Abstract
Enzymatic catalysis has conflicting structural requirements of the enzyme. In order for the enzyme to form a Michaelis complex, the enzyme must be in an open conformation so that the substrate can get into its active center. On the other hand, in order to maximize the stabilization of the transition state of the enzymatic reaction, the enzyme must be in a closed conformation to maximize its interactions with the transition state. The conflicting structural requirements can be resolved by a flexible active center that can sample both open and closed conformational states. For a bisubstrate enzyme, the Michaelis complex consists of two substrates in addition to the enzyme. The enzyme must remain flexible upon the binding of the first substrate so that the second substrate can get into the active center. The active center is fully assembled and stabilized only when both substrates bind to the enzyme. However, the side-chain positions of the catalytic residues in the Michaelis complex are still not optimally aligned for the stabilization of the transition state, which lasts only approximately 10−13 s. The instantaneous and optimal alignment of catalytic groups for the transition state stabilization requires a dynamic enzyme, not an enzyme which undergoes a large scale of movements but an enzyme which permits at least a small scale of adjustment of catalytic group positions. This review will summarize the structure, catalytic mechanism, and dynamic properties of 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase and examine the role of protein conformational dynamics in the catalysis of a bisubstrate enzymatic reaction.
Keywords: Bisubstrate enzyme, enzymatic catalysis, folate biosynthesis, 6-hydroxymethyl-7, 8-dihydropterin pyrophosphokinase, HPPK, NMR, protein dynamics, X-ray crystallography
Introduction
Folate cofactors are essential for life. Mammals obtain folates from their diet, because they cannot synthesize folates de novo but have an active transport system. In contrast, most microorganisms must synthesize folates de novo, because they cannot take folates from their environments due to the lack of an active transport system [1]. Therefore, the folate biosynthetic pathway has been one of the principal targets for developing antimicrobial agents [2, 3]. Among the folate pathway enzymes, the four enzymes in the mid pathway are particularly attractive targets because they are absent in mammals: dihydroneopterin aldolase (DHNA), 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase (HPPK), dihydropteroate synthase (DHPS), and dihydrofolate synthase (DHFS). DHPS is the target of sulfonamides, the clinical use of which marks the beginning of the modern era of antimicrobial chemotherapy [4]. The multiple targets afforded by the pathway also provide opportunities to develop antibiotics with synergetic effects. For example, in clinical use, sulfonamides, which target DHPS, are combined with trimethoprim [4], an antibiotic targeting dihydrofolate reductase, the last enzyme in the folate pathway.
HPPK is a bisubstrate enzyme and catalyzes the transfer of pyrophosphate from ATP to 6-hydroxymethyl-7,8-dihydropterin (HP) to form AMP and 6-hydroxymethyl-7,8-dihydropterin pyrophosphate (HPPP). In E. coli and most other bacteria, HPPK is a monofunctional enzyme. However, in some microorganisms, HPPK is a part of bifunctional (DHNA-HPPK or HPPK-DHPS) or trifunctional (DHNA-HPPK-DHPS) enzymes, encoded by polycistronic genes. It is tempting to speculate that such multifunctional organizations may assist substrate transfer between the active centers, but the recently reported crystal structures of the bifunctional DHNA-HPPK from Streptococcus pneumoniae [5] and the HPPK-DHPS moiety of the trifunctional DHNA-HPPK-DHPS from Saccharomyces cerevisiae [6] do not support such a proposition, as no channel exists for a direct transfer of the product from one active center to another to serve as the substrate for the subsequent reaction in the pathway. Like other enzymes in the folate pathway, HPPK has been explored as a target for developing antimicrobial agents [7, 8].
Conflicting Structural Requirements for Substrate Binding/Product Release and Maximal Transition State Stabilization
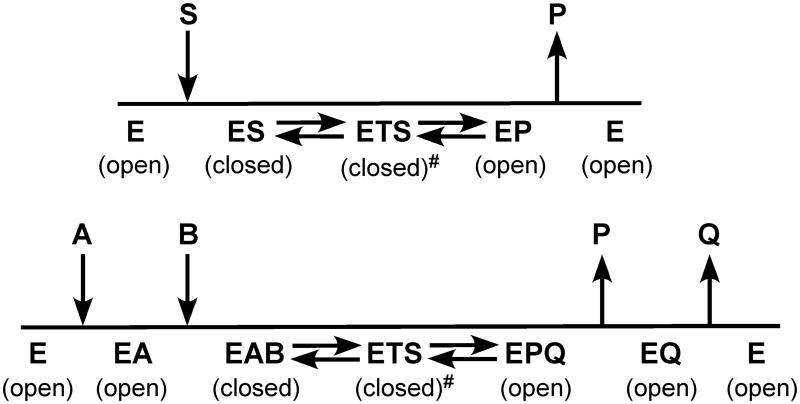
The hallmarks of enzymatic catalysis are formation of Michaelis complex and stabilization of transition state. The two events, however, have different structural requirements as discussed by Wolfenden [9]. In order for the substrate to get into the active center of an enzyme, the active center must be in an open conformation (Fig. 1). On the other hand, in order to maximize transition state stabilization, the active center must be in a closed conformation to maximize interactions with the altered substrate in the transition state. After the chemical reaction, in order for the product to dissociate, the active center must open up again. Consequently, protein dynamics is an important aspect of enzymatic catalysis and plays an important role throughout the catalytic cycle of the enzyme, including substrate binding to form the Michaelis complex, transition state stabilization, and product release. For a bisubstrate enzyme, upon the binding of the first substrate, the enzyme must remain in an open conformation or a partially open conformation so that the second substrate can bind to the enzyme to form the ternary Michaelis complex.
