Abstract
The world population is expected to reach an estimated 9.2 billion by 2050. Therefore, food production globally has to increase by 70% in order to feed the world, while total arable land, which has reached its maximal utilization, may even decrease. Moreover, climate change adds yet another challenge to global food security. In order to feed the world in 2050, biotechnological advances in modern agriculture are essential. Plant genetic engineering, which has created a new wave of global crop production after the first green revolution, will continue to play an important role in modern agriculture to meet these challenges. Plastid genetic engineering, with several unique advantages including transgene containment, has made significant progress in the last two decades in various biotechnology applications including development of crops with high levels of resistance to insects, bacterial, fungal and viral diseases, different types of herbicides, drought, salt and cold tolerance, cytoplasmic male sterility, metabolic engineering, phytoremediation of toxic metals and production of many vaccine antigens, biopharmaceuticals and biofuels. However, useful traits should be engineered via chloroplast genomes of several major crops. This review provides insight into the current state of the art of plastid engineering in relation to agricultural production, especially for engineering agronomic traits. Understanding the bottleneck of this technology and challenges for improvement of major crops in a changing climate are discussed.
Keywords: Plastid engineering, Food security, Climate change, Cereal crops, Chloroplast genome
Introduction
Feeding the world in 2050 has become a main concern worldwide and is at the top of the agenda of the Food and Agriculture Organization (FAO) of the United Nations. Global food production needs to increase by 70% by 2050 in order to feed an expected population of 9.2 billion (High Level Expert Forum, FAO, October 2009; http://www.fao.org). With the success of the Green Revolution and biotechnology advances, agricultural production has been enhanced significantly over the past 40 years. Yield increases for major cereals (wheat, rice, maize) have amounted to 100–200% since the late 1960s (Mann 1999; http://www.fao.org). However, more than one billion people worldwide—about 100 million more than last year and around one-sixth of all of humanity—are still in hunger and undernourished, as shown in Fig. 1 (http://faostat.fao.org/faostat/, 2009). Moreover, yield growth rates have been unequally distributed across crops and regions. In developing countries, 80% of the production increases are projected to come from increases in yields and cropping intensity, and only 20% from expansion of arable land. In land-scarce countries, almost all of the production increases would have to be achieved through yield improvement. Unfortunately, yields of the major cereal crops have been steadily declining (FAO High Level Expert Forum; Rome 12–13 October 2009, http://www.fao.org). Furthermore, the global arable area reached its plateau by the mid-1970s and is now decreasing slowly due to increasing urbanization. Enhancement of crop yield for assuring food supply is thus a very challenging task.
Fig. 1.

Chart showing the prevalence of undernourishment in 102 developing countries in the world. Source The State of Food Insecurity in the World 2009 (www.fao.ord/publications/sofi)
Climate change creates an additional challenge to food security (FAO High Level Expert Forum; Rome 12–13 October 2009, http://www.fao.org). Adaptation of agricultural production and management to climate change will be costly but necessary for food security, reduction of poverty and maintenance of ecosystems (Sharkey et al. 2004; FAO High Level Expert Forum; http://www.fao.org). Reduction and removal of greenhouse gases (mitigation) from agriculture will alsobenecessary,ifglobal mitigation efforts are to be successful. To cope with climate change and secure food production for an increasing world population, the application and advancement of biotechnology are advantageous.
Plant biotechnology has played a significant role in modern agriculture in the past two decades. Since the first genetically modified (GM) crop was commercialized in 1996, global hectarage of biotech crops has continued to grow, reaching 134 million hectares in 2009 (James 2009). This translates to an increase of 9 million hectares over 2008, demonstrating the significance, economic benefits and great potential of GM crops. Of the commercialized GM crops so far, maize represents one of the most successfully engineered grass crops. However, the environmental risks, especially pollen-mediated transgene outflow and unintended genetic and epigenetic effects (Daniell 2002; Filipecki and Malepszy 2006) of the first generation of GM crops produced via nuclear transformation, has restricted its public perception in many countries, especially in Europe. The problem of transgene escape from GM crops to their wild relatives or other cultivated species is due to the presence of the transgene in pollen, which is a consequence of nuclear transformation (Daniell 2002; Grevich and Daniell 2005). Thus, the modification of important crops must be implemented in more sustainable approaches to accelerate crop production in an environmentally friendly manner, i.e. to conserve natural resources and preserve native habitats as well as to adapt cropping systems to climate changes (e.g. high CO2 level in the atmosphere, abiotic stresses created by extreme weather) that threaten crop productivity and food security worldwide (Martino-Catt and Sachs 2008).
Plastid genetic engineering (concept shown in the schematic drawing in Fig. 2; Daniell et al. 2002; Maliga 2004) offers several unique opportunities to plant biotechnologists. Besides the potential for high-level production of foreign proteins (>70% of total soluble protein, Oey et al. 2009; Ruhlman et al. 2010), other advantages of this technology include its effectiveness as a high-precision genetic engineering technique (site-specific transgene integration exclusively via homologous recombination; Verma and Daniell 2007), the absence from plastids of epigenetic effects and gene silencing mechanisms (Verma et al. 2008), the ease with which multiple transgenes can be stacked by linking them together in operons (as shown in Fig. 2, De Cosa et al. 2001; Ruiz et al. 2003; Quesada-Vargas et al. 2005), which is especially useful for metabolic engineering (Bock and Khan 2004; Bock 2007), and the lack of pleiotropic effects due to sub-cellular compartmentalization of even toxic transgene products (Lee et al. 2003; Daniell et al. 2005; Verma et al. 2010). Moreover, to avoid the deleterious effects caused by constitutive expression of transgene in chloroplasts, a nuclear-encoded ethanol-inducible plastid-targeted T7 RNA polymerase that transcribes plastid transgenes from a T7 promoter system was developed (Lössl et al. 2005). This inducible system which triggers transgene expression upon ethanol application further enhances the security and control of production in GM plants. Transgene containment via maternal inheritance is yet another important advantage of chloroplast engineering over nuclear transformation (Daniell et al. 1998, 2005; Daniell 2007; Ruf et al. 2007; Svab and Maliga 2007). Due to the potential for transgenes outcrossing with wild relatives or other crops via pollen, field production of many GM crops engineered via nuclear transformation has encountered strong opposition in several countries. Expression of foreign genes in chloroplasts could confine the pollen transmission because of the maternal inheritance nature of chloroplasts (Hagemann 2010), providing a better solution for engineering of agronomically important crops (Daniell 2007). Transgene containment via maternal inheritance would, however, not be applicable to a few crops that show biparental inheritance of chloroplast genomes (e.g. Medicago sativa, Actinidia sinensis) as well as several other species which are suspected to have biparental inheritance (Sears 1980; Corriveau and Coleman 1988; Harris and Ingram 1991; Zhang et al. 2003; Hu et al. 2008). Chloroplast transformation is ideal for harvesting transgene products expressed in leaves before development of any reproductive structures. However, transgene escape via seed dispersal would be a problem in transplastomic lines. For total transgene containment, reversible cytoplasmic male sterility has also been engineered via the chloroplast genome (Ruiz and Daniell 2005) to address rare events of paternal transmission (Ruf et al. 2007; Svab and Maliga 2007) or seed dispersal.
