Abstract
The ability of the Amber ff99 force field to predict relative free energies of RNA helix formation was investigated. The test systems were three hexaloop RNA hairpins with identical loops and varying stems. The potential of mean force of stretching the hairpins from the native state to an extended conformation was calculated with umbrella sampling. Because the hairpins have identical loop sequence, the differences in free energy changes are only from the stem composition. The Amber ff99 force field was able to correctly predict the order of stabilities of the hairpins, although the magnitude of the free energy change is larger than that determined by optical melting experiments. The two measurements cannot be compared directly because the unfolded state in the optical melting experiments is a random coil, while the end state in the umbrella sampling simulations was an elongated chain. The calculations can be compared to reference data by using a thermodynamic cycle. By applying the thermodynamic cycle to the transitions between the hairpins using simulations and nearest neighbor data, agreement was found to be within the sampling error of simulations, thus demonstrating that ff99 force field is able to accurately predict relative free energies of RNA helix formation.
Keywords: hairpin, hexaloop, molecular dynamics, free energy calculation, umbrella sampling, thermodynamic cycle
INTRODUCTION
RNA plays many important roles in the organism beyond simply carrying genetic information. It catalyzes reactions,1–3 participates in post-translational gene regulation,4 controls protein localization5 and guides post-transcriptional modification.6–7 In addition many RNA sequences are transcribed in genomes, but their functions are not yet known.8
Molecular dynamics (MD) simulations provide insights in RNA structure and dynamics. Beginning with the introduction of Ewald methods for calculation of long range electrostatic interactions, it has been possible to run precise simulations on RNA structures.9 Subsequently, molecular dynamics has been used to gain insight into roles of ions, water and modified nucleotides on the RNA structure.10–18 Other, specialized calculations have been used to understand conformational changes, including targeted molecular dynamics for modeling HIV dimerization,19 nudged elastic band (NEB) for modeling loop conformational changes20–22 and umbrella sampling for calculating free energy of folding.23–24
An important factor in estimating the accuracy of MD simulations is the accuracy of the set of parameters used to describe the potential energy of the system, the force field. There are several force fields commonly used for simulations of RNA, belonging broadly to groups derived for use with CHARMM25 and Amber26 molecular modeling packages. The several commonly used Amber force fields are all derivatives of the original Cornell et al. force field ff94,27 derived using a combination of experimental data and quantum mechanical calculations Force fields ff9828 and ff9929 improved the sugar pucker and glycosidic torsions of nucleic acids in ff94. Later, in 2007, the ff99bsc030 correction introduced an improved description of the alpha and gamma backbone torsions. Finally, two separate sets of parameters have been derived using quantum mechanics to describe glycosidic torsions of all four bases.31–32 Recent CHARMM force fields are derivatives of the CHARMM2733 force field. CHARMM3624, 34 improves on CHARMM27 by reparameterizing the torsions of 2′-hydroxyl group of RNAs and several backbone and sugar pucker torsions to better describe the BI/BII conformation equilibrium of DNAs. On a basic level, the accuracy of force field parameters can be estimated by comparing it with quantum mechanics (QM) calculations. Because of the high computational cost of QM calculations, these computations are confined to calculating the energies of stacking and hydrogen bonding in vacuum and involving only bases.35–40 Comparing these results with the equivalent calculations performed using Amber force fields shows that Amber force fields are able to predict hydrogen bonding and stacking interactions with reasonable accuracy.41 The downside of these comparisons is that they only consider the bases, the calculations are performed in vacuum and the ions are not present.
Ultimately, the accuracy of force field can best be judged by how well it performs in simulations of nucleic acids systems in solution. In this work, the ability of Amber ff99 force field to accurately predict free energy changes of helix formation was examined. Experimental data, in form of the free energy changes of folding calculated from optical melting experiments, and nearest neighbor parameters were used to determine the reference free energy changes.42–43 Nearest neighbor parameters are a set of empirical parameters derived from optical melting experiments that can predict free energy change of RNA secondary structure formation by adding pair-wise contributions. For duplex formation, the nearest neighbor parameters have statistical errors of less than 0.1 kcal/mol each.44–45
Specifically, experimental and nearest neighbor free energies of folding were compared with the free energies calculated using MD simulations. To determine the free energy change of unfolding, potential of mean force (PMF), also called umbrella sampling, calculations were performed.46–48 The PMF method uses a set of equilibrium molecular dynamics calculations, called windows, along a reaction coordinate to determine a free energy change along that coordinate. Individual simulations are forced to sample different regions along the reaction coordinate with harmonic restraints, also called umbrella potentials. Finally, the windows are combined, the effect of the biasing potential is removed and the free energy change along the reaction coordinate is obtained. In this work, the umbrella sampling simulations follow the unfolding coordinate measured by the distance between O5′ atom of the 5′-end nucleotide and the O3′ atom of the 3′-end nucleotide in RNA hairpin loops. This reaction coordinate was chosen to mimic the procedure performed in single molecule stretching experiments. In this study the endpoints are distances of 15 Å (native conformation) and 75 Å, a denatured state that corresponds roughly to the contour length of a 12-mer of a single-stranded RNA, which has a 5.9 Å distance between successive nucleotides.49
The RNA systems used were three hairpin stem-loop structures that have the same loop (hexaloop GUAAUA),42 but differ in the stem region. Therefore, the differences in folding free energies between the hairpin stem-loops were caused by the differences in the double-helical region. The hairpins were chosen instead of simple double helices because of their larger stability compared to double helices with the same number of nucleotides,50 and because the endpoint of a pulling simulation is a well-defined structure that can be sampled on the timescales of molecular dynamics simulations. The GUAAUA sequence is highly conserved in the L11 region of the large subunit ribosomal RNA.51–52 Although tetraloops have been studied extensively using MD simulations,23, 53–56 simulation studies done on hexaloops are fewer,57–58 despite of large amount of experimental data available.42, 59–60 In addition, the choice of hexaloop hairpins as a test system enabled examination of their folding pathway, results of which can be compared to the similar tests performed recently on the tetraloop systems.23–24
The report is organized as follows. In the Materials and Methods, the properties of the hairpins used in the simulations and the details of the umbrella sampling simulations are described. In the Results and Discussion section, the results of the MD simulation are analyzed, the free energy changes are calculated using the results from the simulations and experiments/nearest neighbor data and the difference between the two are discussed. Finally, the Conclusion section summarizes the results of simulations and comparisons with the experimental data.
