Abstract
Self-assembly of misfolded proteins into ordered fibrillar structures is a fundamental property of a wide range of proteins and peptides. This property is also linked with the development of various neurodegenerative diseases such as Alzheimer’s and Parkinson’s. Environmental conditions modulate the misfolding and aggregation processes. We used a peptide, CGNNQQNY, from yeast prion protein Sup35, as a model system to address effects of environmental conditions on aggregate formation. GNNQQNY peptide self-assembles in fibrils with structural features that are similar to amyloidogenic proteins. Atomic Force Microscopy (AFM) and Thioflavin T (ThT) fluorescence assay were employed to follow the aggregation process at various pHs and ionic strengths. We also used single molecule AFM force spectroscopy to probe interactions between the peptides under various conditions. The ThT fluorescence data showed that the peptide aggregates fast at pH values approaching the peptide isoelectric point (pI=5.3) and the kinetics is 10 times slower at acidic pH (pH 2.0) suggesting that electrostatic interactions contribute to the peptide self-assembly into aggregates. This hypothesis was tested by the experiments performed at low (11 mM) and high (150 mM) ionic strengths. Indeed, the aggregation lag time measured at pH 2 at low ionic strength (11 mM) is 195 hours, whereas the lag time decreases ~5 times when ionic strength is increased to 150 mM. At conditions close to the pI value, pH 5.6, the aggregation lag time is 12 ± 6 hours under low ionic strength, and there is minimal change to the lag time at 150 mM NaCl. Ionic strength also influences the morphology of aggregates visualized with AFM. In pH 2.0 and at high ionic strength, the aggregates are twofold taller than those formed at low ionic strength. In parallel, AFM force spectroscopy studies revealed minimal contribution of electrostatics on dissociation of transient peptide dimers.
Keywords: AFM, Force spectroscopy, protein aggregation, electrostatics, nanoimaging
1.Introduction
Misfolding and aggregation of proteins are fundamental phenomena of protein biophysics leading to the development of a number of devastating neurodegenerative diseases, including Alzheimer’s, Parkinson’s, and Huntington’s diseases, amyotrophic lateral sclerosis, frontal temporal dementia, and the human prion diseases [1]. The accumulation of abnormal protein aggregates detected as extracellular or intracellular deposits represents a common pathological signature for these diverse neurodegenerative disorders. Proteins from such deposits can spontaneously assemble into aggregates in test tubes, and this finding dramatically facilitated physical, chemical and structural analysis of protein aggregates. Studies during the past decade, with the use of such techniques as x-ray fibril diffraction [2], electron microscopy (EM) [3], and atomic force microscopy (AFM) [4–6], as well as spectroscopic methods [7, 8], showed that misfolded proteins are assembled into fibrils with a periodic structure, stabilized by β-sheet structural motifs. The breakthrough was made in the laboratory of D. Eisenberg, in which the crystallographic structure of short peptide (GNNQQNY peptide) from the yeast prion protein Sup35 was obtained [9]. These studies have identified several classes of so-called steric zippers. Despite the fundamental similarities in the structure with extended protein strands that are perpendicular to the axis of β-sheets, these classes vary in the basic steric zipper structure. Such variation mainly involves the orientation of peptides and β-sheets with respect to each other. Based on solid state NMR and x-ray crystallography, the underlying structure of prion amyloids was found to share common structural features. They contain similarly arranged parallel β-sheets that are packed in register for a variety of different prion proteins, such as Ure2p1-89 (the basis of the [URE3] prion) [10], Ure2p10-39 [11], Sup35p ([PSI+] prion) [12], Rnq1p ([PIN+] prion) [13]. Generally, β-strands of a protein molecule run perpendicular to the axis of the fibril and hydrogen bonding in such structures occurs along the length of the fibril.
The mechanism of peptide/protein self-assembly into well-ordered structures is one of the fundamental problems in soft condensed matter and biophysics. The ability to control the self-assembly process and understand specific properties of self-assembling systems may enable one to create novel materials with desired properties. Therefore, a deeper understanding of the effect of various interactions such as electrostatic, hydrophobic, conformational, etc. on the mechanism of self-assembly is required. These competing interactions may influence the form and topology of aggregating proteins/peptides, leading to various structures in self-assembling systems which might possess properties that are beneficial (novel materials) or detrimental (cause of neurodegeneration). This study explores the effect of electrostatics on aggregating properties of small amyloidogenic peptide, CGNNQQNY.
It has been previously demonstrated that the effects of pH and ionic strength have a profound impact on aggregation of amyloidogenic proteins [14–21]. Salt concentration/ionic strength has been found to increase the intermolecular interactions between proteins [16, 20]. Although acidity and ionic strength are the two main environmental conditions affecting aggregation, there are also more subtle effects on aggregation that can have a great impact on the aggregate morphology [22, 23]. Here, we look at the effects of electrostatic interactions at four different pHs and two different ionic strengths on aggregating properties and aggregate morphologies of CGNNQQNY peptide. This short amyloidogenic peptide, 7-GNNQQNY-14, is part of the prion-forming N-terminus of the Yeast Prion Protein, sup35. The use of single molecule force spectroscopy allowed us to characterize intermolecular interactions between two single peptides of different charged states. We studied the aggregation kinetics using Thioflavin T (ThT) fluorescence assay and analyzed the morphology of the formed aggregates with atomic force microscopy (AFM). We found that for the charged peptide (pH 2) ionic strength accelerates the aggregation kinetics, whereas at conditions close to the isoelectric point of the peptide, the aggregation kinetics is faster and does not depend on ionic strength as much. The sizes of the aggregates were also affected by pH and ionic strength of solution. The further away from the isoelectric point, the slower the aggregation kinetics, and smaller, more linear aggregates were formed. Increasing ionic strength, on the other hand, consistently accelerated aggregation kinetics, as well as increased size of all aggregates formed, whether they were linear or globular in nature.
2. Methods
2.1 Peptides and Linkers
The peptide CGNNQQNY (PepQ) was synthesized by BioSynthesis, Inc. (Lewisville, TX). A second batch of peptides was also synthesized using a 12-channel automated microwave peptide synthesizer (CEM Corporation, Matthews, NC). All synthesized peptides were purified by C18 reverse-phase HPLC, and their molecular weights were confirmed by MALDI-TOF mass spectrometry. The aminopropyl silatrane (APS) was used for mica surface functionalization as described in [24]. NHS-PEG-MAL (N-hydroxysuccinimide-polyethylene glycol-maleimide, MW=3,400 g/mol) was purchased from Laysan Bio, Inc. (Arab, AL).