Figure 1.
Conflicting structural requirement of enzymatic catalysis. Top, a single substrate system, where E, S, TS, and P represent enzyme, substrate, transition state, and product, respectively. Bottom, an ordered bisubstrate system with the two substrates represented by A and B and the two products by P and Q. The conformation of the enzyme in the transition state is slightly different from that in the Michaelis complex as indicated by #.
Because E. coli HPPK is small (∼18 kDa), stable, and amenable to both X-ray crystallographic and NMR analysis, the enzyme has emerged as an excellent model for studying the role of protein dynamics in enzymatic catalysis. Below we review the role of protein conformational dynamics in the catalysis by E. coli HPPK as revealed by a combination of biochemical, X-ray crystallographic, and NMR studies.
Product Release is Rate-Limiting
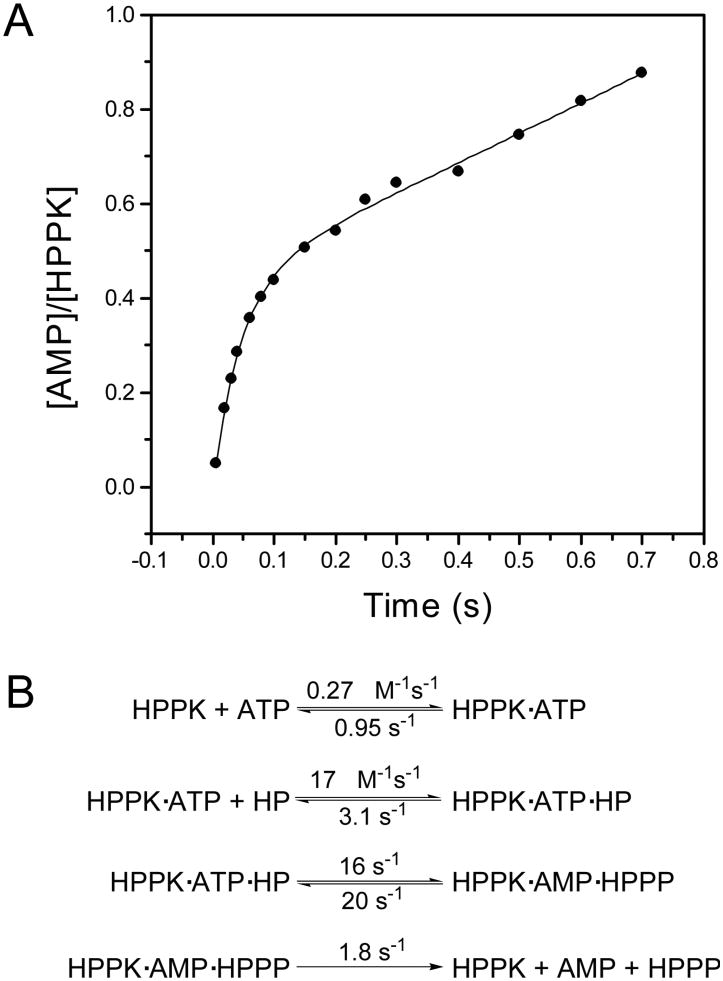
The HPPK-catalyzed reaction follows an apparently ordered kinetic mechanism with MgATP binding to the enzyme first [10, 11]. Bermingham and coworkers showed by Hummel and Dreyer measurements that HPPK forms a binary complex with MgATP or its analogue MgAMPCPP but not with HP; binding of the nucleotides is slow and is followed by the rapid addition of HP [10]. We showed by NMR that HP can bind to the free HPPK, but the affinity of HP for the free HPPK is much lower [12]. The Kd for the binding of HP to the free HPPK is in the mM range [12], whereas the Kd for the binding of HP to the binary nucleotide complex is in the sub-μM range [10, 11]. Furthermore, the cellular ATP concentration is estimated at ∼3 mM [13] and the Kd for the binding of MgATP to HPPK is 2.6-4.5 μM [10, 11, 14], indicating that HPPK is essentially all in the MgATP-bound form, ready for the rapid addition of HP from the upstream of the folate pathway. Using equilibrium binding and transient-state kinetic (both stopped-flow and quench-flow) studies, we derived a set of rate constants for the HPPK-catalyzed reaction. The Kd value for the binding of MgATP was most conveniently measured with a competitive binding assay using the fluorescent ATP analogue Ant-ATP [14]. But the rate constants for the binding of MgATP could be measured by stopped-flow fluorometry based on the small change in the intrinsic tryptophan fluorescence of HPPK [11]. Both the Kd and rate constants for the binding of HP can be measured by fluorometry in the presence of the ATP analogue AMPCPP [11]. The equilibrium binding data provided a consistent test for the kinetic analysis. The rate for the pyrophosphoryl transfer was measured by quench-flow analysis. The reaction was run with radioactive ATP as the tracer in a KinTek quench-flow apparatus and stopped with EDTA. The substrates and products were separated by TLC and quantified by phosphorimage. The pre-steady state experiment showed a classical burst kinetic behavior (Fig. 2A), indicating that product release is the rate-limiting step in the HPPK-catalyzed reaction [11]. The rate constant for the burst phase of the reaction was further confirmed by a single turnover experiment. A set of kinetic constants were derived by global analysis of the transient kinetic data using the program DYNAFIT (Fig. 2B). The slow product release was attributed to the conformational changes that are required to allow the products to exit, as the substrates are buried at the active center of the enzyme [15]. Product release may be random as both products can bind to the free enzyme, but AMP is probably preferentially released first, because HPPP binds to the enzyme much more tightly than AMP [16]. Surprisingly, when HPPP binds to the free HPPK its fluorescence does not change. Stopped-flow fluorometry also showed that after the chemical reaction, there is a conformational change which is slower than the chemical step [16].
