Summary
APPL1 is a multifunctional adaptor protein that binds membrane receptors, signaling proteins and nuclear factors, thereby acting in endosomal trafficking and in different signaling pathways. Here, we uncover a novel role of APPL1 as a positive regulator of transcriptional activity of NF-κB under basal but not TNFα-stimulated conditions. APPL1 was found to directly interact with TRAF2, an adaptor protein known to activate canonical NF-κB signaling. APPL1 synergized with TRAF2 to induce NF-κB activation, and both proteins were necessary for this process and function upstream of the IKK complex. Although TRAF2 was not detectable on APPL endosomes, endosomal recruitment of APPL1 was required for its function in the NF-κB pathway. Importantly, in the canonical pathway, APPL1 appeared to regulate the proper spatial distribution of the p65 subunit of NF-κB in the absence of cytokine stimulation, since its overexpression enhanced and its depletion reduced the nuclear accumulation of p65. By analyzing the patterns of gene transcription upon APPL1 overproduction or depletion we found altered expression of NF-κB target genes that encode cytokines. At the molecular level, overexpressed APPL1 markedly increased the level of NIK, the key component of the noncanonical NF-κB pathway, by reducing its association with the degradative complex containing TRAF2, TRAF3 and cIAP1. In turn, high levels of NIK triggered nuclear translocation of p65. Collectively, we propose that APPL1 regulates basal NF-κB activity by modulating the stability of NIK, which affects the activation of p65. This places APPL1 as a novel link between the canonical and noncanonical machineries of NF-κB activation.
Key words: APPL1, NF-κB pathway, NIK, TRAF2
Introduction
Nuclear factor κB (NF-κB) is a key transcription factor involved in the immune responses, cell survival and proliferation (Hayden and Ghosh, 2004; Hayden and Ghosh, 2008). The mammalian NF-κB family includes five members: RelA/p65, c-Rel, RelB, NF-κB1 (p105/p50) and NF-κB2 (p100/p52) that form homo- or heterodimers to transactivate gene expression (Ghosh and Karin, 2002; Vallabhapurapu and Karin, 2009). The primary regulation of the NF-κB pathway operates through the sequestration of NF-κB dimers in the cytoplasm by IκB inhibitory proteins (Scheidereit, 2006; Vallabhapurapu and Karin, 2009). NF-κB dimers can be activated via canonical or noncanonical signaling cascades, which operate with different kinetics and perform nonredundant physiological functions (Shih et al., 2011).
The activation of the canonical NF-κB pathway is generally rapid and occurs within minutes in response to proinflammatory stimuli, such as tumor necrosis factor α (TNFα) or interleukin 1β (IL-1β). Cytokine stimulation leads to the recruitment of adaptors (such as TRAFs) to the cytoplasmic domain of the receptor. In turn, these adaptors recruit the inhibitor of kappa-B kinase (IKK) complex, which leads to phosphorylation and degradation of the IκBα inhibitor (Naudé et al., 2011; Wajant and Scheurich, 2011). This allows p65 dimers to translocate to the nucleus and activate the transcription of target genes (Vallabhapurapu and Karin, 2009).
In contrast to the rapid canonical NF-κB signaling, the noncanonical pathway operates in the range of hours upon engagement of the selected group of receptors, such as B cell-activating factor receptor (BAFF-R), lymphotoxin β receptor (LTβR), and receptor activator of NF-κB (RANK) (Sun and Ley, 2008). This induces stabilization and accumulation of the NF-κB-inducing kinase (NIK) and subsequent IKK1-dependent p100 phosphorylation, what results in p100 processing to its active derivative p52 and translocation of RelB–p52 dimers to the nucleus (Hoffmann and Baltimore, 2006; Sun, 2011). The central event that leads to the noncanonical NF-κB signaling is the stabilization of NIK. Under resting conditions the stability of NIK protein is negatively regulated by TRAF3, TRAF2 and cellular inhibitor of apoptosis 1 (cIAP1) and cIAP2, which form a degradative complex controlling polyubiquitination and proteasomal targeting of NIK (Zarnegar et al., 2008b). Although the canonical and noncanonical NF-κB pathways were first thought to transduce signals independently (Pomerantz and Baltimore, 2002), several studies provided evidence for interconnections between them. It was noted that p65-containing dimers could be activated through a NIK–IKK2 axis when NIK protein accumulates in cells (Malinin et al., 1997; Zarnegar et al., 2008a).
A growing number of studies show that many proteins known to regulate endocytosis participate also in nuclear signaling and transcriptional regulation, mostly by modulating the activity of various nuclear factors (Borlido et al., 2009; Pyrzynska et al., 2009). Among such multifunctional endocytic proteins is an adaptor protein containing pleckstrin homology domain, phosphotyrosine binding domain, and leucine zipper motif 1 (APPL1). Through its localization to a subpopulation of early endosomes, termed APPL endosomes, APPL1 acts as a dynamic scaffold for endosomal signaling proteins (Miaczynska et al., 2004; Zoncu et al., 2009). APPL1 was reported as an interacting partner for the serine/threonine kinases Akt and LKB1, phosphatidylinositol 3-kinase (PI3K) and inositol 5-phosphatase OCRL (Erdmann et al., 2007; Mitsuuchi et al., 1999; Zhou et al., 2009). In addition, APPL1 associates with a number of transmembrane receptors (including nerve growth factor receptor TrkA, netrin 1 receptor DCC and receptors for adiponectin and follicle stimulating hormone) (Deepa and Dong, 2009). Apart from its role in endocytosis and endosomal transport, APPL1 was reported to undergo nucleocytoplasmic shuttling and participate in transcriptional regulation, e.g. by interactions with histone deacetylases (HDACs) (Banach-Orlowska et al., 2009; Miaczynska et al., 2004). We recently showed that APPL1 modulates the activity of the β-catenin–Reptin–HDAC complex, and is able to increase expression of β-catenin/TCF target genes in the canonical Wnt signaling (Rashid et al., 2009).
Here, we report that APPL1 functions as a novel positive regulator of the NF-κB signaling that operates under basal conditions without cytokine stimulation. We found that overexpression of APPL1 stabilizes NIK, which in turn triggers p65 translocation to the nucleus and induction of selected NF-κB target genes. This indicates that APPL1 may serve as a link between the canonical and noncanonical machineries of NF-κB activation.
Results
APPL1 directly interacts with TRAF2
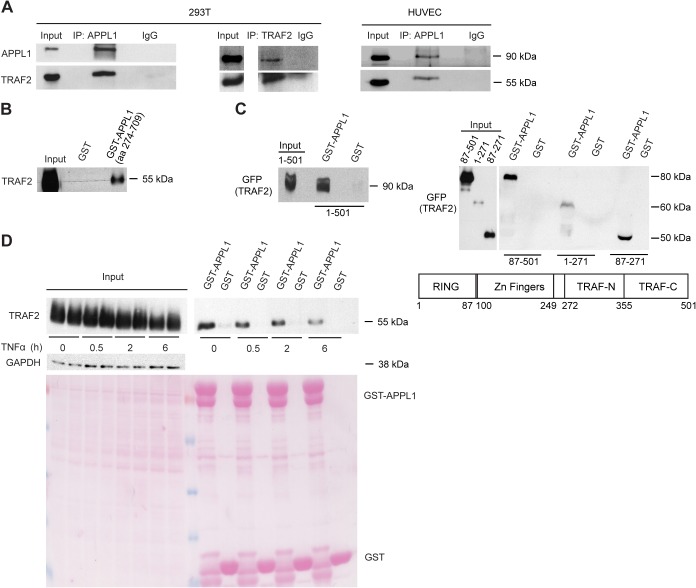
The TRAF2 protein was previously shown to interact with APPL1 in a yeast two-hybrid system (Rual et al., 2005). To test whether these proteins might associate in mammalian cells we performed reciprocal co-immunoprecipitations from lysates of HEK293T cells. We detected the presence of TRAF2 in immunoprecipitates of APPL1 and vice versa, demonstrating an association between the two endogenous proteins, as confirmed also in primary endothelial cells (Fig. 1A). We further verified that APPL1–TRAF2 binding is direct, as the C-terminal fragment of APPL1 fused to GST (a.a. 274–709; containing the PH and PTB domains) purified from bacteria was able to pull down in vitro translated TRAF2 (Fig. 1B). Next, we identified the fragment of APPL1 responsible for TRAF2 binding as the C-terminal PTB domain (a.a. 429–709) which was able to associate with the full-length TRAF2 in GST pull-down experiments (Fig. 1C). By performing converse experiments with the fragments of TRAF2 we determined that its region containing zinc finger motifs (a.a. 87–271) is responsible for the association with APPL1 (Fig. 1C).
Fig. 1.
APPL1 interacts with TRAF2. (A) Endogenous APPL1 and TRAF2 interact in HEK293T and HUVEC cells. Cell lysates from untransfected HEK293T and HUVEC cells were subjected to coimmunoprecipitation with either rabbit anti-APPL1 antiserum or anti-TRAF2 mouse monoclonal antibody. Rabbit and mouse IgG were used as negative controls, respectively. Input represents 10% of total cell lysate used for immunoprecipitation. (B) APPL1 and TRAF2 interact directly. An in vitro translated TRAF2 was incubated with GST alone or APPL1 (a.a. 274–709) fused with GST. The protein complexes were analyzed by western blotting with anti-TRAF2 monoclonal antibody. Input shows 5% of total in vitro translated TRAF2. (C) The zinc finger region of TRAF2 binds to the PTB domain of APPL1. HEK293T cells were transfected with GFP-tagged TRAF2 (full length a.a. 1–501, left panel; or a.a. 87–501, 1–271, 87–271, right panel) 24 h prior to lysis. Cell lysates were incubated with GST alone or GST–APPL1 (a.a. 429–709). GFP–TRAF2 bound to APPL1 was detected with anti-GFP antibody. Input shows 10% of total cell lysate. (D) APPL1–TRAF2 interaction is regulated by TNFα. HEK293T cells were transfected with TRAF2 expression vector and, after 24 h, were stimulated with TNFα for 0.5, 2, 6 h or left untreated. Cell lysates were subjected to pull-down experiments with GST alone or GST–APPL1 (a.a. 274–709) and processed as in B (upper panel). Input shows 10% of total cell lysate. GAPDH level and Ponceau S staining (lower panel) serve as loading controls.
TRAF2 is one of the key factors mediating TNFα signaling (Chung et al., 2007; Wajant and Scheurich, 2011). To determine whether the APPL1–TRAF2 interaction depends on TNFα stimulation, we treated TRAF2-transfected HEK293T cells with TNFα for 0.5, 2 and 6 h. In GST pull-down assays we detected that the interaction between APPL1 and TRAF2 was the strongest in unstimulated cells and was progressively reduced with prolonged times of TNFα treatment (Fig. 1D). This negative regulation by TNFα provided the first indication that APPL1–TRAF2 interaction may not be important for TNFα-stimulated NF-κB signaling.
