Summary
Dynamic interactions with the cytoskeleton drive the movement and positioning of nuclei in many cell types. During muscle cell development, myoblasts fuse to form syncytial myofibers with nuclei positioned regularly along the length of the cell. Nuclear translocation in developing myotubes requires microtubules, but the mechanisms involved have not been elucidated. We find that as nuclei actively translocate through the cell, they rotate in three dimensions. The nuclear envelope, nucleoli and chromocenters within the nucleus rotate together as a unit. Both translocation and rotation require an intact microtubule cytoskeleton, which forms a dynamic bipolar network around nuclei. The plus- and minus-end-directed microtubule motor proteins, kinesin-1 and dynein, localize to the nuclear envelope in myotubes. Kinesin-1 localization is mediated at least in part by interaction with klarsicht/ANC-1/Syne homology (KASH) proteins. Depletion of kinesin-1 abolishes nuclear rotation and significantly inhibits nuclear translocation, resulting in the abnormal aggregation of nuclei at the midline of the myotube. Dynein depletion also inhibits nuclear dynamics, but to a lesser extent, leading to altered spacing between adjacent nuclei. Thus, oppositely directed motors acting from the surface of the nucleus drive nuclear motility in myotubes. The variable dynamics observed for individual nuclei within a single myotube are likely to result from the stochastic activity of competing motors interacting with a complex bipolar microtubule cytoskeleton that is also continuously remodeled as the nuclei move. The three-dimensional rotation of myotube nuclei may facilitate their motility through the complex and crowded cellular environment of the developing muscle cell, allowing for proper myonuclear positioning.
Key words: Dynein, Kinesin, Microtubule, Muscle, Nuclei
Introduction
Nuclear movement and positioning in many systems is controlled by connections between the nuclear envelope and the cytoskeleton (reviewed by Starr & Fridolfsson, 2010). Microtubule-dependent nuclear movement is particularly important for cells as they migrate and differentiate during development. For example, nuclear movement is tightly coupled to neuronal migration during brain development and mutations that disrupt this movement result in severe developmental defects such as lissencephaly (reviewed by Kuijpers and Hoogenraad, 2011). In neurons, nuclear migration requires the minus-end-directed microtubule motor protein, dynein. Dynein walks the nucleus toward the centrosome along a microtubule cage that surrounds the nucleus (Xie et al., 2003; Shu et al., 2004; Tsai et al., 2007). In contrast, as nuclei migrate through hypodermal precursor cells in the developing C. elegans embryo, kinesin-1 is the predominant motor moving the nucleus toward the plus-end of a polarized parallel non-centrosomal bundle of microtubules, with dynein driving small backsteps along this network (Fridolfsson and Starr, 2010). In these examples, both the polarity of the microtubule network and the type of motors present on the nuclear surface determine the overall direction of nuclear translocation.
Proper nuclear positioning is also critical in skeletal muscle cells. Mammalian skeletal muscle fibers are large multinucleated cells formed by the fusion of hundreds of post-mitotic mononucleated myocytes. Adult muscle fibers can extend many centimeters in length and, except for a cluster of specialized nuclei at the neuromuscular junction, the nuclei are found at the periphery of the cell, evenly spaced along the long-axis of the fiber (Bruusgaard et al., 2003; Kummer et al., 2004). This positioning is thought to ensure sufficient transcriptional capacity as well as to minimize transport distances between the nuclei and the cytoplasm in these extraordinarily long cells (Bruusgaard et al., 2003). Abnormally clustered nuclei have been found in patients with autosomal dominant Emery-Dreifuss muscular dystrophy (Mattioli et al., 2011), suggesting that correct nuclear positioning may be required for proper muscle function.
Nuclei in developing chick myotubes have been observed to translocate along the long axis of the cell (Cooper and Konigsberg, 1958; Capers, 1960; Cooper and Konigsberg, 1961). Although this translocation was shown to be dependent on an intact microtubule cytoskeleton (Englander and Rubin, 1987), the mechanisms that drive this translocation have not yet been explored.
Early studies in primary myotubes suggested that nuclei may rotate as they translocate (Cooper and Konigsberg, 1958; Capers, 1960; Cooper and Konigsberg, 1961). Nuclear rotation has been explored in cultured fibroblasts, where nuclei rotate in two dimensions, within the plane of substrate attachment (Ji et al., 2007; Levy and Holzbaur, 2008). This rotation occurs more frequently in migrating cells and is driven by dynein motors (Levy and Holzbaur, 2008). The function of rotation in migrating fibroblasts is still unclear, but has been suggested to be important in maintaining nuclear centrality. Given the length and complexity of the myotube, it is possible that nuclear dynamics during development, including both translocation and rotation, are essential for proper distribution of nuclei in the mature muscle fibers; however, this has not yet been examined.
In this study, we use live cell microscopy to examine the dynamics of nuclear movement in developing C2C12 myotubes, a well-established model system that faithfully replicates most features of early myogenesis and myofibril assembly, with cytoskeletal organization and dynamics closely resembling that of developing myotubes in vivo (reviewed by Sanger et al., 2010). We find that nuclei translocate within myotubes and also display robust three-dimensional rotation. Kinesin-1 and dynein both localize to the nuclear surface, likely mediated by interactions with the klarsicht/ANC-1/Syne homology (KASH) proteins. While both motors contribute to nuclear dynamics, kinesin-1 is the more dominant motor in this system. Loss of either kinesin or dynein causes abnormal aggregation and inappropriate dispersal of nuclei in myotubes, indicating that normal nuclear dynamics are essential for the proper distribution of nuclei in developing muscle cells.
Results
Nuclei both translocate and rotate in three dimensions in developing myotubes
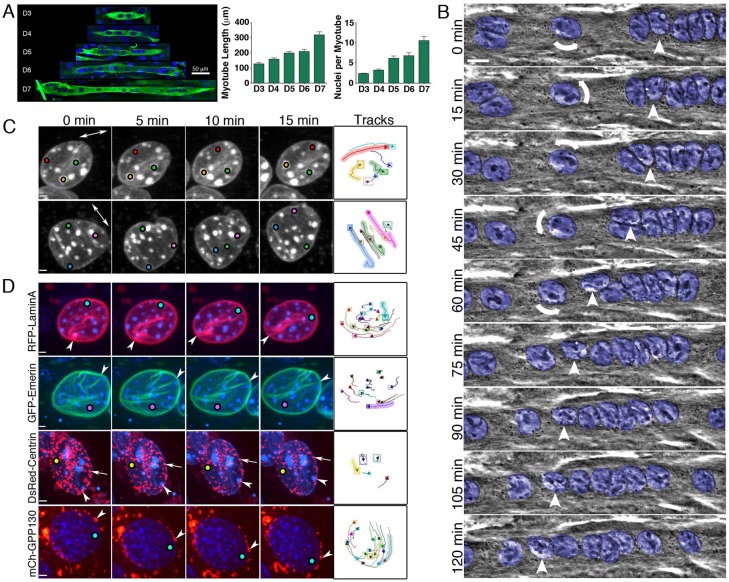
Myotubes are formed as myoblasts fuse to generate multinucleated syncytial cells. As additional myoblasts fuse, the length of the cell increases concomitant with an increase in the number of nuclei per myotube (Fig. 1A). Most myotubes display a relatively even distribution of nuclei throughout the length of the cell (Fig. 1A), with the average distance between nuclei in myotubes 7 days post differentiation (D7) found to be 22.7±0.95 µm (mean ± s.e.m.; n = 630 nuclei in 66 myotubes).
Fig. 1.
Nuclei actively rotate and translocate during myotube differentiation. (A) Representative images of myotubes differentiated for 3–7 days in vitro (D3–D7). Nuclei were stained with Hoechst dye; α-actinin is in green. Images are maximum projections of confocal z-series. Myotube length and number of nuclei per myotube are shown (mean ± s.e.m.; n>30 myotubes). Scale bar: 50 μm. (B) Phase-contrast images from a time-lapse sequence of nuclear migration in a D7 myotube (supplementary material Movie 1). The long-axis of the myotube runs in the horizontal direction; all visible nuclei in the myotube have been pseudo-colored blue. The arrow indicates the counter-clockwise rotation of a nucleus, visible with phase-contrast optics by watching prominent nucleoli; the arrowhead follows the path of a nucleus as it deforms and passes an adjacent nucleus. Scale bar: 10 µm. (C) Representative examples of nuclear rotation (supplementary material Movie 2). DNA was labeled with Hoechst dye and maximum projections of confocal z-stacks are shown over time. Bidirectional arrow indicates the long-axis of the myotube and colored dots label bright chromocenters to aid visualization of rotation. (D) Nuclei in myotubes expressing fluorescent constructs to label the nuclear lamina (LaminA), inner nuclear membrane (emerin), outer nuclear membrane (centrin-2) or Golgi (GPP130) are shown (supplementary material Movie 3). Colored dots label bright chromocenters in each nucleus; fiduciary marks of nuclear envelope-associated structures are demarcated with arrowheads and arrows. Chromocenters were tracked in the X, Y and Z planes over time. The right panels in C and D show representative tracks of individual chromocenters during rotation of the nuclei. Shaded tracks correspond to the chromocenters labeled with colored dots in the time series. Scale bars: 2 µm.
We imaged C2C12 myotubes with time-lapse phase-contrast microscopy and found that nuclei display robust and complex dynamics. Nuclei translocate along the length of the developing cell (Fig. 1B; supplementary material Movie 1) at an average rate of 11.7±7.8 µm/hr (0.2±0.13 µm/min; mean ± s.d.), similar to initial observations of nuclear translocation in primary myotubes (Englander and Rubin, 1987). Average velocities remained relatively constant in myotubes differentiated for 4–7 days in culture. We focused primarily on myotubes differentiated 7 days in order to study dynamics in cells that had undergone substantial myofibrillogenesis; past this time point myotubes begin to form branched networks and contract too strongly for accurate analysis.
