Abstract
The mRNA levels of a set of immune-related genes were analysed with peripheral blood samples from at-risk, new-onset and long-term type 1 diabetes (T1D) patients, in comparison to those from healthy controls. The selected set includes T lymphocyte genes [CD3G and cytotoxic T lymphocyte-associated antigen 4 (CTLA4)], B lymphocyte genes (CD19 and CD20) and myeloid cell-related genes [CD11b, Toll-like receptor (TLR)-9, arginase (ARG1)]. Also included is a subset of the S100 family members that has been documented recently as regulatory elements of innate immunity. Samples from patients with long-term T1D had a reduced level of mRNA for most of selected innate and adaptive immune genes. No such reduction was detected in samples collected from at-risk or new-onset T1D patients. Analyses of regulatory gene expression ratios revealed a dynamic disproportion of CTLA4 versus CD3G expression in samples from at-risk, new-onset and long-term T1D patients. These changes could serve as immunological biomarkers for the status of the immune system during T1D progression and therapeutic interventions.
Keywords: autoimmunity, CTLA4, diabetes, human, gene expression
Introduction
T1D develops as a result of breakdown in immunological tolerance induction mechanisms. However, the pathogenesis of T1D has proved difficult to study in humans because of ethical and practical concerns that limit access to appropriate samples, such as pancreatic tissues. Studies using peripheral blood samples have advantageous accessibility and minimum body invasion compared to tissue samples. Although the gene expression profiles in peripheral blood may not reflect an active disease process taking place in the pancreas, such analyses performed with samples from patients at different stages of T1D may yield valuable biomarker information for the stage and progression of the disease. Indeed, our previous study [1] with peripheral blood samples from T1D patients has identified dynamic profiles of mRNA expression levels for a number of inflammatory cytokines and cytotoxic effector molecules.
In this study, we examined the mRNA levels for a set of immune genes including some essential elements of immune cells as well as newly emerging regulatory components. Quantitative reverse transcription–polymerase chain reaction (qRT–PCR) assays were used to generate robust expression data with a small volume of sample collection. Commercially tested primer and probe sets were utilized for the qPCR assays, purporting to facilitate repetition of the gene expression analyses in independent studies.
We reasoned that the expression level of a gene associated characteristically with an immune-cell subset may mark the abundance and/or activity of the cells. In this regard the expression of CD3G, which encodes for the gamma chain of the CD3–T cell receptor (TCR) complex, was taken to represent T lymphocytes, a major mediator of autoimmune damage in T1D. The expression of CTLA4 was analysed to assess T cell regulation during T1D progression. CTLA4 was selected because its allelic variations have been attributed to T1D risks. Another major subset in the adaptive immune system, B lymphocytes, also plays an important role in human T1D pathogenesis [2]. The expression of CD20 (MS4A1) and CD19 was analysed to assess the abundance and/or activity levels in the B cell compartment.
The response of the adaptive immune system is initiated and co-ordinated by the innate immune system, in which myeloid cells expressing CD11b are a major constituent. A cornerstone premise of immunology is the capacity of the immune system to distinguish ‘self’, the body's own tissue, and ‘non-self’, exemplified by infectious agents. One mechanism to achieve this is thought to be by an innate system recognizing pathogen-associated molecular patterns (PAMPs) of microbial pathogens [3], mediated by Toll-like receptors (TLRs). Some members in the TLR family, including TLR-9 [4], have also been demonstrated as initiators of autoimmunity. In lieu of PAMPs, it has been proposed that tissue damage releases endogenous damage-associated molecular patterns (DAMPs), a collective of innate immune signals that trigger adaptive immune responses [5,6]. Among the DAMP molecules is the S100 family of proteins [7]. Some members of S100 genes, most notably S100A8 and S100A9, were shown recently to play an important role in immunological tolerance by regulating myeloid-derived suppressor cells (MDSCs). MDSCs depend on arginase (ARG1) for their regulatory function [8]. Overall, for most of the S100 family members, their immunological functions remain to be identified. The S100 genes constitute a large family with approximately 20 members. They are absent in invertebrates and highly conserved in mammalian genomes [9]. The appearance of S100 genes at the vertebrate branching point of evolution coincides with specific adaptive immunity, suggesting a role in participating and/or co-ordinating innate and adaptive immune responses.
Therefore, we studied gene expression levels of CD3G, CTLA4, CD20, CD19, CD11b, ARG1, TLR9 and members of the S100 family to identify potential biomarkers during T1D progression.