Fig. 2.
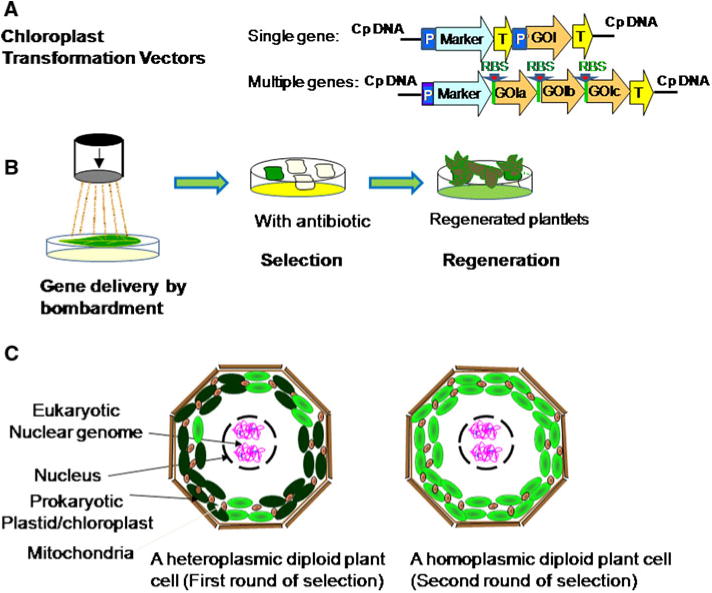
Schematic drawing of chloroplast transformation (Modified from Daniell et al. 2002): a Design and make gene constructs expressing single or multiple genes. Cp (chloroplast), P (promoter), marker (antibiotic selectable marker gene), T (terminator), GOI (gene of interest) and RBS (ribosome binding site); b Chloroplast transformation using particle gun bombardment followed by 2–3 rounds of antibiotic selections and subsequent regeneration of homoplasmic transformants; c heteroplasmic diploid plant cell (first round of selection) versus homoplasmic plant cell (second round of selection). Transgene integration is mediated by site-specific recombination
To date, a number of approaches have been developed to generate marker-free transplastomic plants to reduce the potential impact of antibiotic selectable marker which is a metabolic burden once the homoplastomic state is achieved in plastid transformants (Iamtham and Day 2000; Corneille et al. 2001; Lutz et al. 2006; Lutz and Maliga 2007). One of the widely known marker gene excision systems is the Cre-lox system based on site-specific recombination (Corneille et al. 2001; Lutz et al. 2006; Lutz and Maliga 2007). Another marker gene excision system has been developed by Iamtham and Day (2000), based on the homologous DNA recombination machinery in chloroplasts to excise antibiotic marker genes. This system, in comparison with the Cre-lox system which requires adding foreign recombinases, their target sites as well as subsequent removal of the foreign recombinase genes from the transplastomic plants, can obviate the need to remove foreign recombinase genes. Excision of antibiotic resistance gene once transformed plants have been obtained may simplify the regulatory approvalof GM crops (Daniell 2002; Lutz and Maliga 2007).
The state of the art of plastid biotechnology
Thus far, more than 100 transgenes have been stably integrated and expressed via the chloroplast genome to confer important agronomic traits, as well as to express industrially valuable biomaterials and therapeutic proteins (Bock 2007; Verma and Daniell 2007; Daniell et al. 2009; Bock and Warzecha 2010). Chloroplast transformation via organogenesis has been accomplished in a number of crops including lettuce (Lelivelt et al. 2005; Kanamoto et al. 2006; Ruhlman et al. 2007, 2010), cabbage (Liu et al. 2007), oilseed rape (Cheng et al. 2010), cauliflower (Nugent et al. 2006), poplar (Okumura et al. 2006), sugarbeet (De Marchis et al. 2009), potato (Sidorov et al. 1999; Nguyen et al. 2005; Valkov et al. 2010), tomato (Ruf et al. 2001) and eggplant (Singh et al. 2010). Organogenesis and (somatic) embryogenesis are the two pathways of regeneration in most flowering plants (angiosperms). Organogenesis involves the regeneration of adventitious shoots or roots through the formation of organized, meristematic tissues, whereas somatic embryogenesis involves the formation of embryos or embryo-like structures from somatic tissues. So far, most successful plastid transformation has been achieved through organogenesis using green leaves as explants. Unfortunately, most of the important food crops such as rice, wheat, maize etc. regenerate primarily through the somatic embryogenesis pathway. Successful plastid transformations through somatic embryogenesis were reported for carrot, cotton, soybean and rice (Kumar et al. 2004a, b; Dufourmantel et al. 2004; Grevich and Daniell 2005; Lee et al. 2006). The different types of plastids and regeneration systems (organogenesis or somatic embryogenesis) do affect plastid transformation. Although somatic embryogenesis has been successful in dicots, achieving homoplasmy in monocots has been quite challenging. This is one of the major obstacles to advancing plastid engineering in cereal crops. Chloroplast transformation using non-green cells and high level expression in chromoplasts or proplastids has been shown in carrots demonstrating the broad potential of the technology (Kumar et al. 2004a, b; Daniell et al. 2005). However, in ripe red tomato, expression of vaccine antigens in fruits is hardly detectable when compared to leaves (Zhou et al. 2008). This is due to the fact that ripe red tomatoes contain chromoplasts, in which most plastid genome-encoded genes are down regulated and plastid gene expression is less active than in photosynthetically active chloroplasts (Kahlau and Bock 2008). Furthermore, protein stability is another important factor determining the accumulation level of a foreign protein to be expressed in the plastids. Several studies have shown that major stability determinants are located in the N-ter-minus, contributing to our understanding of the protein stability and determinants in plastids (Apel et al. 2010; Leelavathi and Reddy 2003; Ruhlman et al., 2007; Lee et al. 2011).
Although homoplasmy was not established, success in rice chloroplast transformation (Lee et al. 2006) has provided encouragement to explore this technology in major cereal crops. To date, a number of agronomic traits have been engineered via plastid biotechnology, including insect resistance (McBride et al. 1995; Kota et al. 1999; De Cosa et al. 2001; Dufourmantel et al. 2006; Jin et al. 2011), herbicide tolerance (Daniell et al. 1998; Ye et al. 2001; Lutz et al. 2001), disease resistance against bacterial fungal and viral pathogens (DeGray et al. 2001; Lee et al. 2011; Verma et al. 2010), drought tolerance (Lee et al. 2003), salt tolerance (Kumar et al. 2004a), and cold tolerance (Craig et al. 2008). Table 1 summarizes the achievements of plastid engineering for improvement of agronomic traits. In addition, enhanced nutritional value (Apel and Bock 2009) or the ability to clean the environment from toxic metals like mercury (Ruiz et al. 2003; Hussein et al. 2007) or confer reversible cytoplasmic male sterility (Ruiz and Daniell 2005) have been demonstrated.
