Abstract
Congenital heart disease (CHD) is the most common form of developmental malformation and is the leading noninfectious cause of infant mortality. Emerging evidence indicates that genetic defects are involved in the pathogenesis of CHD. Nevertheless, CHD is genetically heterogeneous, and the molecular basis for CHD in a majority of patients remains unknown. In this study, the whole coding region of GATA6, a gene encoding a zinc-finger transcription factor crucial for normal cardiogenesis, was sequenced in 380 unrelated patients with CHD. The relatives of the index patients harboring the identified mutations and 200 unrelated control individuals were subsequently genotyped. The functional effect of the mutations was characterized using a luciferase reporter assay system. As a result, two novel heterozygous GATA6 mutations, p.D404Y and p.E460X, were identified in two families with ventricular septal defect and tetralogy of Fallot, respectively. The mutations co-segregated with CHD in the families with complete penetrance, and were absent in 400 control chromosomes. Functional analysis demonstrated that the mutated GATA6 proteins were associated with significantly decreased transactivational activity in comparison with their wild-type counterpart. These findings provide novel insight into the molecular mechanism implicated in CHD, suggesting potential implications for the early prophylaxis and personalized treatment of CHD.
The transcription factor GATA6 is a zinc finger DNA-binding protein that has been shown to be essential for cardiac development. In this article, two novel heterologous polymorphisms were associated with two cardiac malformations in families.
Introduction
Congenital heart disease (CHD), characterized by a defect in the structure of the heart and great vessels, represents the most common form of developmental anomaly, with an estimated prevalence of 1% among live births, and is the leading noninfectious cause of infant mortality, with over 29% of neonates who die of a birth defect having a cardiovascular abnormality (Roger et al., 2012). Clinically CHD is classified into at least 21 different types with specific anatomic or hemodynamic lesions, of which ventricular septal defect is the most prevalent type, occurring in 30–60% of all children with various kinds of congenital cardiovascular deformations, and accounts for 14–16% of birth defects that require an invasive procedure within the first year of life (van der Linde et al., 2011; Roger et al., 2012). Tetralogy of Fallot, a tetrad of (1) right ventricular outflow tract obstruction, (2) over-riding of the aortic root, (3) ventricular septal defect, and (4) right ventricular hypertrophy, is the most common form of cyanotic CHD worldwide, occurring in about 3 of every 10,000 live births, and constitutes ∼7–10% of all congenital heart malformations. Left untreated surgically, 25% of patients with severe obstruction die within the first year, 40% die by 3 years, 70% by 10 years, and 95% by 40 years (Starr, 2010). Various congenital cardiac deformities can occur separately or together with each other, such as ventricular septal defect, atrial septal defect, tetralogy of Fallot, double-outlet right ventricle, atrioventricular septal defect, aortic stenosis, transposition of great arteries, and patent ductus arteriosus, leading to cardiac enlargement, ventricular dysfunction or heart failure, pulmonary hypertension, Eisenmenger's syndrome, delayed fetal brain development, arrhythmias, poor quality of life, and even sudden cardiac death in the absence of surgical treatment or catheter-based repair (Cheng et al., 2011; Shedeed and Elfaytouri, 2011; Teixeira et al., 2011; van der Bom et al., 2011; Perry, 2012; Silka and Bar-Cohen, 2012). Despite the high prevalence and the important clinical significance, the molecular basis for CHD remains unknown in an overwhelming majority of patients (Bruneau, 2008; Benson, 2010; Chen et al., 2011; Li et al., 2012; Liu et al., 2012; Rani et al., 2012; Wang et al., 2012b).
Cardiac morphogenesis is a complex biological process, which is controlled by an evolutionarily conserved network of transcription factors that connect signaling pathways with genes for muscle growth, patterning, and contractility (Pikkarainen et al., 2004; Olson, 2006; Bartlett et al., 2010). Mutations in components of the cardiac gene network have been implicated in the pathogenesis of CHD, including GATA and NK transcription factors that are essential for normal cardiogenesis (Schott et al., 1998; Garg et al., 2003).
The GATA transcription factors are a family of zinc-finger-containing DNA-binding proteins, characteristic of the ability to bind the consensus DNA sequence GATA of target gene promoters. In vertebrates, the GATA family consists of six members (GATA1–GATA6), of which GATA4, GATA5, and GATA6 are broadly expressed in various mesoderm- and endoderm-derived tissues, especially in the embryonic heart (Pikkarainen et al., 2004). Of these three cardiac GATA transcription factors, GATA4 has been most frequently probed as a molecular determinant for CHD, and a long list of GATA4 mutations have been detected in cases with a wide variety of CHD, including ventricular septal defect, atrial septal defect, atrioventricular septal defect, tetralogy of Fallot, pulmonary stenosis, and patent ductus arteriosus (Benson, 2010; Butler et al., 2010; Chen et al., 2010a, 2010b; Liu et al., 2011; Salazar et al., 2011; Wang et al., 2011; Yang et al., 2012a, 2012c). The human GATA6 gene is another member of the GATA family, and its expression and function overlap at least partially with that of GATA4 during cardiovascular development (Pikkarainen et al., 2004), which makes GATA6 a logical candidate gene for CHD.
Materials and Methods
Study subjects
A cohort of 380 unrelated patients with CHD was enrolled from the Chinese Han population. The available relatives of the index patients carrying the identified GATA6 mutations were also recruited. Subjects were evaluated by individual and familial histories, review of the medical records, complete physical examination, 12-lead electrocardiogram (ECG), and two-dimensional transthoracic echocardiography with color-flow Doppler. The types of CHD were determined using two-dimensional continuous-wave Doppler and color Doppler techniques on transthoracic echocardiography. Where necessary, transesophageal echocardiography and angiography were used for further clarification of the anatomy. These procedures were performed by skillful echocardiologists. Some patients underwent cardiac catheterization and, if required, cardiac surgery. The patients with known chromosomal abnormalities or syndromic cardiovascular defects were excluded from the study.