MATERIALS AND METHODS
Starting structures
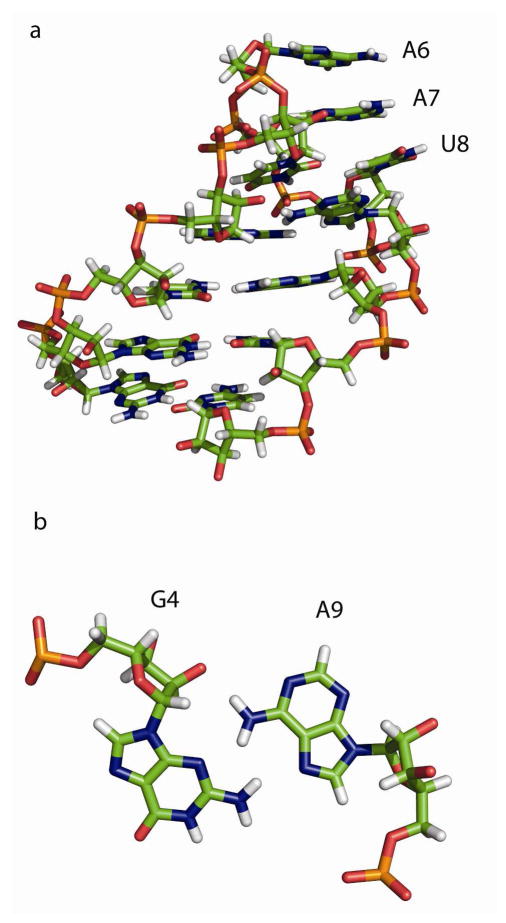
The calculations were done starting with the hexaloop hairpin sequence: 5′-GGCGUAAUAGCC-3′, where unpaired hairpin nucleotides are underlined. The atomic coordinates were taken from an NMR structure.42 The coordinates of the hairpin GCGUAAUAGC were also determined in the E. coli ribosome crystal structure solved to 3.5 Å resolution.43 The mass-weighted conformational root mean square deviation (RMSD) for the 10 nucleotides common to the NMR and x-ray structure is 0.28 Å. The stability of this hairpin is −2.7 ± 0.15 kcal/mol in 0.1 M NaCl at 37 °C, as measured by optical melting experiments.59 Figure 1 illustrates the solution structure of the hairpin. The important structural features include the G4-A9 sheared base pair and the continuous stacking of A6, A7 and U8. NMR data, however, show that there is a large flexibility of the loop region at 35 °C, well below the melting temperature of 63 °C, and that the structure of G4-A9 mismatch is fluxional.42
Figure 1.
Solution structure of native hairpin GGCGUAAUAGCC.42 Figure 1a shows the structure, illustrating the continuous stack of A6, A7 and U8. Figure 1b shows the sheared base pair between G4 and A9. G4 and A9 are bonded via trans Hoogsteen/sugar edge hydrogen bond.87
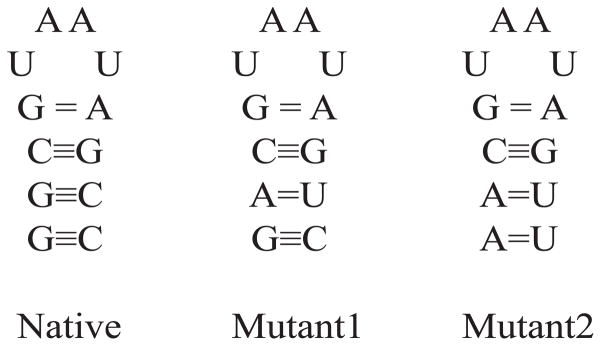
Two additional, mutant hairpins were created by manually modifying the atoms in the stem region of the native hairpin. In mutant1, basepair G2-C11 was replaced with A2-U11. In mutant2, G1-C12 has been replaced with A1-U12 and G2-C11 was replaced with A2-U11. This preserves the loop region, and changes the stem region. The secondary structures of the three hairpins are given in Figure 2.
Figure 2.
Secondary structures of native, mutant1 and mutant2 hairpins. They differ only in the stem region. Mutant1 has the G2-C11 base pair replaced with the A2-U11, and the mutant2 has G1-C12 replaced with A1-U12 and G2-C11 with A2-U11. This diagram shows the number of hydrogen bonds in each pair.
Free simulations MD protocol
Stability of the hairpins was tested by running three independent simulations for each hairpin. Hairpins were first neutralized by adding Na+ ions and then they were immersed in a truncated octahedron box of TIP3P61 water such that the edges of the box were at least 8 Å from the solute molecules. The systems were then equilibrated to temperature of 300 K and pressure of 1 atm using the following procedure. First, a 1000 step minimization of only water molecules was run to relieve the potentially bad contacts between solute and solvent molecules, and then the whole system was minimized during another 1000 steps of minimization. Then the system was gradually warmed to 300 K during 20 ps using a Langevin thermostat62 with a frequency of collision of 1 ps−1. Next, pressure was set to 1 atm using the Berendsen method63 with isotropic position scaling and a 1 ns MD simulation was run. After that, three independent 100 ns simulations were run for each hairpin, with the difference between runs being the random number seeds for the pseudo-random number generator. In all simulations, particle-mesh Ewald (PME) method64–65 was used to calculate the electrostatic interactions. The SHAKE algorithm66 was applied to fix all bonds containing hydrogens, which permits a 2 fs time step. Lennard-Jones interactions were truncated at 8 Å. Simulations were run using the pmemd program of Amber.26
Umbrella sampling protocol
PMF calculations were done using the Amber software package26 and the ff99 force field28–29 using an explicit solvent and the TIP3P61 water model. Umbrella sampling windows were separated by 1 Å. The ends of the RNA hairpin were held at the appropriate distance in each window using a harmonic restraint with a force constant of 5 kcal/mol. Each window simulation was at least 10 ns (and in some cases longer to ensure convergence). Because of the large size of the system, especially in unfolded states, separate starting structures were created for each window. They were obtained by running a series of restrained simulations in implicit solvent using the Generalized Born solvation model67–69 with a 1 ns simulation time per window. One simulation was performed for each umbrella sampling window, by restraining the distance between the O5′ atom at the 5′-end and the O3′ atom at the 3′-end of the hairpin. The end points of implicit solvent simulation simulations were used to make the starting points for the explicit solvent simulations. Each molecule was first neutralized with Na+ ions,70 and then, to reproduce the conditions of the optical melting experiments,42 enough Na+ and Cl− ions71 were added to make the concentration 0.1 M in each window. Each of the structures was then solvated in a cube of TIP3P61 water with water molecules at least 8 Å from the edges of the RNA hairpin. The system was then energy minimized in two stages. First, the RNA molecule was kept frozen and water and ions were minimized and then in the second stage the whole system was allowed to move. The window restraints were then turned on, and the system was slowly heated to 300 K and to pressure of 1 atm during the 100 ps using the Langevin thermostat.62 After that, an NTP production simulation was run for 10 ns or more per window. The Particle-mesh Ewald (PME) method64–65 was used to calculate the electrostatic interactions. The SHAKE algorithm66 was used to fix all bonds containing hydrogen atoms. Simulations were run using the pmemd module of Amber. The first 1 ns of each window simulation were used for equilibration and the data were collected from the remaining time. To obtain an estimate for errors, three independent simulations were run for native and four simulations for mutant hairpins. For all hairpins, first, an “exploratory” simulation was run on windows from 15 to 75 Å end-to-end distance, to establish an exact point of unfolding. This point was set to where all hydrogen bonds in the hairpin were broken (more details in the Results and Discussion), which occurs by the 55 Å window in all simulations. Then an additional two (or three for mutant hairpins) umbrella sampling simulations were run on windows from 15 to 55 Å. The individual runs differ in the random seed for the Langevin thermostat. Distributions of end-to-end distances from all windows were then combined, the biasing potential was removed and the free energy change along the reaction coordinate was calculated using the Weighted Histogram Analysis Method (WHAM)46–48 as implemented in a program by Alan Grossfield.72 In the WHAM calculations the data points were sorted in bins separated by 0.1 Å and the convergence criterion was set to 1×10−6 kcal/mol. The convergence of simulations was tested by comparing the free energy change calculated using the full sampling to that calculated using shorter sampling, the reasoning being that if the simulations have sampled the conformational space properly the results should be similar. The tests for convergence for Native, Mutant1 and Mutant2 hairpins are shown in Figures S3, S4 and S5 respectively in the Supporting Materials section. The total simulation time, which includes free and umbrella sampling simulations was 7.2 Ss.