Materials and methods
Subjects
The sample source and subject profiles are similar to what have been described in detail in a previous report [1], with minor differences. Briefly, peripheral blood samples were collected from four groups of subjects [n, mean age ± standard deviation (s.d.) in years, male : female ratio]: at-risk (n = 19, 15·0 ± 12·0, 10:9), new-onset (n = 33, 15·4 ± 7·6, 16:17) and long-term (n = 59, 38·9 ± 13·6, 31:28) T1D patients and healthy controls (n = 70, 39·7 ± 10·0, 35:35). The long-term diabetic patients and the healthy control group have a similar distribution of age and gender. Subjects in the at-risk group have autoantibodies against at least one of the following specificities: insulin (IAA), glutamic acid decarboxylase-65 (GAD65), islet cell autoantigen (ICA)512/IA2 (IA2) and/or ICA. Risk status of the at-risk patients was stratified based on the protocol for the Type 1 Diabetes TrialNet Natural History Study [10]. Low risk (n = 2) was defined as having one positive autoantibody with a normal oral glucose tolerance test (OGTT). Moderate risk (n = 10) was defined as having two positive autoantibodies with a normal OGTT test. High risk (n = 6) was defined as having three or more positive autoantibodies with a normal OGTT test or one to four positive autoantibodies with an abnormal OGTT test. The small sample size in each subgroup of the at-risk patients precluded us from analysing potential gene expression difference between low-, medium- and high-risk patients. All patients in the new-onset T1D group were diagnosed within a year of sample collection with the mean disease duration of 78 ± 74 days. Long-term T1D patients had a diabetes duration of more than 5 years (22·4 ± 11·0 years) with average haemoglobin A1c (HbA1c) (%) = 7·9 ± 1·3. Control blood samples were obtained from local healthy volunteers negative for any of the islet autoantibodies. Blood samples were collected from participants who were well (afebrile and off antibiotic treatment for at least 2 weeks prior to the visit) and not on steroids or other immunomodulatory therapies for at least 1 month prior to blood sample collection.
RNA isolation, cDNA synthesis, qRT–PCR for assessment of gene expression levels
Peripheral blood was collected into a Paxgene Blood RNA tube (Qiagen, Valencia, CA, USA) and stored at −80°C. We expected that whole blood collected in the Paxgene Blood RNA tube had an advantage of preserving optimal mRNA integrity and facilitating longitudinal comparison of archived samples, in comparison to methods that entail cell purification, such as flow cytometry, which may compromise the cellular viability (and thus mRNA intergrity), even though the latter would be advantageous to pinpoint potential changes to cellular subsets. Total RNA was isolated within 3 months of sample collection using a Paxgene Blood RNA kit (Qiagen); RNA preparation was treated with DNase I (Invitrogen, Carlsbad, CA, USA) to remove genomic DNA contamination. The levels of mRNA expression were determined using qRT–PCR, as described previously [1].
Statistical analyses
Outlier detection
Before computational analyses, log2 transformation was applied to the value of gene expression. The sas robustreg procedure was applied to identify outliers.
Analysis of variance (anova), Student's t-test and regression analysis of gene expressions
anova tests were carried out for each gene. In order to identify genes that are expressed differentially between two groups, a Student's t-test was performed for each gene comparing healthy controls with each T1D group. Before a conclusion about gene expression difference was drawn, potential impact of age and gender difference between groups was considered. Potential effect of age was adjusted statistically. A difference of gene expression or gene-expression ratio between groups was considered statistically significant only if both Student's t-test without age adjustment and the regression analysis with age adjustment indicated thus at a false discovery rate (FDR) < 0·05. In this study the t-test between a T1D group and the healthy group was not regarded as post-anova analysis. anova, t-tests and linear regression with adjustment of age were used in parallel to ensure that consistent results were obtained from different types of analysis. Indeed, for those genes that anova did not identify differential expression, the t-test and linear regression did not identify differential expression either.