The 1.67 mM stock solution of NHS-PEG-MAL was prepared in DMSO (Sigma-Aldrich Inc.) and stored at −20°C. The 10 mM tris(2-carboxyethyl)phosphine (TCEP) hydrochloride (Hampton Research Inc.) was prepared in water. Maleimide polyethylene glycol silatrane (MAS), with five ethylene glycol repeats, was synthesized as described in [25].
2.2. Aggregation studies using ThT fluorescence
The extent of peptide aggregation was followed by Thioflavin T (ThT) fluorescence. First, the lyophilized peptide powder was dissolved in either water or buffer in an initial volume of 100 μl. Then initial concentration was obtained by using NanoDrop (Thermo Scientific, Wilmington, DE), detecting the absorbance of the tyrosine residue at 274 nm, and using an extinction coefficient of 1405 L·mol−1·cm−1 for tyrosine. The peptide solution was diluted down to 0.5 mg/ml, using either water or a buffer as a solvent.
For the ThT fluorescence assay, 10 μL aliquots from the samples were withdrawn periodically and added to 590 μL of 5 μM ThT solution. Fluorescence intensity was measured on a Cary Eclipse spectrofluorimeter (Varian Inc. Palo Alto, CA) at 485 nm while exciting at wavelength 450 nm. Each reading is an average of 15 values determined by simple reads after subtracting out the fluorescence contribution from free ThT.
Initial readings of fluorescence were taken immediately after solution was prepared. Stirring the solution at a regular interval was done in order to accelerate the aggregation process. Magnetic stir bars were placed into the peptide solutions, and the samples were stirred for 20 minutes once every two hours.
2.3. AFM
Once ThT fluorescence levels had reached a plateau, samples of aggregated peptides were deposited on APS modified mica. 2 μl of sample was initially deposited on the mica and allowed to sit for 2 minutes, followed by the addition of 8 μl of distilled water, which was then allowed to sit for 2 minutes. The samples were then dried by 2 minutes of spin coating at 2000 rpm using a Model WS-400BZ-6NPP/LITE spin coater (Laurell Technologies Corporation, North Wales, PA).
Images were acquired in air using a MultiMode SPM NanoScope IV system (Bruker Nano, Santa Barbara, CA) operating in tapping mode. Silicon-etched tapping mode probes (OTESPA) with nominal spring constants of ~42 N/m and a resonant frequency of ~330 Hz were used. Image analysis was done using Femtoscan Online (Advanced Technologies Center, Moscow, Russia). The first step in analyzing the features found on the mica surface was to subtract the background that was less than 1 nm in height. The heights of all features found on the surface were determined by using “enum features” function found in the software used. This function was also used to sort out the elongation factor of the aggregates. For this, we used the form factor parameter (Rs/Rp) obtained from the “enum features” readout. The aggregates were sorted out by their form factor value, with a form factor of 0 to 0.5 interpreted as a fully elongated fibril, 0.5 to 0.8 as a protofibril, and anything above 0.8 was considered to be an oligomer. The mean value for the height was assigned to the maximum of Gaussian fitting for the histogram obtained for the data set not less than 100 molecules for the smallest data set.
2.4. Dynamic Force Spectroscopy
Freshly cleaved mica (Asheville-Schoonmaker Mica Co., Newport News, VA, USA) surfaces were treated with amino-propyl-silatrane (APS) for 30 min according to previously reported protocol [24]. The APS modified mica surfaces were treated with 167 μM NHS-PEG-MAL in DMSO for 3 h followed by rinsing with DMSO to remove non-bound NHS-PEG-MAL, rinsing with water, and drying with argon. Maleimide-functionalized AFM probes and mica were incubated for 1 h in 190 nM of peptide solution (HEPES buffer, pH 7.0). Prior to immobilization of peptide, its solution was treated with 0.25 mM TCEP hydrochloride for 10 min in pH 7.0 buffer to reduce any disulfide bonds. After washing with HEPES (pH 7.0, 10 mM HEPES, 50 mM NaCl) buffer, unreacted maleimide was quenched with 10 mM β-mercaptoethanol for 10 min followed by rinsing with HEPES pH 7.0 buffer.
Silicon nitride (Si3N4) AFM probe tips (MLCT – VEECO, Santa Barbara, CA, USA) were washed in ethanol by immersion for 30 min and then activated by UV treatment for 30 min. Activated probe tips were treated with 167 μM maleimide-silatrane for 3 h followed by rinsing with double distilled water. Alternatively, in a double tether setup, after UV treatment for 30 minutes, the tips were treated similarly to the surfaces after mica cleavage as described above. The modified probe tips and mica were stored in carbonate (NaH2CO3/Na2HCO3) buffer (pH 9.8) overnight.
Force–distance measurements were performed in various buffers: pH 2.0, pH 3.7, pH 5.6 (IS=11 mM) and pH 2.0 (IS=150 mM) at room temperature with the Molecular Force Probe 3D system (Asylum Research, Santa Barbara, CA) or Force Robot 300 (JPK Instruments, Dresden, Germany). The ramp size was 200 nm with various loading rates. An application force was kept at a low value (100 pN). Silicon nitride cantilevers with nominal values of spring constants in the range of 0.04–0.07 N/m were used. Spring constants for each cantilever were obtained using a thermal method with the MFP-3D instrument, regardless of which instrument was ultimately used. In the case of Force Robot, we confirm the spring constant measurement prior to force spectroscopy experiments. The approach velocity was kept at 500 nm/s while the retraction velocity was varied between 80 nm/s and 6,000 nm/s, and the corresponding values of apparent loading rates were between 102 and 105,000 pN/s.
We have established for our experimental system that WLC model consistently provided a better quality fit to the measured force-distance curves at the rupture point in a greater range of rupture forces (pulling rates) compared to Freely Joint Chain model (see figures s1 and s2 in supplementary material). The force curves were analyzed using wormlike chain (WLC) approximation and the Igor Pro 6.04 software package provided by the manufacturer. The unbinding events where interactions between peptides are ruptured upon tip retraction appear at a certain distance in the experimental force–distance curves defined by the length of the flexible PEG linker. The analysis of the unbinding events included fitting the part of the force-distance curve with Worm-Like Chain model describing behavior of polymer linkers under applied external force (see supplemental material for details). The fit provided contour length and persistence length of the linkers in our system. Contour length and persistence length were further used to estimate apparent loading rate as described in [26]. The most probable rupture force was obtained from the Gaussian fit [27] of the force distribution compiled in a statistical histogram. Data analysis in dynamic force spectroscopy was performed as described in [25, 26, 28, 29].
