embryos. We found a noticeable increase in lethality compared with magi-1(zh66) and afd-1(RNAi) alone (Figure S2). These results indicate that although the two proteins can physically interact and colocalize, they may act through partially redundant effector pathways.
MAGI-1 helps to partition junctions during development
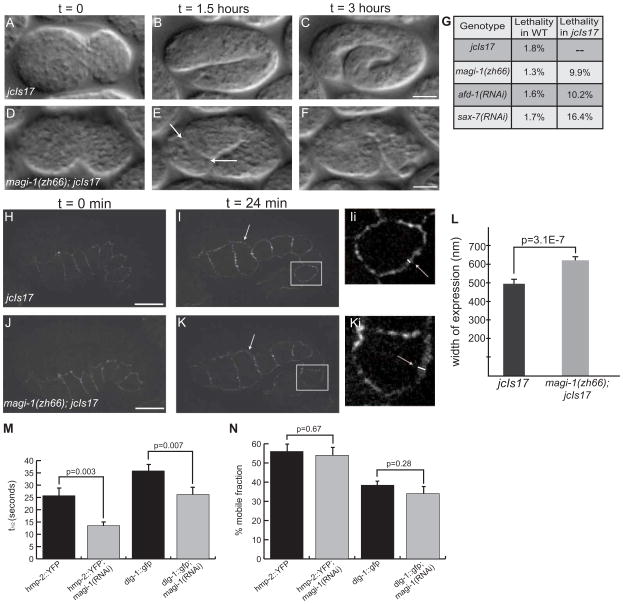
The effects of loss of MAGI-1 on adhesion-dependent processes and its position within the apical junction led us to examine potential roles for MAGI-1 in maintaining spatial ordering of apical junctional components, a theory independently suggested in a recent study [12]. We hypothesized that overexpression of members of apical junctional complexes might lead to more severe phenotypes when MAGI-1 is absent. We therefore generated the strain zh66/zh66; jcIs17 [Phmp-1::hmp-1::gfp, Pdlg-1::dlg-1::dsRed]. In contrast to magi-1(zh66) and jcIs17 embryos (Figure 5A–C), which each display a small amount of lethality (1–2%), depleting magi-1 injcIs17 worms results in ~10% embryonic lethality that is predominantly due to enclosure failure (Figure 5D–F, G; Movie 4). This lethality is most likely due to overexpression of junctional proteins because gfp(RNAi) reduced the lethality in zh66; jcIs17 embryos to 3.25% (n=660). We also knocked down sax-7 and afd-1 in jcIs17 worms and found similar lethality (Figure 5G). These phenotypic similarities provide further evidence that MAGI-1, SAX-7, and AFD-1 can act together during morphogenesis in C. elegans embryos.
Figure 5. MAGI-1 helps to partition junctions during development.
(A–C)jcIs17 embryos enclose (A) and elongate as wild-type embryos do, reaching approximately 4-fold before they hatch (C). (D–F) magi-1(zh66); jcIs17 embryos enclose ventrally, but the anterior epidermis does not complete enclosure. When elongation starts, anterior cells spill out of the opening in the epidermis (arrows, E). Scale bars are 10 μm. (G) Depleting magi-1, afd-1 or sax-7 in jcIs17 embryos causes similar non-additive increases in lethality, also primarily due to anterior enclosure defects. (H–K) At the onset of elongation, jcIs17 (H) and magi-1(zh66); jcIs17 (J) embryos look indistinguishable with respect to HMP-1::GFP. As elongation progresses, HMP-1::GFP expression is expanded at seam-dorsal and seam-ventral boundaries in magi-1(zh66); jcIs17 embryos (arrows, Ii, Ki; white lines indicate width of junctional material). Scale bars are 10 μm. (L) magi-1(zh66); jcIs17 embryos exhibit an expanded area of HMP-1::GFP expression (618±29 nm, mean ± SEM, n=10) in seam cells compared to wild-type embryos (493±49 nm, n=13). Significance was calculated using a two-tailed Student’s t-test. (M) magi-1(RNAi) decreases the half-life (t1/2) of fluorescence recovery after photobleaching in embryos expressing hmp-2::yfp or dlg-1::gfp. (N) magi-1(RNAi) does not significantly affect the mobile fraction of either HMP-2::YFP or DLG-1::GFP.