A total of 200 ethnically matched, unrelated healthy individuals randomly selected from the individuals undergoing routine physical examinations were used as control individuals. As determined by reviews of medical histories and analyses of echocardiographic records, the control individuals had no apparent congenital cardiovascular defects, except for subclinical cardiac aberrations such as bicuspid aortic valve or patent foramen ovale. The ethnic origin of a participant was ascertained by a combination of self-reported ethnicity and a personal questionnaire asking questions about the birthplace, language, religion, and ancestry.
Peripheral venous blood samples from CHD cases and control individuals were acquired. The study protocol was reviewed and approved by the local institutional ethics committee, and written informed consent was obtained from all participants or their guardians before study.
Genetic studies
Genomic DNA from all participants was extracted from blood lymphocytes with a Wizard Genomic DNA Purification Kit (Promega, Madison, WI). The GATA6 gene was initially sequenced in 380 unrelated patients with CHD and subsequently genotyped for the available relatives of the index patients carrying identified mutations and the 200 control individuals. The referential genomic DNA sequence of GATA6 was derived from GenBank (accession No. NT_010966). With the help of on-line Primer 3 program (http://frodo.wi.mit.edu), the primer pairs used to amplify the coding exons and exon/intron boundaries of GATA6 by polymerase chain reaction (PCR) were designed as shown in Table 1. The PCR was carried out using HotStar Taq DNA Polymerase (Qiagen GmbH, Hilden, Germany) on a PE 9700 Thermal Cycler (Applied Biosystems, Foster, CA), with standard conditions and concentrations of reagents. Amplified products were analyzed on 1% agarose gels stained with ethidium bromide and purified with a QIAquick Gel Extraction Kit (Qiagen GmbH). Both strands of each PCR product were sequenced with a BigDye® Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems) under an ABI PRISM 3130 XL DNA Analyzer (Applied Biosystems). The sequencing primers were the same as previously designed for specific region sequencing. The DNA sequences were viewed and analyzed with DNA Sequencing Analysis Software v5.1 (Applied Biosystems). The sequence variation was validated by resequencing an independent PCR-generated amplicon from the subject and met our quality control thresholds with a call rate >99%. Additionally, an identified GATA6 variant was checked in National Center for Biotechnology Information (NCBI) single-nucleotide polymorphism (SNP) database (http://ncbi.nlm.nih.gov/SNP) to confirm the novelty.
Table 1.
The Intronic Primers to Amplify the Coding Exons and Flanking Splice Sites of GATA6
| Exon | Forward primer (5′–3′) | Reverse primer (5′–3′) | Amplicon (bp) |
|---|---|---|---|
| 2-A | ttg, tta, acc, cgt, cga, tct, cc | gcg, agg, gtc, tgg, tac, atc, tc | 543 |
| 2-B | tgc, tgt, tca, ctg, acc, tcg, ac | ctg, gga, gag, tag, ggg, aag, c | 466 |
| 2-C | ccg, aca, gcc, ctc, cat, acg | gaa, aac, agg, gcc, cga, gtg | 539 |
| 3 | ggc, caa, gga, gaa, aag, ctc, ag | gtt, gga, aca, gcc, ggg, aca, g | 485 |
| 4 | tct, tgg, ccc, aga, aaa, gtc, ag | tca, ttt, gct, gat, tct, ttg, taa, ctg | 387 |
| 5–6 | ctg, gga, tta, gag, gcg, tga, gc | ttt, act, aga, gag, cag, ccc, agt | 473 |
| 7 | att, tct, cct, gcc, ctg, ggt, ct | ctg, cac, aaa, agc, aga, cac, ga | 382 |
Alignment of multiple GATA6 protein sequences
Alignment of multiple GATA6 protein sequences across various species was performed using the online program of MUSCLE, version 3.6 (http://ncbi.nlm.nih.gov).
Prediction of the causative potential of a GATA6 sequence variation
The disease-causing potential of a GATA6 sequence variation was assessed by MutationTaster (an online program at http://mutationtaster.org), which automatically gave a probability for the variation to be either a pathogenic mutation or a benign polymorphism. Notably, the p value used here is the probability of the correct prediction rather than the probability of error as used in t-test statistics (i.e., a value close to 1 indicates a high security of the prediction).
Plasmids and site-directed mutagenesis
The recombinant expression plasmid pcDNA3-hGATA6 was kindly provided by Dr. Angela Edwards-Ghatnekar, from the Division of Rheumatology and Immunology, Medical University of South Carolina (Charleston, SC). The atrial natriuretic factor (ANF)–luciferase reporter gene, which contains the 2600-bp 5′-flanking region of the ANF gene, namely ANF(-2600)-Luc, was kindly provided by Dr. Ichiro Shiojima, from the Department of Cardiovascular Science and Medicine, Chiba University Graduate School of Medicine (Chuo-ku, Chiba, Japan). The identified mutation was introduced into the wild-type GATA6 using a QuickChange II XL Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA) with a complementary pair of primers. The mutant was sequenced to confirm the desired mutation and to exclude any other sequence variations.
Reporter gene assay
HEK-293 cells were cultured in the Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum. The HEK 293 cell line is a specific cell line originally generated by transformation of human embryonic kidney cell cultures with sheared adenovirus 5 DNA. This cell line has been widely used in cell biology research for many years mainly due to its extreme transfectability by the various transfection techniques, with efficiencies approaching 100%. The ANF(-2600)-Luc reporter vector and an internal control reporter plasmid pGL4.75 (hRluc/CMV; Promega) were used in transient transfection assays to evaluate the transcriptional activation function of the GATA6 mutants. HEK-293 cells were transfected with 0.4 μg of wild-type or mutant pcDNA3-hGATA6 expression vector, 0.4 μg of ANF(-2600)-Luc reporter construct, and 0.04 μg of pGL4.75 control reporter vector using PolyFect Transfection Reagent (Qiagen GmbH). For cotransfection experiments, 0.2 μg of wild-type pcDNA3-hGATA6, 0.2 μg of mutant pcDNA3-hGATA6, 0.4 μg of ANF(-2600)-Luc, and 0.04 μg of pGL4.75 were used. Firefly luciferase and Renilla luciferase activities were measured with the Dual-Glo luciferase assay system (Promega) 48 h after transfection. The activity of the ANF promoter was presented as fold activation of Firefly luciferase relative to Renilla luciferase. Three independent experiments were performed at minimum for wild-type and mutant GATA6.