RESULTS AND DISCUSSION
Free simulations of hairpins
The stability of the three hairpins (Figure 2) was tested by running three independent, 100 ns explicit solvent simulations using the Amber software package26 and the ff99 force field.29 The RMSD plots of heavy atoms to the NMR structure as a function of time are given in Figure S1 in Supporting Materials. The native hairpin stabilized at around 3.5 Å mass-weighted RMSD from the NMR structure. The RMSD of the stem region is on average 1.04 Å for all three simulations (data not shown), so most of the flexibility is in the loop. Mutant1 shows a somewhat larger RMSD because, in addition to the flexible loop, it has one of the GC basepairs in the stem replaced with the weaker AU pair. In two of the trajectories the stem remained stable, while the third shows a formation of a “ladder-like” structure in the double helical region, a phenomenon related to the inability of ff99 to properly model the glycosidic torsion potential.53 Finally mutant2 shows a relatively high RMSD relative to the NMR structure in all three trajectories. Mutant2 is predicted by nearest neighbor model to be weakly unstable at 300K, with a folding free energy change of 0.8 kcal/mol (Table 2). There is again the formation of a “ladder-like” conformation and also a fraying of the A1-U12 basepair in all three trajectories. To test whether umbrella sampling can be run on a weakly unstable system, a 30 ns MD simulation of mutant2 was run with a 17 Å end-to-end restraint. The 30 ns simulation time is longer than any window time used in the umbrella sampling simulations. Figure S2 of the Supporting Materials shows RMSD as a function of time for both the whole mutant2 hairpin and for its stem region. The average RMSDs to the starting structure for the complete hairpin and the stem region were 2.6 Å and 1.2 Å, respectively. Therefore, the stem region remains stable for the time scales required to run a single window simulation of umbrella sampling measurements because the end-to-end restraints keep the base pairs in the stem at the appropriate positions for pairing. Because of these factors, the flexibility of the mutant hairpins should not have a significant influence on the accuracy of calculations.
Table 2.
Free energy of melting native, mutant1 and mutant2 hairpins calculated using the combination of free energy changes from optical melting experiments and nearest neighbor parameters (from the NNDB).
| Native | Mutant1 | Mutant2 | |
|---|---|---|---|
| ΔGNN (kcal/mol) | −3.5 ± 0.2 | −1.3 ± 0.3 | 0.8 ± 0.3 |
Errors are calculated from the errors given with the parameters in NNDB and by taking into account that the changes in enthalpy and entropy are correlated.45
It has been reported that the ff99 force field performs poorly in describing the structure of the hairpins. This is apparent in this study, as evidenced by the relatively high RMSD of the loop region and the partial distortion of the stem regions of the mutant hairpins. The recently published31–32 improvements to the Amber ff99 force field description of the glycosidic torsion prevent the formation of “ladder-like” structures; their inclusion could be used to improve the stability of hairpins in free simulations.
Free energy change from umbrella sampling simulations
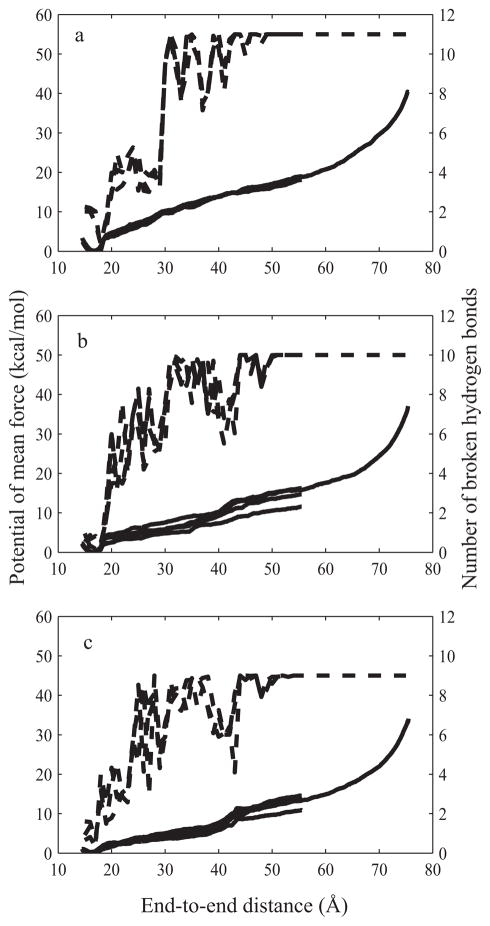
Figure 3 shows free energy change (solid lines, left side y-axis) and the average number of broken hydrogen bonds (dashed lines, right side y-axis) along the reaction coordinate for native, and both mutant hairpins. The free energy change was calculated by applying the WHAM procedure on the end-to-end distances distributions from the individual umbrella sampling simulations. The average number of broken hydrogen bonds was calculated by averaging the number of broken hydrogen bonds in each umbrella simulation window over the course of the simulation. A hydrogen bond was defined as being broken if the distance between the donor and acceptor atoms was larger than 4 Å. The native hairpin has four basepairs, three GC and one GA (sheared) pair for a total of 11 hydrogen bonds; mutant1 has three GC pairs and one each of AU and GA pairs for a total of 10 hydrogen bonds. Finally, mutant2 has two GC pairs, two AU pairs and one GA pair for a total of 9 hydrogen bonds.
Figure 3.
Potential of mean force (PMF) and number of broken hydrogen bonds plotted against the end-to-end distance for native (Figure 3a), mutant1 (Figure 3b) and mutant2 sequences (Figure 3c). The PMF calculation has been run three times for the native hairpin and four times for mutant1 and mutant2 hairpins. Solid lines and the left-hand side y-axis are the PMF plots, dashed lines and the right-hand side y-axis denote data for the number of hydrogen bonds broken.
All PMF calculations showed five distinct regions:
A well-shaped region, spanning from 15 to 19 Å with a minimum at 17 Å, corresponding to the native state. The minima of all three molecules were within several tenths of an angstrom of each other and to the distance between 5′-end O5′ and 3′-end O3′ atoms in the solution structure.42
In the second region, from approximately 19 Å when first hydrogen bond breaks to approximately 42 Å, there was monotonic increase in free energy change that corresponded to an equilibrium between conformations with partially broken hydrogen bonds in the stem region. All hydrogen bonds in the first base pair (1G-12C in native and mutant1 and 1A-12U in the mutant2) are broken by 20 Å. Hydrogen bonds in the second basepair (2G-11C in native and 2A-11U in both mutants) break by around 30 Å. The last basepair in the stem region (3C-10G in all three hairpins) breaks by around 42 Å.
In the third distinct region, starting around 42 Å end-to-end distance, there was a steep increase in the slope of free energy change curve as the last hydrogen bond in the stem was broken. These first three regions correspond approximately to the three regions described in work by Deng et al.23 with tetraloop sequences.
The fourth region, spanning from 50 Å to approximately 55 Å, was the region where the sheared hydrogen bond in the loop (4G-9A) was gradually broken in all hairpins, and the hairpin stretched to an almost linear form after that.
The region after 55 Å end-to-end distance was characterized by a steeper slope of the free energy curve. At this point all hydrogen bonds were broken and the faster increase in free energy was a consequence of stretching the angles and bonds that requires more potential energy than the breaking of hydrogen bonds.