We filmed magi-1(zh66); jcIs17 embryos to analyze their defects. During enclosure and at the comma stage, HMP-1::GFP is evenly distributed along junctions; localization does not change significantly at the 1.5 fold stage in jcIs17 embryos (Figure 5H, I, Ii). magi-1(zh66); jcIs17 embryos look indistinguishable from jcIs17 embryos during enclosure (Figure 5J) through the comma stage. However, at the 1.5 fold stage, just before embryos start to elongate, magi-1(zh66); jcIs17 embryos display an expanded area of HMP-1::GFP expression at the borders between lateral (seam) and ventral epidermal cells, and at seam-dorsal borders (Figure 5K, Ki). We quantified the expansion of HMP-1::GFP width and found a significant increase in magi-1(zh66); jcIs17 embryos compared to jcIs17 embryos (Figure 5L).
Previous work has shown apical junction formation occurs in two steps: assembly, during which proteins are targeted to the membrane, and establishment, when junctions segregate into domains along the apicobasal axis that are maintained throughout development [5]. Because enclosure and comma stage jcIs17 and zh66/zh66; jcIs17 embryos look indistinguishable, our results are consistent with a role for MAGI-1 in spatial compaction of the apical junction or maintenance of its compacted state. Loss of compaction may in turn weaken cell-cell adhesion.
In addition to aiding apicobasal compaction of the apical junction, MAGI-1 could also stabilize junctional components by restricting their movement. To address this question, we examined the mobility of junctional proteins in wild-type and magi-1(RNAi) treated embryos using Fluorescence Recovery After Photobleaching (FRAP). In hmp-2::yfp embryos, depleting MAGI-1 significantly reduced the half-life of recovery after photobleaching, t1/2, from 25.7±6.2 sec to 13.5±2.8 sec (n=6, significantly different in a two-tailed Student’s t-test, p=0.003) (Figure 5M). In dlg-1::gfp embryos, magi-1(RNAi) significantly reduced the half-life of recovery from t1/2 = 35.8±5.1 sec to t1/2 = 26.2±5.6 sec (n=6, p=0.007) (Figure 5M). magi-1(RNAi) treatment did not significantly change the percent mobile fraction for either protein (hmp-2::yfp vs. hmp-2::yfp; magi-1(RNAi): 56±7.4% vs. 54±7.8%, n=6, p=0.673; dlg-1::gfp vs. dlg-1::gfp; magi-1(RNAi): 38.4±4% vs. 34±7.3%, n=6, p=0.28) (Figure 5N). These results suggest that MAGI-1 normally stabilizes proteins once they arrive and assemble at the apical junction.
Discussion
An enhancer screen identifies putative adhesion modulators during C. elegans morphogenesis
C. elegans is a useful system for identifying additional conserved modulators of cell-cell adhesion in metazoans [2]. Because forward genetic approaches do not always identify modifiers or redundant genes, we used genome-wide RNAi in a hypomorphic hmp-1/α-catenin mutant background to uncover 55 novel regulators of cell junctions in vivo. We focused on magi-1, a critical regulator of junctional stability in vertebrate tissue culture [26, 27]. We subsequently confirmed the genetic interaction between magi-1 and the CCC using magi-1 and hmp-2/β-catenin mutants, providing further support for their functional interaction.