Statistics
Data are expressed as means±standard deviations. Continuous variables were tested for normality of distribution, and Student's unpaired t-test was used for comparison of numeric variables between two groups. Pearson's χ2 test or Fisher's exact test was used, when appropriate, to compare the categorical variables between two groups. A two-tailed p<0.05 was considered to be statistically significant.
Results
Baseline characteristics of the study population
A cohort of 380 unrelated patients with CHD was enrolled and clinically evaluated in contrast to a total of 200 ethnically matched, unrelated healthy individuals used as controls. None of them had overt environmental risk factors for CHD, such as maternal illness and drug use in the first trimester of pregnancy, parental smoking, and chronic exposure to toxicants and ionizing radiation. The baseline clinical characteristics of the 380 unrelated CHD patients are summarized in Table 2.
Table 2.
Clinical Characteristics of the 380 Unrelated Patients with Congenital Heart Disease
| Parameter | Number or mean value | Percentage or range |
|---|---|---|
| Male | 182 | 48 |
| Age at the initial diagnosis of CHD (year) | 3.18 | 0–16 |
| Age at the present study (year) | 6.24 | 1–35 |
| Positive family history of CHD | 65 | 17 |
| Distribution of various types of CHD | ||
| Isolated CHD | 258 | 68 |
| VSD | 84 | 22 |
| ASD | 61 | 16 |
| PDA | 30 | 8 |
| TGA | 19 | 5 |
| DORV | 19 | 5 |
| RVOTO | 11 | 3 |
| Others | 34 | 9 |
| Complex CHD | 122 | 32 |
| TOF | 42 | 11 |
| VSD and ASD | 30 | 8 |
| VSD and PDA | 15 | 4 |
| ASD and PDA | 11 | 3 |
| VSD and TGA | 5 | 1 |
| Others | 19 | 5 |
| Incidence of arrhythmias | ||
| Atrial fibrillation | 23 | 6 |
| Atrioventricular block | 15 | 4 |
| Treatment | ||
| Surgical repair | 247 | 65 |
| Catheter-based closure | 121 | 32 |
| Follow-up | 12 | 3 |
ASD, atrial septal defect; CHD, congenital heart disease; DORV, double-outlet right ventricle; PDA, patent ductus arteriosus; RVOTO, right ventricular outflow tract obstruction; TGA, transposition of great arteries; TOF, tetralogy of Fallot; VSD, ventricular septal defect.
GATA6 sequence variations
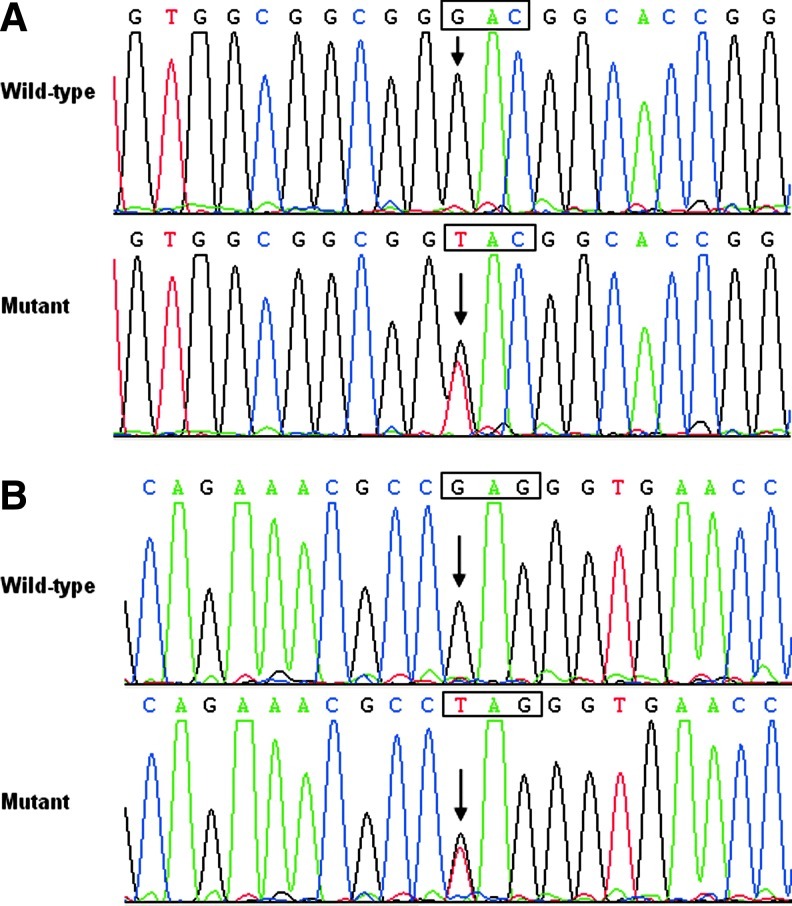
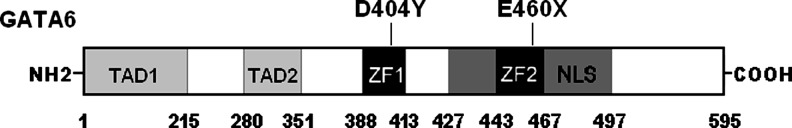
The whole coding region of the GATA6 gene was sequenced in the 380 unrelated CHD patients. Two heterozygous sequence variations in GATA6 were identified in 2 out of 380 patients. The total population prevalence of GATA6 variation based on the cohort patients was ∼0.53%. Specifically, a substitution of thymine for guanine in the first nucleotide of codon 404 of the GATA6 gene (c.1210G>T), equivalent to the transition of aspartic acid to tyrosine at amino acid position 404 (p.D404Y), was identified in the proband from family 1. A replacement of guanine by thymine at coding nucleotide 1378 (c.1378G>T), predicting the truncated protein without C-terminal 136 amino acids (p.E460X), was identified in the index patient from family 2. The sequence chromatograms showing the detected heterozygous GATA6 variations in contrast to corresponding control sequences are shown in Figure 1. A schematic diagram of GATA6 showing the structural domains and the locations of the identified mutations is presented in Figure 2. The variants were neither observed in 400 control alleles nor found in the SNP database at NCBI, which was consulted again on July 19, 2012.