Optical melting experiments, which were used as a reference, interpret the melting data using a two-state model.73–74 The two-state model assumes that the molecule exists in either completely paired or completely unpaired state, and is a reasonably good approximation for small hairpins and duplexes such are the ones used in this study. This assumption is supported by isothermal titration calorimetry experiments.75 The free energy change of unfolding for the two state model can be estimated from the probabilities of finding the molecule in the two states, folded or unfolded. The native state was defined as the area between 15 and 19 Å end-to-end distances and the unfolded state as the area beyond 55 Å, which is the point at which all hydrogen bonds are broken. Then the free energy of unfolding23 is:
| (1) |
where R is the gas constant, T=300 K is the absolute temperature, the numerator is the sum of probabilities of folded states and the denominator is the sum of probabilities of the unfolded states. Probabilities for folded and unfolded states were obtained from the WHAM procedure.
The exact positions of folded and unfolded regions do not have a large effect on the relative free energies of folding. For the native region, the areas where end-to-end distances were smaller than 15 Å and larger than 19 Å had small probabilities so their contribution to the folded probability was negligible. Similarly, the area beyond 55 Å contributed little to the total probability of the unfolded state because the probabilities decrease rapidly in this region. The end point was determined by following the breaking of hydrogen bonds during the umbrella sampling simulations (Figure 3). There were three basepairs in the stem region and one (sheared) basepair in the loop. At the 55 Å window, all hydrogen bonds of the four basepairs had broken in all three hairpins and in all simulations. This point was chosen as the transition to unfolded state.
The free energy changes of folding are given in Table 1. The errors are calculated as the standard deviation of the three (or four for mutant hairpins) independent calculations. As can be seen from Table 1, ff99 was able to predict the correct order of stabilities of the three hairpins. The native hairpin has three GC basepairs, in mutant1 one of the GC pair is replaced by a weaker AU and mutant2 has two of the GCs replaced by weaker AU pairs, therefore the native hairpin is more stable than mutant1 which in turn is more stable than mutant2.
Table 1.
Free energy change between native and stretched RNA hairpins calculated using the umbrella sampling simulations in units of kcal/mol.
| Native | Mutant1 | Mutant2 | |
|---|---|---|---|
| Run1 | −20.5 | −17.7 | −15.0 |
| Run2 | −19.7 | −17.8 | −15.4 |
| Run3 | −20.6 | −13.2 | −16.3 |
| Run4 | −16.3 | −12.5 | |
|
| |||
| Average | −20.2 ± 0.5 | −16.2 ± 2.1 | −14.8 ± 1.6 |
Three independent calculations were performed for native hairpin and four for mutant1 and mutant2. The reported error of the average values is the standard deviation over the independent calculations.
Calculating the free energy of unfolding using the nearest neighbor parameters
The nearest neighbor methodology was developed to predict the folding stability of nucleic acid secondary structure.45, 76–77 It is based on sets of empirical rules derived from optical melting experiments. Stability of the secondary structure elements depends on the sequence of the motif and the sequence of adjacent basepairs. The overall folding free energy change of a motif is a sum of contributions of all individual basepair increments. All the parameters have been gathered in a web-based resource for easy access.50
Nearest neighbor parameters are accurate for predicting the stabilities of Watson-Crick helices, with the errors in the individual increments on the order of 0.1 kcal/mol.45 The error in other structural elements, such as loops, is generally larger, at about 0.5 kcal/mol.77 The loop region is common to all three hairpins so to avoid including the larger error terms in the calculation, the free energy change of melting of native hairpin59 and the nearest neighbor rules for the free energy change of helices were used to calculate the free energy changes of melting the mutant hairpins stem-loops. From the definition of the nearest neighbor model, free energy change of forming a hairpin at a temperature T is:
| (2) |
so the free energy change of the loop region is independent of the nearest neighbor free energy change of the stem region. The free energy changes of the mutant hairpins were calculated by subtracting the stem free energy change of the native hairpin from the experimentally determined free energy change and then adding the appropriate free energy changes for stem formations for mutant1 or mutant2.
The nearest neighbor parameters were derived using 1 M NaCl, while the folding free energy change of the umbrella sampling calculations and the optical melting of the native hairpin were measured in 0.1M NaCl. The salt correction for the free energy changes calculated using the nearest neighbor parameters, however, is sequence independent.77–78 Therefore, the salt correction cancels when using the thermodynamic cycle and the nearest neighbor parameters can be used directly.
Table 2 lists the free energy changes of folding for the three hairpins calculated using the nearest neighbor rules. The errors were calculated using the errors values provided with the parameters in the nearest neighbor database50 and taking into account that the errors of enthalpy, entropy and free energy are correlated (see Supporting Information).44
Comparison between simulations and nearest neighbor model free energies
The denatured state of optical melting experiments is a random coil conformation, while the umbrella sampling simulations produce an extended conformation similar to the final product of single molecule pulling experiments. Therefore, the free energy changes calculated using the two methods cannot be directly compared without accounting for the different end states. The free energy changes determined by optical melting are an order of magnitude smaller than the free energy changes from umbrella sampling simulations. One reason for this difference comes from the work that would be required to stretch a random coil (melted RNA in the optical melting experiments) with its many different conformations into a linear form (final state of RNA in the umbrella sampling simulations, and single molecule pulling experiments), thus reducing the molecule’s entropy. Replica exchange molecular dynamics79 would sample the same end states as the optical melting experiments, but was not used here because of the difficulties of adequately sampling the random coil state. An approximation for the free energy of stretching can be obtained from the polymer theory of flexible chains, specifically from the worm-like chain (WLC) model.80 This model is commonly used for comparing single molecule pulling experiments using optical tweezers to optical melting experiments.49, 81 According to the WLC model free energy of stretching can be approximated as:81–82
| (3) |
where R is the gas constant, T is temperature, L is the contour length of the polymer, X is the end-to-end distance and P is the persistence length. Free energy of stretching is directly proportional to the contour length and inversely proportional to the persistence length.49, 81 Using 5.9Å as a contour length per nucleotide, and P=10 Å, values commonly used for single stranded RNA molecules,49, 81 the predicted free energy change to stretch a 12 nucleotide hairpin to the end-to-end distance of 55 Å is 4.1 kcal/mol. This value is clearly much smaller than the difference between simulation and melting experiments, which are on the order of 10 kcal/mol (see Tables 1 and 2). So while the polymer theory can give a quantitative explanation of the processes involved, the predictive values are poor.
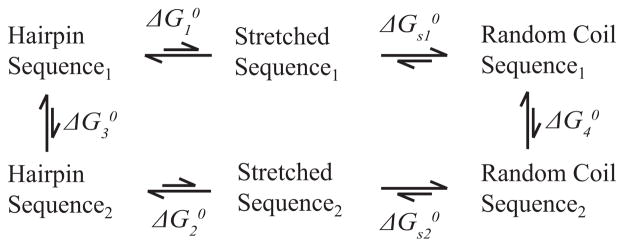
In order to make a meaningful comparison between the simulations and optical melting, a thermodynamic cycle can be used. Figure 4 shows a thermodynamic cycle between two hairpins. Separate umbrella sampling calculations were performed for each sequence yielding and . Then, by the nature of the thermodynamic cycle:
| (4) |
where ΔΔG0 is the free energy difference in stability for a change in sequence. If the stretching free energy change between the stretched state and the random coil are identical, then the ΔΔG0 for the chemical transformation calculated by nearest neighbor rules is the same as the differences calculated by molecular dynamics:
| (5) |
Figure 4.
Thermodynamic cycle for transition between two hairpins. The difference in and determined by umbrella sampling, is equal to the difference between and , which can be accurately predicted using nearest neighbor parameters if the free energy changes required to stretch the hairpins from the random coil to an extended conformation ( and ) are sequence-independent, an approximation which is routinely made in single molecule stretching experiments.
