Abstract
In recent years, quantification of absolute protein numbers in cellular structures using fluorescence microscopy has become a reality. Two popular methods are available to a broad range of researchers with minimal equipment and analysis requirements: stepwise photobleaching to count discrete changes in intensity from a small number of fluorescent fusion proteins, and comparing the fluorescence intensity of a protein to a known in vivo or in vitro standard. This review summarizes the advantages and disadvantages of each method, and gives recent examples of each that answer important questions in their respective fields. We also highlight new counting methods that could become widely available in the future.
Live-cell protein quantification
The discovery and utilization of green fluorescent protein (GFP) from Aequorea victoria for cell imaging is a watershed achievement in biological research. Fluorescence occurs when light is emitted from the fluorophore in response to an absorbed light, and GFP is capable of fluorescence without enzymatic modification or a cofactor, allowing expression of a single gene to result in detectable emission in any organism. Fluorescence imaging has since become a powerful tool to answer many questions in biology.
Cell biology is becoming increasingly quantitative. Many researchers are interested in counting protein molecules in live cells to define stoichiometry of functional protein complexes and to build models of cellular structures [1–23]. As technology and equipment improve, quantitative fluorescence microscopy is becoming more accurate. Genome-wide studies might miss information about low abundance proteins or local protein concentrations [24–26], stressing the need for single-cell and even single-molecule experiments.
Although various methods for counting proteins in vivo exist, this review focuses on two fluorescence microscopy methods that are currently the most accessible to most researchers: stepwise photobleaching and ratio comparison to fluorescent standards. Specific details of the methods have been reported elsewhere [27–29], so this review focuses on the advantages and disadvantages of both methods and some applications of each. Both methods can utilize standard imaging equipment and fluorescent fusion proteins (see glossary box), without requiring specialized systems or analysis software. This review also touches on some new methods that will likely be useful for protein quantification in the future.
Glossary Box.
- Blinking
reversible loss of emission intensity from FPs due to transition to a nonemissive triplet state more likely to occur at higher excitation intensities
- Diffraction limit
the best resolution that can be obtained by a light microscope, given by optical emission wavelength (λ) divided by two times the numerical aperture (N.A.) of the objective lens (λ/2N.A.); ~200 nm at best.
- Flow cytometry
a process by which cells or microscopic particles in suspension flow past a detector one at a time and the detector counts the number and records the fluorescence intensity and other parameters
- Fluorescence correlation spectroscopy (FCS)
a technique in which fluctuations of fluorescence intensity are measured within a small volume and physical properties (e.g. rate of diffusion, concentration of molecules, interactions) of the fluorescent molecules passing through that volume can be mathematically extracted using autocorrelation functions
- Fluorescent fusion protein
the gene for a fluorescent protein, such as GFP, is inserted in frame up- or downstream of the gene for a protein of interest, so that when transcribed and translated, the resulting protein of interest is fused to GFP
- Förster resonance energy transfer (FRET)
energy transfer from a donor fluorophore to an acceptor fluorophore in close proximity (<10 nm and depending on the alignment of the fluorophores with respect to one another) when the donor emission wavelength overlaps the acceptor excitation wavelength
- Full width at half maximum (FWHM)
on a Gaussian curve, the width of the curve at a height that is half the maximum height. The FWHM of the point spread function approximates Z-axis or axial resolution
- Maturation efficiency
the time it takes for a fluorophore, such as GFP, to mature to its fluorescent state via rearrangements and chemical reactions among amino acids
- Noise
inconstant imprecise output above and below a real signal that disturbs or interferes with detection of the signal, usually referred to as ‘snow’ on a television screen when the broadcast signal is lost
- Photobleaching
irreversible loss of fluorescence due to exposure to an excitation light source
- Point spread function (PSF)
the apparent blurring of intensity from a point source of light, such as a fluorescent bead or protein, due to diffraction of light by the lens
- Super-resolution microscopy
any technique that breaks the diffraction limit of fluorescence microscopy (~200 nm) by pinpointing the exact location of point sources and representing the image using those locations rather than the additive point spread functions of all point sources in an imaging field
- Zinc finger nuclease (ZFN)
a protein constructed by fusing a zinc finger, which is a DNA binding motif, to a restriction enzyme usually FokI is used because it has a non-specific cleavage site. Zinc fingers can be combined to recognize specific sequences of DNA on either side of a desired cleavage site and FokI dimerizes in the middle and creates a double-stranded break. Existing DNA repair mechanisms can utilize homologous sequences supplied exogenously to insert DNA while repairing the break
Counting protein molecules by stepwise photobleaching
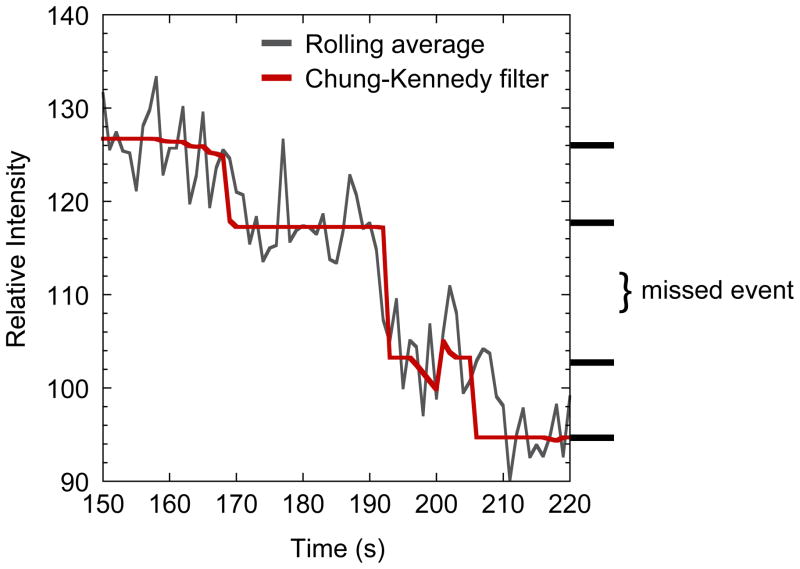
One fluorescence microscopy method for counting protein molecules is stepwise photobleaching. This approach relies on the irreversible and stochastic loss of fluorescence from repeated exposure of fluorescent proteins (FPs) to a light source. The method involves continuous exposure of the sample to the excitation light at low enough intensity that the sample is slowly bleached until its emission intensity reaches background level. The appropriate light intensity and exposure time differ depending on the number of fluorescent molecules in the structure of interest. The key to optimizing these conditions is to minimize missed bleaching events that occur when two (or more) fluorophores are bleached simultaneously or too closely to be resolved, resulting in a step approximately twice the size of other steps (Figure 1). Because the likelihood of missed events increases exponentially with the number of molecules in a structure [28], the bleaching method is only useful for low protein numbers. Das et al. estimate that a maximum of 15 bleaching steps can be directly detected without mathematical extrapolation, although they detected no more than seven steps in their experiments [30].
