Abstract
Wnt5a is an extracellular glycoprotein that activates Wnt signaling pathways, important in development and tissue homeostasis. Wnt5a expression is often misregulated during cancer progression. In this study, we analyzed the transcriptional regulation of two of the Wnt5a alternative promoters, termed A and B. Transient transfection of promoter A and B luciferase reporter constructs in to NIH3T3 and Caco-2 cells indicated that the separated promoters are both functional and that 300–450 base pair (bp) of upstream sequence is sufficient for activity. Promoter B constructs displayed distinct patterns of expression in the two cell types. The endogenous levels of promoter A-derived transcripts were found to be greater than the promoter B transcripts by four- to sixfold in fibroblast cells. Treatment of NIH3T3 cells with tumor necrosis factor (TNF)-alpha leads to an increase in both promoter A and B activities, but promoter B was more responsive. Using inhibitors of TNF-alpha effector proteins, we provide evidence that the transcription factor nuclear factor-kappaB and the MEK1/2 and p38 kinases have distinct roles in determining the activity levels of promoters, A and B. These results support the conclusion that Wnt5a promoters, A and B, are differentially regulated and provide a model for complex transcriptional regulation of Wnt5a.
In two cell lines, the promoters of Wnt5a was examined using a luciferase reporter. Gene expression was greater for the A promoter than for the B promoter. The latter promoter was more responsive to induction by the cytokine TNF-α.
Introduction
Wnt5a is a lipid-modified glycoprotein belonging to the Wnt family of secreted protein ligands. Wnt5a binds to distinct membrane receptors of the Fz family, along with the Ror2 coreceptor, and activates the noncanonical Wnt signaling pathways, PCP/CE and Ca2+ (reviewed in Kikuchi et al., 2011). In some cells, depending on the receptor context, Wnt5a has also been shown to block or activate the canonical β-catenin pathway (Mikels and Nusse, 2006; McDonald and Silver, 2009). Wnt5a has important functions during development, particularly in events that require cell movement and rearrangements (Yamaguchi et al., 1999; Hardy et al., 2008; Blakely et al., 2011). Wnt5a also functions in postnatal cellular differentiation and tissue homeostasis. In particular, Wnt5a has a role in hematopoiesis (Liang et al., 2003) and mesenchymal stem cell differentiation (Yang et al., 2003; Bilkovski et al., 2010). Mechanistically, Wnt5a-initiated cell signaling leads to altered cell function through rearrangements of the cytoskeleton and/or changes in gene expression (reviewed in Kikuchi et al., 2011).
Significantly, WNT5A expression is altered in numerous cancers, including lung (Huang et al., 2005), breast (Iozzo et al., 1995; Pukrop et al., 2006), thyroid (McCall et al., 2007), osteosarcoma (Nakano et al., 2003; Enomoto et al., 2009), gastric (Saitoh et al., 2002), colorectal (Ying et al., 2008; Hibi et al., 2009), leukemias (Martin et al., 2010; Deng et al., 2011), skin (Taki et al., 2003; Weeraratna et al., 2010), pancreatic (Ripka et al., 2007), melanoma (Iozzo et al., 1995; Weeraratna et al., 2002), esophageal (Li et al., 2010), and prostate (Wang et al., 2007). WNT5A has been found to be both overexpressed and downregulated. WNT5A overexpression has been associated with metastatic behavior in various cancers (Weeraratna et al., 2002; Kurayoshi et al., 2006; Ripka et al., 2007; Enomoto et al., 2009).
And, upregulation of WNT5A expression has been found to induce an epithelial–mesenchymal transition in melanoma cells (Dissanayake et al., 2007). Clearly, understanding of the mechanisms regulating WNT5A expression is of particular importance.
Current published data indicate that changes in WNT5A expression during cancer progression do not involve genetic (DNA) changes, such as gene mutation and rearrangements, but rather nongenetic changes. Hypo- and hypermethylation of the WNT5A gene has been detected in tissue derived from diverse tumor types. Hypermethylation of Wnt5a is more common and has been detected in the early stages of colorectal cancer (Ying et al., 2008; Hibi et al., 2009), myeloid and acute lymphoblastic leukaemia (Roman-Gomez et al., 2007; Martin et al., 2009) and esophageal squamous cell carcinoma (Li et al., 2010). In contrast, Wnt5a was found to be hypomethylated in prostate cancer tissue (Wang et al., 2007).
Alternatively, there is evidence that WNT5A upregulation in certain cancers involves changes in specific signaling pathways and transcription factors. In two studies, the drug phenylmethimazole, which inhibits the Toll-like receptor 3, also coordinately decreased the level of WNT5A mRNA in papillary thyroid carcinoma (McCall et al., 2007) and pancreatic cancer and melanoma (Schwartz et al., 2009), leading to decreased cell proliferation and migration. Similarly, the CUTL1 transcription factor, which is a target of transforming growth factor β and when overexpressed enhances cancer cell motility and invasiveness (Michl et al., 2005), increases WNT5A transcription (Ripka et al., 2007). The nuclear factor (NF)-kappaB has also been implicated in the regulation of WNT5A transcription (Saitoh and Katoh, 2002; Ge et al., 2011; Rauner et al., 2011). NF-kappaB is known to mediate the expression of many genes that influence growth and inflammation and is often upregulated in cancer (Basseres and Baldwin, 2006).
The human and mouse Wnt5a gene region generates transcript variants derived from distinct transcription start sites and alternative splicing (see Table 1 and Katoh and Katoh, 2009). Little is known regarding the differential regulation of the Wnt5a alternative promoters. It is likely, however, that distinct patterns of Wnt5a expression can be achieved through the activity of gene regulatory proteins that affect the transcription from one, but not the other promoter and that altered Wnt5a transcript levels, particularly in cancer cells, can be achieved via multiple pathways involving these distinct promoters. Indeed, the alternative promoters of various mammalian genes have been found to display distinct activities at particular developmental stages, in specific tissue types, and in cancer cells (Liang et al., 2005; Oh et al., 2005; Banday et al., 2011; Bee et al., 2011; Demura et al., 2011). In this study, we focused on two of the Wnt5a alternative promoters that are common in mouse and human to address the question of Wnt5a differential promoter regulation. The genomic upstream regions of these promoters were separately cloned for individual analysis and promoter-specific transcript levels were quantified by quantitative real-time (qRT)-PCR. We focused on the general activity of the separated promoters and on the response of each promoter to tumor necrosis factor (TNF)-alpha-induced cell signaling and the role of NF-kappaB and other effector proteins in the response to TNF-alpha.
Table 1.
Comparison of Mouse and Human Wnt5a Genes
| Mouse | Human | |
|---|---|---|
| Location | Chromosome 14 | Chromosome 3 |
| 29,317,936–29,340,633 | 55,499,743–55,523,973 | |
| Forward | Reverse | |
| Number of transcriptsa | 6 | 8 |
| Transcript size (name and ID)b | 7009 bp (Wnt5a-001;ID-063465) | 6042 bp (WNT5A-201;ID474267) |
| 3650 bp (Wnt5a-002;ID-112272) | 1299 bp (WNT5A-005;ID497027) | |
| Proteins produceda | 2 | 6 |
| Protein Length (AA) and IDc | 380 residues (ID-064878) | 380 residues (ID-417310) |
| 360 residues (ID-107891) | 365 residues (ID-420104) | |
| N-terminus of proteinb,d | MKKPIGILSPGVALGTAGGA (20) | MKKSIGILSPGVALG (15) |
| MSSKFFLMALATFFSFAQVV… | MAGSAMSSKFFLVALAIFFS… | |
| Exons and introns (bp)b,e | Wnt5a-001 | WNT5A-201 |
| Exons: 1365, 134, 251, 293, 4966 | Exons: 324, 134, 251, 293, 4835 | |
| Introns: 5706, 1184, 4894, 3903 | Introns: 6061, 1220, 4684, 3786 | |
| Wnt5a-002 | WNT5A-005 | |
| Exons: #1–19, #5–2953 | Exons: #1b-63, #5–558 | |
| Intron: #1–399 | Introns: #1–412 |
Source: Ensembl Mouse Wnt5a ENSMUSG00000021994 and Human WNT5A ENSG00000114251.
Total number of transcripts or proteins generated from the Wnt5a genomic region.
For the two mouse and human transcripts analyzed in this study, Transcript ID is preceded by ENSMUST000000 for mouse or ENST00000 for human.
Derived from the two transcripts analyzed. ID is preceded by ENSMUSO00000 for mouse or ENSP00000 for human.
Italicized AA sequence and the number in parenthesis indicate the additional AA's and N-terminus on the longer transcript (b). The AA sequence of the longer transcript includes all the AA's shown and is continuous. Bottom sequence includes the N-terminus and first 20 AA's of the shorter transcript (b).
Only the unique exons and introns for the shorter transcript are included; all others are identical to the longer transcript.
Materials and Methods
Cell lines and culture
NIH3T3 mouse fibroblasts were cultured in the Dulbecco's Modified Eagle's Medium (DMEM) and supplemented with 10% calf serum (Atlanta Biologicals, Lawrenceville, GA) and 1% penicillin/streptomycin (5000 I.U./mL and 5000 μg/mL). Caco-2, a human colorectal adenocarcinoma-derived cell line, was grown in the RPMI 1640 medium containing 10% fetal bovine serum (Gibco, Carlsbad, CA) and 1% penicillin/streptomycin. GM03349 cells are normal human dermal fibroblast cells obtained from the National Institute of General Medical Sciences (NIGMS) Human Genetic Mutant Cell Repository (Corriell, Camden, NJ). They were grown in the Minimum Essential Medium (MEM; Mediatech, Inc., Manassas, VA), supplemented with 10% fetal bovine serum (Gibco), 2 mM L-glutamine, and 1% penicillin/streptomycin. All cell types were cultured in a 37°C and 5% carbon dioxide humidified incubator.