The approximation that stretching entropies are sequence-independent is commonly used in single molecule stretching experiments.49, 81
Similar thermodynamic cycle calculations were performed for all possible combinations, i.e. between native and mutant1, native and mutant2 hairpins and between mutant1 and mutant2 hairpins. In addition to removing the influence of stretching entropy, the thermodynamic cycle also reduces the effect of approximations in the molecular dynamics force field. For example, inaccuracies in the backbone torsional parameters, that may be prominent in the loop regions,31–32 might cancel by taking the difference between two potential of mean force calculations.
Table 3 shows the comparison of free energy changes when going from native to mutant1, native to mutant2 and mutant1 to mutant2 hairpin obtained by applying the thermodynamic cycle on the free energies of folding of all three hairpins calculated using either umbrella sampling simulations or nearest neighbor data. The differences between the simulation and the reference data were 1.8, 1.2 and −0.6 kcal/mol for the mutant1 to native, mutant2 to native and mutant2 to mutant1 transitions, respectively. All three values were within error bounds of umbrella sampling results.
Table 3.
Free energy change in kcal/mol for transition from native to mutant1, native to mutant2 and mutant1 to mutant2 hairpins calculated using a thermodynamic cycle.
| Native ← Mutant1 | Native ← Mutant2 | Mutant1 ← Mutant2 | |
|---|---|---|---|
| Simulation | −4.0 ± 2.2 | −5.5 ± 1.7 | −1.5 ± 2.7 |
| Nearest Neighbor | −2.2 ± 0.2 | −4.3 ± 0.2 | −2.1 ± 0.2 |
|
| |||
| Difference | 1.8 | 1.2 | −0.6 |
CONCLUSION
RNA was discovered to play many important roles in the biology, far beyond simply being the carrier of genetic information. Molecular dynamics can play an important role in examining its structure and dynamics, so it is important to assess the accuracy of various MD force fields that are commonly used. In this, work the ability of Amber ff99 force field to predict relative free energies of RNA helix formation was examined. Three hexaloop hairpins, with differing closing helices, were used as our test systems. The native hairpin was a solution structure42 and two mutants were made by replacing the GC basepairs of the stem region of native hairpin with AU pairs. Multiple umbrella sampling simulations on all hairpins were performed to determine the change in free energy change as the hairpins were stretched from a native form to an elongated conformation. Stretching the hexaloop hairpin produces several distinct areas along the free energy change curve. First, there was a native conformation basin, then a region where the hydrogen bonds in the stem are gradually broken. The third region was characterized by a sudden increase as the last hydrogen bonds in the stem were broken. In the fourth region the hydrogen bonds in the loop region were gradually broken. Finally, the fifth region is characterized by a steep increase of free energy slope due to the stretching of bonds and angles. These areas generally correspond to the areas observed in stretching the tetraloop hairpins23 although the transitions are not as pronounced due to the larger stiffness of the tetraloops.50 In this work, the unfolded state was considered to be only those structures that do not have any of the hydrogen bonds found in the native structure. This facilitates the comparison of the folding free energy changes to optical melting experiments, where only the native and random coil states are considered.83
The Amber ff99 force field was able to predict the correct order of helix formation stabilities as shown in Tables 1 and 2. Absolute values were, however, different from the values determined from optical melting experiments and nearest neighbor rules. This was a consequence of different end states in the two approaches. The end state in optical melting is a random coil, while umbrella sampling produces extended conformations. There was a large entropic cost to extend a random coil to a linear form. Polymer theory was used to try to estimate this effect, but the predictions are only qualitative, in part because the polymer model probably only works well once the strands are relatively long, such as those used as adapters in optical tweezer experiments.49, 81 The stretching entropy was not considered in prior unfolding studies of tetraloops.23–24 Another reason for the discrepancy between the free energy calculated from umbrella sampling simulations and the sum of nearest neighbor and stretching free energy could be the inaccuracy of the ff99 force field. Free simulations of the hairpins show a relatively large RMSD of the loop region. This can affect the accuracy of absolute free energies of folding calculated from umbrella sampling. All three hairpins, however, have the same loop region, and the inaccuracies in free energy coming from this region should cancel when relative free energies are calculated. This is indeed the case as can be seen from Table 3. There is also an advantage to performing molecular pulling computations; the required sampling time is significantly less than other methods, such as replica exchange, that require sampling of a random coil conformation.
The calculations were compared to experiments via a thermodynamic cycle (Table 3). With this approach, Amber’s ff99 was able to predict relative free energies of RNA helix formation with accuracy within the error bounds of the simulations. This is the first demonstration that molecular mechanics can reproduce nearest neighbor parameters. QM calculations84–85 have been used to predict the stacking energies of different di-nucleotide steps. Parameters for stacking interactions have also been extracted from the PDB database and shown to be of comparable accuracy to the thermodynamic derived parameters.86
The agreement with experiment is a testament to the Amber ff99 force field’s28–29 ability to accurately predict free energy difference of RNA helix formation. A few comments are in order regarding these results. The magnitude of errors of the free energy changes from the simulations for the mutant hairpins are much larger than the native hairpin error (see Table 1). This is a consequence of the larger flexibility of the mutant hairpins. The native hairpin has three GC basepairs in the stem region, mutant1 replaces the middle GC with the AU pair and mutant2 replaces both middle and first GC with an AU pairs. More flexible molecules can explore larger areas of conformational space during the sampling and therefore take longer to converge. This was addressed by running an additional umbrella sampling run for mutant1 and mutant2 hairpins, but it appears that to get higher precision, even more simulations were needed.
Recently the improvements to α/γ backbone torsions derived for DNAs30 were shown to work for RNAs as well,32, 53 and also two new sets of glycosidic torsion31–32 parameters have been derived. These new parameters, when replacing the respective ff99 parameters, showed an improved agreement with the experimental structures in long simulations.31–32, 53 This work was started before the publication of these findings, so the revised force fields were not included in these calculations. The total change to the force field ff99 was not large, however, and their apparent effect would be further reduced in our results due to the fact that relative free energy changes are determined. Still, based on the already published tests of the new parameters, a better agreement might be expected in predicting the relative free energies of helix formation.
Despite these recent improvements, the Amber RNA force fields still have documented deficiencies, such as the inability to properly describe the structural characteristics of loop regions of hairpins,53 or to properly predict the free energy changes of conformational changes.21 Therefore, it is important that this work shows agreement in free energy changes for helices. As shown here, Amber’s ff99 can accurately predict the relative free energies of helix formation. This suggests that, by comparing the relative free energies, inaccuracies can cancel out and still produce accurate final results. This also suggests that the modeling of A-form helices by the Amber force field may be more accurate than the modeling of loop structures.
Supplementary Material
Acknowledgments
This work was funded in part by National Institutes of Health Grant R01HG004002 to D.H.M. Computer time was provided by the University of Rochester Center for Integrated Research Computing.
