Summary
Assembly of contractile apparatuses in striated muscle requires precisely regulated reorganization of the actin cytoskeletal proteins into sarcomeric organization. Regulation of actin filament dynamics is one of the essential processes of myofibril assembly, but the mechanism of actin regulation in striated muscle is not clearly understood. Actin depolymerizing factor (ADF)/cofilin is a key enhancer of actin filament dynamics in striated muscle in both vertebrates and nematodes. Here, we report that CAS-1, a cyclase-associated protein in Caenorhabditis elegans, promotes ADF/cofilin-dependent actin filament turnover in vitro and is required for sarcomeric actin organization in striated muscle. CAS-1 is predominantly expressed in striated muscle from embryos to adults. In vitro, CAS-1 binds to actin monomers and enhances exchange of actin-bound ATP/ADP even in the presence of UNC-60B, a muscle-specific ADF/cofilin that inhibits the nucleotide exchange. As a result, CAS-1 and UNC-60B cooperatively enhance actin filament turnover. The two proteins also cooperate to shorten actin filaments. A cas-1 mutation is homozygous lethal with defects in sarcomeric actin organization. cas-1-mutant embryos and worms have aggregates of actin in muscle cells, and UNC-60B is mislocalized to the aggregates. These results provide genetic and biochemical evidence that cyclase-associated protein is a critical regulator of sarcomeric actin organization in striated muscle.
Key words: Actin turnover, ADF/cofilin, Cyclase-associated protein, Myofibril, Muscle
Introduction
In striated muscle, actin filaments are organized in sarcomeric structures in which length and polarity of the filaments are highly ordered, so that coordinated actin–myosin interaction can efficiently produce contractile forces (Clark et al., 2002). Assembly and maintenance of sarcomeric actin filaments require specific mechanisms for controlling actin dynamics, which is different from highly dynamic actin-regulatory systems in non-muscle cells (Ono, 2010). However, the mechanism by which turnover of sarcomeric actin filaments is enhanced or suppressed is not clearly understood. Actin depolymerizing factor (ADF)/cofilin is an essential factor in striated muscle for enhancing actin filament dynamics (Ono, 2007; Ono, 2010). In mammalian striated muscle, cofilin-2, a muscle-specific ADF/cofilin, is predominantly expressed (Ono et al., 1994; Thirion et al., 2001; Vartiainen et al., 2002), and a mutation in human cofilin-2 causes nemaline myopathy (Agrawal et al., 2007). Knockdown of cofilin in cultured rat cardiomyocytes causes disorganization of myofibrils (Skwarek-Maruszewska et al., 2009). In the nematode Caenorhabditis elegans, UNC-60B, a muscle-specific ADF/cofilin, is required for proper assembly of sarcomeric actin filaments in striated muscle (Ono et al., 2003; Ono et al., 1999). Thus, ADF/cofilin is a conserved enhancer of actin filament dynamics in striated muscle and functions together with other actin regulators for proper assembly and maintenance of sarcomeric actin organization.
Although ADF/cofilin by itself can enhance actin filament turnover by filament severing (Andrianantoandro and Pollard, 2006; Ichetovkin et al., 2000; Pavlov et al., 2007) and monomer dissociation from the pointed ends (Carlier et al., 1997; Maciver et al., 1998), the effect of ADF/cofilin can be enhanced or suppressed by other factors (Van Troys et al., 2008). Studies in C. elegans striated muscle show that tropomyosin (LEV-11) (Ono and Ono, 2002; Yu and Ono, 2006) and UNC-87, a calponin-like protein (Yamashiro et al., 2007), protect actin filaments from severing by ADF/cofilin, while actin-interacting protein 1 (AIP1) enhances disassembly of ADF/cofilin-bound actin filaments (Mohri et al., 2006; Mohri and Ono, 2003; Ono, 2001; Ono et al., 2011). Tropomodulin (UNC-94/TMD-1) caps the pointed ends of actin filaments and protects them from ADF/cofilin-mediated depolymerization (Yamashiro et al., 2008). In vertebrate striated muscle, actin filaments are stabilized by tropomyosin (Mudry et al., 2003), tropomodulin (Littlefield et al., 2001), and nebulin (Pappas et al., 2010). Although functional relationships between these proteins and ADF/cofilin have not been determined in vertebrates, the regulatory mechanism of actin filament stability is expected to be similar in vertebrates and nematodes.
Cyclase-associated protein (CAP) is an enhancer of ADF/cofilin-dependent actin turnover, which has been characterized mostly in non-muscle cells. Vertebrates have two CAP isoforms, CAP1 and CAP2 (Yu et al., 1994), and CAP2 is predominantly expressed in heart and skeletal muscle (Bertling et al., 2004; Peche et al., 2007; Wolanski et al., 2009). However, the function of CAP2 in striated muscle is not clearly understood. CAP (also known as Srv2) was originally identified in yeast as a component of the Ras–cAMP signaling (Fedor-Chaiken et al., 1990; Field et al., 1990) and has been shown to bind to actin monomers (Freeman et al., 1995). Homologs of CAP have been subsequently found in other eukaryotes, and their activities to regulate actin cytoskeleton are conserved (Hubberstey and Mottillo, 2002). Biochemical studies by Moriyama and Yahara showed that human CAP1 enhances actin filament turnover in vitro in the presence of ADF/cofilin by promoting dissociation of ADF/cofilin from actin monomers and enhancing exchange of actin-bound ATP/ADP (Moriyama and Yahara, 2002). These activities are consistent with the cooperative roles of CAP and ADF/cofilin in the regulation of actin dynamics in yeast (Balcer et al., 2003) and mammalian cultured cells (Bertling et al., 2004). A similar function was previously proposed for profilin (Blanchoin and Pollard, 1998; Didry et al., 1998). However, relatively high concentrations of profilin are required to enhance actin dynamics because it preferentially binds to ATP–G-actin rather than ADP–G-actin, which is the predominant form of ADF/cofilin-bound actin monomers. In contrast, yeast CAP preferentially binds to ADP–G-actin (Mattila et al., 2004) and efficiently enhances actin turnover at low concentrations in the presence of ADF/cofilin (Chaudhry et al., 2007; Quintero-Monzon et al., 2009).
The C. elegans genome has two genes that encode CAP homologs, but their functions have not been characterized. In C. elegans muscle, mutations in profilins cause only minor phenotypes in sarcomeric actin organization (Polet et al., 2006), and they do not appear to cooperate strongly with ADF/cofilin in vivo (Yamashiro et al., 2008). These observations led us to hypothesize that CAP is involved in ADF/cofilin-dependent actin dynamics in C. elegans muscle. We found that one of the C. elegans CAP genes, cas-1, is expressed in striated muscle and required for viability and sarcomeric actin organization, providing genetic evidence that CAP is an essential factor for myofibril assembly in striated muscle.
Results
CAS-1 is expressed in the body wall muscle and localizes to the M-lines
cah-1 was previously reported as a gene that encodes a homolog of cyclase-associated protein (CAP), which is adjacent to mec-4 on the X chromosome (Lai et al., 1996). Later, this gene was renamed as cas-1 (cyclase-associated protein-1) because the gene name ‘cah’ was suited better to represent the carbonic anhydrase gene family. As a result, currently known cah-1 is a gene that encodes a carbonic anhydrase (Fasseas et al., 2011) and unrelated to cas-1, which is the major subject of the current study. Although ‘CAP’ is the common name for this protein family, the gene name ‘cap’ has already been used for designation of C. elegans genes encoding heterodimeric actin capping protein subunits (cap-1 and cap-2) (Waddle et al., 1993).
The amino acid sequence of CAS-1 has a conserved domain structure (supplementary material Fig. S1A) and shows 32–41% identity with CAPs in other organisms. In particular, sequence identity of CAS-1 with human CAP1, human CAP2, and yeast Srv2/CAP is 37, 38 and 32%, respectively. The C. elegans genome sequencing project identified a second CAP gene with 41% identity with CAS-1, and we designated this gene as cas-2. Functional analysis of cas-2 is currently in progress and will be reported elsewhere. Vertebrates have two CAP isoforms: CAP1 (a non-muscle isoform) and CAP2 (a muscle isoform) (Yu et al., 1994) (supplementary material Fig. S1B). However, phylogenetic analysis of CAP sequences suggested that the two C. elegans isoforms have evolved separately from vertebrate isoforms (supplementary material Fig. S1B). Prediction of secondary structures of CAS-1 by Jpred 3 (Cole et al., 2008) indicated that the N-terminal region (residues 3–231) is enriched in α-helices, and that the C-terminal region (residues 342–492) is primarily composed of β-strands (supplementary material Fig. S1A). These are consistent with known structures of the N-terminal helical folded domain (HFD) of Dictyostelium CAP (Ksiazek et al., 2003; Mavoungou et al., 2004; Yusof et al., 2005) and the C-terminal β-sheet domain of yeast Srv2/CAP and human CAP1 (Dodatko et al., 2004). However, alignment of the N-terminal sequences shows that C. elegans CAS-1 as well as human CAP1 and CAP2 contain gaps as compared with yeast Srv2/CAP (supplementary material Fig. S1C). This region of yeast Srv2/CAP is predicted to form a coiled coil (Nishida et al., 1998), but this feature may not be conserved in C. elegans CAS-1 (see Discussion). The central region of C. elegans CAS-1 has a proline-rich region (P1) and a WASP-homology 2 (WH2) domain (supplementary material Fig. S1A,D). P1 of yeast Srv2/CAP is known to bind to profilin (Bertling et al., 2007). However, CAS-1 lacks a second proline-rich region (P2), which is present in yeast Srv2/CAP (supplementary material Fig. S1D). Proline residues are present in this region of CAS-1, but they are scattered and not clustered as they are in yeast Srv2/CAP (supplementary material Fig. S1D). Human CAP-1 and CAP-2 also have scattered prolines in this region (supplementary material Fig. S1D). Thus, C. elegans CAS-1 shows greater similarity to mammalian CAPs than yeast Srv2/CAP.
Since there was no functional information on cas-1, we first determined expression patterns of cas-1 in C. elegans. A promoter–reporter analysis using a 3-kb-upstream sequence as a promoter and green fluorescent protein (GFP) as a reporter resulted in no detectable GFP expression, suggesting that another genetic element is required for expression of cas-1 (K.O., D. L. Baillie and S.O., unpublished observations). Therefore, we generated antibody against the CAS-1 protein and determined localization patterns of CAS-1 by immunofluorescence microscopy. The antibody was raised in rabbits using a C-terminal fragment of CAS-1 (residues 225–495) as an immunogen. On western blots, affinity-purified anti-CAS-1 antibody reacted specifically with a band corresponding to the size of CAS-1 in whole worm lysates (supplementary material Fig. S2, lane 13) and with purified recombinant full-length CAS-1 protein and the C-terminal fragment of CAS-1 (supplementary material Fig. S2, lanes 9 and 11), but it did not react with CAS-2, a second CAP isoform in C. elegans (supplementary material Fig. S2, lanes 8 and 12).