Table 1. Expression of agronomic trait-encoding genes via the plastid genome.
| Agronomic traits | Gene | Plastid genome | Expression level (%) total soluble protein (tsp) or activity | Reference |
|---|---|---|---|---|
| Insect resistance | Cry1A(c) | Tobacco | 3–5% | McBride et al. (1995) |
| Cry2Aa2 | Tobacco | 2–3%, killed resistant insects | Kota et al. (1999) | |
| Cry2Aa2 operon | Tobacco | 46%, killed cotton bollworm, beet armyworm | De Cosa et al. (2001) | |
| Cry1Ab protoxin | Soybean | Not quantified but adequate | Dufourmantel et al. (2006) | |
| Bgl1 | Tobacco | Killed aphids and whiteflies | Jin et al. (2011) | |
| Herbicide resistance | EPSPS (petunia) | Tobacco | Up to 5 mM glyphosate | Daniell et al. (1998) |
| Bar | Tobacco | Not quantified | Iamtham and Day (2000) | |
| Bar | Tobacco | More than 7% | Lutz et al. (2001) | |
| Herbicide tolerance | hppd | Tobacco Soybean | Ca 5% | Dufourmantel et al. (2007) |
| Disease resistance | MSI-99 | Tobacco | 21–43%, antibacterial, antifungal activities | DeGray et al. (2001) |
| Pectate lyase | Tobacco | Resistance against Erwinia soft rot | Verma et al. (2010) | |
| RC101 | Tobacco | Resistance against Erwinia soft rot and tobacco mosaic virus | Lee et al. (2011) | |
| Protegrin | Tobacco | Resistance against Erwinia soft rot | Lee et al. (2011) | |
| Drought tolerance | tps1 from yeast | Tobacco | 169-fold more tps 1 transcript than from nuclear transformants | Lee et al. (2003) |
| Salt tolerance | Badh | Carrot | Up to 400 mM salt tolerance | Kumar et al. (2004a) |
| Cold tolerance | Δ9desaturase | Tobacco | Not quantified | Craig et al. (2008) |
Nevertheless, despite the significant progress in plastid genetic engineering and the unique advantages of this technology over nuclear transformation, plastid transformation is still limited to dicotyledon species (as shown in Table 1). The development of this technology for major cereal crops (e.g. maize, rice, wheat, barley) to enhance their productivity in the era of climate change is of significance for future agriculture worldwide. Increasing the photosynthetic capacity of plants to cope with the elevated CO2 level and improving adaptation of plants to extreme climatic stresses such as increasingly severe droughts, heat or floods are the challenges ahead, and solutions are likely to be found with the help of biotechnology.
Significance of crop chloroplast genome sequences for plastid engineering
The chloroplast genome is highly conserved within most land plants (Jansen et al. 2005), composed of a single circular chromosome with a quadripartite structure that includes two copies of an inverted repeat (IR) that separate the large and small single copy regions (LSC and SSC). The size of the circular genome varies from 35 to 217 kb, but among photosynthetic organisms most are between 115 and 165 kb (Jansen et al. 2005). The chloroplast genome information is essential for successful chloroplast transformation. To achieve efficient homologous recombination and integration of transgenes into chloroplast genomes, flanking sequences with the intergenic spacer regions are essential. Endogenous regulatory sequences are required for optimal expression of foreign genes within chloroplasts (Verma and Daniell 2007). In a recent study in which lettuce chloroplast flanking sequence was used to integrate transgenes into the tobacco chloroplast genome, all sequences unique for lettuce chloroplast genome (including single nucleotide changes) were eliminated by the tobacco chloroplast genome during homologous recombination events. Increase in the number of homologous recombination events significantly decreased the efficiency of transformation (Ruhlman et al. 2010). The site of integration may also play a significant role in transgene expression. In early studies of transgene integration into the chloroplast genome, transcriptionally silent spacer regions were used. However, the transcriptionally active spacer regions between the trnI and trnA genes located within the ribosomal operon resulted in the highest levels of transgene expression (De Cosa et al. 2001; Ruhlman et al. 2010) and this is the most commonly used site for transgene integration (Verma and Daniell 2007; Verma et al. 2008). While the trnI/trnA site has been used to engineer all agronomic traits including insect or herbicide resistance, drought or salt tolerance, phyto-remediation and cytoplasmic male sterility, other sites have been used only for insect resistance or the bar gene. Likewise, a lot more vaccine antigens and biopharmaceuticals have been engineered via the trnI/trnA site than any other site (Daniell et al. 2009). The trnI-trnA site has several advantages over other integration sites: location within the inverted repeat regions of the chloroplast genome doubles the copy number of transgenes; the copy correction mechanism, and presence of replication origin and introns for accurate splicing of foreign transcripts (Verma and Daniell 2007). A recent study showed that transgene integration into the trnI-trnA site increased expression of the lux operon (driven by the same regulatory sequences) 25-fold more than the transcriptionally silent spacer region (rps12-trnV), under-scoring the importance of the site of transgene integration (Krichevsky et al. 2010).
The lack of chloroplast crop genome information has in the past limited the application of plastid transformation technology in many crops (Daniell et al. 2006; Ruhlman et al. 2010). Although the tobacco chloroplast genome was sequenced in 1986, until 2004 only five other crop chloroplast genomes were sequenced, in spite of hundreds of chloroplast genomes of weedy species having been sequenced for phylogenetic studies. Species specific flanking sequences and endogenous regulatory sequences are needed to make chloroplast vectors (see Ruhlman et al. 2010).
When the soybean complete chloroplast genome was published (Saski et al. 2005), there were less than ten crop chloroplast genomes sequenced, almost 20 years after the first chloroplast genome was published. However, in the past few years about thirty crop chloroplast genomes have been published but several hundred non-crop chloroplast genomes have been sequenced. For a summarized list of sequenced crop chloroplast genomes, see Verma et al. (2008). Recently, the complete chloroplast genome sequence of perennial ryegrass (Lolium perenne L.) was reported, providing valuable information for the future plastid engineering of the forage species (Diekmann et al. 2009). As pointed out above, utilization of heterologous intergenic spacer regions that have low sequence identities between the target genome and the flanking sequences in the chloroplast transformation vectors resulted in substantially lower frequencies of transformants (Verma and Daniell 2007). Use of heterologous flanking sequences have clearly shown that even single nucleotide changes are eliminated during recombination via multiple recombination events, thereby significantly reducing transformation efficiency. In this study, the same tissue culture system was used to eliminate the contribution of other factors (Ruhlman et al. 2010). Similar results were observed in plastid transformation of Arabidopsis, potato and tomato, where tobacco chloroplast genome flanking or regulatory sequences were used; the efficiency was much lower than in tobacco (Sikdar et al. 1998; Sidorov et al. 1999; Ruf et al. 2001). Furthermore, when Petunia and Solanum flanking sequences were used in the chloroplast transformation of tobacco, a similar situation occurred (DeGray et al. 2001; Zubko et al. 2004). Moreover, published studies show that not a single intergenic spacer region is conserved among nine grass chloroplast genomes (Saski et al. 2007) or only four spacer regions are conserved among Solanaceous crops (Daniell et al. 2006). Because transgenes are integrated into intergenic spacer regions, their sequence information is essential for making chloroplast vectors. It is therefore unlikely that a single, highly conserved intergenic spacer region will be appropriate throughout the grass family which consists of the principal crops rice, wheat, maize, sorghum, barley etc. These examples demonstrate the significance of complete chloroplast genome sequences in successful plastid engineering and the importance of utilization of species-specific flanking sequences for assuring efficient homologous recombination within the intergenic spacer regions.