Footnotes
Supporting Information Available. The supporting information contains the procedure describing calculation of the errors in the nearest neighbor free energy calculations, figures of RMSD vs. time for the free simulations of native and mutant hairpins, figures of RMSD vs. time that show the stability of mutant conformations in individual windows of umbrella sampling simulations and figures showing the convergence of umbrella sampling simulations. This information is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Doudna JA, Cech TR. The chemical repertoire of natural ribozymes. Nature. 2002;418:222–228. doi: 10.1038/418222a. [DOI] [PubMed] [Google Scholar]
- 2.Nissen P, Hansen J, Ban N, Moore PB, Steitz TA. The structural basis of ribosome activity in peptide bond synthesis. Science. 2000;289:920–930. doi: 10.1126/science.289.5481.920. [DOI] [PubMed] [Google Scholar]
- 3.Scott WG. Ribozymes. Curr Opin Struct Biol. 2007;17:280–286. doi: 10.1016/j.sbi.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 4.Tucker BJ, Breaker RR. Riboswitches as versatile gene control elements. Curr Opin Struct Biol. 2005;15:342–348. doi: 10.1016/j.sbi.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 5.Walter P, Blobel G. Signal recognition particle contains a 7s RNA essential for protein translocation across the endoplasmicreticulum. Nature. 1982;299:691–698. doi: 10.1038/299691a0. [DOI] [PubMed] [Google Scholar]
- 6.Bachellerie JP, Cavaille J, Huttenhofer A. The expanding snoRNA world. Biochimie. 2002;84:775–790. doi: 10.1016/s0300-9084(02)01402-5. [DOI] [PubMed] [Google Scholar]
- 7.Wahl MC, Will CL, Luhrmann R. The spliceosome: Design principles of a dynamic RNP machine. Cell. 2009;136:701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 8.David L, Huber W, Granovskaia M, Toedling J, Palm CJ, Bofkin L, Jones T, Davis RW, Steinmetz LM. A high-resolution map of transcription in the yeast genome. Proc Natl Acad Sci USA. 2006;103:5320–5325. doi: 10.1073/pnas.0601091103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee H, Darden T, Pedersen L. Accurate crystal molecular dynamics simulations using particle-mesh-Ewald - RNA dinucleotides - ApU and GpC. Chem Phys Lett. 1995;243:229–235. [Google Scholar]
- 10.Auffinger P, Louise-May S, Westhof E. Molecular dynamics simulations of solvated yeast tRNA(Asp) Biophys J. 1999;76:50–64. doi: 10.1016/S0006-3495(99)77177-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Auffinger P, Bielecki L, Westhof E. The Mg2+ binding sites of the 5S rRNA loop E motif as investigated by molecular dynamics simulations. Chem Biol. 2003;10:551–561. doi: 10.1016/s1074-5521(03)00121-2. [DOI] [PubMed] [Google Scholar]
- 12.Krasovska MV, Sefcikova J, Reblova K, Schneider B, Walter NG, Sponer J. Cations and hydration in catalytic RNA: Molecular dynamics of the hepatitis delta virus ribozyme. Biophys J. 2006;91:626–638. doi: 10.1529/biophysj.105.079368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rhodes MM, Reblova K, Sponer J, Walter NG. Trapped water molecules are essential to structural dynamics and function of a ribozyme. Proc Natl Acad Sci USA. 2006;103:13380–13385. doi: 10.1073/pnas.0605090103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Auffinger P, Westhof E. Water and ion binding around RNA and DNA (C,G) oligomers. J Mol Biol. 2000;300:1113–1131. doi: 10.1006/jmbi.2000.3894. [DOI] [PubMed] [Google Scholar]
- 15.Auffinger P, Westhof E. RNA solvation: A molecular dynamics simulation perspective. Biopolymers. 2000;56:266–274. doi: 10.1002/1097-0282(2000)56:4<266::AID-BIP10027>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 16.Sklenovsky P, Florova P, Banas P, Reblova K, Lankas F, Otyepka M, Sponer J. Understanding RNA Flexibility Using Explicit Solvent Simulations: The Ribosomal and Group I Intron Reverse Kink-Turn Motifs. J Chem Theory Comput. 2011;7:2963–2980. doi: 10.1021/ct200204t. [DOI] [PubMed] [Google Scholar]
- 17.Noy A, Soteras I, Luque FJ, Orozco M. The impact of monovalent ion force field model in nucleic acids simulations. Phys Chem Chem Phys. 2009;11:10596–10607. doi: 10.1039/b912067j. [DOI] [PubMed] [Google Scholar]
- 18.Wong KY, Lee TS, York DM. Active Participation of the Mg(2+) Ion in the Reaction Coordinate of RNA Self-Cleavage Catalyzed by the Hammerhead Ribozyme. J Chem Theory Comput. 2011;7:1–3. doi: 10.1021/ct100467t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aci S, Mazier S, Genest D. Conformational pathway for the kissing complex - Extended dimer transition of the SL1 stem-loop from genomic HIV-1 RNA as monitored by targeted molecular dynamics techniques. J Mol Biol. 2005;351:520–530. doi: 10.1016/j.jmb.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 20.Mathews DH, Case DA. Nudged elastic band calculation of minimal energy paths for the conformational change of a GG noncanonical pair. J Mol Biol. 2006;357:1683–1693. doi: 10.1016/j.jmb.2006.01.054. [DOI] [PubMed] [Google Scholar]
- 21.Van Nostrand KP, Kennedy SD, Turner DH, Mathews DH. Molecular mechanics investigation of an adenine-adenine noncanonical pair conformational change. J Chem Theory Comput. 2011;7:3779–3792. doi: 10.1021/ct200223q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reblova K, Strelcova Z, Kulhanek P, Besseova I, Mathews DH, Van Nostrand K, Yildirim I, Turner DH, Sponer J. An RNA molecular switch: Intrinsic flexibility of 23S rRNA helices 40 and 68 5′-UAA/5′-GAN internal loops studied by molecular dynamics methods. J Chem Theory Comput. 2010;6:910–929. [PMC free article] [PubMed] [Google Scholar]
- 23.Deng NJ, Cieplak P. Free energy profile of RNA hairpins: A molecular dynamics simulation study. Biophys J. 2010;98:627–636. doi: 10.1016/j.bpj.2009.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Denning EJ, Priyakumar UD, Nilsson L, Mackerell AD., Jr Impact of 2′-hydroxyl sampling on the conformational properties of RNA: Update of the CHARMM all-atom additive force field for RNA. J Comput Chem. 2011;32:1929–1943. doi: 10.1002/jcc.21777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brooks BR, Brooks CL, Mackerell AD, Nilsson L, Petrella RJ, Roux B, Won Y, Archontis G, Bartels C, Boresch S, Caflisch A, Caves L, Cui Q, Dinner AR, Feig M, Fischer S, Gao J, Hodoscek M, Im W, Kuczera K, Lazaridis T, Ma J, Ovchinnikov V, Paci E, Pastor RW, Post CB, Pu JZ, Schaefer M, Tidor B, Venable RM, Woodcock HL, Wu X, Yang W, York DM, Karplus M. CHARMM: The Biomolecular Simulation Program. J Comput Chem. 2009;30:1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Case DA, Darden TA, Cheatham TE, Simmerling CL, Wang JM, Duke RE, Luo R, Walker RC, Zhang W, Merz KM, Roberts B, Wang B, Hayik S, Roitberg A, Seabra G, Kolossvai I, Wong KF, Paesani F, Vanicek J, Liu J, Wu X, Brozell SR, Steinbrecher T, Gohlke H, Cai Q, Ye X, Wang J, Hsieh MJ, Cui G, Roe DR, Mathews DH, Seetin MG, Sagui C, Babin V, Luchko T, Gusarov S, Kovalenko A, Kollman PA. AMBER. Vol. 11. University of California; San Francisco, CA: 2010. [Google Scholar]
