Abstract
At least eight families of mammalian patatin-like phospholipase domain-containing proteins (PNPLA) (E.C. 3.1.1.3) catalyse the hydrolysis of triglycerides, including PNPLA4 (alternatively PLPL4 or GS2), which also acts as a retinol transacylase and participates in retinol-ester metabolism in the body. Bioinformatic methods were used to predict the amino acid sequences, secondary and tertiary structures and gene locations for PNPLA4 genes and encoded proteins using data from several vertebrate genome projects. PNPLA4 genes were located on the X-chromosome for the eutherian mammalian genomes examined. Opossum (marsupial), chicken, anole lizard, clawed toad, zebrafish and lancelet PNPLA4 genes were also identified. Most vertebrate PNPLA4 genes typically contained six coding exons whereas the lancelet PNPLA4 gene contained five coding exons. PNPLA4 subunits were the smallest among the PNPLA-like proteins examined containing 252–255 residues, shared >64 % sequence identities and key amino acid residues and predicted motifs, including ‘patatin’ (residues 6–176); putative catalytic dyad active site residues, Ser43 and Asp163; oxy-anion ‘hole’ residues (10–15); and conserved serine residues, which may perform structural roles for this enzyme. Predicted tertiary structures for PNPLA4 ‘patatin’ were similar to those reported for potato ‘patatin’, suggesting that it is strongly conserved during evolution. Human PNPLA4 contained a CpG49 island within the gene promoter, a miRNA-186 binding site within the mRNA 3′-noncoding region for the PNPLA4b isoform and exhibited wide tissue expression at a higher than average level. These and previous studies of vertebrate PNPLA-like gene families have suggested that PNPLA4 is an ancient gene in evolution which has resulted from a duplication of an ancestral invertebrate ATGL-like gene (encoding adipose triglyceride lipase).
Electronic supplementary material
The online version of this article (doi:10.1007/s13205-012-0063-7) contains supplementary material, which is available to authorized users.
Keywords: Patatin-like phospholipase domain containing proteins, Vertebrate PNPLA4, PNPLA4, X-chromosome, Comparative studies
Introduction
At least eight mammalian patatin-like phospholipase domain-containing proteins (PNPLA-like) (E.C. 3.1.1.3) and genes have been reported which encode patatin-motif containing lipases (Wilson et al. 2006; Kienesberger et al. 2009; Holmes 2012). Human PNPLA4 (also designated as PLPL4 or GS2) is localized on the X-chromosome at Xp22.3, between the genes for steroid sulfatase (STS) and Kallman syndrome (KAL) (Lee et al. 1994). Other human PNPLA-like genes are separately localized on the human genome, including PNPLA1 (chromosome 6) (Mungall et al. 2003), ATGL (also called PNPLA2 or adipose triglyceride lipase) (chromosome 11), PNPLA3 (PLPL3) and PNPLA5 (PLPL5) (chromosome 22) (Schoenborn et al. 2006), PNPLA6 (PLPL6 or NTE) (neuropathy target esterase) (chromosome 19) (Lush et al. 1998; Grimwood et al. 2004), PNPLA7 (PLPL7) (chromosome 9) (Humphray et al. 2009) and PNPLA8 (PLPL8 or IPLA2G) (calcium-independent phospholipase A2-gamma) (chromosome 7) (Tanaka et al. 2000; Mancuso et al. 2000; Scherer et al. 2005).