APPL1 positively regulates the NF-κB pathway and enhances the activation of NF-κB induced by TRAF2
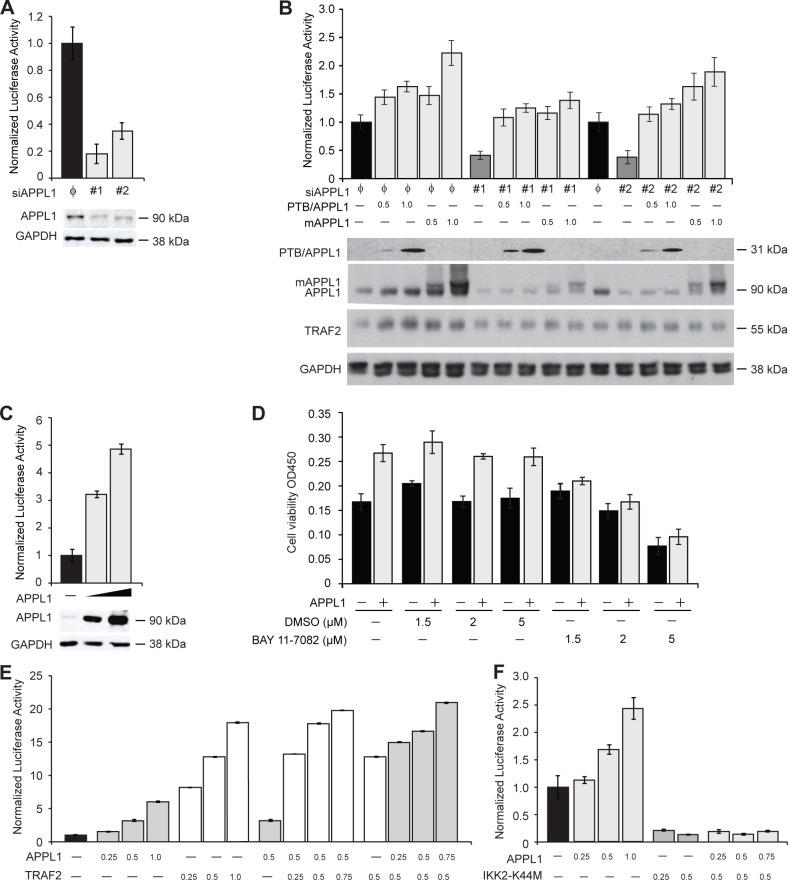
Considering a well-established role of TRAF2 in the NF-κB pathway, we hypothesized that APPL1 may regulate the NF-κB activity via its interaction with TRAF2. First we examined the effect of APPL1 on the transcriptional activation of NF-κB in serum-grown HEK293T cells, using a luciferase reporter gene under control of the human κB promoter. We found that depletion of APPL1 with two independent siRNAs significantly reduced the basal transcriptional activity of NF-κB (Fig. 2A). This effect was rescued by overexpression of siRNA-resistant mouse APPL1, as well as by the PTB domain of APPL1 (Fig. 2B). These data confirm that the PTB domain, responsible for TRAF2 binding, is crucial for mediating the effects of APPL1 in the NF-κB pathway. In parallel, we found that APPL1 expression potently activated the reporter gene in a dose-dependent manner, with the maximal 5-fold induction over the empty vector control (Fig. 2C). The effects of APPL1 depletion and overexpression consistently demonstrated that APPL1 can positively regulate the NF-κB signaling pathway. In agreement with a previous observation that silencing of APPL1 reduces cell proliferation (Miaczynska et al., 2004), we confirmed that overexpression of APPL1 potentiated cell viability (Fig. 2D). Importantly, this APPL1-driven increase was specifically abolished by the use of IKK inhibitor BAY 11-7082 at low concentrations (1.5–2 µM) which do not yet affect the overall viability of cells (Fig. 2D). This argues that the APPL1 function in the regulation of cell viability is NF-κB-dependent.
Fig. 2.
APPL1 enhances activity of the NF-κB pathway and acts synergistically with TRAF2. (A) APPL1 knockdown inhibits the NF-κB pathway. HEK293T cells were cotransfected with reporter vectors and APPL1 siRNA #1 and #2 or non-targeting siRNA (φ). The luciferase reporter activity was measured as described in the Materials and Methods. Knockdown of the APPL1 level was confirmed by immunoblotting (lower panel) with GAPDH as a loading control. (B) Overexpression of APPL1 or its PTB domain overcomes the inhibitory effect of its depletion on the NF-κB reporter. HEK293T cells were cotransfected with reporter vectors together with APPL1 siRNA #1 and #2 or non-targeting siRNA (φ) alone or in combination with vectors encoding the HA-tagged PTB of human APPL1 (PTB/APPL1) or full-length mouse HA–APPL1 (mAPPL1), as indicated (all values are in µg of DNA). The luciferase assay was performed as in A (upper panel). The levels of PTB/APPL1, mAPPL1, TRAF2 and GAPDH were analyzed by western blotting (bottom panel). mAPPL1 and PTB/APPL1 were detected with anti-APPL1 and anti-HA antibodies, respectively. (C) Dose–response effect of APPL1 overexpression on the NF-κB reporter activation. HEK293T cells were cotransfected with reporter vectors as well as 0.5 or 1 µg of APPL1 expression vector. APPL1 and GAPDH (loading control) levels were analyzed by western blotting (bottom panel). (D) APPL1-induced increase in cell viability is dependent on NF-κB. HEK293T cells were transfected with APPL1 expression vector and treated with different concentrations of BAY 11-7082 or DMSO, as a control, for 48 h. Cell viability was measured as described in the Materials and Methods, and expressed as OD450. (E) APPL1 and TRAF2 act synergistically to induce the NF-κB reporter. HEK293T cells were cotransfected with reporter vectors, together with the different combinations of APPL1 and TRAF2 expression vectors, as indicated (all values are in µg of DNA). (F) APPL1 acts upstream of IKK2. HEK293T cells were cotransfected with reporter vectors and different combinations of APPL1 and IKK2-K44M expression vectors, as indicated (all values are in µg of DNA).
We further tested whether APPL1 and TRAF2 synergize in the activation of NF-κB. TRAF2 overexpression resulted in up to 20-fold reporter gene induction, as previously reported (Takeuchi et al., 1996). Coexpression of both APPL1 and TRAF2 potentiated NF-κB activation, with the maximum observed for high amounts of APPL1 expression vector, indicating that APPL1 and TRAF2 activate NF-κB synergistically (Fig. 2E). We also analyzed the TRAF2 mutants deficient in the E3 ligase activity for their ability to synergize with APPL1 in NF-κB activation. Three previously characterized TRAF2 mutants were used: deletion of the N-terminal RING domain (deltaRING, dR) and two quaternary point mutants (TRAF2-Rm mutated on the RING domain at C49A, H51A, C54A and C57A; TRAF2-RmZm mutated at C49A, H51A within the RING and at C209A and C212A within the fourth zinc finger domains) (Habelhah et al., 2004). The ability of all three mutants to stimulate the NF-κB reporter was significantly reduced in comparison to the wild-type TRAF2 and their ability to synergize with APPL1 was proportionally diminished (supplementary material Fig. S1A). However, none of the mutants had any dominant-negative inhibitory effects on the NF-κB activation induced by APPL1 overexpression.
To determine if APPL1 is required for TRAF2 to activate the NF-κB pathway, we measured the reporter activity in cells overexpressing TRAF2 but depleted for APPL1. Knockdown of APPL1 inhibited both the basal and TRAF2-stimulated activation of NF-κB (supplementary material Fig. S1B). In a converse experiment, APPL1 overexpression could not overcome the reporter inhibition caused by knockdown of TRAF2, suggesting that APPL1-induced NF-κB activation is mediated via TRAF2 (supplementary material Fig. S1C). These data indicate that both APPL1 and TRAF2 are necessary for the full activation of NF-κB.
To further analyze at which step of the NF-κB signaling pathway APPL1 acts we used the kinase-deficient mutant of IKK2 (IKK2-K44M), previously reported to inhibit NF-κB activation (Mercurio et al., 1997). The stimulatory effect of APPL1 on the NF-κB reporter was abolished upon coexpression of the kinase-defective mutant of IKK2 (Fig. 2F), arguing that APPL1 acts upstream of the IKK complex, similarly to TRAF2 (Wajant and Scheurich, 2011). Consistently, APPL1 depletion had no effect on the NF-κB activation induced by overexpression of an NF-κB component p65 (supplementary material Fig. S1D). Cumulatively, the reporter assay data indicate that APPL1 positively regulates the NF-κB pathway, is required for TRAF2-mediated NF-κB activation and functions upstream of the IKK complex.
Endosomal recruitment of APPL1 is necessary for its function in the NF-κB pathway
APPL1 is recruited to a subpopulation of early endosomes, termed APPL endosomes, due to its interactions with Rab5 (Miaczynska et al., 2004) and Rab21 (Zhu et al., 2007). We tested whether APPL1 point mutants, unable to associate with Rab5 or Rab21 and thus deficient in binding to endosomal membranes, can stimulate the NF-κB activity as the wild-type protein does. We used three previously described mutants, unable to bind Rab5 (APPL1-K280E/Y283C/G319R and APPL1-A318D) or Rab21 (APPL1-G320A) (Miaczynska et al., 2004; Zhu et al., 2007). When tested in the reporter assay, none of these mutants could stimulate the NF-κB activity. Instead, they reduced the basal NF-κB activity, possibly due to unproductive sequestration of some signaling components (Fig. 3A).
Fig. 3.
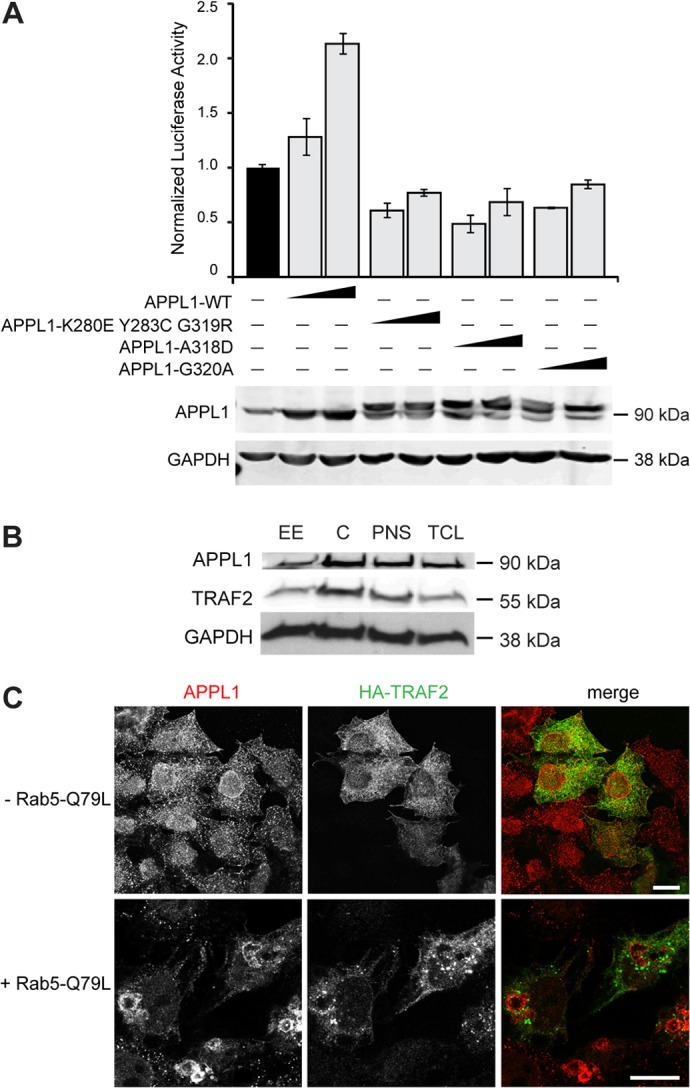
Endosomal recruitment of APPL1 is necessary for its function in the NF-κB pathway. (A) APPL1 endosomal localization is necessary for NF-κB activation. HEK293T cells were cotransfected with reporter vectors together with 0.5 and 1 µg of untagged wild-type APPL1 or APPL1 mutants unable to bind Rab5 (myc-APPL1-K280E,Y283C,G319R and myc-APPL1-A318D) or Rab21 (myc-APPL1-G320A), and measurements of the luciferase activity were made (upper panel). The overexpression levels of APPL1 variants were tested by immunoblotting with anti-APPL1 antibody (lower panel). (B) TRAF2 is localized on endosomal membranes. Early endosomal (EE) and cytoplasmic (C) fractions, postnuclear supernatant (PNS) and total cell lysate (TCL) from HeLa cells were analyzed by western blotting with anti-APPL1, -TRAF2 and -GAPDH (loading control). (C) TRAF2 is not recruited to the enlarged APPL1-positive endosomes. Twenty-four hours after transfection with HA–TRAF2 alone (upper panel) or in combination with Rab5-Q79L (bottom panel), HeLa cells were fixed and stained with antibodies against APPL1 (red) and HA (green). Scale bars: 20 µm.