- 27.Cornell WD, Cieplak P, Bayly CI, Gould IR, Merz KM, Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, Kollman PA. A second generation force field for the simulation of proteins, nucleic acids, and organic molecules. J Am Chem Soc. 1995;117:5179–5197. [Google Scholar]
- 28.Cheatham TE, Cieplak P, Kollman PA. A modified version of the Cornell et al. force field with improved sugar pucker phases and helical repeat. J Biomol Struct Dyn. 1999;16:845–862. doi: 10.1080/07391102.1999.10508297. [DOI] [PubMed] [Google Scholar]
- 29.Wang JM, Cieplak P, Kollman PA. How well does a restrained electrostatic potential (RESP) model perform in calculating conformational energies of organic and biological molecules? J Comput Chem. 2000;21:1049–1074. [Google Scholar]
- 30.Perez A, Marchan I, Svozil D, Sponer J, Cheatham TE, Laughton CA, Orozco M. Refinenement of the AMBER force field for nucleic acids: Improving the description of alpha/gamma conformers. Biophys J. 2007;92:3817–3829. doi: 10.1529/biophysj.106.097782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yildirim I, Stern HA, Kennedy SD, Tubbs JD, Turner DH. Reparameterization of RNA chi torsion parameters for the AMBER force field and comparison to NMR spectra for cytidine and uridine. J Chem Theory Comput. 2010;6:1520–1531. doi: 10.1021/ct900604a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zgarbova M, Otyepka M, Sponer J, Mladek A, Banas P, Cheatham TE, Jurecka P. Refinement of the Cornell et al. nucleic acids force field based on reference quantum chemical calculations of glycosidic torsion profiles. J Chem Theory Comput. 2011;7:2886–2902. doi: 10.1021/ct200162x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Foloppe N, MacKerell AD. All-atom empirical force field for nucleic acids: I. Parameter optimization based on small molecule and condensed phase macromolecular target data. J Comput Chem. 2000;21:86–104. [Google Scholar]
- 34.Hart K, Foloppe N, Baker CM, Denning EJ, Nilsson L, MacKerell AD. Optimization of the CHARMM Additive Force Field for DNA: Improved Treatment of the BI/BII Conformational Equilibrium. J Chem Theory Comput. 2012;8:348–362. doi: 10.1021/ct200723y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sponer J, Jurecka P, Hobza P. Accurate interaction energies of hydrogen-bonded nucleic acid base pairs. J Am Chem Soc. 2004;126:10142–10151. doi: 10.1021/ja048436s. [DOI] [PubMed] [Google Scholar]
- 36.Reha D, Kabelac M, Ryjacek F, Sponer J, Sponer JE, Elstner M, Suhai S, Hobza P. Intercalators. 1. Nature of stacking interactions between intercalators (ethidium, daunomycin, ellipticine, and 4′,6-diaminide-2-phenylindole) and DNA base pairs. Ab initio quantum chemical, density functional theory, and empirical potential study. J Am Chem Soc. 2002;124:3366–3376. doi: 10.1021/ja011490d. [DOI] [PubMed] [Google Scholar]
- 37.Sponer JE, Leszczynski J, Sychrovsky V, Sponer J. Sugar edge/sugar edge base pairs in RNA: Stabilities and structures from quantum chemical calculations. J Phys Chem B. 2005;109:18680–18689. doi: 10.1021/jp053379q. [DOI] [PubMed] [Google Scholar]
- 38.Sponer JE, Spackova N, Leszczynski J, Sponer J. Principles of RNA base pairing: Structures and energies of the trans Watson-Crick/sugar edge base pairs. J Phys Chem B. 2005;109:11399–11410. doi: 10.1021/jp051126r. [DOI] [PubMed] [Google Scholar]
- 39.Jurecka P, Sponer J, Cerny J, Hobza P. Benchmark database of accurate (MP2 and CCSD(T) complete basis set limit) interaction energies of small model complexes, DNA base pairs, and amino acid pairs. Phys Chem Chem Phys. 2006;8:1985–1993. doi: 10.1039/b600027d. [DOI] [PubMed] [Google Scholar]
- 40.Sharma P, Sponer JE, Sponer J, Sharma S, Bhattacharyya D, Mitra A. On the role of the cis Hoogsteen:sugar-edge family of base pairs in platforms and triplets-quantum chemical insights into RNA structural biology. J Phys Chem B. 2010;114:3307–3320. doi: 10.1021/jp910226e. [DOI] [PubMed] [Google Scholar]
- 41.Kolar M, Berka K, Jurecka P, Hobza P. On the reliability of the AMBER force field and its empirical dispersion contribution for the description of noncovalent complexes. ChemPhysChem. 2010;11:2399–2408. doi: 10.1002/cphc.201000109. [DOI] [PubMed] [Google Scholar]
- 42.Fountain MA, Serra MJ, Krugh TR, Turner DH. Structural features of a six-nucleotide RNA hairpin loop found in ribosomal RNA. Biochemistry. 1996;35:6539–6548. doi: 10.1021/bi952697k. [DOI] [PubMed] [Google Scholar]
- 43.Schuwirth BS, Borovinskaya MA, Hau CW, Zhang W, Vila-Sanjurjo A, Holton JM, Cate JH. Structures of the bacterial ribosome at 3.5 angstrom resolution. Science. 2005;310:827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- 44.Bevington PR, Robinson DK. Data reduction and error analysis for the physical sciences. 3. McGraw-Hill; Boston, MA: 2003. pp. 36–50. [Google Scholar]
- 45.Xia T, SantaLucia J, Jr, Burkard ME, Kierzek R, Schroeder SJ, Jiao X, Cox C, Turner DH. Thermodynamic parameters for an expanded nearest-neighbor model for formation of RNA duplexes with Watson-Crick base pairs. Biochemistry. 1998;37:14719–14735. doi: 10.1021/bi9809425. [DOI] [PubMed] [Google Scholar]
- 46.Kumar S, Bouzida D, Swendsen RH, Kollman PA, Rosenberg JM. The weighted histogram analysis method for freeenergy calculations on biomolecules. 1. The method. J Comput Chem. 1992;13:1011–1021. [Google Scholar]
- 47.Kumar S, Rosenberg JM, Bouzida D, Swendsen RH, Kollman PA. Multidimensional free-energy calculations using the weighted histogram analysis method. J Comput Chem. 1995;16:1339–1350. [Google Scholar]
- 48.Roux B. The calculation of the potential of mean force using computer simulations. Comput Phys Commun. 1995;91:275–282. [Google Scholar]
- 49.Bustamante C. Unfolding single RNA molecules: bridging the gap between equilibrium and non-equilibrium statistical thermodynamics. Q Rev Biophys. 2005;38:291–301. doi: 10.1017/S0033583506004239. [DOI] [PubMed] [Google Scholar]
- 50.Turner DH, Mathews DH. NNDB: the nearest neighbor parameter database for predicting stability of nucleic acid secondary structure. Nucleic Acids Res. 2010;38:D280–D282. doi: 10.1093/nar/gkp892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xing YY, Draper DE. Stabilization of a ribosomal-RNA tertiary structure by ribosomal-protein L11. J Mol Biol. 1995;249:319–331. doi: 10.1006/jmbi.1995.0299. [DOI] [PubMed] [Google Scholar]
- 52.Gutell RR, Gray MW, Schnare MN. A compilation of large subunit (23s and 23s-like) ribosomal RNA structures: 1993. Nucleic Acids Res. 1993;21:3055–3074. doi: 10.1093/nar/21.13.3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Banas P, Hollas D, Zgarbova M, Jurecka P, Orozco M, Cheatham TE, Sponer J, Otyepka M. Performance of molecular mechanics force fields for RNA simulations: Stability of UUCG and GNRA hairpins. J Chem Theory Comput. 2010;6:3836–3849. doi: 10.1021/ct100481h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gong Z, Zhao YJ, Xiao Y. RNA stability under different combinations of Amber force fields and solvation models. J Biomol Struct Dyn. 2010;28:431–441. doi: 10.1080/07391102.2010.10507372. [DOI] [PubMed] [Google Scholar]