PNPLA4 catalyses the hydrolysis of triglycerides and participates in retinol-ester metabolism in the body, with a specific role reported for this enzyme in the epidermis in regulating access to retinol from retinol-ester storage depots (Kienesberger et al. 2009; Gao and Simon 2005; Gao et al. 2009). Retinol and related retinoid compounds play key roles in the body including supporting vision (Palczewski 2011), regulating epithelial cell growth and differentiation (Long et al. 2010), contributing to the growth of bone tissue (Oki et al. 2008), immune function (Pino-Lagos et al. 2010) and the activation of tumor suppressor genes (Ye et al. 2009). This retinol-ester metabolic role is in contrast to functions reported for other PNPLA-like enzymes including ATGL (or adipose triglyceride lipase) in triglyceride hydrolysis in adipocyte and non-adipocyte lipid droplets (Zimmermann et al. 2004; Haemmerle et al. 2011); PNPLA3 in contributing to hepatic fat metabolism and non-alcoholic fatty liver disease (Romeo et al. 2008); PNPLA6 (or neuropathy target esterase) which contributes to membrane lipid homeostasis and assists in maintaining axonal integrity (Zaccheo et al. 2004; Rainier et al. 2008); and PNPLA8 which serves as a calcium-independent phospholipase A2 and catalyzes the hydrolysis of membrane phospholipids (Tanaka et al. 2000; Mancuso et al. 2000).
PNPLA4 and other members of the PNPLA-like enzymes belong to the patatin family of acyl hydrolases whose proteins are characterized by a conserved amino acid sequence of Gly-X-Ser-X-Gly at their active sites, a Ser-Asp catalytic dyad (Ser43/Asp163 for human PNPLA4) (Rydel et al. 2003; Holmes 2012) instead of the Ser-His-Asp/Glu triad reported for other lipases (Cygler and Schrag 1997) and an oxy-anion ‘hole’ providing access to the active site (Rydel et al. 2003). Although three-dimensional structural analyses have not been reported for mammalian PNPLA4, the crystal structure for human PNPLA8 (also IPLA2G or cytosolic phospholipase A2) has been described (Dessen et al. 1999) showing structural similarity to potato patatin (Rydel et al. 2003).
This paper reports the predicted gene structures and amino acid sequences for PNPLA4 genes and proteins, including primate (human [Homo sapiens], chimpanzee [Pan troglodytes], orang-utan [Pongo abelii], rhesus monkey [Rhesus mulatta], marmoset [Callithrix jacchus]), other eutherian mammals (rat[Rattus norvegicus], horse [Equus caballus], cow [Bos taurus], dog [Canis familiaris]), a marsupial mammal (opossum) [Monodelphis domestica] and other vertebrates, including chicken [Gallus gallus], lizard [Anolis carolensis], frog [Xenopustropicalis], zebrafish [Danio rerio] and lancelet [Branchiostoma floridae]. Predicted secondary and tertiary structures for PNPLA4 protein subunits are also described, as well as the structural relationships of these genes and enzymes with other PNPLA-like gene families.
Methods
PNPLA4 and other PNPLA-like gene and protein identification
Basic Local Alignment Search Tool (BLAST) studies were undertaken using web tools from the National Center for Biotechnology Information (NCBI) (http://blast.ncbi.nlm.nih.gov/Blast.cgi) (Altschul et al. 1990). Protein BLAST analyses used the human PNPLA4 (Gao and Simon 2005) and PNPLA-like amino acid sequences deduced from reported sequences for these genes (Schoenborn et al. 2006; Dunham et al. 1999; Lush et al. 1998; Grimwood et al. 2004; Humphray et al. 2009; Tanaka et al. 2000; Mancuso et al. 2000). Non-redundant protein sequence databases for several mammalian and other vertebrate genomes were examined using the blastp algorithm, including human (Homo sapiens) (International Genome Sequencing Consortium 2001); chimpanzee (Pan troglodytes) (Chimpanzee Sequencing and Analysis Consortium 2005); orang-utan (Pongo abelii) (Locke et al. 2011); rhesus monkey (Mucaca mulatta) (Gibbs et al. 2007), marmoset (Callithrix jacchus) (http://genome.ucsc.edu/cgi-bin/hgGateway?db=calJac1); horse (Equus caballus) (Wade et al. 2009), cow (Bos taurus) (The Bovine Genome Sequencing and Analysis Consortium et al. 2009); mouse (Mus musculus) (Mouse Genome Sequencing Consortium 2002); rat (Rattus norvegicus) (Rat Genome Sequencing Project Consortium 2004); dog (Canis familiaris) (Lindblad-Toh et al. 2005); opossum (Monodelphis domestica) (Mikkelsen et al. 2007); chicken (Gallus gallus) (International Chicken Genome Sequencing Consortium 2004); lizard (Anolis carolensis) (Alfoldi et al. 2011); frog (Xenopus tropicalis) (Hellsten et al. 2010); zebrafish (Danio rerio) (Sprague et al. 2005); sea squirt (Ciona intestinalis) (Dehal et al. 2002); and lancelet (Branchiostoma floridae) (Putnam et al. 2008). This procedure produced multiple BLAST ‘hits’ for each of the protein databases which were individually examined and retained in FASTA format, and a record kept of the sequences for predicted encoded PNPLA-like proteins. These records were derived from annotated genomic sequences using the gene prediction method: GNOMON (http://www.ncbi.nlm.nih.gov/genome/guide/gnomon.shtml) and predicted sequences with high similarity scores generated.