Most nuclei in myotubes are mobile and exhibit complex dynamics, characterized by episodic, rather than continuous, movement. Translocation velocities of individual nuclei can vary greatly over the course of >3 hours of imaging (Fig. 1B; supplementary material Movie 1). Nuclei migrate in either direction along the long-axis of the cell; individual nuclei will occasionally change direction. Nuclei within the same myotube can move individually, or in groups, and nuclei can pass one another as they translocate (Fig. 1B, arrowhead), suggesting that the dynamics of each nucleus is influenced, but not dependent, on those nearby.
Remarkably, nuclei also rotate in three dimensions, as assessed by monitoring the movement of nucleoli in time-lapse phase-contrast imaging (Fig. 1B, arrow; supplementary material Movie 1). To monitor this rotation more precisely, we labeled the DNA with a live-cell Hoechst dye, and used confocal microscopy to track the positions of the brightly stained chromocenters in focal stacks over time (Fig. 1C; supplementary material Movie 2). Within an individual nucleus, the positions of chromocenters remain fixed relative to one another over the time course of observation (15 min), allowing accurate tracking of their trajectories in XYZ space.
Nuclei rotate clockwise or counterclockwise about the axes perpendicular to the long axis of the myotube. Occasionally individual nuclei change their direction of rotation (supplementary material Movie 1). The extent and rate of rotation varies considerably, with 50–60% of nuclei rotating more than 2°/min, some reaching velocities as high as 14.4°/min (Fig. 1C, bottom panel). Isolated nuclei, as well as nuclei moving in concert with others, can rotate; the rate and direction of rotation for individual nuclei appear to be independent of the dynamics of neighboring nuclei. Rotation is usually accompanied by translocation. The ability of nuclei to pass one another closely correlates with nuclear rotation. Analysis of time series from 75 myotubes indicates that in 29 pass events, all involved rotation of at least one nucleus, and in 93% of these events, both nuclei rotated. The observed rotation during passing is often accompanied by deformation of nuclear shape (Fig. 1B; supplementary material Movie 1).
When nuclei rotate in two dimensions in fibroblasts, nucleoli within the interior of the nucleus rotate with the nuclear lamina and nuclear envelope as an intact structure (Ji et al., 2007; Levy and Holzbaur, 2008). To determine whether this is also true for nuclei rotating in three dimensions, we transfected myotubes with RFP–laminA to label the nuclear lamina, co-labeled the DNA with Hoechst dye to observe chromocenters and examined nuclear rotation using time-lapse confocal microscopy. Comparison of chromocenter movements with fiduciary marks in the RFP–laminA labeling show that the nuclear lamina is rotating along with the chromatin (Fig. 1D; supplementary material Movie 3). Expression of a GFP-tagged emerin (GFP–emerin) construct reveals that the inner membrane of the nucleus also rotates in tandem with the Hoechst-labeled chromocenters.
To determine whether structures associated with the outer nuclear membrane also move in conjunction with the envelope, we expressed either a DsRed-Centrin-2 or mCherry-GPP130 construct and observed the dynamics of the labeled nuclei. We found that the centrosomal protein centrin-2 localizes in puncta both in the cytoplasm and at the outer surface of the nucleus, as described for other centrosomal proteins in myotubes (Bugnard et al., 2005; Srsen et al., 2009). DsRed–Centrin-2 likely identifies sites of microtubule nucleation, known to occur from the surface of the nuclei in these cells (Tassin et al., 1985; Bugnard et al., 2005; Zaal et al., 2011). Tracking shows that individual centrin-2 puncta rotate with the nucleus (Fig. 1D; supplementary material Movie 3). We also found that the Golgi, labeled with mCherry-tagged Golgi matrix protein, GPP130 (Linstedt et al., 1997), rotates with the nucleus (Fig. 1D; supplementary material Movie 3). Together, these experiments reveal that the nuclear interior and nuclear envelope rotate together as a unit in three dimensions, along with associated organelles including the Golgi.
As myotubes develop in culture, we noted a dramatic change in the shape of nuclei in myoblasts compared to those in myotubes (Fig. 2A; supplementary material Fig. S1). While nuclei in mononucleated myoblasts resemble the disc-shaped structures seen in fibroblasts, in myotubes the nuclei are more spherical (supplementary material Fig. S1). This change in shape correlates with changes in nuclear rotation. The flattened nuclei in myoblasts occasionally exhibit two-dimensional rotation in the plane of the substratum, which resembles the nuclear rotation described in fibroblasts (Ji et al., 2007; Levy and Holzbaur, 2008), and is distinct from the three-dimensional rotation observed in more fully developed myotubes.
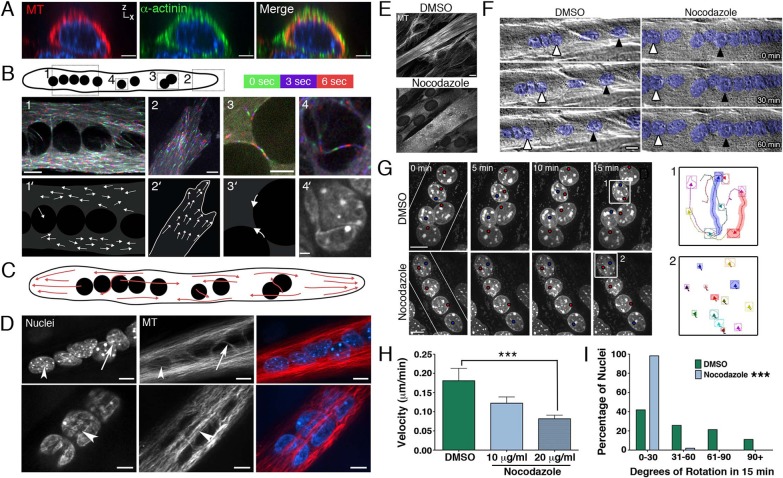
Fig. 2.
The microtubule cytoskeleton forms a dynamic network around nuclei in myotubes and is required for both nuclear translocation and rotation. (A) Image of a myotube in cross section stained with antibodies to tubulin (MT, red) and α-actinin (green). Nuclei are labeled with Hoechst dye (blue). Microtubules in myotubes run parallel to the developing myofibril network, forming a tube around the centrally located nuclei. Shown is a single Y plane in XZ. Scale bars: 5 µm. (B) Myotubes were transfected with GFP-EB3 to label the growing plus-ends of microtubules (supplementary material Movies 4, 5). Images were obtained every 3 seconds and three sequential frames were pseudo-colored with green, purple and red, respectively, and overlaid (1–4) to show the direction of microtubule growth. 1′–3′ depict the direction of representative microtubule growth. 4′ shows the invagination in the nuclear surface. The DNA was labeled with Hoechst dye. Single Z plane in XY. Scale bars: 10 μm (panel 1); 5 μm (panels 2 and 3); 2 μm (panel 4′). (C) Model of microtubule orientation within a myotube. (D) Microtubules lie between adjacent nuclei (arrow) and within invaginations (arrowheads) on the nuclear surface. Cells were stained for tubulin (red) and nuclei were labeled with Hoechst dye (blue). Single Z plane in XY. Scale bars: 10 µm. (E) Representative images of myotubes and surrounding myoblasts treated with DMSO or nocodazole (10 µg/ml) for 30 min at 37°C. Cells were stained with antibodies to tubulin. Scale bar: 20 µm. (F) Phase-contrast images from time-lapse sequences of nuclear migration in myotubes treated with DMSO (left) or 10 µg/ml nocodazole (right). The long-axis of the myotubes run in the horizontal direction and all visible nuclei in myotubes have been pseudo-colored blue. Arrowheads follow the migration of representative nuclei. Scale bar: 10 µm. (G) Representative examples of nuclear rotation in myotubes treated with DMSO (top panel) or 10 µg/ml nocodazole (bottom panel: supplementary material Movie 6). DNA was labeled with Hoechst dye and maximum projections of confocal z-stacks are shown over time. White lines depict the orientation of the myotube and colored dots label bright chromocenters to aid visualization. Right panel shows representative chromocenter tracks during rotation of the boxed nuclei on the left. Shaded tracks correspond to the chromocenters labeled with colored dots in the time series. Scale bars: 10 µm. (H) Velocity of nuclear translocation following treatment of myotubes with DMSO or nocodazole (mean ± s.e.m.; ***P<0.001, ANOVA; n>30 nuclei in five to seven myotubes). (I) Quantification of the degree of nuclear rotation in treated myotubes (***P<0.001, χ2-test; shown are pooled data for n>100 nuclei in 20–22 myotubes).
Microtubules are necessary for both nuclear translocation and rotation in myotubes
During myogenesis, the microtubule cytoskeleton is reorganized from the radial, centrosomal array found in mononucleated myoblasts to a linear array oriented along the long-axis of multinucleated myotubes (Warren, 1974; Pizon et al., 2005). In cross section, both the microtubules and the myofibrils surround the centrally located nuclei (Fig. 2A), with the microtubule array located more toward the interior, consistent with studies showing that nascent myofibrils form initially at the cell cortex and move inward (reviewed by Sanger et al., 2010).
To explore the polarity and dynamics of the microtubule array, we labeled the plus-tips of microtubules with a fluorescently-tagged end-binding protein (GFP–EB3) that tracks with the growing end of the microtubule (Akhmanova and Steinmetz, 2008). At the ends of the myotube, microtubules are generally oriented with their plus-ends toward the cortex (Fig. 2B2,2′,C; supplementary material Movie 4). In contrast, in the center of the cell, we observe an anti-parallel microtubule organization along the long axis of the cell (Fig. 2B1,1′,C; supplementary material Movie 4), also described by (Pizon et al., 2005; Zhang, T. et al., 2009). Most cells showed a small overall bias in microtubule growth toward one end of the myotube (58.6±2.4 vs 41.4±2.4; mean ± s.e.m.) for dominant versus non-dominant direction, respectively). Often there were areas within a myotube that exhibited highly polarized growth, raising the possibility that local differences in microtubule polarity may influence microtubule-dependent nuclear movement.