Figure 2.
Transient kinetic analysis. A, burst kinetics of the HPPK-catalyzed reaction. The reaction mixture contained 10 μM HPPK, 100 μM ATP, 55 μM HP, 25 mM DTT, 0.5 mM EDTA, and 10 mM MgCl2. The solid lines were obtained by nonlinear least square fit to an exponential equation. Reproduced with permission from [11]. B, kinetic scheme with the rate constants for the wild-type HPPK obtained by transient kinetic studies by Li and coworkers [11].
HPPK Undergoes Dramatic Conformational Changes through its Catalytic Cycle
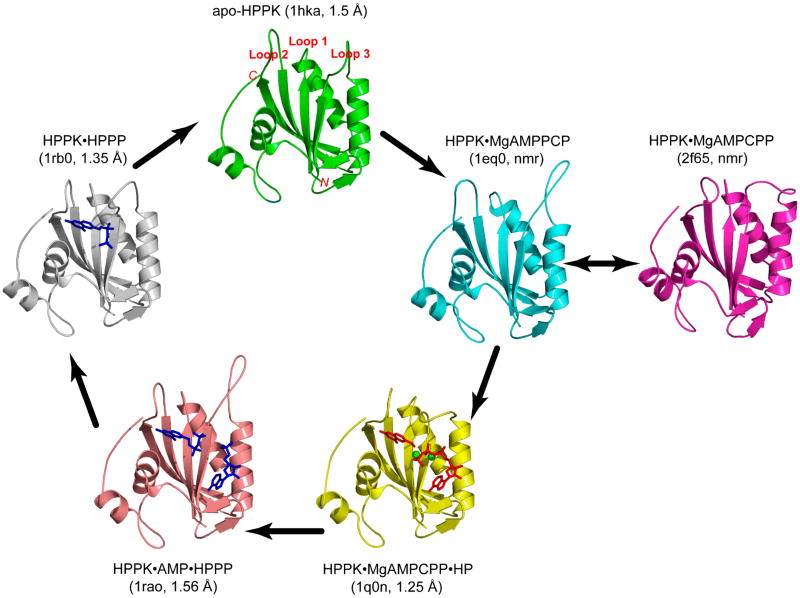
The structure of HPPK has been determined by X-ray crystallography and NMR for various stages of the catalytic cycle using substrates, substrate analogues, and products (Fig. 3). The crystal structure of apo HPPK from E. coli was determined by multiwavelength anomalous diffraction at 1.5-Å resolution (PDB code: 1HKA) [12]. The structure reveals a three-layered αβα fold formed by six β-strands and four α-helices (Fig. 5 in [12]). The fold of the HPPK molecule creates a valley that is approximately 26-Å long, 10-Å wide, and 10-Å deep. Three flexible loops, β1-α1 (loop 1), β2-β3 (loop 2), and α2-β4 (loop 3), form one wall of the valley. The other wall of the valley is relatively rigid and is constructed by the structural motif β6-loop-α3, which is part of the protein's hydrophobic core. ATP- and HP-binding sites were initially identified by NMR spectroscopy and molecular modeling [12]. Binding of ATP causes significant changes in the chemical shifts of many backbone amide resonances, not only in ATP-binding site but also in HP-binding site, suggesting that the binding of ATP induces significant conformational changes. The side chain positions of several conserved residues such as R82 and R92 also suggest conformational changes upon substrate binding, because these side chains point away from the active center of the enzyme as observed in the crystal structure of apo HPPK.
Figure 3.
Snapshots of the catalytic cycle of the HPPK-catalyzed reaction. The HPPK molecule is illustrated as a ribbon diagram (arrows, β-strands; spirals, helices; tubes, loops), the ligands in the crystal structures as stick models (substrates in red, products in blue), and the two Mg2+ ions as spheres (in green). In parenthesis are the PDB accession codes and the resolution of the structures (when applicable). The black arrows indicate the conformational changes of HPPK during the catalytic cycle.
The assignment of substrate-binding sites and suggested substrate-induced conformational changes were confirmed by the crystal structure of the ternary complex of HPPK with AMPCPP, HP, and two Mg2+ ions at 1.25-Å resolution (PDB code: 1Q0N) [15]. We also determined the NMR solution structure of HPPK in complex with AMPCPP, 7,7-dimethyl-6-hydroxymethylpterin, and Mg2+ ions (PDB code: 2F63) [17], which is very similar to the crystal structure of the ternary complex. In the crystal structure, HP is sandwiched between two aromatic rings of F123 and Y53 and forms six hydrogen bonds with residues T42, P43, L45, and N55 (Fig. 5(a) in [15]). Two Mg2+ ions are found in the ternary complex, one between α- and β-phosphate and the other between β- and γ-phosphate, and both are six-coordinated (Fig. 6(a) in [15]). Twelve residues are involved in the binding of AMPCPP, including Q74, E77, R84, R88, W89, R92, I98, R110, T112, H115, Y116, and R121, among which E77, R92, H115, and R121 are conserved. For I98, R110, and T112, the functional groups involved in AMPCPP binding are amides and/or carbonyls. The γ-phosphate group of AMPCPP is tethered by the side chains of H115, Y116, and R121, whereas the β- and α-phosphate groups form hydrogen bonds with the guanidinium groups of R92 and R84. The ribose forms hydrogen bonds with the side chain of Q74 and the carbonyl group of R110. The backbone amide and carbonyl groups of I98 and T112 are responsible for the recognition of the adenine moiety.