Results
Reduced gene expression in both innate and adaptive immune branches in long-term T1D groups but not in at-risk or new-onset T1D patients
Relative mRNA expression levels were determined with triplicate, qRT–PCR for the selected set of 13 innate and adaptive immune genes, with peripheral blood samples from 59 long-term (LT) T1D patients and 33 new-onset (NO) T1D patients, 19 subjects at risk (AT) for T1D and 70 healthy (HT) controls. Log2 transformation was applied to the expression values of all genes before further computational analyses. Four statistical outliers, out of the 2353 expression values for all 13 genes determined for all samples, were excluded from further analyses. These four outliers included one ARG1 value in the HT group, one ARG1 value in the NO group, one S100A10 value in the AT group and one S100A13 in the HT group,
The relative expression levels of the 13 innate and adaptive genes in healthy control samples were compared with those of at-risk, new-onset and long-term diabetic groups. Compared to healthy controls, the AT and NO T1D groups did not exhibit any decrease of expression in any of the 13 genes. Conversely, the LT T1D group showed a significantly decreased expression for most (10 of 13) of the selected genes. As shown in Fig. 1, affected genes include basic indicators for lymphocytes and myeloid cells, CD3G, CD20, CD11b and TLR9. Among the six S100 genes tested (Fig. 2), the LT T1D patient samples had reduced expression level for S100A6, S100A10, S100A11 and S100A13, with the most significant difference seen in S100A13 expression. No significant difference was detected for expression of S100A8 and S100A9. Taken together with the previous findings that cytokine mRNA levels were reduced in the LT T1D samples, but not in the AT and NO patients [1], the results suggest that there could be a mild immune gene expression insufficiency across innate and adaptive immune branches in the LT T1D group. However, such an insufficiency did not occur before or at the onset of diabetes. This pattern of gene expression in different stages of T1D is summarized in Table 1.
Fig. 1.
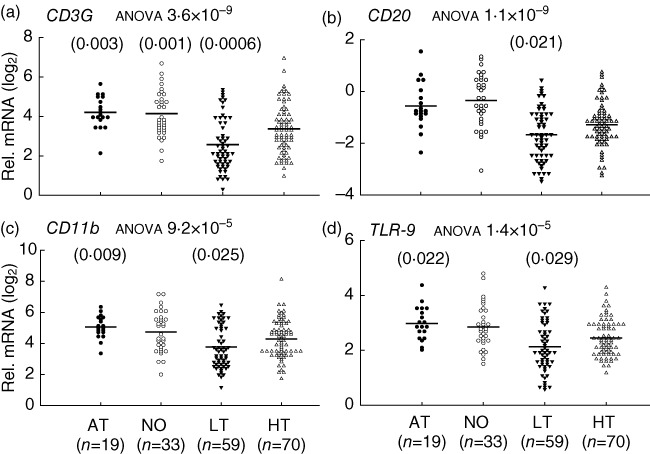
Innate and adaptive gene expression insufficiencies in long-term (LT), but not new-onset (NO) and at-risk (AT), type 1 diabetes (T1D) patients, compared to healthy (HT) controls. Relative mRNA expression levels are calculated as fold change against a common control and plotted in a log2 scale. The line indicates the average value in a group. The P-value from analysis of variance (anova) for each gene is indicated after the title of each plot. The P-value for comparison between a T1D group and healthy controls is presented on top of the gene expression value plots if it was less than 0·05 in both regression analyses with age adjustment, and Student's t-test without age adjustment. Shown is the higher (more nullifying) P-value from the regression analyses or Student's t-tests. Cut-off P-values for the t-test and regression analysis are 0·026 and 0·029, respectively, determined with the Benjamini–Hochberg procedure at a false discovery rate (FDR) <0·05 for a family of 13 hypothesis tests (13 genes). However, P-values less than 0·05 but above 0·026 or 0·029 are still presented to shown potential marginal significance.
Fig. 2.
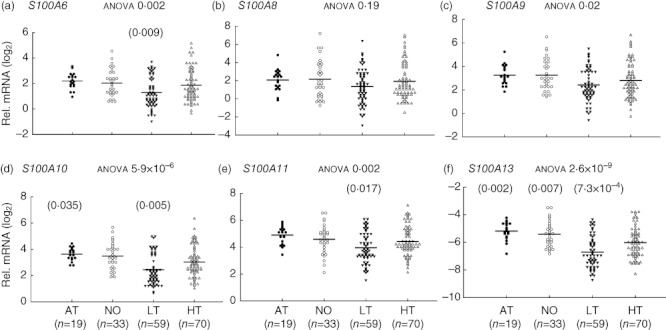
S100 gene expression insufficiencies in long-term (LT), but not new-onset (NO) and at-risk (AT), type 1 diabetes (T1D) patients, compared to healthy (HT) controls. Relative gene expression levels were calculated as fold change against a common control and plotted in the log2 scale. The line indicates the average value in a group. The P-value from analysis of variance (anova) for each gene is indicated after the title of each plot. The P-value for comparison between a T1D group and healthy control is presented on top of the gene expression value plots if it is less than 0·05. Shown is the higher (more nullifying) P-value from two different statistical tests: regression analysis with age adjustment or Student's t-test without age adjustment. Cut-off P-values for the t-test and regression analysis are 0·026 and 0·029, respectively, determined with the Benjamini–Hochberg procedure at a false discovery rate (FDR) <0·05 for a family of 13 hypothesis tests (13 genes). However, P-values less than 0·05 but above 0·026 or 0·029 are still presented to show potential marginal significance.