Having determined that Rab binding and endosomal localization of APPL1 is important for its function in the NF-κB pathway, we tested whether TRAF2 might be present with APPL1 on endosomal membranes. A small pool of TRAF2 could be detected by western blotting of cellular fractions enriched in early endosomes (Fig. 3B). However, microscopy analyses failed to detect an unequivocal localization of endogenous or overexpressed TRAF2 to APPL endosomes in untreated or TNFα-treated HEK293T or HeLa cells (Fig. 3C; supplementary material Fig. S2; data not shown). Colocalization of APPL1 and TRAF2 was not observed even in the presence of overexpressed Rab5-Q79L mutant, which enhances membrane fusion and causes the formation of enlarged endosomes (Stenmark et al., 1994) containing Rab5 effectors such as APPL1 (Miaczynska et al., 2004). Under these conditions, APPL1-positive large vacuoles were negative for TRAF2, independently of TNFα treatment (Fig. 3C; supplementary material Fig. S2), although overexpressed TRAF2 seemed to be concentrated in punctate structures in the vicinity of the vacuoles (Fig. 3C, lower panel). Cumulatively, these data indicate that TRAF2 is not a stable component of APPL endosomes, however it is still possible that APPL1 and TRAF2 interact on endosomal membranes in a transient, dynamic manner.
APPL1 triggers p65 translocation to the nucleus
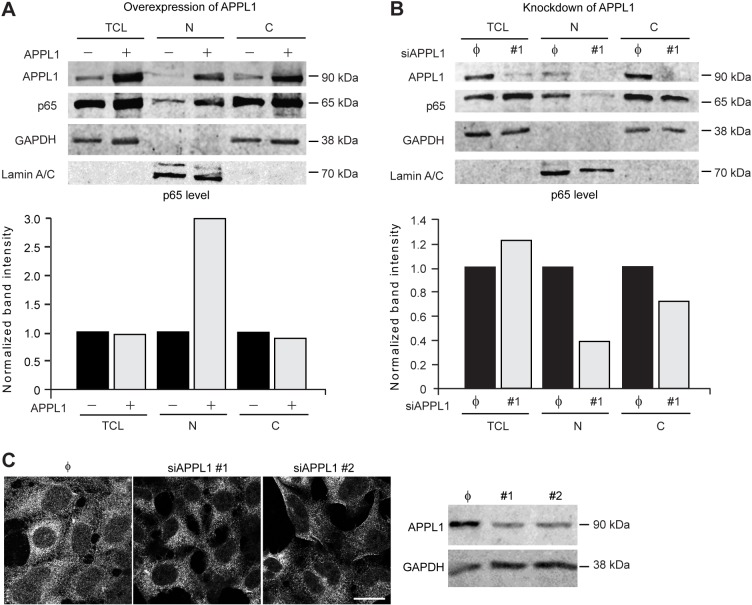
One of the final events in the canonical activation of NF-κB pathway is the translocation of p65 to the nucleus. To further confirm the role of APPL1 in the regulation of NF-κB activity we overexpressed APPL1 and monitored the nuclear localization of p65 by cell fractionation and by immunofluorescence microscopy. Fractionation experiments from HEK293T cells revealed that APPL1 overexpression resulted in a strong increase of p65 level in the nucleus (Fig. 4A). These results are in full agreement with the stimulatory effects of APPL1 overexpression on the NF-κB luciferase reporter (Fig. 2; supplementary material Fig. S1). Conversely, upon APPL1 knockdown the nuclear fraction contained less p65 than in control cells (Fig. 4B), again confirming the results of the reporter assays. In agreement with the biochemical data, immunofluorescence microscopy revealed that p65 was predominantly sequestered in the cytoplasm in control HEK293T cells. The nuclear pool of p65 was further reduced upon depletion of APPL1 (Fig. 4C) and increased upon its overexpression, although the level of p65 nuclear staining did not directly correlate with the level of APPL1 overexpression (supplementary material Fig. S3). Taken together, these data indicate that APPL1 activates the NF-κB pathway by increasing p65 translocation to the nucleus.
Fig. 4.
APPL1 induces nuclear accumulation of p65. (A) APPL1 overexpression induces translocation of p65 to the nucleus. HEK293T cells were transfected with APPL1-expressing vector or empty vector as a control. Forty-eight hours after transfection total cell lysate (TCL), cytoplasmic (C) and nuclear (N) fractions were analyzed by western blotting with anti-APPL1, -p65, -GAPDH and -lamin-A/C antibodies (upper panel), and the intensity of the p65 band was quantified (lower panel). A representative result from four independent experiments is shown. (B) APPL1 knockdown prevents nuclear accumulation of p65. HEK293T cells were transfected with APPL1 siRNA #1 or non-targeting siRNA (φ). Forty-eight hours after transfection the cell fractions were prepared and analyzed as in A. (C) Immunofluorescence microscopy confirms inhibition of translocation of p65 to the nucleus upon APPL1 knockdown. Forty-eight hours after transfection with non-targeting siRNA (φ) or APPL1 siRNAs (#1 or #2), HEK293T cells were fixed and stained with anti-p65 antibody (left panel). Scale bar: 20 µm. Knockdown of APPL1 level was confirmed by immunoblotting (right panel) with GAPDH level as a loading control.
We further validated these finding in cancer cell lines H1299 (small lung cancer), SKBR3 (breast cancer) and DLD1 (colon cancer). Levels of APPL1 and IκBα in these cell lines differ by up to two-fold, although they do not correlate with each other (supplementary material Fig. S4A). Depletion of APPL1 in these cells inhibited the activity of the NF-κB reporter, concomitant with the reduced nuclear accumulation of p65 (supplementary material Fig. S4B,C). However, overexpression of APPL1 enhanced only weekly the reporter activity and the nuclear accumulation of p65 in H1299 and SKBR3 cells (supplementary material Fig. S4D,E). These data argue that although in these cell lines APPL1 is necessary for the basal NF-κB activity, its increased levels are not sufficient to potentiate the pathway activity further. Nevertheless, taken together these findings confirm a contribution of APPL1 to the control of basal NF-κB activation in different cell types.
In contrast, we could not detect any role for APPL1 in the regulation of TNFα-stimulated NF-κB activity. We did not observe any changes in the distribution of p65 upon overexpression or depletion of APPL1 in TNFα-stimulated cells (supplementary material Fig. S5A,B). Similarly, IκBα degradation upon TNFα stimulation was not changed upon depletion of APPL1 (supplementary material Fig. S5C). Collectively, these results demonstrate that APPL1 does not play a role in the regulation of TNFα signaling and operates in the NF-κB pathway mainly under basal conditions.
APPL1 levels modulate the expression of NF-κB target genes
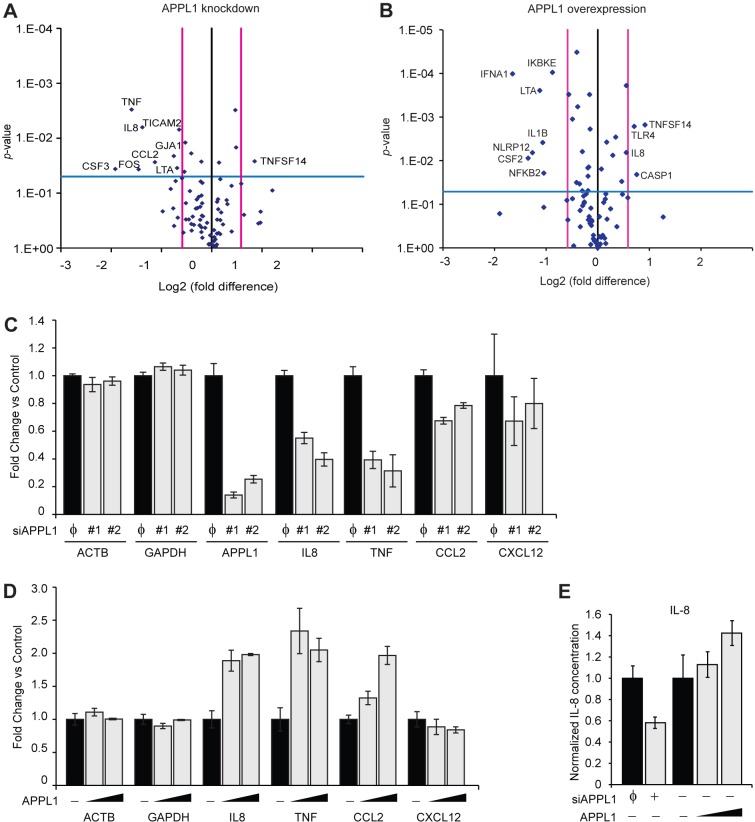
To determine which NF-κB target genes may be regulated by APPL1 we performed RT-PCR analysis and measured the expression patterns of 84 NF-κB-responsive genes in HEK293T cells depleted of APPL1 or overexpressing it (Fig. 5A,B). In parallel, we profiled expression of the same genes in cells treated with TNFα (supplementary material Fig. S5D). In general, the manipulation of APPL1 levels and the stimulation with TNFα resulted in the induction of different sets of genes, additionally confirming that APPL1-mediated and TNFα-induced NF-κB activation represent two distinct mechanisms.
Fig. 5.
APPL1 regulates expression of NF-κB target genes. Analysis of gene expression upon APPL1 knockdown (A) or overexpression (B) in HEK293T cells. A predesigned 96-well array containing primer pairs for detection of 84 genes involved in NF-κB signaling was used. The vertical lines indicate the 1.5-fold change in gene expression threshold. The horizontal line indicates the P = 0.05 of the t-test threshold. Only names of the genes significantly down- or upregulated are mentioned on the charts. Expression of the genes encoding IL-8, TNF, CCL2 and CXCL12 upon APPL1 knockdown (C) or overexpression (D) was validated by qRT-PCR in HEK293T cells 2 days after transfection with non-targeting siRNA (φ) or APPL1 siRNAs (#1 or #2) (C) or APPL1-expressing vector (D). (E) APPL1 level regulates the amounts of secreted IL-8. ELISA analyses of IL-8 production by HEK293T cells 48 h after transfection with non-targeting siRNA (φ), APPL1 siRNA #1 (+) or APPL1-expressing vector.