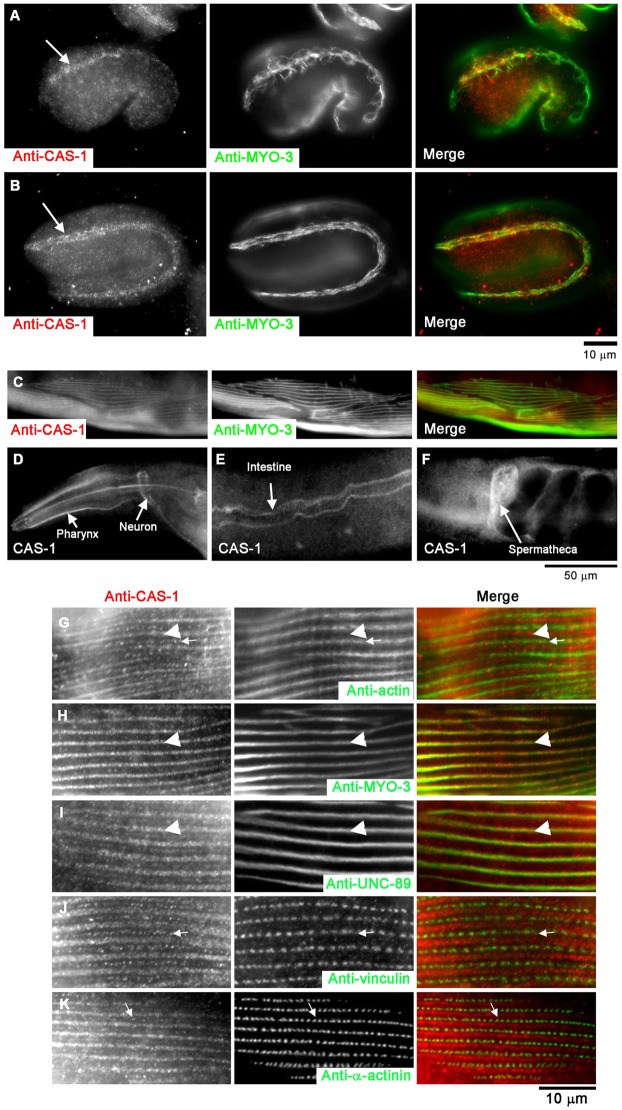
In embryos, CAS-1 was specifically expressed in the body wall muscle as early as the 1.5-fold stage (∼420 min old; Fig. 1A). MYO-3 is a muscle-specific myosin heavy chain and used as a marker for embryonic body wall muscle (Fig. 1A). At this stage, MYO-3 was not completely assembled into myofibrils (Fig. 1A), indicating that this is an early stage of myofibril assembly. Nonetheless, CAS-1 was concentrated to a region where MYO-3 had been assembled into myofibrils (Fig. 1A). At the twofold stage (∼450 min old), CAS-1 remained associated with well-organized myofibrils (Fig. 1B), as indicated by the linear arrangement of MYO-3 (Fig. 1B). Throughout the embryonic stages, expression of CAS-1 was specific in the body wall muscle, and no other embryonic tissues expressed detectable levels of CAS-1 (Fig. 1A,B).
Fig. 1.
Expression and localization patterns of CAS-1 in C. elegans embryos and adult worms. (A,B) Expression patterns of CAS-1 in embryos at the 1.5-fold (∼420-min old; A) and 2-fold (∼450-min old; B) stages. Embryos were fixed and stained with anti-CAS-1 (left panels) and anti-MYO-3 (a marker for the body wall muscle; middle panels). Merged images are shown in the right panels (CAS-1 in red and MYO-3 in green). CAS-1 was expressed predominantly in the body wall muscle as indicated by arrows in A and B. (C–F) Expression patterns of CAS-1 in adult worms. (C) Adult worms were fixed and stained with anti-CAS-1 (left panel) and anti-MYO-3 (middle panel), and a region of the body wall muscle is shown. A merged image is shown in the right panel (CAS-1 in red and MYO-3 in green). Immunostaining of CAS-1 was also detected in the pharynx (D), the neurons (D), the intestinal lumen (E), and the spermatheca of the somatic gonad (F). (G–K) Sarcomeric localization of CAS-1 in C. elegans adult body wall muscle. Adult worms were fixed and stained with anti-CAS-1 (left panels) and anti-actin (G, middle), anti-MYO-3 (H, middle), anti-UNC-89 (I, middle), anti-vinculin (J, middle) or anti-α-actinin (K, middle) antibodies. Merged images are shown in the right panels (CAS-1 in red, and actin, MYO-3, UNC-89, vinculin and α-actinin in green). Arrowheads in G–I indicate positions of the M-lines, and arrows in G, J and K indicate positions of CAS-1 spots in the I-bands.
In adult worms, CAS-1 was expressed in the body wall muscle (Fig. 1C), the pharynx (Fig. 1D), the neurons (Fig. 1D), the intestine (Fig. 1E), and the spermatheca (Fig. 1F). In the adult body wall muscle, CAS-1 localized in a striated pattern indicating that it was associated with sarcomeres (Fig. 1C). Interestingly, CAS-1 only partially overlapped with bands of actin (Fig. 1G, arrows). Instead, strong striated staining of CAS-1 overlapped with MYO-3 (Fig. 1H, arrowheads) that is enriched in the M-lines within the thick filaments (Miller et al., 1983). We confirmed the enrichment of CAS-1 in the M-line by colocalization with UNC-89 (Fig. 1I, arrowheads), a specific marker of the M-lines (Benian et al., 1996). Relatively weakly stained spots of CAS-1 were present (Fig. 1G,J,K, arrows), but these did not colocalize with vinculin (Fig. 1J) or α-actinin (Fig. 1K), indicating that CAS-1 did not localize to dense bodies, which are equivalent to the Z-discs in mammalian striated muscle. It should be noted that mammalian CAP2 also localizes to the M-line in human skeletal muscle (Peche et al., 2007), suggesting that the M-line localization is a common feature of CAP in striated muscle across different species.
CAS-1 promotes actin filament turnover in the presence of ADF/cofilin in vitro
We produced recombinant CAS-1 proteins and characterized their biochemical properties. Biochemical studies on yeast Srv2/CAP (Balcer et al., 2003; Mattila et al., 2004) and human CAP1 (Moriyama and Yahara, 2002) have shown that the N-terminal and C-terminal halves of CAPs have distinct functions. Therefore, we produced full-length CAS-1 (residues 1–495) as well as the N-terminal half (CAS-1N; residues 1–224) containing the putative helical-folded domain and the C-terminal half (CAS-1C; residues 225–495) containing the proline-rich sequence, the WH2 domain, and the putative G-actin-binding domain (supplementary material Fig. S1A). We used a maltose-binding-protein (MBP) fusion system that is reported to enhance solubility and proper folding of recombinant proteins (Fox and Waugh, 2003). The MBP fusion system significantly improved solubility of the CAS-1 protein in Escherichia coli, and we were successful in purifying full-length CAS-1, CAS-1N and CAS-1C as MBP fusion proteins (supplementary material Fig. S2, lanes 2–4). Cleavage of MBP by TEV protease was very inefficient. Therefore, we used MBP fusion proteins in the following experiments and included purified MBP (supplementary material Fig. S2, lane 1) in control experiments.
We tested whether the CAS-1 protein binds to G-actin as reported for CAP proteins from other organisms using three methods. First, we examined effects of MBP–CAS-1, MBP–CAS-1N and MBP–CAS-1C on spontaneous actin polymerization from G-actin (Fig. 2A–D). MBP–CAS-1 (full-length) slowed down the initial phase of actin polymerization in a concentration-dependent manner (Fig. 2A,D), indicating that CAS-1 bound to G-actin and inhibited actin nucleation. MBP–CAS-1C had a similar inhibitory effect on actin polymerization to full-length CAS-1 (Fig. 2C,D). However, MBP–CAS-1N moderately slowed down polymerization only when CAS-1N was present at a high concentration (15 µM CAS-1N to 5 µM actin; Fig. 2B,D). MBP alone had no effect on the rate of actin polymerization at 15 µM (Fig. 2D, black circle). These results indicate that the N-terminal and C-terminal halves of CAS-1 independently bind to G-actin, and that the C-terminal half binds to G-actin more strongly than the N-terminal half.
Fig. 2.
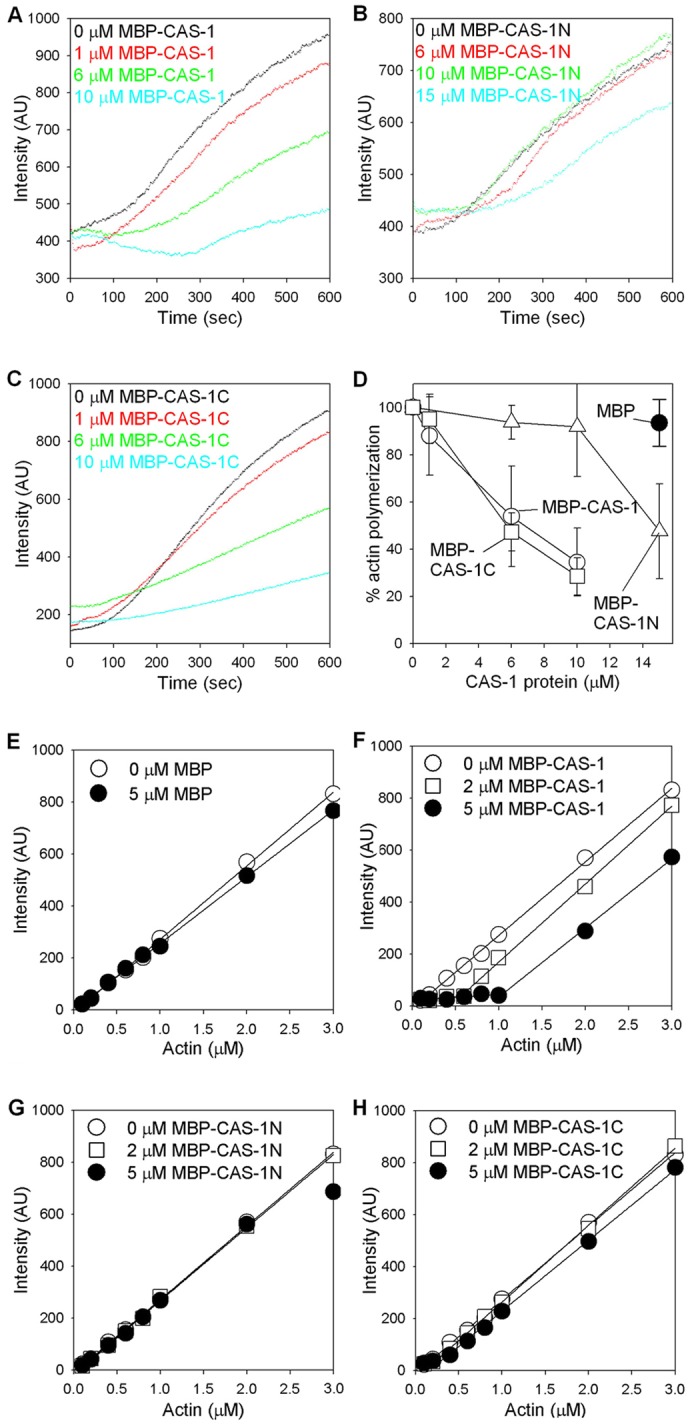
Effects of CAS-1 on actin polymerization. (A–D) Effects of CAS-1 on the initial phase of actin polymerization. G-actin (5 µM, 20% pyrene labeled) was polymerized by addition of salt at time 0 in the presence of 0–15 µM MBP–CAS-1 (A), MBP–CAS-1N (B) or MBP–CAS-1C (C), and intensity of the pyrene fluorescence (arbitrary units; AU) was monitored over time. (D) Relative rates of actin polymerization in the presence of MBP–CAS-1 (white circles), MBP–CAS-1N (white triangles), MBP–CAS-1C (white squares), and MBP (black circle) were determined as described in Materials and Methods. Data are means ± s.d. of three independent experiments. (E–H) Effects of CAS-1 on the steady state of actin polymerization. Varying concentrations of actin (20% pyrene labeled) were polymerized in the presence of 0–5 µM MBP (E), MBP–CAS-1 (F), MBP–CAS-1N (G) or MBP–CAS-1C (H) for 18 hr, and the intensity of the pyrene fluorescence (arbitrary units) was measured.
However, at the steady state, full-length CAS-1 sequestered actin monomers, but either CAS-1N or CAS-1C was not effective in sequestering actin monomers. Various concentrations of actin monomers were polymerized in the presence of 2 or 5 µM MBP, MBP–CAS-1, MBP–CAS-1N or MBP–CAS-1C, and the amounts of polymerized actin were quantified (Fig. 2E–H). MBP or MBP–CAS-1N did not affect actin polymerization (Fig. 2E,G). MBP–CAS-1 shifted the critical concentration from 0.13 µM by actin alone to 0.55 and 1.0 µM in the presence of 2 and 5 µM MBP–CAS-1, respectively (Fig. 2F). From these values, dissociation constant for binding of MBP–CAS-1 to actin was estimated as 1.4 µM (2 µM MBP–CAS-1) and 0.62 µM (5 µM MBP–CAS-1). Interestingly, although MBP–CAS-1C had a strong inhibitory effect on the initial phase of actin polymerization (Fig. 2C), it shifted the critical concentration only weakly to 0.20 and 0.27 µM in the presence of 2 and 5 µM MBP–CAS-1C, respectively (Fig. 2H). From these values, dissociation constant for binding of MBP–CAS-1 to actin was estimated as 9.2 µM (2 µM MBP–CAS-1C) and 4.5 µM (5 µM MBP–CAS-1C). These suggest that CAS-1C is sufficient to inhibit actin nucleation and needs to cooperate with CAS-1N for sequestration of actin monomers.