3. Results
3.1. Aggregation in water
Initially, aggregation of CGNNQQNY (PepQ) peptide was tested in distilled water, at conditions typically used for structural studies of aggregates formed by a similar peptide [30]. Specifically, we wanted to know in what extent N-terminal cysteine changes the aggregation kinetics. AFM images of aggregates at the plateau of ThT fluorescence intensity showed that this peptide readily forms fibrils in water (figure 1). Here, it is clearly seen that long and straight fibrils have formed. Similar fibrils were observed for PepQ peptide [30, 31], suggesting that the addition of cysteine did not change the aggregation kinetics of the peptide. The aggregation kinetic curve for the peptide (figure s3) monitored with ThT fluorescence has a typical sigmoidal shape consistent with nucleation-polymerization model of fibril formation [32]. A lag phase with no increase in ThT fluorescence is followed by an exponential growth phase wherein ThT fluorescence increases until a plateau is reached, indicating the end of aggregate formation. The fluorescence curve was fitted using the model described in [32] and lag time of aggregation was found to be ~2.8 hours.
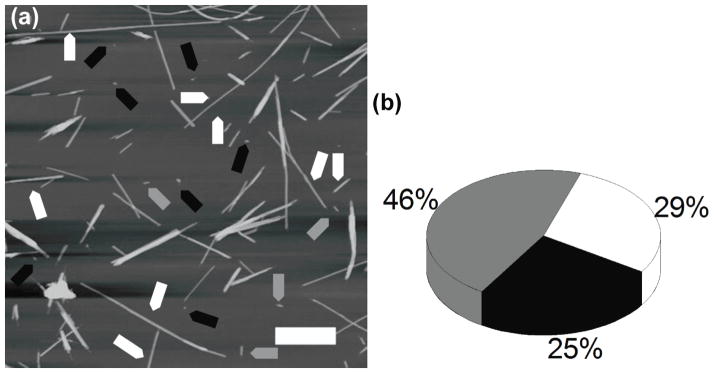
Figure 1.
Fibrils grown in water. (a) AFM images of fibrils grown in water; full length fibrils, short fibrils and globular aggregates are indicated with white, gray and black arrows respectively. (b) Yield of aggregates formed with oligomers (black), protofibrils (gray), and full length fibrils (white).
The morphology of the aggregates type formed by PepQ peptide in water was analyzed by AFM (Fig. 1(a)). The aggregate morphologies were classified in three major groups according to their form factor value (see Methods section). They are (figure 1(a)) (i) fully elongated fibrils (white arrows) with a form factor of 0 to 0.5; (ii) short fibrils (gray arrows) with a form factor 0.5 to 0.8, and (iii) oligomers (black arrows) with form factor above 0.8. The yield of the aggregate morphology (Fig. 1(b)) shows that, generally, these fibrils appear to be quite uniform in morphology, with an even divide between globular oligomers and long, mature fibrils (29% and 25%, respectively).
3.2. Effect of pH on aggregation kinetics
The effect of pH on aggregation of PepQ peptide was investigated at four different pH values: 2.0, 3.7, 5.6 and 7.0. The pI value for the peptide is 5.3. The kinetic curves of the peptide aggregation in these four pH buffers were obtained by ThT fluorescence and are shown in figure s4. Fluorescence curves exhibit distinct growth patterns at different pH values. The fastest kinetics was observed at pH 5.6, represented with blue diamonds, with a lag time of 12± 6 hours, while an increase or decrease in pH resulted in larger lag times. The longest lag time occurred with peptide incubated in pH 2.0, black squares, with a lag time of 196 ± 24 hours. The other two pHs, pH 3.7 (red circles) and 7.0 (green triangles), had intermediate lag times of 41 ± 21 h and 47 ± 5 h, respectively. It is clear that aggregation kinetics gets faster at pH values closer to pI value of the peptide. Because of this phenomenon of differing aggregation rates as a result of pH, we hypothesize that electrostatic interactions play a significant role in the self-assembly of protein aggregates.
3.3. Effect of pH on aggregate morphology
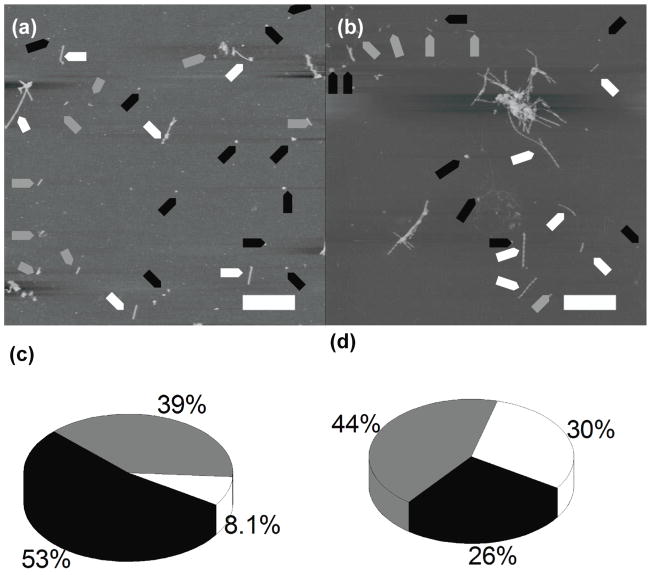
With the use of AFM, we then tested the peptide aggregation in four different pHs: 2.0, 3.7, 5.6, and 7.0 at a low ionic strength of 11 mM. Two typical AFM images for samples obtained in pH 5.6 and pH 2.0, are shown in Fig. 2(a) and Fig. 2(b) respectively. Regardless of the pH, the peptide is able to form all three types of morphologies indicated by arrows. However, according to Figs. 2(c) and D, the proportion of these morphologies vary varies depending on the pH. Oligomers were generally favored at pH 5.6, as seen in the AFM image in figure 2(a), and the pie chart in figure 2(c), which is close to the peptide’s pI of 5.3. Longer fibrils were generally favored at pH 2.0, as seen in AFM image in figure 2(b) and the pie chart in figure 2(d). More data are shown in Fig. s5 (supplemental figure) and the proportion of oligomers in pH 5.6 was 53%, with fibrils making up only 8% of all aggregates. There were also intermediates that made up 39% of the total. On the other hand, pH 2.0 had 26% of all aggregates as globular, 30% were fully formed fibrils, and the remaining 44% being intermediates. The other two pHs are shown in AFM images (i) and pie charts (ii) of figures s5(b) and s5(d). The two pHs represented here had similar proportions of oligomers and fibrils (ii), with a greater than twofold proportion of oligomers to fibrils (ii), and with a similar proportion of protofibrils (ii). The proportion of oligomers is 38%, fibrils comprise of 17%, and the remaining 45% are intermediates at pH 3.7 (figure s5(b ii)). These proportions remain similar at pH 7.0, with 39% of aggregates being in the globular state, 16% in full-length fibrils, and the remaining 45% being in an intermediate state between globular and fibrillar aggregates (figure s5(d ii)).