FIG. 1.
Sequence electropherograms showing the GATA6 mutations in contrast to their corresponding controls. The arrow indicates the heterozygous nucleotides of G/T in the probands from families 1 (A) and 2 (B), respectively (mutant), or the homozygous nucleotides of G/G in the corresponding control individuals (wild-type). The square denotes the nucleotides comprising a codon of GATA6.
FIG. 2.
Schematic representation of GATA6 protein structure with the congenital heart disease (CHD)-related mutations indicated. The mutations identified in patients with CHD are shown above the structural domains. NH2 means aminoterminus; TAD, transcriptional activation domain; ZF, zinc finger; NLS, nuclear localization signal; and COOH, carboxylterminus.
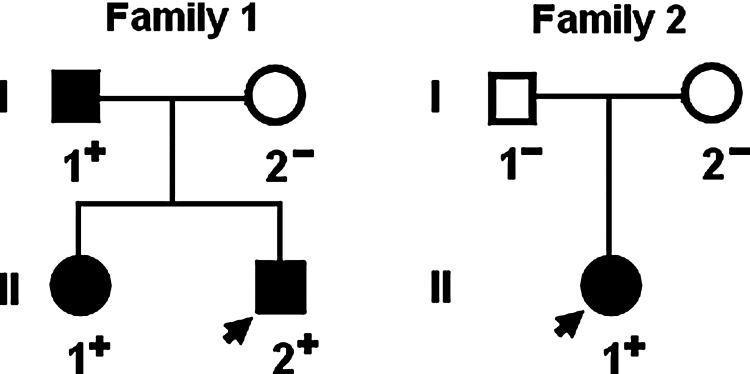
A genetic screen of the mutation carriers' family members demonstrated that the variations were present in all affected family members available, but absent in unaffected family members examined. Analysis of the pedigree showed that in each family, the variation co-segregated with CHD with complete penetrance. The pedigree structures of the two families are illustrated in Figure 3. All the affected family members from the family 1 had the same anatomic type of subarterial ventricular septal defect, and the proband's father (I-1) and elder sister (II-1) had also patent ductus arteriosus and pulmonary stenosis, respectively. Besides, the proband's father (I-1) had also ECG documented atrial fibrillation. In the family 2, the proband had tetralogy of Fallot, and none of his relatives is affected. The phenotypic characteristics and results of genetic screening of the affected pedigree members are listed in Table 3.
FIG. 3.
Pedigree structures of the families with CHD. Families are designated as family 1 and family 2, respectively. Family members are identified by generations and numbers. Squares indicate male family members; circles, female members; closed symbols, affected members; open symbols, unaffected members; arrows, probands; ‘‘+’’, carriers of the heterozygous mutations; and ‘‘−’’, noncarriers.
Table 3.
Phenotypic Characteristics and Status of GATA6 Mutations of the Affected Pedigree Members
|
Subject information |
Phenotypes |
Genotypes |
|||
|---|---|---|---|---|---|
| Identity | Gender | Age at time of present study (years) | Age at initial diagnosis of CHD (years) | Cardiovascular structural defects | GATA6 mutations |
| Family 1 | D404Y | ||||
| I-1 | M | 32 | 12 | VSD, PDA | +/− |
| II-1 | F | 6 | 5 | VSD, PS | +/− |
| II-2 | M | 3 | 2 | VSD | +/− |
| Family 2 | E460X | ||||
| II-1 | F | 2 | 0 | TOF | +/− |
“+” Indicates present and “−”denotes absent; F, female; M, male; PDA, patent ductus arteriosus; PS, pulmonary stenosis.
Multiple-sequence alignments
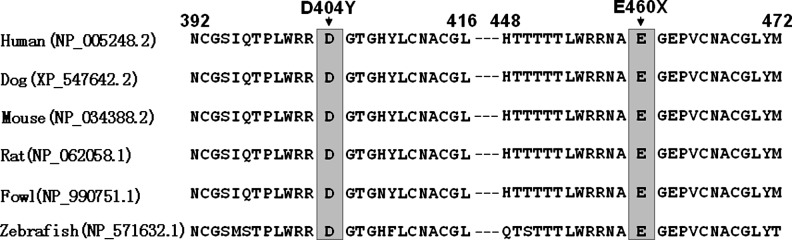
A cross-species alignment of multiple GATA6 protein sequences indicated that the affected amino acids were highly conserved evolutionarily, as shown in Figure 4, indicating that the amino acids are functionally important.
FIG. 4.
Alignment of multiple GATA6 protein sequences across species. The altered amino acids of p.D404 and p.E460 are completely conserved evolutionarily.
Disease-causing potential of sequence variations
The GATA6 sequence variations of c.1210G>T and c.1378G>T were both automatically predicted to be disease causing, with p values of 0.99995 and 1.00000, respectively. No SNPs in the altered regions were found in the MutationTaster database.