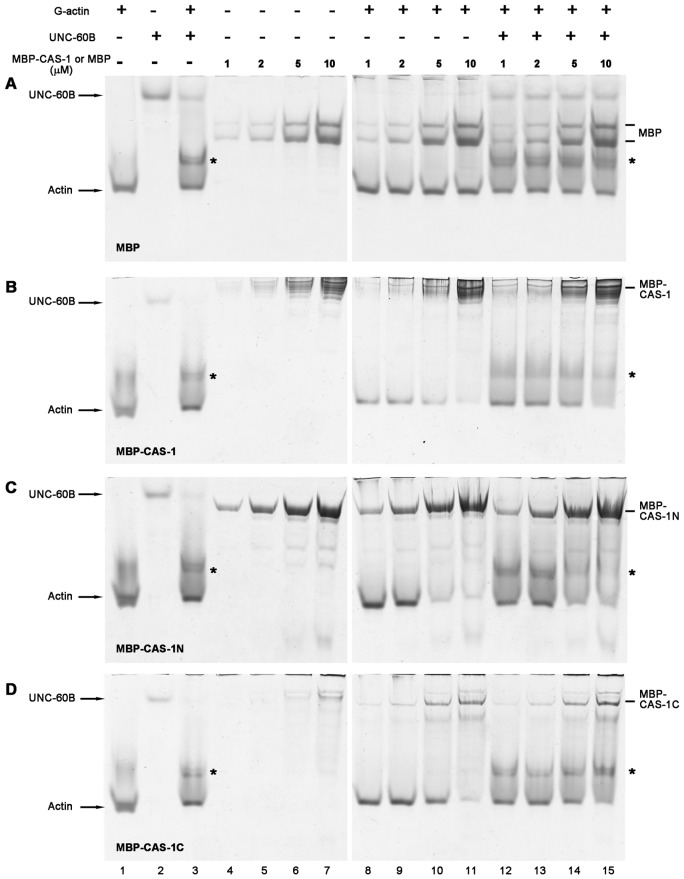
Second, we tested G-actin binding of CAS-1 by nondenaturing polyacrylamide gel electrophoresis (PAGE) (Fig. 3). Under nondenaturing conditions, G-actin alone migrated in a major band (Fig. 3A–D, lane 1). As a control, increasing concentrations of MBP did not affect the mobility of the G-actin band when MBP and G-actin were mixed (Fig. 3A, lanes 8–11), indicating that MBP did not interact with G-actin. However, in mixtures of G-actin and MBP–CAS-1, the G-actin band was gradually diminished as the concentration of MBP–CAS-1 was increased (Fig. 3B, lanes 8–11), indicating that the G-actin band was shifted due to its binding with MBP–CAS-1. Similar band shift of G-actin was observed in the presence of MBP–CAS-1N (Fig. 3C, lanes 8–11) or MBP–CAS-1C (Fig. 3D, lanes 8–11). Thus, these results also indicate that the N-terminal and C-terminal halves of CAS-1 independently bind to G-actin.
Fig. 3.
Binding of CAS-1 to G-actin and the UNC-60B–actin complex, examined by nondenaturing polyacrylamide gel electrophoresis. Various concentrations (1–10 µM) of MBP (A), MBP–CAS-1 (B), MBP–CAS-1N (C) or MBP–CAS-1C (D) were incubated with buffer only (lanes 4–7) or buffer with 10 µM G-actin (lanes 8–11) or 10 µM G-actin and 10 µM UNC-60B (lanes 12–15) and examined by nondenaturing acrylamide gel electrophoresis. G-actin alone (lane 1), UNC-60B alone (lane 2), and mixtures of G-actin and UNC-60B (lane 3) were also applied to determine positions of free G-actin, free UNC-60B and the actin–UNC-60B complex (asterisks).
Nondenaturing PAGE also allowed us to determine whether CAS-1 interacts with UNC-60B–actin complex. UNC-60B alone migrated much slower than G-actin (Fig. 3A–D, lane 2), and the UNC-60B–G-actin complex appeared above the G-actin band (Fig. 3A–D, lane 3, asterisks). MBP did not affect the band of the UNC-60B–G-actin complex (Fig. 3A, lanes 12–15), while it was gradually diminished as increasing concentrations of MBP–CAS-1 was added (Fig. 3B, lanes 12–15, asterisk) indicating that CAS-1 bound to the UNC-60B–G-actin complex or that CAS-1 promoted dissociation of UNC-60B from G-actin. MBP–CAS-1N similarly reduced the band of the UNC-60B–G-actin complex (Fig. 3C, lanes 12–15, asterisk), whereas MBP–CAS-1C did not affect the mobility of the band of the UNC-60B–G-actin complex (Fig. 3D, lanes 12–15, asterisk). These results show that the N-terminal half of CAS-1 is necessary and sufficient for interacting with the UNC-60B–G-actin complex.
Since nondenaturing PAGE was performed under low ionic conditions, we also tested binding of CAS-1 with actin and UNC-60B under physiological ionic conditions by pull-down assays in which MBP–CAS-1 (or CAS-1 fragments) was pulled down by beads with conjugated anti-MBP antibody (supplementary material Fig. S3). We also tested if CAS-1 differentially binds to ATP– or ADP–actin, as yeast Srv2/CAP binds to ADP–actin with higher affinity than ATP–actin (Mattila et al., 2004). The results indicated that ATP–actin and ADP–actin equally bound to MBP–CAS-1 (supplementary material Fig. S3A,C, lane 3) and MBP–CAS-1C (Fig. 4A,C, lane 7). However, the amounts of ATP– or ADP–actin that co-precipitated with MBP–CAS-1N (supplementary material Fig. S3A,C, lane 5) were not significantly different from those with MBP (supplementary material Fig. S3A,C, lane 1), suggesting that binding of MBP–CAS-1N with actin is too weak to be detectable under these experimental conditions. We also found that UNC-60B did not co-precipitate with MBP–CAS-1, MBP–CAS-1N, or MBP–CAS-1C (supplementary material Fig. S3A,C, lanes 4, 6 and 8) and that UNC-60B did not affect the amount of precipitated actin (supplementary material Fig. S3A,C, lanes 4, 6 and 8). Based on our previous studies (Yamashiro et al., 2005), UNC-60B is expected to bind to actin under these conditions. Therefore, lack of UNC-60B in the precipitates suggests that UNC-60B was dissociated from actin in the presence of MBP–CAS-1 or MBP–CAS-1C.
Fig. 4.
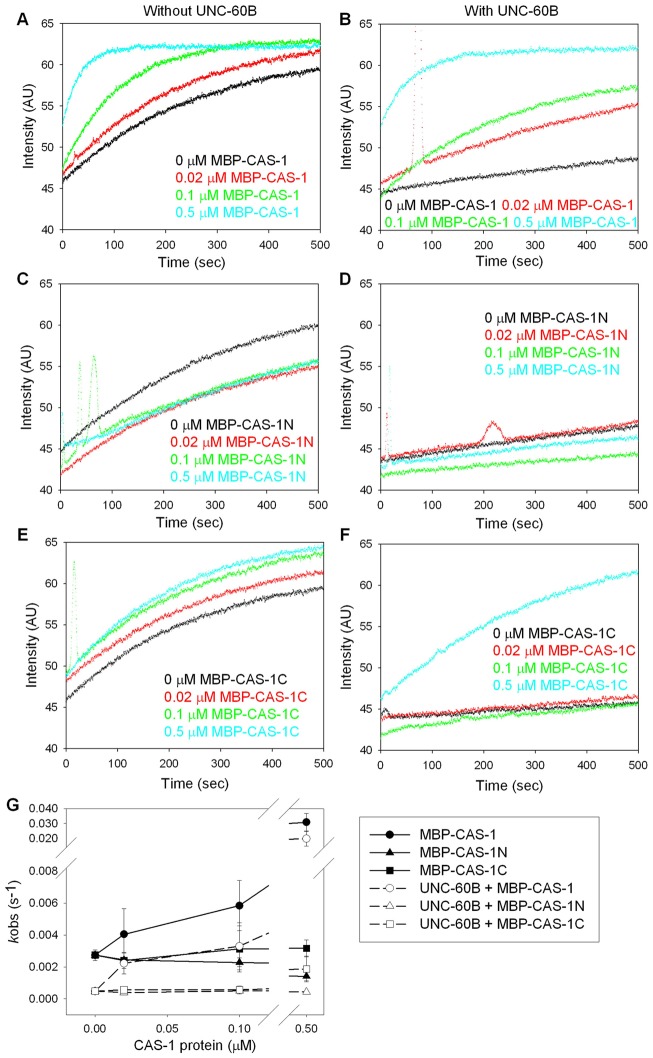
Effects of CAS-1 on exchange of actin-bound nucleotides in the absence and presence of UNC-60B. ATP–G-actin (1 µM) was incubated with etheno–ATP in the presence of 0–0.5 µM MBP–CAS-1 (A,B), MBP–CAS-1N (C,D), or MBP–CAS-1C (E,F) without UNC-60B (A,C,E) or with 1 µM UNC-60B (B,D,F), and the fluorescence of etheno–ATP (arbitrary units) was monitored over time. (G) Rates of exchange of nucleotides (kobs) were determined from the data and plotted as a function of concentrations of the CAS-1 variants. Data are means ± s.d. of three independent experiments.
Next, we tested whether CAS-1 has activity to enhance exchange of actin-bound nucleotides, as demonstrated for CAPs in other organisms (Moriyama and Yahara, 2002; Quintero-Monzon et al., 2009) (Fig. 4). ATP-bound G-actin was incubated with various concentrations of CAS-1 in the presence of etheno–ATP whose fluorescence is increased upon binding to actin, and changes in its fluorescence were monitored. MBP–CAS-1 strongly enhanced the rate of nucleotide exchange (Fig. 4A). In the presence of 0.5 µM MBP–CAS-1, the exchange rate was enhanced more than 10-fold (Fig. 4G, black circles). Substoichiometric concentrations of MBP–CAS-1 (0.02–0.5 µM MBP–CAS-1 to 1 µM G-actin) had significant effects. However, neither MBP–CAS-1N (Fig. 6C,G, black triangles) nor MBP–CAS-1C (Fig. 4E, black squares) enhanced nucleotide exchange. MBP–CAS-1N even had a weak inhibitory effect on nucleotide exchange (Fig. 4G, black triangles). MBP–CAS-1C slightly enhanced overall fluorescence (Fig. 4E), which might be due to its binding to G-actin as observed for human CAP1 (Moriyama and Yahara, 2002). These results indicate that both N- and C-terminal halves of CAS-1 are required for this activity. We also tested effects of CAS-1 on nucleotide exchange of ADP-bound actin (supplementary material Fig. S4A,C,E). However, ADP–actin exchanged nucleotides much faster than ATP–actin, as reported previously (Kinosian et al., 1993), and we were unable to determine the effects of CAS-1.
Fig. 6.