Microtubules are often found in close apposition to the nucleus. Microtubules lie between adjacent nuclei (Fig. 2D, top panel, arrow) as well as within invaginations, or grooves, on the nuclear surface (Fig. 2D, arrowheads). The deeper, wider grooves contain both microtubules (Fig. 2D, bottom panel) and myofibrils (not shown), whereas only microtubules are found in smaller invaginations (Fig. 2D, top panel, arrowhead). Microtubules in close contact with the nuclear surface are dynamic. Microtubules can polymerize along the curve of the nuclear surface (Fig. 2B3,3′, top arrow; supplementary material Movie 5), between two nuclei (bottom arrow), and within invaginations (Fig. 2B4,4′; supplementary material Movie 5). Together, these observations indicate that the microtubule network is in close association with the nuclei in myotubes, forming a complex and constantly changing network around each nucleus.
Microtubule depolymerization effectively inhibits nuclear translocation in primary myotubes, whereas the inhibition of actin dynamics had no effect (Englander and Rubin, 1987). In C2C12 cells, we found that treatment of myotubes with concentrations of nocodazole that depolymerize the microtubule network (Fig. 2E) significantly reduced the rate of nuclear translocation when compared to the DMSO-treated control myotubes (0.082±0.009 µm/min vs 0.181±0.032 µm/min, respectively; mean ± s.e.m., P<0.001; Fig. 2F,H). Additionally, nuclear rotation was abolished in myotubes treated with 10 µg/ml nocodazole (Fig. 2G,I; supplementary material Movie 6). Thus, microtubules are necessary for both nuclear translocation and rotation.
While an intact microtubule network is necessary for nuclear dynamics, nuclei are still able to translocate and rotate in myotubes treated with a lower dose of nocodazole (100 ng/ml; supplementary material Movie 7). This concentration inhibits microtubule growth (supplementary material Movie 7, central panel), but does not eliminate the microtubule network, suggesting that dynamic microtubules are not essential for nuclear translocation or rotation.
Kinesin-1 drives nuclear translocation and rotation in myotubes
In other systems displaying microtubule motor-dependent nuclear translocation and/or rotation such as migrating neurons and fibroblasts, the motors act from the surface of the nucleus (Levy and Holzbaur, 2008; Zhang, X. et al., 2009). To determine whether the kinesin-1 motor localized to the nuclear envelope, we performed immunofluorescence on D7 C2C12 myotubes. Antibodies directed against kinesin light chains 1 and 2 (KLC1/2) and kinesin heavy chain (KHC, mAb 1614) abundantly decorate the nuclear surface (representative images are shown in Fig. 3A). A similar pattern was found when myotubes were stained with a second pan-KHC antibody (SUK4 mAb, not shown). We also found that transfection of myotubes with a GFP-tagged KIF5C tail domain construct, which binds to KHC partner proteins, including KLC1/2 (Bi et al., 1997; Konishi and Setou, 2009), led to uniform decoration of the nucleus envelope (Fig. 3A).
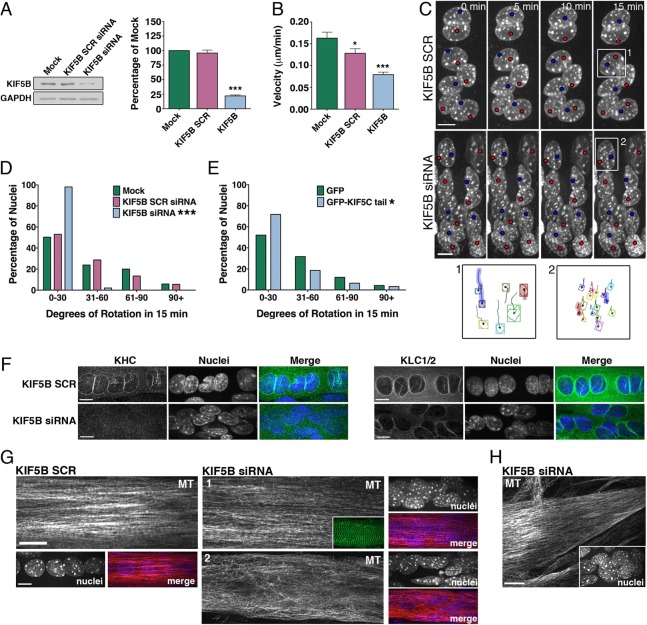
Fig. 3.
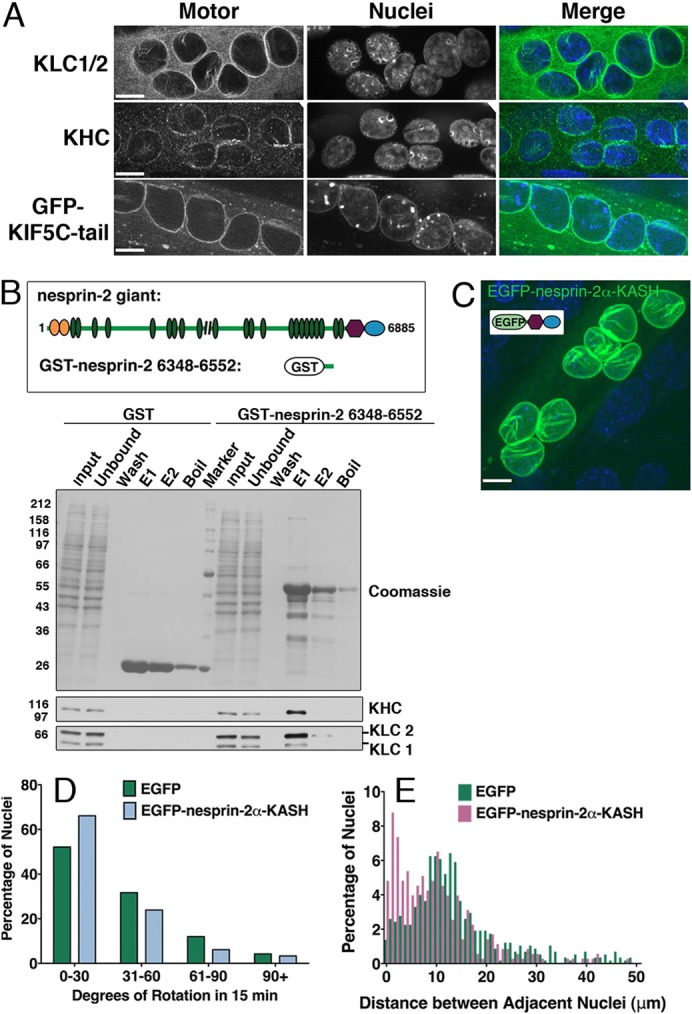
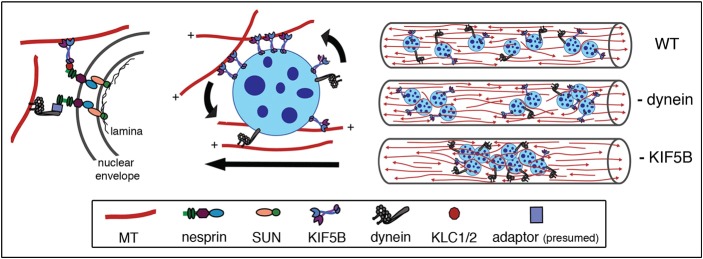
Kinesin-1 localizes to the nuclear envelope in myotubes and binds to the LINC protein, nesprin-2. (A) Myotubes were stained for subunits of the plus-end-directed microtubule motor kinesin-1 [kinesin light chains 1/2 (KLC1/2); kinesin heavy chain (KHC)]. Also shown is a myotube transfected with EGFP-KIF5C tail, the domain of KHC that interacts with KLC1/2. Myotubes were fixed with methanol for KLC1/2 and KHC. DNA was labeled with Hoechst dye. Scale bar: 10 µm. (B) Schematic of nesprin-2giant and GST–nesprin-26348–6552 (orange, green, purple and blue shapes represent the actin-binding domain, spectrin repeats, transmembrane domain and KASH domain, respectively). Pull-down assays with GST–nesprin26348–6552 or GST beads incubated with C2C12 myotube lysate. Bound protein was eluted first with 80 mM glutathione (two rounds: E1,E2); beads were then boiled in denaturing buffer (Boil). Blots were probed with antibodies to KLC1/2 (63–90 mAb) and KHC (SUK4 mAb); blot shown is representative of three independent replicates. (C) Representative image of a myotube expressing EGFP–nesprin-2α–KASH (maximum projection of a confocal z-series; DNA was labeled with Hoechst dye). Scale bar: 10 µm. (D) Quantification of the degree of nuclear rotation in myotubes treated with EGFP or EGFP–nesprin2α–KASH (χ2-test; P = 0.18; shown are pooled data for n>140 nuclei from 19–21 myotubes). (E) Frequency distributions of the distance between adjacent nuclei in myotubes treated with EGFP–nesprin-2α–KASH (1 µm bin width; less than 8% of the data lie above 50 µm, so distribution is truncated at 50 µm for clarity, n>382 nuclei in >29 myotubes).
KASH proteins mediate links between the nucleus and the cytoskeleton in many systems (reviewed by Starr and Fridolfsson, 2010), and the KASH protein nesprin-2 has recently been shown to bind to KLC (Schneider et al., 2011). To examine this interaction in myotubes, we performed GST pulldown assays from C2C12 myotube lysate with the minimal KLC binding domain of nesprin-2, residues 6348–6552 and observed specific binding of KLC1, KLC2 and KHC to the nesprin-2 construct (Fig. 3B). This result suggests that nesprin-2 mediates, at least in part, the localization of kinesin-1 to the nuclear envelope in myotubes.
To further test this possibility, we expressed the KASH domain of nesprin-2α (EGFP–nesprin-2α–KASH) in myotubes and assessed its effect on nuclear rotation. This domain localizes specifically to the nuclear envelope in myotubes (Fig. 3C) and acts as a dominant negative by displacing the endogenous nesprins from the nuclear membrane (Zhen et al., 2002; Grady et al., 2005). We found that expression of EGFP–nesprin-2α–KASH partially displaced kinesin-1 from the nuclear envelope in myotubes (supplementary material Fig. S2). Although this displacement was not complete, we found that EGFP–nesprin-2α–KASH-positive nuclei rotated less than those in control cells (Fig. 3D). Furthermore, expression of the dominant negative construct induced aggregation of nuclei along the myotube, with a decreased mean distance between adjacent nuclei (13.0±0.9 µm vs 19.3±0.9 µm in EGFP–nesprin-2α–KASH-positive myotubes as compared to control myotubes expressing EGFP, respectively, mean ± s.e.m., P<0.0001; mean myotube length was not different; Fig. 3E).