APPL1 depletion or overproduction affected expression of several NF-κB-responsive genes, mainly those encoding extracellular ligands and receptors involved in inflammatory response (Fig. 5A,B). We specifically followed and further validated the changes in expression of three genes encoding cytokines: interleukin-8 (IL-8), TNFα and CCL2. Their expression was reduced upon knockdown of APPL1 with two independent siRNA oligonucleotides, with the strongest effects observed for IL-8 and TNF (Fig. 5C). Consistently, overexpression of APPL1 exerted an opposite effect, inducing expression of the three genes (Fig. 5D). As the investigated cytokines are targets of the canonical NF-κB pathway, we included in our analysis a gene encoding CXCL12 cytokine which is primarily regulated by the noncanonical NF-κB pathway (Madge et al., 2008). While we observed a weak reduction in the CXCL12 expression upon depletion of APPL1, no changes were detected upon APPL1 overexpression (Fig. 5C,D), arguing that CXCL12 is not a direct target of APPL1 action. We further verified that the changes observed at the level of gene expression were reflected by altered protein levels. In agreement with the RT-PCR data, ELISA-based analysis demonstrated that the amount of secreted IL-8 is reduced upon depletion of APPL1 and increased upon APPL1 overexpression (Fig. 5E). Collectively, these results indicate that the levels of APPL1 protein can modulate the expression of NF-κB-dependent genes encoding cytokines.
APPL1 stabilizes NIK
In search for potential mechanisms by which APPL1 could regulate the nuclear accumulation of p65 and expression of the canonical NF-κB target genes independently of the classical TNFα-stimulated machinery, we turned our attention to the noncanonical NF-κB pathway. It was previously reported that accumulation of NIK, a key noncanonical kinase, can also stimulate the canonical NF-κB signaling (Malinin et al., 1997; Zarnegar et al., 2008a). Importantly, in addition to its activating role in the canonical NF-κB signaling, TRAF2 functions as an inhibitor of the noncanonical pathway by promoting degradation of NIK (Grech et al., 2004).
In the alternative pathway, following activation of NIK, p100 is proteolytically processed to its transcriptionally active form p52 (Hayden and Ghosh, 2004; Hayden and Ghosh, 2008). We first confirmed that endogenous p100 is detectable in HEK293T cells, arguing that the noncanonical NF-κB pathway can operate in this experimental system. Strikingly, we observed that overexpression of APPL1 resulted in an increase in p100 processing to p52, compared with control cells (Fig. 6A). These findings indicated that APPL1 may act in the regulation of the noncanonical NF-κB pathway.
Fig. 6.
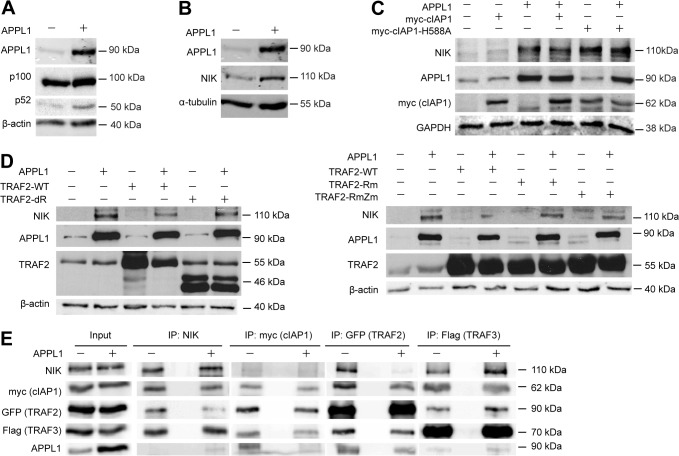
APPL1 stabilizes NIK. (A) APPL1 overexpression promotes p100 processing to p52. HEK293T cells were transfected with APPL1-expression vector or an empty vector for 48 h. Cell lysates were analyzed by western blotting for APPL1, p100, p52 and β-actin (loading control). (B) APPL1 overexpression regulates NIK protein level. The levels of APPL1, NIK and α-tubulin (loading control) were analyzed by western blotting upon APPL1 overexpression for 48 h. (C) APPL1 overexpression stabilizes NIK. HEK293T cells were transfected with APPL1, myc–cIAP1 and myc–cIAP1-H588A expression vectors either alone or in combination. The levels of APPL1, NIK, myc (cIAP1) and GAPDH (loading control) were analyzed 48 h after transfection. (D) E3 ligase-deficient TRAF2 mutants do not affect APPL1-induced NIK stability. HEK293T cells were transfected with APPL1, TRAF2, TRAF2-Rm, TRAF2-RmZm and TRAF2-dR expression vectors either alone or in combination. The levels of APPL1, NIK, TRAF2 and β-actin (loading control) were analyzed by western blotting 48 h after transfection. (E) APPL1 overexpression modulates the composition of the NIK degradative complex. Cell lysate from HEK293T cells transfected with NIK, myc–cIAP1, GFP–TRAF2, Flag–TRAF3 together with or without APPL1 were subjected to coimmunoprecipitation with rabbit anti-NIK, anti-myc, anti-GFP or mouse anti-Flag antibodies. Input represents 10% of total cell lysate used for immunoprecipitation.
The noncanonical NF-κB signaling depends on the levels of NIK (Senftleben et al., 2001; Sun, 2011; Thu and Richmond, 2010) which under resting conditions are very low, due to constant degradation by an ubiquitination-dependent mechanism involving cIAP E3 ligases complexed to TRAF2 and TRAF3 (Varfolomeev et al., 2007; Zarnegar et al., 2008b). We found that overexpression of APPL1 increased markedly the level of NIK compared with control cells (Fig. 6B), providing a possible explanation for the enhanced p100 processing. It was therefore plausible that APPL1 may also regulate the levels of cIAP1/2 or related E3 ligases, responsible for NIK degradation. However, we detected only a moderate increase of cIAP1 protein level upon knockdown of APPL1 but no reduction upon APPL1 overexpression (supplementary material Fig. S6). This argued against modulation of cIAP levels being the main mechanism for APPL1-induced NIK stabilization.
We further tested whether APPL1 could act through inhibition of cIAP-mediated NIK degradation. It was previously shown that coexpression of NIK with cIAP1 leads to degradation of NIK whereas its coexpression with a RING domain mutant of cIAP1 deficient in the E3 ligase activity (cIAP1-H588A) does not (Varfolomeev et al., 2007). We therefore overexpressed APPL1, cIAP1 and cIAP1-H588A alone or in combination and determined the levels of NIK. As expected, upon overexpression of cIAP1 the level of NIK protein was hardly detectable. On the contrary, overexpression of cIAP1-H588A showed upregulation of NIK to the similar degree as upon overexpression of APPL1 (Fig. 6C). Importantly, coexpression of APPL1 along with cIAP1 prevented NIK degradation (Fig. 6C), indicating that excess of APPL1 protein interferes with cIAP1-mediated NIK degradation. In agreement with the known role of TRAF2 as an inhibitor of the noncanonical pathway by promoting NIK degradation, co-overexpression of TRAF2 along with APPL1 partly counteracted the APPL1-induced stabilization of NIK. This effect was not observable when the E3 ligase deficient mutants of TRAF2 were co-overexpressed along with APPL1 (Fig. 6D).
In order to get further mechanistic insights into how APPL1 may prevent cIAP1-mediated NIK degradation, we tested the impact of APPL1 overexpression on the composition of the NIK degradative complex consisting of TRAF2, TRAF3, cIAP1 and NIK itself. Within this complex, TRAF2 recruits cIAP1, while TRAF3 binds NIK (Zarnegar et al., 2008b). By performing a series of reciprocal co-immunoprecipitations we could observe that APPL1 overexpression reduced the amounts of TRAF2 in NIK immunoprecipitations and of NIK in TRAF2 immunoprecipitations (Fig. 6E). These data suggest that through its interaction with TRAF2, APPL1 either reduces formation or promotes dissociation of the NIK degradative complex, thus inducing NIK stabilization.
APPL1-mediated stabilization of NIK activates the canonical NF-κB pathway
Finally, we wished to determine whether NIK stabilization is necessary for the APPL1-induced activation of the canonical NF-κB pathway and the nuclear accumulation of p65. To this end we overexpressed kinase-defective mutant of NIK (NIK-KD, K429A-K430A) (Malinin et al., 1997) along with APPL1 or the empty vector, and we assessed NF-κB activation by luciferase reporter assay. As shown in Fig. 7A, transient overexpression of dominant-negative NIK inhibited the NF-κB pathway in APPL1-expressing cells. This argues that NIK activity is required for APPL1-mediated NF-κB activation. Moreover, fractionation experiments revealed that APPL1 overexpression was not sufficient to increase p65 level in the nucleus when NIK was depleted (Fig. 7B). Interestingly, APPL1 protein levels were decreased upon knockdown of NIK in both total lysate and the nuclear fraction (Fig. 7B), what might additionally favor low NF-κB activity. These results support the conclusion that APPL1-induced NF-κB activation occurs via NIK. Consistently, knockdown of APPL1 did not affect the nuclear localization of p65 induced by overexpression of NIK (Fig. 7C), arguing that the primary role of APPL1 in NF-κB activation is the stabilization of NIK.
Fig. 7.