BLAT analyses were subsequently undertaken for each of the predicted PNPLA4 and other PNPLA-like amino acid sequences using the UC Santa Cruz web browser (Kent et al. 2003) with the default settings to obtain the predicted locations for each of the vertebrate PNPLA-like genes, including predicted exon boundary locations and gene sizes (Table 1; Supplementary Table 1). Structures for human PNPLA4 isoforms were obtained using the AceView website to examine predicted gene and protein structures to interrogate this database of human mRNA sequences (Thierry-Mieg and Thierry-Mieg 2006).
Table 1.
PNPLA4 and other PNPLA-like lipase genes and proteins 1RefSeq: the reference amino acid sequence; 2predicted Ensembl amino acid sequence; and 3scaffold IDs are shown; GenBank IDs are derived from NCBI sources http://www.ncbi.nlm.nih.gov/genbank/; UNIPROT refers to UniprotKB/Swiss-Prot IDs for individual PNPLA4 and other PNPLA-like lipase subunits (see http://kr.expasy.org); gene size refers to base pairs of nucleotide sequences; pI refers to theoretical isoelectric points; the number of coding exons are listed; ‘na’ means data not available
| Animal | Species | PNPLA gene (other name) | Chromosome coordinates | Gene size | Exon strand | Subunit (MW) | Amino acids | pI | GenBank ID | UNIPROT ID | ¹NCBI reference ID ²NCBI predicted ID |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Human | Homo sapiens | PNPLA4 (PLPL4) | X:7,866,804–7,895,475 | 29,493 | 6 –ve | 27,980 | 253 | 9.0 | BC020746 | P41247 | ¹NM_001142389.1 |
| Chimpanzee | Pan troglodytes | PNPLA4 (PLPL4) | X:7,736,089–7,762,317 | 26,229 | 6 –ve | 27,964 | 253 | 9.2 | na | na | ²XP_001139947.1 |
| Orangutan | Pongo abelii | PNPLA4 (PLPL4) | X:7,601,921–7,627,933 | 26,013 | 6 –ve | 28,292 | 255 | 9.3 | na | na | ²XP_002831412.1 |
| Gibbon | Nomascus leucogenys | PNPLA4 (PLPL4) | ³GL3937281:5,083,830–5,109,334 | 25,505 | 6 –ve | 27,994 | 253 | 9.2 | na | na | ²XP_003261040.1 |
| Rhesus monkey | Macaca mulatta | PNPLA4 (PLPL4) | X:5,489,556–5,514,987 | 25,432 | 6 –ve | 28,105 | 253 | 9.1 | na | na | ²NP_001180773 |
| Marmoset | Callithrix jacchus | PNPLA4 (PLPL4) | X:5,725,477–5,753,451 | 27,975 | 6 –ve | 28,194 | 253 | 9.1 | na | na | ²XP_002762646.1 |
| Rat | Rattus norvegicus | Pnpla4 (Plpl4) | X:64,019,691–64,022,515 | 2,825 | 6 –ve | 27,439 | 252 | 9.1 | FQ216301 | na | ²XP_343791.1 |
| Guinea pig | Cavia porcellus | PNPLA4 (PLPL4) | ³121:3,377,779–3,414,755 | 36,977 | 6 –ve | 28,052 | 253 | 9.1 | na | na | ²XP_003462801.1 |
| Horse | Equus caballus | PNPLA4 (PLPL4) | X:4,394,005–4,419,747 | 25,743 | 6 –ve | 28,109 | 253 | 8.9 | na | F6R9V1 | ²XP_001488340.1 |