Figure 1.
Missed bleaching events. A blown up image of a photobleaching curve with two bleaching events too close to be counted separately. The dark gray trace is a rolling average of every three data points from raw bleaching data, whereas the red trace is Chung-Kennedy filtered. See text for a description of the filter. The location of plateaus is marked on the right side, and the 2× drop marked as a missed event. Modified from the MotB data in ©Coffman et al., 2011. Originally published in J. Cell Biol. doi: 10.1083/jcb.201106078.
The upper limit for the number of countable molecules, using the photobleaching method, can be extended using mathematical aids. Several groups have simulated bleaching events to estimate the likely number of visible bleach steps [28,30,31]. This approach is essential when using dyes that have <100% labeling efficiency, or fluorophores that have <100% maturation efficiency. Another way to estimate the number of bleach steps is to measure the steps that are visible (the difference of consecutive plateau means), calculate the step size of bleaching a single fluorophore using a gamma distribution, and divide the starting intensity by the step size [2,10,32,33]. The histogram of photobleaching step sizes can include steps that are >1 bleaching event, therefore the mode of the gamma distribution (μ − SD2/μ, where μ = mean and SD = standard deviation) represents the most probable step size from bleaching one molecule [2,33]. We do not recommend a Fourier (or power) spectral analysis of the step sizes to determine the size that occurs with the highest frequency, because it is technically more difficult than calculating the mode of the gamma distribution and does not significantly improve the accuracy [10,32]. Even with mathematical aids, the largest reported number counted by photobleaching is ~30 molecules [32].
Whether the bleaching events are counted directly or the step size is calculated, analysis of the data is more complex than the ratio imaging method. A background correction is necessary to remove fluorescence from diffused proteins and other background sources from the starting intensity. For photobleaching data, regions of interest (ROIs) should be selected to minimize the chance of emission overlap between multiple structures [28,30]. Because the raw data is noisy, it is also necessary to filter the data to reveal the discrete drops. Three filters were recently evaluated for quantification of the bacterial replisome [15] (Figure 2). An edge-preserving running average, known as the Chung-Kennedy filter, is the most appropriate of those tested. Briefly, it calculates the mean and SD in two consecutive sets within the data from one photobleaching ROI, and reports the mean of the set with the lower SD. If a set includes points along two plateaus, the SD will be higher, and it will not be reported. Thus the reported values lie along single plateaus, and the steps are preserved [2,32] (Figure 1). It is similar to a running median but produces an SD ~30% lower [15]. The number of data points in the set that is averaged should be large enough to reduce the noise but small enough to ensure that few steps are missed [15]. Leake et al. used a modified photobleaching approach to quantify mobile particles at the cell membrane in Escherichia coli, which included automated tracking of particles during bleaching [11]. ImageJ plug-ins are available for automated particle tracking, and the Chung-Kennedy filter can be applied using formulas in a spreadsheet application, thus both analyses are executable by most researchers.
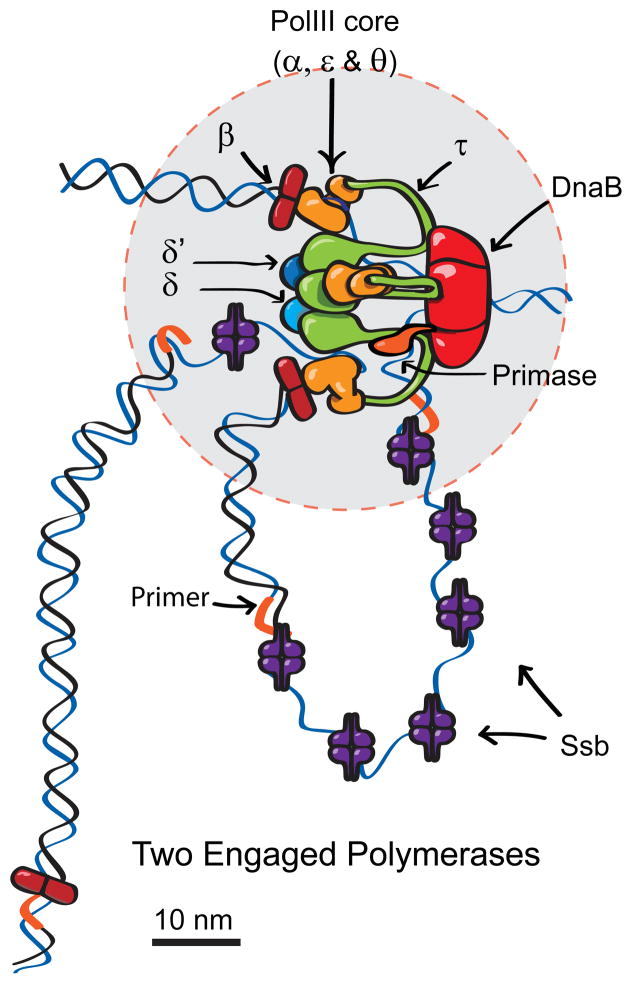
Figure 2.
A revised model of the bacterial DNA polymerase based on stoichiometry data obtained using stepwise photobleaching. Three Pol III polymerase cores (orange) exist in each replisome, two of which are usually associated with β clamps, and the third might be poised to extend the next lagging strand primer. A third β clamp is distant to the core replisome (50 nm gray circle) in 75% of replisomes. Previous models included only two polymerases coordinately replicating the leading and lagging DNA strands. The presence of a third Pol III may help lagging strand synthesis keep pace with leading strand synthesis. From Reyes-Lamothe et al., Science, 2010. Reprinted with permission from AAAS.