In order to more directly determine whether kinesin-1 drives translocation and/or rotation in myotubes, we reduced the expression of the motor with siRNA and evaluated the resulting effects on nuclear dynamics. Myotubes were transfected with siRNA against KIF5B, which is the only kinesin-1 isoform expressed in skeletal muscle (Kanai et al., 2000), on day 4 of differentiation and nuclear dynamics were assessed 72 hours later in D7 myotubes. We were able to consistently reduce the levels of KIF5B to ∼20% of expression levels in mock siRNA-treated myotubes (Fig. 4A).
Fig. 4.
KIF5B acts from the nuclear surface to drive nuclear translocation and rotation in myotubes. Myotubes were treated with KIF5B siRNA or with scrambled control oligos (SCR). (A) Representative immunoblots and quantification of KIF5B in myotubes treated with siRNA and SCR siRNA for KIF5B; GAPDH served as a loading control (mean ± s.e.m.; ANOVA, n = 3 independent replicates). (B) Mock- or siRNA-treated myotubes were imaged with phase-contrast microscopy for 60 min and absolute velocity was quantified (mean ± s.e.m.; ANOVA; n>50 nuclei in 10–12 myotubes). (C) Representative examples of nuclear rotation in myotubes treated with KIF5B SCR siRNA or KIF5B siRNA (supplementary material Movie 8). DNA was labeled with Hoechst dye and maximum projections of confocal z-stacks are shown over time. Red and blue dots label chromocenters to aid visualization. The panels below show representative chromocenter tracks during rotation of the boxed nuclei. Shaded tracks correspond to the chromocenters labeled with colored dots in the time series. Scale bars: 10 µm. (D,E) Quantification of the degree of nuclear rotation in myotubes treated with siRNA (D) or GFP-KIF5C tail dominant-negative construct to disrupt KIF5B localization and/or function (E) (χ2-test on pooled data for n>120 nuclei from 20–27 myotubes). (F) Treated myotubes were stained for subunits of the plus-end-directed microtubule motor kinesin-1 [kinesin heavy chain (KHC); kinesin light chains 1/2 (KLC1/2)]. DNA was labeled with Hoechst dye. Scale bars: 10 µm. (G,H) Treated myotubes were stained for tubulin; DNA was labeled with Hoechst dye. Representative images from the central region of the myotubes are shown in G (inset in siRNA panel 1 shows α-actinin staining). Representative image from the end of a KIF5B-siRNA-treated myotube is shown in H (inset shows the aggregation of nuclei from the center region of this myotube, which is to the left of the region of the myotube shown). Scale bars: 10 µm. For all panels, *P<0.05, **P<0.01, ***P<0.001 vs Mock or GFP.
The mean nuclear translocation velocity in KIF5B-deficient cells was reduced to 48.7±3.5% (mean ± s.e.m.) of mock-treated myotubes (Fig. 4B). Although we did note a small decrease in the mean rate of translocation in the KIF5B scrambled control siRNA-treated (SCR) myotubes, the dynamics of nuclear movement in KIF5B-deficient myotubes were distinctly different than those in either the mock or KIF5B SCR control myotubes. Nuclei tended to pile on top of one another and squeeze together as a unit in the KIF5B-deficient myotubes rather than exhibiting persistent, independent translocation.
We found that depletion of KIF5B also led to a striking reduction in nuclear rotation (Fig. 4C,D; supplementary material Movie 8), similar to the effect of nocodazole (Fig. 2I). In a parallel experiment, we found that inhibiting kinesin-1 using a dominant negative GFP-KIF5C tail-domain construct, which acts to displace endogenous motors from cargos (Bi et al., 1997; Konishi and Setou, 2009), also induced a decrease in nuclear rotation (Fig. 4E).
Together, these observations suggest that KIF5B drives nuclear dynamics by acting from the surface of the nucleus. Consistent with this hypothesis, we found that KHC is no longer localized to the nucleus in cells treated with siRNA-treated cells, although some residual signal for KLC1/2 persists (Fig. 4F). However, it is also possible that loss of the motor alters microtubule organization in myotubes, which may in turn impact on the dynamics of the nuclear movement. In >50% of KIF5B-deficient myotubes, the microtubule network was not different from that seen in control cells (Fig. 4G, SCR and siRNA panel 1; 58% of myotubes, n = 50). In 42% of KIF5B-depleted cells, microtubules tended to form a more complex network in the center of the myotube, but only in the vicinity of nuclei. In these myotubes, microtubules appeared to wrap more frequently around the nuclei, with the direction of GFP–EB3 comets relative to the long-axis of the myotube more variable (Fig. 4G, siRNA panel 2; supplementary material Movie 9). However, in regions devoid of nuclei such as myotube ends, microtubule organization and polarity were indistinguishable from control myotubes (Fig. 4H; supplementary material Movie 9). We also noted KIF5B-deficient myotubes can develop organized myofibrils, as judged by YFP–α-actinin localization in D7 C2C12 cells (Fig. 4G, siRNA panel 1 inset). Kinesin-1-depleted myotubes were observed to contract spontaneously, again suggesting that these cells develop functional sarcomeres despite the mis-organization of the nuclei.
Dynein also contributes to nuclear translocation and rotation
Antibodies to both dynein and dynactin reveal that the minus end-directed microtubule motor complex is also enriched at the nuclear periphery of myotubes, in addition to a strong cytosolic localization of the motor consistent with a broad range of cellular functions (Fig. 5A). The localization of dynein and dynactin on the envelope was more irregular than that observed for KHC and KLC1/2 (Fig. 3A). This may reflect a limitation of epitope accessibility, or may suggest that the dynein complex is less homogenously distributed across the nuclear surface than the kinesin-1 motor.
Fig. 5.
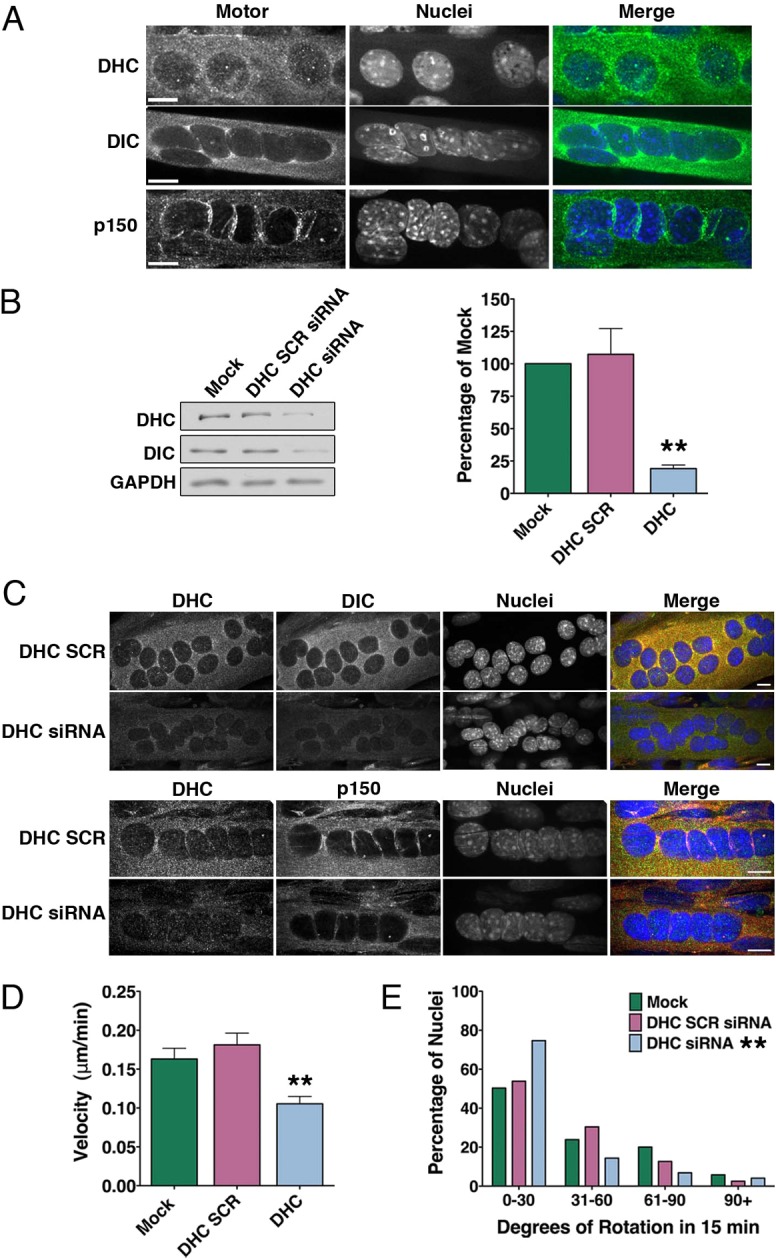
Dynein localizes to the nuclear envelope in myotubes and drives nuclear translocation and rotation. (A) Myotubes were stained for components of the minus-end-directed dynein/dynactin complex [dynein heavy chain (DHC); dynein intermediate chain (DIC); p150Glued subunit of dynactin]. Myotubes were fixed with methanol for p150 analysis, and with 1:1 acetone:methanol for DHC and DIC analysis. DNA was labeled with Hoechst dye. Scale bars: 10 µm. (B–D) Myotubes were treated with siRNA to DHC or with scrambled control oligos (SCR). (B) Representative immunoblots (left) and quantification (right) of dynein heavy chain (DHC) and dynein intermediate chain (DIC) in myotubes treated with siRNA for DHC; GAPDH served as a loading control (mean ± s.e.m.; ANOVA, n = 3 independent replicates). (C) Myotubes treated with DHC siRNA or DHC SCR siRNA were stained for DHC, DIC and p150Glued. DNA was labeled with Hoechst dye. Scale bars: 10 µm. (D) Mock- or siRNA-treated myotubes were imaged with phase-contrast microscopy for 60 min and absolute velocity was quantified (mean ± s.e.m.; ANOVA; n>50 nuclei in 10–12 myotubes). (E) Quantification of the degree of nuclear rotation in myotubes treated with DHC siRNA. Data for siRNA is compared to the same Mock data as in Fig. 4D (χ2-test; pooled data for n>120 nuclei from 20–27 myotubes). For all panels, **P<0.01.