The utilization of endogenous regulatory sequences is another important factor which should be considered in the vector construction for monocot plastid transformation. Use of lettuce psbA promoter and UTRs in tobacco or tobacco psbA regulatory sequences in lettuce resulted in >90% reduction in the levelof transgene expression (Ruhlmanet al. 2010). Therefore, in addition to use of endogenous flanking sequences for homologous recombination, it is important to use endogenous regulatory sequences for efficient transgene expression. These studies point out the need for construction of species specific chloroplast vectors, the foundation of successful plastid transformation.
Plastid engineering in cereal crops for food security
Unfortunately, most plastid engineering has been restricted to tobacco and a few other dicot crops. Future plastid engineering should focus on the major cereal crops in order to enhance crop production and improve the nutritional content necessary to feed the world. The encouraging development in soybean plastid engineering has showed the potential of this technology for major food crops (Dufourmantel et al. 2006; Grevich and Daniell 2005). Engineering of carotenoid biosynthesis in transplastomic tomatoes has demonstrated the potential of plastid genome engineering for the nutritional enhancement of food crops (Apel and Bock 2009). Nevertheless, plastid genome engineering is still restricted largely to dicot plants, especially tobacco. The most important food crops in the world, such as rice, wheat, maize, sorghum, and barley, are still not amenable to plastid transformation. With the urgent need to increase food crop yield in order to feed 9.2 billion people in the world in 2050, breakthroughs in plastid engineering of cereal crops are of great importance for enabling the genetic manipulation of these crops in a contained and environmentally friendly manner (Daniell 2002; Daniell et al. 2005). Better understanding of the plastid genetics and genomes of cereal crops, efficient tissue culture and regeneration systems for these crops, improvement of selection systems (with new selectable marker genes), identification of intergenic spacer regions for transgene integration, and understanding of the possible factors/proteins affecting the process from heteroplasmy to homoplasmy and subsequent promotion of homoplasmy, are the challenges ahead. Nonetheless, encouraging progress has been made in these areas. For instance, a new selectable marker, chloramphenicol acetyltransferase gene (cat), for plastid transformation was recently reported by Li et al. (2010). Authors showed that the cat gene allows the transplastomic tobacco lines to quickly reach the homoplasmic state, an advantage for plastid transformation of plants.
Major obstacles of plastid engineering in cereal crops include the difficulty of expressing transgenes in non-green plastids, in which gene expression and gene regulation systems are quite distinct from those of chloroplasts. When proplastids, which are about five-fold smaller in size than chloroplasts, are used as the transformation target, irreversible physical plastids due to biolistic bombardment might be greater. It may also be necessary to develop new selection markers for a monocot-specific selection scheme. In addition, instability of DNA in cereal chloroplasts is yet another challenge (Oldenburg and Bendich 2004a, b). It has been recently shown by Zheng et al. (2011) that when dark grown maize seedlings are transferred to light, the DNA content per plastid declines rapidly to a minimum at 24 h during greening while chlorophyll fluorescence continues to increase. Therefore, the authors concluded that the increase in chloroplast DNA that accompanies plastid enlargement is a consequence of cell and leaf growth, whereas light stimulates photosynthetic capacity and chloroplast DNA instability. This may explain one of the challenges in achieving homoplasmy in cereal crop plastid transformation. It was also proposed that the loss of DNA from chloroplasts causes leaf senescence in coleoptiles and young leaves of rice (Sodmergen et al. 1991), but this seems not to be the case in maize where degradation of cpDNA during maize chloroplast maturation may benefit the plants (Oldenburg and Bendich 2004b). Therefore, the stability issue of cpDNA in cereal crops is of great importance.
Despite failure to obtain homoplasmic transformants, the rice plastid transformation by Lee et al. (2006) has enriched our understanding of the challenges and difficulties in rice and other monocot plastid transformation. We know that rice plastid transformation is quite inefficient and no homoplasmy has ever been achieved, when compared to chloroplast transformation in dicots, which is well established in many labs. There are several possible reasons for the difficulty in monocot plastid transformation: (1) lack of an efficient regeneration system in monocotyledonous cereal crops, including rice; (2) the target plastids are smaller than chloroplasts, leading to the possibility of physical damage due to biolistic bombardment with 0.6 μm gold particles possibly being greater in the proplastids. The attempt to use smaller particle sizes (0.4 μm) has resulted in three- to four-fold increases in plastid transformation frequency during the transformation of proplastids in dark-grown tobacco suspension cells (Langbecker et al. 2004); (3) transcription and translation levels are lower in proplastids than in chloroplasts in accordance with previous reports by Mullet (1993) and Silhavy and Maliga (1998) so the recovery of the newly transformed cells is attenuated further during the initial selection phase; (4) the effect of the Prrn plastid-encoded RNA polymerase PEP and NEP promoters in leucoplasts; (5) utilization of a variety of 5′ UTRs; (6) all of the homoplastomic plants produced thus far have been selected under light conditions, through a repeated process of subculturing of transformed leaf tissues; however, this subculturing process cannot be applied to cereal crops; (7) the necessity of developing new selection markers for a monocot-specific selection scheme, because the use of suitable selection markers has been shown to be critical for success in the transformation of plastids in tobacco, lettuce and other dicots (see reviews by Bock 2007; Daniell et al. 2009). With efforts from the global research community, these hurdles may be overcome in the near future.
Plastid engineering of biosynthetic pathways for improvement of the nutritional value of food crops
Climate change introduces new challenges to agricultural production, not only in yield (quantity) but also in its nutritional aspect (food quality). Plastid engineering has become a valuable tool for metabolic engineering of pathways such as photosynthetic carbon reduction, starch and lipid biosyntheses etc., as it can express several transgenes by linking them in operons, making it possible to engineer several enzymatic steps, in some cases the entire pathway, simultaneously (Bock and Khan 2004; Bock 2007; Wurbs et al. 2007; Apel and Bock 2009). Furthermore, in plastid genome engineering, the expression level of foreign genes in plastids can be regulated by using desirable promoters and/or 5′ and 3′ untranslated regions (Bock 2007). A recent study on manipulation of the carotenoid biosynthetic pathway in tomato using plastid transformation has demonstrated the advantages of plastid genome manipulation for metabolic engineering of complex pathways, as well as the potential of the technology for improvement of nutritional content in food crops (Apel and Bock 2009). By introducing the lycopene β- cyclase genes from Erwinia herbicola and daffodil (Narcissus pseudonarcissus) into the tomato plastid genome, the authors have reported a significant high accumulation of total carotenoid with an increase of over 50%, and most of the lycopene was converted into β-carotene with provitamin A levels reaching 1 mg per gram dry weight in fruits (Apel and Bock 2009). Another study by Craig et al. (2008) has demonstrated the feasibility of using plastid transformation to engineer lipid metabolic pathways in both vegetative and reproductive tissues. Transplastomic tobacco plants expressing a fatty acid D9 desaturase gene exhibited altered fatty acid profiles and improved cold tolerance compared with the control (Craig et al. 2008).