Figure 2.
Fibrils grown in low salt buffers. AFM images of fibrils grown in (a) pH 5.6 buffer and (b) pH 2.0 buffer; full length fibrils, short fibrils and globular aggregates are indicated with white, gray and black arrows, respectively. Yield of aggregates formed in (c) pH 5.6 and (d) pH 2.0, with globular aggregates (black), protofibrils (gray), and full length fibrils (white).
The aggregates were also analyzed for their height. Here, we see that the difference in heights depends mainly on pH rather than the specific type of aggregate. At pH 2.0, the majority of all aggregates had heights around 6.5 nm (figures s5(a, iii–v)). At pH 3.7, the aggregates had a larger range of heights from 5 nm, 8 nm, and 12 nm for fibrils, short fibrils, and oligomers, respectively (figures s5(b) iii–v). At pH 5.6, there was a shift toward higher aggregates, with the center of the distribution of 9 nm, with an additional distribution maximum around 5 nm (figure s5C iii–v). At pH 7.0, there was a distribution of 7 nm in height in oligomers and fibrils, as well as a distribution at around 4 nm (figures s5D iii–v). There were also many aggregates at all pHs, mainly protofibrils and oligomers, that were around 2 nm, which may represent an early stage of aggregation.
3.4. Effect of ionic strength on the aggregation kinetics
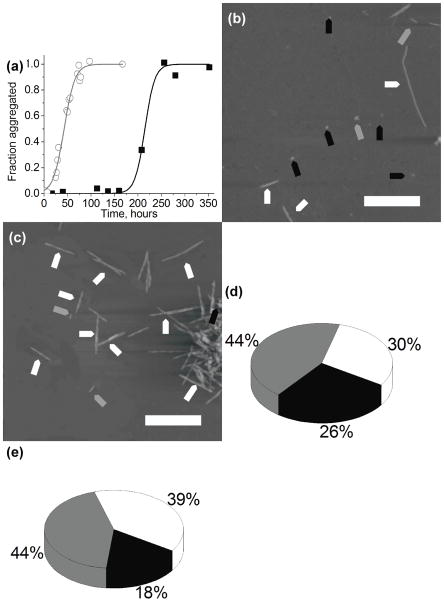
The influence of ionic strength on the PepQ peptide aggregation was studied at pH 2.0 two ionic strengths - 11 mM and 150 mM. The ThT aggregation data are shown in figure 3(a) in which black squares represent aggregation in 11mM salt concentration, and the open gray circles represent aggregation in 150 mM salt concentration. The data show that the addition of salt increases the rate of aggregation most dramatically, as indicated by a shorter lag time of 36 ± 15 hours, compared to 196 ± 24 hours under low salt conditions. All of the other pHs also show an effect of ionic strength on their lag times, where the change in lag time was the smallest at pH 5.6. The increase in ionic strength at pH 5.6 resulted in a small shift from 12 ± 6 hours to 7 ± 3 hours. The other two pHs showed an intermediate shift from 41 ± 21 to 22 ± 11 hours at pH 3.7, and 47 ± 5 to 33 ± 16 hours at pH 7.0.
Figure 3.
Fibrils grown in low salt buffers. (a) Kinetic curves of the peptide aggregation at pH 2.0 at low salt (black squares) and high salt (open gray circles). AFM images of fibrils grown in (b) pH 2.0 low salt buffer and (c) pH 2.0 high salt buffer; fibrils, protofibrils and globular aggregates are indicated with white, gray, and black arrows, respectively. Yield of aggregates formed in (d) pH 2.0 low salt buffer and (e) pH 2.0 high salt buffer, with globular aggregates, protofibrils and full length fibrils shown in black, gray, and white, respectively.
3.5. Effect of ionic strength on aggregate morphology
We used AFM to study the effect of ionic strength on the morphology of aggregates (figures 3(b) and 3(c) and supplemental figures s5(a i) and s6(a i). At pH 2.0, increasing ionic strength resulted in an 8% decrease in oligomers, and a 9% increase in elongated fibrils (Fig. 3(d) and 3(e), Fig. s5(a ii), and Fig. s6(a ii). At pH 3.7, oligomers increased from 38% to 49%, with a reduction in fibrils from 17% to 5.4% (Fig. s5(b ii), and Fig. s6(b ii)). At pH 5.6, the proportion of globular oligomers slightly increased and fibrils slightly decreased, while short fibrils remained the same (Fig. s5(c ii), and Fig. s6(c ii)). There was a reverse trend at pH 7.0, where there was a reduction in oligomers from 39% to 18%, and an increase in fibrils from 16% to 39%, with protofibrils barely changing (Fig. s5(d ii), and Fig. s6(d ii)). One interesting result to note is that protofibrils have a range from 39% to 46% across all pHs and ionic strength. This is the most stable percentage, and it does not appear to be affected by ionic strength. In pH 2.0, the oligomers formed under high ionic strength conditions have heights of around 10 nm, shown in figure s6 (a iii), while aggregates formed under lower ionic strength are smaller, have heights of around 5 nm (figure s5(a iii)). The distributions of aggregates also have larger sizes under high ionic strength as compared to low ionic strength, as seen in figure s5(a–d, iii–v). We have also observed association of the fibrils into bundles at higher ionic strength. The fibrils themselves have a similar height of around 5.5 nm and 6.3 nm in low and high ionic strengths, respectively; along with some larger fibrils that (11.2 nm) under high ionic strength, shown in figure s5(a) and s6(a), respectively. At pH 3.7 and high ionic strength, the aggregates have a large spread of heights with maxima of distributions at 5 nm, 9 nm, and 17 nm (figure s6(b iii–v)). At pH 5.6, there was a shift toward higher aggregates, with the center of the distribution at 9 nm, with an additional maximum at around 27 nm (figure s6(c iii–v). The formation of very large aggregates up to 60 nm in height was also observed at pH 5.6. At pH 7.0, there was a wide distribution of heights, with the maxima occurring at 15 nm and 40 nm in all aggregates (figure s5(b iii–v), as well as the heights of 9 nm and 48 nm in the case of oligomers (figure s5(b iii)).