Transcriptional activity of the GATA6 mutants
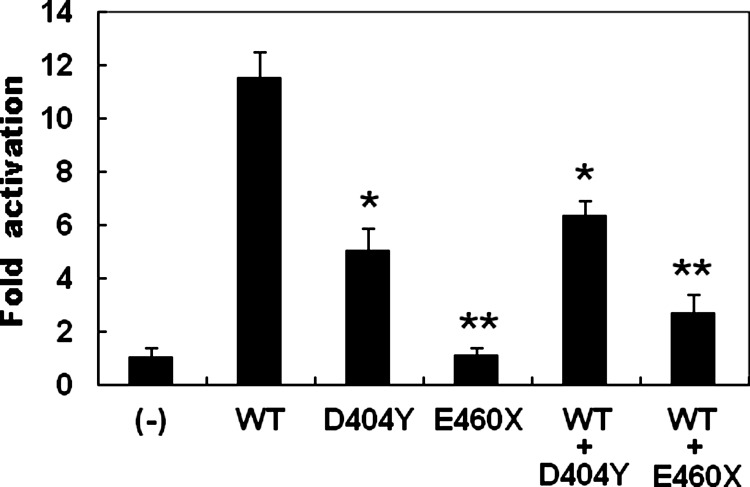
The wild-type GATA6, the D404Y-mutant, and the D460X-mutant GATA6 activated the ANF promoter by ∼11-fold, ∼5-fold, and ∼1-fold, respectively. When wild-type GATA6 was coexpressed with the same amount of D404Y-mutant or E460X-mutant GATA6, the induced activation of the ANF promoter was ∼6-fold or ∼3-fold, respectively. These results reveal that both GATA6 mutants are associated with significantly reduced activation activity compared with their wild-type counterpart (Fig. 5).
FIG. 5.
Functional impairments resulted from GATA6 mutations. Activation of atrial natriuretic factor–luciferase reporter in HEK-293 cells by GATA6 wild-type (WT), mutant D404Y, or mutant E460X, alone or in combination, demonstrated significantly decreased transactivational activity by mutant proteins. Experiments were performed in triplicate, and mean and standard deviations are shown. ** and * represent p<0.001 and p<0.005, respectively, when compared with wild-type GATA6.
Discussion
In the present study, two novel heterozygous mutations of GATA6 (p.D404Y and p.E460X) were identified in two families with CHD. The mutant alleles were present in all the affected family members available, but absent in unaffected relatives examined and 400 normal chromosomes from an ethnically matched control population. A cross-species alignment of GATA6 protein sequences showed that the altered amino acids were completely conserved evolutionarily. The two variants were both predicted to be causative, and the functional analysis substantiated that the mutant GATA6 proteins were associated with significantly decreased transactivational activity. Therefore, functionally impaired GATA6 may predispose to CHD in these families.
The GATA6 gene maps to human chromosome 18q11.1-q11.2 by fluorescence in situ hybridization, which encodes a protein with 595 amino acids (Suzuki et al., 1996). By alignment of GATA6 with GATA4, the functional domains of GATA6 are predicted to be two transcriptional activation domains (TAD1, amino acids 1–215; TAD2, amino acids 280–351), two adjacent zinc fingers (ZF1, amino acids 388–413; ZF2, amino acids 443–467), and one nuclear localization signal (NLS, amino acids 427–497). The 2 TADs are both pivotal for the transcriptional activity of GATA6. The C-terminal ZF1 is responsible for DNA sequence recognition and binding to the consensus motif, whereas the N-terminal ZF2 is required for sequence specificity and stability of protein–DNA binding. The NLS is required for the subcellular trafficking and distribution of GATA6. The GATA6 mutations p.D404Y and p.E460X identified in this study are located in ZF1 and ZF2, respectively, and the p.E460X mutation eliminates the partial ZF2, NLS, and the entire C-terminus, thus may be expected to endanger the transcriptional activation of GATA6 by interfering with the specific binding of GATA6 with target gene promoter or subcellular trafficking.
GATA6 is reported to be an upstream transcriptional regulator of multiple genes expressed during embryogenesis and cardiac morphogenesis, including the genes that encode ANF, brain natriuretic peptide, α-myosin heavy chain, β-myosin heavy chain, and c gap-junction protein (Rémond et al., 2011). Hence, the functional effect of the GATA6 mutation may be assessed by analysis of the transcriptional activity of the ANF promoter in tool cells. In this study, the functional characteristics of the novel GATA6 mutations identified in our CHD patients were delineated by transcriptional activation assay, and the results demonstrated a significantly decreased transcriptional activity on a downstream gene. These findings suggest that dysfunctional GATA6 resulting from loss-of-function mutations is potentially an alternative pathogenic mechanism involved in CHD.
Association of mutated GATA6 with increased susceptibility to CHD has been reported previously. Kodo et al. (2009) sequenced the GATA6 gene in 21 unrelated patients with persistent truncus arteriosus, and two heterozygous mutations of p.E486GfsX10 and p.N466H were identified in two patients, respectively, with a prevalence of 9.52%. It is notable that the proband harboring the frameshift mutation also had atrial septal defect, but his father only had pulmonary stenosis, and his sister had pulmonary stenosis, atrial septal defect, and patent ductus arteriosus. Functional analysis demonstrated both GATA6 mutations resulted in defects in nuclear localization and the transactivation activity (Kodo et al., 2009). Lin et al. (2010) made genetic analysis of GATA6 in 270 unrelated patients with CHD, and discovered a novel heterozygous mutation of p.S184N in three unrelated patients, including one with tetralogy of Fallot and two with atrial septal defect, with a prevalence of 1.11%. Biochemical analysis showed that the mutation had remarkably decreased transcriptional activity (Lin et al., 2010). Maitra et al. (2010) genotyped GATA6 in 310 unrelated patients with CHD, and two heterozygous mutations of p.A178V and p.L198V were identified in two patients, respectively, with a prevalence of 0.65%. The p.L198V carrier was affected with tetralogy of Fallot, whereas the p.A178V carrier was affected with atrioventricular septal defect, hypoplastic left ventricle, and ventricular septal defect. Functional studies revealed the p.A178V mutation had increased transcriptional activity, whereas the p.L198V mutation had no effect on the transcriptional activity (Maitra et al., 2010). Recently, Zheng et al. (2012) carried out mutational analysis of GATA6 in 130 unrelated patients with ventricular septal defect, and found a mutation p.G220S in a patient, with a mutation prevalence of 0.77%. Additionally, the proband's father carrying the p.G220S mutation had also atrial septal defect (Zheng et al., 2012). To date, six distinct GATA6 mutations identified in eight of 731 index patients with CHD have been reported, with a mutation prevalence of 1.09% (Kodo et al., 2009; Lin et al., 2010; Maitra et al., 2010; Zheng et al., 2012). Similarly, a GATA6 mutation prevalence of 0.53% (2/380) was observed in our CHD population, implying that the GATA6 mutations could be an uncommon cause of CHD.