WNT5A promoter A and B luciferase reporter constructs
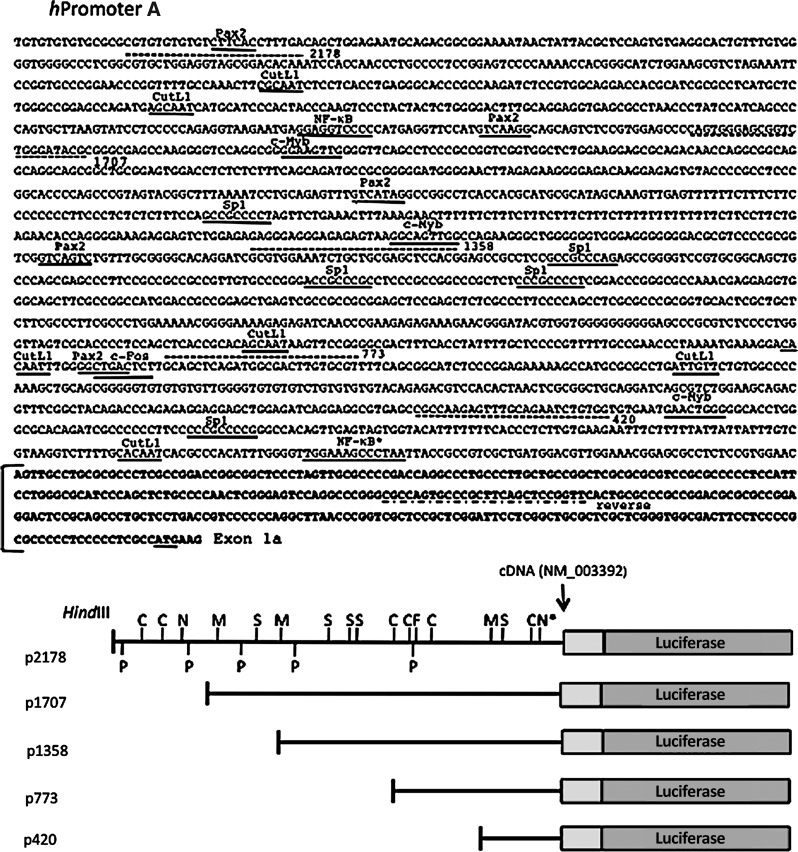
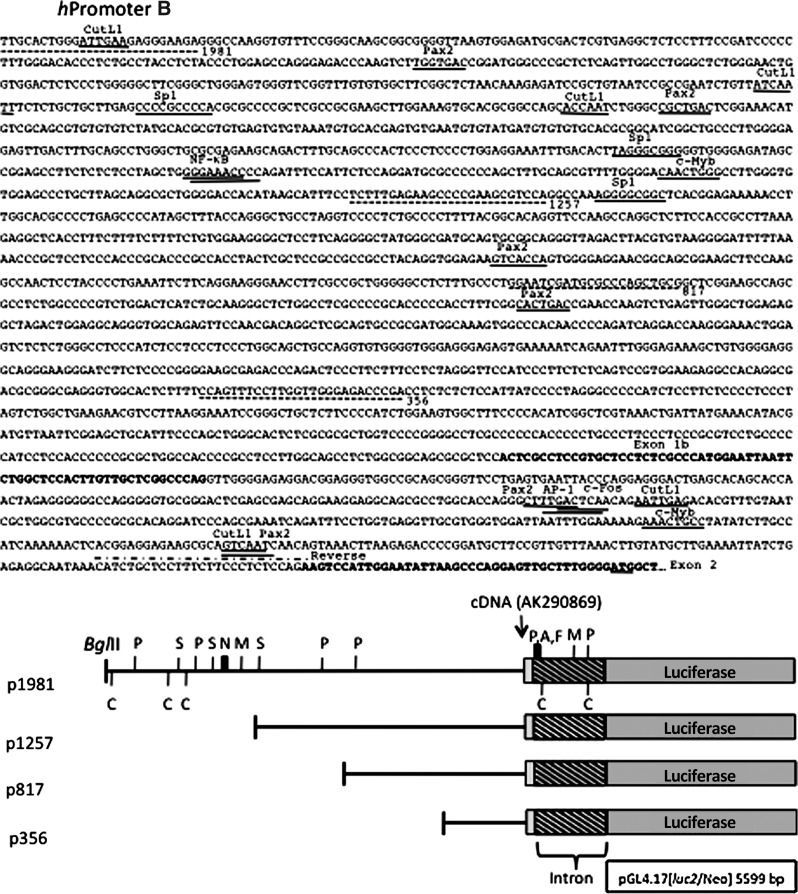
Sequences upstream of the human WNT5A alternative transcription start sites referred to as hpromoter A and hpromoter B (see Fig. 2) were PCR amplified from purchased human genomic DNA (Promega, Madison WI). HindIII restriction sites were included on the primers for the hpromoter A sequences and BglII sites for the hpromoter B sequences and used for cloning of the PCR fragments into the vector, pGL4.17 [luc2/Neo] (Promega). Five constructs were generated for the hpromoter A containing 2178, 1707, 1358, 773, 420 bp upstream of the end of the cDNA as defined in NM_003392 (Fig. 2). Four constructs were generated for the hpromoter B containing 1981, 1257, 817, 356 bp upstream of the cDNA defined in AK290869 (Fig. 2). The primers used to generate the hpromoter A and B constructs are shown in Table 2. The clones were verified by DNA sequencing.
FIG. 2.
Human WNT5A hpromoter A and hpromoter B luciferase reporter constructs. Genomic sequences: The included genomic sequences for the p2178 and p1981 constructs are shown. Putative transcription factor binding sites with names are underlined. Some sites are on the opposite strand. Dotted lines indicate the positions of the PCR primers used to generate the luciferase reporter constructs. Exon sequences are shown in bold and the ATG start codons are underlined. Luciferase reporter constructs: The WNT5A upstream sequences are represented by the black line. The numbers are base-pair (bp) upstream from the first nucleotide of the cDNA, indicated with the NCBI accession number. The gray boxes are sequences downstream of the first nucleotide. The indicated HindIII and BglII sites were used for cloning. The hpromoter B constructs include a 412-bp intron, unique to the hpromoter B-derived transcript. The vector sequences are not to scale. Positions of the selected transcription factor binding sites are indicated. A, AP-1; C, CutL1; F, c-Fos; M, c-Myb; N, nuclear factor (NF)-kappaB; P, Pax2; S, Sp1. NF-κB* is the site identified in Ge et al., (2011).
Table 2.
Primer and Probe Sequences for PCR and qRT-PCR
| Primers for Wnt5a reporter constructs | Primer sequences |
|---|---|
| hpromoter A | |
| p2178 | F-GCTAAGCTT/GCGTGTGTGTGTCTTCACCTTG |
| p1707 | F-GCTAAGCTT/CAGTGGGAGCGGTCTGGGATACGC |
| p1358 | F-GCTAAGCTT/GAGGGAGAGAGTAAGGCAGTTGG |
| p773 | F-GCTAAGCTT/CTCACCGCACAGCAATAAGTTCCGG |
| p420 | F-GCTAAGCTT/CGCCAAGAGTTTGCAGAATCTGTGG |
| R-GCTAAGCTT/GAACCGGAGCTGAAGCGGGCACTGGCG | |
| hpromoter B | |
| p1981 | F-GCTAGATCT/TTGCACTGGGATTGAAGAGGGAAGA |
| p1257 | F-GCTAGATCT/TCTTTGAGAAGCCCCGAAGCGTCCA |
| p817 | F-GCTAGATCT/GGAATCGATGCGCCCAGCTGCGGCTCG |
| p356 | F-GCTAGATCT/CCAGTTTCCTTGGTTGGGAGACCCGA |
| R-GCTAGATCT/TTGATTGACTGCGCTTCTCCTCCGT |
| Primers and probes for qRT-PCR | Primer sequences |
|---|---|
| hpromoter A | |
| Forward | TCGGGTGGCGACTTCCT |
| Reverse | CAACTCCTGGGCTTAATATTCCAAT |
| Probe | CGCCCCCTCCCCCTCGCCATGAAG |
| hpromoter B | |
| Forward | CCTCTCGCCCATGGAATT |
| Reverse | GGGCTTAATATTCCAATGGACTTC |
| Probe | CTGGCTCCACTTGTTGCTCGGCC |
| mpromoter A | |
| Forward | GTGGCGACTTCCTCTCCGT |
| Reverse | CGGTCCCCAAAGCCACT |
| Probe | CCCCTCGCCATGAAGAAGCCCA |
| mpromoter B | |
| Forward | ACTTGTTGCTCCGGCCC |
| Reverse | CGGTCCCCAAAGCCACT |
| Probe | AGAAGCCCATTGGAATATTAAGCCCGG |
All primers are shown 5′ to 3′. The diagonal lines demarcate the added extra nucleotides with restriction sites from actual primer sequence. Reverse primers (R) were paired with each of the associated forward (F) primers.