3.6. Dynamic Force Spectroscopy
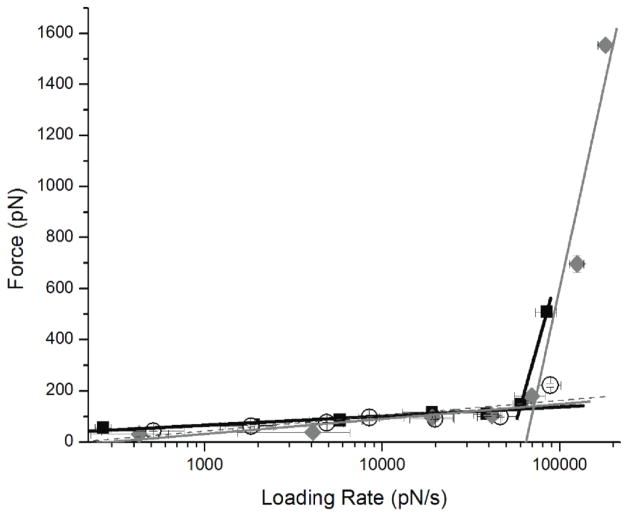
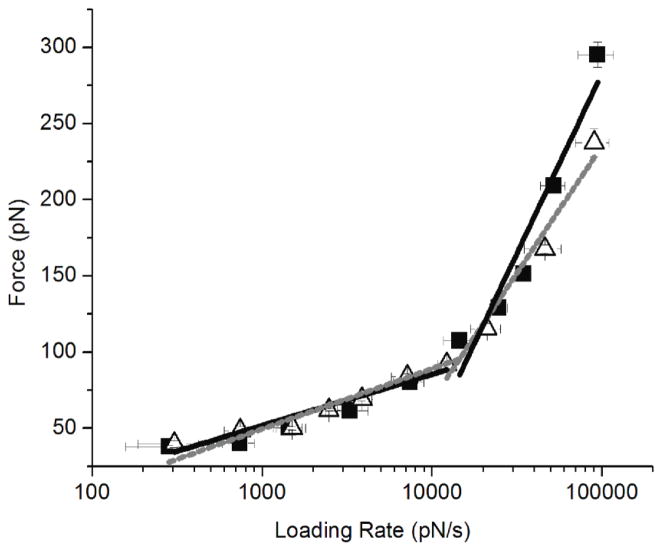
In order to understand what effect electrostatics has on the interactions between PepQ peptides, we have performed force spectroscopy measurements at various pH values. Individual peptide molecules were covalently tethered to the AFM tip and mica surface via two PEG (MW 3,400) flexible linkers on the tip and the surface, and their interactions were probed by multiple approach-retraction cycles as described in [17, 26, 28, 29, 33, 34]. A double tether approach for force spectroscopy was used in order to give enough rupture distance in the analysis of the force curves to distinguish specific peptide-peptide interactions over short range non-specific interactions [26, 28, 29, 33, 34]. The experiments were performed at different pulling rates to generate dynamic force spectra [17, 26, 28, 29, 33–36]. Figure 4 shows DFS spectra measured in pH 2.0, pH 3.7, and pH 5.6 buffers. The plot for pH 2.0 have two slopes (figure 4). The data points for pH 3.7 fall on one line (figure 4). The plot for pH 5.6 has two slopes; note the second of which is quite steep (figure 4), suggesting that two different dissociation pathways exist for the dimer dissociation at these conditions [26, 28, 29, 33, 34].
Figure 4.
Dynamic Force Spectroscopy plots for PepQ interactions measured at pH 2.0 (black squares), pH 3.7 (open circles), and pH 5.6 (gray diamonds).
We have also performed DFS study at two ionic strengths for pH 2.0 shown in figure 5. These experiments were also performed at different loading rates to generate dynamic force spectra [17, 26, 28, 29, 33–36]. The parameters extracted from the fit of the spectra with equation 1 are summarized in Table 2. Both sets of slopes are nearly identical, with similar lifetimes of 3.2 s and 1.9 s for low salt and high salt, respectively, for the first slope. The second set of slopes also show similar lifetimes of 0.40 s and 0.60 s, respectively.
Figure 5.
Dynamic Force Spectroscopy plots of PepQ interactions obtained at pH 2.0 at high salt (open triangles and gray lines) and low salt concentrations (black squares and lines).
Table 2.
Parameters obtained from dynamic force spectroscopy at pH 2.0 in low and high ionic strengths (IS).
| IS (mM) | koff(1)(s−1) | T1 (s) | ΔG (KBT) | x(1)(nm) | koff(2)(s−1) | T2 (s) | ΔG (KBT) | x(2)(nm) |
|---|---|---|---|---|---|---|---|---|
| 10 | 3.3 ± 1.7 | 0.30 | 28.3 | 0.24 ± 0.04 | 62.0 ± 9.4 | 0.02 | 25.3 | 0.04 ± 0.01 |
| 150 | 2.0 ± 0.8 | 0.50 | 28.8 | 0.28 ± 0.03 | 54.0 ± 8.9 | 0.02 | 25.5 | 0.06 ± 0.01 |
koff – dissociation rate constant for the peptide-peptide pair, x – the position of energy barrier. The indices 1 and 2 in the subscript of the parameters refer to the outer and inner barriers of the energy landscape, respectively.