Table 1.
Relative gene expression pattern in at-risk, new-onset and long-term type 1 diabetes (T1D) patients compared to healthy controls (HT)
| Gene | At-risk (AT) | New-onset (NO) | Long-term (LT) |
|---|---|---|---|
| CD3G | ↑versus HT | ↑versus HT | ↓versus HT |
| S100A10 | ↓versus HT | ||
| S100A13 | ↑versus HT | ↑versus HT | ↓versus HT |
| CD11b | ↑versus HT | ↓versus HT | |
| TLR-9 | ↑versus HT | ↓versus HT | |
| CD20 | ↓versus HT | ||
| S100A6 | ↓versus HT | ||
| S100A11 | ↓versus HT | ||
| CTLA4 | ↓versus HT | ||
| CTLA4/CD3G | ↓versus HT | ↓versus HT | ↑versus HT |
Increase and decrease, in gene expression or gene expression ratio relative to controls. CTLA4: cytotoxic T lymphocyte-associated antigen 4; HT: healthy; TLR: Toll-like receptor.
Altered ratios of cytotoxic T lymphocyte-associated antigen 4 (CTLA4) versus CD3G expression in T1D
The development of T1D is influenced strongly by genetic factors. One of the genetic determinants of T1D risk is the polymorphisms of the CTLA4 locus, associated with T1D in various ethnic populations by studies from many groups. Genetic studies indicate that it is not the qualitative change of mature CTLA4 protein, but subtle quantitative variations of CTLA4 expression that are attributable to T1D risk [11]. The association is consistent with the function of CTLA4 as a master regulator of T lymphocytes [12,13].
We analysed CTLA4 mRNA levels in peripheral blood samples from the AT, NO and LT T1D patients in comparison to HT controls. No difference was detected between the HT controls and AT subjects or NO patients, but CTLA4 mRNA levels were decreased significantly in the LT diabetic patients compared to HT controls (Fig. 3a). However, a different pattern emerged when CTLA4 expression was examined in reference to the CD3G expression level in the same samples. The ratios of CTLA4 : CD3G expression were decreased substantially in both the AT and NO groups, but increased significantly in the LT diabetic group (Fig. 3b, Table 1).
Fig. 3.
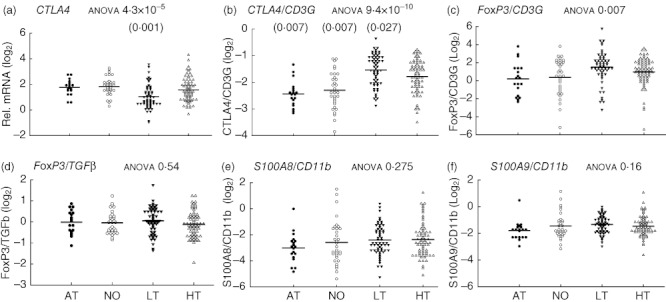
Dynamics and equilibrium of innate and adaptive regulatory gene expression ratios in type 1 diabetes (T1D) patients at different stages of the disease. Relative mRNA levels, or the expression ratios of two genes, are plotted in a log2 scale. The line indicates the average value in a group. The P-value from analysis of variance (anova) for each gene expression value or ratio is indicated after the title of each plot. The P-value for comparison between a T1D group and healthy controls is presented according to the same description in Fig. 2. (a) Cytotoxic T lymphocyte-associated antigen 4 (CTLA4) expression levels in healthy and T1D subjects. (b–f) Gene expression ratios of CTLA4 versus CD3G, forkhead box protein 3 (FoxP3) versus CD3G, FoxP3 versus transforming growth factor (TGF)-β, S100A8 versus CD11b and S100A9 versus CD11b, respectively. Sample numbers are the same as in Fig. 2, except for (c) and (d), LT (n = 55) and HT (n = 69).