Putative transcription factor binding site identification
Genomic sequences included in the luciferase reporter construct, p2178 and p1981 (see Fig. 2), were analyzed for a select group of transcription factors (NF-kappaB, CutL1, Pax2, AP-1, STAT3, c-Fos, Sp1, and c-Myb) with the online analysis program PROMO, which uses the TRANSFAC database, and MacVector (MacVector, Inc., Cary, NC) nucleic acid subsequence analysis. Only those sites that were at least 95% similar to the consensus sequence were mapped on to the sequence. Transcription factors with shorter consensus sequence binding sites generated a greater number of potential sites than those with longer sequences.
Transient transfections and dual luciferase assays
NIH3T3 cells (6×104 cells/well) and Caco-2 cells (4×104 cells/well) were plated in 24-well plates and incubated for 24 h. The hpromoter A and B constructs (0.5 μg) and the parental plasmid pGL4.17 (0.5 μg) were transiently transfected into NIH3T3 cells using TransIT-LT (Mirus, Madison, WI) and Caco-2 using Nanojuice (Novagen, San Diego, CA), along with 10 ng of a Renilla control vector (phRL-SV40; Promega, Madison, WI) according to instructions. Each construct was transfected into four separate wells of cells. After 48 h, the media was removed and the cells were washed with 1× phosphate-buffered saline (PBS). 1× Passive Lysis Buffer (Promega) was added and the samples incubated at 37°C for 15 min and stored at −80°C. Samples were assayed for firefly and Renilla luciferase activity on a Synergy 2 multimode microplate reader (BioTek, Winooski, Vermont) using the Dual-Luciferase Assay System (Promega). Promoter activity was expressed as the average value of the ratio of firefly luciferase to Renilla luciferase activity for the four replicas. The transfection was repeated three independent times. The fold-change relative to the longest construct was determined for each independent trial. These values were averaged and the standard error and statistical significance determined using the Student t-test.
Stable WNT5A hpromoter A and B cells and compound treatments
NIH3T3 cells were cotransfected with WNT5A hpromoters A p2178 or hpromoter B p1981 reporter constructs and the selection vector pMC1neo Poly A (Agilent Technologies, Santa Clara, CA) at a mass ratio of 4:1 using the GenPORTER transfection reagent (Genlantis, San Diego, CA). Three days after transfection 500 μg of geneticin bisulfate (G418) was added. Following initial cell death, the resistant cells were replated in a medium with G418 and after 3 or 4 weeks, cells were collected as a group and expanded once more before being stored in liquid nitrogen.
For treatment experiments, p2178 and p1981 NIH3T3 cell lines were plated in 48-well plates at 2×104 cells per well and allowed to grow 1 day. The cells were treated with 5 ng/mL TNF-alpha, with and without a NF-kappaB inhibitor (JSH-23, 30 μM), MAP (mitogen-activated protein) kinase/ERK kinase (MEK) 1/2 inhibitor (U0126, 10 μM), mitogen-activated protein kinase p38 inhibitor (SB203850, 10 μM), and Jun N-terminal kinase (JNK) inhibitor (SP600125, 20 μM) for 6 and 12 h. Cells were also treated with the same concentration of inhibitors, but without TNF-alpha. Control cells were treated with dimethyl sulfoxide (DMSO) only. All wells contained the same concentration of DMSO. The cells were collected by removing the media and washing with 1× PBS. 100 μL of 1× Cell Lysis Buffer (Promega) for 15 min at 37°C. 20 μL of lysate from each sample was added to a black-sided 96-well plate (Corning, Inc., Corning, NY) to be assayed for luciferase activity on a Synergy 2 multimode microplate reader using the Luciferase Assay System (Promega). Each study was independently replicated three to four times. Standard error values were determined and significance using the Student t-test (p<0.05) for replicas.
Verification and amplification efficiencies of custom TaqMan primer–probe sets
Amplification (PCR) products from TaqMan qRT-PCR using the mouse and human promoter A and B custom primer–probe sets were analyzed on a 1% DNA agarose gel to determine the product size. The PCR products were then gel purified and quantified by reading the absorbance at 260 nm. An initial qRT-PCR amplification was conducted to adjust the cycle numbers. A 5-log dilution series of the purified PCR products, including five points, was prepared based on the test amplification such that the resulting cycle numbers were between 10 and 32. Each dilution was amplified in triplicate using the appropriate primer–probe set. The replica values for each concentration were averaged. A graph of concentration versus the average cycle number was generated and the slope of the line determined. The efficiency value (E) was calculated using the equation E=10(−1/slope)−1. Efficiency values between 90% and 100% with an established standard deviation value of±10% are considered acceptable (Applied Biosystems, 2006).
RNA isolation and quantitative real-time PCR
For comparison of promoter A and B transcript levels in mouse and human fibroblast cells, NIH3T3 cells and GM03349 cells were grown to near 80% confluency in 100-mm plates. RNA was isolated from the cells as described below. For analysis of transcript levels in TNF-alpha-treated cells, NIH3T3 cells were plated in 100-mm plates and either untreated or treated with TNF-alpha and various inhibitors as previously described. The total RNA was isolated from the cells using the SV Total RNA Isolation Kit (Promega). 1–3 μg of RNA was converted to cDNA using the QuantiTect Reverse Transcription kit (Qiagen, Valencia, CA) or the Maxima Reverse Transcriptase (Fermentas, Glen Burnie, Maryland). qRT-PCR was conducted using the Applied Biosystem TaqMan Assay (Applied Biosystems, Foster City, CA). For NIH3T3 cells, primers used for amplification included mouse cyclin D1 (Mm00432358) and custom-made primer–probes that amplified mouse Wnt5a transcripts derived specifically from mpromoter A and mpromoter B (Table 1). For the human GM03349 cells, human cyclin D1 (Hs00765553) and custom-human Wnt5a primer–probes that amplified transcripts derived from hpromoter A and hpromoter B were used (Table 1). Each amplification set included the internal control primer for GAPDH (mouse-Mm03302249_g1; human- Hs99999905). qRT-PCR was conducted on the Applied Biosystems StepOne Real-Time PCR System thermal cycling block. Amplification conditions were 95°C for 15 s and 60°C for 1 min for 40 cycles. Amplifications were in triplicate for each sample and primer set. The CT (ΔΔCT) value was determined for each cDNA sample and primer set, in comparison to the internal control, and relative change determined.
Results
Comparison of mouse and human Wnt5a genes
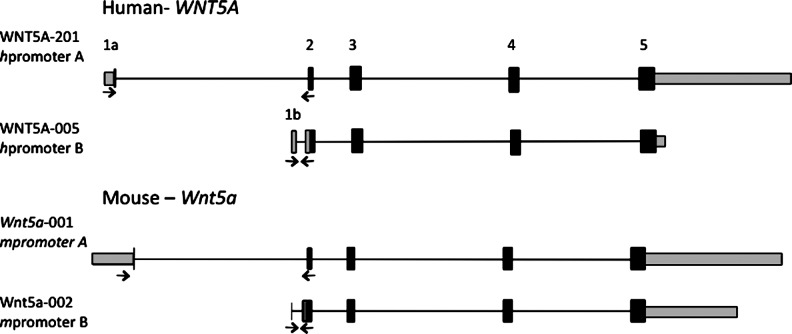
We compared the mouse and human Wnt5a gene regions using data provided by Ensembl (Flicek et al., 2012) (Table 2 and Fig. 1). Multiple transcripts are generated from the mouse and human Wnt5a genes. Of the 6 mouse transcripts, only two generate proteins and these transcripts were selected for analysis (Fig. 1). In contrast, 6 of the 8 human WNT5A transcripts produce proteins. These 6 transcription units have unique transcription start sites (Ensembl ENSG000001142510).
FIG. 1.
Genomic structure of selected human and mouse Wnt5a transcripts. These structures were derived from Ensembl: Mouse Wnt5a ENSMUSG00000021994 and Human WNT5A ENSG00000114251. Coding exon regions are shown as black boxes. Gray boxes are 5′ or 3′ untranslated regions. Lines are intron sequences. The sizes are proportional to sequence length. The exons are numbered for the human transcripts. The arrows indicate the relative position of custom primers for quantitative real time (qRT)-PCR used to detect the specific transcripts.
We selected for analysis the two human transcripts that share similarity to the mouse transcripts (Fig. 1). The different transcripts include distinct first exons; the longer human transcript (WNT5A-201) includes exon 1a, whereas the shorter transcript (WNT5A-005) includes exon 1b. These transcripts share exons 2, 3, 4, and a portion of 5. WNT5A-201 has a longer 3′ untranslated regions. The WNT5A-201 and -005 transcripts have different starts of translation. The alternative transcription start site for the shorter mouse transcript (002) is in a similar relative genomic location, in comparison to the human transcript (005). One distinction is that the mouse transcript (001) has a larger exon 1 in comparison to the human exon 1a. The proteins generated from the longer transcripts include an extra 20 AA's on the N-terminus for the mouse and 15 AA's for the human (Table 2). It is important to note that the longer human transcript WNT5A-201 is associated with the cDNA RefSeq NM_003392, which is the transcript most often studied.
In this study, we refer to gene regulatory sequences associated with the long and short Wnt5a transcripts as promoter A (long) and promoter B (short) and a qualifying h for the human gene and m for the mouse. The promoter A and B designation was adopted by Katoh and Katoh (2009) in their comparison of the Wnt5a genomic regions of human, chimpanzee, mouse, and rat.