Quantification by ratio comparison to fluorescent standards
Another straightforward approach for counting protein molecules is to measure the ratio of the fluorescence intensities of a protein of interest to a standard with a known molecule number (discussed later). In essence, this method uses a series of images of cells that express either the protein of interest or the standard, each fused to an FP that has suitable fluorescent properties (see the section on properties of fluorescent proteins). If the standard can be distinguished from the protein of interest, it is desirable to include cells that express the standard and experimental fusion proteins on the same slide to ensure comparable illumination. If the standard is not distinguishable, images can be taken consecutively or another marker can be imaged separately to distinguish the control cells [27,34] as long as the two fluorophores are sufficiently distant to eliminate Förster resonance energy transfer (FRET). One advantage of this method is that a much greater number of protein molecules can be counted than with the photobleaching method.
Several corrections might be necessary to achieve accurate measurements. Correcting for uneven illumination in the microscope system is needed if the whole field is used, but this is optional if measurements are only taken near the center of a field [2,27,29,35]. If the molecules of interest are at different depths relative to the coverslip, the effect of depth on intensity should be calibrated and corrected using fluorescent beads [27,36]. This correction is different for each system but is essential if an in vitro standard is measured using total internal reflection fluorescence (TIRF) microscopy in which the sample is much closer to the coverslip compared to the molecules in a cell. Fluorescence intensity and exposure time are linearly proportional within a certain range of exposure times for each camera, so different exposure times can be utilized and corrected to keep the signal to noise ratio high and avoid saturation [29,36]. Exposure times below or above the linear range will skew the intensity by camera noise or photobleaching, respectively [29]. Changes to excitation intensity might result in nonlinear changes to photon counts due to the increased likelihood of blinking molecules as excitation intensity increases [37]; therefore excitation intensity should be kept constant between samples. After appropriate corrections, the ROI should be chosen to include >90% of the signal in the structure [2,27]. Complex analysis to determine the centroids of fluorescent spots is not always necessary as it yields similar data compared to simply fitting an ROI centered on the brightest pixel [27,28]. Similar to the bleaching method, intracellular background must be subtracted. The background should be taken from a concentric area unless there are overlapping neighboring signals or an inhomogeneous cytoplasmic intensity [9,18,27,35].
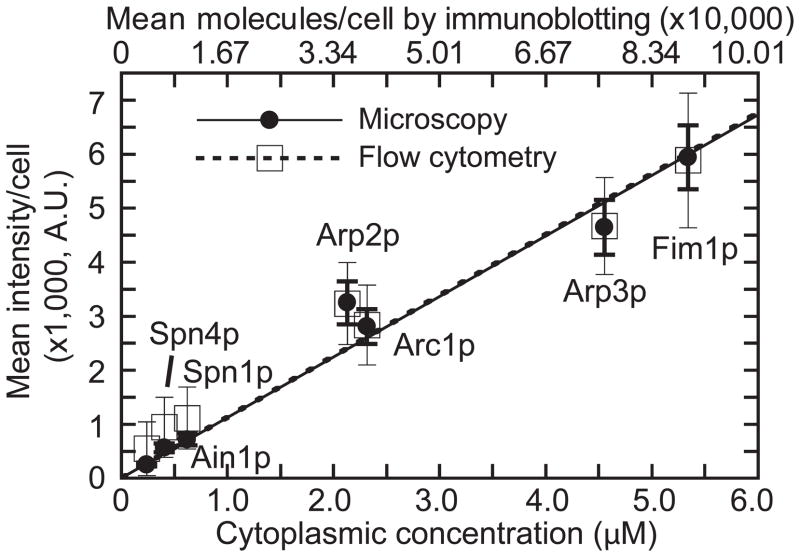
The most important factor for the ratio comparison method is a trustworthy standard. Some commonly used standards were recently evaluated [2,36,38], resulting in a wide variety of reliable standards that can now be used to generate a standard curve without the need for immunoblotting or flow cytometry [29] (Figure 3). The standards used were: 1) MotB, a protein from E. coli that has 22 molecules per motor [10]; 2) purified enhanced GFP (EGFP with F64L and S65T mutations) diluted to view single molecules [36]; and 3) viral particles that have 120 EGFP per assembled capsid [39]. Purified EGFP and viral particles are extracellular, and MotB is intracellular, making it necessary to verify equivalent fluorescence in different conditions (see the section on properties of fluorescent proteins).
Figure 3.
Standard curve for counting protein molecules. Mean numbers of mYFP-tagged protein molecules per cell and cellular concentrations, both from immunoblotting, correlate linearly with cell-size corrected mean fluorescence intensity per cell from microscopy (solid line and filled circles) and flow cytometry (dashed line and open squares). The fission yeast cytokinesis proteins shown are small to medium sized (377–621 amino acids) and cover a range of cellular concentrations (0.22–5.34 μM). Ain1: α-actinin-like actin crosslinking protein; Spn1 and Spn4: septin GTPases; Arc1, Arp2, and Arp3: subunits of the actin filament-nucleating Arp2/3 complex; and Fim1: actin crosslinker fimbrin. From Wu and Pollard, Science, 2005. Reprinted with permission from AAAS.
When comparing structures of different sizes, or proteins of the same structure but with very different intensities, it can be necessary to use the sum intensity of multiple z-sections rather than the intensity of the best focal plane [2,18]. The full width at half maximum (FWHM) of the Gaussian-fitted z-axis point spread function (PSF) of fluorescent bead or quantum dot images is the appropriate spacing of z-sections for sum intensity [2,18,40,41]. The percentage of total intensity of a protein in the best focal plane differs, depending on the density/distribution of the protein or size/shape of the structure [2,18]. To ensure the accuracy of the numbers measured, additional verifications are recommended. For example, one might need to sequence genomic DNA to make sure that there are no gene duplications or mutations in the tags. If the excitation power is constant, similar results should be obtained by mixing the cells or imaging consecutively, and measuring ratios on different days [2,3,18].
The ratio method of quantification can be extended to count proteins over time during their accumulation at a specific location in the cell [3,9,12,18]. Time-lapse quantification requires correction for the photobleaching over time. In two studies, the intensity at the best focal plane was corrected to represent the total intensity based on the percentage calculated from single stacks at FWHM spacing [9,18]. This correction can introduce some error if the positioning of the slice is off-centered compared to the real location of the best focal plane. In the fission yeast Schizosaccharomyces pombe, observation of protein numbers over time revealed a two-step accumulation of proteins, which was not obvious when taking a static measurement [2,9]. Thus, the number of molecules of each protein and their relative stoichiometries can be obtained using the ratio method at one or many time points.