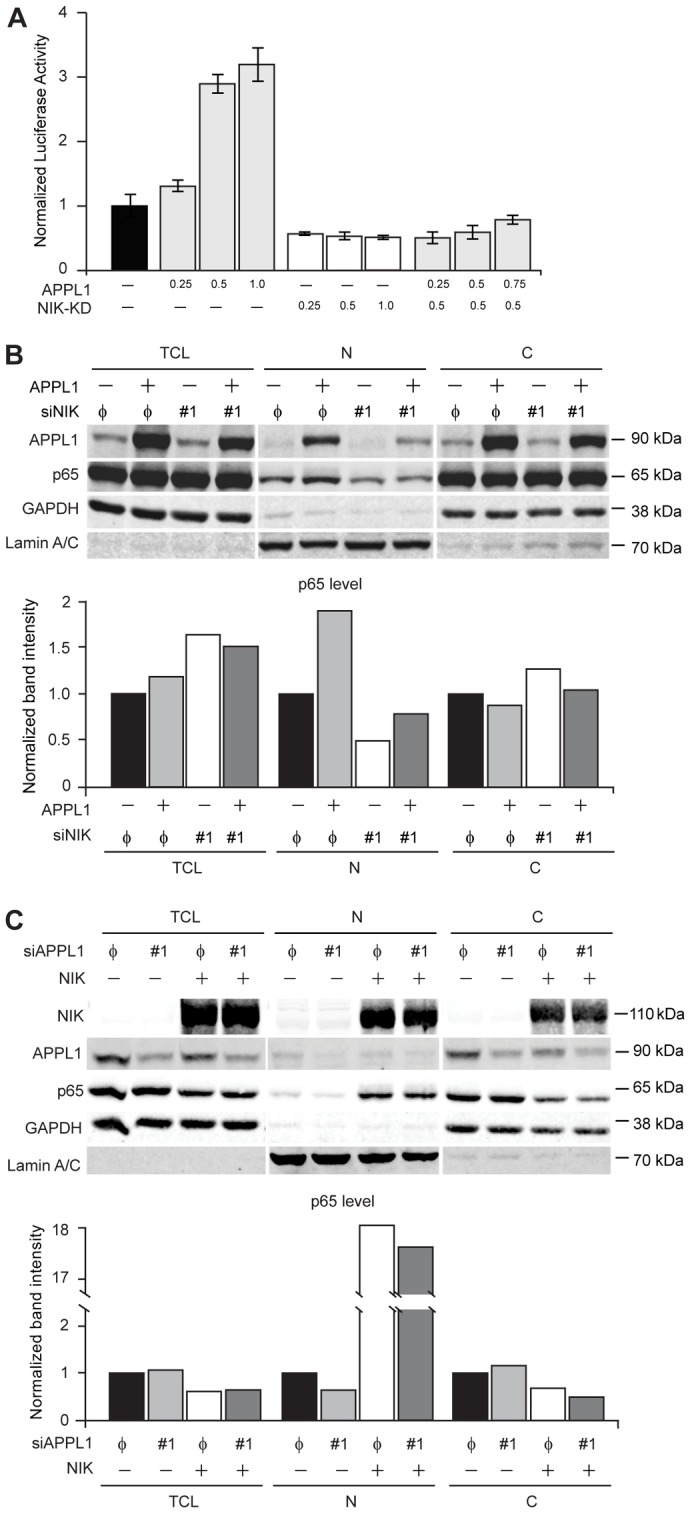
APPL1-induced NIK stability activates the canonical NF-κB pathway. (A) A kinase-deficient mutant of NIK abolishes APPL1-induced activation of the NF-κB reporter. HEK293T cells were cotransfected with reporter plasmids along with the different combinations of APPL1 and NIK-KD expression vectors, as indicated (all values are in µg of DNA). The luciferase reporter activity was measured as described in the Materials and Methods. (B) APPL1 overexpression does not trigger nuclear translocation of p65 upon NIK depletion. HEK293T cells were transfected with APPL1-expressing vector or siRNAs (non-targeting φ or NIK #1; efficiency of NIK knockdown was verified by RT-PCR). Forty-eight hours after transfection total cell lysate (TCL), cytoplasmic (C) and nuclear (N) fractions were analyzed by western blotting with anti-APPL1, -p65, -GAPDH and -lamin-A/C antibodies (upper panel) and the intensity of p65 band was quantified (lower panel). (C) NIK overexpression enhances nuclear localization of p65 upon knockdown of APPL1. HEK293T cells were transfected with NIK-expressing vector or siRNAs (non-targeting φ or APPL1 #1). Forty-eight hours after transfection, the cell fractions were prepared and analyzed as in B. Representative results from three independent experiments are shown in B and C.
Discussion
In this report, we reveal a novel role for an endocytic protein APPL1 in the regulation of NF-κB activity in the absence of cytokine stimulation. We uncover that APPL1 can positively affect basal transcriptional activity of the NF-κB, inducing nuclear accumulation of p65 and expression of specific target genes. Mechanistically, we propose that through its interaction with TRAF2, APPL1 prevents NIK degradation by reducing its association with the degradative complex. The resulting NIK stabilization then triggers p65 translocation to the nucleus. In this way, APPL1 serves as a link between the canonical and noncanonical machineries of NF-κB activation, as previously reported for TRAF2.
Up to date, protein interaction studies identified several proteins involved in endocytosis that play additional functions in NF-κB signaling. Most of them act as inhibitors of the pathway, including Tom1 (target of Myb1) and Tollip (Toll-interacting protein) that form a complex regulating endocytosis of IL-1 receptor (IL-1R) (Brissoni et al., 2006; Yamakami and Yokosawa, 2004). Additionally, Tollip is found in a complex with IRAK (IL-1R associated kinase) to block its phosphorylation and prevent IKK and NF-κB activation upon stimulation of IL-1R (Burns et al., 2000). Moreover, β-arrestins inhibit NF-κB activity by binding to IκBα and preventing its phosphorylation and subsequent degradation by proteasome (Gao et al., 2004). However, little is known about the role of endocytic proteins in the activation of the NF-κB pathway. Previously, yeast two hybrid tests revealed that APPL1 interacts with the adaptor protein TRAF2 (Rual et al., 2005). Our biochemical data clearly confirm that this interaction occurs at the level of endogenous proteins and is negatively regulated by TNFα. In addition to its soluble form in the cytoplasm and in the nucleus, APPL1 protein is localized to a subpopulation of early endosomes which participate in intracellular trafficking of receptors and cargo sorting (Miaczynska et al., 2004; Zoncu et al., 2009). By microscopy we could not detect stable recruitment of TRAF2 to endosomes which may indicate that it either interacts with cytoplasmic APPL1 or binds endosomal APPL1 in a transient manner. The latter option would be favored by the fact that the ability of APPL1 to associate with Rab proteins on endosomes is required for its function as an activator of NF-κB. This could be due to potential posttranslational modifications or other interactions of APPL1 occurring on endosomes and critical for its activity. Moreover, our data would not exclude a possibility that APPL endosomes could act as a platform for a dynamic assembly of complexes involved in NF-κB signaling, although a direct proof for it is currently missing.
It is an emerging notion that many endocytic proteins play additional or alternative roles in intracellular signaling, either related to or independent of their functions in endocytosis. In agreement with this, APPL1 was reported to act as an adaptor protein in different signaling pathways, including PI3K/Akt, insulin, adiponectin, Wnt and, as presented in this study, NF-κB (Chandrasekar et al., 2008; Cheng et al., 2009; Mao et al., 2006; Rashid et al., 2009; Saito et al., 2007; Schenck et al., 2008). These functions require different interacting partners of APPL1. It is possible that depending on the cell type, extracellular factors or abundance of interacting proteins, APPL1 may help integrating multiple signal transduction cascades within the cell. In particular, adiponectin signal transduction was previously linked to NF-κB signaling (Heilbronn and Campbell, 2008; Ruan et al., 2003). Depending on the cell type, adiponectin was shown to either suppress NF-κB activity, like in endothelial cells and adipocytes (Ajuwon and Spurlock, 2005; Ouchi et al., 2000) or to activate the NF-κB pathway in synovial or cardiac fibroblasts (Hattori et al., 2007; Tang et al., 2007). By interacting with adiponectin receptors, APPL1 mediates activation of the downstream kinases, including Akt, LKB1 and AMPK (Mao et al., 2006; Saito et al., 2007). Moreover, APPL1 directly interacts with Akt and LKB1 (Cheng et al., 2009; Mitsuuchi et al., 1999; Tan et al., 2010; Zhou et al., 2009). Akt-mediated IKK phosphorylation was shown to activate the IκB–NF-κB pathway (Ozes et al., 1999). These reports and our data indicate that APPL1 can contribute to the regulation of NF-κB signaling at different levels, e.g. via Akt activation and/or NIK stabilization, possibly in a cell type-dependent manner.
In contrast to the canonical activation of NF-κB signaling via cytokine stimulation, the noncanonical pathway is less investigated, particularly in non-immune cells. Here we show that in HEK293T cells the noncanonical pathway can be activated via NIK stabilization caused by APPL1 overexpression, in the absence of cytokine stimulation. This argues that the noncanonical pathway can be induced in non-immune cells by intrinsic cues. Under resting conditions the protein level of NIK is kept very low because of its constant association with the TRAF2–TRAF3–cIAP1/2 degradative complex (Liao et al., 2004; Qing et al., 2005). TRAF2 exhibits diverse roles in NF-κB signaling, as it activates the canonical NF-κB pathway and inhibits the noncanonical pathway as a component of the NIK degradation machinery (Au and Yeh, 2007). In support of the latter function, TRAF2 deletion was shown to stabilize NIK, activate IKKα, and induce processing of p100 to the active p52 form which results in the constitutive noncanonical NF-κB activity (Grech et al., 2004). Analyzing the composition of the NIK degradative complex we observed that overexpression of APPL1 limits the association between TRAF2 and NIK which is a prerequisite for NIK degradation. We propose that due to its ability to interact with TRAF2, APPL1 upon overexpression can compete off TRAF2 from the NIK degradative complex, thus leading to NIK stabilization.
We demonstrate that NIK-mediated activation of the noncanonical NF-κB by overexpressed APPL1 can further stimulate the canonical NF-κB signaling defined by p65 nuclear translocation. However, the subset of genes expressed under these conditions differs from those induced by acute TNFα signaling in the canonical pathway, consistent with our proposal that APPL1 regulates basal NF-κB activity in the absence of cytokine stimulation. This further suggests that in unstimulated non-immune cells there are mechanisms linking canonical and noncanonical pathways, of which APPL1-mediated NIK stabilization represents one example. More generally, taking into account other known cellular functions of APPL1 it is conceivable that APPL1 may help coordinating basal NF-κB activity with other signaling or metabolic pathways.
Materials and Methods
Cell culture and transfection
HEK293T (human embryonic kidney 293T), SKBR3 (human breast carcinoma) and DLD1 (human colon carcinoma) and HeLa cells were maintained in DMEM (Dulbecco's modified Eagle's medium) supplemented with 10% fetal calf serum and 2 mM L-glutamine (Sigma, St Louis, MO). H1299 (human non-small cell lung carcinoma) cells were grown in RPMI-1640 medium (Sigma), supplemented as above. HUVEC (human umbilical vein endothelial cells) were purchased from Invitrogen (Carlsbad, CA) and grown in EBM-2 basal medium supplemented with the EGM-2 SingleQuots kit (Lonza, Basel, Switzerland). HEK293T cells were treated with 20 ng/ml of human TNFα (PeproTech, Rocky Hill, NJ) in a starvation medium (serum-free medium containing 0.2% bovine serum albumin). Transient transfection was done using either Lipofectamine2000 (Invitrogen) or Fugene6 (Promega, Madison, WI) for DNA delivery or HiPerFect (Qiagen, Hilden, Germany) for siRNA delivery (at final concentration of 10 nM for siRNA and 33 nM for esiRNA). SureFECT (SABiosciences, Frederick, MD) was used for simultaneous transfection of DNA and siRNA. All transfections were performed according to manufacturers' instructions and analyzed after 24 or 48 h. In all experiments DNA and siRNA amount was kept constant by contransfection either with empty vector or non-targeting siRNA.
Antibodies
The following antibodies were used: mouse monoclonal against IKK2 (611254, BD, San Jose, CA), IκBα (4814, Cell Signaling, Beverly, MA), Lamin A/C (sc-7292, Santa Cruz, CA), TRAF2 (558890, BD), β-actin (A5441, Sigma), α-tubulin (T5168, Sigma), Myc (05–419, Upstate, Lake Placid, NY), Flag (F3165, Sigma), GFP (G1546, Sigma), rat monoclonal against HA (3F10, Roche), rabbit polyclonal against GAPDH (sc-25778), cIAP1 (sc-7943), cIAP2 (sc-7944), TRAF3 (sc-947), XIAP and livin (9770, Cell Signaling), secondary horseradish peroxidase-conjugated goat anti-mouse and anti-rabbit antibodies (Jackson ImmunoResearch, West Grove, PA), IRDye 800 CW donkey anti-mouse and IRDye 680 anti-rabbit (LI-COR Biosciences, Lincoln, NE), Alexa Fluor 488-conjugated anti-rat and Alexa Fluor 555-conjugated anti-rabbit antibodies (Invitrogen). Anti-APPL1 (4428) antibody was generated using peptide SKKRENDKVKYEVTE+C that was synthesized based on the sequence of APPL1 followed by immunization of rabbits (Eurogentec, Seraing, Belgium). Purified donkey anti-mouse (715-005-150) and anti-rabbit (711-005-152) IgGs were purchased from Jackson ImmunoResearch.