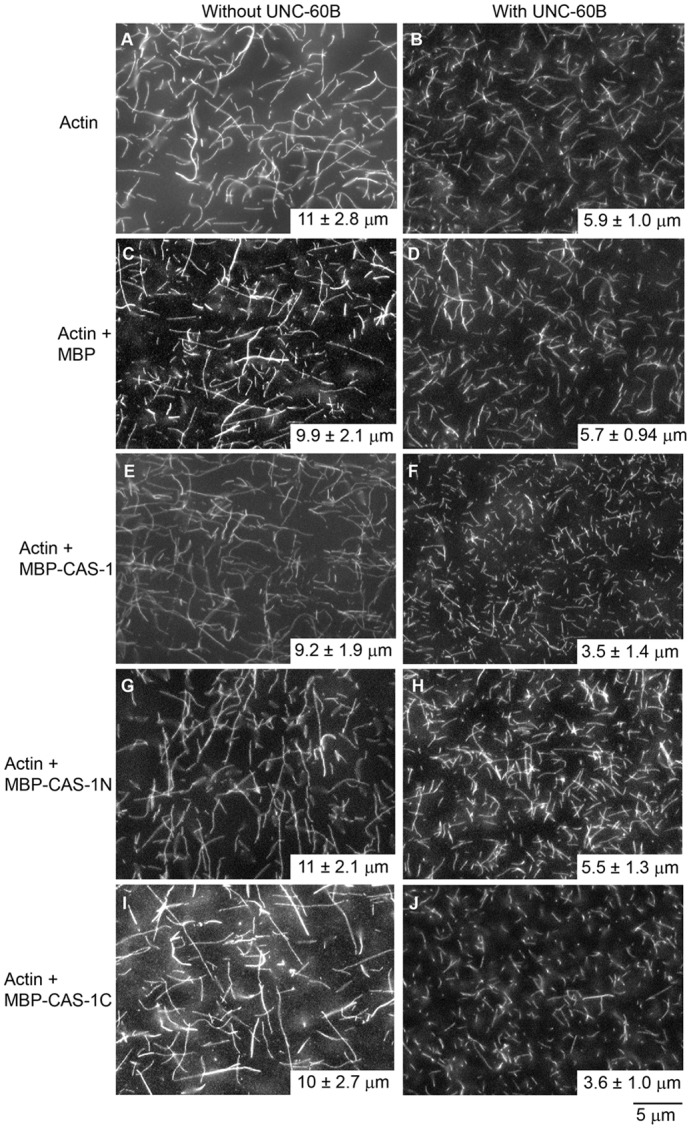
Effects of CAS-1 and UNC-60B on the lengths of actin filaments. Actin (2 µM, 20% DyLight549-labeled) was polymerized without an MBP fusion protein (A,B), or with 0.5 µM MBP (C,D), MBP–CAS-1 (E,F), MBP–CAS-1N (G,H) or MBP–CAS-1C (I,J) in the absence (A,C,E,G,I) or presence of 1 µM UNC-60B (B,D,F,H,J) and observed by fluorescence microscopy. Filament lengths (averages ± s.d., n = 50) are shown on the lower right corner of each panel.
UNC-60B, in contrast, bound to G-actin and inhibited exchange of actin-bound ATP (Fig. 4B, black curve) as demonstrated previously (Yamashiro et al., 2005). However, this inhibitory effect of UNC-60B was relieved by MBP–CAS-1 (Fig. 4B,G, white circles). Again, substoichiometric concentrations of MBP–CAS-1 (0.02–0.5 µM MBP–CAS-1) strongly enhanced nucleotide exchange even in the presence of 1 µM UNC-60B. MBP–CAS-1N had no effect on UNC-60B-inhibited nucleotide exchange (Fig. 4D), whereas 0.5 µM MBP–CAS-1C weakly relieved the effect of UNC-60B (Fig. 4F). This might be due to dissociation of UNC-60B from G-actin, as the C-terminal actin-binding site of yeast SRV2/CAP1 is known to compete with cofilin for G-actin binding (Mattila et al., 2004). Similar experiments using ADP-bound actin showed similar effects of CAS-1 on exchange of actin-bound nucleotides (supplementary material Fig. S4B,D,F), suggesting that CAS-1 interacts equally with ATP– and ADP–actin, as reported for plant CAP (Chaudhry et al., 2007). Thus, both N- and C-terminal halves of CAS-1 are required for efficient exchange of actin-bound nucleotides in the presence of ADF/cofilin.
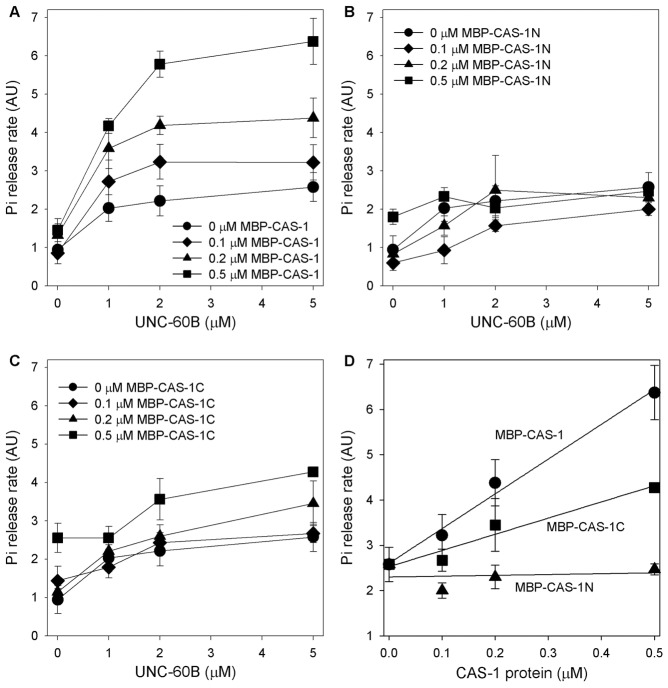
To determine effects of CAS-1 on actin filament turnover, we measured the rate of inorganic phosphate (Pi) release from actin filaments. The Pi release rate is an appropriate parameter for actin filament turnover or actin monomer recycling because it is enhanced after completion of essential turnover processes: monomer dissociation from F-actin, nucleotide exchange on G-actin, incorporation into F-actin, and ATP hydrolysis on F-actin (Quintero-Monzon et al., 2009). Alternatively Pi release can be enhanced if F-actin-binding of ADF/cofilin is enhanced (Blanchoin and Pollard, 1999). In the absence of MBP–CAS-1, UNC-60B by itself moderately enhanced Pi release (Fig. 5A, black circles). MBP–CAS-1 further enhanced UNC-60B-dependent Pi release in a dose-dependent manner (Fig. 5A,D, circles). The Pi release was enhanced by substoichiometric amounts of MBP–CAS-1, suggesting that CAS-1 enhanced actin turnover in a catalytic manner. MBP–CAS-1N did not significantly enhance Pi release (Fig. 5B,D, triangles), whereas MBP–CAS-1C moderately enhanced Pi release (Fig. 5C,D, squares). These results indicate that the C-terminal half of CAS-1 is required for enhancement of actin filament turnover, and that both N- and C-terminal halves are required for strong enhancement of this activity.
Fig. 5.
Effects of CAS-1 and UNC-60B on actin filament turnover as determined by the rate of phosphate release. Final 5 µM F-actin was mixed with various concentrations of UNC-60B and MBP–CAS-1 (A), MBP–CAS-1N (B) or MBP–CAS-1C (C), and the rate of Pi release (arbitrary units) was determined and plotted as a function of concentrations of UNC-60B. (D) Rate of Pi release from 5 µM F-actin with 5 µM UNC-60B in the presence of 0–0.5 µM MBP–CAS-1 (circles), MBP–CAS-1N (triangles) or MBP–CAS-1C (squares) were plotted as a function of the concentrations of the CAS-1 variants. Data are means ± s.d. of three independent experiments.
Effects of CAS-1 and UNC-60B were also examined by direct observation of actin filaments (Fig. 6). Fluorescently labeled actin was polymerized in the presence or absence of CAS-1 and/or UNC-60B for 30 min, and the filaments were observed by fluorescence microscopy. MBP–CAS-1 by itself does not have significant effect on lengths of actin filaments (Fig. 6, compare A and E). UNC-60B (1 µM) shortened actin filaments to ∼50% of the control (Fig. 6, compare A and B) due to its severing effect (Ono et al., 2008; Ono et al., 2004). Interestingly, in the presence of UNC-60B, MBP–CAS-1 further shortened filaments by 40% (Fig. 6, compare B and F). MBP (Fig. 6C,D) and MBP–CAS-1N (Fig. 6G,H) did not have a significant effect on filament lengths, while MBP–CAS-1C had a similar effect on filament lengths to MBP–CAS-1 (Fig. 6I,J). Although the precise mechanism of this effect is not clear, CAS-1 and UNC-60B can cooperate to keep actin filaments short and maintain a dynamic state of actin filaments.
cas-1 is essential for viability and sarcomeric actin organization
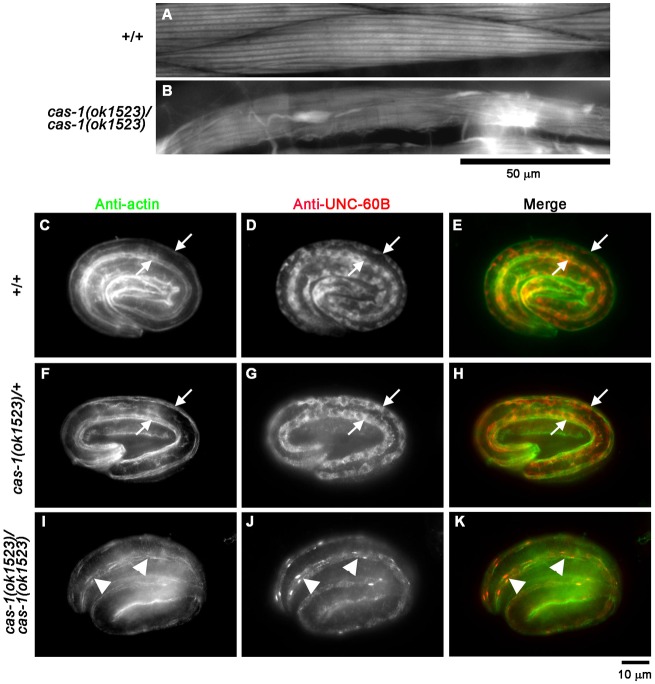
To determine in vivo roles of CAS-1 in C. elegans, we characterized cas-1 mutant phenotypes. cas-1(ok1523) is a deletion of 2.7 kb, which removes second and third exons and induces a frame-shift. This results in potential production of an mRNA coding for only 39 amino acids from the N-terminus, which is unlikely to be functional. Therefore, cas-1(ok1523) is a putative null allele. Most of cas-1(ok1523) homozygotes were arrested at variable larval stages and became immobile, while heterozygous animals were indistinguishable from wild type. Very few cas-1(ok1523) homozygotes became adults but failed to reproduce. Examination of cas-1(ok1523) mutant worms by phalloidin staining showed that sarcomeric actin filaments in the body wall muscle of arrested larvae and survived adults were severely disorganized with formation of a number of F-actin aggregates (Fig. 7B). In comparison, wild-type worms had highly ordered sarcomeric actin structures in the body wall muscle (Fig. 7A). Abnormalities of F-actin organization were not detected in other tissues, suggesting that cas-1 primarily functions in the body wall muscle and is required for organized assembly of sarcomeric actin filaments.
Fig. 7.
cas-1 mutation causes disorganization of actin and UNC-60B in the body wall muscle. (A,B) F-actin organization in late larval body wall muscle. Wild-type (+/+; A) or cas-1(ok1523) homozygous (cas-1(ok1523)/cas-1(ok1523); B) worms were stained with tetramethylrhodamine–phalloidin, and regions of the body wall muscle are shown. Scale bar: 50 µm. (C–K) Localization of UNC-60B and actin in embryos. Wild-type (+/+; C–E), cas-1 heterozygous (cas-1(ok1523)/+; F–H) or cas-1 homozygous (cas-1(ok1523)/cas-1(ok1523); I–K) embryos were fixed and immunostained for actin (C,F,I) and UNC-60B (D,G,J). Merged images are shown in E, H and K (actin in green and UNC-60B in red). Arrows in C–H indicate positions of the body wall muscle. Arrowheads in I–K indicate abnormal aggregates of actin and UNC-60B.
Abnormalities of actin filament organization were also detected in embryonic muscle of the cas-1(ok1523) mutant, suggesting that CAS-1 is required for an early stage of myofibril assembly (Fig. 7C–K). In wild-type and cas-1/+ heterozygous embryos, actin was assembled into linearly organized myofibrils in the body wall muscle (Fig. 7C,F, arrows). However, in the cas-1 homozygous embryos, actin became discontinuous and was often concentrated into aggregates (Fig. 7I, arrowheads). Thus, CAS-1 is required for organized assembly of actin filaments in embryonic muscle.