The studies described above have demonstrated the feasibility and potential of plastid biotechnology for manipulation of plants for future food production, with adaptation to biotic and abiotic stresses under the changing climate.
Conclusions and future perspectives
Adaptation to climate change—including the ability to mitigate exposure to, and cope with, extreme climate changes—will be necessary to ensure global food security in both the short and long term. With better understanding and technological advancement of plastid genome engineering, this technology should be able to contribute to the improvement of crop productivity as well as other important agronomical traits and industrial applications (Table 2). The sobering fact that globally the rate of growth in yields of the major cereal crops has been steadily declining is an unfortunate reality. The rate of growth in global cereal yields, for example, dropped from 3.2% per year in 1960 to 1.5% in 2000. The challenge for biotechnology is to assist in reversing this decline in order to meet global food needs.
Table 2. Aspirational goals for plastid biotechnology.
| Examples | Future directions |
|---|---|
| Agronomic importance | Except for insect, herbicide and salt tolerance engineered in soybean and carrot, all others traits should be engineered in useful crops. Nitrogen fixation should be first demonstrated in a suitable model plant |
| Insect resistance | |
| Herbicide resistance | |
| Disease resistance | |
| Drought and salt tolerance | |
| Cold tolerance | |
| Nitrogen fixation | |
| Metabolic pathways | Metabolic engineering has been demonstrated in tomato; should be advanced in major crops |
| Provitamin A | |
| Engineering vitamin B12 | |
| Synthesis of long-chain PUFAs | |
| Biopharmaceuticals and vaccines for human health | Tobacco is not a suitable system for oral delivery, a major advantage of plant molecular farming. To date, only a few vaccine antigens have been expressed in lettuce chloroplasts. More efforts should be made to express biopharmaceuticals and vaccine antigens in useful crops suitable for oral delivery. Antigen expression in tomato fruit should be improved |
| Human growth hormones | |
| Human growth factors | |
| Human blood proteins | |
| Vaccines for animal health | Tobacco is not suitable for animal feed. More efforts should be made to develop animal vaccines in crops or edible algae which can be used as animal feed |
| Bacterial antigens | |
| Viral antigens | |
| Phyto-remediation | Only phyto-remediation to clean up mercury in soil has been demonstrated so far. Many other toxic metals and persistent organic pollutants should be addressed. Phyto-remediation systems should be developed to clean up water |
| Heavy metals | |
| Organomercury compounds | |
| Biofuels and industrial enzymes | Cocktails for two biomasses (pine wood, citrus peel) were developed; more work is needed for developing new enzymes for several other biomasses. In addition, valuable compounds and secondary metabolites could be engineered |
| Cell wall degrading enzymes | |
| Biopolymers | |
| Compounds for cosmetic application | |
| Photosynthesis | Photosynthetic efficiency in major crops could be improved to enhance food production. Performance of Rubisco under elevated atmospheric CO2 and abiotic stresses could be addressed |
| Rubisco | |
| C3 and C4 plants |
It is evident that plastid genome engineering holds great promise for the improvement of crop productivity. Efforts should in the future be directed to plastid genome engineering of monocots so as to facilitate the manipulation and improvement of many agronomically and economically important crops in order to feed the world in 2050.
Acknowledgments
We thank the Research Council of Norway for grant BILAT-174998/D15 and Bioforsk core funding to Jihong Liu Clarke and USDA-CSREES, USDA-NIFA and NIH 2 R01 GM 063879 to Henry Daniell. The authors thank Nicholas Clarke and Sonja Klemsdal for critical reading and Shuai Guo for his help with references.
Contributor Information
Jihong Liu Clarke, Email: jihong.liu-clarke@bioforsk.no, Plant Health and Protection Division, Bioforsk- Norwegian, Institute for Agricultural and Environmental Research, Hoegskoleveien 7, 1432 Aas, Norway.
Henry Daniell, Email: daniell@mail.ucf.edu, Department of Molecular Biology and Microbiology, College of Medicine, University of Central Florida, 336 Biomolecular Science Building, Orlando, FL 32816-2364, USA.
References
- Apel W, Bock R. Enhancement of carotenoid biosynthesis in transplastomic tomatoes by induced lycopene-to-provitamin A conversion. Plant Physiol. 2009;151:59–66. doi: 10.1104/pp.109.140533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apel W, Schulze WX, Bock R. Identification of protein stability determinants in chloroplasts. Plant J. 2010;63:636–650. doi: 10.1111/j.1365-313X.2010.04268.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock R. Plastid biotechnology: prospects for herbicide and insect resistance, metabolic engineering and molecular farming. Curr Opin Biotechnol. 2007;18:100–106. doi: 10.1016/j.copbio.2006.12.001. [DOI] [PubMed] [Google Scholar]
- Bock R, Khan MS. Taming plastids for a green future. Trends Biotechnol. 2004;22:311–318. doi: 10.1016/j.tibtech.2004.03.005. [DOI] [PubMed] [Google Scholar]
- Bock R, Warzecha H. Solar-powered factories for new vaccines and antibiotics. Trends Biotechnol. 2010;28(5):246–252. doi: 10.1016/j.tibtech.2010.01.006. [DOI] [PubMed] [Google Scholar]
- Cheng L, Li HP, Qu B, Huang T, Tu JX, Fu TD, Liao YC. Chloroplast transformation of rapeseed (Brassica napus) by particle bombardment of cotyledons. Plant Cell Rep. 2010;29:371–381. doi: 10.1007/s00299-010-0828-6. [DOI] [PubMed] [Google Scholar]
- Corneille S, Lutz K, Svab Z, Maliga P. Efficient elimination of selectable markergenes from the plastid genome by the CRE-lox site-specific recombination system. Plant J. 2001;72:171–178. doi: 10.1046/j.1365-313x.2001.01068.x. [DOI] [PubMed] [Google Scholar]
- Corriveau JL, Coleman AW. Rapid screening method to detect potential biparental inheritance of plastid DNA and results for over 200 angiosperm species. Amer J Bot. 1988;75:1443–1458. [Google Scholar]
- Craig W, Lenzi P, Scotti N, et al. Transplastomic tobacco plants expressing a fatty acid desaturase gene exhibit altered fatty acid profiles and improved cold tolerance. Transgenic Res. 2008;17:769–782. doi: 10.1007/s11248-008-9164-9. [DOI] [PubMed] [Google Scholar]