Plasmids
APPL1-encoding plasmids (pcDNA3-APPL1, pcDNA3HA-APPL1, pcDNA3HA-mAPPL1, pGEX-6P-3-APPL1 a.a. 274–709, pGEX-6P-3-APPL1 and pcDNA3HA-APPL1 a.a. 429–709, myc-APPL1-K280E,Y283C,G319R, myc-APPL1-A318D, myc-APPL1-G320A) and pCMV-Rab5-Q79L were previously described (Miaczynska et al., 2004; Stenmark et al., 1994). The pcDNA3-IKK2-K44M construct was provided by Dr Frank Mercurio (Addgene plasmid 11104) (Mercurio et al., 1997). pcDNA3myc-cIAP1, pcDNA3myc-cIAP1-H588A, pcDNA3-NIK-KD were a kind gift from Dr Krishna Rajalingam (Institute of Biochemistry II, Goethe University, Frankfurt). pcDNA3Flag-Strawberry-p65 construct was a gift from Dr Tomasz Lipniacki (Institute of Fundamental Technological Research, Warsaw). pNF-κB-Luc and pRL-TK-luc plasmids for NF-κB luciferase reporter assay were obtained from Stratagene (La Jolla, CA). Full-length human TRAF2 was amplified from the clone IRAKp961L0764Q (obtained from the Source BioScience ImaGenes, Berlin, Germany) by PCR using the oligonucleotides pairs: 5′-GGCGCGGCCGCTATGGCTGCAGCTAGC-3′ (forward) and 5′-GTCTAGACGTTAGAGCCCTGTCAGGTC-3′ (reverse) (for cloning into pcDNA3 or pcDNA3HA via NotI/XbaI digestion), 5′-GCGAAGCTTCTCATGGCTGCAGCTAGCG-3′ (forward) and 5′-CGGTCGACGCAGTTAGAGCCCTGTCAG-3′ (reverse) (for cloning into pEGFP-C3 via HindIII/SalI digestion). Expression vectors for GFP–TRAF2 deletion mutants were constructed by PCR amplification of TRAF2 coding sequences using oligonucleotide pairs as listed below and cloning them into HindIII/SalI-digested pEGFP-C3 vector: for 87–501, 5′-GCGAAGCTTCTCATGAGCAGTTCGGCC-3′ (forward) and 5′-CGGTCGACGCAGTTAGAGCCCTGTCAG-3′ (reverse); for 1–271, 5′-GCGAAGCTTCTCATGGCTGCAGCTAGCG-3′ (forward) and 5′-CGGTCGACGCAGTTACCGGTTCAGGACGC-3′ (reverse); for 87–271, 5′-GCGAAGCTTCTCATGAGCAGTTCGGCC-3′ (forward) and 5′-CGGTCGACGCAGTTACCGGTTCAGGACGC-3′ (reverse). Mutations in TRAF2 expression vector were introduced using the QuikChange Site-Directed Mutagenesis Kit (Stratagene). TRAF2-Rm was mutated at C49A, H51A, C54A and C57A; TRAF2-RmZm was mutated at C49A, H51A, C209A and C212A; HA-TRAF2-dR was constructed by ligating the 87–501 fragment into the pcDNA3HA. Constructs were verified by sequencing.
Small interfering RNA
The following Stealth RNAi oligonucleotides (Invitrogen) were used: against APPL1#1 (ID#HSS178039), APPL1#2 (ID#HSS119759), NIK#1 (ID#VHS40827) and non-specific siRNAs (ID#12935-200 and ID#12935-400) as controls.
Production and purification of endoribonuclease-prepared siRNA (esiRNA)
Optimal esiRNA target regions with a length of 400–600 bp were selected using the DEQOR web server (http://deqor.mpi-cbg.de). In brief, T7 promoter sequence was added to the selected regions of a gene of interest by two PCRs. The first PCR reaction was carried out with gene-specific primer pairs that were tagged at 5′ ends with a part of the T7 promoter (underlined). During the second PCR, primers specific to T7 promoter were used to amplify the whole T7 sequence. The sequences for these primers are as follows: for TRAF2-#1, 5′-TCACTATAGGGAGAGGGGTCTTCATCTGGAAGATC-3′ (forward primer) and 5′-TCACTATAGGGAGACCGTCCCGCACGTAGGAATTC-3′ (reverse primer); for TRAF2-#2, 5′-TCACTATAGGGAGAGACGGGGGAAGGAGCCACCAG-3′ (forward primer) and 5′-TCACTATAGGGAGACAGATTTGAATGGTTTAATAAC-3′ (reverse primer), for control mRFP, 5′-TCACTATAGGGAGAGCGACTACTTGAAGCTG-3′ (forward primer) and 5′-TCACTATAGGGAGACGCGCGCTCGTACTGTTC-3′ (reverse primer), for control β-Gal, 5′-TCACTATAGGGAGAGGCTGGCGTAATAGCGAAGAG-3′ (forward primer) and 5′-TCACTATAGGGAGACCATTAAAGCGAGTGGCAACA-3′ (reverse primer), for T7 promoter, 5′-GCTAATACGACTCACTATAGGGAGAG-3′ (forward primer) and 5′-GCTAATACGACTCACTATAGGGAGAC-3′ (reverse primer). Further esiRNA synthesis was carried out as described previously (Kittler et al., 2005). Average of two esiRNA controls is presented on the graphs (φ).
Immunofluorescence
Cells grown on coverslips were fixed with 3% paraformaldehyde for 15 min and then permeabilized in 0.1% Triton X-100 for 2 min at room temperature. After washing in PBS, free aldehyde groups were quenched by 15 min incubation with 50 mM NH4Cl. Washed coverslips were blocked in 10% fetal bovine serum for 0.5 h and incubated with primary antibodies diluted in 5% fetal bovine serum for 1 h in a humid chamber. The coverslips were washed, and Alexa Fluor-tagged secondary antibodies were added for 0.5 h. The coverslips were washed and mounted onto glass slides using Moviol (Sigma). Images were acquired on a laser-scanning confocal microscope (Leica TCS SP2 AOBS) using a ×63/1.4 numerical aperture oil immersion objective. The presented microscopy images were assembled using Adobe Photoshop CS5.
Western blot analysis
Cells were extracted in ice-cold lysis buffer (50 mM Hepes pH 7.5, 150 mM NaCl, 1 mM EGTA, 1 mM EDTA, 1% Triton X-100, 10% glycerol) supplemented with 5 µg/ml DNase and protease inhibitor cocktail. Protein concentration was measured with BCA Protein Assay Kit (Pierce, Rockford, IL) and samples of 15–20 µg total proteins were subjected to SDS-PAGE. Resolved proteins were transferred to nitrocellulose membrane, which was blocked with 5% skim milk, and then incubated with the specific antibodies, and detected with either enhanced chemiluminescence or Odyssey infrared imaging system (LI-COR Biosciences, Lincoln, NE).
Immunoprecipitation and GST pull-down assay
HEK293T cells were lysed in ice-cold lysis buffer containing 5 µg/ml DNase and protease inhibitor cocktail. If required, antibodies were cross-linked to Protein-G–agarose (Roche) by using dimethyl pimelimidate (DMP) (Sigma) and incubated with cell lysates overnight at 4°C with rotation. When no cross-linker was used, proteins were immunoprecipitated by overnight incubation of lysates with an appropriate antibody, followed by 2 h incubation with Protein-G–agarose beads. In all cases immune complexes were washed six times in a lysis buffer before elution (with 100 mM glycine pH 2.5 or Laemmli buffer) and SDS-PAGE.
GST, GST–APPL1 (a.a. 274–709) and GST–APPL1 (a.a. 429–709) fusion proteins used in pull-down assays as bait were expressed in E. coli and purified according to the manufacturer's instructions (GE Healthcare, Uppsala, Sweden). Isopropyl β-D-thiogalactoside (Sigma) at a concentration of 0.5 mM was used to induce the expression. The purified GST fusion proteins bound to the glutathione–Sepharose beads (GE Healthcare) were incubated overnight with rotation at 4°C with either lysates of HEK293T cells transfected with GFP–TRAF2 or TRAF2 or with in vitro translated TRAF2 (synthesized using TNT T7 Coupled Reticulocyte Lysate System from Promega; according to the manufacturer's protocol). Beads were washed with the lysis buffer and elution of bound proteins was done with 10 mM glutathione in 50 mM Tris/HCl pH 8.0 for 15 min at room temperature. Eluates were resuspended in Laemmli buffer, subjected to SDS-PAGE and immunoblotted for the proteins of interest.
Luciferase assays
HEK293T cells were transiently transfected with the pNF-κB-luc and pRL-TK-luc reporter vectors (100 ng each when cotransfected with plasmids and 25 ng pNF-κB-luc and 5 ng pRL-TK-luc when cotransfected with siRNA) and the different combinations of plasmids or siRNA. In all assays, the amount of DNA transfected was kept constant for a total 1.25 µg by cotransfection with empty pcDNA3.1 vector. Forty-eight hours after transfection cells were collected and lysed with passive lysis buffer (Promega). Cell lysates were assayed using the dual luciferase assay kit according to the manufacturer's instructions (Promega). The firefly luciferase activity derived from the NF-κB-responsive reporter was normalized to its respective Renilla luciferase activity as a control for the transfection efficiency. Results are expressed as the fold induction relative to the basal level measured in cells transfected with control vector or siRNA. Values are mean ± s.d. from three or four independent transfections performed in parallel and are representative of at least three experiments.
Preparation of cell fractions
Fractions enriched in early endosomes, cytosol and PNS were prepared from HeLa cells as described (Urbanska et al., 2011). For nuclear and cytoplasmic fractions HEK293T cells were trypsinized, centrifuged (800 g, 3 min, 4°C) and resuspended in a lysis buffer (20 mM Hepes pH 7.9, 20 mM NaF, 1 mM Na3VO4, 1 mM sodium pyrophosphate, 1 mM EDTA, 1 mM EGTA, 1 mM DTT, DNase and protease inhibitor cocktail). After 15 min of lysis on ice, Nonidet P40 was added to the cell extracts to a final concentration of 0.2% for further 15 min. Afterwards cell lysates were applied on the top of 6 ml of sucrose buffer (0.7 M sucrose, 60 mM KCl, 15 mM NaCl, 15 mM Tris/HCl pH 7.5, 2 mM EDTA, 0.5 mM EGTA, 14 mM 2-mercaptoethanol and 0.1% Triton X-100) and centrifuged (1300 g, 10 min, 4°C). After centrifugation, the cytoplasmic fraction was harvested from the top of the sucrose buffer, and the nuclear pellet was lysed in RIPA buffer (50 mM Tris-HCl pH 7.4, 150 mM NaCl, 0.5 mM EDTA, 1% Triton X-100, 0.5% deoxycholate, 0.1% SDS, 5 µg/ml DNase and protease inhibitors). Both fractions were centrifuged for 15 min at 20,000 g to remove insoluble complexes. The purity of fractions was tested by immunoblotting for GAPDH as a cytoplasmic marker and Lamin A/C as a nuclear marker. The band intensity was determined using the LI-COR Odyssey Infrared Imaging System. p65 levels were normalized to GAPDH for total cell lysates and cytoplasmic fractions, or to Lamin A/C for nuclear fractions. Data are shown as the fold change to the respective controls, which were normalized to 1.