Association of functionally impaired GATA6 with increased susceptibility to congenital cardiovascular defects has been established in animals. Homozygous GATA6 knockout mice die after implantation due to defects in visceral endoderm function and extraembryonic development (Morrisey et al., 1998; Zhao et al., 2008). Although the mice heterozygous for either GATA4 or GATA6 deletion are viable without obvious cardiovascular defects, the mice that are compound heterozygous for both GATA4 and GATA6 die by E13.5 with 100% penetrance, showing a phenotypic spectrum of cardiovascular defects, including ventricular septal defect, persistent truncus arteriosis, myocardial hypoplasia, reduced myocardial proliferation, and impaired differentiation of vascular smooth muscle cells (Xin et al., 2006). Similarly, a compound null of a GATA5 and GATA6 allele also leads to double-outlet right ventricle and ventricular septal defect in mice (Laforest and Nemer, 2011). These experimental results indicate an exquisite sensitivity of the developing cardiovascular system to the levels of GATA4, GATA5, and GATA6 and suggest that these GATA factors act cooperatively to regulate downstream target genes.
The identification of novel GATA6 mutations associated with CHD helps to make an early diagnosis and timely treatment, which is of potential importance in clinical practice, because uncorrected CHD may give rise to such complications as congestive heart failure, pulmonary hypertension, arrhythmias, and even death. As the mortality rate of CHD operations is very low in patients without cardiac failure at an early age, timely surgical repair or transcatheter closure of CHD is feasible. Especially, this information will be very useful for genetic counseling for these families, because genetic testing for p.D404Y and p.E460X in newly born children can be followed by careful medical examination and preventive intervention if CHD is observed.
Interestingly, atrial fibrillation was documented in one CHD patient (I-1) harboring the p.D404Y mutation, consistent with our previous reports on the link of GATA6 with atrial fibrillation (Yang et al., 2012b, 2012e). Similarly, mutations in other cardiac transcriptional factor genes, such as GATA5 and GATA4, were also associated in atrial fibrillation (Posch et al., 2010; Jiang et al., 2011; Yang et al., 2011; Wang et al., 2012a; Yang et al., 2012d). These observations suggest that atrial fibrillation may share a common genetic origin with CHD.
Conclusion
The findings expand the mutational spectrum of GATA6 linked to CHD and provide a novel insight into the molecular pathogenesis of CHD, implying the potential implications for the early diagnosis and personalized treatment of CHD.
Acknowledgments
We are greatly thankful to the participants for their devotion to the study. This work was supported in part by the grants from the National Natural Science Fund of China (81070153 and 30570768), the Personnel Development Foundation of Shanghai, China (2010019), the Natural Science Fund of Shanghai, China (10ZR1428000), and the Key Program of Basic Research of Shanghai, China (10JC1414002).
Disclosure Statement
No competing financial interests exist.
References
- Bartlett H. Veenstra G.J. Weeks D.L. Examining the cardiac NK-2 genes in early heart development. Pediatr Cardiol. 2010;31:335–341. doi: 10.1007/s00246-009-9605-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson D.W. Genetic origins of pediatric heart disease. Pediatr Cardiol. 2010;31:422–429. doi: 10.1007/s00246-009-9607-y. [DOI] [PubMed] [Google Scholar]
- Bruneau B.G. The developmental genetics of congenital heart disease. Nature. 2008;451:943–948. doi: 10.1038/nature06801. [DOI] [PubMed] [Google Scholar]
- Butler T.L. Esposito G. Blue G.M. Cole A.D. Costa M.W. Waddell L.B., et al. GATA4 mutations in 357 unrelated patients with congenital heart malformation. Genet Test Mol Biomarkers. 2010;14:797–802. doi: 10.1089/gtmb.2010.0028. [DOI] [PubMed] [Google Scholar]
- Chen M.W. Pang Y.S. Guo Y. Pan J.H. Liu B.L. Shen J., et al. GATA4 mutations in Chinese patients with congenital cardiac septal defects. Pediatr Cardiol. 2010a;31:85–89. doi: 10.1007/s00246-009-9576-1. [DOI] [PubMed] [Google Scholar]
- Chen Y. Mao J. Sun Y. Zhang Q. Cheng H.B. Yan W.H., et al. A novel mutation of GATA4 in a familial atrial septal defect. Clin Chim Acta. 2010b;411:1741–1745. doi: 10.1016/j.cca.2010.07.021. [DOI] [PubMed] [Google Scholar]
- Chen Y. Zhou B. Li H. Peng Y. Wang Y. Rao L. Analysis of RTN4 3′UTR insertion/deletion polymorphisms in ventricular septal defect in a Chinese Han population. DNA Cell Biol. 2011;30:323–327. doi: 10.1089/dna.2010.1116. [DOI] [PubMed] [Google Scholar]