Transient transfection of WNT5A hpromoter A and B constructs
To examine the activity and regulation of the separated human WNT5A alternative hpromoters A and B, different amounts of upstream regions for each promoter were cloned into a luciferase reporter vector (Fig. 2). The genomic sequences included in the longest constructs (p2178 and p1981) are shown. Putative binding sites for a select group of transcription factors are positioned on the sequence and mapped on the constructs. These may or may not represent functional sites. The hpromoter B construct p1981 includes 2337 bp of the hpromoter A 6061 bp intron 1.
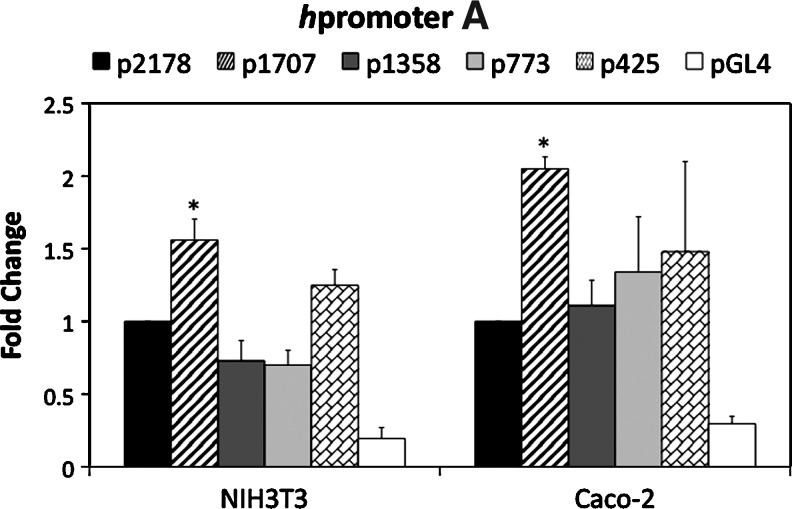
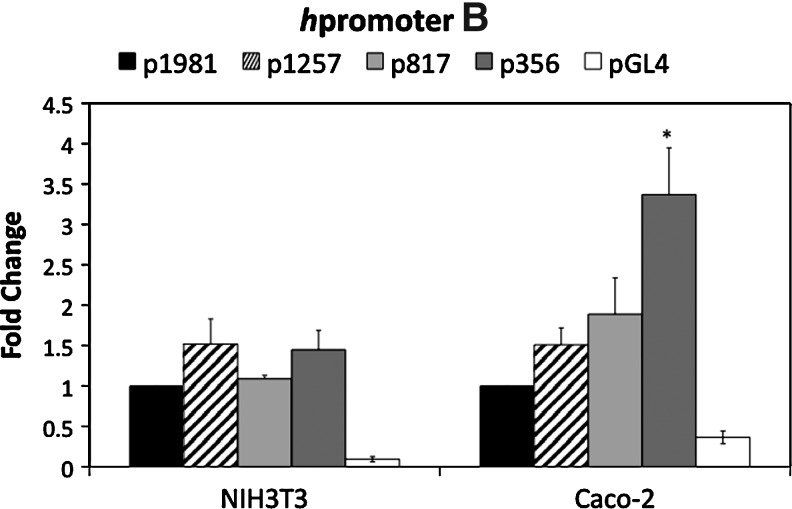
The constructs in Figure 2 were transiently transfected into NIH3T3 and Caco-2 cells, along with a Renilla luciferase control vector. After 48 h, the cells were assayed for both firefly and Renilla luciferase activity. The fold-change relative to the longest construct (p2178 for hpromoter A and p1981 for hpromoter B) was graphed (Fig. 3). hPromoter A and B constructs are active in both cell types at relatively the same level, although the hpromoter B appears more active in Caco-2 cells. A low level of activity was not detected for any of the hpromoter A and B constructs, indicating that 425 bp (hpromoter A) and 356 bp (hpromoter B) of sequence are sufficient for transcriptional initiation and a relatively high level of activity.
FIG. 3.
Transient transfection of hpromoter A and hpromoter B constructs into NIH3T3 and Caco-2 cells. The constructs shown in Figure 2 and the empty vector pGL4-17 were transiently transfected into the cells, along with a Renilla control vector. The ratio of firefly to Renilla luciferase was determined for four replicas and fold-change determined relative to the longest constructs, p2178 and p1981. The mean fold-change for three or four independent transfections is shown. The bars are±S.E.M. The asterisks indicate significance (p<0.05) in comparison to the longest construct.
The hpromoter A constructs displayed a similar pattern of activity in both NIH3T3 and Caco-2 cells (Fig. 3). A significant increase in activity was measured comparing the p2178 and p1707 constructs. This indicates that a negative acting sequence is contained in the removed sequences. A putative NF-kappaB site is missing from the p1707 construct (Fig. 2). Activity dropped for the p1358 and p773 constructs, both in NIH3T3 and Caco-2 cells, suggesting removal of positive acting sequences. In NIH3T3 cells, an increase in activity was observed for the p425 construct, suggesting removal of a negative acting sequence.
The hpromoter B constructs displayed distinctive patterns of activity in NIH3T3 and Caco-2 cells (Fig. 3). In NIH3T3 cells, the expression levels from the four constructs were not significantly different. In the Caco-2 cells, there was increasing activity as more of the hpromoter B upstream sequences were removed with maximal activity (about a 3.5-fold increase) measured with the shortest construct, p356. These data indicate the presence of negative acting sequences that are functional in Caco-2 cells, but not NIH3T3.
Endogenous promoters A and B are active in human and mouse fibroblastic cells
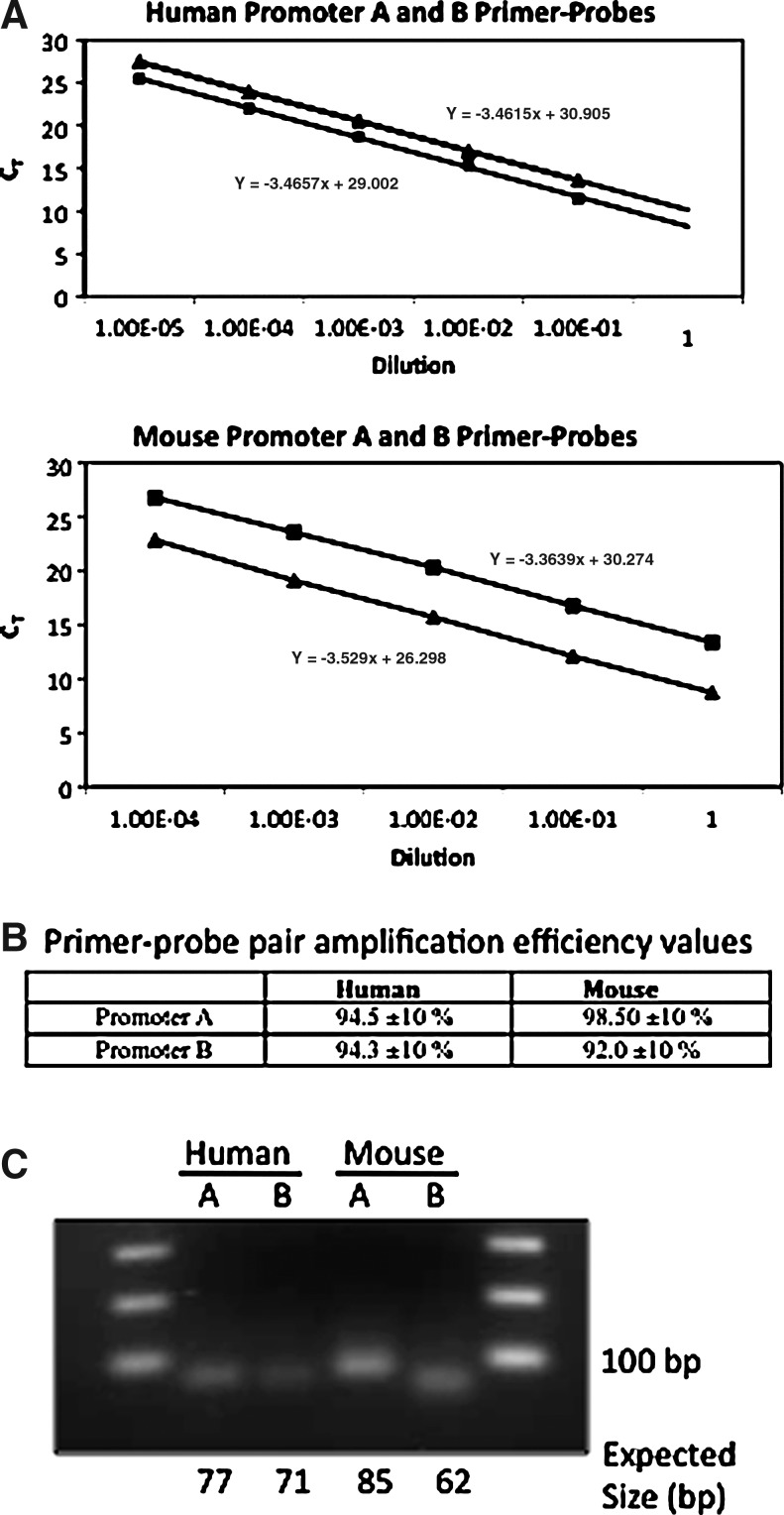
The transient transfection data show that both hpromoter A and hpromoter B are active when assayed separately. We designed TaqMan primer–probe sets specific to the human and mouse promoter A- and B-derived transcripts to determine the level of endogenous promoter-specific transcripts (Table 1). The amplified product flanks the exon 1 and exon 2 splice junction. The TaqMan forward primer and probes for the hpromoter A and hpromoter B transcripts are homologous to sequences in the unique exon 1a and 1b, respectively, and the reverse primer in exon 2 (Fig. 1). The mouse primer–probe sets are similar with the exception that for the mpromoter B transcript, the probe is located entirely in exon 2. Amplification with these primer–probe sets generated products of the expected length (Fig. 4C). The amplification efficiencies of the primer–probe sets were determined to be between 90% and 100% (Fig. 4A, B), as recommended by Applied Biosystems (2006).