To test the role of dynein in nuclear dynamics, we used siRNA to deplete the motor. We were able to consistently reduce the levels of DHC to ∼20% of expression levels in mock siRNA-treated myotubes after 72 hours of knock-down (Fig. 5B). DHC siRNA concomitantly reduces levels of DIC, suggesting that the dynein complex is destabilized, as has been observed in other cell types (Caviston et al., 2007; Levy and Holzbaur, 2008). Immunofluorescence analysis of myotubes treated with DHC siRNA demonstrate a depletion of DHC and DIC throughout the cell (Fig. 5C). Although western blot analysis indicates that overall expression levels of p150Glued were not reduced in cells treated with DHC siRNA (not shown), immunostaining for p150Glued suggests that dynactin may also be depleted from the nuclear envelope in dynein-depleted myotubes (Fig. 5C).
Knockdown of DHC reduced the average rate of nuclear translocation and rotation (Fig. 5D,E). However, the effect of dynein depletion on nuclear dynamics was not as striking as observed for the kinesin-1 motor. Mean nuclear translocation velocity in dynein-deficient cells was reduced to 64.7% of mock-treated myotubes (Fig. 5C), with 25% of the nuclei continuing to rotate more than 30° in 15 min, as compared to only 2% of nuclei in KIF5B-deficient myotubes (Fig. 5E, Fig. 4D). These data suggest that while both dynein and KIF5B contribute to nuclear dynamics, the plus-end-directed KIF5B is the more dominant motor.
Motor-dependent nuclear dynamics are necessary for proper nuclear distribution in myotubes
After 7 days of differentiation, myotubes display a relatively even distribution of nuclei along the length of control cells (Fig. 1A, Fig. 6A,B). However, we noted that the nuclei in myotubes deficient for KIF5B tended to densely aggregate at the midline of the cell (Fig. 6A,B). This aggregation was also observed in myotubes expressing the GFP–KIF5C tail construct. In myotubes treated with siRNA for DHC, we observed some nuclear aggregation, but these aggregates were not enriched toward the myotube center as seen with kinesin-1 disruption. Instead, clusters of nuclei were observed along the length of the myotube in dynein-depleted cells (Fig. 6A,B).
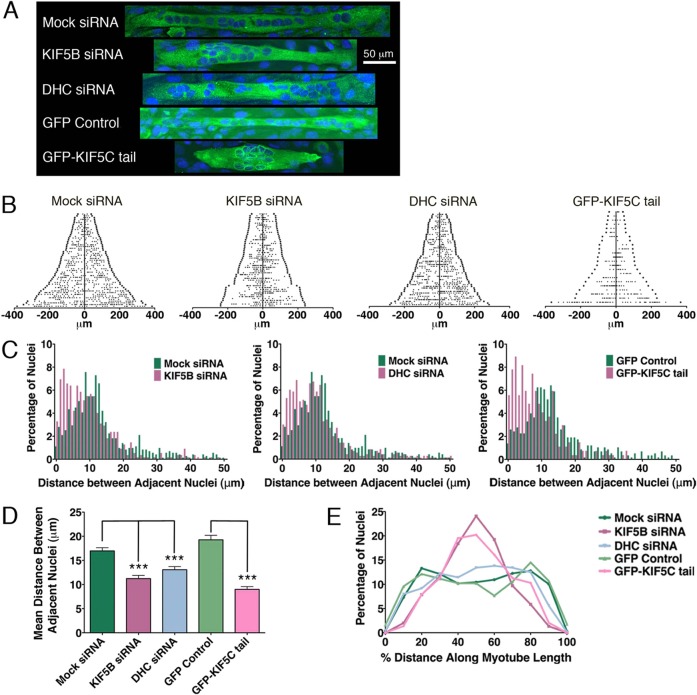
Fig. 6.
Motor-dependent nuclear dynamics are necessary for proper distribution of nuclei in myotubes. Myotubes were transfected with either siRNA + GFP, GFP or GFP-KIF5C tail. (A) Representative images of treated myotubes. Nuclei were stained with Hoechst dye; images are maximum projections of confocal z-series. (B) Distribution of nuclei in myotubes. Each line on the y-axis represents an individual myotube, organized according to length (n = 42–55 siRNA myotubes; 23 myotubes GFP-KIF5C tail). The ends of the myotube are marked with a dark square; data points represent individual nuclei. (C) Frequency distributions of the distance between adjacent nuclei in myotubes treated with siRNA or GFP-KIF5C tail (1 µm bin width; less than 8% of the data lies above 50 µm so distributions are truncated at 50 µm for clarity). (D) Mean distance between adjacent nuclei (mean ± s.e.m.; ANOVA, ***P<0.001). (E) Histogram depicting the position of nuclei as a percentage of the distance along the myotube length (bin width = 10%).
Mean myotube length was not different between control and treated myotubes, nor did we detect a difference in the number of nuclei per myotube. However, the distance between nuclei was noticeably reduced when either kinesin or dynein function was disrupted (Fig. 6C,D). The mean distance between adjacent nuclei was significantly smaller in KIF5B-depleted or dynein-depleted myotubes (Fig. 6D), and disruption of kinesin function using a dominant negative approach (expression of GFP–KIF5C tail) also significantly reduced the average distance between nuclei (Fig. 6D). These data indicate that proper nuclear dynamics are necessary to prevent aggregation of nuclei. Live cell data suggest that nuclei need to rotate in order to pass one another. As nuclei in myotubes treated with siRNA to KIF5B or dynein do not rotate normally (Figs 4, 5), reduced rotation may be sufficient to induce nuclear aggregation.
To further compare the effects of motor depletion or inhibition on nuclear localization, we plotted the position of nuclei as a function of distance along the myotube length (Fig. 6E). Nuclei in KIF5B-deficient myotubes clustered near the midline as compared to control myotubes. This same pattern was seen for cells treated with GFP–KIF5C tail. Conversely, despite their increased propensity to aggregate, the nuclei in dynein-deficient myotubes were found along the full myotube length, similar to the distribution of nuclei in the control cells (Fig. 6E). This suggests, that in addition to driving rotation and preventing aggregation, KIF5B is also necessary for proper translocation of the nuclei out toward the ends of these elongated cells.
Discussion
Nuclei in developing muscle cells are very mobile (Capers, 1960). We propose that this mobility is necessary to achieve an even distribution of nuclei along the length of mature myofibers. Consistent with this hypothesis, we find that the microtubule motor proteins kinesin-1 and dynein are required for normal nuclear dynamics in developing myotubes. Depletion or inhibition of these motors leads to nuclear aggregation and abnormal nuclear distribution. The motor proteins localize to the nuclear envelope, likely mediated at least in part by interactions with the KASH proteins of the LINC complex, and drive both nuclear translocation and rotation along the microtubule network.
The nuclear dynamics we observe in C2C12 myotubes are consistent with initial observations in primary chick myotubes (Cooper, 1958; Capers, 1960; Cooper and Konigsberg, 1961; Englander and Rubin, 1987). We find that nuclei translocate along the long-axis of the myotube, rotate in all dimensions and can even pass one another. Both primary and C2C12 myotubes exhibit similar developmental patterns to what has been described in vivo, including microtubule arrangement and myofibril assembly (Warren, 1974; Sanger et al., 2010). Thus, the nuclear dynamics described here likely inform on the dynamics that occur during vertebrate myotube development.
These nuclear dynamics require an intact microtubule network (Fig. 2). Nuclear rotation was abolished by microtubule depolymerization. While translocation was inhibited in nocodazole-treated cells, some residual nuclear movement suggests that there could be additional forces involved, such as contractions of the myofibril network. Myofibrils are often in close contact with nuclei and their contractions might also influence nuclear movement (supplementary material Movie 10). To investigate this possibility, we inhibited contraction with N-benzyl-ptoluene sulphonamide (BTS), a specific inhibitor of skeletal muscle myosin (Cheung et al., 2002), and found that the nuclei were still able to translocate and rotate (supplementary material Movie 11). Thus, myofibril contraction is likely to play only a minor role. During our phase-contrast imaging of myotube dynamics, we often observed bulk fluid dynamics within the cell that resemble the cytoplasmic streaming described in plants and Drosophila oocytes (Serbus et al., 2005; Shimmen, 2007) (supplementary material Movie 1). This flow may also contribute to nuclear dynamics and organelle distribution in myotubes, either directly, or indirectly by influencing the polarity of the microtubule network.
Our observations indicate that kinesin-1 is the primary motor driving nuclear dynamics in myotubes (Fig. 4), while dynein contributes to a lesser extent. Though dynein is the primary driver of nuclear movement in migrating neurons (Shu et al., 2004; Tsai et al., 2007; Vallee et al., 2009) and nuclear rotations in fibroblasts (Levy and Holzbaur, 2008), in C. elegans hypodermal precursor cells, kinesin-1 activity predominates (Fridolfsson and Starr, 2010). Kinesin has a significantly higher stall force than mammalian dynein (∼5–6 pN for kinesin vs 1.1 pN for dynein) (Hendricks et al., 2010). Thus, within the complex cellular environment of the myotube, which is densely packed with developing myofibrils, kinesin-1 may be a more effective driver of the motility of this large (∼10 µm diameter) organelle. The localization of both kinesin and dynein to the nucleus suggests that a population of motors acts from the nuclear surface to drive both nuclear translocation and rotation. Reduced nuclear rotation observed in myotubes expressing either the GFP–KIF5C tail or EGFP–nesprin-2α–KASH dominant-negative constructs also argues for nuclear-based motor activity, since the localization of these constructs to the nuclear envelope can displace endogenous motors.