- Cheng H.H. Almodovar M.C. Laussen P.C. Wypij D. Polito A. Brown D.W., et al. Outcomes and risk factors for mortality in premature neonates with critical congenital heart disease. Pediatr Cardiol. 2011;32:1139–1146. doi: 10.1007/s00246-011-0036-3. [DOI] [PubMed] [Google Scholar]
- Garg V. Kathiriya I.S. Barnes R. Schluterman M.K. King I.N. Butler C.A., et al. GATA4 mutations cause human congenital heart defects and reveal an interaction with TBX5. Nature. 2003;424:443–447. doi: 10.1038/nature01827. [DOI] [PubMed] [Google Scholar]
- Jiang J.Q. Shen F.F. Fang W.Y. Liu X. Yang Y.Q. Novel GATA4 mutations in lone atrial fibrillation. Int J Mol Med. 2011;28:1025–1032. doi: 10.3892/ijmm.2011.783. [DOI] [PubMed] [Google Scholar]
- Kodo K. Nishizawa T. Furutani M. Arai S. Yamamura E. Joo K., et al. GATA6 mutations cause human cardiac outflow tract defects by disrupting semaphorin-plexin signaling. Proc Natl Acad Sci U S A. 2009;106:13933–13938. doi: 10.1073/pnas.0904744106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laforest B. Nemer M. GATA5 interacts with GATA4 and GATA6 in outflow tract development. Dev Biol. 2011;358:368–378. doi: 10.1016/j.ydbio.2011.07.037. [DOI] [PubMed] [Google Scholar]
- Li H. Chen Y. Zhou B. Peng Y. Bai W. Rao L. RNT4 3′-UTR insertion/deletion polymorphisms are not associated with atrial septal defect in Chinese Han population: a brief communication. DNA Cell Biol. 2012;31:1121–1124. doi: 10.1089/dna.2011.1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X. Huo Z. Liu X. Zhang Y. Li L. Zhao H., et al. A novel GATA6 mutation in patients with tetralogy of Fallot or atrial septal defect. J Hum Genet. 2010;55:662–667. doi: 10.1038/jhg.2010.84. [DOI] [PubMed] [Google Scholar]
- Liu H. Dai L. Mao M. Wang X. Hua Y. Xie L. Absence of association between length variation of an intronic region in the NFATC1 gene and congenital heart defects in a Han Chinese population. DNA Cell Biol. 2012;31:88–91. doi: 10.1089/dna.2011.1286. [DOI] [PubMed] [Google Scholar]
- Liu X.Y. Wang J. Zheng J.H. Bai K. Liu Z.M. Wang X.Z., et al. Involvement of a novel GATA4 mutation in atrial septal defects. Int J Mol Med. 2011;28:17–23. doi: 10.3892/ijmm.2011.638. [DOI] [PubMed] [Google Scholar]
- Maitra M. Koenig S.N. Srivastava D. Garg V. Identification of GATA6 sequence variants in patients with congenital heart defects. Pediatr Res. 2010;68:281–285. doi: 10.1203/PDR.0b013e3181ed17e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrisey E.E. Tang Z. Sigrist K. Lu M.M. Jiang F. Ip H.S., et al. GATA6 regulates HNF4 and is required for differentiation of visceral endoderm in the mouse embryo. Genes Dev. 1998;12:3579–3590. doi: 10.1101/gad.12.22.3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson E.N. Gene regulatory networks in the evolution and development of the heart. Science. 2006;313:1922–1927. doi: 10.1126/science.1132292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry J.C. Sudden cardiac death and malignant arrhythmias: the scope of the problem in adult congenital heart patients. Pediatr Cardiol. 2012;33:484–490. doi: 10.1007/s00246-012-0171-5. [DOI] [PubMed] [Google Scholar]
- Pikkarainen S. Tokola H. Kerkelä R. Ruskoaho H. GATA transcription factors in the developing and adult heart. Cardiovasc Res. 2004;63:196–207. doi: 10.1016/j.cardiores.2004.03.025. [DOI] [PubMed] [Google Scholar]
- Posch M.G. Boldt L.H. Polotzki M. Richter S. Rolf S. Perrot A., et al. Mutations in the cardiac transcription factor GATA4 in patients with lone atrial fibrillation. Eur J Med Genet. 2010;53:201–203. doi: 10.1016/j.ejmg.2010.03.008. [DOI] [PubMed] [Google Scholar]
- Rani D.S. Nallari P. Dhandapany P.S. Tamilarasi S. Shah A. Archana V., et al. Cardiac Troponin T (TNNT2) mutations are less prevalent in Indian hypertrophic cardiomyopathy patients. DNA Cell Biol. 2012;31:616–624. doi: 10.1089/dna.2011.1366. [DOI] [PubMed] [Google Scholar]
- Rémond M.C. Iaffaldano G. O'Quinn M.P. Mezentseva N.V. Garcia V. Harris B.S., et al. GATA6 reporter gene reveals myocardial phenotypic heterogeneity that is related to variations in gap junction coupling. Am J Physiol Heart Circ Physiol. 2011;301:H1952–H1964. doi: 10.1152/ajpheart.00635.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roger V.L. Go A.S. Lloyd-Jones D.M. Benjamin E.J. Berry J.D. Borden W.B., et al. American Heart Association Statistics Committee and Stroke Statistics Subcommittee. Heart disease and stroke statistics—2012 update: a report from the American Heart Association. Circulation. 2012;125:e2–e220. doi: 10.1161/CIR.0b013e31823ac046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salazar M. Consoli F. Villegas V. Caicedo V. Maddaloni V. Daniele P., et al. Search of somatic GATA4 and NKX2.5 gene mutations in sporadic septal heart defects. Eur J Med Genet. 2011;54:306–309. doi: 10.1016/j.ejmg.2011.01.004. [DOI] [PubMed] [Google Scholar]