Unlike the long-term T1D group, the age distribution of the AT and NO patients is different from that of HT controls. The variations of CTLA4 : CD3G ratios among the groups could be contributed by gender and age. A commonly encountered difficulty in T1D studies is the lack of samples from gender- and age-matched healthy children. Given this obstacle, we could not rule out experimentally the possibility that the gender- and age-difference between groups may contribute to the altered CTLA4 : CD3G ratios in the T1D groups. We conducted regression analyses of gene expression in each group with respect to age and examined the potential effect of gender by Student's t-test. We did not detect dependence of CTLA4 : CD3G ratio on gender. Regression analyses indicated that CTLA4 : CD3G ratios was significantly (P = 0·0002) dependent on age in the AT group, but not in the NO or LT T1D group or in healthy individuals. With adjustment for age difference between the groups, regression analyses detected a significant difference in CTLA4 : CD3G ratios between healthy and each T1D group (Fig. 3b). Using a different method, Student's t-test, without adjusting for age, the ratios of CTLA4 : CD3G expression between the healthy and each T1D group were also found to be statistically significant. CTLA4 critically mediates the suppressive function of forkhead box protein 3 (FoxP3+) regulatory T cells [14]. However, no significant difference was detected for FoxP3 : CD3G ratios between healthy controls and any one of the T1D subject groups (Fig. 3c). It should be noted that whereas a number of genetic studies have associated T1D with genetic variations of the CTLA4 locus that affect CTLA4 expression [11], polymorphisms of the FOXP3 gene were not associated with common T1D cases [15].
Marginal correlation of gene expression changes with glycaemic controls in T1D patients
To examine the potential impact of glycaemic control on the immune gene expression, fasting glucose, fasting c-peptide or HbA1c measurements were tested for correlation with gene expression levels at a significance level of 0·05. The analysis was conducted for the expression values of 25 genes, including 12 genes reported in a previous study [1] (not shown) and 13 genes in this study, as well as the CTLA4 : CD3G ratio. In the AT group, no significant correlation was detected for HbA1c or fasting glucose with expression values of any of the selected genes. However, the expression of ARG1 correlated with fasting c-peptide [correlation (r) = 0·47; P = 0·043]. In the NO group, no significant correlation was detected between fasting glucose or c-peptide and the expression level of any of the selected genes. However, the HbA1c values correlated with expression of ARG1 (r = −0·77; P = 0·004) and S100A8 (r = −0·59; P = 0·041). In the LT group, correlation was detected between HbA1c values and CD3G (r = 0·31; P = 0·003), CD19 (r = 0·31, P = 0·018), CD20 (r = 0·39; P = 0·003) and ARG1 (r = 0·29; P = 0·028), interferon (IFN)-γ (r = 0·27, P = 0·042) and granzyme B (r = −0·36, P = 0·006).
In the AT group, no correlation was detected between the CTLA4 : CD3G ratio and fasting glucose, fasting c-peptide or HbA1c measurements. In the NO group, the CTLA4 : CD3G ratio was not correlated with fasting glucose or fasting c-peptide values, but a correlation between HbA1c values and CTLA4 : CD3G ratios was detected (r = 0·71; P = 0·01). In the LT group, HbA1c values were correlated weakly with the CTLA4 : CD3G ratio (r = −0·31; P = 0·021).
However, if we applied the Benjamini–Hochberg procedure to control the FDR at a level <0·05 for each family of 26 hypothesis tests (for the 25 genes and one ratio), none of the correlations was significant. Hence, the correlations detected at a significance level of 0·05 described above should be regarded as only marginally significant.
Stable ratios of some innate and adaptive regulatory genes throughout different stages of T1D
The dynamic change of CTLA4 : CD3G ratios at different stages of T1D prompted us to examine the expression relationship of another two genes that play key roles in T cell regulation, transforming growth factor (TGF)-β and FoxP3. TGF-β is essential for generation of induced FoxP3+ regulatory T (Treg) cells [16]. Evidence gathered from a recent study indicated that a defect in TGF-β-induced Treg cells may contribute to the development of autoimmune diabetes in the non-obese diabetic (NOD) mouse model of T1D [17]. Interestingly, although the mRNA expression levels of both TGF-β and FoxP3 were reduced substantially in peripheral blood samples from the LT T1D patients [1], the average ratios of FoxP3 : TGF-β mRNA were maintained at a virtually identical level among all four groups (Fig. 3d) (mean ± s.d., HT 1·03 ± 0·47, AT 1·06 ± 0·39, NO 1·03 ± 0·42, LT 1·10 ± 0·51).
S100A8, S100A9 and ARG1 are thought to be important regulatory genes for myeloid cell function [8]. Those regulatory mechanisms was identified originally in tumour-induced immune suppression, but has been implicated in non-malignant inflammatory pathology including autoimmune damage [8]. No significant variation of S100A8 : CD11b or S100A9 : CD11b expression ratios was identified between healthy controls and any of the T1D groups (Fig. 3e,f).