Subcellular localization of UNC-60B was significantly altered in the cas-1 mutant muscle (Fig. 7J). In wild-type and cas-1/+ heterozygous embryos, UNC-60B was specifically expressed in the body wall muscle and localized to the diffuse cytoplasm (Fig. 7D,G, arrows). However, in the cas-1 homozygous embryos, diffuse localization of UNC-60B was diminished, and UNC-60B was concentrated into aggregates where actin was also accumulated (Fig. 7J,K, arrowheads). The formation of UNC-60B–actin aggregates suggests that UNC-60B and actin remain in a stable complex in the absence of CAS-1, and that proper recycling of UNC-60B and actin for persistent actin filament turnover is not operated in the cas-1 mutant. This is consistent with our in vitro observations that CAS-1 promotes UNC-60B-dependent actin filament turnover.
In contrast, localization of CAS-1 was not disturbed in an unc-60B-null mutant (Fig. 8). unc-60B(su158) is a null allele, and unc-60B(su158) homozygotes had severely disorganized actin filaments with formation of actin aggregates in the body wall muscle (Fig. 8B), as described previously (Ono et al., 2003). Nonetheless, CAS-1 remained associated with striated myofibrils (Fig. 8A, arrows). Striations of CAS-1 did not overlap with those of actin (Fig. 8A–C, arrows), indicating that CAS-1 remained localized to the M-lines in the unc-60B mutant. Furthermore, CAS-1 was absent from the actin aggregates (Fig. 8A–C, arrowheads). These results suggest that myofibril localization of CAS-1 is independent of UNC-60B and actin, and that CAS-1 does not bind to actin filaments in the absence of UNC-60B-mediated actin filament turnover.
Fig. 8.
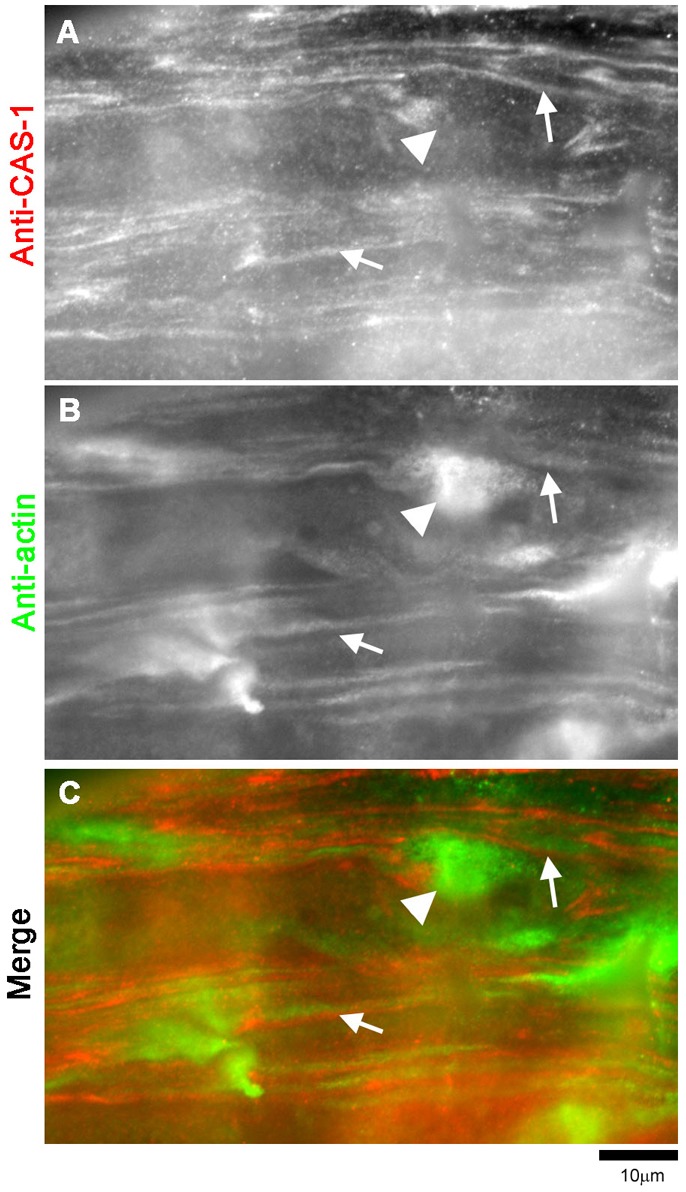
UNC-60B deficiency does not affect myofibril localization of CAS-1. Immunolocalization of CAS-1 (A) and actin (B) in the body wall muscle of unc-60B(su158) adult worms. A merged image is shown in C with CAS-1 in red and actin in green. Arrows indicate positions of the M-lines, where CAS-1 remained localized. Arrowheads indicate actin aggregates where CAS-1 did not localize.
Discussion
In this study, we demonstrate that CAS-1 is a C. elegans homolog of CAP, which is essential for viability and sarcomeric actin organization in striated muscle. In vitro, CAS-1 binds to monomeric actin, enhances exchange of actin-bound nucleotides, and, promotes actin filament turnover in the presence of UNC-60B (a muscle-specific ADF/cofilin). Previous studies have shown that UNC-60B is an essential regulator of sarcomeric actin organization in the body wall muscle (McKim et al., 1994; Ono et al., 2003; Ono et al., 1999). These results strongly suggest that enhancement of actin filament turnover by CAS-1 and UNC-60B is essential for assembly of sarcomeric actin filaments in the C. elegans body wall muscle. Mammals also have muscle-specific isoforms of ADF/cofilin (cofilin-2) (Ono et al., 1994; Thirion et al., 2001; Vartiainen et al., 2002) and CAP (CAP2) (Peche et al., 2007; Swiston et al., 1995; Wolanski et al., 2009; Yu et al., 1994), suggesting that collaboration of ADF/cofilin and CAP is a conserved mechanism of the regulation of actin dynamics in striated muscle.
The sequence and domain structure of C. elegans CAS-1 are conserved with CAPs from other species with some differences (supplementary material Fig. S1). In addition to the lack of a second proline-rich region, we noticed a potential difference between CAS-1 and other CAPs in the N-termini. The N-terminal end of yeast Srv2/CAP is predicted to form a coiled coil and implicated in its association with adenylate cyclase (Nishida et al., 1998) or oligomerization (Quintero-Monzon et al., 2009). Analysis by COILS, a prediction tool for coiled coils (Lupas et al., 1991), indicates that the N-terminal region of yeast Srv2/CAP exhibits high probability of forming a coiled coil, but an equivalent region of C. elegans CAS-1 shows very low probability of forming a coiled coil (data not shown). However, analysis by Multicoil2, a more recently published tool for prediction of coiled coils (Trigg et al., 2011), predicted that both yeast Srv2/CAP and C. elegans CAS-1 do not form coiled coils (data not shown). The N-terminal 50 amino acids of Dictyostelium CAP are flexible and unstructured as revealed by NMR (Mavoungou et al., 2004). Therefore, the N-terminal region of CAP may become structured only when it binds to a ligand, but this property still needs to be verified by biochemical or biophysical methods.
CAS-1 has similar biochemical activities to CAPs from other species, but we detected two unique properties of CAS-1. First, the N-terminal half of CAS-1 (CAS-1N) weakly bound to G-actin in the absence of UNC-60B. An equivalent region of human CAP1 also weakly binds to G-actin (Moriyama and Yahara, 2002), but that of yeast Srv2/CAP only binds to the actin–cofilin complex (Quintero-Monzon et al., 2009). G-actin binding of the CAS-1N resulted in slight retardation of polymerization and weak inhibition of nucleotide exchange. However, their functional significance is not clear, since these effects are weak and require high concentrations of CAS-1N. Second, the C-terminal half of CAS-1 (CAS-1C) alone had much weaker activity to enhance exchange of G-actin-bound nucleotide than full-length CAS-1. In yeast and human CAPs, the C-terminal halves promote nucleotide exchange as strongly as full-length proteins (Moriyama and Yahara, 2002; Quintero-Monzon et al., 2009). CAS-1C contained WH2 and putative β-sheet domains, and bound to G-actin, suggesting that G-actin binding is not sufficient to enhance nucleotide exchange. Therefore, our CAS-1C construct may not have an essential sequence for nucleotide exchange or may include an inhibitory sequence. To clarify this problem, additional experiments are required to determine regulatory sequences for promotion of nucleotide exchange.
Our biochemical studies showed that CAS-1 strongly promoted exchange of G-actin-bound nucleotides antagonistically to UNC-60B and enhanced actin filament turnover in collaboration with UNC-60B. These observations agree with recently reported properties of CAPs in mammals (Moriyama and Yahara, 2002), yeast (Balcer et al., 2003; Quintero-Monzon et al., 2009) and plants (Chaudhry et al., 2007). Although CAP sequesters actin monomers in vitro (Freeman et al., 1995; Gieselmann and Mann, 1992), stoichiometric amounts of CAP are required for effective actin monomer sequestration. In contrast, enhancement of actin filament turnover can be achieved by substoichiometric concentrations of CAP in the presence of ADF/cofilin. Therefore, the latter function is more likely to be a physiologically important function of CAP. The role of CAP in actin filament turnover is also consistent with our observed CAS-1 knockout phenotypes and previously reported essential roles of CAP in lamellipodial extension (Rogers et al., 2003), stress fiber formation (Freeman and Field, 2000), cell polarity (Baum et al., 2000) and plant development (Barrero et al., 2002; Deeks et al., 2007), which cannot be explained simply by the actin-monomer sequestering activity. CAS-1 is also expressed in several non-muscle cells (Fig. 1D–F) and might be involved in the regulation of other actin-dependent events.
Functional significance of localization of CAS-1 to the M-lines in sarcomeres is still unclear. However, it is intriguing that mammalian CAP2 is predominantly expressed in striated muscle and also localizes to the M-lines (Peche et al., 2007). Apparently, this localization pattern is independent of UNC-60B and actin (Fig. 8). Given that CAS-1 can enhance actin turnover at very low concentrations, the M-line might be a temporary anchoring or storage site. Therefore, dynamic nature of CAS-1 at the M-lines is important information to understand its significance. Proline-rich regions of CAP are known to bind to Src-homology 3 (SH3) domains (Freeman et al., 1996; Lila and Drubin, 1997). Intriguingly, C. elegans UNC-89, an obscurin-like protein, is a component of the M-lines in the body wall muscle and has an SH3 domain near its N-terminus (Benian et al., 1996). Whether UNC-89 binds to CAS-1 and is required for M-line localization of CAS-1 will be determined in the future. The M-lines are closely located to the pointed ends of sarcomeric actin filaments, where actin monomers are actively exchanged (Littlefield et al., 2001). We have previously shown that ADF/cofilin (UNC-60B) cooperates with tropomodulin, an actin pointed-end capping protein, to promote sarcomeric actin organization (Yamashiro et al., 2008). If the pointed ends of sarcomeric actin filaments are sites of ADF/cofilin-induced actin turnover, accumulation of CAS-1 to the M-lines may facilitate cooperation between ADF/cofilin and CAS-1.
Our study provides biochemical and genetic evidence that CAS-1 is an essential regulator of actin filament dynamics for sarcomeric actin organization in striated muscle. We hypothesize that C. elegans CAS-1 is functionally homologous to vertebrate CAP2 based on their similarities in muscle expression and M-line localization. Previous studies have shown that muscle-specific ADF/cofilin isoforms are essential for sarcomeric actin assembly in both C. elegans and mammals (Ono, 2010). Therefore, cooperative enhancement of actin filament turnover by ADF/cofilin and CAP might be a conserved mechanism for organized assembly of sarcomeres in striated muscle. Requirement of CAP for muscle actin organization and viability in C. elegans strongly suggests that a deficiency in vertebrate CAP2 also causes severe muscle defects or lethality. Additional analysis of the function of CAS-1 in C. elegans and functional studies on CAP2 in vertebrate muscle should be important for advancing our knowledge on the mechanism of assembly and organization of sarcomeric actin filaments in striated muscle.
Materials and Methods
Nematode strains
Nematodes were grown at 20°C as described previously (Brenner, 1974). Wild-type strain N2 was obtained from the Caenorhabditis Genetics Center (Minneapolis, MN). cas-1(ok1523)X was isolated as the strain VC1144 by the C. elegans Gene Knockout Consortium (Vancouver, British Columbia, Canada and Oklahoma, OH, USA), outcrossed four times, and maintained in the strain ON169 +/szT1[lon-2(e678)] I; cas-1(ok1523)/szT1 X. unc-60B(su158)V was described previously (Ono et al., 2003; Zengel and Epstein, 1980).