KIF5B binds to the nuclear envelope in myotubes, at least in part, through interactions with nesprins (Fig. 3), outer nuclear membrane KASH proteins known to connect the nuclear lamina with the cytoskeleton (Starr and Fridolfsson, 2010). Our data confirm the interaction between kinesin-1 and nesprin-2, mediated by binding to kinesin light chain (Schneider et al., 2011); however, nesprin-1 may also contribute to kinesin recruitment to the nucleus. Dynein and dynactin have been shown to precipitate with nesprin-1 and -2 from mouse brain lysate (Zhang, X. et al., 2009), although dynein did not bind to the nesprin construct used here. The interaction between dynein and nesprins may be mediated through a different domain, or may be recruited to the nuclear envelope through other binding partners. Mature skeletal muscle fibers and C2C12 myotubes express both nesprin-1 and nesprin-2, with the relative expression of alternatively spliced isoforms modulated over the course of muscle development (Apel et al., 2000; Zhang et al., 2001; Zhang et al., 2005; Randles et al., 2010). Evidence from knock-out mice suggests that nesprin-1 and nesprin-2 are both critical in muscle for nuclear positioning and anchorage (Zhang, X. et al., 2007; Zhang et al., 2010). Moreover, mutations in both nesprin-1 and nesprin-2 have been found in patients with Emery-Dreifuss muscular dystrophy, suggesting that nuclear positioning and/or anchorage in skeletal muscle is essential for proper muscle function (Zhang, Q. et al., 2007).
We propose that nuclear-bound motors influence nuclear dynamics by exerting force on the local microtubule network. The net sum of all forces would dictate the direction and speed of nuclear rotation and translocation. Kinesin distribution at the nuclear envelope appears patchy, whereas the GFP–KIF5C tail construct decorates the envelope more evenly, suggesting that under normal conditions not all of the kinesin binding sites on the envelope are occupied. A non-uniform distribution of KIF5B on the envelope could create asymmetrical areas of force generation that may lead to nuclear rotation. The dynein motor complex also appears to be non-uniformly distributed, best illustrated by the staining for the dynactin subunit p150Glued, which shows enhanced accumulation between adjacent nuclei (Fig. 5A). An uneven distribution of oppositely directed motors is also likely to lead to force imbalances that further influence nuclear dynamics.
The microtubule network in myotubes forms a dynamic bidirectional array along the long-axis of the cell, surrounding the centrally located nuclei (Warren, 1974; Tassin et al., 1985) (see Fig. 2). However, microtubules are also found between adjacent nuclei and lie within invaginations on the nuclear surface. Most or all of these microtubules are dynamic and therefore the network surrounding a given nucleus is always changing, which will significantly influence how the motors on the surface of an individual nucleus contribute to nuclear dynamics. Most myotubes exhibited a slight overall bias in microtubule growth directed toward one end of the myotube. We also noticed that in many myotubes, the net direction of nuclear movement was toward one end of the cell. It is possible that a bias in the direction of overall microtubule growth correlates with the net direction of nuclear movement, especially given the recent finding that even subtle biases in microtubule polarity are sufficient to direct cargo transport (Parton et al., 2011). Therefore, it is likely that the nuclear dynamics are influenced both by motors bound to the nuclear envelope, and by dynamics and polarity of the microtubule network (see model, Fig. 7).
Fig. 7.
Model depicting the roles of opposing kinesin and dynein motors in the nuclear dynamics in developing myotubes. Motors are bound to the nucleus through nesprins or other links. The number and distribution of opposing motors on a nucleus and the polarity of the local microtubule network determine the direction and speed of rotation and translocation. Depletion of either dynein or KIF5B from myotubes causes abnormal aggregation and inappropriate dispersal of nuclei along the length of the myotube.
The distribution of nuclei along the length of C2C12 myotubes was most severely disrupted upon depletion of KIF5B, leading to nuclear aggregation near the midline of the myotube (Fig. 6). Similarly, loss of kinesin-1 function in Drosophila embryos also caused central aggregation of myonuclei in a recent study (Metzger et al., 2012). This is in line with our observation that loss of KIF5B more substantially inhibited nuclear translocation and rotation. We hypothesize that the rotation of nuclei is essential to fluidize the dynamics of these large organelles. The decreased mobility in KIF5B-deficient myotubes may inhibit nuclei from moving around obstacles in their path, including other nuclei, myofibrils, or even the dense microtubule network surrounding the nucleus. In live cell recordings, one or both of the nuclei involved in passing events rotated as they moved past one another, consistent with this hypothesis. Thus, loss of nuclear rotation in the kinesin-depleted myotubes may be a critical factor leading to nuclear aggregation at the cell center. Loss of dynein also caused aggregation of nuclei, although the nuclei were still able to distribute along the length of the myotube. This suggests that although kinesin may be the primary driver, dynein is also needed for proper nuclear spacing. Loss of dynein-driven mobility might cause nuclear traffic jams, which appear as localized aggregates at discrete sites along the cell length.
While motor-driven rotation and translocation from the nuclear surface appears to be the dominant driver of movement in this system, the process may have a feed-forward component. Microtubules in the myotube are nucleated in part from the surface of nuclei (Tassin et al., 1985; Zaal et al., 2011), polymerizing in all directions (Tassin et al., 1985). Thus nuclei actively contribute to the development of a bidirectional microtubule array in myotubes. Since loss of kinesin activity would be expected to be most significant at myotube ends where microtubules are polarized with plus-ends oriented outward, as kinesin is depleted, the nuclei begin to aggregate toward the center of the myotube, and microtubule organization in cell areas depleted of nuclei becomes more unipolar and less bipolar (Fig. 4H; supplementary material Movie 9). Further, the local microtubule network surrounding the aggregated nuclei becomes increasingly disordered; we observed a direct correlation between degree of nuclear clumping and local disorganization of the microtubule cytoskeleton, consistent with this model (Fig. 4G and data not shown). Together, these processes create a progressively more complex cytoskeletal network for the nuclei to navigate through, thereby further reducing nuclear translocation and rotation and further enhancing aggregation over time.
Thus, we propose that the dynamics of the nuclei in this system are stochastic, dictated by the organization and dynamics of the microtubule network surrounding an individual nucleus as well as the complement of motors on its surface. This allows for a fluid distribution of nuclei in the myotube; as differentiation continues, the distributed nuclei will become anchored in place under the sarcolemma in myofibers. It has been proposed that proper nuclear positioning is necessary to ensure sufficient transcriptional capacity and minimize transport distances in the myofiber (Bruusgaard et al., 2003), which likely affects muscle function. Indeed, nesprin-1 null mice, exhibit abnormal distribution of nuclei, which correlates with significantly decreased exercise capacity (Zhang et al., 2010). Similarly, Drosophila mutant larvae expressing mutant MAP7 display nuclear positioning defects during muscle development, which correlates with decreased locomotion (Metzger et al., 2012), again suggesting that normal nuclear distribution is required for normal muscle function.
Nuclear dynamics may also contribute to the ability of skeletal muscle fibers to repair themselves after injury. During fiber repair, myogenic cells fuse with the injured fiber, contributing a new nucleus (Chargé and Rudnicki, 2004). These nuclei are positioned at the periphery of the myofiber (Rich and Lichtman, 1989; Terada et al., 2010; Li et al., 2011), and it is likely that both nuclear translocation and rotation are required for this positioning. In diseases like the muscular dystrophies, where mechanically induced muscle damage occurs frequently, nuclear dynamics may be essential for proper fiber repair and subsequent muscle function.
Materials and Methods
Reagents
The following constructs were generously provided: pRFP-Lmna and pGFP-emerin (Howard Worman, Columbia University), mCherry-GPP130 (Adam Linstedt, Carnegie Mellon University), DsRed1-Centrin-2 (Joseph Gleeson, University of California San Diego), GFP-KIF5C tail (Mitsutoshi Setou, Hamamatsu University School of Medicine), EGFP-Nesprin2α-KASH (residues 483–542) (Catherine Shanahan, King's College London), YFP-α-actinin (Joeseph Sanger, SUNY Upstate Medical University), and GFP-EB3 (Anna Akhmanova, University Utrecht). siRNA oligos were directed against mouse KIF5B (Gene ID 16573; 5′-CAACAGACAUGUCGCAGUU-3′, scrambled control: 5′-GAACGAAUGUCGCUUACCAUU-3′), and mouse dynein heavy chain (GenBank NM_030238; 5′-GAAAUCAACUUGCCCGAUAUU-3′, scrambled control: 5′-GACCGAUAAUCAAACCUGUUU-3′). Primary antibodies include anti-α-tubulin (YL1/2, AdD Serotec), anti-GFP (GFP-1020, Aves Labs, Inc.), anti-DHC (R-325, Santa Cruz Biotechnology, Inc.), anti- KIF5B (ab15705, Abcam) anti-α-actinin (clone EA-53, Sigma-Aldrich), anti-KLC 1/2 (63–90, from Scott Brady), anti-KHC (MAB1614, clone H2, Millipore; SUK4, Abcam), anti-DIC (MAB1618, Millipore), anti-p150Glued (BD Biosciences), and anti-GAPDH (mAb 9484, Abcam). Alexa fluorophore-conjugated secondary antibodies were from Molecular Probes (Invitrogen) and Cy2-conjugated goat anti-chicken antibodies were from Jackson ImmunoResearch Laboratories, Inc.
Cell culture, transfections and drug treatment
Mouse C2C12 myoblasts (American Type Culture Collection, Manassas, VA, CRL-1772) were maintained at 37°C/5% CO2 in growth medium [Dulbecco's modified Eagle's medium (DMEM) with glutamax and 10% fetal bovine serum]. To induce myogenic differentiation, myoblasts were grown to ∼80% confluence and switched to DMEM with glutamax and 10% horse serum for 3–7 days. Medium was replaced every 24 hours. For live-cell imaging, cells were grown on 50 mm glass-bottomed dishes (FluorDish, World Precision Instruments) coated with 5 µg/cm2 rat tail collagen, type 1 (BD Biosciences). For immunofluorescence analyses, cells were grown on collagen-coated 1-cm2 squares of ACLAR embedding film (Ted Pella, Inc.). DNA transfections were performed with Lipofectamine 2000 (Invitrogen). For RNAi knockdown, cells were transfected with siRNA duplexes (Thermo Scientific Dharmacon) at a final concentration of 50 nM using Lipofectamine RNAiMax (Invitrogen). Unless otherwise noted, cells were transfected 4 days after induction of differentiation and imaged 72 hours post-transfection. Myotubes were treated with nocodazole at 10 or 20 µg/ml (Sigma-Aldrich) or DMSO control for 30 min at 37°C. Myotubes were imaged in the presence of 50 µM N-benzyl-ptoluene sulphonamide (BTS; Tocris Bioscience) in DMSO to inhibit myofibril contraction.