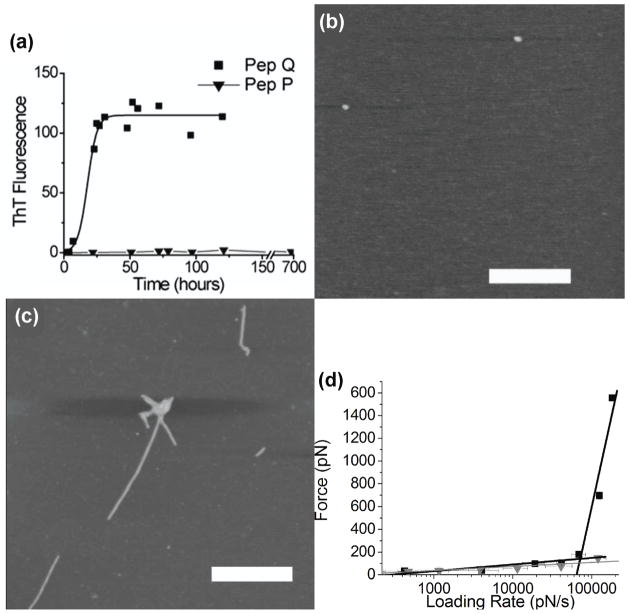
We proposed earlier [37] that different slopes on the DFS plots corresponded to different conformations of misfolded dimers that might lead to different morphologies of aggregates. Here, we tested this assumption by performing parallel DFS analysis of Pep Q and CGNNPQNY (Pep P); the first glutamine (Q) in PepQ was replaced with proline (P) to yield PepP. The difference in the aggregation of these two peptides (Fig. 6(a)) is the lack of fibrils for PepP as tested with ThT fluorescence. Figures 6(b) and 6(c) show AFM images obtained for PepQ (48 hours when plateau of fluorescence was reached) and PepP (600 hours – the longest time point we have observed). These images show that PepP does not form any aggregates while images of PepQ sample show fibrillar type aggregates. AFM force spectroscopy results are summarized in Fig. 6(d) in which DFS data for both peptides are shown. The major difference between these two peptides is the lack of large forces at high loading rates for PepP, so its DFS plot is presented with one line (black squares). On the contrary, the data for PepQ have large forces at high loading rates leading to a steep second linear regime in the DFS spectrum (blue triangles). From these findings, we suggest that the second steep slope of DFS plot characterizes the formation of fibrillar aggregates, suggesting that the dimers with large lifetimes (small off rates) lead to the formation of aggregates different from fibrils.
Figure 6.
(a) Kinetic curves of aggregation of PepQ peptide (black squares) and CGNNPQNY (PepP, gray triangles) obtained with ThT fluoresence. AFM images of aggregates formed by PepP (b) and PepQ (c) peptides in pH 5.6 buffer. The scale bars are 500 nm. (D) Dynamic force spectroscopy plots for PepQ (black squares) and PepP (gray triangles).
4. Discussion and Conclusions
The data presented here show a strong effect of electrostatics on the aggregation of PepQ peptide. The aggregation rate is the fastest at pH 5.6, which is close to pI of the peptide (5.3). At this pH, the peptide chain has the positive and negative charges neutralizing each other and therefore there is less repulsion, which contributes to faster aggregation. There are also hydrophobic interactions that take place between peptides, which is most readily seen at pH 5.6 due to the neutralization of overall charge on the peptide chain. At pH 2.0, however, the overall charge is highly positive, which then leads to greater repulsion. This repulsion has slowed down the aggregation kinetics at this pH, as this repulsion needs to be overcome before there is interaction between charged peptides. However, the fact that this interaction does eventually take place demonstrates that this is not an entirely strong effect. When increasing salt concentrations, we can effectively screen this electrostatic repulsion. This has the result of accelerating aggregation kinetics, with the repulsion lessened from the ion screening. AFM topographic analysis revealed the effect of electrostatics on the aggregation morphologies. The overall sizes of all aggregates formed at low salt concentration are smaller compared to samples grown in high salt aggregation. The heights of the aggregates had two populations in the high salt aggregates, corresponding to the low salt heights, as well as twice as that.
Surprisingly, the effect of electrostatics was not that pronounced in the force spectroscopy experiments in which interactions between the peptides within the transiently formed dimer were probed. According to figure 5, the increase of ionic strength at pH 2.0 slightly decreased the interactions at high loading rates with no effect on the interactions at slow loading rates. These findings suggest that intermolecular rather than electrostatic interactions with the transiently formed dimer define the dimer stability. Indeed, the distances between the peptide monomers within in crystal structures [38] are relatively short (~0.5 nm), so the contribution of the long-range electrostatic interactions on the dimers dissociation at such distances is small. However, electrostatic repulsion prevents the formation of such dimers, initially slowing down the kinetics of the peptide aggregation as we observed in the experiments with ThT fluorescence. The primary role of intermolecular interactions rather than electrostatics on the dimers stability is confirmed by our comparative studies of PepP and PepQ, which differed in only one amino acid.
The DFS data for these PepQ and PepP are dramatically different. The results with these two mutants provide additional insight into the mechanism of protein aggregation. ThT is a widely used fluorescent probe for detecting amyloids [32, 39–42]. In our experiments (Fig. 6(a)), we did not see any increase of ThT fluorescence for PepP peptide, suggesting that no amyloid type aggregates are formed. These data are in sharp contrast with similar probing of PepQ aggregation (Fig. s4), which is typical for aggregation of amyloid proteins. These data suggest that PepP peptide should not form amyloid aggregates. Indeed, we did not observe aggregates for PepP with AFM (Figs. 6(b) and 6(c)). The DFS data (Fig. 6(d)) obtained for PepQ and PepP are very different, too. PepQ forms dimers, dissociating at the rupture forces as large as ~1.6 nN, whereas the stability of PepP dimers probed at the same loading rates is more than 10 times less. The DFS plot for peptide PepQ has two well-defined linear plots, whereas the DFS plot for PepP has only one line with a small slope similar to the one for PepQ. Comparison of these data leads us to the hypothesis that the second linear plot on the DFS plots is the signature for the formation of fibrils and the linear plot with a small slope describe the properties of dimers that are not involved into the fibrils formation and may be responsible for the formation of small aggregates (oligomers). We have recently applied DFS analysis to Aβ peptide [28] and α-synuclein [26, 29]. The two-slope DFS plots are the signatures for these amyloid proteins and they also form aggregates of different morphologies, including fibrils. However, the values of rupture forces obtained at high loading rates for these systems are considerably smaller than those for PepQ, suggesting that fibrils formed by this peptide, Aβ and α-synucelin are structurally different. Additional data are needed to test this assumption.