The conformational changes upon the formation of the ternary complex are illustrated in Fig. 3. The valley created by the fold of the enzyme has both ends and the front open to the solvent when the enzyme is ligand-free (Fig. 3(a) in [15]). In the ternary complex, one end and the front of the valley are sealed, leaving only one end open (Fig. 3(a) in [15]). If the apo enzyme looks like a half-closed right hand, the ternary complex appears to be a tightly closed fist. The most significant conformational differences reside in the three aforementioned flexible loops. A network of hydrogen bonds that couples the three flexible loops and helps to stabilize the complex and seal the active center where the pyrophosphoryl transfer occurs has been identified (Fig. 4 in [15]). The HP-binding site in the free HPPK is not blocked. Rather, the hydrogen bond partners for the binding of HP are not in place for hydrogen bonding. Consequently, HP has a very low affinity for the free HPPK. The critical part of the active center of HPPK is assembled only when both substrates bind to the enzyme.
Stammer and coworkers determined the crystal structure of a ternary complex of HPPK with MgATP and an HP analogue at 2-Å resolution (PDB code: 1DY3) [18]. In comparison with our ternary structure, the main differences are the conformations of loops 2 and 3 of the protein. The pterin moiety of the bound HP analogue is displaced somewhat, but the hydrogen bonds between the pterin and the protein are maintained. ATP and AMPCPP superimpose very well, and the coordination chemistry of the two Mg2+ ions is the same between the two structures, indicating that AMPCPP is an excellent analogue for ATP for HPPK. The differences in the conformations of loops 2 and 3 are caused by the two substituents of the HP analogue, particularly the bulky phenethyl group. As a result, the side-chains of R82 and R92 have quite different conformations and interact with the nucleotide triphosphate differently. Both guanidinium groups of R82 and R92 move inwards with R82 interacting with both α- and β-phosphate and R92 interacting with β-phosphate only. Both of the two distinct conformations for each of the two arginine residues revealed by the two structures were observed in the crystal structure of HPPK in complex with 6-hydroxymethylpterin/6-carboxylpterin, two Mg2+ ions, and AMPCPP at 0.89-Å resolution (PDB code: 1F9Y) [19], suggesting that the roles of R82 and R92 are rather dynamic. R92 first binds to the α-phosphate group of ATP and then shifts to interact with the β-phosphate as R82, which initially does not bind to ATP, moves in and binds to α-phosphate when the pyrophosphoryl transfer is about to occur.
After the chemical step, HPPK undergoes another conformational change as revealed by the crystal structure of HPPK in complex with the products HPPP and AMP at 1.56-Å resolution (PDB code: 1RAO) (Fig. 3) [20]. In particular, loop 3 moves away from the active center and adopts a conformation even more open than in the apo HPPK. In comparison with the ternary complex with a substrate (HP) and a substrate analogue (AMPCPP), the Cα atom of E87 moves by ∼23 Å, the guanidinium group of R82 by ∼8 Å, and the guanidinium group of R84 by ∼23 Å. The conformational change not only opens up the active center but also weakens the interactions between the products and the enzyme. The phosphate of AMP and the pyrophosphate of HPPP are disordered, suggesting that the two products are rather dynamic. AMP is more exposed to the solvent than HPPP, in accordance with the preferential release of AMP. After the departure of AMP, HPPK undergoes yet another conformational change and adopts a conformation similar to that of the apo HPPK. HPPP becomes less exposed to solvent and is well fixed in the binary product complex with a single conformation, suggesting that the dissociation of HPPP is the rate-liming step in the HPPK-catalyzed reaction. HPPP interacts with not only loop 2 but also loop 3. The dynamics of the loops, particularly loop 3, may facilitate the release of HPPP.
The crystallization of HPPK with MgATP has not been successful, because of the low level of ATPase activity of the enzyme [21]. The published structures of HPPK in complex with a nucleotide include the crystal structure of HPPK in complex with MgADP (PDB code: 1EQM; 1.5-Å resolution) [21], which was obtained because of the hydrolysis of ATP to ADP during the crystallization process, and two NMR solution structures, one in complex with MgAMPPCP (PDB code: 1EQ0) [21] and the other with MgAMPCPP (PDB code: 2F65) [17]. Loop 3 in both the HPPK•MgADP crystal structure and the HPPK•MgAMPPCP NMR solution structure adopts a conformation more open than in the apo HPPK, very similar to that in the ternary product structure. The fact that loop 3 can also adopt a “super open” conformation in solution indicates that the “super open” conformation observed in the crystal structures are not due to crystal packing [21]. In comparison with the structures of the apo HPPK and the ternary complex with the substrates and substrate analogues, it appears that such an unusual conformational change is required for the assembly of the active center. In particular, the side chains of catalytically important R82 and R92 cannot move into the active center without first opening loop 3 [15].
The conformational properties of HPPK in complex with MgAMPCPP are quite different from those of the other two binary nucleotide complexes described above. In particular, both loops 2 and 3 can assume multiple conformations, which are generally in-between those of the apo HPPK and the ternary substrate complex. Because AMPCPP is a better ATP analogue for HPPK than ADP or AMPPCP in terms of both binding affinity and bound conformation, the conformational properties of the binary substrate complex HPPK•MgATP may be better represented by those of HPPK•MgAMPCPP. The structures of HPPK•MgADP and HPPK•MgAMPPCP may represent intermediate conformations before the formation of the stable binary substrate complex.