Cell viability assays
Cells grown in 96-well plates (5000 cells/ well) were transfected and treated with BAY 11–7082 (Sigma) in a final volume of 100 µl medium containing 0.5% serum. After 48 h, 10 µl Cell Counting Kit-8 solution (Sigma) was added to the plates for 2 h at 37°C. The absorbance was measured at 450 nm. Values are mean ± s.d. from six independent transfections performed in parallel and are representative of at least three experiments.
Quantitative PCR
Total RNA was extracted from HEK293T cells 48 h upon APPL1 overexpression or knockdown or 5 h after treatment with TNFα using High Pure Isolation Kit (Roche). For cDNA synthesis random nonamers, oligo(dT)23 and M-MLV reverse transcriptase (Sigma) were used according to manufacturer's instructions. For estimation of gene expression, a 96-well PCR array designed for the NF-κB signaling pathway and containing primer pairs for detection of 84 genes of interest was employed (PAHS-025, SABiosciences). Quantitative PCR was performed according to manufacturer's recommendations, using a 7900HT Fast Real-Time PCR thermocycler (Applied Biosystems, Foster City, CA). The Ct based fold-change calculations were performed using RT2 PCR Data Analysis Software (SABiosciences). For estimation of the expression of NIK, APPL1, IL-8, CCL2 and CXCL12 the following pairs of primers were used: NIK 5′-GACTTTGGCCATGCTGTGTGT-3′ (forward), 5′-GGATGTAGTCCCCTGTGAGCAA-3′ (reverse) and 5′-CCTGCACATCCGGGAGTT-3′ (forward) and 5′-GATGCCAGTGGCGATGTCT-3′ (reverse), APPL1 5′-GAGGACAGCCCGCAGACA-3′ (forward), 5′-TCCGATGCATAGCTTGATACAACT-3′ (reverse), IL-8 5′-GCTCTCTTGGCAGCCTTCCTGA-3′ (forward), 5′-TTTCCTTGGGGTCCAGACAGAGC-3′ (reverse), CCL2 5′-GAAGAATCACCAGCAGCAAG-3′ (forward), 5′-CTTGGCCACAATGGTCTTGA-3′ (reverse), CXCL12 5′-CAAACTGTGCCCTTCAGATTG-3′ (forward), 5′-CGGGTCAATGCACACTTGTC-3′ (reverse), ACTB 5′-CAGGTCATCACCATTGGCAAT-3′ (forward), 5′-TCTTTGCGGATGTCCACGT-3′ (reverse), GAPDH 5′-CATGTTCGTCATGGGTGTGAACCA-3′ (forward), 5′-GTGATGGCATGGACTGTGGTCAT-3′ (reverse). The PCR reaction was performed with the Kapa Sybr Fast ABI Prism qPCR Kit (KapaBiosystems, Boston MA). For estimation of TNF expression the TaqManR Assays (#4448892) and the TaqMan Gene Expression Master Mix (Applied Biosystems) were used. The expression values were obtained at least in duplicate from 3 biological replicates. Relative quantification (RQ) method and Data Assist software (Applied Biosystems) were used to estimate fold change of gene expression. Data were normalized according to the level of ACTB and GAPDH. Only samples with efficient silencing (at least 70% reduction in APPL1 mRNA level) were used in these experiments.
IL-8 protein estimation
The concentration of human IL-8 protein in cell culture supernatants was estimated using the Quantikine ELISA (R&D Systems, Minneapolis, MN) according to the manufacturer's recommendations. Values are mean ± s.d. from two independent transfections performed in parallel and are representative of at least three experiments.
Statistical analysis
The statistical significance was assessed by the Mann Whitney test. The P-values relative to controls were marked with the asterisks on the charts (**P<0.005).
Supplementary Material
Acknowledgments
We are grateful to Krishna Rajalingam (Goethe University, Frankfurt) for advice on the project and for providing the cIAP1 and NIK constructs. We thank Iwona Pilecka and Kamil Jastrzebski for critical reading of the manuscript, and Tomasz Lipniacki (IFTR, Warsaw) for the p65 construct.
Footnotes
Funding
This work was supported by a Senior Research Fellowship from the Wellcome Trust [grant number 076469/Z/05/Z]; the European Union [grant numbers LSHG-CT-2006-019050 (EndoTrack), GA No 229676 (HEALTH-PROT)]; and by the Polish-Norwegian Research Fund [grant number PNRF-27-AI-1/07]. Deposited in PMC for immediate release.
Supplementary material available online at http://jcs.biologists.org/lookup/suppl/doi:10.1242/jcs.105171/-/DC1
References
- Ajuwon K. M., Spurlock M. E. (2005). Adiponectin inhibits LPS-induced NF-kappaB activation and IL-6 production and increases PPARgamma2 expression in adipocytes. Am. J. Physiol. Regul. Integr. Comp. Physiol. 288, R1220–R1225 10.1152/ajpregu.00397.2004 [DOI] [PubMed] [Google Scholar]
- Au P. Y., Yeh W. C. (2007). Physiological roles and mechanisms of signaling by TRAF2 and TRAF5. Adv. Exp. Med. Biol. 597, 32–47 10.1007/978-0-387-70630-6_3 [DOI] [PubMed] [Google Scholar]
- Banach–Orlowska M., Pilecka I., Torun A., Pyrzynska B., Miaczynska M. (2009). Functional characterization of the interactions between endosomal adaptor protein APPL1 and the NuRD co-repressor complex. Biochem. J. 423, 389–400 10.1042/BJ20090086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borlido J., Zecchini V., Mills I. G. (2009). Nuclear trafficking and functions of endocytic proteins implicated in oncogenesis. Traffic 10, 1209–1220 10.1111/j.1600-0854.2009.00922.x [DOI] [PubMed] [Google Scholar]
- Brissoni B., Agostini L., Kropf M., Martinon F., Swoboda V., Lippens S., Everett H., Aebi N., Janssens S., Meylan E.et al. (2006). Intracellular trafficking of interleukin-1 receptor I requires Tollip. Curr. Biol. 16, 2265–2270 10.1016/j.cub.2006.09.062 [DOI] [PubMed] [Google Scholar]
- Burns K., Clatworthy J., Martin L., Martinon F., Plumpton C., Maschera B., Lewis A., Ray K., Tschopp J., Volpe F. (2000). Tollip, a new component of the IL-1RI pathway, links IRAK to the IL-1 receptor. Nat. Cell Biol. 2, 346–351 10.1038/35014038 [DOI] [PubMed] [Google Scholar]
- Chandrasekar B., Boylston W. H., Venkatachalam K., Webster N. J., Prabhu S. D., Valente A. J. (2008). Adiponectin blocks interleukin-18-mediated endothelial cell death via APPL1-dependent AMP-activated protein kinase (AMPK) activation and IKK/NF-kappaB/PTEN suppression. J. Biol. Chem. 283, 24889–24898 10.1074/jbc.M804236200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng K. K., Iglesias M. A., Lam K. S., Wang Y., Sweeney G., Zhu W., Vanhoutte P. M., Kraegen E. W., Xu A. (2009). APPL1 potentiates insulin-mediated inhibition of hepatic glucose production and alleviates diabetes via Akt activation in mice. Cell Metab. 9, 417–427 10.1016/j.cmet.2009.03.013 [DOI] [PubMed] [Google Scholar]
- Chung J. Y., Lu M., Yin Q., Wu H. (2007). Structural revelations of TRAF2 function in TNF receptor signaling pathway. Adv. Exp. Med. Biol. 597, 93–113 10.1007/978-0-387-70630-6_8 [DOI] [PubMed] [Google Scholar]
- Deepa S. S., Dong L. Q. (2009). APPL1: role in adiponectin signaling and beyond. Am. J. Physiol. Endocrinol. Metab. 296, E22–E36 10.1152/ajpendo.90731.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdmann K. S., Mao Y., McCrea H. J., Zoncu R., Lee S., Paradise S., Modregger J., Biemesderfer D., Toomre D., De Camilli P. (2007). A role of the Lowe syndrome protein OCRL in early steps of the endocytic pathway. Dev. Cell 13, 377–390 10.1016/j.devcel.2007.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H., Sun Y., Wu Y., Luan B., Wang Y., Qu B., Pei G. (2004). Identification of beta-arrestin2 as a G protein-coupled receptor-stimulated regulator of NF-kappaB pathways. Mol. Cell 14, 303–317 10.1016/S1097-2765(04)00216-3 [DOI] [PubMed] [Google Scholar]
- Ghosh S., Karin M. (2002). Missing pieces in the NF-kappaB puzzle. Cell 109 Suppl., S81–S96 10.1016/S0092-8674(02)00703-1 [DOI] [PubMed] [Google Scholar]
- Grech A. P., Amesbury M., Chan T., Gardam S., Basten A., Brink R. (2004). TRAF2 differentially regulates the canonical and noncanonical pathways of NF-kappaB activation in mature B cells. Immunity 21, 629–642 10.1016/j.immuni.2004.09.011 [DOI] [PubMed] [Google Scholar]
- Habelhah H., Takahashi S., Cho S. G., Kadoya T., Watanabe T., Ronai Z. (2004). Ubiquitination and translocation of TRAF2 is required for activation of JNK but not of p38 or NF-kappaB. EMBO J. 23, 322–332 10.1038/sj.emboj.7600044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori Y., Hattori S., Akimoto K., Nishikimi T., Suzuki K., Matsuoka H., Kasai K. (2007). Globular adiponectin activates nuclear factor-kappaB and activating protein-1 and enhances angiotensin II-induced proliferation in cardiac fibroblasts. Diabetes 56, 804–808 10.2337/db06-1405 [DOI] [PubMed] [Google Scholar]
- Hayden M. S., Ghosh S. (2004). Signaling to NF-kappaB. Genes Dev. 18, 2195–2224 10.1101/gad.1228704 [DOI] [PubMed] [Google Scholar]
- Hayden M. S., Ghosh S. (2008). Shared principles in NF-kappaB signaling. Cell 132, 344–362 10.1016/j.cell.2008.01.020 [DOI] [PubMed] [Google Scholar]
- Heilbronn L. K., Campbell L. V. (2008). Adipose tissue macrophages, low grade inflammation and insulin resistance in human obesity. Curr. Pharm. Des. 14, 1225–1230 10.2174/138161208784246153 [DOI] [PubMed] [Google Scholar]
- Hoffmann A., Baltimore D. (2006). Circuitry of nuclear factor kappaB signaling. Immunol. Rev. 210, 171–186 10.1111/j.0105-2896.2006.00375.x [DOI] [PubMed] [Google Scholar]
- Kittler R., Heninger A. K., Franke K., Habermann B., Buchholz F. (2005). Production of endoribonuclease-prepared short interfering RNAs for gene silencing in mammalian cells. Nat. Methods 2, 779–784 10.1038/nmeth1005-779 [DOI] [PubMed] [Google Scholar]
- Liao G., Zhang M., Harhaj E. W., Sun S. C. (2004). Regulation of the NF-kappaB-inducing kinase by tumor necrosis factor receptor-associated factor 3-induced degradation. J. Biol. Chem. 279, 26243–26250 10.1074/jbc.M403286200 [DOI] [PubMed] [Google Scholar]