| Dog | Canis familiaris | PNPLA4 (PLPL4) | X:4,847,874–4,875,987 | 28,114 | 6 –ve | 28,111 | 253 | 8.7 | na | E2R3S8 | ²XP_548849.3 |
| Cow | Bos taurus | PNPLA4 (PLPL4) | Un.004.9:49,923–92,708 | 42,786 | 6 –ve | 28,117 | 253 | 8.7 | BT021623 | na | ²XP_590366.2 |
| Panda | Ailuropoda melanoleuca | PNPLA4 (PLPL4) | ³GL194268.1:23,365–49,508 | 26,144 | 6 +ve | 28,227 | 253 | 9.1 | na | na | ²XP_002929669.1 |
| Elephant | Loxodonta africana | PNPLA4 (PLPL4) | ³94:3,450,626–3,485,515 | 34,890 | 6 –ve | 28,113 | 253 | 8.8 | na | na | ²XP_003420679.1 |
| Pig | Sus scrofa | PNPLA4 (PLPL4) | X:3,428,888–3,458,816 | 29,929 | 6 –ve | 28,452 | 256 | 9.1 | na | na | ¹AC071250.1 |
| Opossum | Monodelphis domestica | PNPLA4 (PLPL4) | 7:35,933,455–36,004,661 | 71,207 | 6 +ve | 28,263 | 253 | 9.3 | na | F7F1B0 | ²XP_001365352.1 |
| Chicken | Gallus gallus | PNPLA4 (PLPL4) | 1:130,120,480–130,133,840 | 13,361 | 6 +ve | 28,377 | 253 | 8.6 | EU419877 | B3TZB7 | ²NP_001124212.1 |
| Lizard | Anolis carolensis | PNPLA4 (PLPL4) | 3:117,647,108–117,660,289 | 13,182 | 6 –ve | 27,759 | 253 | 8.2 | na | na | ²XP_003218860.1 |
| Frog | Xenopus tropicalis | PNPLA4 (PLPL4) | ³430:395,676–405,924 | 10,249 | 6 +ve | 28,623 | 255 | 9.0 | na | F6SLH7 | ²XP_002939012.1 |
| Zebrafish | Danio rerio | PNPLA4 (PLPL4) | 1:31,551,478–31,565,593 | 14,116 | 6 –ve | 27,769 | 252 | 9.3 | BC133946 | B8JKG7 | ¹NM_001089482.1 |
| Lancelet | Branchiostoma floridae | PNPLA4 (PLPL4) | 4Un:621,873,136–621,877,356 | 4,221 | 5 –ve | 30,817 | 273 | 7.7 | na | na | ²XP_002595239.1 |
| Human | Homo sapiens | PNPLA1 (PLPL1) | 6:36,238,237–36,275,490 | 37,254 | 8 +ve | 57,875 | 532 | 8.4 | BC103905 | Q8N8W4 | ²NP_001139189 |
| Rat | Rattus norvegicus | Pnpla1 (Plpl1) | 20:7,139,384–7,171,885 | 32,502 | 9 +ve | 63,589 | 589 | 6.3 | na | na | ¹NM_001191841.1 |
| Mouse | Mus musculus | Pnpla1 (Plpl1) | 17:28,995,812–29,023,893 | 28,082 | 9 +ve | 65,171 | 592 | 8.6 | AK132521 | Q3V1D5 | ¹NM_001034885.3. |
| Opossum | Monodelphis domestica | PNPLA1 (PLPL1) | 2:275,536,646–275,586,990 | 50,345 | 9 +ve | 60,027 | 540 | 6.3 | na | na | ²XP_001378816.2 |
| Chicken | Gallus gallus | PNPLA1 (PLPL1) | 26:1,321,888–1,327,134 | 5,247 | 8 –ve | 41,612 | 376 | 8.5 | na | na | ²XP_425818.2 |
| Human | Homo sapiens | ATGL (PNPLA2) | 11:819,719–824,859 | 5,141 | 9 +ve | 55,316 | 504 | 6.7 | BC011958 | Q96AD5 | ¹NM_023376 |
| Rat | Rattus norvegicus | Atgl (Pnpla2) | 1:201,642,058–201,646,343 | 4,286 | 9 +ve | 52,567 | 478 | 6.2 | AC109542 | P0C548 | ¹NM_001108509.2. |