Discussion
The overall immunological status during human T1D initiation and progression remains a challenge to characterize. This study, with peripheral blood samples from T1D patients at different stages of diabetes, examined levels and ratios of innate and adaptive immune gene expression. The study revealed reduction of mRNA levels in long-term T1D patient samples for most of the selected genes in both the innate and adaptive branches. However, this type of reduction was not detected in at-risk or new-onset T1D groups. Therefore, such evidence does not suggest any causal role for immune insufficiencies in autoimmunity. Whereas it remains unknown what causes the reduction of mRNA levels in the samples from long-term T1D patients, reduced mRNA levels for common immune genes might serve as a biomarker of immune changes in the process of diabetes progression. It should be noted, however, that the reduction of immune gene expression in long-term T1D patients was subtle, and its effect remains unknown. Conversely, subtle deficiencies in expression of some immune genes, including TLRs, have been documented to affect human immune defence to infections [18].
Aside from the gene-expression level changes, this study uncovered a dynamic profile for CTLA4 : CD3G expression ratios in samples from different stages of T1D. CTLA4, a master regulator in T cell tolerance [12], was expressed at a higher proportion relative to CD3G in the long-term T1D population. In contrast to long-term diabetic patients, at-risk and new-onset T1D cohorts had a lower ratio of CTLA4 : CD3G mRNA expression in peripheral blood. It should be emphasized that the results remain not fully conclusive, due to lack of control samples from age-matched healthy children, and that we had to resort to a statistical adjustment for age-difference. However, the observed difference is consistent with the CTLA4 genetic predisposition associated with human T1D pathogenesis. Besides the MHC locus, the CTLA4 gene is shown to be one of the loci carrying a strong association with T1D. The linkage disequilibrium of T1D risk with the single nucleotide polymorphisms (SNPs) at the CTLA4 locus has been established by independent studies from several different groups. The exact attribution of each CTLA4 SNP to CTLA4 expression level, as well as T1D association, remains a debate. For example, a well-known report showed that unstimulated CD4 T cells from 14 healthy subjects had about two- to threefold lower levels of soluble (s) CTLA4 splice variants associated with the T1D-risk +6230G allele [19]. However, a later study with 11 non-diabetic subjects, most of whom were parents of T1D children, found that neither sCTLA4 nor the full-length (f) CTLA4 splice variant levels were linked to the +6230G>A SNP if the subjects had the same –318C SNP in the promoter region, but the –318C T1D-risk allele was associated with lower levels of both fCTLA4 and sCTLA4 transcripts [20].
What might cause a distinct change of CTLA4 : CD3G ratios in different stages of T1D remains to be determined. Whereas the decreased ratios in the AT and NO groups could be consistent with genetic predisposition, the higher ratio of CTLA4 : CD3G in the LT group probably reflects an immunological process that is associated with diabetes but has no causative role in diabetes per se. It remains to be identified whether such changes reflect altered CD4 or CD8 T cell lineages, or effector T (Teff) cell or Treg cell subsets. There seems to be a marginal correlation between HbA1c measurements and CTLA4 : CD3G gene expression ratios but, intriguingly, the weak correlation appears positive in the new-onset group but negative in the long-term T1D group. Further studies are needed to examine this observation, but it by no means excludes the potential impact of imperfect glycaemic controls on immune gene expression. None the less, those changes might be useful as immune biomarkers during T1D progression. The variations were subtle, but could have an immunological impact. In animal models, a modest reduction of CTLA4 through transgenic shRNA provoked autoimmune diabetes, not only in the standard NOD mouse model but also in mice with a mixed genetic background that were otherwise free from autoimmune damage of pancreatic islets [21]. However, whether and how a change in CTLA4 levels relative to CD3G expression affects immune function needs to be determined experimentally. In mouse models of anti-tumour immunity, subtle reduction of CTLA4 promoted Teff cell activity without compromising Treg cell function, indicating an exquisite sensitivity of Teff cells to CTLA4 changes [22].