Proteins
Rabbit muscle actin was purified from acetone powder (Pel-Freez Biologicals) as described previously (Pardee and Spudich, 1982). Pyrene-labeled rabbit muscle actin was prepared as described (Kouyama and Mihashi, 1981). ADP–G-actin was prepared as described previously (Moriyama and Yahara, 2002) with some modifications. ATP–G-actin (15 µM) in G-buffer was incubated with 20 units/ml hexokinase (Worthington Biochemical Corp.) in the presence of 50 µM MgCl2, 0.2 mM EGTA, and 0.3 mM glucose at 4°C for 2 hr. UNC-60B was expressed in E. coli and purified as described previously (Ono and Benian, 1998).
Recombinant CAS-1 proteins
The full-length protein coding sequence of CAS-1 was amplified from a cDNA clone yk1657b05 (kindly provided by Yuji Kohara, Mishima, Shizuoka, Japan), and cloned into the pGEX-2T vector. Originally, we attempted bacterial expression of CAS-1 using a glutathione S-transferase fusion system, but solubility of recombinant fusion proteins was very poor (our unpublished observations). Then, full-length CAS-1, CAS-1N or CAS-1C sequences were re-cloned into the pDEST-HisMBP, a vector for bacterial expression as fusion proteins with MBP with an N-terminal histidine tag as described previously (Nallamsetty and Waugh, 2007) using the Gateway® technology with ClonaseTM II (Invitrogen). pDEST-HisMBP was developed by the group of David Waugh (Nallamsetty et al., 2005) and obtained through Addgene. The protein coding sequences were verified by DNA sequencing. E. coli BL21(DE3) was transformed with the expression vectors, and protein expression was inducted by adding 1 mM isopropyl-β-D-1-thiogalactopyranoside for 3 hr at room temperature. The cells were harvested by centrifugation at 5000 g for 10 min and disrupted by a French pressure cell at 360–580 kg/cm2 in phosphate-buffered saline. The homogenates were cleared at 20,000 g for 15 min and applied to a His60 Ni SuperflowTM column (Clontech) and washed with 150 mM NaCl, 3 mM imidazole, 50 mM sodium phosphate buffer, pH 7.9 to remove unbound proteins. Bound proteins were eluted with 150 mM NaCl, 250 mM imidazole, 50 mM sodium phosphate buffer, pH 7.9. MBP–CAS-1 (full-length) was further purified with a HiPrep 26/60 Sephacryl S-300 column (GE Healthcare), while MBP–CAS-1N and MBP–CAS-1C were further purified with a Resource Q column (GE Healthcare). MBP was used without additional chromatographic purification. Finally, purified proteins were dialyzed against F-buffer [0.1 M KCl, 20 mM Hepes-NaOH, pH 7.5, 2 mM MgCl2 and 0.2 mM dithiothreitol (DTT)] containing 50% glycerol and stored at −20°C.
Preparation of anti-CAS-1 antibody
Anti-CAS-1 antisera were raised in rabbit using GST-CAS-1C as an immunogen at Lampire Biological Laboratories. Anti-CAS-1 antibody was affinity-purified by adsorbing to MBP–CAS-1 that had been immobilized on nitrocellulose membranes and eluting with 0.1 M glycine–HCl, pH 2.5.
Fluorescence microscopy
Immunofluorescent staining of adult nematodes was performed as described previously (Finney and Ruvkun, 1990). Immunofluorescent staining of embryos was performed as described previously (Ono et al., 2003). Briefly, worm embryos were collected by cutting gravid adults and picking from agar plates on polylysine-coated slides, permeabilized by a freeze-crack method (Epstein et al., 1993), and fixed with methanol at −20°C for 5 min. They were washed by phosphate-buffered saline (PBS) for 10 min and stained with primary antibodies diluted in 1% bovine serum albumin in PBS. They were visualized by staining with Alexa-Fluor-488-conjugated goat anti-mouse IgG (Invitrogen) and Cy3-conjugated donkey anti-rabbit IgG (Jackson ImmunoResearch). Primary antibodies used were rabbit anti-CAS-1 polyclonal (this study), mouse anti-MYO-3 monoclonal (5–6) (Miller et al., 1983), mouse anti-UNC-89 monoclonal (MH42) (Benian et al., 1996), mouse anti-vinculin monoclonal (MH24) (Francis and Waterston, 1985), mouse anti-α-actinin monoclonal (MH40) (Francis and Waterston, 1985), mouse anti-actin monoclonal (C4; MP Biomedicals) and rabbit anti-UNC-60B (Ono et al., 1999) antibodies.
Staining of whole worms with tetramethylrhodamine–phalloidin (Sigma-Aldrich) was performed as described previously (Ono, 2001).
Samples were mounted with ProLong Gold (Invitrogen) and observed by epifluorescence using a Nikon Eclipse TE2000 inverted microscope with a CFI Plan Fluor ELWD 40× (Dry; NA 0.60) or Plan Apo 60× (oil; NA 1.40) objective. Images were captured by a SPOT RT monochrome CCD camera (Diagnostic Instruments) and processed by IPLab imaging software (BD Biosciences) and Adobe Photoshop CS3.
Monitoring kinetics of actin polymerization
Kinetics of actin polymerization was monitored by measuring fluorescence of pyrene-labeled actin. Pyrene-labeled G-actin (final concentration of 5 µM; referred to as final 5 µM; 20% labeled) was mixed with MBP–CAS-1, MBP–CAS-1N, MBP–CAS-1C or MBP in G buffer (0.2 mM ATP, 0.2 mM CaCl2, 0.2 mM DTT, 2 mM Tris-HCl, pH 8.0), and polymerization was initiated by adding salts to final concentrations of 0.1 M KCl, 2 mM MgCl2, 1 mM EGTA, 20 mM Hepes-NaOH, pH 7.5. Fluorescence of pyrene (excitation at 366 nm and emission at 384 nm) was monitored for 10 min with an LS50B fluorescence spectrophotometer (PerkinElmer). Relative rates of actin polymerization were calculated by the increase of the pyrene fluorescence from time 0 to 350 sec (ΔF), when approximately 50% of control actin (no CAS-1 protein) was polymerized, as ΔFCAS-1/ΔFcontrol×100 (%).
Determination of critical concentration of actin
Varying concentrations of pyrene-labeled G-actin (20% labeled) were polymerized in 0.1 M KCl, 2 mM MgCl2, 1 mM EGTA, 20 mM Hepes-NaOH, 0.2 mM DTT, 0.2 mM ATP, pH 7.5 for 18 hr at room temperature. Fluorescence of pyrene (excitation at 366 nm and emission at 384 nm) was measured with a LS50B fluorescence spectrophotometer (PerkinElmer). Equilibrium dissociation constant for CAS-1 binding to G-actin was calculated from the apparent change in the critical concentration as described (Carlier et al., 1993).
Non-denaturing polyacrylamide gel electrophoresis
Non-denaturing polyacrylamide gel electrophoresis was performed as described previously (Ono et al., 2001; Safer, 1989). Briefly, protein samples except for MBP or CAS-1 proteins were pre-incubated in G buffer for 30 min at room temperature, followed by addition of MBP or CAS-1 proteins for 30 min, supplemented with 0.25 volume of a loading buffer (50% glycerol and 0.05% Bromophenol Blue), and electrophoresed using a Bicine/triethanolamine buffer system (50 mM Bicine, 40 mM triethanolamine, 0.2 mM ATP, 0.5 mM EGTA) at 4°C. The proteins were visualized by staining with Coomassie Brilliant Blue R-250 (National Diagnostic).
Assay for exchange of actin-bound nucleotides
Effects of CAS-1 variants and UNC-60B on the exchange rate of actin-bound nucleotides were examined by monitoring increase in the fluorescence of etheno–ATP that is associated with G-actin binding (Wang and Taylor, 1981). G-actin (1.5 µM) was prepared in 100 µl of G-buffer without ATP, and then 50 µl of etheno–ATP (120 µM) with or without CAS-1 variants and UNC-60B was mixed. Final concentrations of G-actin and UNC-60B were both 1 µM. Then, fluorescence of etheno–ATP (excitation at 350 nm and emission at 410 nm) was monitored for 10 min with a HITACHI F-4500 fluorescence spectrophotometer. The exponential rates (kobs) were calculated by curve fitting using SigmaPlot 10 (Systat Software).
Measurements of actin turnover by phosphate release
Phosphate release during actin filament turnover was measured using an EnzChek Pi assay kit (Invitrogen). Final 5 µM F-actin was mixed with various concentrations of UNC-60B and CAS-1 variants in the presence of 0.1 mM 2-amino-6-mercapto-7-methylpurine riboside and 0.4 unit/ml purine nucleoside phosphorylase in F-buffer (0.1 M KCl, 2 mM MgCl2, 0.5 mM ATP, 0.1 mM DTT, 20 mM Hepes-NaOH, pH 7.5), and absorbance at 360 nm was monitored over time using a DU640 spectrophotometer (Beckman Coulter). Phosphate release was initially enhanced due to dilution-induced actin depolymerization. Therefore, a steady-state rate of Pi release was determined by a slope of a linear phase followed by the initial burst of Pi release (see supplementary material Fig. S5).
Direct observation of actin filaments by fluorescence microscopy
DyLight549-labeled actin was prepared as described previously (Liu et al., 2010). G-actin (2 µM, 20% DyLight549 labeled) was mixed with 0.5 µM MBP, MBP–CAS-1, MBP–CAS-1N, or MBP–CAS-1C in the presence or absence of 1 µM UNC-60B in G-buffer and polymerized for 30 min at room temperature by addition of final 0.1 M KCl, 2 mM MgCl2, 1 mM EGTA, and 20 mM Hepes-NaOH, pH 7.5. The samples were mounted on nitrocellulose-coated coverslips and filaments observed by fluorescence microscopy using a Nikon TE2000 microscope with a Plan Apo 60× (oil; NA 1.40) objective. Images were captured by a SPOT RT monochrome CCD camera and processed by IPLab imaging software and Adobe Photoshop CS3. Filament lengths were measured using a length-measurement tool of IPLab after manually tracing filaments on captured images.
Supplementary Material
Acknowledgments
Some C. elegans strains were provided by the Caenorhabditis Genetics Center (Minneapolis, MN), which is supported by the National Institutes of Health National Center for Research Resources. Monoclonal antibodies 5-6 and MH24 were developed by Henry Epstein (University of Texas Medical Branch, Galveston) and Robert Waterston (University of Washington), respectively, and obtained from the Developmental Studies Hybridoma Bank developed under auspices of the National Institute of Child Health and Human Development and maintained by the Department of Biological Sciences, University of Iowa, Iowa City, IA.
Footnotes
Funding
This work was supported by the National Institutes of Health [grant number R01 AR48615 to S.O.]. Deposited in PMC for release after 12 months.