| Mouse | Mus musculus | Atgl (Pnpla2) | 7:148,641,186–148,645,564 | 4,379 | 9 +ve | 53,657 | 486 | 6.1 | BC064747 | Q8BJ56 | ¹NR_028142 |
| Opossum | Monodelphis domestica | ATGL (PNPLA2) | 4Un:45,368,040–45,372,831 | 4,792 | 9 +ve | 53,547 | 490 | 6.8 | na | na | ²XP_001380646.2 |
| Chicken | Gallus gallus | ATGL (PNPLA2) | 5:16,838,493–16,868,610 | 30,118 | 9 –ve | 53,610 | 483 | 6.7 | EU419874 | A8WEN5 | ¹NM_001113291.1 |
| Zebrafish | Danio rerio | ATGL (PNPLA2) | ³3,512:30,775–45,374 | 14,600 | 10 +ve | 52,253 | 473 | 6.8 | BC075928 | na | ¹NM_001002338.1 |
| Sea squirt | Ciona intestinalis | ATGL (PNPLA2) | ³127:40,380–41,927 | 1,548 | 1 +ve | 57,387 | 516 | 8.1 | AK112234 | na | na |
| Fruit fly | Drosophila melanogaster | ATGL (BRUMMER) | 3L:14,770,298–14,779,178 | 8,881 | 7 –ve | 57,227 | 507 | 5.9 | AY051668 | Q9VUH7 | NM_140466.1 |
| Human | Homo sapiens | PNPLA3 (PLPL3) | 22:44,319,792–44,342,259 | 22,468 | 9 +ve | 52,865 | 481 | 6.3 | BC014449 | Q9NST1 | ¹NM_025225.2 |
| Rat | Rattus norvegicus | Pnpla3 (Plpl3) | 7:122,152,145–122,171,991 | 19,847 | 9 +ve | 45,908 | 414 | 6.8 | EDM15609 | na | na |
| Mouse | Mus musculus | Pnpla3 (Plpl3) | 15:83,998,304–84,016,512 | 18,209 | 9 +ve | 45,772 | 413 | 6.6 | BC028792 | Q91WW7 | ¹NM_054088.3 |
| Opossum | Monodelphis domestica | PNPLA3 (PLPL3) | 8:16,368,181–16,401,317 | 33,137 | 9 –ve | 51,235 | 460 | 7.5 | na | na | ²XP_001367550.1 |
| Chicken | Gallus gallus | PNPLA3 (PLPL3) | 1:71,223,761–71,268,760 | 45,000 | 9 +ve | 56,429 | 509 | 8.9 | MGC86401 | na | ²XP_416457.2 |
| Human | Homo sapiens | PNPLA5 (PLPL5) | 22:44,276,678–44,287,760 | 12,299 | 9 –ve | 47,912 | 429 | 6.3 | BC031820 | Q7Z6Z6 | ¹NM_138814 |
| Rat | Rattus norvegicus | Pnpla5 (Plpl5) | 7:122,105,840–122,115,990 | 10,151 | 9 –ve | 50,408 | 453 | 8.5 | na | D3ZXU1 | ¹NM_001130497.1 |
| Mouse | Mus musculus | Pnpla5 (Plpl5) | 15:83,943,618–83,953,543 | 9,926 | 9 –ve | 48,480 | 432 | 9.0 | BC109360 | Q32LZ8 | ¹NM_029427.1 |
| Cow | Bos taurus | PNPLA5 (PLPL5) | 5:115,038,715–115,060,288 | 21,574 | 9 –ve | 50,457 | 455 | 7.5 | na | na | ²XP_001253781.3 |
Predicted structures and properties of vertebrate PNPLA4 subunits
Alignments of predicted PNPLA4 amino acid sequences were undertaken using a ClustalW method (http://www.ebi.ac.uk/Tools/msa/clustalw2/) (Chenna et al. 2003). Predicted secondary and tertiary structures for vertebrate PNPLA4 subunits were obtained using PSIPRED (McGuffin et al. 2000) and SWISS MODEL web tools, respectively (Guex and Peitsch 1997; Kopp and Schwede 2004). The reported tertiary structure for potato patatin (Rydel et al. 2003) served as the reference for the predicted PNPLA4 tertiary structures, with