Important considerations in counting proteins
Several important factors affect both protein counting methods. Because some dyes label inefficiently, mathematical simulations of expected results or binomial fits of the data are necessary when those dyes are used [30,31]. Thus, genetically encoded FPs should be used where possible, because they will be in a 1:1 stoichiometry with the protein of interest if the endogenous locus is disrupted or replaced with the fusion protein [27,29]. The maturation efficiency or proportion of unfolded FPs can affect this ratio in some cases [27–29]. We also note that in vivo fluorescence quantification can include both molecules that are incorporated into a structure and molecules that are close to the intended structure but within the diffraction limit, and fluorescence recovery after photobleaching (FRAP) can be used to reveal the different fractions [2,7,9,42–44].
Properties of fluorescent proteins
The properties of FPs are important considerations for both counting methods. Before constructing fusion proteins, the folding and maturation efficiency, brightness, and photostability of the FPs used in the fusions should be considered [27,45,46]. It is important for researchers to be aware of recently developed FPs, and use the best one(s) available to maximize the signal-to-noise ratio, especially for less abundant proteins [35]. Because GFP can dimerize at high concentrations [46], GFP and YFP variants with the monomeric mutation (A206K) are preferred. Red FPs such as mCherry have inferior physical properties, but can still be used as a second label if needed [47]. Cellular autofluorescence should be reduced as much as possible, especially when quantifying proteins that have low levels. Thus, FPs that have excitation/emission wavelengths for which autofluorescence is low, such as mYFP and mECitrine in fission yeast, are preferred [29]. It remains unclear whether maturation or folding efficiency of FPs is a major source of error. For example, 75–80% of YFP was found to be active in a single-molecule pull-down assay and in Xenopus laevis oocytes that were injected with mRNA for a tagged gene [28,47]. However, in the budding yeast Saccharomyces cerevisiae, γ-tubulin had a similar ratio to the standard, whether they were both tagged with EGFP or a super-folding variant, suggesting that the ratio method is insensitive to the folding efficiency [3]. Additionally, all YFP is folded in E. coli cell extracts, and the total fluorescence in the cell was changed by <10% after protein translation was stopped [10,11]. The discrepancy in maturation or folding efficiency in these reports might be attributed to the use of different FPs or different organisms, and might need to be evaluated for individual cases. Together, these data suggest that YFP maturation and folding efficiency are not major issues for counting proteins, especially for proteins with low turnover rates.
Functionality of fluorescent fusion proteins
The functionality and expression level of a fusion protein should be similar to wild type. One of the advantages of using yeasts is that the native protein can be replaced by a fluorescent fusion protein using homologous recombination, and the functionality of the fusion protein can be easily checked. A flexible linker between the FP and protein of interest might improve the functionalities for some proteins [2]. To circumvent the inability to replace endogenous genes with tagged versions, several options to count proteins exist; however, these methods might require assumptions about the relative incorporation of tagged and untagged proteins. These assumptions should be stated clearly so that interpretations can be evaluated. For example, a small fraction of actin was tagged because the tagged version could not replace the endogenous actin in fission yeast [40]. To quantify local actin abundance in actin patches, the data was corrected after measuring the ratio of tagged and untagged protein by immunoblotting. This correction is only accurate if the assumption that tagged and untagged actin are utilized with similar efficiency in actin patches is true. The tagged actin cannot incorporate into the contractile ring because of the properties of formins [48]. Thus, actin concentration in the ring was estimated from electron microscope images [40]. If the tagged protein must be added exogenously, it might be necessary to express proteins near the wild type level to minimize adverse effects of overexpression [8]. Another strategy was employed by Engel et al.; they counted exogenous tagged proteins in green algae Chlamydomonas reinhardtii flagella using the stepwise photobleaching method in a mutant background in which the endogenous protein does not localize, thus eliminating the need for assumptions about the ratio of tagged and untagged proteins [32]. With the recent introduction of a novel ‘genome editing’ technique, endogenous genes can be tagged in any model organism in which the zinc finger nuclease genes can be introduced [49]. Thus quantitative fluorescence microscopy will be easier in more organisms in the future.
In vivo vs. in vitro standards and quenching
Another source of concern in counting proteins is the environment in which the proteins are being measured. In early studies [41,50–56], in vitro standards were common, but it was unknown how the environment affects the fluorescence intensity, leading researchers to use immunoblotting or internal standards to calibrate fluorescence intensity inside the cell [27,29]. However, it was recently reported that the intensity of a single YFP variant YPet in vitro is equal to the intensity drop from photobleaching each YPet in E. coli [15]. In addition, ratio measurements between E. coli and S. pombe agree with the numbers obtained by bleaching S. pombe proteins, suggesting that the two intracellular environments have similar effects on GFP fluorescence [2]. Together, these data suggest that in vitro YFP/GFP is comparable to YFP/GFP in bacteria or yeast. The effects of other cell types on fluorescence intensity have not been tested, although molecule numbers have been reported [8,23]. Some fluorophores are sensitive to changes in pH and halide ion concentrations [46], which could be different in some cell types or in different compartments within cells.
Fluorescence quenching might occur if FPs are packed into very dense structures. Purified EGFP and EGFP-tagged viral particles yield different values for a single EGFP, suggesting that quenching occurs in the viral particle where 120 tagged coat proteins are packed into a small volume [36]. However, an earlier study reported that there was no quenching based on extinction coefficients [39]. Moreover, evidence for quenching inside fission yeast cells is lacking so far [2,40]. For example, after disrupting actin patches using Latrunculin A, the total cellular fluorescence of the actin crosslinker fimbrin was unchanged, indicating that the close proximity of >500 fimbrin molecules in an actin patch did not generate obvious quenching [40]. Thus, the effects of quenching on the accuracy of protein quantification should be addressed individually for specific structures of interest. One way to measure quenching due to environment changes is by fluorescence lifetime imaging, but this technique requires specialized equipment and analysis [57,58]. Moreover, the relationship between fluorescence lifetime (in nanoseconds) and the fluorescence intensity measured on >100 millisecond time-scales is unclear, making it difficult to correct fluorescence intensity using differences in fluorescence lifetime.