Despite the essential role of CTLA4 for the function of FoxP3+ Treg cells [14], unlike the substantial disproportion of CTLA4 : CD3G expression, FoxP3 : CD3G expression ratios did not differ significantly comparing healthy controls and any of the T1D groups. FoxP3+ Treg cells can be generated naturally in the thymus, or induced in the periphery through a mechanism dependent on TGF-β. The defect of TGF-β-induced Treg cells has been implicated in the pathogenesis of autoimmune diabetes in the NOD mouse model [17]. Interestingly, the ratios of TGF-β : FoxP3 were stably maintained to such an extent that the average ratios were virtually identical in all groups of T1D patients and healthy controls. Therefore, the balance of these two key regulatory elements of adaptive immune tolerance was not apparently perturbed even in the condition of long-term diabetes. In the same vein, for innate immune regulation, no significant change of S100A9 : CD11b ratios was detected between healthy controls and any of the T1D or at-risk groups. Thus, the expression relationships of those key immunoregulatory genes were maintained throughout T1D progression, whereas CTLA4 : CD3G expression ratios exhibited a dynamic variation.
Gene expression analyses using whole blood samples, instead of purified cell populations, were conducted in this study. Whole blood assays require less manipulation, and contain cell populations that may be lost in the cell isolation process. In addition, the whole blood samples used in this study were collected into a Paxgene Blood RNA tube, which contains RNA-stabilizing reagents to protect RNA molecules from degradation and minimize induction of gene expression changes after blood collection. However, this approach could not differentiate if the changes in gene expression were caused by altered cellular activation and/or subset composition, which requires further studies using cell-based analyses. Conversely, this approach could circumvent potential artificial variability introduced by extensive in-vitro handling that would be required for cell sorting by flow cytometry or magnetic beads. qRT–PCR has the advantages of high sensitivity and flexibility over flow cytometry assay, as for this assay blood samples do not need to be processed immediately, but instead can be frozen in the Paxgene tube for a few months before processing, which enables examination of a large numbers of samples, including samples collected longitudinally from the same patients in the same experiment.
Overall, the profiles of immune gene expression ratios, together with gene expression levels, could be useful as biomarkers to assess immune status during progression of T1D, and assist monitoring of immune intervention. These parameters might provide some practical and reproducible biomarker indicators for whether a substantial change might have occurred in the patient's immune system, especially if a longitudinal analysis is conducted to compare samples collected at different stages of T1D development. The information could be important for T1D patients given the key role of the immune system in maintaining a patient's health. Further studies are needed to identify biomarker patterns that could precisely stage T1D pathogenesis, and predict diabetes development and progression, or lack thereof, in at-risk and new-onset individuals.
Acknowledgments
This work was supported by the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK, award DP3DK085696). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIDDK or NIH. X.C. was supported partly by the National Science Foundation (award 0746882, Stochastic Modeling, Analysis and Simulation of Gene Networks). This work was also supported, in part, by TrialNet Coordinating Center (HHSN267200800019C) and the University of Miami TrialNet Clinical Center (1U01-DK085499-04). TrialNet is a clinical trials network funded by the National Institutes of Health through the National Institute of Diabetes and Digestive and Kidney Diseases, the National Institute of Allergy and Infectious Diseases, the Eunice Kennedy Shriver National Institute of Child Health and Human Development, the National Center for Research Resources, the Juvenile Diabetes Research Foundation International and the American Diabetes Association. The authors gratefully acknowledge the expert support of R. Alejandro, D. Mineo and D. M. Berman. The authors thank Dr A. Pugliese for designing the IRB protocol and critically reviewing the manuscript.
Disclosures
The authors declare that there are no conflicts of interest.