HPPK Dynamics as Revealed by NMR
While X-ray crystallography has captured the dramatic conformational changes of HPPK through its catalytic cycle at atomic resolution, NMR has revealed the internal motions of HPPK on a wide range of timescales. First, it appears that the catalytic loops of the apo enzyme can assume a range of conformations. Sixteen residues show no or very weak cross peaks in the 1H-15N HSQC spectrum of the apo enzyme [22], seven of which are located in loops 2 and 3, suggesting that both loops assume multiple conformations and undergo conformational transitions in an intermediate exchange regime on the ms timescale. Second, the binary substrate complex of HPPK can assume multiple conformations on the slow NMR chemical shift timescale, as evidenced by multiple sets of NMR signals for several residues in loops 2 and 3 and the very weak or missing NH cross peaks for several residues in loops 1 and 3 [17]. Third, the ternary complex shows only one set of NMR signals and the cross peak intensities are rather uniform, suggesting that the binding of the second substrate shifts the multiple conformations of the binary complex to an apparently single conformation of the ternary complex [17].
The dynamic properties of HPPK from the apo form to the binary substrate complex with MgATP (represented by MgAMPCPP) to the Michaelis complex (ternary substrate complex) with MgATP (represented by MgAMPCPP) and HP (represented by 7,7-dimethyl-6-hydroxypterin, an HP analogue) have been investigated by 15N NMR relaxation studies [23]. The dynamic parameters S2 and Rex obtained are summarized in Fig. 8 in [23]. S2 is the squared-order parameter, reflecting the amplitude of internal motion on the ps-ns timescale and Rex is the chemical exchange contribution to transverse relaxation, reflecting conformational exchange on the μs-ms timescale. The results indicate that the binding of the nucleotide to HPPK does not cause major changes in the dynamic properties of the enzyme on both ps-ns and μs-ms timescales, as measured by S2 and Rex, respectively. The internal mobility of HPPK is not reduced and is even moderately increased in the binary complex, particularly in the catalytic loops. The internal mobility of the catalytic loops is significantly quenched upon the formation of the ternary complex, but some mobility remains. The enhanced motions in the catalytic loops of the binary substrate complex may be required for the assembling of the ternary complex. On the other hand, some degrees of mobility in the catalytic loops of the ternary complex may be required for the optimal stabilization of the transition state, which may need the instantaneous adjustment and alignment of the side-chain positions of catalytic residues.
These experimental observations about the dynamic properties of HPPK have been corroborated by computational studies. Correlated motions involving loops-2 and 3 have been deduced from a coarse-grained model of HPPK, harmonically constrained according to its crystal structure [24]. The two distinct conformations of loop 1 revealed by crystallography have been also observed in a molecular dynamics simulation study [25]. Essential dynamics analysis of the apo HPPK and the binary substrate complex has revealed three major modes of motion involving the three catalytic loops [25].
Mechanism of Pyrophosphoryl Transfer
The HPPK-catalyzed pyrophosphoryl transfer reaction is proposed to follow a concerted mechanism (ANDN in the IUPAC nomenclature) with a loose transition state [15, 20]. The hydroxyl group of HP is well positioned for attacking the β-phosphorus atom of ATP according to the HPPK•MgAMPCPP•HP structure [15]. The angle formed by the hydroxyl oxygen of HP, the β-phosphorus of AMPCPP, and the bridging methylene carbon of AMPCPP is 174.5°; and the distance between the hydroxyl oxygen of HP and the β-phosphorus of AMPCPP is 3.2 Å. A model for the transition state of the proposed concerted mechanism has been built [20] (Fig. 9 in [15]). In this model, the angle formed by the hydroxyl oxygen of HP, the β-phosphorus of ATP, and the bridging oxygen of α-phosphate of ATP is 180°; and the distance between the hydroxyl oxygen of HP and the β-phosphorus of ATP and that between the β-phosphorus and the bridging oxygen of α-phosphate of ATP are both 2.5 Å. As described earlier, HPPK has a low level of ATPase activity. Because the hydrolytic products are ADP and phosphate rather than AMP and pyrophosphate, it is very unlikely that the HPPK-catalyzed pyrophosphoryl transfer follows a dissociative mechanism (DN + AN) involving the formation of a substituted metaphosphate reaction intermediate and pyrophosphate.
Two Mg2+ ions are required for the catalysis of HPPK and other pyrophosphoryl transfer enzymes [26]. As observed in HPPK•MgAMPCPP•HP [15], one Mg2+ ion is coordinated with both α- and β-phosphates of AMPCPP and the other with both β- and γ-phosphates of AMPCPP as well as the hydroxyl oxygen of HP; both Mg2+ ions are coordinated with the carboxyl groups of D95 and D97. The two Mg2+ ions may play several important roles in catalysis. First, the Mg2+ ions may help to induce the conformational changes necessary to bring many catalytic residues into the active site. Second, these ions may orient the hydroxyl oxygen of HP, the β-phosphorus, and the bridging oxygen between the α- and β-phosphorus atoms into an appropriate arrangement for the reaction. Third, the coordination of both Mg2+ ions with β-phosphate may activate the β-phosphorus for the nucleophilic attack. Fourth, the coordination of Mg2+ with the hydroxyl oxygen of HP may reduce the pK a of the hydroxyl group and therefore facilitates the reaction. Fifth, the Mg2+ ions may stabilize the negative charge developed in the transition state of the reaction.