- Daniell H. Molecular strategies for gene containment in transgenic crops. Nat Biotechnol. 2002;20:581–586. doi: 10.1038/nbt0602-581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniell H. Transgene containment by maternal inheritance: effective or elusive? Proc Natl Acad Sci USA. 2007;104:6879–6880. doi: 10.1073/pnas.0702219104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniell H, Datta R, Varma S, Gray S, Lee SB. Containment of herbicide resistance through genetic engineering of the chloroplast genome. Nat Biotechnol. 1998;16:345–348. doi: 10.1038/nbt0498-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniell H, Khan M, Allison L. Milestones in chloroplast genetic engineering: an environmentally friendly era in biotechnology. Trends Plant Sci. 2002;7(2):84–91. doi: 10.1016/s1360-1385(01)02193-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniell H, Kumar S, Dufourmantel N. Breakthrough in chloroplast genetic engineering of agronomically important crops. Trends Biotechnol. 2005;23:238–245. doi: 10.1016/j.tibtech.2005.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniell H, Lee SB, Grevich J, et al. Complete chloroplast genome sequences of Solanum bulbocastanum, Solanum lycopersicum and comparative analyses with other Solanaceae genomes. Theor Appl Genet. 2006;112:1503–1518. doi: 10.1007/s00122-006-0254-x. [DOI] [PubMed] [Google Scholar]
- Daniell H, Singh ND, Mason H, Streatfield SJ. Plant-made vaccines and biopharmaceuticals. Trends Plant Sci. 2009;14:669–679. doi: 10.1016/j.tplants.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Cosa B, Moar W, Lee SB, Miller M, Daniell H. Overexpression of the Bt cry2Aa2 operon in chloroplasts leads to formation of insecticidal crystals. Nat Biotechnol. 2001;19:71–74. doi: 10.1038/83559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Marchis F, Wang YX, Stevanato P, Arcioni S, Bellucci M. Genetic transformation of the sugar beet plastome. Transgenic Res. 2009;18:17–30. doi: 10.1007/s11248-008-9193-4. [DOI] [PubMed] [Google Scholar]
- DeGray G, Rajasekaran K, Smith F, Sanford J, Daniell H. Expression of an antimicrobial peptide via the chloroplast genome to control phytopathogenic bacteria and fungi. Plant Physiol. 2001;127:852–862. [PMC free article] [PubMed] [Google Scholar]
- Diekmann K, Hodkinson TR, Wolfe KH, Van Den Bekerom R, Dix PJ, Barth S. Complete chloroplast genome sequence of a major allogamous forage species perennial ryegrass (Lolium perenne L.) DNA Res. 2009;16:165–176. doi: 10.1093/dnares/dsp008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufourmantel N, Pelissier B, Garcon F, Peltier G, Ferullo JM, Tissot G. Generation of fertile transplastomic soybean. Plant Mol Biol. 2004;55:479–489. doi: 10.1007/s11103-004-0192-4. [DOI] [PubMed] [Google Scholar]
- Dufourmantel N, Tissot G, Goutorbe F, et al. Generation and analysis of soybean plastid transformants expressing Bacillus thuringiensis Cry1Ab protoxin. Plant Mol Biol. 2006;58:659–668. doi: 10.1007/s11103-005-7405-3. [DOI] [PubMed] [Google Scholar]
- Dufourmantel N, Dubald M, Matringe M, Canard H, Garcon F, Job C, Kay E, Wisniewski JP, Ferullo JM, Pelissier B, Sailland A, Tissot G. Generation and characterization of soybean and marker-free tobacco plastid transformants over-expressing a bacterial 4-hydroxyphenylpyruvate dioxygenase which provides strong herbicide tolerance. Plant Biotechnol J. 2007;5:118–133. doi: 10.1111/j.1467-7652.2006.00226.x. [DOI] [PubMed] [Google Scholar]
- Filipecki M, Malepszy S. Unintended consequences of plant transformation: a molecular insight. J Appl Genet. 2006;47:277–286. doi: 10.1007/BF03194637. [DOI] [PubMed] [Google Scholar]
- Grevich JJ, Daniell H. Chloroplast genetic engineering: recent advances and future perspectives. Crit Rev Plant Sci. 2005;24:83–107. [Google Scholar]
- Hagemann R. The foundation of extranuclear inheritance: plastid and mitochondrial genetics. Mol Genet Genomics. 2010;283:199–209. doi: 10.1007/s00438-010-0521-z. [DOI] [PubMed] [Google Scholar]
- Harris SA, Ingram R. Chloroplast DNA and biosystematics: the effects of intraspecific diversity and plastid transmission. Taxon. 1991;40:393–412. [Google Scholar]
- Hu Y, Zhang Q, Rao G, Sodmergen Occurrence of plastids in the sperm cells of Caprifoliaceae: biparental plastid inheritance in Angiosperms is unilaterally derived from maternal inheritance. Plant Cell Physiol. 2008;49:958–968. doi: 10.1093/pcp/pcn069. [DOI] [PubMed] [Google Scholar]
- Hussein SH, Ruiz ON, Terry N, Daniell H. Phytoremediation of mercury and organomercurials in chloroplast transgenic plants: enhanced root uptake, translocation to shoots and volatilization. Environm Sci Technol. 2007;41:8439–8446. doi: 10.1021/es070908q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iamtham S, Day A. Removal of antibiotic resistance genes from transgenic tobacco plastids. Nat Biotechnol. 2000;18:1172–1176. doi: 10.1038/81161. [DOI] [PubMed] [Google Scholar]
- James C. ISAAA Brief No 41. ISAAA; NY: 2009. Global status of commercialized biotech/GM Crops: 2009. [Google Scholar]
- Jansen RK, Raubeson LA, Boore JL, et al. Methods for obtaining and analyzing chloroplast genome sequences. Meth Enzym. 2005;395:348–384. doi: 10.1016/S0076-6879(05)95020-9. [DOI] [PubMed] [Google Scholar]
- Jin S, Kanagaraj A, Verma D, Lange T, Daniell H. Release of hormones from conjugates: chloroplast expression of B-glucosidase results in elevated phytohormone levels with significant increase in biomass and protection from aphids and whiteflies conferred by sucrose esters. Plant Physiol. 2011;155:222–235. doi: 10.1104/pp.110.160754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahlau S, Bock R. Plastid transcriptomics and translatomics of tomato fruit development and chloroplast-to-chromoplast differentiation: chromoplast gene expression largely serves the production of a single protein. Plant Cell. 2008;20:856–874. doi: 10.1105/tpc.107.055202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanamoto H, Yamashita A, Asao H, Okumura S, Takase H, Hattori M, Yokota A, Tomizawa K. Efficient and stable transformation of Lactuca sativa L. cv. Cisco (lettuce) plastids. Transgenic Res. 2006;15:205–217. doi: 10.1007/s11248-005-3997-2. [DOI] [PubMed] [Google Scholar]