Validation of protein quantification by complementary approaches
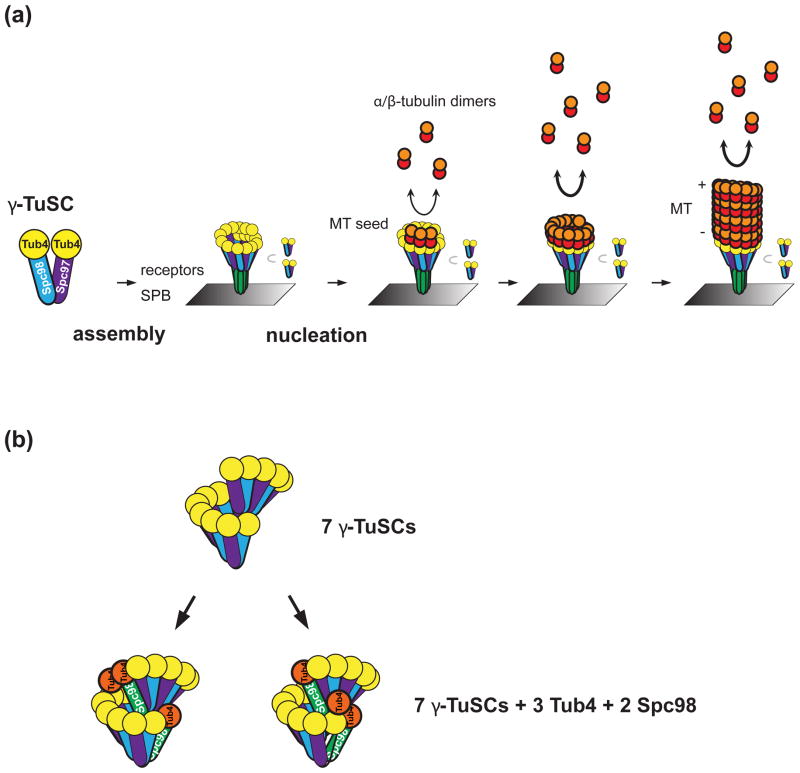
As alluded to previously, validating quantification results with complementary techniques is important. Global cellular concentrations can be validated by flow cytometry for abundant proteins [40] (Figure 3) or by fluorescence correlation spectroscopy for sparse proteins at higher spatial and temporal resolution [58,59]. The quantification of E. coli MotB is consistent with electron micrographs and biochemical data [10]. Similarly, formin speckles have about two molecules, and this is consistent with the strong dimerization of formins [2,60,61]. Biochemical characterization or other findings should guide the interpretation of results, especially for experiments in which native untagged protein is still present in the cell [8]. Protein concentrations from fluorescence microscopy should also be consistent with quantitative immunoblotting [29,62] (Figure 3). The function of the protein can also be informative. In a recent paper about the microtubule organizing center, 17 molecules of γ-tubulin were counted per microtubule [3]. The authors assumed that all the γ-tubulin was involved in microtubule nucleation and that each nucleation site was equivalent (Figure 4). Microtubules exhibit 13 protofilaments in electron micrographs, therefore an explanation for four additional γ-tubulins was offered [3].
Figure 4.
A revised model of the γ-tubulin ring in S. cerevisiae based on the ratio of γ-tubulin to fluorescent standards. Previously it was expected that 13 γ-tubulins should form a ring that serves as a template for the 13 protofilaments in each microtubule, but budding yeast lack many of the components that other eukaryotes use to stabilize the ring structure. (a) Microtubule (MT) nucleation at a spindle pole body (SPB, gray rectangle) from a nucleation platform consisting of 17 γ-tubulins (Tub4). α- and β-tubulin dimers (red and orange circles) interact with γ-tubulin more strongly than with the MT plus (+) end so that nucleation can occur. (b) Excess γ-tubulin small complexes (γ-TuSC) might stabilize the γ-tubulin ring by overlapping in a spiral. Two possible locations for the Tub4 and Spc98 in excess of the 7 complexes (orange circle and green rod, respectively) are shown. ©Erlemann et al., 2012. Originally published in J. Cell Biol. doi: 10.1083/jcb.201111123.
A protein counting experiment should utilize suitable fluorescent protein genes, ideally fused to the gene of interest at the native locus, producing fusion proteins that accomplish wild type functions. Suitable standards and controls for environmental changes or the possibility of quenching will ensure appropriate interpretations of the data, which should be validated with complementary experiments if possible. Ultimately, the need for such standards might be superseded by super-resolution and single molecule techniques.
The future of counting proteins using fluorescence microscopy
Super-resolution microscopy techniques, such as photoactivated localization microscopy (PALM) and stochastic optical reconstruction microscopy (STORM), can produce much higher resolution images of intracellular structures than confocal microscopy by pinpointing exact locations of individual fluorescent molecules [63,64]. Quantitative information from PALM imaging has recently been derived [65–67]. The main hindrance to quantification is that the contribution of blinking fluorophores to the final image is unknown. Super-resolution techniques have been made more accessible to researchers (although at a lower resolution) with the introduction of bleaching/blinking assisted localization microscopy (BaLM). BaLM requires no specialized equipment, uses standard dyes and FPs, and uses freely available ImageJ software and plug-ins to analyze the data [68]. The disadvantage of this method is that it has not been done in live cells and fixation could affect quantification [35,68]. The key to counting proteins with super-resolution microscopy techniques will be to simplify the analysis of high density images of FPs and minimize errors (caused by blinking or failure to photobleach) to ensure that each FP is counted only once [65–67]. Although algorithms to eliminate duplicate counting have been reported, a proof of principle experiment showing that these algorithms can reproduce numbers that are known by other methods is lacking. In a recent study, PALM data subjected to such an algorithm was compared to chromatin immunoprecipitation results [67], but disagreed with previous fluorescence quantification [2], suggesting that improvements are needed to reconcile these methods.
One of the difficulties with current microscopy methods is to balance excitation and bleaching of the molecules. As a result most images are collected with >200 ms exposures. Slimfield fluorescence microscopy uses a specialized system to generate a compact excitation field 100-times the intensity of a normal field, allowing a 3 ms exposure that can be used in combination with stepwise photobleaching to count proteins [15]. Such characteristics are ideal for live cell imaging to quantify very dynamic structures, and this will likely be a useful technique for many applications, including single-molecule microscopy.