- Madge L. A., Kluger M. S., Orange J. S., May M. J. (2008). Lymphotoxin-alpha 1 beta 2 and LIGHT induce classical and noncanonical NF-kappa B-dependent proinflammatory gene expression in vascular endothelial cells. J. Immunol. 180, 3467–3477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinin N. L., Boldin M. P., Kovalenko A. V., Wallach D. (1997). MAP3K-related kinase involved in NF-kappaB induction by TNF, CD95 and IL-1. Nature 385, 540–544 10.1038/385540a0 [DOI] [PubMed] [Google Scholar]
- Mao X., Kikani C. K., Riojas R. A., Langlais P., Wang L., Ramos F. J., Fang Q., Christ–Roberts C. Y., Hong J. Y., Kim R. Y.et al. (2006). APPL1 binds to adiponectin receptors and mediates adiponectin signalling and function. Nat. Cell Biol. 8, 516–523 10.1038/ncb1404 [DOI] [PubMed] [Google Scholar]
- Mercurio F., Zhu H., Murray B. W., Shevchenko A., Bennett B. L., Li J., Young D. B., Barbosa M., Mann M., Manning A.et al. (1997). IKK-1 and IKK-2: cytokine-activated IkappaB kinases essential for NF-kappaB activation. Science 278, 860–866 10.1126/science.278.5339.860 [DOI] [PubMed] [Google Scholar]
- Miaczynska M., Christoforidis S., Giner A., Shevchenko A., Uttenweiler–Joseph S., Habermann B., Wilm M., Parton R. G., Zerial M. (2004). APPL proteins link Rab5 to nuclear signal transduction via an endosomal compartment. Cell 116, 445–456 10.1016/S0092-8674(04)00117-5 [DOI] [PubMed] [Google Scholar]
- Mitsuuchi Y., Johnson S. W., Sonoda G., Tanno S., Golemis E. A., Testa J. R. (1999). Identification of a chromosome 3p14.3-21.1 gene, APPL, encoding an adaptor molecule that interacts with the oncoprotein-serine/threonine kinase AKT2. Oncogene 18, 4891–4898 10.1038/sj.onc.1203080 [DOI] [PubMed] [Google Scholar]
- Naudé P. J., den Boer J. A., Luiten P. G., Eisel U. L. (2011). Tumor necrosis factor receptor cross-talk. FEBS J. 278, 888–898 10.1111/j.1742-4658.2011.08017.x [DOI] [PubMed] [Google Scholar]
- Ouchi N., Kihara S., Arita Y., Okamoto Y., Maeda K., Kuriyama H., Hotta K., Nishida M., Takahashi M., Muraguchi M.et al. (2000). Adiponectin, an adipocyte-derived plasma protein, inhibits endothelial NF-kappaB signaling through a cAMP-dependent pathway. Circulation 102, 1296–1301 10.1161/01.CIR.102.11.1296 [DOI] [PubMed] [Google Scholar]
- Ozes O. N., Mayo L. D., Gustin J. A., Pfeffer S. R., Pfeffer L. M., Donner D. B. (1999). NF-kappaB activation by tumour necrosis factor requires the Akt serine-threonine kinase. Nature 401, 82–85 10.1038/43466 [DOI] [PubMed] [Google Scholar]
- Pomerantz J. L., Baltimore D. (2002). Two pathways to NF-kappaB. Mol. Cell 10, 693–695 10.1016/S1097-2765(02)00697-4 [DOI] [PubMed] [Google Scholar]
- Pyrzynska B., Pilecka I., Miaczynska M. (2009). Endocytic proteins in the regulation of nuclear signaling, transcription and tumorigenesis. Mol. Oncol. 3, 321–338 10.1016/j.molonc.2009.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qing G., Qu Z., Xiao G. (2005). Stabilization of basally translated NF-kappaB-inducing kinase (NIK) protein functions as a molecular switch of processing of NF-kappaB2 p100. J. Biol. Chem. 280, 40578–40582 10.1074/jbc.M508776200 [DOI] [PubMed] [Google Scholar]
- Rashid S., Pilecka I., Torun A., Olchowik M., Bielinska B., Miaczynska M. (2009). Endosomal adaptor proteins APPL1 and APPL2 are novel activators of beta-catenin/TCF-mediated transcription. J. Biol. Chem. 284, 18115–18128 10.1074/jbc.M109.007237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rual J. F., Venkatesan K., Hao T., Hirozane–Kishikawa T., Dricot A., Li N., Berriz G. F., Gibbons F. D., Dreze M., Ayivi–Guedehoussou N.et al. (2005). Towards a proteome-scale map of the human protein-protein interaction network. Nature 437, 1173–1178 10.1038/nature04209 [DOI] [PubMed] [Google Scholar]
- Ruan H., Pownall H. J., Lodish H. F. (2003). Troglitazone antagonizes tumor necrosis factor-alpha-induced reprogramming of adipocyte gene expression by inhibiting the transcriptional regulatory functions of NF-kappaB. J. Biol. Chem. 278, 28181–28192 10.1074/jbc.M303141200 [DOI] [PubMed] [Google Scholar]
- Saito T., Jones C. C., Huang S., Czech M. P., Pilch P. F. (2007). The interaction of Akt with APPL1 is required for insulin-stimulated Glut4 translocation. J. Biol. Chem. 282, 32280–32287 10.1074/jbc.M704150200 [DOI] [PubMed] [Google Scholar]
- Scheidereit C. (2006). IkappaB kinase complexes: gateways to NF-kappaB activation and transcription. Oncogene 25, 6685–6705 10.1038/sj.onc.1209934 [DOI] [PubMed] [Google Scholar]
- Schenck A., Goto–Silva L., Collinet C., Rhinn M., Giner A., Habermann B., Brand M., Zerial M. (2008). The endosomal protein Appl1 mediates Akt substrate specificity and cell survival in vertebrate development. Cell 133, 486–497 10.1016/j.cell.2008.02.044 [DOI] [PubMed] [Google Scholar]
- Senftleben U., Cao Y., Xiao G., Greten F. R., Krähn G., Bonizzi G., Chen Y., Hu Y., Fong A., Sun S. C.et al. (2001). Activation by IKKalpha of a second, evolutionary conserved, NF-kappa B signaling pathway. Science 293, 1495–1499 10.1126/science.1062677 [DOI] [PubMed] [Google Scholar]
- Shih V. F., Tsui R., Caldwell A., Hoffmann A. (2011). A single NFκB system for both canonical and non-canonical signaling. Cell Res. 21, 86–102 10.1038/cr.2010.161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenmark H., Parton R. G., Steele–Mortimer O., Lütcke A., Gruenberg J., Zerial M. (1994). Inhibition of rab5 GTPase activity stimulates membrane fusion in endocytosis. EMBO J. 13, 1287–1296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S. C. (2011). Non-canonical NF-κB signaling pathway. Cell Res. 21, 71–85 10.1038/cr.2010.177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S. C., Ley S. C. (2008). New insights into NF-kappaB regulation and function. Trends Immunol. 29, 469–478 10.1016/j.it.2008.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi M., Rothe M., Goeddel D. V. (1996). Anatomy of TRAF2. Distinct domains for nuclear factor-kappaB activation and association with tumor necrosis factor signaling proteins. J. Biol. Chem. 271, 19935–19942 10.1074/jbc.271.33.19935 [DOI] [PubMed] [Google Scholar]
- Tan Y., You H., Wu C., Altomare D. A., Testa J. R. (2010). Appl1 is dispensable for mouse development, and loss of Appl1 has growth factor-selective effects on Akt signaling in murine embryonic fibroblasts. J. Biol. Chem. 285, 6377–6389 10.1074/jbc.M109.068452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang C. H., Chiu Y. C., Tan T. W., Yang R. S., Fu W. M. (2007). Adiponectin enhances IL-6 production in human synovial fibroblast via an AdipoR1 receptor, AMPK, p38, and NF-kappa B pathway. J. Immunol. 179, 5483–5492 [DOI] [PubMed] [Google Scholar]
- Thu Y. M., Richmond A. (2010). NF-κB inducing kinase: a key regulator in the immune system and in cancer. Cytokine Growth Factor Rev. 21, 213–226 10.1016/j.cytogfr.2010.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbanska A., Sadowski L., Kalaidzidis Y., Miaczynska M. (2011). Biochemical characterization of APPL endosomes: the role of annexin A2 in APPL membrane recruitment. Traffic 12, 1227–1241 10.1111/j.1600-0854.2011.01226.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallabhapurapu S., Karin M. (2009). Regulation and function of NF-kappaB transcription factors in the immune system. Annu. Rev. Immunol. 27, 693–733 10.1146/annurev.immunol.021908.132641 [DOI] [PubMed] [Google Scholar]
- Varfolomeev E., Blankenship J. W., Wayson S. M., Fedorova A. V., Kayagaki N., Garg P., Zobel K., Dynek J. N., Elliott L. O., Wallweber H. J.et al. (2007). IAP antagonists induce autoubiquitination of c-IAPs, NF-kappaB activation, and TNFalpha-dependent apoptosis. Cell 131, 669–681 10.1016/j.cell.2007.10.030 [DOI] [PubMed] [Google Scholar]
- Wajant H., Scheurich P. (2011). TNFR1-induced activation of the classical NF-κB pathway. FEBS J. 278, 862–876 10.1111/j.1742-4658.2011.08015.x [DOI] [PubMed] [Google Scholar]
- Yamakami M., Yokosawa H. (2004). Tom1 (target of Myb 1) is a novel negative regulator of interleukin-1- and tumor necrosis factor-induced signaling pathways. Biol. Pharm. Bull. 27, 564–566 10.1248/bpb.27.564 [DOI] [PubMed] [Google Scholar]
- Zarnegar B., Yamazaki S., He J. Q., Cheng G. (2008a). Control of canonical NF-kappaB activation through the NIK-IKK complex pathway. Proc. Natl. Acad. Sci. USA 105, 3503–3508 10.1073/pnas.0707959105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarnegar B. J., Wang Y., Mahoney D. J., Dempsey P. W., Cheung H. H., He J., Shiba T., Yang X., Yeh W. C., Mak T. W.et al. (2008b). Noncanonical NF-kappaB activation requires coordinated assembly of a regulatory complex of the adaptors cIAP1, cIAP2, TRAF2 and TRAF3 and the kinase NIK. Nat. Immunol. 9, 1371–1378 10.1038/ni.1676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L., Deepa S. S., Etzler J. C., Ryu J., Mao X., Fang Q., Liu D. D., Torres J. M., Jia W., Lechleiter J. D.et al. (2009). Adiponectin activates AMP-activated protein kinase in muscle cells via APPL1/LKB1-dependent and phospholipase C/Ca2+/Ca2+/calmodulin-dependent protein kinase kinase-dependent pathways. J. Biol. Chem. 284, 22426–22435 10.1074/jbc.M109.028357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu G., Chen J., Liu J., Brunzelle J. S., Huang B., Wakeham N., Terzyan S., Li X., Rao Z., Li G.et al. (2007). Structure of the APPL1 BAR-PH domain and characterization of its interaction with Rab5. EMBO J. 26, 3484–3493 10.1038/sj.emboj.7601771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoncu R., Perera R. M., Balkin D. M., Pirruccello M., Toomre D., De Camilli P. (2009). A phosphoinositide switch controls the maturation and signaling properties of APPL endosomes. Cell 136, 1110–1121 10.1016/j.cell.2009.01.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.