FIG. 4.
Verification and amplification efficiencies of Wnt5a promoter A and B TaqMan primer–probe sets. (A) Purified human and mouse promoter A- and B-specific PCR products were diluted as indicated and used for TaqMan amplification with each primer–probe set. CT (cycle number) values were plotted. Squares are promoter A primer–probes; triangles are promoter B primer–probes. The slopes of the lines were determined and used to calculate the amplification efficiencies shown in (B). (C) DNA agarose gel of mouse and human promoter A and promoter B PCR products, showing the expected product sizes.
The level of endogenous promoter A- and B-specific transcripts were determined by qRT-PCR using our verified primer–probe sets in mouse (NIH3T3) and human (GM03349) fibroblast cells. The threshold cycle numbers for the promoter A- and promoter B-specific amplifications were compared. Cycle numbers for NIH3T3 were 26.34, 28.31, and 29.27 for total Wnt5a, mpromoter A, and mpromoter B transcripts, respectively. Cycle numbers for GM03349 cells were 28.59, 28.90, and 30.90 for the same target transcripts. The cycle numbers for human and mouse promoter B were less by 2 and 2.7, respectively, than those for promoter A, indicating 4 and 6.5 times the number of transcripts derived from promoter A. These results indicate that both mouse and human promoter A and B are active in fibroblast cells, but there are a greater number of promoter A-specific transcripts than B.
Involvement of TNF signaling pathway in the regulated activity of the Wnt5a alternative promoters A and B
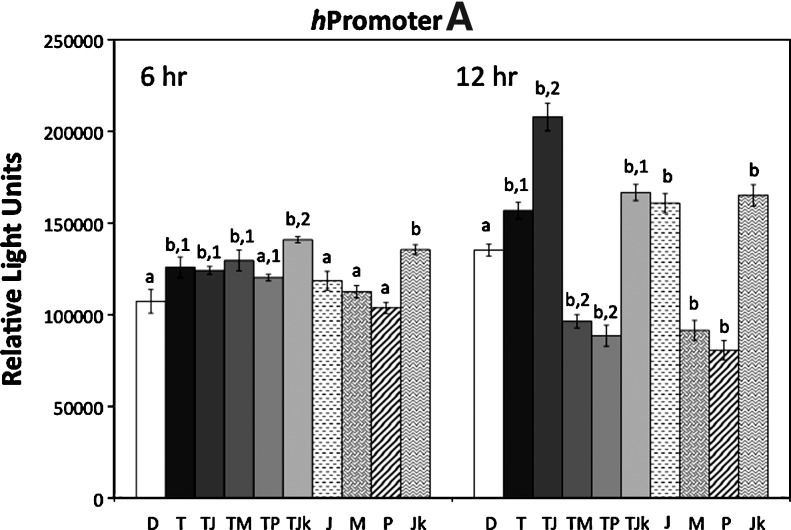
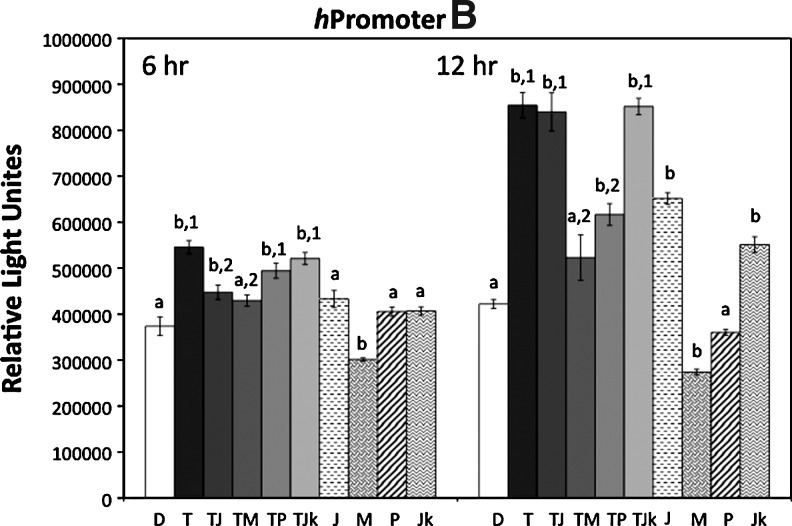
There is evidence indicating that the TNF-alpha signaling pathway affects Wnt5a transcription. However, these studies are limited and it is not clear which of the TNF-alpha effector proteins (NFkappa-B, p38, ERK, or JNK) are involved and if Wnt5a alternative promoters A and B are differentially affected. Using our stable lines of NIH3T3 containing the WNT5A hpromoter A p2178 and hpromoter B p1981 luciferase reporter constructs, the effect of TNF-alpha on promoter activity was analyzed. The cells were treated with 5 ng/mL TNFalpha and inhibitors of NF-kappaB, p38, MEK1/2 (hence ERK), and JNK for 6 and 12 h (Fig. 5). Luciferase activity increased in TNF-alpha-treated cells at 6 h for both hpromoter B and hpromoter A, but less for hpromoter A. At 12 h, there was a significant increase in activity of both hpromoters A and B, as measured by luciferase activity. The hpromoter B showed a greater increase of ∼1.5- to 2.25-fold at 12 h, compared to 1.2- to 1.5-fold for the hpromoter A. These results indicate that TNF-alpha increases activity of both hpromoters A and B, but the effect is greater for the hpromoter B.
FIG. 5.
Increase in hpromoter A and hpromoter B activity in response to TNF-alpha. NIH3T3 cells stably transfected with hpromoter A p2178 and hpromoter B p1981 reporter constructs were treated for 6 and 12 h with 5 ng/mL TNF-alpha only (T) and the following inhibitors with (solid bars) and without (patterned bars) TNF-alpha: NF-kappaB inhibitor (JSH-23, 30 μM) (TJ and J), MEK1/2 inhibitor (U0126, 10 μM) (TM and M), p38 inhibitor (SB203850, 10 μM) (TP and P), Jun N-terminal kinase inhibitor (SP600125, 20 μM) (TJk and Jk). Control cells were treated with DMSO (D) (open bar). Cells were collected and assayed for luciferase activity, expressed as relative light units. Bars are±S.E.M. N=4. For statistical analyses, letters are pair-wise comparisons of each value to the DMSO control. Numbers are statistical pair-wise comparisons between TNF-alpha plus inhibitor and the TNF-alpha only values. A different letter or number indicates significance (p<0.05).
The effects of the various inhibitors are shown in Figure 5. At 6 h, inhibitors of NF-kappaB, MEK1/2, and p38 had no affect on the slight increase in hpromoter A activity due to TNF-alpha treatment. A small, but significant increase in luciferase above the DMSO control and TNF-alpha-treated cells was detected in the TNF-alpha plus JNK inhibitor-treated cells. This is likely due to an effect of JNK independent of TNF-alpha as the JNK inhibitor, alone, increased activity. At 12 h, hpromoter A activity was increased by TNF-alpha; however, the NF-kappaB inhibitor increased activity in comparison to both the control- and TNF-alpha-treated cells. The MEK1/2 and p38 inhibitors reduced activity to below DMSO control levels, with and without TNF-alpha. Again, the JNK inhibitor increased activity alone, but had no effect on TNF-alpha-treated cells. These results indicate that for the hpromoter A, the TNF-alpha-induced increase in activity at 12 h involves MEK1/2 and p38, but that these kinases also contribute to the level of constitutive hpromoter A activity in untreated cells. The increase in reporter activity in cells treated with TNF-alpha plus NF-kappaB inhibitor and NF-kappaB inhibitor only, suggest that NF-kappaB has a negative effect on promoter A activity, at least at 12 h.
At 6 h for the hpromoter B, both the NF-kappaB and MEK1/2 inhibitors decreased activity, relative to the TNF-alpha-treated cells. P38 and JNK inhibitors had no effect on the TNF-alpha increase in activity. NF-kappaB, p38, and JNK inhibitors, alone, did not alter activity, whereas the MEK1/2 inhibitor slightly, but significantly decreased activity relative to the DMSO control. At 12 h, only the MEK1/2 and p38 inhibitors significantly reduced activity of the hpromoter B relative to the TNF-alpha-treated cells, whereas the NF-kappaB and JNK inhibitors had no effect. The NF-kappaB and JNK inhibitors, alone, increased activity, whereas the MEK1/2 inhibitor decreased activity. The p38 inhibitor, alone, slightly decreased activity, but not significantly. These results suggest that NF-kappaB is involved in the early response (6 h) of hpromoter B to TNF-alpha, but at 12 h, p38 is more important. MEK1/2 kinase likely has a role in both the TNF-alpha increase in hpromoter B activity and the constitutive level of hpromoter B activity.