Average population behaviors can hide stochastic events, therefore single-molecule techniques are becoming more popular [69]. Single-molecule microscopy has been developed for proteins that have a very low copy number, especially in bacteria, and the overall picture of single-molecule experiments agrees with the bulk assays while providing a specific look at stochastic events [33]. Two similar in vitro methods utilizing single molecules are promising for obtaining stoichiometric information by photobleaching. Colocalization single-molecule spectroscopy (CoSMoS) with dye-conjugated protein tags was used in vitro to examine the stoichiometry and assembly of the spliceosome [70]. Single-molecule pull-down (SiMPull) allows quantitative determination of stoichiometry and mutually exclusive interaction partners [47]. Single-molecule biology is an actively growing field that might become a standard technique in counting proteins.
One advantage of single molecule and super-resolution techniques would be the ability to count molecules directly rather than as part of a collective image and possibly even distinguish different protein complexes that are within a diffraction limited area. Especially super-resolution imaging holds promise for counting higher numbers of proteins once the analysis has been improved.
Concluding remarks
Counting protein molecules globally in cells and locally in specific complexes is an important step toward generating structural models and numerical simulations of protein complex functions. Protein numbers are also useful to determine the reaction rates in cellular processes and reconstitute multiprotein complexes in vitro. Here we have given an overview of two popular methods for counting proteins using fluorescence microscopy: stepwise photobleaching and ratio comparison to a known standard. Both methods are suitable for any laboratory with a fluorescence microscope that wishes to know the stoichiometry of a favorite protein complex. With the zinc finger nuclease technology, even previously difficult or impossible quantifications have become simple in organisms that had no efficient gene targeting methods. Each counting method has advantages and disadvantages depending on the application, and the properties of FPs and functionality of fusion proteins are important considerations for either method. Regardless of the method, protein quantities should be validated by biochemical methods or electron microscopy to ensure the accuracy of interpretations. One interesting outcome of such experiments will be determining if protein complexes have absolute numbers or tolerate a range of binding. Counting proteins using fluorescence microscopy is already an active field, and super-resolution and single-molecule techniques are promising directions for future improvements.
Box 1. Sources of variation in counting proteins.
All quantification techniques necessitate the same question: how can one distinguish between biological variation and variation from other sources? Other sources include systematic error, due to the microscope system itself; measurement error, due to the limitations of resolution and techniques; and noise, arising from stochasticity of photon emission and collection [35]. For many applications it would be useful to know the true biological variation in a structure. In some cases, biochemistry can tell us that a structure should have a fixed number of molecules [10,28,30,31]. In these instances the error likely arises from sources other than biological variation. But what if biological variation is a key feature of the structure? For example, cytokinesis nodes, which are precursors of the contractile ring in fission yeast, display a large variation in numbers of a single protein from one node to another [2,9] and pairs of node proteins do not always colocalize in individual nodes [9,61,71] suggesting that their varied composition might be significant. Interestingly, only a few proteins in actin patches have a large variation in their peak intensity [18]. In these cases, how much variation can be attributed to biology?
A partial answer to the aforementioned question is revealed using the coefficient of variation that equals the SD divided by the mean. Photobleaching data on single molecules of Venus FP have a 27% variation [33], which is only attributable to non-biological sources. Interestingly, coefficients of variation for MotB [2,10], kinetochore components [2,8,27], and most peak numbers in actin patches [18] were ≤ 27%, suggesting that these structures might tolerate less biological variation. By contrast, proteins in fission yeast cytokinesis nodes exhibited 35–60% variation at their plateaus of accumulation [2,9], suggesting that ~5–30% of the variation could be biological, although some less abundant proteins at nodes might exhibit greater variation due to lower signal-to-noise ratios. These examples suggest that a coefficient of variation from non-biological sources, such as from a single molecule bleaching experiment, could be used to assess the amount of biological variation in a set of data from the same system.
Acknowledgments
We thank Edward Salmon and Nicole Neuman for critical reading of the manuscript and members of the Wu laboratory for helpful discussions. V.C. Coffman is supported by an American Heart Association predoctoral fellowship and The Ohio State University Elizabeth Clay Howald Presidential Fellowship. This work is supported by the National Institutes of Health (NIH) grant R01GM086546 to J.-Q. Wu.
Abbreviations
- BaLM
blinking/bleaching assisted localization microscopy
- FP
fluorescent protein
- FWHM
full width at half maximum
- GFP
green fluorescent protein
- PALM
photoactivated localization microscopy
- PSF
point spread function
- ROI
region of interest
- SD
standard deviation
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Anderson M, et al. Function and assembly of DNA looping, clustering, and microtubule attachment complexes within a eukaryotic kinetochore. Mol Biol Cell. 2009;20:4131–4139. doi: 10.1091/mbc.E09-05-0359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coffman VC, et al. CENP-A exceeds microtubule attachment sites in centromere clusters of both budding and fission yeast. J Cell Biol. 2011;195:563–572. doi: 10.1083/jcb.201106078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Erlemann S, et al. An extended gamma-tubulin ring functions as a stable platform in microtubule nucleation. J Cell Biol. 2012;197:59–74. doi: 10.1083/jcb.201111123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gao Q, et al. A non-ring-like form of the Dam1 complex modulates microtubule dynamics in fission yeast. Proc Natl Acad Sci USA. 2010;107:13330–13335. doi: 10.1073/pnas.1004887107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gardner MK, et al. Chromosome congression by Kinesin-5 motor-mediated disassembly of longer kinetochore microtubules. Cell. 2008;135:894–906. doi: 10.1016/j.cell.2008.09.