References
- 1.Han D, Leyva CA, Matheson D, et al. Immune profiling by multiple gene expression analysis in patients at-risk and with type 1 diabetes. Clin Immunol. 2011;139:290–301. doi: 10.1016/j.clim.2011.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pescovitz MD, Greenbaum CJ, Krause-Steinrauf H, et al. Rituximab, B-lymphocyte depletion, and preservation of beta-cell function. N Engl J Med. 2009;361:2143–52. doi: 10.1056/NEJMoa0904452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Janeway CA, Jr, Medzhitov R. Innate immune recognition. Annu Rev Immunol. 2002;20:197–216. doi: 10.1146/annurev.immunol.20.083001.084359. [DOI] [PubMed] [Google Scholar]
- 4.Leadbetter EA, Rifkin IR, Hohlbaum AM, et al. Chromatin–IgG complexes activate B cells by dual engagement of IgM and Toll-like receptors. Nature. 2002;416:603–7. doi: 10.1038/416603a. [DOI] [PubMed] [Google Scholar]
- 5.Oppenheim JJ, Yang D. Alarmins: chemotactic activators of immune responses. Curr Opin Immunol. 2005;17:359–65. doi: 10.1016/j.coi.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 6.Bianchi ME. DAMPs, PAMPs and alarmins: all we need to know about danger. J Leukoc Biol. 2007;81:1–5. doi: 10.1189/jlb.0306164. [DOI] [PubMed] [Google Scholar]
- 7.Foell D, Wittkowski H, Roth J. Mechanisms of disease: a ‘DAMP’ view of inflammatory arthritis. Nat Clin Pract Rheumatol. 2007;3:382–90. doi: 10.1038/ncprheum0531. [DOI] [PubMed] [Google Scholar]
- 8.Gabrilovich DI, Nagaraj S. Myeloid-derived suppressor cells as regulators of the immune system. Nat Rev Immunol. 2009;9:162–74. doi: 10.1038/nri2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marenholz I, Heizmann CW, Fritz G. S100 proteins in mouse and man: from evolution to function and pathology (including an update of the nomenclature) Biochem Biophys Res Commun. 2004;322:1111–22. doi: 10.1016/j.bbrc.2004.07.096. [DOI] [PubMed] [Google Scholar]
- 10.Mahon JL, Sosenko JM, Rafkin-Mervis L, et al. The TrialNet natural history study of the development of type 1 diabetes: objectives, design, and initial results. Pediatr Diabetes. 2009;10:97–104. doi: 10.1111/j.1399-5448.2008.00464.x. [DOI] [PubMed] [Google Scholar]
- 11.Kristiansen OP, Larsen ZM, Pociot F. CTLA-4 in autoimmune diseases – a general susceptibility gene to autoimmunity? Genes Immun. 2000;1:170–84. doi: 10.1038/sj.gene.6363655. [DOI] [PubMed] [Google Scholar]
- 12.Teft WA, Kirchhof MG, Madrenas J. A molecular perspective of CTLA-4 function. Annu Rev Immunol. 2006;24:65–97. doi: 10.1146/annurev.immunol.24.021605.090535. [DOI] [PubMed] [Google Scholar]
- 13.Shevach EM. Immunology. Regulating suppression. Science. 2008;322:202–3. doi: 10.1126/science.1164872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wing K, Onishi Y, Prieto-Martin P, et al. CTLA-4 control over Foxp3+ regulatory T cell function. Science. 2008;322:271–5. doi: 10.1126/science.1160062. [DOI] [PubMed] [Google Scholar]
- 15.Zavattari P, Deidda E, Pitzalis M, et al. No association between variation of the FOXP3 gene and common type 1 diabetes in the Sardinian population. Diabetes. 2004;53:1911–4. doi: 10.2337/diabetes.53.7.1911. [DOI] [PubMed] [Google Scholar]
- 16.Chen W, Jin W, Hardegen N, et al. Conversion of peripheral CD4+CD25– naive T cells to CD4+CD25+ regulatory T cells by TGF-beta induction of transcription factor Foxp3. J Exp Med. 2003;198:1875–86. doi: 10.1084/jem.20030152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.D’Alise AM, Ergun A, Hill JA, et al. A cluster of coregulated genes determines TGF-{beta}-induced regulatory T-cell (Treg) dysfunction in NOD mice. Proc Natl Acad Sci USA. 2011;108:8737–42. doi: 10.1073/pnas.1105364108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Turvey SE, Hawn TR. Towards subtlety: understanding the role of Toll-like receptor signaling in susceptibility to human infections. Clin Immunol. 2006;120:1–9. doi: 10.1016/j.clim.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 19.Ueda H, Howson JM, Esposito L, et al. Association of the T-cell regulatory gene CTLA4 with susceptibility to autoimmune disease. Nature. 2003;423:506–11. doi: 10.1038/nature01621. [DOI] [PubMed] [Google Scholar]
- 20.Anjos SM, Shao W, Marchand L, et al. Allelic effects on gene regulation at the autoimmunity-predisposing CTLA4 locus: a re-evaluation of the 3′+6230G>A polymorphism. Genes Immun. 2005;6:305–11. doi: 10.1038/sj.gene.6364211. [DOI] [PubMed] [Google Scholar]
- 21.Chen Z, Stockton J, Mathis D, et al. Modeling CTLA4-linked autoimmunity with RNA interference in mice. Proc Natl Acad Sci USA. 2006;103:16400–5. doi: 10.1073/pnas.0607854103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miska J, Bas E, Devarajan P, et al. Autoimmunity-mediated antitumor immunity: tumor as an immunoprivileged self. Eur J Immunol. 2012 doi: 10.1002/eji.201242590. doi: 10.1002/eji.201242590. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]