Supplementary material available online at http://jcs.biologists.org/lookup/suppl/doi:10.1242/jcs.104950/-/DC1
References
- Agrawal P. B., Greenleaf R. S., Tomczak K. K., Lehtokari V. L., Wallgren–Pettersson C., Wallefeld W., Laing N. G., Darras B. T., Maciver S. K., Dormitzer P. R.et al. (2007). Nemaline myopathy with minicores caused by mutation of the CFL2 gene encoding the skeletal muscle actin-binding protein, cofilin-2. Am. J. Hum. Genet. 80, 162–167 10.1086/510402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrianantoandro E., Pollard T. D. (2006). Mechanism of actin filament turnover by severing and nucleation at different concentrations of ADF/cofilin. Mol. Cell 24, 13–23 10.1016/j.molcel.2006.08.006 [DOI] [PubMed] [Google Scholar]
- Balcer H. I., Goodman A. L., Rodal A. A., Smith E., Kugler J., Heuser J. E., Goode B. L. (2003). Coordinated regulation of actin filament turnover by a high-molecular-weight Srv2/CAP complex, cofilin, profilin, and Aip1. Curr. Biol. 13, 2159–2169 10.1016/j.cub.2003.11.051 [DOI] [PubMed] [Google Scholar]
- Barrero R. A., Umeda M., Yamamura S., Uchimiya H. (2002). Arabidopsis CAP regulates the actin cytoskeleton necessary for plant cell elongation and division. Plant Cell 14, 149–163 10.1105/tpc.010301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum B., Li W., Perrimon N. (2000). A cyclase-associated protein regulates actin and cell polarity during Drosophila oogenesis and in yeast. Curr. Biol. 10, 964–973 10.1016/S0960-9822(00)00640-0 [DOI] [PubMed] [Google Scholar]
- Benian G. M., Tinley T. L., Tang X., Borodovsky M. (1996). The Caenorhabditis elegans gene unc-89, required for muscle M-line assembly, encodes a giant modular protein composed of Ig and signal transduction domains. J. Cell Biol. 132, 835–848 10.1083/jcb.132.5.835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertling E., Hotulainen P., Mattila P. K., Matilainen T., Salminen M., Lappalainen P. (2004). Cyclase-associated protein 1 (CAP1) promotes cofilin-induced actin dynamics in mammalian nonmuscle cells. Mol. Biol. Cell 15, 2324–2334 10.1091/mbc.E04-01-0048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertling E., Quintero–Monzon O., Mattila P. K., Goode B. L., Lappalainen P. (2007). Mechanism and biological role of profilin-Srv2/CAP interaction. J. Cell Sci. 120, 1225–1234 10.1242/jcs.000158 [DOI] [PubMed] [Google Scholar]
- Blanchoin L., Pollard T. D. (1998). Interaction of actin monomers with Acanthamoeba actophorin (ADF/cofilin) and profilin. J. Biol. Chem. 273, 25106–25111 10.1074/jbc.273.39.25106 [DOI] [PubMed] [Google Scholar]
- Blanchoin L., Pollard T. D. (1999). Mechanism of interaction of Acanthamoeba actophorin (ADF/Cofilin) with actin filaments. J. Biol. Chem. 274, 15538–15546 10.1074/jbc.274.22.15538 [DOI] [PubMed] [Google Scholar]
- Brenner S. (1974). The genetics of Caenorhabditis elegans. Genetics 77, 71–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlier M. F., Jean C., Rieger K. J., Lenfant M., Pantaloni D. (1993). Modulation of the interaction between G-actin and thymosin β 4 by the ATP/ADP ratio: possible implication in the regulation of actin dynamics. Proc. Natl. Acad. Sci. USA 90, 5034–5038 10.1073/pnas.90.11.5034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlier M. F., Laurent V., Santolini J., Melki R., Didry D., Xia G. X., Hong Y., Chua N. H., Pantaloni D. (1997). Actin depolymerizing factor (ADF/cofilin) enhances the rate of filament turnover: implication in actin-based motility. J. Cell Biol. 136, 1307–1322 10.1083/jcb.136.6.1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhry F., Guérin C., von Witsch M., Blanchoin L., Staiger C. J. (2007). Identification of Arabidopsis cyclase-associated protein 1 as the first nucleotide exchange factor for plant actin. Mol. Biol. Cell 18, 3002–3014 10.1091/mbc.E06-11-1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark K. A., McElhinny A. S., Beckerle M. C., Gregorio C. C. (2002). Striated muscle cytoarchitecture: an intricate web of form and function. Annu. Rev. Cell Dev. Biol. 18, 637–706 10.1146/annurev.cellbio.18.012502.105840 [DOI] [PubMed] [Google Scholar]
- Cole C., Barber J. D., Barton G. J. (2008). The Jpred 3 secondary structure prediction server. Nucleic Acids Res. 36, W197–W201 10.1093/nar/gkn238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deeks M. J., Rodrigues C., Dimmock S., Ketelaar T., Maciver S. K., Malhó R., Hussey P. J. (2007). Arabidopsis CAP1 – a key regulator of actin organisation and development. J. Cell Sci 120, 2609–2618 10.1242/jcs.007302 [DOI] [PubMed] [Google Scholar]
- Didry D., Carlier M. F., Pantaloni D. (1998). Synergy between actin depolymerizing factor/cofilin and profilin in increasing actin filament turnover. J. Biol. Chem. 273, 25602–25611 10.1074/jbc.273.40.25602 [DOI] [PubMed] [Google Scholar]
- Dodatko T., Fedorov A. A., Grynberg M., Patskovsky Y., Rozwarski D. A., Jaroszewski L., Aronoff–Spencer E., Kondraskina E., Irving T., Godzik A.et al. (2004). Crystal structure of the actin binding domain of the cyclase-associated protein. Biochemistry 43, 10628–10641 10.1021/bi049071r [DOI] [PubMed] [Google Scholar]
- Epstein H. F., Casey D. L., Ortiz I. (1993). Myosin and paramyosin of Caenorhabditis elegans embryos assemble into nascent structures distinct from thick filaments and multi-filament assemblages. J. Cell Biol. 122, 845–858 10.1083/jcb.122.4.845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasseas M. K., Tsikou D., Flemetakis E., Katinakis P. (2011). Molecular and biochemical analysis of the α class carbonic anhydrases in Caenorhabditis elegans. Mol. Biol. Rep. 38, 1777–1785 10.1007/s11033-010-0292-y [DOI] [PubMed] [Google Scholar]
- Fedor–Chaiken M., Deschenes R. J., Broach J. R. (1990). SRV2, a gene required for RAS activation of adenylate cyclase in yeast. Cell 61, 329–340 10.1016/0092-8674(90)90813-T [DOI] [PubMed] [Google Scholar]
- Field J., Vojtek A., Ballester R., Bolger G., Colicelli J., Ferguson K., Gerst J., Kataoka T., Michaeli T., Powers S.et al. (1990). Cloning and characterization of CAP, the S. cerevisiae gene encoding the 70 kd adenylyl cyclase-associated protein. Cell 61, 319–327 10.1016/0092-8674(90)90812-S [DOI] [PubMed] [Google Scholar]
- Finney M., Ruvkun G. (1990). The unc-86 gene product couples cell lineage and cell identity in C. elegans. Cell 63, 895–905 10.1016/0092-8674(90)90493-X [DOI] [PubMed] [Google Scholar]
- Fox J. D., Waugh D. S. (2003). Maltose-binding protein as a solubility enhancer. Methods Mol. Biol. 205, 99–117 [DOI] [PubMed] [Google Scholar]
- Francis G. R., Waterston R. H. (1985). Muscle organization in Caenorhabditis elegans: localization of proteins implicated in thin filament attachment and I-band organization. J. Cell Biol. 101, 1532–1549 10.1083/jcb.101.4.1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman N. L., Field J. (2000). Mammalian homolog of the yeast cyclase associated protein, CAP/Srv2p, regulates actin filament assembly. Cell Motil. Cytoskeleton 45, 106–120 [DOI] [PubMed] [Google Scholar]
- Freeman N. L., Chen Z., Horenstein J., Weber A., Field J. (1995). An actin monomer binding activity localizes to the carboxyl-terminal half of the Saccharomyces cerevisiae cyclase-associated protein. J. Biol. Chem. 270, 5680–5685 10.1074/jbc.270.10.5680 [DOI] [PubMed] [Google Scholar]
- Freeman N. L., Lila T., Mintzer K. A., Chen Z., Pahk A. J., Ren R., Drubin D. G., Field J. (1996). A conserved proline-rich region of the Saccharomyces cerevisiae cyclase-associated protein binds SH3 domains and modulates cytoskeletal localization. Mol. Cell. Biol. 16, 548–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gieselmann R., Mann K. (1992). ASP-56, a new actin sequestering protein from pig platelets with homology to CAP, an adenylate cyclase-associated protein from yeast. FEBS Lett. 298, 149–153 10.1016/0014-5793(92)80043-G [DOI] [PubMed] [Google Scholar]
- Hubberstey A. V., Mottillo E. P. (2002). Cyclase-associated proteins: CAPacity for linking signal transduction and actin polymerization. FASEB J. 16, 487–499 10.1096/fj.01-0659rev [DOI] [PubMed] [Google Scholar]
- Ichetovkin I., Han J., Pang K. M., Knecht D. A., Condeelis J. S. (2000). Actin filaments are severed by both native and recombinant dictyostelium cofilin but to different extents. Cell Motil. Cytoskeleton 45, 293–306 [DOI] [PubMed] [Google Scholar]
- Kinosian H. J., Selden L. A., Estes J. E., Gershman L. C. (1993). Nucleotide binding to actin. Cation dependence of nucleotide dissociation and exchange rates. J. Biol. Chem. 268, 8683–8691 [PubMed] [Google Scholar]
- Kouyama T., Mihashi K. (1981). Fluorimetry study of N-(1-pyrenyl)iodoacetamide-labelled F-actin. Local structural change of actin protomer both on polymerization and on binding of heavy meromyosin. Eur. J. Biochem. 114, 33–38 10.1111/j.1432-1033.1981.tb06167.x [DOI] [PubMed] [Google Scholar]
- Ksiazek D., Brandstetter H., Israel L., Bourenkov G. P., Katchalova G., Janssen K. P., Bartunik H. D., Noegel A. A., Schleicher M., Holak T. A. (2003). Structure of the N-terminal domain of the adenylyl cyclase-associated protein (CAP) from Dictyostelium discoideum. Structure 11, 1171–1178 10.1016/S0969-2126(03)00180-1 [DOI] [PubMed] [Google Scholar]
- Lai C. C., Hong K., Kinnell M., Chalfie M., Driscoll M. (1996). Sequence and transmembrane topology of MEC-4, an ion channel subunit required for mechanotransduction in Caenorhabditis elegans. J. Cell Biol. 133, 1071–1081 10.1083/jcb.133.5.1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lila T., Drubin D. G. (1997). Evidence for physical and functional interactions among two Saccharomyces cerevisiae SH3 domain proteins, an adenylyl cyclase-associated protein and the actin cytoskeleton. Mol. Biol. Cell 8, 367–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littlefield R., Almenar–Queralt A., Fowler V. M. (2001). Actin dynamics at pointed ends regulates thin filament length in striated muscle. Nat. Cell Biol. 3, 544–551 10.1038/35078517 [DOI] [PubMed] [Google Scholar]
- Liu Z., Klaavuniemi T., Ono S. (2010). Distinct roles of four gelsolin-like domains of Caenorhabditis elegans gelsolin-like protein-1 in actin filament severing, barbed end capping, and phosphoinositide binding. Biochemistry 49, 4349–4360 10.1021/bi100215b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupas A., Van Dyke M., Stock J. (1991). Predicting coiled coils from protein sequences. Science 252, 1162–1164 10.1126/science.252.5009.1162 [DOI] [PubMed] [Google Scholar]
- Maciver S. K., Pope B. J., Whytock S., Weeds A. G. (1998). The effect of two actin depolymerizing factors (ADF/cofilins) on actin filament turnover: pH sensitivity of F-actin binding by human ADF, but not of Acanthamoeba actophorin. Eur. J. Biochem. 256, 388–397 10.1046/j.1432-1327.1998.2560388.x [DOI] [PubMed] [Google Scholar]
- Mattila P. K., Quintero–Monzon O., Kugler J., Moseley J. B., Almo S. C., Lappalainen P., Goode B. L. (2004). A high-affinity interaction with ADP-actin monomers underlies the mechanism and in vivo function of Srv2/cyclase-associated protein. Mol. Biol. Cell 15, 5158–5171 10.1091/mbc.E04-06-0444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavoungou C., Israel L., Rehm T., Ksiazek D., Krajewski M., Popowicz G., Noegel A. A., Schleicher M., Holak T. A. (2004). NMR structural characterization of the N-terminal domain of the adenylyl cyclase-associated protein (CAP) from Dictyostelium discoideum. J. Biomol. NMR 29, 73–84 10.1023/B%3AJNMR.0000019513.86120.98 [DOI] [PubMed] [Google Scholar]