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Joglekar AP, et al. Molecular architecture of the kinetochore-microtubule attachment site is conserved between point and regional centromeres. J Cell Biol. 2008;181:587–594. doi: 10.1083/jcb.200803027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Joglekar AP, et al. Molecular architecture of a kinetochore-microtubule attachment site. Nat Cell Biol. 2006;8:581–585. doi: 10.1038/ncb1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johnston K, et al. Vertebrate kinetochore protein architecture: protein copy number. J Cell Biol. 2010;189:937–943. doi: 10.1083/jcb.200912022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Laporte D, et al. Assembly and architecture of precursor nodes during fission yeast cytokinesis. J Cell Biol. 2011;192:1005–1021. doi: 10.1083/jcb.201008171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leake MC, et al. Stoichiometry and turnover in single, functioning membrane protein complexes. Nature. 2006;443:355–358. doi: 10.1038/nature05135. [DOI] [PubMed] [Google Scholar]
- 11.Leake MC, et al. Variable stoichiometry of the TatA component of the twin-arginine protein transport system observed by in vivo single-molecule imaging. Proc Natl Acad Sci USA. 2008;105:15376–15381. doi: 10.1073/pnas.0806338105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin MC, et al. Overlapping and distinct functions for cofilin, coronin and Aip1 in actin dynamics in vivo. J Cell Sci. 2010;123:1329–1342. doi: 10.1242/jcs.065698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Markus SM, et al. Motor- and tail-dependent targeting of dynein to microtubule plus ends and the cell cortex. Curr Biol. 2009;19:196–205. doi: 10.1016/j.cub.2008.12.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moore JK, et al. Dynactin function in mitotic spindle positioning. Traffic. 2008;9:510–527. doi: 10.1111/j.1600-0854.2008.00710.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reyes-Lamothe R, et al. Stoichiometry and architecture of active DNA replication machinery in Escherichia coli. Science. 2010;328:498–501. doi: 10.1126/science.1185757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ribeiro SA, et al. Condensin regulates the stiffness of vertebrate centromeres. Mol Biol Cell. 2009;20:2371–2380. doi: 10.1091/mbc.E08-11-1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shimogawa MM, et al. Bir1 is required for the tension checkpoint. Mol Biol Cell. 2009;20:915–923. doi: 10.1091/mbc.E08-07-0723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sirotkin V, et al. Quantitative analysis of the mechanism of endocytic actin patch assembly and disassembly in fission yeast. Mol Biol Cell. 2010;21:2894–2904. doi: 10.1091/mbc.E10-02-0157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tang X, et al. A CAAX motif can compensate for the PH domain of Num1 for cortical dynein attachment. Cell Cycle. 2009;8:3182–3190. doi: 10.4161/cc.8.19.9731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thorpe PH, et al. Kinetochore asymmetry defines a single yeast lineage. Proc Natl Acad Sci USA. 2009;106:6673–6678. doi: 10.1073/pnas.0811248106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vizcay-Barrena G, et al. Subcellular and single-molecule imaging of plant fluorescent proteins using total internal reflection fluorescence microscopy (TIRFM) J Exp Bot. 2011;62:5419–5428. doi: 10.1093/jxb/err212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yeh E, et al. Pericentric chromatin is organized into an intramolecular loop in mitosis. Curr Biol. 2008;18:81–90. doi: 10.1016/j.cub.2007.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schittenhelm RB, et al. Detrimental incorporation of excess Cenp-A/Cid and Cenp-C into Drosophila centromeres is prevented by limiting amounts of the bridging factor Cal1. J Cell Sci. 2010;123:3768–3779. doi: 10.1242/jcs.067934. [DOI] [PubMed] [Google Scholar]
- 24.Ghaemmaghami S, et al. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 25.Arava Y, et al. Genome-wide analysis of mRNA translation profiles in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2003;100:3889–3894. doi: 10.1073/pnas.0635171100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huh WK, et al. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 27.Joglekar AP, et al. Counting kinetochore protein numbers in budding yeast using genetically encoded fluorescent proteins. Methods Cell Biol. 2008;85:127–151. doi: 10.1016/S0091-679X(08)85007-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ulbrich MH, Isacoff EY. Subunit counting in membrane-bound proteins. Nat Methods. 2007;4:319–321. doi: 10.1038/NMETH1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu JQ, et al. Chapter 9: Counting proteins in living cells by quantitative fluorescence microscopy with internal standards. Methods Cell Biol. 2008;89:253–273. doi: 10.1016/S0091-679X(08)00609-2. [DOI] [PubMed] [Google Scholar]
- 30.Das SK, et al. Membrane protein stoichiometry determined from the step-wise photobleaching of dye-labelled subunits. Chembiochem. 2007;8:994–999. doi: 10.1002/cbic.200600474. [DOI] [PubMed] [Google Scholar]
- 31.Shu D, et al. Counting of six pRNAs of phi29 DNA-packaging motor with customized single-molecule dual-view system. EMBO J. 2007;26:527–537. doi: 10.1038/sj.emboj.7601506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Engel BD, et al. Intraflagellar transport particle size scales inversely with flagellar length: revisiting the balance-point length control model. J Cell Biol. 2009;187:81–89. doi: 10.1083/jcb.200812084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taniguchi Y, et al. Quantifying E. coli proteome and transcriptome with single-molecule sensitivity in single cells. Science. 2010;329:533–538. doi: 10.1126/science.1188308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee IJ, Wu JQ. Characterization of Mid1 domains for targeting and scaffolding in fission yeast cytokinesis. J Cell Sci. 2012 doi: 10.1242/jcs.102574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Waters JC. Accuracy and precision in quantitative fluorescence microscopy. J Cell Biol. 2009;185:1135–1148. doi: 10.1083/jcb.200903097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lawrimore J, et al. Point centromeres contain more than a single centromere-specific Cse4 (CENP-A) nucleosome. J Cell Biol. 2011;195:573–582. doi: 10.1083/jcb.201106036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Garcia-Parajo MF, et al. Real-time light-driven dynamics of the fluorescence emission in single green fluorescent protein molecules. Proc Natl Acad Sci USA. 2000;97:7237–7242. doi: 10.1073/pnas.97.13.7237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Short B. Setting a new standard for kinetochores. J Cell Biol. 2011;195:539. [Google Scholar]
- 39.Charpilienne A, et al. Individual rotavirus-like particles containing 120 molecules of fluorescent protein are visible in living cells. J Biol Chem. 2001;276:29361–29367. doi: 10.1074/jbc.M101935200. [DOI] [PubMed] [Google Scholar]