- Schott J.J. Benson D.W. Basson C.T. Pease W. Silberbach G.M. Moak J.P., et al. Congenital heart disease caused by mutations in the transcription factor NKX2–5. Science. 1998;281:108–111. doi: 10.1126/science.281.5373.108. [DOI] [PubMed] [Google Scholar]
- Shedeed S.A. Elfaytouri E. Brain maturity and brain injury in newborns with cyanotic congenital heart disease. Pediatr Cardiol. 2011;32:47–54. doi: 10.1007/s00246-010-9813-7. [DOI] [PubMed] [Google Scholar]
- Silka M.J. Bar-Cohen Y. A contemporary assessment of the risk for sudden cardiac death in patients with congenital heart disease. Pediatr Cardiol. 2012;33:452–460. doi: 10.1007/s00246-012-0165-3. [DOI] [PubMed] [Google Scholar]
- Starr J.P. Tetralogy of Fallot: yesterday and today. World J Surg. 2010;34:658–668. doi: 10.1007/s00268-009-0296-8. [DOI] [PubMed] [Google Scholar]
- Suzuki E. Evans T. Lowry J. Truong L. Bell D.W. Testa J.R., et al. The human GATA-6 gene: structure, chromosomal location, and regulation of expression by tissue-specific and mitogen-responsive signals. Genomics. 1996;38:283–290. doi: 10.1006/geno.1996.0630. [DOI] [PubMed] [Google Scholar]
- Teixeira F.M. Coelho R.M. Proença C. Silva A.M. Vieira D. Vaz C., et al. Quality of life experienced by adolescents and young adults with congenital heart disease. Pediatr Cardiol. 2011;32:1132–1138. doi: 10.1007/s00246-011-0039-0. [DOI] [PubMed] [Google Scholar]
- van der Bom T. Zomer A.C. Zwinderman A.H. Meijboom F.J. Bouma B.J. Mulder B.J. The changing epidemiology of congenital heart disease. Nat Rev Cardiol. 2011;8:50–60. doi: 10.1038/nrcardio.2010.166. [DOI] [PubMed] [Google Scholar]
- van der Linde D. Konings E.E. Slager M.A. Witsenburg M. Helbing W.A. Takkenberg J.J., et al. Birth prevalence of congenital heart disease worldwide: a systematic review and meta-analysis. J Am Coll Cardiol. 2011;58:2241–2247. doi: 10.1016/j.jacc.2011.08.025. [DOI] [PubMed] [Google Scholar]
- Wang J. Fang M. Liu X.Y. Xin Y.F. Liu Z.M. Chen X.Z., et al. A novel GATA4 mutation responsible for congenital ventricular septal defects. Int J Mol Med. 2011;28:557–564. doi: 10.3892/ijmm.2011.715. [DOI] [PubMed] [Google Scholar]
- Wang J. Sun Y.M. Yang Y.Q. Mutation spectrum of the GATA4 gene in patients with idiopathic atrial fibrillation. Mol Biol Rep. 2012a;39:8127–8135. doi: 10.1007/s11033-012-1660-6. [DOI] [PubMed] [Google Scholar]
- Wang Y. Liu Y. Peng W. Wang M. Sun J. Zhang Z., et al. ECE1 polymorphisms may contribute to the susceptibility of sporadic congenital heart disease in a Chinese population. DNA Cell Biol. 2012b;31:1425–1430. doi: 10.1089/dna.2012.1626. [DOI] [PubMed] [Google Scholar]
- Xin M. Davis C.A. Molkentin J.D. Lien C.L. Duncan S.A. Richardson J.A., et al. A threshold of GATA4 and GATA6 expression is required for cardiovascular development. Proc Natl Acad Sci U S A. 2006;103:11189–11194. doi: 10.1073/pnas.0604604103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y.Q. Li L. Wang J. Liu X.Y. Chen X.Z. Zhang W., et al. A novel GATA4 loss-of-function mutation associated with congenital ventricular septal defect. Pediatr Cardiol. 2012a;33:539–546. doi: 10.1007/s00246-011-0146-y. [DOI] [PubMed] [Google Scholar]
- Yang Y.Q. Li L. Wang J. Zhang X.L. Li R.G. Xu Y.J., et al. GATA6 loss-of-function mutation in atrial fibrillation. Eur J Med Genet. 2012b doi: 10.1016/j.ejmg.2012.06.007. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Yang Y.Q. Wang J. Liu X.Y. Chen X.Z. Zhang W. Wang X.Z., et al. Novel GATA4 mutations in patients with congenital ventricular septal defects. Med Sci Monit. 2012c;18:CR344–CR350. doi: 10.12659/MSM.882877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y.Q. Wang J. Wang X.H. Wang Q. Tan H.W. Zhang M., et al. Mutational spectrum of the GATA5 gene associated with familial atrial fibrillation. Int J Cardiol. 2012d;157:305–307. doi: 10.1016/j.ijcard.2012.03.132. [DOI] [PubMed] [Google Scholar]
- Yang Y.Q.Wang M.Y.Zhang X.L.Tan H.W.Shi H.F.Jiang W.F., et al.2011GATA4 loss-of-function mutations in familial atrial fibrillation Clin Chim Acta 4121825–1830. [DOI] [PubMed] [Google Scholar]
- Yang Y.Q. Wang X.H. Tan H.W. Jiang W.F. Fang W.Y. Liu X. Prevalence and spectrum of GATA6 mutations associated with familial atrial fibrillation. Int J Cardiol. 2012e;155:494–496. doi: 10.1016/j.ijcard.2011.12.091. [DOI] [PubMed] [Google Scholar]
- Zhao R. Watt A.J. Battle M.A. Li J. Bondow B.J. Duncan S.A. Loss of both GATA4 and GATA6 blocks cardiac myocyte differentiation and results in acardia in mice. Dev Biol. 2008;317:614–619. doi: 10.1016/j.ydbio.2008.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng G.F. Wei D. Zhao H. Zhou N. Yang Y.Q. Liu X.Y. A novel GATA6 mutation associated with congenital ventricular septal defect. Int J Mol Med. 2012;29:1065–1071. doi: 10.3892/ijmm.2012.930. [DOI] [PubMed] [Google Scholar]