Endogenous Wnt5a promoters affected by TNF-alpha
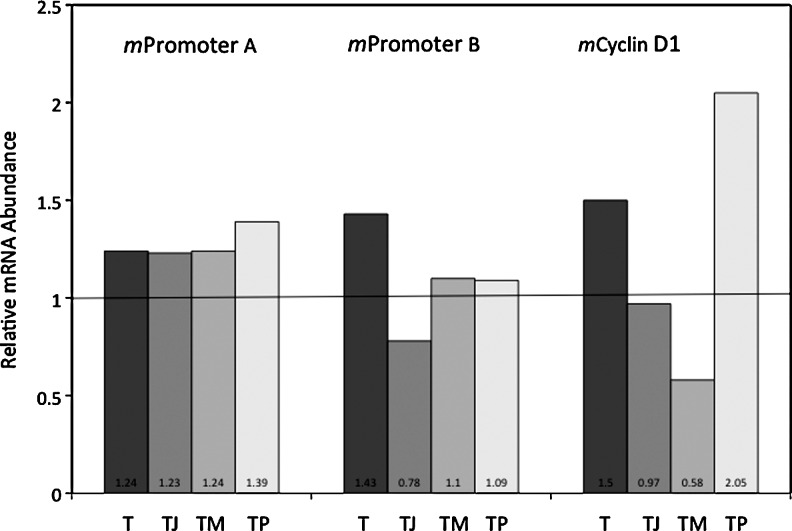
To determine if the genomic promoters A and B are also responding to TNF-alpha stimulation, NIH3T3 cells were treated with TNF-alpha for 6 h and RNA isolated. qRT-PCR analysis was conducted using mpromoter A- and B-specific TaqMan primer–probes and commercial cyclin D1 primer–probes (Fig. 6). Cyclin D1 transcription is known to increase in TNF-alpha-stimulated cells. A relative increase in all three target sequences was detected at 6 h. mPromoter A transcripts increased by 1.24×, whereas mpromoter B increased by1.43× and cyclin D1 transcript levels by 1.5×. Treatment with the NF-kappaB inhibitor, JSH-23, had no effect on mpromoter A transcript levels in TNF-alpha-treated cells, but caused a decrease in mpromoter B and cyclin D1 transcripts. These results suggest that NF-kappaB has a role in TNF-alpha stimulation of mpromoter B and cyclin D1 activity at 6 h in mouse cells. The data in Figure 6 also confirms that at 6 h MEK1/2 has a positive effect on mpromoter B and cyclin D1 transcription. A p38 inhibitor reduced mpromoter B transcript levels, but increased levels for cyclin D1 transcripts. These results suggest that MEK1/2 and p38 have a role in the level of mpromoter B activity.
FIG. 6.
qRT-PCR analysis of endogenous mpromoter A, mpromoter B, and mcyclin D1-specific transcripts in TNF-alpha-treated cells. NIH3T3 cells were treated for 6 h with 5 ng/mL TNF-alpha (T), with and without a NF-kappaB inhibitor (JSH-23, 30 μM) (TJ), MEK1/2 inhibitor (U0126, 10 μM) (TM), and p38 inhibitor (SB203850, 10 μM) (TP). Control cells were treated with DMSO (D). The relative level of mpromoter A, mpromoter B, and mcyclin D1 transcripts were determined in comparison to the DMSO-treated cells, using GAPDH as the internal control.
Discussion
The human WNT5A upstream sequences from the alternative transcription start sites termed hpromoter A and hpromoter B were individually cloned and analyzed to determine relative levels of transcriptional activity and to map potential regulatory sequences by deletion analysis. Our results suggest that hpromoters A and B have similar transcriptional potential when assayed separately, but that the hpromoter A is more active in human and mouse fibroblast cells. Deletion analysis of the hpromoter A and B upstream sequences indicate that both positive and negative acting regulatory sequences are located within the ∼2000 bp of upstream sequence of both hpromoter A and B and that 356 to 425 bp of upstream sequence is sufficient for a high level of expression. The relative expression levels varied only two- to fourfold for the different constructs, indicating that any removed regulatory sequences have only a minor effect on WNT5A activity at least in NIH3T3 and Caco-2 cells. These data support the conclusion that WNT5A is constitutively expressed in these cell types and that its regulation involves subtle modulations in transcriptional activity. Alternatively, it is possible that WNT5A regulation is primarily centered on the most proximal promoter region. This may involve hpromoter B intron sequences, which are included on the reporter construct. hPromoter A intron 1 sequences are not required for activity of the promoter as they were not included in the constructs, although possible regulatory sequences may be located within these sequences.
The patterns of expression for the different constructs were similar for the hpromoter A in both a fibroblastic (NIH3T3) and epithelial cell (Caco2). In contrast, the hpromoter B displayed a distinctive pattern of expression in Caco-2 in comparison to NIH3T3. We found that in Caco-2 cells removal of hpromoter B upstream sequences lead to a continual increase in expression. In NIH3T3, the levels of expression were similar for the different constructs. This finding suggests that negative regulatory sequences are being removed between the 1981- and 356-bp region of hpromoter B that are functional in Caco-2 cells, but not in NIH3T3 cells. The different expression patterns of the hpromoter B constructs in Caco-2 are likely a consequence of cell-specific regulatory factors. In general, this finding suggests that hpromoters A and B are differentially regulated.
Figure 2 displays potential transcription factor binding sites. These sites were selected for analysis based on their previous involvement in WNT5A expression and link to TNF-alpha and MAPK signaling. hpromoters A and B include multiple binding sites within the sequences contained in the shortest construct (p420 and p356). Common to both are c-Myb and CUTL1 sites. Wnt5a was found to be a target of CUTL1 and overexpression of CUTL1 increased total WNT5A transcripts and protein levels (Ripka et al., 2007). It is not known if both hpromoters A and B are affected by CUTL1 and which of the CUTL1 sites are functional. The hpromoter B harbors overlapping sites for Pax-2, AP-1, and c-Fos within its unique first intron. There is support for the involvement of Pax-2 in WNT5A transcription; however, the identified binding site was located 100-kb upstream of the promoter A (Tamimi et al., 2008). Pax-2 is unlikely to be an essential transcription factor for the hpromoter A, as a consensus Pax-2 binding site is lacking from the p420 construct. STAT3 has been implicated, but not proven to be involved in WNT5A regulation and a putative STAT3 site was identified in exon 4, common to both promoter A and B transcripts (Katoh and Katoh, 2007). No STAT3 sites were identified in the promoter A and B sequences we analyzed. The involvement of these and other transcription factors in promoter A and B activities will require overexpressing and RNAi knockdown of the transcription factors, and then quantifying the levels of promoter A- and B-derived transcripts using our costume-designed TaqMan primer–probes. One difficulty is the possibility that the activity levels of promoters A and B are dependent on the combinational inputs of a diverse group of transcription factors and that the contribution of anyone factor is limited.
Collectively, our results using stable lines of NIH3T3 cells containing the WNT5A hpromoter A and B luciferase reporter constructs and qRT-PCT analysis of endogenous mpromoter A and B transcripts support the conclusion that activation of the TNF-alpha signaling pathway increases both promoter A and B activity, but that promoter B is more strongly affected. We used inhibitors to block the activity of downstream effectors of TNF signaling, including the transcription factor NF-kappaB, and the kinases MEK 1/2, p38, and JNK. The results indicate that at 6 h, NF-kappaB is responsible for some of the increased promoter B activity, but not for promoter A. At 12 h, the NF-kappaB inhibitor (without TNF-alpha treatment) increased hpromoters A and B activities. This finding suggests that NF-kappaB is active in NIH3T3 cells not treated with any inducer of NF-kappaB and that it is having a negative effect on hpromoters A and B. Potential NF-kappaB binding sites were identified in the human promoter A and B sequences contained in the p2178 and p1981 reporter vectors (Fig. 2). The NF-kappaB site in promoter B is the same as the one identified by Katoh and Katoh (2009). In promoter A, the NF-kappaB site nearest the beginning of the transcription start is the same as the site identified by Ge et al. (2011). In that study, an increase in WNT5A mRNA and protein levels was detected in primary condylar chondrocyte cells treated with interleukin-1β (an inducer of TNF signaling pathway), which was reduced by an inhibitor of NF-kappaB activation. Moreover, using ChIP analysis, Ge et al., provided evidence that NF-kappaB binding is enriched at this sequence. In another study, WNT5A transcript levels increased in bone marrow stromal cells treated with TNF-alpha for 48 h and this induction was decreased by treatment with an inhibitor of NF-kappaB (Rauner et al., 2011). In both studies, however, total WNT5A transcripts were measured, thus it is not possible to know if the detected increase was due to hpromoter A- or B-derived transcripts.
We also found that the kinases p38 and ERK contribute to the activity levels of promoters A and B, particularly at 12 h. For promoter A, activity levels were reduced by the p38 and MEK1/2 kinase inhibitors to the same level as in cells treated with and without TNF-alpha. For promoter B, the kinase inhibitors reduced promoter activity in both TNF-alpha-treated and untreated cells, but activity was greater in the TNF-alpha-treated cells. Hence, for both promoters A and B, the p38 and MEK1/2 kinases appear to be involved in the constitutive activity of the promoters and likely have some role in TNF-alpha stimulation of activity. This appears to be the first report of p38 involvement in WNT5A transcriptional regulation. WNT5A stimulation of the noncanonical Ca2+ pathway has been shown to result in p38 kinase activation (Ma and Wang, 2007). This and our finding suggest a positive feedback loop with p38 kinase leading to increased WNT5A expression, and then WNT5A receptor binding leading to further p38 kinase activation.