Overall, the results of this study demonstrate that the aggregating properties of short amyloidogeneic peptide are strongly affected by the environment. We show here that electrostatics plays an important role in the process of self-assembly of short amyloidogenic peptide, PepQ. Electrostatic interactions define both aggregation kinetics and final morphology of aggregated species. Either neutralization of the positively charged peptide by high ionic strength or compensation of the overall peptide charge by pH leads to a drop of the kinetics of the aggregates assembly. The decrease of the electrostatic repulsion of the peptides facilitates the assembly of thick fibrils and the formation of large aggregates. At the same time, electrostatic interactions do not play that strong effect in the stability of already assembled aggregates, as the effect of electrostatics is small in the dissociation of dimers. A finding of the role of the sequence on the interpeptide interactions suggested that fibrils and oligomers are assembled by different pathways characterized by different interactions within the dimers. This is a novel finding that shed a new light into the mechanism of protein misfolding and self-assembly in nanoaggregates.
Supplementary Material
Table 1.
Parameters obtained from dynamic force spectroscopy in different pHs.
| pH | koff(1)(s−1) | T1 (s) | ΔG (KBT) | x(1)(nm) | koff(2)(s−1) | T2 (s) | ΔG (KBT) | x(2)(nm) |
|---|---|---|---|---|---|---|---|---|
| 2.0 | 3.3 ± 1.7 | 0.30 | 28.3 | 0.24 ± 0.04 | 62.0 ± 9.4 | 0.02 | 25.3 | 0.04 ± 0.01 |
| 3.7 | 7.5 ± 8.0 | 0.13 | 27.4 | 0.16 ± 0.05 | ||||
| 5.6 | 11.2 ± 11.5 | 0.09 | 27.0 | 0.16 ±0.05 | 46.9 ±7.6 | 0.02 | 25.6 | 0.003 ± 0.001 |
koff – dissociation rate constant for the peptide-peptide pair, x – the position of energy barrier. The indices 1 and 2 in the subscript of the parameters refer to the outer and inner barriers of the energy landscape, respectively.
Acknowledgments
We thank B. H. Kim, J. Yu, Z. Lv., Y. Zhang, L. Shlyakhtenko, and other members of the Lyubchenko lab for insightful discussions. We also thank J. Garrison for use of his equipment and training for peptide synthesis. The work was supported by grants from NIH (R01 GM096039), DOE (DE-FG02-08ER64579), Nebraska Research Initiative (NRI) and NATO (983204) to YLL.
Footnotes
Classification Numbers: 87.14.E- Proteins
87.64.-t Spectroscopic and microscopic techniques in biophysics and medical physics
87.64. Dz Scanning tunneling and atomic force microscopy
82.37. Gk STM and AFM manipulations of a single molecule
References
- 1.Dobson CM. Protein folding and misfolding. Nature. 2003;426(6968):884–90. doi: 10.1038/nature02261. [DOI] [PubMed] [Google Scholar]
- 2.Madine J, et al. Structural insights into the polymorphism of amyloid-like fibrils formed by region 20–29 of amylin revealed by solid-state NMR and X-ray fiber diffraction. J Am Chem Soc. 2008;130(45):14990–5001. doi: 10.1021/ja802483d. [DOI] [PubMed] [Google Scholar]
- 3.Schmidt M, et al. Comparison of Alzheimer Abeta(1–40) and Abeta(1–42) amyloid fibrils reveals similar protofilament structures. Proc Natl Acad Sci U S A. 2009;106(47):19813–8. doi: 10.1073/pnas.0905007106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ono K, Condron MM, Teplow DB. Structure-neurotoxicity relationships of amyloid beta-protein oligomers. Proc Natl Acad Sci U S A. 2009;106(35):14745–50. doi: 10.1073/pnas.0905127106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maurstad G, et al. Dehydration stability of amyloid fibrils studied by AFM. Eur Biophys J. 2009;38(8):1135–40. doi: 10.1007/s00249-009-0526-x. [DOI] [PubMed] [Google Scholar]
- 6.Lyubchenko YL, et al. Nanoimaging for protein misfolding and related diseases. J Cell Biochem. 2006;99(1):52–70. doi: 10.1002/jcb.20989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fernandez-Busquets X, et al. Recent structural and computational insights into conformational diseases. Curr Med Chem. 2008;15(13):1336–49. doi: 10.2174/092986708784534938. [DOI] [PubMed] [Google Scholar]
- 8.Munishkina LA, et al. Conformational behavior and aggregation of alpha-synuclein in organic solvents: modeling the effects of membranes. Biochemistry. 2003;42(9):2720–30. doi: 10.1021/bi027166s. [DOI] [PubMed] [Google Scholar]
- 9.Sawaya MR, et al. Atomic structures of amyloid cross-beta spines reveal varied steric zippers. Nature. 2007;447(7143):453–7. doi: 10.1038/nature05695. [DOI] [PubMed] [Google Scholar]
- 10.Baxa U, et al. In Sup35p filaments (the [PSI+] prion), the globular C-terminal domains are widely offset from the amyloid fibril backbone. Mol Microbiol. 79(2):523–32. doi: 10.1111/j.1365-2958.2010.07466.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chan JC, et al. Parallel beta-sheets and polar zippers in amyloid fibrils formed by residues 10–39 of the yeast prion protein Ure2p. Biochemistry. 2005;44(31):10669–80. doi: 10.1021/bi050724t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shewmaker F, Wickner RB, Tycko R. Amyloid of the prion domain of Sup35p has an in-register parallel beta-sheet structure. Proc Natl Acad Sci U S A. 2006;103(52):19754–9. doi: 10.1073/pnas.0609638103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wickner RB, Dyda F, Tycko R. Amyloid of Rnq1p, the basis of the [PIN+] prion, has a parallel in-register beta-sheet structure. Proc Natl Acad Sci U S A. 2008;105(7):2403–8. doi: 10.1073/pnas.0712032105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gsponer J, Haberthur U, Caflisch A. The role of side-chain interactions in the early steps of aggregation: Molecular dynamics simulations of an amyloid-forming peptide from the yeast prion Sup35. Proc Natl Acad Sci U S A. 2003;100(9):5154–9. doi: 10.1073/pnas.0835307100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hess S, Lindquist SL, Scheibel T. Alternative assembly pathways of the amyloidogenic yeast prion determinant Sup35-NM. EMBO Rep. 2007;8(12):1196–201. doi: 10.1038/sj.embor.7401096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoyer W, et al. Dependence of alpha-synuclein aggregate morphology on solution conditions. J Mol Biol. 2002;322(2):383–93. doi: 10.1016/s0022-2836(02)00775-1. [DOI] [PubMed] [Google Scholar]