Our screen also identified the junctional proteins jac-1/p120-catenin [14], zoo-1/ZO-1 [15] and srgp-1/srGAP [28], whose synergy with hmp-1(fe4) we have previously reported. Additionally, we identified cytoskeletal modulators including unc-34/Enabled [29], TMD-1/tropomodulin [30], and FLI-1/Flightless, which has been implicated in numerous non-epithelial processes in C. elegans [31]. We also identified proteins involved in vesicular trafficking. These include a component of the exocyst complex (SEC-8) and components of the AP-2 clathrin adaptor complex (APS-2, APA-2), which could be involved in trafficking CCC components. We also found genes involved in protein turnover, including EEL-1, an E3 ligase that has been implicated in indirect regulation of hemidesmosome-like structures in C. elegans [32]. In summary, our screen is the first genome-wide approach to identifying proteins that act alongside the CCC during morphogenesis and should provide the basis for future experiments to clarify the roles of the CCC interactome during morphogenesis.
MAGI-1 aids collective cell migration during epidermal morphogenesis
We identified a role for MAGI-1 in modulating cell-cell contact during ventral enclosure, consistent with work in vertebrate tissue culture, where loss of MAGI-1 weakens cell-cell adhesion, leading to dismantling of cell junctions [26, 27]. Loss of C. elegans magi-1 function could similarly weaken cell junctions, thereby impairing normal coordination of cell movements during ventral enclosure. Because defects in anterior epidermal cell migration occur before contralateral cells make nascent junctional connections, the non-uniform shape of ventral cells during enclosure may reflect weakened adhesion along lateral edges of cells when MAGI-1 is absent. Similarly, over-reaching of contralateral cells at the ventral midline may reflect lack of suppression of motility upon ventral midline contact. Similar defects in collective cell migration have been observed in cultured cells during healing of scratch wounds in the presence of function-blocking anti-E-cadherin antibodies [33].
MAGI-1 localizes between the CCC and the DLG-1/AJM-1 complex at the junction
While MAGI-1 had been reported to be at cell-cell junctions [10], we determined that MAGI-1 localizes to a unique junctional domain between the CCC and the DLG-1/AJM-1 complex. Our results are contrast with a recent paper that placed MAGI-1 apical to both the CCC and the DLG-1/AJM-1 complex [12]. This difference could possibly be due to localization differences between endogenous MAGI-1, which we examined in our antibody staining experiments, and fluorescently tagged MAGI-1, used in the previous study. The embryo presented in [12] was also significantly older than the embryos examined here; however, we examined embryos of various ages and found no changes in apicobasal ordering of proteins at the apical junction as development proceeded. Our results are internally consistent and reflect numerous permutations of pair-wise and triple labeling.
Localization of MAGI-1 to junctions partly depends on SAX-7/L1CAM
We failed to find any candidates whose removal completely prevented localization of MAGI-1 at junctions. This is not surprising since multiple domains of the protein are sufficient to target it to junctions [12]. We did, however, see significant effects on MAGI-1 localization due to loss of sax-7 function. A yeast two-hybrid assay further demonstrated that SAX-7 and MAGI-1 can physically interact, suggesting that localization of MAGI-1 to junctions depends in part on SAX-7 binding. The expanded area of expression of AJM-1 in sax-7 null embryos relative to wild-type also supports this view, because expansion of the DLG-1/AJM-1 complex had been previously demonstrated in magi-1 null embryos [12].
MAGI-1 interacts downstream with AFD-1/afadin/AF6 and regulates the actin cytoskeleton at junctions
We showed that MAGI-1 and AFD-1, another screen enhancer, can interact in a directed yeast two-hybrid assay via the RA domains and the C-terminus of AFD-1. Moreover, AFD-1 localization is significantly disrupted in magi-1 null embryos. Importantly, afd-1(RNAi) has no discernable effect on the ability of MAGI-1::GFP to localize to junctions (data not shown). To our knowledge, this is the first report linking MAGI-1 and AFD-1/afadin. Afadin has previously been linked to ZO-1, another MAGUK, in both vertebrates and Drosophila [34, 35], but not to MAGIs. While we cannot rule out an interaction between AFD-1 and the C. elegans ZO ortholog, ZOO-1, our antibody staining shows that AFD-1 and MAGI-1 colocalize at cell junctions, while ZOO-1 localizes predominantly with the CCC [15]. We conclude that AFD-1 and MAGI-1 are part of the same junctional subdomain.