Myotubes grown on ACLAR film were fixed with 4% paraformaldehyde (PFA) and permeabilized with 0.1% Triton X-100. Alternatively, myotubes were fixed with ice-cold methanol with 1 mM EDTA or 1:1 acetone:methanol as noted. Fixed cells were incubated with primary and secondary antibodies as noted then stained with Hoechst 33342 (0.5 µg/ml) and mounted on 40 mm glass coverslips with ProLong Gold antifade reagent (Invitrogen).
siRNA knockdown was assessed by lysing cells in HEM lysis buffer with 1% Triton X-100 and protease inhibitors (Roche); total protein was measured using the BCA protein assay kit (Pierce), equal total protein was separated by SDS-PAGE and subjected to immunoblot analysis. Chemiluminescence was enhanced by SuperSignal West Pico Chemiluminescent Substrate (Thermo Scientific) and exposed to film. Densitometry was performed with ImageJ (NIH, Rockville, MD).
GST-pulldown assays
GST-Nesprin2 constructs from Angelika Noegel and Iakowos Karakesisoglou (Schneider et al. 2011) were expressed in BL21 DE3 E. coli and purified by binding to glutathione–Sepharose 4B (Amersham). C2C12 myotube lysates were prepared using HEM lysis buffer [50 mM HEPES, 25 mM NaCl, 1 mM EDTA, 1 mM MgCl2, pH 7.0 plus 0.1% Triton X-100 and protease inhibitors (Roche)]. Lysates were incubated overnight at 4°C with equal amounts of purified GST fusion proteins coupled to GST–Sepharose beads. Interacting proteins were eluted with 40 mM glutathione, pH 7.5, and analyzed by western blotting.
Microscopy
Live cell imaging was performed in Phenol-Red-free DMEM with 25 mM HEPES (Gibco), 10% horse serum, and 2 mM Glutamax, overlaid with mineral oil (Sigma-Aldrich). For nuclear translocation analyses, imaging was performed on a Leica DMI6000B inverted epifluorescence microscope using an Apochromatic 63× 1.4 NA oil-immersion objective with a 1.6× magnifier (Leica Microsystems) in an environmental chamber at 37°C. Digital images were acquired with a Hamamatsu ORCA-R2 charge-coupled device camera using LAS-AF software (Leica Microsystems). Phase-contrast images were taken every 30 sec for 60 or 180 min.
Fluorescence imaging was performed on a Perkin Elmer UltraView Vox Spinning Disk Confocal with a Nikon Ti Microscope equipped with PFS, a motorized stage, and 40×/1.30 NA, 60×/1.49 NA, and 100×/1.49 NA oil-immersion apochromatic objectives (Nikon) in an environmental chamber set to 37°C. Digital images were acquired with a Hamamatsu EMCCD C9100-50 camera and Volocity 3D Image Analysis Software (Improvision/Perkin Elmer). Where noted, DNA was labeled with Hoechst 33342 dye (0.5 µg/ml) for 20 min prior to imaging. Z-series, encompassing the depth of each myotube (∼15–30 µm), were obtained with a 0.5 µm step size. Images were taken at a rate of 1 z-series per minute for 15 minutes. For dual-color movies, z-series of each fluorophore were taken consecutively to minimize exposure times. GFP–EB3 analyses were performed using an Apochromatic 100× 1.49 NA oil-immersion objective (Nikon). Images were taken in a single z-plane at a rate of 1 frame every 3 seconds. Images of fixed myotubes were acquired at 40×, 60× or 100×. Z-series encompassing the entire depth of each myotube (∼15–30 µm) were taken at a step-size of 0.5 µm. For analysis of nuclear distribution, z-series (1 µm z-step) of adjacent areas were obtained at 40× with 20% overlap. Images were stitched together digitally in Volocity to obtain composite images of ∼1000×1000 µm.
Image analysis
To assess nuclear translocation, the centroid of each nucleus was manually tracked using LAS-AF software (Leica Microsystems); mean velocity reported reflects both translocating and non-moving nuclei. To quantify nuclear rotation, bright Hoechst-dye-positive chromocenters in individual nuclei were automatically tracked in XYZ over time using Volocity 3D Image Analysis Software (Improvision/Perkin Elmer). Principal component analysis was used to find the angular velocity and translational speed from the tracked chromocenter positions. Analysis routines were implemented in Matlab. For analyses of nuclear rotation in treated myotubes, angular velocity was first quantified in a subset of nuclei exhibiting a wide range of rotation. These nuclei then served as standards for comparison as rotation of the remaining nuclei was visually assessed and binned into 30° categories. The direction of GFP–EB3 comets was quantified from the resulting overlay images of three sequential pseudo-colored frames (ImageJ). Comet direction was evaluated with regard to the long-axis of the myotubes. Nuclear distribution was assessed by defining the position of the myotube ends and the centroid of each nucleus in the myotube in XY; only unbranched myotubes were included in the analysis. Nuclear position was linearized along the myotube length and distances between adjacent nuclei and nuclear position as a percent of the distance along the myotube length were calculated. All graphs and statistical analyses were performed using GraphPad Prism V5.0 (GraphPad Software, Inc.).
Supplementary Material
Acknowledgments
We gratefully acknowledge Mariko Tokito for technical assistance, and Adam G. Hendricks, Matthew G. Bray and Michael F. Howland for development of Matlab scripts.
Footnotes
Funding
This work was supported by the National Institutes of Health [grant number PO1 GM087253 to E.L.F.H., T32 GM-07229 and T32 AR-053461 to M.H.W.]. Deposited in PMC for release after 12 months.
Supplementary material available online at http://jcs.biologists.org/lookup/suppl/doi:10.1242/jcs.108688/-/DC1
References
- Akhmanova A., Steinmetz M. O. (2008). Tracking the ends: a dynamic protein network controls the fate of microtubule tips. Nat. Rev. Mol. Cell Biol. 9, 309–322 10.1038/nrm2369 [DOI] [PubMed] [Google Scholar]
- Apel E. D., Lewis R. M., Grady R. M., Sanes J. R. (2000). Syne-1, a dystrophin- and Klarsicht-related protein associated with synaptic nuclei at the neuromuscular junction. J. Biol. Chem. 275, 31986–31995 10.1074/jbc.M004775200 [DOI] [PubMed] [Google Scholar]
- Bi G. Q., Morris R. L., Liao G., Alderton J. M., Scholey J. M., Steinhardt R. A. (1997). Kinesin- and myosin-driven steps of vesicle recruitment for Ca2+-regulated exocytosis. J. Cell Biol. 138, 999–1008 10.1083/jcb.138.5.999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruusgaard J. C., Liestøl K., Ekmark M., Kollstad K., Gundersen K. (2003). Number and spatial distribution of nuclei in the muscle fibres of normal mice studied in vivo. J. Physiol. 551, 467–478 10.1113/jphysiol.2003.045328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugnard E., Zaal K. J., Ralston E. (2005). Reorganization of microtubule nucleation during muscle differentiation. Cell Motil. Cytoskeleton 60, 1–13 10.1002/cm.20042 [DOI] [PubMed] [Google Scholar]
- Capers C. R. (1960). Multinucleation of skeletal muscle in vitro. J. Biophys. Biochem. Cytol. 7, 559–566 10.1083/jcb.7.3.559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caviston J. P., Ross J. L., Antony S. M., Tokito M., Holzbaur E. L. (2007). Huntingtin facilitates dynein/dynactin-mediated vesicle transport. Proc. Natl. Acad. Sci. USA 104, 10045–10050 10.1073/pnas.0610628104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chargé S. B., Rudnicki M. A. (2004). Cellular and molecular regulation of muscle regeneration. Physiol. Rev. 84, 209–238 10.1152/physrev.00019.2003 [DOI] [PubMed] [Google Scholar]
- Cheung A., Dantzig J. A., Hollingworth S., Baylor S. M., Goldman Y. E., Mitchison T. J., Straight A. F. (2002). A small-molecule inhibitor of skeletal muscle myosin II. Nat. Cell Biol. 4, 83–88 10.1038/ncb734 [DOI] [PubMed] [Google Scholar]
- Cooper W. G., Konigsberg I R. (1958). Behavior of myoblasts in tissue culture. Program of the 9th Annual Meeting, Tissue Culture Association, Philadelphia, April 9–10 . 27 [Google Scholar]
- Cooper W. G., Konigsberg I. R. (1961). Dynamics of myogenesis in vitro. Anat. Rec. 140, 195–205 10.1002/ar.1091400305 [DOI] [PubMed] [Google Scholar]
- Englander L. L., Rubin L. L. (1987). Acetylcholine receptor clustering and nuclear movement in muscle fibers in culture. J. Cell Biol. 104, 87–95 10.1083/jcb.104.1.87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridolfsson H. N., Starr D. A. (2010). Kinesin-1 and dynein at the nuclear envelope mediate the bidirectional migrations of nuclei. J. Cell Biol. 191, 115–128 10.1083/jcb.201004118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grady R. M., Starr D. A., Ackerman G. L., Sanes J. R., Han M. (2005). Syne proteins anchor muscle nuclei at the neuromuscular junction. Proc. Natl. Acad. Sci. USA 102, 4359–4364 10.1073/pnas.0500711102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendricks A. G., Perlson E., Ross J. L., Schroeder H. W., 3rd, Tokito M., Holzbaur E. L. (2010). Motor coordination via a tug-of-war mechanism drives bidirectional vesicle transport. Curr. Biol. 20, 697–702 10.1016/j.cub.2010.02.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji J. Y., Lee R. T., Vergnes L., Fong L. G., Stewart C. L., Reue K., Young S. G., Zhang Q., Shanahan C. M., Lammerding J. (2007). Cell nuclei spin in the absence of lamin b1. J. Biol. Chem. 282, 20015–20026 10.1074/jbc.M611094200 [DOI] [PubMed] [Google Scholar]