- 17.McAllister C, et al. Protein interactions and misfolding analyzed by AFM force spectroscopy. J Mol Biol. 2005;354(5):1028–42. doi: 10.1016/j.jmb.2005.10.012. [DOI] [PubMed] [Google Scholar]
- 18.Richt JA, et al. Production of cattle lacking prion protein. Nat Biotechnol. 2007;25(1):132–8. doi: 10.1038/nbt1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Uversky VN, Kabanov AV, Lyubchenko YL. Nanotools for megaproblems: probing protein misfolding diseases using nanomedicine modus operandi. J Proteome Res. 2006;5(10):2505–22. doi: 10.1021/pr0603349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yun S, et al. Role of electrostatic interactions in amyloid beta-protein (A beta) oligomer formation: a discrete molecular dynamics study. Biophys J. 2007;92(11):4064–77. doi: 10.1529/biophysj.106.097766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chernoff YO. Identity determinants of infectious proteins. Proc Natl Acad Sci U S A. 2008;105(36):13191–2. doi: 10.1073/pnas.0806234105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.DePace AH, Weissman JS. Origins and kinetic consequences of diversity in Sup35 yeast prion fibers. Nat Struct Biol. 2002;9(5):389–96. doi: 10.1038/nsb786. [DOI] [PubMed] [Google Scholar]
- 23.Xu S, Bevis B, Arnsdorf MF. The assembly of amyloidogenic yeast sup35 as assessed by scanning (atomic) force microscopy: an analogy to linear colloidal aggregation? Biophys J. 2001;81(1):446–54. doi: 10.1016/S0006-3495(01)75712-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shlyakhtenko LS, et al. Silatrane-based surface chemistry for immobilization of DNA, protein-DNA complexes and other biological materials. Ultramicroscopy. 2003;97(1–4):279–87. doi: 10.1016/S0304-3991(03)00053-6. [DOI] [PubMed] [Google Scholar]
- 25.Atarashi R, et al. Prion strain-dependent differences in conversion of mutant prion proteins in cell culture. J Virol. 2006;80(16):7854–62. doi: 10.1128/JVI.00424-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu J, Malkova S, Lyubchenko YL. alpha-Synuclein misfolding: single molecule AFM force spectroscopy study. J Mol Biol. 2008;384(4):992–1001. doi: 10.1016/j.jmb.2008.10.006. [DOI] [PubMed] [Google Scholar]
- 27.Abid K, Soto C. The intriguing prion disorders. Cell Mol Life Sci. 2006;63(19–20):2342–51. doi: 10.1007/s00018-006-6140-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim BH, et al. Single-Molecule Atomic Force Microscopy Force Spectroscopy Study of Abeta-40 Interactions. Biochemistry. 50(23):5154–5162. doi: 10.1021/bi200147a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu J, Lyubchenko YL. Early Stages for Parkinson’s Development: α-Synuclein Misfolding and Aggregation. J Neuroimmune Pharmacol. 2008 doi: 10.1007/s11481-008-9115-5. [DOI] [PubMed] [Google Scholar]
- 30.Balbirnie M, Grothe R, Eisenberg DS. An amyloid-forming peptide from the yeast prion Sup35 reveals a dehydrated beta-sheet structure for amyloid. Proc Natl Acad Sci U S A. 2001;98(5):2375–80. doi: 10.1073/pnas.041617698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krasnoslobodtsev AV, et al. Nanoimaging for prion related diseases. Prion. 4(4):265–74. doi: 10.4161/pri.4.4.13125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nielsen L, et al. Effect of environmental factors on the kinetics of insulin fibril formation: elucidation of the molecular mechanism. Biochemistry. 2001;40(20):6036–46. doi: 10.1021/bi002555c. [DOI] [PubMed] [Google Scholar]
- 33.Kransnoslobodtsev AV, et al. Nanomedicine and protein misfolding diseases. Nanomedicine. 2005;1(4):300–5. doi: 10.1016/j.nano.2005.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krasnoslobodtsev AV, Shlyakhtenko LS, Lyubchenko YL. Probing Interactions within the synaptic DNA-SfiI complex by AFM force spectroscopy. J Mol Biol. 2007;365(5):1407–16. doi: 10.1016/j.jmb.2006.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kuhner F, et al. LexA-DNA bond strength by single molecule force spectroscopy. Biophys J. 2004;87(4):2683–90. doi: 10.1529/biophysj.104.048868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zheng P, et al. Single Molecule Force Spectroscopy Reveals that Electrostatic Interactions Affect the Mechanical Stability of Proteins. Biophys J. 100(6):1534–41. doi: 10.1016/j.bpj.2011.01.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lyubchenko YL, et al. Nanoimaging for protein misfolding diseases. Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2(5):526–43. doi: 10.1002/wnan.102. [DOI] [PubMed] [Google Scholar]
- 38.Eisenberg An amyloid-forming peptide from the yeast prion Sup35 reveals a dehydrated b-sheet structure for amyloid. PNAS. 2001;98(5):2375–2380. doi: 10.1073/pnas.041617698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Narayanan S, Walter S, Reif B. Yeast prion-protein, sup35, fibril formation proceeds by addition and substraction of oligomers. Chembiochem. 2006;7(5):757–65. doi: 10.1002/cbic.200500382. [DOI] [PubMed] [Google Scholar]
- 40.Ban T, et al. Direct observation of amyloid fibril growth monitored by thioflavin T fluorescence. J Biol Chem. 2003;278(19):16462–5. doi: 10.1074/jbc.C300049200. [DOI] [PubMed] [Google Scholar]
- 41.Biancalana M, et al. Molecular mechanism of thioflavin-T binding to the surface of beta-rich peptide self-assemblies. J Mol Biol. 2009;385(4):1052–63. doi: 10.1016/j.jmb.2008.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Picotti P, et al. Amyloid fibril formation and disaggregation of fragment 1–29 of apomyoglobin: insights into the effect of pH on protein fibrillogenesis. J Mol Biol. 2007;367(5):1237–45. doi: 10.1016/j.jmb.2007.01.072. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.