There are only a few studies linking MAPK signaling pathway to WNT5A transcriptional regulation. In one, SW480 cells (human colon cancer cell line) were subjected to amino acid deprivation to induce the MAPK pathway (Wang and Chen, 2009). An increase in ERK1/2 phosphorylation in these cells was associated with a decrease in WNT5A mRNA levels. These data are contrary to our findings as we observed a decrease in Wnt5a transcription (of both promoter A and B transcripts) when cells, with and without TNF-alpha, are treated with the MEK1/2 inhibitor. Consistent with our findings, Rauner et al. (2011), measured an increase in Wnt5a transcript levels due to stimulation with TNF-alpha, which was reduced by treatment with a MAPK inhibitor. In neither of these studies were the individual promoter A- and B-derived transcripts analyzed.
Our finding that the Wnt5a alternative promoters A and B are differentially regulated is not unexpected. Alternative promoter usage giving rise to multiple transcripts appears to be a common feature of mammalian genes; it is estimated that 50% of mammalian genes have alternative promoters (Kimura et al., 2006; Baek et al., 2007). The idea that alternative promoters provide for more complex differential gene regulation is supported by numerous studies. For example, the two promoters of the gene coding for the transcription factor, Runx1, are differentially utilized during hematopoiesis, allowing for fine control of overall Runx1 levels at the appropriate developmental stage (Bee et al., 2010). Three alternative promoters have been identified in the gene coding for acetyl-CoA carboxylase β, which show unique tissue-specific activity in both human and rat (Oh et al., 2005). And, the human microsomal epoxide hydrolase gene (EPHX1) has an alternative promoter that is utilized only for liver-specific expression of the gene (Liang et al., 2005). Alternative promoter usage has also been associated with human disease. In one study, the five alternative promoters of the aromatase (CYP19) gene were found to have distinct patterns of activity in nonsmall cell lung cancer cells, in comparison to the surrounding normal tissue (Demura et al., 2011). And, a genome-wide analysis of genes associated with disease susceptibility showed a positive association with the presence of alternative promoters (Liu, 2010). The misregulation of WNT5A in numerous cancers involves both increases and decreases in transcript levels, depending on the cancer type. It is likely that this altered expression involves differential utilization of the WNT5A alternative promoters, including promoters A and B. Distinct promoter A and B expression patterns are likely to occur in normal cells as well. In both cases, the expected outcome would be a change in the ratio of WNT5A protein isoforms A and B. We are currently using our promoter A and B TaqMan primer–probes to analyze the levels of promoter-specific transcripts in different cancer cells and analyzing the functional distinctions, if any, between protein isoforms A and B.
Conclusion
We provide evidence for differential regulation of two of the Wnt5a alternative transcription start sites in different cell types and as a consequence of TNF signaling pathway activation. The promoter B was found to be more sensitive to TNF-alpha. The transcription factor NF-kappaB and the MEK1/2 and p38 kinases have distinct roles in determining the activity levels of promoters A and B. These findings indicate a mechanism of more complex and subtle regulation of Wnt5a expression involving its alternative promoters. Further studies are necessary to determine if and to what degree the amounts of promoter-specific transcripts vary during development, in different cell types, and during cellular transformation.
Acknowledgments
This work was funded by an NIH R15 grant (DK070581-01A1) to KSK. We thank the various undergraduates in the laboratory who assisted with some of this research.
Disclosure Statement
No competing financial interests exist.
References
- Applied Biosystems. Application Note: TaqMan Gene Expression Assay. Publication 127APO5-03; Foster City, CA: 2006. [Google Scholar]
- Baek D. Davis C. Ewing B. Gordon D. Green P. Characterization and predictive discovery of evolutionarily conserved mammalian alternative promoters. Genome Res. 2007;17:125–155. doi: 10.1101/gr.5872707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banday A.R. Azim S. Tabish M. Alternative promoter usage and differential expression of multiple transcripts of mouse Prkar1a gene. Mol Cell Biochem. 2011;357:263–274. doi: 10.1007/s11010-011-0897-z. [DOI] [PubMed] [Google Scholar]
- Basseres D.S. Baldwin A.S. Nuclear factor-kappaB and inhibitor of kappaB kinase pathways in oncogenic initiation and progression. Oncogene. 2006;25:6817–6830. doi: 10.1038/sj.onc.1209942. [DOI] [PubMed] [Google Scholar]
- Bee T. Swiers G. Muroi S. Nottingham W. Santos A.C. Li P.S., et al. Nonredundant roles for Runx1 alternative promoters reflect their activity at discrete stages of developmental hematopoiesis. Blood. 2010;115:3042–3050. doi: 10.1182/blood-2009-08-238626. [DOI] [PubMed] [Google Scholar]
- Bilkovski R. Schulte D.M. Oberhauser F. Gomolka M. Udelhoven M. Hettich M.M., et al. Role of WNT-5a in the determination of human mesenchymal stem cells into preadipocytes. J Biol Chem. 2010;285:6170–6178. doi: 10.1074/jbc.M109.054338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blakely B.D. Bye C.R. Fernando C.V. Horne M.K. Macheda M.L. Stacker S.A., et al. Wnt5a regulates midbrain dopaminergic axon growth and guidance. PLoS One. 2011;6:e18373. doi: 10.1371/journal.pone.0018373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demura M. Demura Y. Ameshima S. Ishizaki T. Sasaki M. Miyamori I., et al. Changes in aromatase (CYP19) gene promoter usage in non-small cell lung cancer. Lung Cancer. 2011;73:289–293. doi: 10.1016/j.lungcan.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng G. Li Z.Q. Zhao C. Yuan Y. Niu C.C. Pan J., et al. WNT5A expression is regulated by the status of its promoter methylation in leukaemia and can inhibit leukemic cell malignant proliferation. Oncol Rep. 2011;25:367–376. doi: 10.3892/or.2010.1108. [DOI] [PubMed] [Google Scholar]
- Dissanayake S.K. Wade M. Johnson C.E. O'Connell M.P. Leotlela P.D. French A.D., et al. The Wnt5A/protein kinase C pathway mediates motility in melanoma cells via the inhibition of metastasis suppressors and initiation of an epithelial to mesenchymal transition. J Biol Chem. 2007;282:17259–17271. doi: 10.1074/jbc.M700075200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enomoto M. Hayakawa S. Itsukushima S. Ren D.Y. Matsuo M. Tamada K., et al. Autonomous regulation of osteosarcoma cell invasiveness by Wnt5a/Ror2 signaling. Oncogene. 2009;28:3197–3208. doi: 10.1038/onc.2009.175. [DOI] [PubMed] [Google Scholar]
- Flicek P. Amode M.R. Barrell D. Beal K. Brent S. Carvalho-Silva D., et al. Ensembl 2012. Nucleic Acids Res. 2012;40:D84–D90. doi: 10.1093/nar/gkr991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge X.P. Gan Y.H. Zhang C.G. Zhou C.Y. Ma K.T. Meng J.H., et al. Requirement of the NF-kappaB pathway for induction of Wnt-5A by interleukin-1beta in condylar chondrocytes of the temporomandibular joint: functional crosstalk between the Wnt-5A and NF-kappaB signaling pathways. Osteoarthritis Cartilage. 2011;19:111–117. doi: 10.1016/j.joca.2010.10.016. [DOI] [PubMed] [Google Scholar]
- Hardy K.M. Garriock R.J. Yatskievych T.A. D'Agostino S.L. Antin P.B. Krieg P.A. Non-canonical Wnt signaling through Wnt5a/b and a novel Wnt11 gene, Wnt11b, regulates cell migration during avian gastrulation. Dev Biol. 2008;320:391–401. doi: 10.1016/j.ydbio.2008.05.546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibi K. Mizukami H. Goto T. Kitamura Y. Sakata M. Saito M., et al. WNT5A gene is aberrantly methylated from the early stages of colorectal cancers. Hepatogastroenterology. 2009;56:1007–1009. [PubMed] [Google Scholar]
- Huang C.L. Liu D. Nakano J. Ishikawa S. Kontani K. Yokomise H., et al. Wnt5a expression is associated with the tumor proliferation and the stromal vascular endothelial growth factor—an expression in non-small-cell lung cancer. J Clin Oncol. 2005;23:8765–8773. doi: 10.1200/JCO.2005.02.2871. [DOI] [PubMed] [Google Scholar]
- Iozzo R.V. Eichstetter I. Danielson K.G. Aberrant expression of the growth factor Wnt-5A in human malignancy. Cancer Res. 1995;55:3495–3499. [PubMed] [Google Scholar]