- 55.Riccardi L, Nguyen PH, Stock G. Free-energy landscape of RNA hairpins constructed via dihedral angle principal component analysis. J Phys Chem B. 2009;113:16660–16668. doi: 10.1021/jp9076036. [DOI] [PubMed] [Google Scholar]
- 56.DePaul AJ, Thompson EJ, Patel SS, Haldeman K, Sorin EJ. Equilibrium conformational dynamics in an RNA tetraloop from massively parallel molecular dynamics. Nucleic Acids Res. 2010;38:4856–4867. doi: 10.1093/nar/gkq134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Joli F, Hantz E, Hartmann B. Structure and dynamics of phosphate linkages and sugars in an abasic hexaloop RNA hairpin. Biophys J. 2006;90:1480–1488. doi: 10.1529/biophysj.105.070862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Joli F, Bouchemal N, Laigle A, Hartmann B, Hantz E. Solution structure of a purine rich hexaloop hairpin belonging to PGY/MDR1 mRNA and targeted by antisense oligonucleotides. Nucleic Acids Res. 2006;34:5740–5751. doi: 10.1093/nar/gkl617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Serra MJ, Axenson TJ, Turner DH. A model for the stabilities of RNA hairpins based on a study of the sequence dependence of stability for hairpins of 6 nucleotides. Biochemistry. 1994;33:14289–14296. doi: 10.1021/bi00251a042. [DOI] [PubMed] [Google Scholar]
- 60.Vecenie CJ, Morrow CV, Zyra A, Serra MJ. Sequence dependence of the stability of RNA hairpin molecules with six nucleotide loops. Biochemistry. 2006;45:1400–1407. doi: 10.1021/bi051750u. [DOI] [PubMed] [Google Scholar]
- 61.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 1983;79:926–935. [Google Scholar]
- 62.Chandrasekhar S. Stochastic problems in physics and astronomy. Rev Mod Phys. 1943;15:1. [Google Scholar]
- 63.Berendsen HJC, Postma JPM, Vangunsteren WF, Dinola A, Haak JR. Molecular dynamics with coupling to an external bath. J Chem Phys. 1984;81:3684–3690. [Google Scholar]
- 64.Darden T, York D, Pedersen L. Particle mesh Ewald: An N.Log(N) method for Ewald sums in large systems. J Chem Phys. 1993;98:10089–10092. [Google Scholar]
- 65.Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LG. A smooth particle mesh Ewald method. J Chem Phys. 1995;103:8577–8593. [Google Scholar]
- 66.Ryckaert JP, Ciccotti G, Berendsen HJC. Numericalintegration of cartesian equations of motion of a system with constraints - molecular-dynamics of N-alkanes. J Comput Phys. 1977;23:327–341. [Google Scholar]
- 67.Hawkins GD, Cramer CJ, Truhlar DG. Pairwise solute descreening of solute charges from a dielectric medium. Chem Phys Lett. 1995;246:122–129. [Google Scholar]
- 68.Hawkins GD, Cramer CJ, Truhlar DG. Parametrized models of aqueous free energies of solvation based on pairwise descreening of solute atomic charges from a dielectric medium. J Phys Chem. 1996;100:19824–19839. [Google Scholar]
- 69.Tsui V, Case DA. Theory and applications of the generalized Born solvation model in macromolecular simulations. Biopolymers. 2000;56:275–291. doi: 10.1002/1097-0282(2000)56:4<275::AID-BIP10024>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 70.Aqvist J. Ion Water Interaction Potentials Derived from Free- Energy Perturbation Simulations. J Phys Chem. 1990;94:8021–8024. [Google Scholar]
- 71.Dang LX. Mechanism and Thermodynamics of Ion Selectivity in Aqueous-Solutions of 18-Crown-6 Ether - a Molecular-Dynamics Study. J Am Chem Soc. 1995;117:6954–6960. [Google Scholar]
- 72.Grossfield A. WHAM: the weighted histogram analysis method. Vol. 205. Grossfield Lab, University of Rochester Medical Center; Rochester, NY: http://membrane.urmc.rochester.edu/content/wham. [Google Scholar]
- 73.Cantor CR, Schimmel PR. Biophysical Chemistry. W.H. Freeman and Company; New York: 1980. pp. 1109–1182. [Google Scholar]
- 74.Bloomfield VA, Crothers DM, Tinoco IJ. Nucleic Acids: Structures, Properties and Fuctions. University Science Books; Sausalito, CA: 2000. pp. 259–334. [Google Scholar]
- 75.Diamond JM, Turner DH, Mathews DH. Thermodynamics of three-way multibranch loops in RNA. Biochemistry. 2001;40:6971–6981. doi: 10.1021/bi0029548. [DOI] [PubMed] [Google Scholar]
- 76.Mathews DH, Sabina J, Zuker M, Turner DH. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J Mol Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- 77.Mathews DH, Disney MD, Childs JL, Schroeder SJ, Zuker M, Turner DH. Incorporating chemical modification constraints into a dynamic programming algorithm for prediction of RNA secondary structure. Proc Natl Acad Sci USA. 2004;101:7287–7292. doi: 10.1073/pnas.0401799101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.SantaLucia J. A unified view of polymer, dumbbell, and oligonucleotide DNA nearest-neighbor thermodynamics. Proc Natl Acad Sci USA. 1998;95:1460–1465. doi: 10.1073/pnas.95.4.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mitsutake A, Sugita Y, Okamoto Y. Generalized-ensemble algorithms for molecular simulations of biopolymers. Biopolymers. 2001;60:96–123. doi: 10.1002/1097-0282(2001)60:2<96::AID-BIP1007>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 80.Flory PJ. Statistical Mechanics of Chain Molecules. Interscience Publishers; New York: 1969. pp. 401–410. [Google Scholar]
- 81.Tinoco I, Li PTX, Bustamante C. Determination of thermodynamics and kinetics of RNA reactions by force. Q Rev Biophys. 2006;39:325–360. doi: 10.1017/S0033583506004446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bustamante C, Marko JF, Siggia ED, Smith S. Entropic elasticity of lambda-phage DNA. Science. 1994;265:1599–1600. doi: 10.1126/science.8079175. [DOI] [PubMed] [Google Scholar]
- 83.Schroeder SJ, Turner DH. Optical melting measurements of nucleic acid thermodynamics. Methods Enzymol. 2009;468:371–387. doi: 10.1016/S0076-6879(09)68017-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Svozil D, Hobza P, Sponer J. Comparison of intrinsic stacking energies of ten unique dinucleotide steps in A-RNA and BDNA duplexes. Can we determine correct order of stability by quantum-chemical calculations? J Phys Chem B. 2010;114:1191–1203. doi: 10.1021/jp910788e. [DOI] [PubMed] [Google Scholar]
- 85.Johnson CA, Bloomingdale RJ, Ponnusamy VE, Tillinghast CA, Znosko BM, Lewis M. Computational model for predicting experimental RNA and DNA nearest-neighbor free energy rankings. J Phys Chem B. 2011;115:9244–9251. doi: 10.1021/jp2012733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dima RI, Hyeon C, Thirumalai D. Extracting stacking interaction parameters for RNA from the data set of native structures. J Mol Biol. 2005;347:53–69. doi: 10.1016/j.jmb.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 87.Leontis NB, Westhof E. Geometric nomenclature and classification of RNA base pairs. RNA. 2001;7:499–512. doi: 10.1017/s1355838201002515. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.