- Kota M, Daniell H, Varma S, Garczynski SF, Gould F, Moar WJ. Overexpression of the Bacillus thuringiensis (Bt) Cry2Aa2 protein in chloroplasts confers resistance to plants against susceptible and Bt-resistant insects. Proc Natl Acad Sci USA. 1999;96:1840–1845. doi: 10.1073/pnas.96.5.1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krichevsky A, Meyers B, Vainstein A, Maliga P, Citovsky V. Autoluminescent plants. PLoS One. 2010;5:215461. doi: 10.1371/journal.pone.0015461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Dhingra A, Daniell H. Plastid expressed betaine aldehyde dehydrogenase gene in carrot cultured cells, roots and leaves confers enhanced salt tolerance. Plant Physiol. 2004a;136:2843–2854. doi: 10.1104/pp.104.045187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Dhingra A, Daniell H. Stable transformation of the cotton plastid genome and maternal inheritance of transgenes. Plant Mol Biol. 2004b;56:203–216. doi: 10.1007/s11103-004-2907-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langbecker C, Ye GN, Broyles DL, Duggan LL, Xu CW, Hajdukiewicz PTJ, Armstrong CL, Staub JM. High-frequency transformation of undeveloped plastids in tobacco suspension cells. Plant Physiol. 2004;135:39–46. doi: 10.1104/pp.103.035410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SB, Kwon HB, Kwon SJ, Park SC, Jeong MJ, Han SE, Byun MO, Daniell H. Accumulation of trehalose within transgenic chloroplasts confers drought tolerance. Mol Breed. 2003;11:1–13. [Google Scholar]
- Lee SM, Kang KS, Chung H, Yoo SH, Xu XM, Lee SB, Cheong JJ, Daniell H, Kim M. Plastid transformation in the monocotyledonous cereal crop, rice (Oryza sativa) and transmission of transgenes to their progeny. Mol Cells. 2006;21:401–410. [PMC free article] [PubMed] [Google Scholar]
- Lee SB, Li B, Jin S, Daniell H. Expression and characterization of antimicrobial peptides Retrocyclin-101 and Protegrin-1 in chloroplasts to control viral and bacterial infections. Plant Biotechnol J. 2011;9:100–115. doi: 10.1111/j.1467-7652.2010.00538.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leelavathi S, Reddy VS. Chloroplast expression of His-tagged GUS-fusions: a general strategy to overproduce and purify foreign proteins using transplastomic plants as bioreactors. Mol Breed. 2003;11:49–58. [Google Scholar]
- Lelivelt CLC, McCabe MS, Newell CA, deSnoo CB, van Dun KMP, Birch-Machin I, Gray JC, Mills KHG, Nugent JM. Stable plastid transformation in lettuce (Lactuca sativa L.) Plant Mol Biol. 2005;58:763–774. doi: 10.1007/s11103-005-7704-8. [DOI] [PubMed] [Google Scholar]
- Li W, Ruf S, Bock R. Chloramphenicol acetyltransferase as selectable marker for plastid transformation. Plant Mol Biol. 2010 doi: 10.1007/s11103-010-9678-4. [DOI] [PubMed] [Google Scholar]
- Liu CW, Lin CC, Chen JJW, Tseng MJ. Stable chloroplast transformation in cabbage (Brassica oleracea L. var. capitata L.) by particle bombardment. Plant Cell Rep. 2007;26:1733–1744. doi: 10.1007/s00299-007-0374-z. [DOI] [PubMed] [Google Scholar]
- Lössl A, Bohmert K, Harloff H, Eibl C, Mühlbauer S, Koop HU. Inducible trans-activation of plastid transgenes: expression of the Reutropha phb Operon in Transplastomic Tobacco. Plant Cell Physiol. 2005;46(9):1462–1471. doi: 10.1093/pcp/pci157. [DOI] [PubMed] [Google Scholar]
- Lutz KA, Maliga P. Construction of marker-free transplastomic plants. Curr Opin Biotechnol. 2007;18:107–114. doi: 10.1016/j.copbio.2007.02.003. [DOI] [PubMed] [Google Scholar]
- Lutz KA, Knapp JE, Maliga P. Expression of bar in the plastid genome confers herbicide resistance. Plant Physiol. 2001;125:1585–1590. doi: 10.1104/pp.125.4.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz KA, Svab Z, Maliga P. Construction of marker-free transplastomic tobacco using the Cre-loxP site-specific recombination system. Nat Protoc. 2006;1:900–910. doi: 10.1038/nprot.2006.118. [DOI] [PubMed] [Google Scholar]
- Maliga P. Plastid transformation in higher plants. Ann Rev Plant Biol. 2004;55:289–313. doi: 10.1146/annurev.arplant.55.031903.141633. [DOI] [PubMed] [Google Scholar]
- Mann CC. Crop scientists seek a new revolution. Science. 1999;283:310–314. [Google Scholar]
- Martino-Catt SJ, Sachs ES. Editor's choice series: the next generation of biotech crops. Plant Physiol. 2008;147:3–5. doi: 10.1104/pp.104.900256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride KE, Svab Z, Schaaf DJ, Hogan PS, Stalker DM, Maliga P. Amplification of a chimeric Bacillus gene in chloroplasts leads to an extraordinary level of an insecticidal proteinin tobacco. Bio-Technol. 1995;13:362–365. doi: 10.1038/nbt0495-362. [DOI] [PubMed] [Google Scholar]
- Mullet J. Dynamic regulation of chloroplast transcription. Plant Physiol. 1993;103:309–313. doi: 10.1104/pp.103.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen TT, Nugent G, Cardi T, Dix PJ. Generation of homoplasmic plastid transformants of a commercial cultivar of potato (Solanum tuberosum L.) Plant Sci. 2005;168:1495–1500. [Google Scholar]
- Nugent GD, Coyne S, Nguyen TT, Kavanagh TA, Dix PJ. Nuclear and plastid transformation of Brassica oleracea var. botrytis (cauliflower) using PEG-mediated uptake of DNA into protoplasts. Plant Sci. 2006;170:135–142. [Google Scholar]
- Oey M, Lohse M, Kreikemeyer B, Bock R. Exhaustion of the chloroplast protein synthesis capacity by massive expression of a highly stable protein antibiotic. Plant J. 2009;57:436–445. doi: 10.1111/j.1365-313X.2008.03702.x. [DOI] [PubMed] [Google Scholar]
- Okumura S, Sawada M, Park YW, Hayashi T, Shimamura M, Takase H, Tomizawa KI. Transformation of poplar (Populus alba) plastids and expression of foreign proteins in tree chloroplasts. Transgenic Res. 2006;15:637–646. doi: 10.1007/s11248-006-9009-3. [DOI] [PubMed] [Google Scholar]
- Oldenburg DJ, Bendich AJ. Changes in the structure of DNA molecules and the amount of DNA per plastid during chloroplast development in maize. J Mol Biol. 2004a;344:1311–1330. doi: 10.1016/j.jmb.2004.10.001. [DOI] [PubMed] [Google Scholar]
- Oldenburg DJ, Bendich AJ. Most chloroplast DNA of maize seedlings in linear molecules with defined ends and branched forms. J Mol Biol. 2004b;335:953–970. doi: 10.1016/j.jmb.2003.11.020. [DOI] [PubMed] [Google Scholar]
- Quesada-Vargas T, Ruiz ON, Daniell H. Characterization of heterologous multigene operons in transgenic chloroplasts. Transcription, processing, and translation. Plant Physiol. 2005;138:1746–1762. doi: 10.1104/pp.105.063040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruf S, Hermann M, Berger IJ, Carrer H, Bock R. Stable genetic transformation of tomato plastids and expression of a foreign protein in fruit. Nat Biotechnol. 2001;19:870–875. doi: 10.1038/nbt0901-870. [DOI] [PubMed] [Google Scholar]