Catalytic and Dynamic Roles of Arginine 82 and Arginine 92
As the pyrophosphoryl transfer reaction takes place in β-phosphoryl group with AMP as the leaving group, the residues that interact with the α- and β-phosphoryl groups are likely to be most important for catalysis, which include R82, R84, and R92 according to the crystal structures. The roles of R82 and R92 in HPPK catalysis have been investigated by site-directed mutagenesis, biochemical analysis, and X-ray crystallography [19, 27]. A similar study of R84 will be described in the next section, as R84 is on loop 3. Substitution of R82 with alanine causes a decrease in the rate constant for the chemical step by a factor of ∼380 with no significant changes in the binding energy or binding kinetics of either substrate. Substitution of R92 with alanine causes a decrease in the rate constant for the chemical step by a factor of ∼3.5×104. The mutation causes no significant changes in the binding energy or binding kinetics of MgATP and significant change in the binding energy of HP, either, but it causes a decrease in the association rate constant for the binding of HP by a factor of ∼4.5 and in the dissociation rate constant by a factor of ∼10. The overall structures of the ternary complexes of both mutants are very similar to the corresponding structure of wild-type HPPK. The results suggest that R82 does not contribute to the binding of either substrate, and R92 is dispensable for the binding of MgATP but plays a role in facilitating the binding of HP. Both R82 and R92 are important for catalysis, and R92 plays a critical role in the transition state stabilization. The mutations also change the rate-limiting step from product release to pyrophosphoryl transfer. Structural analysis reveal that the guanidinium groups of both R82 and R92 are rather dynamic, moving in and out the active center and assuming two conformations even in the ternary substrate complex [19].
Loop 3 is Important for Assembling the Catalytic Center and Catalysis
Loop 3, which begins with R82 and ends with R92, undergoes the most dramatic conformational changes during the catalytic cycle of HPPK. To investigate the roles of loop 3 in catalysis, we have made a deletion mutant by removing most part of loop 3 and characterized the mutant by biochemical and X-ray crystallographic analysis [28]. The deletion mutation does not have any significant effects on the dissociation constants nor the rate constants for the binding of the first substrate MgATP or its analogues. The Kd of HP for the mutant enzyme increases by a factor of ∼100. The association rate constant for the binding of HP to the mutant is the same as that to the wild-type enzyme. The large increase in the Kd of HP for the mutant is due to a large increase in the dissociation rate constant. The deletion mutation causes a shift of the rate-limiting step in the reaction. No burst kinetics is observed as for the wild-type enzyme, indicating that the chemical step is rate-limiting in the mutant-catalyzed reaction. The deletion of loop 3 causes a decrease in the rate constant for the chemical step by a factor of ∼1.1×105. Two crystal structures have been determined for the deletion mutant, all at high resolution [28]. The structure of the apo mutant superimposes well with that of the apo wild-type enzyme, indicating that the deletion mutation does not affect the folding of the enzyme. Although the core structures of the three crystal structures of the ternary complex of the deletion mutant are the same as that of the wild-type enzyme, none of the three structures of the ternary complex of the mutant has a fully assembled active center (Fig. 2 in [28]). The results together suggest that loop 3 is dispensable for the folding of the protein and the binding of the first substrate MgATP, but is required for the assembling of the catalytically competent active center. The loop plays an important role in the stabilization of the ternary complex and is critical for catalysis.
On loop 3, there are two residues with hydrogen bonds to the bound nucleotides in the ternary complexes [18, 29]. In the crystal structure of HPPK•HP•MgAMPCPP [15], the guanidinium group of R84 is hydrogen bonded to the α-phosphate and 3′-hydroxyl group of the nucleotide. The indole ring of W89 is hydrogen bonded to the γ-phosphate of the nucleotide and covers one edge of the bound HP. Whether the functional importance of loop 3 is due to the interactions of R84 and W89 with the substrates has been addressed by site-directed mutagenesis, and in-depth biochemical analysis of the mutants. Substitution of R84 with alanine causes little changes in either dissociation constants or kinetic constants of the HPPK-catalyzed reaction except that the rate constant for the chemical step of the forward reaction decreases by a factor of 4. Substitution of W89A with alanine does not have any significant effects on the Kd and the rate constants for the binding of MgATP, but the Kd for HP of the mutant increases by a factor of 6.5, due to the increase in the dissociation rate constant. The mutation decreases the rate constant for the chemical step of the forward reaction by a factor of ∼23 and the rate constant for the chemical step of the reverse reaction by a factor of ∼33. The results show that R84 plays only a minor role in catalysis and W89 plays a minor role in the binding of HP and a more significant role in catalysis, and suggest that the critical role of loop 3 in HPPK catalysis is not due to the interactions of the guanidinium group of R84 and the indole ring of W89 with the substrates.
Concluding Remarks
The multidisciplinary studies of E. coli HPPK have provided important insights to the structure, dynamics, and catalytic mechanism of the enzyme. Equilibrium binding and transient kinetic analyses have laid a solid thermodynamic and kinetic foundation for dissecting the structure-function relationship. The HPPK-catalyzed reaction shows classical burst kinetics, indicating that product release is rate-limiting. High-resolution X-ray crystallography and NMR have provided snapshots of the enzyme throughout its catalytic cycle. The snapshots reveal not only the interactions of the enzyme with the substrates and products but also dramatic conformational changes in the enzyme action. The catalytically competent active center is assembled only after both substrates are bound. The assembling of the active center involves the coupling of three catalytic loops. The enzyme, particularly loop 3, undergoes dramatic conformational changes during the assembling of the active center and after the chemical reaction. R82 and R92, which demarcate the beginning and the end of loop 3, are critical for catalysis, and loop 3 is important for the positioning of R82 and R92 for catalysis.
Because the apo enzyme and the binary substrate complex both assume multiple conformations, the conformational transitions in HPPK cannot be described by the classical induced fit model of one conformation to another. The conformational transitions do exhibit features of the population shift model, but it is not certain whether the conformations of the ligand-bound forms exist in the absence of the ligands, as the solution conformations of the apo enzyme have not been determined and the solution conformations of the binary complex HPPK•MgAMPCPP are only those of the predominant species.