The disorganized actin phenotypes seen in both hmp-1(fe4); magi-1(RNAi) and hmp-1(fe4); afd-1(RNAi) embryos raise the possibility that MAGI-1 influences the actin cytoskeleton through AFD-1. Since MAGI-1 and AFD-1 are spatially distinct from the CCC, our results further suggest that MAGI-1 and AFD-1 modulate the actin cytoskeleton in ways that normally act redundantly with the CCC. While the C. elegans afadin ortholog is missing a conserved actin-binding domain at the C-terminus, the remainder of the C-terminus is intact, so it is possible that the AFD-1 C-terminus can interact with actin. AFD-1 also possesses two RA domains, which are capable of interacting with Rap1, a known cytoskeletal modulator in other systems [36]. Knocking down either magi-1 or afd-1 could affect RAP activity at junctions, contributing to the actin organization defects seen in hmp-1(fe4); magi-1(RNAi) and hmp-1(fe4); afd-1(RNAi) embryos. Determining if AFD-1 can bind actin and interact with RAPs could address these possibilities in the future.
MAGI-1 may act to separate the CCC and DLG-1/AJM-1 complexes
A recent paper suggested that MAGI-1 has a role in maintaining the spatial separation of complexes at the apical junction [12]. Our examination of fluorescent protein expression in magi-1 null mutants overexpressing HMP-1 and DLG-1 supports this experimentally. HMP-1::GFP expression was expanded at seam-ventral and seam-dorsal boundaries in 1.5-fold magi-1 null embryos, immediately before embryos commence elongation, but not during enclosure, when nascent junctions are being formed. Elongation places stress on cell junctions, as the contractile forces are transmitted to CFBs anchored at epidermal cell-cell junctions. The expansion of the domain containing HMP-1::GFP in magi-1(zh66) embryos suggests loss of magi-1 causes a weakening of junctions at this time. Our FRAP analysis further supports this idea. The increased rates of recovery after photobleaching in embryos expressing either DLG-1::GFP or HMP-2::YFP following magi-1 knockdown suggest that once they are incorporated into mature junctions, MAGI-1 may stabilize both the CCC and DLG-1/AJM-1 complex proteins.
Given its localization between the two complexes in our hands, MAGI-1 may separate the two complexes, and stabilize them into a more structurally rigid complex. This model is consistent with the expansion of AJM-1 expression in sax-7 null embryos, the expansion of HMP-1::GFP in magi-1 null embryos, and the mixing of junctional complexes previously reported by others [12]. It also explains the increases in lethality seen in jcIs17 embryos, which over-express HMP-1::GFP and DLG-1::dsRed, following knockdown of magi-1, sax-7, or afd-1. Excess proteins from the two complexes may further exacerbate mixing of the two subdomains, further disorganizing the junction.
Conclusion
We have performed the first genome-wide functional screen for proteins that act alongside the CCC during morphogenesis. MAGI-1, along with SAX-7/L1CAM and AFD-1/afadin, are part of a functional interactome that includes the core CCC. We favor a model in which MAGI-1 serves as a partition between the CCC and DLG-1/AJM-1 complex, acting both to keep the complexes spatially separated, and to stabilize them into a more rigid structure.
Our results also suggest that MAGI-1 is anchored at the plasma membrane in part through an interaction with SAX-7. A connection to the actin cytoskeleton via AFD-1 could stabilize the MAGI-1 partition at its cytoplasmic interface. Such a connection to the actin cytoskeleton could act in parallel to HMP-1, which could explain the enhancement of hmp-1(fe4) we see following loss of function for either magi-1 or afd-1. Our results expand our understanding of the in vivo roles of MAGI-1 in cell-cell adhesion, and underscore the utility of C. elegans as a model system for identifying conserved components of a functionally interacting network of proteins that facilitate cell-cell adhesion in a living embryo.
Supplementary Material
Highlights.