- Ruf S, Karcher D, Bock R. Determining the transgene containment level provided by chloroplast transformation. Proc Natl Acad Sci USA. 2007;104:6998–7002. doi: 10.1073/pnas.0700008104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruhlman T, Ahangari R, Devine A, Samsam M, Daniell H. Expression of cholera toxin B-proinsulin fusion protein in lettuce and tobacco chloroplasts—oral administration protects against development of insulitis in non-obese diabetic mice. Plant Biotechnol J. 2007;5:495–510. doi: 10.1111/j.1467-7652.2007.00259.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruhlman T, Verma D, Samson N, Daniell H. The role of heterologous chloroplast sequence elements in transgene integration and expression. Plant Physiol. 2010;152:2088–2104. doi: 10.1104/pp.109.152017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz ON, Daniell H. Engineering cytoplasmic male sterility via the chloroplast genome by expression of beta-ketothiolase. Plant Physiol. 2005;138:1232–1246. doi: 10.1104/pp.104.057729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz ON, Hussein HS, Terry N, Daniell H. Phytoremediation of organomercurial compounds via chloroplast genetic engineering. Plant Physiol. 2003;132:1344–1352. doi: 10.1104/pp.103.020958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saski C, Lee SB, Daniell H, Wood TC, Tomkins J, Kim HG, Jansen RK. Complete chloroplast genome sequence of Glycine max and comparative analyses with other legume genomes. Plant Mol Biol. 2005;59(2):309–322. doi: 10.1007/s11103-005-8882-0. [DOI] [PubMed] [Google Scholar]
- Saski C, Lee SB, Fjellheim S, Guda C, Jansen RK, Luo H, Tomkins J, Rognli OA, Daniell H, Clarke JL. Complete chloroplast genome sequences of Hordeum vulgare, Sorghum bicolor and Agrostis stolonifera, and comparative analyses with other grass genomes. Theor Appl Genet. 2007;115:571–590. doi: 10.1007/s00122-007-0567-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sears BB. Elimination of plastids during spermatogenesis and fertilization in the plant kingdom. Plasmid. 1980;4:233–255. doi: 10.1016/0147-619x(80)90063-3. [DOI] [PubMed] [Google Scholar]
- Sharkey TD, Laporte M, Lu Y, Weise S, Weber APM. Engineering plants for elevated CO2: a relationship between starch degradation and sugar sensing. Plant Biol. 2004;6:280–288. doi: 10.1055/s-2004-817911. [DOI] [PubMed] [Google Scholar]
- Sidorov VA, Kasten D, Pang SZ, Hajdukiewicz PTJ, Staub JM, Nehra NS. Stable chloroplast transformation in potato: use of green fluorescent protein as a plastid marker. Plant J. 1999;19:209–216. doi: 10.1046/j.1365-313x.1999.00508.x. [DOI] [PubMed] [Google Scholar]
- Sikdar SR, Serino G, Chaudhuri S, Maliga P. Plastid transformation in Arabidopsis thaliana. Plant Cell Rep. 1998;18:20–24. [Google Scholar]
- Silhavy D, Maliga P. Plastid promoter utilization in a rice embryogenic cell culture. Curr Genet. 1998;34:67–70. doi: 10.1007/s002940050367. [DOI] [PubMed] [Google Scholar]
- Singh AK, Verma SS, Bansal KC. Plastid transformation in eggplant (Solanum melongena L.) Transgenic Res. 2010;19:113–119. doi: 10.1007/s11248-009-9290-z. [DOI] [PubMed] [Google Scholar]
- Sodmergen, Kawano S, Tano S, Kuroiwa T. Degradation of chloroplast DNA in second leaves of rice (Oryza sativa) before leaf yellowing. Protoplasma. 1991;160:89–98. [Google Scholar]
- Svab Z, Maliga P. Exceptional transmission of plastids and mitochondria from the transplastomic pollen parent and its impact on transgene containment. Proc Natl Acad Sci USA. 2007;104:7003–7008. doi: 10.1073/pnas.0700063104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valkov V, Gargano D, Manna C, Formisano G, Dix PJ, Gray JC, Scotti N, Cardi T. High efficiency plastid transformation in potato and regulation of transgene expression in leaves and tubers by alternative 5′ and 3′ regulatory sequences. Transgenic Res. 2010 doi: 10.1007/s11248-010-9402-9. [DOI] [PubMed] [Google Scholar]
- Verma D, Daniell H. Chloroplast vector systems for biotechnology applications. Plant Physiol. 2007;145:1129–1143. doi: 10.1104/pp.107.106690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma D, Samson NP, Koya V, Daniell H. A protocol for expression of foreign genes in chloroplasts. Nat Protoc. 2008;3:739–758. doi: 10.1038/nprot.2007.522. [DOI] [PubMed] [Google Scholar]
- Verma D, Kanagaraj A, Jin S, Singh ND, Kolattukudy PE, Daniell H. Chloroplast-derived enzyme cocktails hydrolyse lignocellulosic biomass and release fermentable sugars. Plant Biotechnol J. 2010;8:332–350. doi: 10.1111/j.1467-7652.2009.00486.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurbs D, Ruf S, Bock R. Contained metabolic engineering in tomatoes by expression of carotenoid biosynthesis genes from the plastid genome. Plant J. 2007;49:276–288. doi: 10.1111/j.1365-313X.2006.02960.x. [DOI] [PubMed] [Google Scholar]
- Ye GN, Hajdukiewicz PTJ, Broyles D, Rodriguez D, Xu CW, Nehra N, Staub JM. Plastid-expressed 5-enolpyruvylshikimate-3-phosphate synthase genes provide high level glyphosate tolerance in tobacco. Plant J. 2001;25:261–270. doi: 10.1046/j.1365-313x.2001.00958.x. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Liu Y, Sodmergen Examination of the cytoplasmic DNA in male reproductive cells to determine the potential for cytoplasmic inheritance in 295 angiosperm species. Plant Cell Physiol. 2003;44:941–951. doi: 10.1093/pcp/pcg121. [DOI] [PubMed] [Google Scholar]
- Zheng Q, Oldenburg DJ, Bendich AJ. Independent effects of leaf growth and light on the development of the plastid and its DNA content in Zea species. J Exp Bot. 2011 doi: 10.1093/jxb/erq441. [DOI] [PubMed] [Google Scholar]
- Zhou F, Badillo-Corona JA, Karcher D, Gonzalez-Rabade N, Piepenburg K, Borchers AMI, Maloney AP, Kavanagh TA, Gray JC, Bock R. High-level expression of human immunodeficiency virus antigens from the tobacco and tomato plastid genomes. Plant Biotechnol J. 2008;6:897–913. doi: 10.1111/j.1467-7652.2008.00356.x. [DOI] [PubMed] [Google Scholar]
- Zubko M, Zubko E, Zuilen K, Meyer P, Day A. Stable transformation of petunia plastids. Transgenic Res. 2004;13:523–530. doi: 10.1007/s11248-004-2374-x. [DOI] [PubMed] [Google Scholar]