- 40.Wu JQ, Pollard TD. Counting cytokinesis proteins globally and locally in fission yeast. Science. 2005;310:310–314. doi: 10.1126/science.1113230. [DOI] [PubMed] [Google Scholar]
- 41.Hirschberg K, et al. Kinetic analysis of secretory protein traffic and characterization of Golgi to plasma membrane transport intermediates in living cells. J Cell Biol. 1998;143:1485–1503. doi: 10.1083/jcb.143.6.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Henikoff S, Henikoff JG. ‘Point’ Centromeres of Saccharomyces Harbor Single Centromere-Specific Nucleosomes. Genetics. 2012;190:1575–1577. doi: 10.1534/genetics.111.137711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Howell BJ, et al. Spindle checkpoint protein dynamics at kinetochores in living cells. Curr Biol. 2004;14:953–964. doi: 10.1016/j.cub.2004.05.053. [DOI] [PubMed] [Google Scholar]
- 44.Pelham RJ, Chang F. Actin dynamics in the contractile ring during cytokinesis in fission yeast. Nature. 2002;419:82–86. doi: 10.1038/nature00999. [DOI] [PubMed] [Google Scholar]
- 45.Shaner NC, et al. A guide to choosing fluorescent proteins. Nat Methods. 2005;2:905–909. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
- 46.Chudakov DM, et al. Fluorescent proteins and their applications in imaging living cells and tissues. Physiol Rev. 2010;90:1103–1163. doi: 10.1152/physrev.00038.2009. [DOI] [PubMed] [Google Scholar]
- 47.Jain A, et al. Probing cellular protein complexes using single-molecule pull-down. Nature. 2011;473:484–488. doi: 10.1038/nature10016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen Q, et al. Formins filter modified actin subunits during processive elongation. J Struct Biol. 2012;177:32–39. doi: 10.1016/j.jsb.2011.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Urnov FD, et al. Genome editing with engineered zinc finger nucleases. Nat Rev Genet. 2010;11:636–646. doi: 10.1038/nrg2842. [DOI] [PubMed] [Google Scholar]
- 50.Chiu CS, et al. Single -molecule measurements calibrate green fluorescent protein surface densities on transparent beads for use with ‘knock-in’ animalsand other expression systems. J Neurosci Methods. 2001;105:55–63. doi: 10.1016/s0165-0270(00)00354-x. [DOI] [PubMed] [Google Scholar]
- 51.Dundr M, et al. Quantitation of GFP-fusion proteins in single living cells. J Struct Biol. 2002;140:92–99. doi: 10.1016/s1047-8477(02)00521-x. [DOI] [PubMed] [Google Scholar]
- 52.Hirschberg K, et al. Kinetic analysis of intracellular trafficking in single living cells with vesicular stomatitis virus protein G-green fluorescent protein hybrids. Methods Enzymol. 2000;327:69–89. doi: 10.1016/s0076-6879(00)27268-6. [DOI] [PubMed] [Google Scholar]
- 53.Khakh BS, et al. Activation-dependent changes in receptor distribution and dendritic morphology in hippocampal neurons expressing P2X2-green fluorescent protein receptors. Proc Natl Acad Sci USA. 2001;98:5288–5293. doi: 10.1073/pnas.081089198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Patterson GH, et al. Quantitative imaging of TATA-binding protein in living yeast cells. Yeast. 1998;14:813–825. doi: 10.1002/(SICI)1097-0061(19980630)14:9<813::AID-YEA280>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 55.Piston DW, et al. Qu antitative imaging of the green fluorescent protein (GFP) Methods Cell Biol. 1999;58:31–48. doi: 10.1016/s0091-679x(08)61947-0. [DOI] [PubMed] [Google Scholar]
- 56.Xu C, et al. Kinetic analysis of receptor-activated phosphoinositide turnover. J Cell Biol. 2003;161:779–791. doi: 10.1083/jcb.200301070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Morgan CG, Mitchell AC. Fluorescence lifetime imaging: an emerging technique in fluorescence microscopy. Chromosome Res. 1996;4:261–263. doi: 10.1007/BF02263674. [DOI] [PubMed] [Google Scholar]
- 58.Shivaraju M, et al. Cell-cycle-coupled structural oscillation of centromeric nucleosomes in yeast. Cell. 2012;150:304–316. doi: 10.1016/j.cell.2012.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kim SA, et al. Fluorescence correlation spectroscopy in living cells. Nat Methods. 2007;4:963–973. doi: 10.1038/nmeth1104. [DOI] [PubMed] [Google Scholar]
- 60.Xu Y, et al. Crystal structures of a Formin Homology-2 domain reveal a tethered dimer architecture. Cell. 2004;116:711–723. doi: 10.1016/s0092-8674(04)00210-7. [DOI] [PubMed] [Google Scholar]
- 61.Coffman VC, et al. Roles of form in nodes and myosin motor activity in Mid1p-dependent contractile-ring assembly during fission yeast cytokinesis. Mol Biol Cell. 2009;20:5195–5210. doi: 10.1091/mbc.E09-05-0428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Emanuele MJ, et al. Measuring the stoichiometry and physical interactions between components elucidates the architecture of the vertebrate kinetochore. Mol Biol Cell. 2005;16:4882–4892. doi: 10.1091/mbc.E05-03-0239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Huang B, et al. Super-resolution fluorescence microscopy. Annu Rev Biochem. 2009;78:993–1016. doi: 10.1146/annurev.biochem.77.061906.092014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gitai Z. New fluorescence microscopy methods for microbiology: sharper, faster, and quantitative. Curr Opin Microbiol. 2009;12:341–346. doi: 10.1016/j.mib.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sengupta P, Lippincott-Schwartz J. Quantitative analysis of photoactivated localization microscopy (PALM) datasets using pair-correlation analysis. Bioessays. 2012;34:396–405. doi: 10.1002/bies.201200022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Annibale P, et al. Quantitative photo activated localization microscopy: unraveling the effects of photoblinking. PLoS One. 2011;6(7):e22678. doi: 10.1371/journal.pone.0022678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lando D, et al. Quantitative single -molecule microscopy reveals that CENP-ACnp1 deposition occurs during G2 in fission yeast. Open Biol. 2012;2:120078. doi: 10.1098/rsob.120078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Burnette DT, et al. Bleaching/blinking assisted localization microscopy for superresolution imaging using standard fluorescent molecules. Proc Natl Acad Sci USA. 2011;108:21081–21086. doi: 10.1073/pnas.1117430109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chiu SW, Leake MC. Functioning nanomachines seen in real-time in living bacteria using single-molecule and super-resolution fluorescence imaging. Int J Mol Sci. 2011;12:2518–2542. doi: 10.3390/ijms12042518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hoskins AA, et al. Ordered and dynamic assembly of single spliceosomes. Science. 2011;331:1289–1295. doi: 10.1126/science.1198830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wu JQ, et al. Assembly of the cytokinetic contractile ring from a broad band of nodes in fission yeast. J Cell Biol. 2006;174:391–402. doi: 10.1083/jcb.200602032. [DOI] [PMC free article] [PubMed] [Google Scholar]