Our genome-wide screen identified functional interactors with the CCC
MAGI-1 physically binds AFD-1/afadin and loss of either disrupts the actin cytoskeleton
MAGI-1 accumulation at junctions depends in part on SAX-7/L1CAM
MAGI-1 helps to partition and stabilize junctions during morphogenesis
Acknowledgments
This work was supported by NIH grant R01 GM58038 to J.H, a Genetics program predoctoral training grant NIH 5T32GM07133 to A.M.L., NIH grant NRSA GM067410 and grant R15 HD059952 to E.A.C., and an American Cancer Society Postdoctoral Fellowship awarded to T.G. We thank Yuji Kohara for providing cDNAs, A. Stetak (Univ. of Basel) for sharing the MAGI-1::GFP strain, T. Loveless (Univ. of Wisconsin) and L. Chen (Univ. of Minnesota) for providing Y2H constructs, and C. Rongo (Rutgers Univ) for the MAGI-1 antibody.
Abbreviations
- CeAJ
C. elegans apical junction
- CCC
cadherin-catenin complex
- CFB
circumferential filament bundle
- MAGUK
membrane-associated guanylate kinase
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Niessen CM, Leckband D, Yap AS. Tissue organization by cadherin adhesion molecules: dynamic molecular and cellular mechanisms of morphogenetic regulation. Physiol Rev. 2011;91:691–731. doi: 10.1152/physrev.00004.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lynch AM, Hardin J. The assembly and maintenance of epithelial junctions in C. elegans. Front Biosci. 2009;14:1414–1432. doi: 10.2741/3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Costa M, Raich W, Agbunag C, Leung B, Hardin J, Priess JR. A putative catenin-cadherin system mediates morphogenesis of the Caenorhabditis elegans embryo. J Cell Biol. 1998;141:297–308. doi: 10.1083/jcb.141.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koppen M, Simske JS, Sims PA, Firestein BL, Hall DH, Radice AD, Rongo C, Hardin JD. Cooperative regulation of AJM-1 controls junctional integrity in Caenorhabditis elegans epithelia. Nat Cell Biol. 2001;3:983–991. doi: 10.1038/ncb1101-983. [DOI] [PubMed] [Google Scholar]
- 5.McMahon L, Legouis R, Vonesch JL, Labouesse M. Assembly of C. elegans apical junctions involves positioning and compaction by LET-413 and protein aggregation by the MAGUK protein DLG-1. J Cell Sci. 2001;114:2265–2277. doi: 10.1242/jcs.114.12.2265. [DOI] [PubMed] [Google Scholar]
- 6.Heasman J, Ginsberg D, Geiger B, Goldstone K, Pratt T, Yoshida-Noro C, Wylie C. A functional test for maternally inherited cadherin in Xenopus shows its importance in cell adhesion at the blastula stage. Development. 1994;120:49–57. doi: 10.1242/dev.120.1.49. [DOI] [PubMed] [Google Scholar]
- 7.Larue L, Ohsugi M, Hirchenhain J, Kemler R. E-cadherin null mutant embryos fail to form a trophectoderm epithelium. Proc Natl Acad Sci U S A. 1994;91:8263–8267. doi: 10.1073/pnas.91.17.8263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Muller HA, Wieschaus E. armadillo, bazooka, and stardust are critical for early stages in formation of the zonula adherens and maintenance of the polarized blastoderm epithelium in Drosophila. J Cell Biol. 1996;134:149–163. doi: 10.1083/jcb.134.1.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Funke L, Dakoji S, Bredt DS. Membrane-associated guanylate kinases regulate adhesion and plasticity at cell junctions. Annu Rev Biochem. 2005;74:219–245. doi: 10.1146/annurev.biochem.74.082803.133339. [DOI] [PubMed] [Google Scholar]