- McKim K. S., Matheson C., Marra M. A., Wakarchuk M. F., Baillie D. L. (1994). The Caenorhabditis elegans unc-60 gene encodes proteins homologous to a family of actin-binding proteins. Mol. Gen. Genet. 242, 346–357 10.1007/BF00280425 [DOI] [PubMed] [Google Scholar]
- Miller D. M., 3rd, Ortiz I., Berliner G. C., Epstein H. F. (1983). Differential localization of two myosins within nematode thick filaments. Cell 34, 477–490 10.1016/0092-8674(83)90381-1 [DOI] [PubMed] [Google Scholar]
- Mohri K., Ono S. (2003). Actin filament disassembling activity of Caenorhabditis elegans actin-interacting protein 1 (UNC-78) is dependent on filament binding by a specific ADF/cofilin isoform. J. Cell Sci. 116, 4107–4118 10.1242/jcs.00717 [DOI] [PubMed] [Google Scholar]
- Mohri K., Ono K., Yu R., Yamashiro S., Ono S. (2006). Enhancement of actin-depolymerizing factor/cofilin-dependent actin disassembly by actin-interacting protein 1 is required for organized actin filament assembly in the Caenorhabditis elegans body wall muscle. Mol. Biol. Cell 17, 2190–2199 10.1091/mbc.E05-11-1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriyama K., Yahara I. (2002). Human CAP1 is a key factor in the recycling of cofilin and actin for rapid actin turnover. J. Cell Sci. 115, 1591–1601 [DOI] [PubMed] [Google Scholar]
- Mudry R. E., Perry C. N., Richards M., Fowler V. M., Gregorio C. C. (2003). The interaction of tropomodulin with tropomyosin stabilizes thin filaments in cardiac myocytes. J. Cell Biol. 162, 1057–1068 10.1083/jcb.200305031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nallamsetty S., Waugh D. S. (2007). A generic protocol for the expression and purification of recombinant proteins in Escherichia coli using a combinatorial His6-maltose binding protein fusion tag. Nat. Protoc. 2, 383–391 10.1038/nprot.2007.50 [DOI] [PubMed] [Google Scholar]
- Nallamsetty S., Austin B. P., Penrose K. J., Waugh D. S. (2005). Gateway vectors for the production of combinatorially-tagged His6-MBP fusion proteins in the cytoplasm and periplasm of Escherichia coli. Protein Sci. 14, 2964–2971 10.1110/ps.051718605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida Y., Shima F., Sen H., Tanaka Y., Yanagihara C., Yamawaki–Kataoka Y., Kariya K., Kataoka T. (1998). Coiled-coil interaction of N-terminal 36 residues of cyclase-associated protein with adenylyl cyclase is sufficient for its function in Saccharomyces cerevisiae ras pathway. J. Biol. Chem. 273, 28019–28024 10.1074/jbc.273.43.28019 [DOI] [PubMed] [Google Scholar]
- Ono S. (2001). The Caenorhabditis elegans unc-78 gene encodes a homologue of actin-interacting protein 1 required for organized assembly of muscle actin filaments. J. Cell Biol. 152, 1313–1319 10.1083/jcb.152.6.1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono S. (2007). Mechanism of depolymerization and severing of actin filaments and its significance in cytoskeletal dynamics. Int. Rev. Cytol. 258, 1–82 10.1016/S0074-7696(07)58001-0 [DOI] [PubMed] [Google Scholar]
- Ono S. (2010). Dynamic regulation of sarcomeric actin filaments in striated muscle. Cytoskeleton (Hoboken) 67, 677–692 10.1002/cm.20476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono S., Benian G. M. (1998). Two Caenorhabditis elegans actin depolymerizing factor/cofilin proteins, encoded by the unc-60 gene, differentially regulate actin filament dynamics. J. Biol. Chem. 273, 3778–3783 10.1074/jbc.273.6.3778 [DOI] [PubMed] [Google Scholar]
- Ono S., Ono K. (2002). Tropomyosin inhibits ADF/cofilin-dependent actin filament dynamics. J. Cell Biol. 156, 1065–1076 10.1083/jcb.200110013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono S., Minami N., Abe H., Obinata T. (1994). Characterization of a novel cofilin isoform that is predominantly expressed in mammalian skeletal muscle. J. Biol. Chem. 269, 15280–15286 [PubMed] [Google Scholar]
- Ono S., Baillie D. L., Benian G. M. (1999). UNC-60B, an ADF/cofilin family protein, is required for proper assembly of actin into myofibrils in Caenorhabditis elegans body wall muscle. J. Cell Biol. 145, 491–502 10.1083/jcb.145.3.491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono S., McGough A., Pope B. J., Tolbert V. T., Bui A., Pohl J., Benian G. M., Gernert K. M., Weeds A. G. (2001). The C-terminal tail of UNC-60B (actin depolymerizing factor/cofilin) is critical for maintaining its stable association with F-actin and is implicated in the second actin-binding site. J. Biol. Chem. 276, 5952–5958 10.1074/jbc.M007563200 [DOI] [PubMed] [Google Scholar]
- Ono K., Parast M., Alberico C., Benian G. M., Ono S. (2003). Specific requirement for two ADF/cofilin isoforms in distinct actin-dependent processes in Caenorhabditis elegans. J. Cell Sci. 116, 2073–2085 10.1242/jcs.00421 [DOI] [PubMed] [Google Scholar]
- Ono S., Mohri K., Ono K. (2004). Microscopic evidence that actin-interacting protein 1 actively disassembles actin-depolymerizing factor/Cofilin-bound actin filaments. J. Biol. Chem. 279, 14207–14212 10.1074/jbc.M313418200 [DOI] [PubMed] [Google Scholar]
- Ono K., Yamashiro S., Ono S. (2008). Essential role of ADF/cofilin for assembly of contractile actin networks in the C. elegans somatic gonad. J. Cell Sci. 121, 2662–2670 10.1242/jcs.034215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono S., Nomura K., Hitosugi S., Tu D. K., Lee J. A., Baillie D. L., Ono K. (2011). The two actin-interacting protein 1 genes have overlapping and essential function for embryonic development in Caenorhabditis elegans. Mol. Biol. Cell 22, 2258–2269 10.1091/mbc.E10-12-0934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pappas C. T., Krieg P. A., Gregorio C. C. (2010). Nebulin regulates actin filament lengths by a stabilization mechanism. J. Cell Biol. 189, 859–870 10.1083/jcb.201001043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardee J. D., Spudich J. A. (1982). Purification of muscle actin. Methods Enzymol. 85 Pt B, 164–181 10.1016/0076-6879(82)85020-9 [DOI] [PubMed] [Google Scholar]
- Pavlov D., Muhlrad A., Cooper J., Wear M., Reisler E. (2007). Actin filament severing by cofilin. J. Mol. Biol. 365, 1350–1358 10.1016/j.jmb.2006.10.102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peche V., Shekar S., Leichter M., Korte H., Schröder R., Schleicher M., Holak T. A., Clemen C. S., Ramanath–Y B., Pfitzer G.et al. (2007). CAP2, cyclase-associated protein 2, is a dual compartment protein. Cell. Mol. Life Sci. 64, 2702–2715 10.1007/s00018-007-7316-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polet D., Lambrechts A., Ono K., Mah A., Peelman F., Vandekerckhove J., Baillie D. L., Ampe C., Ono S. (2006). Caenorhabditis elegans expresses three functional profilins in a tissue-specific manner. Cell Motil. Cytoskeleton 63, 14–28 10.1002/cm.20102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintero–Monzon O., Jonasson E. M., Bertling E., Talarico L., Chaudhry F., Sihvo M., Lappalainen P., Goode B. L. (2009). Reconstitution and dissection of the 600-kDa Srv2/CAP complex: roles for oligomerization and cofilin-actin binding in driving actin turnover. J. Biol. Chem. 284, 10923–10934 10.1074/jbc.M808760200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers S. L., Wiedemann U., Stuurman N., Vale R. D. (2003). Molecular requirements for actin-based lamella formation in Drosophila S2 cells. J. Cell Biol. 162, 1079–1088 10.1083/jcb.200303023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safer D. (1989). An electrophoretic procedure for detecting proteins that bind actin monomers. Anal. Biochem. 178, 32–37 10.1016/0003-2697(89)90351-5 [DOI] [PubMed] [Google Scholar]
- Skwarek–Maruszewska A., Hotulainen P., Mattila P. K., Lappalainen P. (2009). Contractility-dependent actin dynamics in cardiomyocyte sarcomeres. J. Cell Sci. 122, 2119–2126 10.1242/jcs.046805 [DOI] [PubMed] [Google Scholar]
- Swiston J., Hubberstey A., Yu G., Young D. (1995). Differential expression of CAP and CAP2 in adult rat tissues. Gene 165, 273–277 10.1016/0378-1119(95)00522-8 [DOI] [PubMed] [Google Scholar]
- Thirion C., Stucka R., Mendel B., Gruhler A., Jaksch M., Nowak K. J., Binz N., Laing N. G., Lochmüller H. (2001). Characterization of human muscle type cofilin (CFL2) in normal and regenerating muscle. Eur. J. Biochem. 268, 3473–3482 10.1046/j.1432-1327.2001.02247.x [DOI] [PubMed] [Google Scholar]
- Trigg J., Gutwin K., Keating A. E., Berger B. (2011). Multicoil2: predicting coiled coils and their oligomerization states from sequence in the twilight zone. PLoS ONE 6, e23519 10.1371/journal.pone.0023519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Troys M., Huyck L., Leyman S., Dhaese S., Vandekerkhove J., Ampe C. (2008). Ins and outs of ADF/cofilin activity and regulation. Eur. J. Cell Biol. 87, 649–667 10.1016/j.ejcb.2008.04.001 [DOI] [PubMed] [Google Scholar]
- Vartiainen M. K., Mustonen T., Mattila P. K., Ojala P. J., Thesleff I., Partanen J., Lappalainen P. (2002). The three mouse actin-depolymerizing factor/cofilins evolved to fulfill cell-type-specific requirements for actin dynamics. Mol. Biol. Cell 13, 183–194 10.1091/mbc.01-07-0331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddle J. A., Cooper J. A., Waterston R. H. (1993). The alpha and beta subunits of nematode actin capping protein function in yeast. Mol. Biol. Cell 4, 907–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. L., Taylor D. L. (1981). Exchange of 1,N6-etheno-ATP with actin-bound nucleotides as a tool for studying the steady-state exchange of subunits in F-actin solutions. Proc. Natl. Acad. Sci. USA 78, 5503–5507 10.1073/pnas.78.9.5503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolanski M., Khosrowshahian F., Jerant L., Jap I. S., Brockman J., Crawford M. J. (2009). Expression of CAP2 during early Xenopus embryogenesis. Int. J. Dev. Biol. 53, 1063–1067 10.1387/ijdb.062158mw [DOI] [PubMed] [Google Scholar]
- Yamashiro S., Mohri K., Ono S. (2005). The two Caenorhabditis elegans actin-depolymerizing factor/cofilin proteins differently enhance actin filament severing and depolymerization. Biochemistry 44, 14238–14247 10.1021/bi050933d [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashiro S., Gimona M., Ono S. (2007). UNC-87, a calponin-related protein in C. elegans, antagonizes ADF/cofilin-mediated actin filament dynamics. J. Cell Sci. 120, 3022–3033 10.1242/jcs.013516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashiro S., Cox E. A., Baillie D. L., Hardin J. D., Ono S. (2008). Sarcomeric actin organization is synergistically promoted by tropomodulin, ADF/cofilin, AIP1 and profilin in C. elegans. J. Cell Sci. 121, 3867–3877 10.1242/jcs.040477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G., Swiston J., Young D. (1994). Comparison of human CAP and CAP2, homologs of the yeast adenylyl cyclase-associated proteins. J. Cell Sci. 107, 1671–1678 [DOI] [PubMed] [Google Scholar]
- Yu R., Ono S. (2006). Dual roles of tropomyosin as an F-actin stabilizer and a regulator of muscle contraction in Caenorhabditis elegans body wall muscle. Cell Motil. Cytoskeleton 63, 659–672 10.1002/cm.20152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusof A. M., Hu N. J., Wlodawer A., Hofmann A. (2005). Structural evidence for variable oligomerization of the N-terminal domain of cyclase-associated protein (CAP). Proteins 58, 255–262 10.1002/prot.20314 [DOI] [PubMed] [Google Scholar]
- Zengel J. M., Epstein H. F. (1980). Identification of genetic elements associated with muscle structure in the nematode Caenorhabditis elegans. Cell Motil. 1, 73–97 10.1002/cm.970010107 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.