The dynamic properties of HPPK may be characteristic of bisubstrate enzymes. The formation of enzyme-substrate complexes and the maximization of transition-state stabilization have conflicting structural requirements. The formation of an enzyme-substrate complex requires an open active center so that the substrate(s) can get in. On the other hand, the maximization of transition-state stabilization requires a closed active center that maximizes the favorable interactions between the enzyme and the transition state. The conflicting structural requirements can be resolved by a flexible active center that can sample both open and closed conformational states. For a bisubstrate enzyme like HPPK, the Michaelis complex consists of two substrates in addition to the enzyme. The enzyme must remain flexible upon the binding of the first substrate so that the second substrate can get into the active center. The active center is fully assembled and stabilized only when both substrates bind to the enzyme. However, the side-chain positions of the catalytic residues in the Michaelis complex are still not optimally aligned for the stabilization of the transitions state, which lasts only approximately 10−13 s. The instantaneous and optimal alignment of catalytic groups for the transition state stabilization requires a dynamic enzyme, not an enzyme which undergoes a large scale of movements but an enzyme which permits at least a small scale of adjustment of catalytic group positions, resulting in a closed conformation that are slightly different from that of the Michaelis complex as indicated by # in Fig. 1. The dynamic properties of the three forms of HPPK as determined by the 15N relaxation measurements are consistent with the conformational transitions for a bisubstrate enzyme from the apo form to the binary substrate complex and the ternary Michaelis complex.
Acknowledgments
Our work was supported in part by NIH grant GM51901 (H.Y.) and the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research. We thank our collaborators, particularly Drs. Yue Li, Jaroslaw Blaszczyk, Genbin Shi, Bing Xiao, Ewen Lescop, and Changwen Jin, for most fruitful collaborations.
Abbreviations
- AMPCPP
α,β-methyleneadenosine triphosphate
- AMPPCP
β,γ-methyleneadenosine triphosphate
- Ant-ATP
3′(2′)-O-anthraniloyladenosine 5′-triphosphate
- DHFS
dihydrofolate synthase
- DHNA
dihydroneopterin aldolase
- DHPS
dihydropteroate synthase
- HPPK
6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase
- HP
6-hydroxymethyl-7,8-dihydropterin
- HPPP
6-hydroxymethyl-7,8-dihydropterin pyrophosphate
- HSQC
heteronuclear single-quantum coherence
References
- 1.Hitchings GH, Burchall JJ. Inhibition of folate biosynthesis and function as a basis for chemotherapy. Adv Enzymol Relat Areas Mol Biol. 1965;27:417–468. doi: 10.1002/9780470122723.ch9. [DOI] [PubMed] [Google Scholar]
- 2.Walsh C. Where will new antibiotics come from? Nat Rev Microbiol. 2003;1:65–70. doi: 10.1038/nrmicro727. [DOI] [PubMed] [Google Scholar]
- 3.Bermingham A, Derrick JP. The folic acid biosynthesis pathway in bacteria: evaluation of potential for antibacterial drug discovery. Bioessays. 2002;24:637–648. doi: 10.1002/bies.10114. [DOI] [PubMed] [Google Scholar]
- 4.Zinner SH, Mayer KH. Sulfonamides and trimethoprim. In: Mandell GL, Bennett JE, Dolin R, editors. Principles and Practice of Infectious Diseases. 6th. Vol. 1. Churchill Livingstone; Philadelphia: 2005. pp. 440–451. [Google Scholar]
- 5.Garçon A, Levy C, Derrick JP. Crystal structure of the bifunctional dihydroneopterin aldolase/6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase from Streptococcus pneumoniae. J Mol Biol. 2006;360:644–653. doi: 10.1016/j.jmb.2006.05.038. [DOI] [PubMed] [Google Scholar]
- 6.Lawrence MC, Iliades P, Fernley RT, Berglez J, Pilling PA, Macreadie IG. The three-dimensional structure of the bifunctional 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase/dihydropteroate synthase of Saccharomyces cerevisiae. J Mol Biol. 2005;348:655–670. doi: 10.1016/j.jmb.2005.03.021. [DOI] [PubMed] [Google Scholar]
- 7.Al Hassan SS, Cameron R, Curran AWC, Lyall WJS, Nicholson S, Robinson D, Stuart A, Suckling C, Stirling I, Wood HCS. Specific Inhibitors in Vitamin Biosynthesis .7. Syntheses of Blocked 7,8-Dihydropteridines Via Alpha-Amino Ketones. J Chem Soc Perkin Trans. 1985;1:1645–1659. [Google Scholar]
- 8.Shi G, Blaszczyk J, Ji X, Yan H. Bisubstrate analogue inhibitors of 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase: Synthesis and biochemical and crystallographic studies. J Med Chem. 2001;44:1364–1371. doi: 10.1021/jm0004493. [DOI] [PubMed] [Google Scholar]
- 9.Wolfenden R. Thermodynamic and extrathermodynamic requirements of enzyme catalysis. Biophys Chem. 2003;105:559–572. doi: 10.1016/s0301-4622(03)00066-8. [DOI] [PubMed] [Google Scholar]
- 10.Bermingham A, Bottomley JR, Primrose WU, Derrick JP. Equilibrium and kinetic studies of substrate binding to 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase from Escherichia coli. J Biol Chem. 2000;275:17962–17967. doi: 10.1074/jbc.M000331200. [DOI] [PubMed] [Google Scholar]