- 10.Emtage L, Chang H, Tiver R, Rongo C. MAGI-1 modulates AMPA receptor synaptic localization and behavioral plasticity in response to prior experience. PLoS One. 2009;4:e4613. doi: 10.1371/journal.pone.0004613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stetak A, Horndli F, Maricq AV, van den Heuvel S, Hajnal A. Neuron-specific regulation of associative learning and memory by MAGI-1 in C. elegans. PLoS One. 2009;4:e6019. doi: 10.1371/journal.pone.0006019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stetak A, Hajnal A. The C. elegans MAGI-1 protein is a novel component of cell junctions that is required for junctional compartmentalization. Dev Biol. 2010;350:24–31. doi: 10.1016/j.ydbio.2010.10.026. [DOI] [PubMed] [Google Scholar]
- 13.Kamath RS, Ahringer J. Genome-wide RNAi screening in Caenorhabditis elegans. Methods. 2003;30:313–321. doi: 10.1016/s1046-2023(03)00050-1. [DOI] [PubMed] [Google Scholar]
- 14.Pettitt J, Cox EA, Broadbent ID, Flett A, Hardin J. The Caenorhabditis elegans p120 catenin homologue, JAC-1, modulates cadherin-catenin function during epidermal morphogenesis. J Cell Biol. 2003;162:15–22. doi: 10.1083/jcb.200212136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lockwood C, Zaidel-Bar R, Hardin J. The C. elegans zonula occludens ortholog cooperates with the cadherin complex to recruit actin during morphogenesis. Curr Biol. 2008;18:1333–1337. doi: 10.1016/j.cub.2008.07.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chisholm AD, Hardin J. WormBook. 2005. Epidermal morphogenesis; pp. 1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gally C, Wissler F, Zahreddine H, Quintin S, Landmann F, Labouesse M. Myosin II regulation during C. elegans embryonic elongation: LET-502/ROCK, MRCK-1 and PAK-1, three kinases with different roles. Development. 2009;136:3109–3119. doi: 10.1242/dev.039412. [DOI] [PubMed] [Google Scholar]
- 18.Cox EA, Tuskey C, Hardin J. Cell adhesion receptors in C. elegans. J Cell Sci. 2004;117:1867–1870. doi: 10.1242/jcs.01177. [DOI] [PubMed] [Google Scholar]
- 19.Xu Z, Peng AW, Oshima K, Heller S. MAGI-1, a candidate stereociliary scaffolding protein, associates with the tip-link component cadherin 23. J Neurosci. 2008;28:11269–11276. doi: 10.1523/JNEUROSCI.3833-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hirabayashi S, Tajima M, Yao I, Nishimura W, Mori H, Hata Y. JAM4, a junctional cell adhesion molecule interacting with a tight junction protein, MAGI-1. Mol Cell Biol. 2003;23:4267–4282. doi: 10.1128/MCB.23.12.4267-4282.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wegmann F, Ebnet K, Du Pasquier L, Vestweber D, Butz S. Endothelial adhesion molecule ESAM binds directly to the multidomain adaptor MAGI-1 and recruits it to cell contacts. Exp Cell Res. 2004;300:121–133. doi: 10.1016/j.yexcr.2004.07.010. [DOI] [PubMed] [Google Scholar]
- 22.Patrie KM, Drescher AJ, Goyal M, Wiggins RC, Margolis B. The membrane-associated guanylate kinase protein MAGI-1 binds megalin and is present in glomerular podocytes. J Am Soc Nephrol. 2001;12:667–677. doi: 10.1681/ASN.V124667. [DOI] [PubMed] [Google Scholar]
- 23.Ide N, Hata Y, Nishioka H, Hirao K, Yao I, Deguchi M, Mizoguchi A, Nishimori H, Tokino T, Nakamura Y, et al. Localization of membrane-associated guanylate kinase (MAGI)-1/BAI-associated protein (BAP) 1 at tight junctions of epithelial cells. Oncogene. 1999;18:7810–7815. doi: 10.1038/sj.onc.1203153. [DOI] [PubMed] [Google Scholar]