- Kanai Y., Okada Y., Tanaka Y., Harada A., Terada S., Hirokawa N. (2000). KIF5C, a novel neuronal kinesin enriched in motor neurons. J. Neurosci. 20, 6374–6384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konishi Y., Setou M. (2009). Tubulin tyrosination navigates the kinesin-1 motor domain to axons. Nat. Neurosci. 12, 559–567 10.1038/nn.2314 [DOI] [PubMed] [Google Scholar]
- Kuijpers M., Hoogenraad C. C. (2011). Centrosomes, microtubules and neuronal development. Mol. Cell. Neurosci. 48, 349–358 10.1016/j.mcn.2011.05.004 [DOI] [PubMed] [Google Scholar]
- Kummer T. T., Misgeld T., Lichtman J. W., Sanes J. R. (2004). Nerve-independent formation of a topologically complex postsynaptic apparatus. J. Cell Biol. 164, 1077–1087 10.1083/jcb.200401115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy J. R., Holzbaur E. L. (2008). Dynein drives nuclear rotation during forward progression of motile fibroblasts. J. Cell Sci. 121, 3187–3195 10.1242/jcs.033878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Lee Y., Thompson W. J. (2011). Changes in aging mouse neuromuscular junctions are explained by degeneration and regeneration of muscle fiber segments at the synapse. J. Neurosci. 31, 14910–14919 10.1523/JNEUROSCI.3590-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linstedt A. D., Mehta A., Suhan J., Reggio H., Hauri H. P. (1997). Sequence and overexpression of GPP130/GIMPc: evidence for saturable pH-sensitive targeting of a type II early Golgi membrane protein. Mol. Biol. Cell 8, 1073–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattioli E., Columbaro M., Capanni C., Maraldi N. M., Cenni V., Scotlandi K., Marino M. T., Merlini L., Squarzoni S., Lattanzi G. (2011). Prelamin A-mediated recruitment of SUN1 to the nuclear envelope directs nuclear positioning in human muscle. Cell Death Differ. 18, 1305–1315 10.1038/cdd.2010.183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger T., Gache V., Xu M., Cadot B., Folker E. S., Richardson B. E., Gomes E. R., Baylies M. K. (2012). MAP and kinesin-dependent nuclear positioning is required for skeletal muscle function. Nature 484, 120–124 10.1038/nature10914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parton R. M., Hamilton R. S., Ball G., Yang L., Cullen C. F., Lu W., Ohkura H., Davis I. (2011). A PAR-1-dependent orientation gradient of dynamic microtubules directs posterior cargo transport in the Drosophila oocyte. J. Cell Biol. 194, 121–135 10.1083/jcb.201103160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizon V., Gerbal F., Diaz C. C., Karsenti E. (2005). Microtubule-dependent transport and organization of sarcomeric myosin during skeletal muscle differentiation. EMBO J. 24, 3781–3792 10.1038/sj.emboj.7600842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randles K. N., Lam, le T., Sewry C. A., Puckelwartz M., Furling D., Wehnert M., McNally E. M., Morris G. E. (2010). Nesprins, but not sun proteins, switch isoforms at the nuclear envelope during muscle development. Dev. Dyn. 239, 998–1009 10.1002/dvdy.22229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rich M., Lichtman J. W. (1989). Motor nerve terminal loss from degenerating muscle fibers. Neuron 3, 677–688 10.1016/0896-6273(89)90236-5 [DOI] [PubMed] [Google Scholar]
- Sanger J. W., Wang J., Fan Y., White J., Sanger J. M. (2010). Assembly and dynamics of myofibrils. J. Biomed. Biotechnol. 2010, 858606 10.1155/2010/858606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider M., Lu W., Neumann S., Brachner A., Gotzmann J., Noegel A. A., Karakesisoglou I. (2011). Molecular mechanisms of centrosome and cytoskeleton anchorage at the nuclear envelope. Cell. Mol. Life Sci. 68, 1593–1610 10.1007/s00018-010-0535-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serbus L. R., Cha B. J., Theurkauf W. E., Saxton W. M. (2005). Dynein and the actin cytoskeleton control kinesin-driven cytoplasmic streaming in Drosophila oocytes. Development 132, 3743–3752 10.1242/dev.01956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimmen T. (2007). The sliding theory of cytoplasmic streaming: fifty years of progress. J. Plant Res. 120, 31–43 10.1007/s10265-006-0061-0 [DOI] [PubMed] [Google Scholar]
- Shu T., Ayala R., Nguyen M. D., Xie Z., Gleeson J. G., Tsai L. H. (2004). Ndel1 operates in a common pathway with LIS1 and cytoplasmic dynein to regulate cortical neuronal positioning. Neuron 44, 263–277 10.1016/j.neuron.2004.09.030 [DOI] [PubMed] [Google Scholar]
- Srsen V., Fant X., Heald R., Rabouille C., Merdes A. (2009). Centrosome proteins form an insoluble perinuclear matrix during muscle cell differentiation. BMC Cell Biol. 10, 28 10.1186/1471-2121-10-28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr D. A., Fridolfsson H. N. (2010). Interactions between nuclei and the cytoskeleton are mediated by SUN-KASH nuclear-envelope bridges. Annu. Rev. Cell Dev. Biol. 26, 421–444 10.1146/annurev-cellbio-100109-104037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tassin A. M., Maro B., Bornens M. (1985). Fate of microtubule-organizing centers during myogenesis in vitro. J. Cell Biol. 100, 35–46 10.1083/jcb.100.1.35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terada M., Lan Y. B., Kawano F., Ohira T., Higo Y., Nakai N., Imaizumi K., Ogura A., Nishimoto N., Adachi Y.et al. (2010). Myonucleus-related properties in soleus muscle fibers of mdx mice. Cells Tissues Organs 191, 248–259 10.1159/000240245 [DOI] [PubMed] [Google Scholar]
- Tsai J. W., Bremner K. H., Vallee R. B. (2007). Dual subcellular roles for LIS1 and dynein in radial neuronal migration in live brain tissue. Nat. Neurosci. 10, 970–979 10.1038/nn1934 [DOI] [PubMed] [Google Scholar]
- Vallee R. B., Seale G. E., Tsai J. W. (2009). Emerging roles for myosin II and cytoplasmic dynein in migrating neurons and growth cones. Trends Cell Biol. 19, 347–355 10.1016/j.tcb.2009.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren R. H. (1974). Microtubular organization in elongating myogenic cells. J. Cell Biol. 63, 550–566 10.1083/jcb.63.2.550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z., Sanada K., Samuels B. A., Shih H., Tsai L. H. (2003). Serine 732 phosphorylation of FAK by Cdk5 is important for microtubule organization, nuclear movement, and neuronal migration. Cell 114, 469–482 10.1016/S0092-8674(03)00605-6 [DOI] [PubMed] [Google Scholar]
- Zaal K. J., Reid E., Mousavi K., Zhang T., Mehta A., Bugnard E., Sartorelli V., Ralston E. (2011). Who needs microtubules? Myogenic reorganization of MTOC, Golgi complex and ER exit sites persists despite lack of normal microtubule tracks. PLoS ONE 6, e29057 10.1371/journal.pone.0029057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Felder A., Liu Y., Guo L. T., Lange S., Dalton N. D., Gu Y., Peterson K. L., Mizisin A. P., Shelton G. D.et al. (2010). Nesprin 1 is critical for nuclear positioning and anchorage. Hum. Mol. Genet. 19, 329–341 10.1093/hmg/ddp499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Skepper J. N., Yang F., Davies J. D., Hegyi L., Roberts R. G., Weissberg P. L., Ellis J. A., Shanahan C. M. (2001). Nesprins: a novel family of spectrin-repeat-containing proteins that localize to the nuclear membrane in multiple tissues. J. Cell Sci. 114, 4485–4498 [DOI] [PubMed] [Google Scholar]
- Zhang Q., Ragnauth C. D., Skepper J. N., Worth N. F., Warren D. T., Roberts R. G., Weissberg P. L., Ellis J. A., Shanahan C. M. (2005). Nesprin-2 is a multi-isomeric protein that binds lamin and emerin at the nuclear envelope and forms a subcellular network in skeletal muscle. J. Cell Sci. 118, 673–687 10.1242/jcs.01642 [DOI] [PubMed] [Google Scholar]
- Zhang Q., Bethmann C., Worth N. F., Davies J. D., Wasner C., Feuer A., Ragnauth C. D., Yi Q., Mellad J. A., Warren D. T.et al. (2007). Nesprin-1 and -2 are involved in the pathogenesis of Emery Dreifuss muscular dystrophy and are critical for nuclear envelope integrity. Hum. Mol. Genet. 16, 2816–2833 10.1093/hmg/ddm238 [DOI] [PubMed] [Google Scholar]
- Zhang T., Zaal K. J., Sheridan J., Mehta A., Gundersen G. G., Ralston E. (2009). Microtubule plus-end binding protein EB1 is necessary for muscle cell differentiation, elongation and fusion. J. Cell Sci. 122, 1401–1409 10.1242/jcs.039255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Xu R., Zhu B., Yang X., Ding X., Duan S., Xu T., Zhuang Y., Han M. (2007). Syne-1 and Syne-2 play crucial roles in myonuclear anchorage and motor neuron innervation. Development 134, 901–908 10.1242/dev.02783 [DOI] [PubMed] [Google Scholar]
- Zhang X., Lei K., Yuan X., Wu X., Zhuang Y., Xu T., Xu R., Han M. (2009). SUN1/2 and Syne/Nesprin-1/2 complexes connect centrosome to the nucleus during neurogenesis and neuronal migration in mice. Neuron 64, 173–187 10.1016/j.neuron.2009.08.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhen Y. Y., Libotte T., Munck M., Noegel A. A., Korenbaum E. (2002). NUANCE, a giant protein connecting the nucleus and actin cytoskeleton. J. Cell Sci. 115, 3207–3222 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.