- 11.Li Y, Gong Y, Shi G, Blaszczyk J, Ji X, Yan H. Chemical transformation is not rate-limiting in the reaction catalyzed by Escherichia coli 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase. Biochemistry. 2002;41:8777–8783. doi: 10.1021/bi025968h. [DOI] [PubMed] [Google Scholar]
- 12.Xiao B, Shi G, Chen X, Yan H, Ji X. Crystal structure of 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase, a potential target for the development of novel antimicrobial agents. Structure. 1999;7:489–496. doi: 10.1016/s0969-2126(99)80065-3. [DOI] [PubMed] [Google Scholar]
- 13.Neuhard J, Nygaard P. Purines and pyrimidines. In: Neidhardt FC, Ingraham JL, Low KB, Magasanik B, Schaechter M, Umbarger HE, editors. Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology. Vol. 1. ASM Press; Washington, D.C.: 1987. pp. 445–473. [Google Scholar]
- 14.Shi G, Gong Y, Savchenko A, Zeikus JG, Xiao B, Ji X, Yan H. Dissecting the nucleotide binding properties of Escherichia coli 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase with fluorescent 3′(2)′-O-anthraniloyladenosine 5′-triphosphate. Biochim Biophys Acta. 2000;1478:289–299. doi: 10.1016/s0167-4838(00)00043-1. [DOI] [PubMed] [Google Scholar]
- 15.Blaszczyk J, Shi G, Yan H, Ji X. Catalytic center assembly of HPPK as revealed by the crystal structure of a ternary complex at 1.25 angstrom resolution. Structure. 2000;8:1049–1058. doi: 10.1016/s0969-2126(00)00502-5. [DOI] [PubMed] [Google Scholar]
- 16.Garçon A, Bermingham A, Lian LY, Derrick JP. Kinetic and structural characterization of a product complex of 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase from Escherichia coli. Biochem J. 2004;380:867–873. doi: 10.1042/BJ20031850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li G, Felczak K, Shi G, Yan H. Mechanism of the conformational transitions in 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase as revealed by NMR spectroscopy. Biochemistry. 2006;45:12573–12581. doi: 10.1021/bi061057m. [DOI] [PubMed] [Google Scholar]
- 18.Stammers DK, Achari A, Somers DO, Bryant PK, Rosemond J, Scott DL, Champness JN. 2.0 Å X-ray structure of the ternary complex of 7,8-dihydro-6-hydroxymethylpterin pyrophosphokinase from Escherichia coli with ATP and a substrate analogue. FEBS Lett. 1999;456:49–53. doi: 10.1016/s0014-5793(99)00860-1. [DOI] [PubMed] [Google Scholar]
- 19.Blaszczyk J, Li Y, Shi G, Yan H, Ji X. Dynamic roles of arginine residues 82 and 92 of Escherichia coli 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase: Crystallographic studies. Biochemistry. 2003;42:1573–1580. doi: 10.1021/bi0267994. [DOI] [PubMed] [Google Scholar]
- 20.Blaszczyk J, Shi G, Li Y, Yan H, Ji X. Reaction trajectory of pyrophosphoryl transfer catalyzed by 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase. Structure. 2004;12:467–475. doi: 10.1016/j.str.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 21.Xiao B, Shi G, Gao J, Blaszczyk J, Liu Q, Ji X, Yan H. Unusual conformational changes in 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase as revealed by X-ray crystallography and NMR. J Biol Chem. 2001;276:40274–40281. doi: 10.1074/jbc.M103837200. [DOI] [PubMed] [Google Scholar]
- 22.Shi G, Gao J, Yan H. 1H, 13C and 15N resonance assignments of Escherichia coli 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase and its complex with MgAMPPCP. J Biomol NMR. 1999;14:189–190. doi: 10.1023/a:1008309801414. [DOI] [PubMed] [Google Scholar]
- 23.Lescop E, Lu Z, Liu Q, Xu H, Li G, Xia B, Yan H, Jin C. Dynamics of the Conformational Transitions in the Assembling of the Michaelis Complex of a Bisubstrate Enzyme: A N-15 Relaxation Study of Escherichia coli 6-Hydroxymethyl-7,8-dihydropterin Pyrophosphokinase. Biochemistry. 2009;48:302–312. doi: 10.1021/bi8016262. [DOI] [PubMed] [Google Scholar]
- 24.Keskin O, Ji X, Blaszcyk J, Covell DG. Molecular motions and conformational changes of HPPK. Proteins. 2002;49:191–205. doi: 10.1002/prot.10205. [DOI] [PubMed] [Google Scholar]
- 25.Yang R, Lee MC, Yan H, Duan Y. Loop conformation and dynamics of the Escherichia coli HPPK apo-enzyme and its binary complex with MgATP. Biophys J. 2005;89:95–106. doi: 10.1529/biophysj.105.061556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mildvan AS, Weber DJ, Abeygunawrdana C. Solution structure and mechanism of the MutT pyrophosphohydrolase. Ad Enzymol Rel Areas Mol Biol. 1999;73:183–207. doi: 10.1002/9780470123195.ch6. [DOI] [PubMed] [Google Scholar]
- 27.Li Y, Wu Y, Blaszczyk J, Ji X, Yan H. Catalytic roles of arginine residues 82 and 92 of Escherichia coli 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase: Site-directed mutagenesis and biochemical studies. Biochemistry. 2003;42:1581–1588. doi: 10.1021/bi026800z. [DOI] [PubMed] [Google Scholar]
- 28.Blaszczyk J, Li Y, Wu Y, Shi G, Ji X, Yan H. Essential roles of a dynamic loop in the catalysis of 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase. Biochemistry. 2004;43:1469–1477. doi: 10.1021/bi036053l. [DOI] [PubMed] [Google Scholar]
- 29.Bissantz C, Folkers G, Rognan D. Protein-based virtual screening of chemical databases. 1. Evaluation of different docking/scoring combinations. J Med Chem. 2000;43:4759–4767. doi: 10.1021/jm001044l. [DOI] [PubMed] [Google Scholar]