- 24.Zhou S, Opperman K, Wang X, Chen L. unc-44 Ankyrin and stn-2 gamma-syntrophin regulate sax-7 L1CAM function in maintaining neuronal positioning in Caenorhabditis elegans. Genetics. 2008;180:1429–1443. doi: 10.1534/genetics.108.091272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sawyer JK, Harris NJ, Slep KC, Gaul U, Peifer M. The Drosophila afadin homologue Canoe regulates linkage of the actin cytoskeleton to adherens junctions during apical constriction. J Cell Biol. 2009;186:57–73. doi: 10.1083/jcb.200904001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gregorc U, Ivanova S, Thomas M, Guccione E, Glaunsinger B, Javier R, Turk V, Banks L, Turk B. Cleavage of MAGI-1, a tight junction PDZ protein, by caspases is an important step for cell-cell detachment in apoptosis. Apoptosis. 2007;12:343–354. doi: 10.1007/s10495-006-0579-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kranjec C, Banks L. A systematic analysis of HPV E6 PDZ substrates identifies MAGI-1 as a major target of HPV-16 and HPV-18 whose loss accompanies disruption of Tight Junctions. J Virol. 2010 doi: 10.1128/JVI.01756-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zaidel-Bar R, Joyce MJ, Lynch AM, Witte K, Audhya A, Hardin J. The F-BAR domain of SRGP-1 facilitates cell-cell adhesion during C. elegans morphogenesis. J Cell Biol. 2010;191:761–769. doi: 10.1083/jcb.201005082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sheffield M, Loveless T, Hardin J, Pettitt J. C. elegans Enabled exhibits novel interactions with N-WASP, Abl, and cell-cell junctions. Curr Biol. 2007;17:1791–1796. doi: 10.1016/j.cub.2007.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cox-Paulson EA, Walck-Shannon E, Lynch AM, Yamashiro S, Zaidel-Bar R, Eno CC, Ono S, Hardin J. Tropomodulin Protects alpha-Catenin-Dependent Junctional-Actin Networks under Stress during Epithelial Morphogenesis. Curr Biol. 2012 doi: 10.1016/j.cub.2012.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deng H, Xia D, Fang B, Zhang H. The Flightless I homolog, fli-1, regulates anterior/posterior polarity, asymmetric cell division and ovulation during Caenorhabditis elegans development. Genetics. 2007;177:847–860. doi: 10.1534/genetics.107.078964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zahreddine H, Zhang H, Diogon M, Nagamatsu Y, Labouesse M. CRT-1/calreticulin and the E3 ligase EEL-1/HUWE1 control hemidesmosome maturation in C. elegans development. Curr Biol. 2010;20:322–327. doi: 10.1016/j.cub.2009.12.061. [DOI] [PubMed] [Google Scholar]
- 33.Lorger M, Moelling K. Regulation of epithelial wound closure and intercellular adhesion by interaction of AF6 with actin cytoskeleton. J Cell Sci. 2006;119:3385–3398. doi: 10.1242/jcs.03027. [DOI] [PubMed] [Google Scholar]
- 34.Takahashi K, Matsuo T, Katsube T, Ueda R, Yamamoto D. Direct binding between two PDZ domain proteins Canoe and ZO-1 and their roles in regulation of the jun N-terminal kinase pathway in Drosophila morphogenesis. Mech Dev. 1998;78:97–111. doi: 10.1016/s0925-4773(98)00151-8. [DOI] [PubMed] [Google Scholar]
- 35.Yamamoto T, Harada N, Kano K, Taya S, Canaani E, Matsuura Y, Mizoguchi A, Ide C, Kaibuchi K. The Ras target AF-6 interacts with ZO-1 and serves as a peripheral component of tight junctions in epithelial cells. J Cell Biol. 1997;139:785–795. doi: 10.1083/jcb.139.3.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kooistra MR, Dube N, Bos JL. Rap1: a key regulator in cell-cell junction formation. J Cell Sci. 2007;120:17–22. doi: 10.1242/jcs.03306. [DOI] [PubMed] [Google Scholar]
- 37.Cram EJ, Shang H, Schwarzbauer JE. A systematic RNA interference screen reveals a cell migration gene network in C. elegans. J Cell Sci. 2006;119:4811–4818. doi: 10.1242/jcs.03274. [DOI] [PubMed] [Google Scholar]
- 38.Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, Kanapin A, Le Bot N, Moreno S, Sohrmann M, et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.