- Katoh M. Katoh M. STAT3-induced WNT5A signaling loop in embryonic stem cells, adult normal tissues, chronic persistent inflammation, rheumatoid arthritis and cancer. Int J Mol Med. 2007;19:273–278. [PubMed] [Google Scholar]
- Katoh M. Katoh M. Transcriptional mechanisms of WNT5A based on NF-kappaB, Hedgehog, TGFbeta, and Notch signaling cascades. Int J Mol Med. 2009;23:763–769. doi: 10.3892/ijmm_00000190. [DOI] [PubMed] [Google Scholar]
- Kikuchi A. Yamamoto H. Sato A. Matsumoto S. Wnt5a: its signaling, functions and implication in diseases. Acta Physiol (Oxf) 2011;204:17–33. doi: 10.1111/j.1748-1716.2011.02294.x. [DOI] [PubMed] [Google Scholar]
- Kimura K. Wakamatsu A. Suzuki Y. Ota T. Nishikawa T., et al. Diversification of transcriptional modulation: Large-scale identification and characterization of putative alternative promoters of human genes. Genome Res. 2006;16:55–65. doi: 10.1101/gr.4039406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurayoshi M. Oue N. Yamamoto H. Kishida M. Inoue A. Asahara T., et al. Expression of Wnt-5a is correlated with aggressiveness of gastric cancer by stimulating cell migration and invasion. Cancer Res. 2006;66:10439–10448. doi: 10.1158/0008-5472.CAN-06-2359. [DOI] [PubMed] [Google Scholar]
- Li J. Ying J. Fan Y. Wu L. Ying Y. Chan A.T., et al. WNT5A antagonizes WNT/beta-catenin signaling and is frequently silenced by promoter CpG methylation in esophageal squamous cell carcinoma. Cancer Biol Ther. 2010;10:617–624. doi: 10.4161/cbt.10.6.12609. [DOI] [PubMed] [Google Scholar]
- Liang H. Chen Q. Coles A.H. Anderson S.J. Pihan G. Bradley A., et al. Wnt5a inhibits B cell proliferation and functions as a tumor suppressor in hematopoietic tissue. Cancer Cell. 2003;4:349–360. doi: 10.1016/s1535-6108(03)00268-x. [DOI] [PubMed] [Google Scholar]
- Liang S.H. Hassett C. Omiecinski C.J. Alternative promoters determine tissue-specific expression profiles of the human microsomal epoxide hydrolase gene (EPHX1) Mol Pharm. 2005;67:220–230. doi: 10.1124/mol.104.005579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S. Increasing alternative promoter repertories is positively associated with differential expression and disease susceptibility. PLoS One. 2010;5:e9482. doi: 10.1371/journal.pone.0009482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L. Wang H.Y. Mitogen-activated protein kinase p38 regulates the Wnt/cyclic GMP/Ca2+non-canonical pathway. J Biol Chem. 2007;282:28980–28990. doi: 10.1074/jbc.M702840200. [DOI] [PubMed] [Google Scholar]
- Martin V. Valencia A. Agirre X. Cervera J. San Jose-Eneriz E. Vilas-Zornoza A., et al. Epigenetic regulation of the non-canonical Wnt pathway in acute myeloid leukemia. Cancer Sci. 2009;101:425–432. doi: 10.1111/j.1349-7006.2009.01413.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCall K.D. Harii N. Lewis C.J. Malgor R. Kim W.B. Saji M., et al. High basal levels of functional toll-like receptor 3 (TLR3) and noncanonical Wnt5a are expressed in papillary thyroid cancer and are coordinately decreased by phenylmethimazole together with cell proliferation and migration. Endocrinology. 2007;148:4226–4237. doi: 10.1210/en.2007-0459. [DOI] [PubMed] [Google Scholar]
- McDonald S.L. Silver A. The opposing roles of Wnt-5a in cancer. Br J Cancer. 2009;101:209–214. doi: 10.1038/sj.bjc.6605174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michl P. Ramjaun A. Pardo O. Warne P. Wagner M. Poulsom R. D'Arrigo C. Ryder K. Menke A. Gress T. Downward J. CUTL1 is a Target of TGF(beta) signaling that enhances cancer cell motility and invasiveness. Cancer Cell. 2005;7:521–532. doi: 10.1016/j.ccr.2005.05.018. [DOI] [PubMed] [Google Scholar]
- Mikels A.J. Nusse R. Purified Wnt5a protein activates or inhibits beta-catenin-TCF signaling depending on receptor context. PLoS Biol. 2006;4:e115. doi: 10.1371/journal.pbio.0040115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano T. Tani M. Ishibashi Y. Kimura K. Park Y.B. Imaizumi N., et al. Biological properties and gene expression associated with metastatic potential of human osteosarcoma. Clin Exp Metastasis. 2003;20:665–674. doi: 10.1023/a:1027355610603. [DOI] [PubMed] [Google Scholar]
- Oh S.Y. Lee M.Y. Kim J.M. Yoon S. Shin S. Park Y.N., et al. Alternative usages of multiple promoters of the acetyl-CoA carboxylase b gene are related to differential transcriptional regulation in human and rodent tissues. J Biol Chem. 2005;280:5909–5916. doi: 10.1074/jbc.M409037200. [DOI] [PubMed] [Google Scholar]
- Pukrop T. Klemm F. Hagemann T. Gradl D. Schulz M. Siemes S., et al. Wnt 5a signaling is critical for macrophage-induced invasion of breast cancer cell lines. Proc Natl Acad Sci U S A. 2006;103:5454–5459. doi: 10.1073/pnas.0509703103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauner M. Stein N. Winzer M. Goettsch C. Zwerina J. Schett G., et al. WNT5A is induced by inflammatory mediators in bone marrow stromal cells and regulates cytokine and chemokine production. J Bone Miner Res. 2012;27:575–585. doi: 10.1002/jbmr.1488. [DOI] [PubMed] [Google Scholar]
- Ripka S. Konig A. Buchholz M. Wagner M. Sipos B. Kloppel G., et al. WNT5A—target of CUTL1 and potent modulator of tumor cell migration and invasion in pancreatic cancer. Carcinogenesis. 2007;28:1178–1187. doi: 10.1093/carcin/bgl255. [DOI] [PubMed] [Google Scholar]
- Roman-Gomez J. Jimenez-Velasco A. Cordeu L. Vilas-Zornoza A. San Jose-Eneriz E. Garate L., et al. WNT5A, a putative tumour suppressor of lymphoid malignancies, is inactivated by aberrant methylation in acute lymphoblastic leukaemia. Eur J Cancer. 2007;43:2736–2746. doi: 10.1016/j.ejca.2007.10.004. [DOI] [PubMed] [Google Scholar]
- Saitoh T. Katoh M. Expression and regulation of WNT5A and WNT5B in human cancer: up-regulation of WNT5A by TNFalpha in MKN45 cells and up-regulation of WNT5B by beta-estradiol in MCF-7 cells. Int J Mol Med. 2002;10:345–349. [PubMed] [Google Scholar]
- Saitoh T. Mine T. Katoh M. Frequent up-regulation of WNT5A mRNA in primary gastric cancer. Int J Mol Med. 2002;9:515–519. [PubMed] [Google Scholar]
- Schwartz A.L. Malgor R. Dickerson E. Weeraratna A.T. Slominski A. Wortsman J., et al. Phenylmethimazole decreases Toll-like receptor 3 and noncanonical Wnt5a expression in pancreatic cancer and melanoma together with tumor cell growth and migration. Clin Cancer Res. 2009;15:4114–4122. doi: 10.1158/1078-0432.CCR-09-0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taki M. Kamata N. Yokoyama K. Fujimoto R. Tsutsumi S. Nagayama M. Down-regulation of Wnt-4 and up-regulation of Wnt-5a expression by epithelial-mesenchymal transition in human squamous carcinoma cells. Cancer Sci. 2003;94:593–597. doi: 10.1111/j.1349-7006.2003.tb01488.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamimi Y. Ekuere U. Laughton N. Grundy P. WNT5A is regulated by PAX2 and may be involved in blastemal predominant Wilms tumorigenesis. Neoplasia. 2008;10:1470–1480. doi: 10.1593/neo.08442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q. Williamson M. Bott S. Brookman-Amissah N. Freeman A. Nariculam J., et al. Hypomethylation of WNT5A, CRIP1 and S100P in prostate cancer. Oncogene. 2007;26:6560–6565. doi: 10.1038/sj.onc.1210472. [DOI] [PubMed] [Google Scholar]
- Wang Z. Chen H. Amino acid limitation induces down-regulation of WNT5a at transcriptional level. Biochem Biophys Res Commun. 2009;378:789–794. doi: 10.1016/j.bbrc.2008.11.124. [DOI] [PubMed] [Google Scholar]
- Weeraratna A.T. Houben R. O'Connell M.P. Becker J.C. Lack of Wnt5A expression in Merkel cell carcinoma. Arch Dermatol. 2010;146:88–89. doi: 10.1001/archdermatol.2009.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeraratna A.T. Jiang Y. Hostetter G. Rosenblatt K. Duray P. Bittner M., et al. Wnt5a signaling directly affects cell motility and invasion of metastatic melanoma. Cancer Cell. 2002;1:279–288. doi: 10.1016/s1535-6108(02)00045-4. [DOI] [PubMed] [Google Scholar]
- Yamaguchi T.P. Bradley A. McMahon A.P. Jones S. A Wnt5a pathway underlies outgrowth of multiple structures in the vertebrate embryo. Development. 1999;126:1211–1223. doi: 10.1242/dev.126.6.1211. [DOI] [PubMed] [Google Scholar]
- Yang Y. Topol L. Lee H. Wu J. Wnt5a and Wnt5b exhibit distinct activities in coordinating chondrocyte proliferation and differentiation. Development. 2003;130:1003–1015. doi: 10.1242/dev.00324. [DOI] [PubMed] [Google Scholar]
- Ying J. Li H. Yu J. Ng K.M. Poon F.F. Wong S.C., et al. WNT5A exhibits tumor-suppressive activity through antagonizing the Wnt/beta-catenin signaling, and is frequently methylated in colorectal cancer. Clin Cancer Res. 2008;14:55–61. doi: 10.1158/1078-0432.CCR-07-1644. [DOI] [PubMed] [Google Scholar]