a modeling range of residues 6–173. Theoretical isoelectric points and molecular weights for vertebrate PNPL4 and PNPLA-like subunits were obtained using Expasy web tools (http://web.expasy.org/compute_pi/) (Gasteiger et al. 2005). Predicted trans-membrane helices for PNPLA-like sequences were obtained using CBS web tools (Center for Biological Sequence Analysis, Technical University of Denmark) (http://www.cbs.dtu.dk/services/TMHMM/) (Moller et al. 2001). Patatin-motifs were identified for PNPLA-like sequences using web tools from the National Center for Biotechnology Information (NCBI) (http://blast.ncbi.nlm.nih.gov/Blast.cgi).
Human PNPLA4 gene expression and predicted gene regulation sites
The human genome browser (http://genome.ucsc.edu) (Kent et al. 2003) was used to examine GNF Expression Atlas 2 data using various expression chips for the human PNPLA4 gene (http://biogps.gnf.org) (Su et al. 2004). Predicted CpG islands and microRNA (miRNA) binding sites for human PNPLA4 were obtained using the UC Santa Cruz Genome Browser (http://genome.ucsc.edu).
Results and discussion
Alignments and biochemical features of PNPLA4 amino acid sequences
Amino acid sequence alignments for 14 previously unreported vertebrate PNPLA4 amino acid sequences are shown in Fig. 1, together with the reported sequence for human PNPLA4 (Gao and Simon 2005; Gao et al. 2009). The PNPLA4 sequences exhibited >60 % identities, suggesting that these protein subunits are products of the same gene family, whereas the sequences for the predicted vertebrate PNPLA1, ATGL, PNPLA3 and PNPLA5 subunits were 27–37 % identical with the PNPLA4 sequences, indicating that these are members of distinct, but related PNPLA-like gene families (Supplementary Table 2). The sequences for the vertebrate PNPLA6, PNPLA7 and PNPLA8 subunits examined were even more distantly related with vertebrate PNPLA4 sequences with identities of <16 % (Supplementary Table 2). Two of these sequences (PNPLA6 and PNPLA7), however, showed comparatively high sequence identities (58–61 %), suggesting that these are closely related gene families. Amino acid sequences for the eight human PNPLA-like proteins examined contained 253 (PNPLA4), 429–532 (PNPLA1, PNPLA2, PNPLA3 and PNPLA5), 782 (PNPLA8) and 1,317–1,366 (PNPLA6 or NTE and PNPLA7) residues (Table 1; Supplementary Table 1). Consequently, vertebrate PNPLA4 is the smallest among these PNPLA-like proteins with an average molecular weight of ~28,000, while others exhibited MWs which are ~2 (PNPLA1, PNPLA2, PNPLA3 and PNPLA5), ~3 (PNPLA8) or ~5 times larger (PNPLA6 and PNPLA7) than PNPLA4.
Fig. 1.
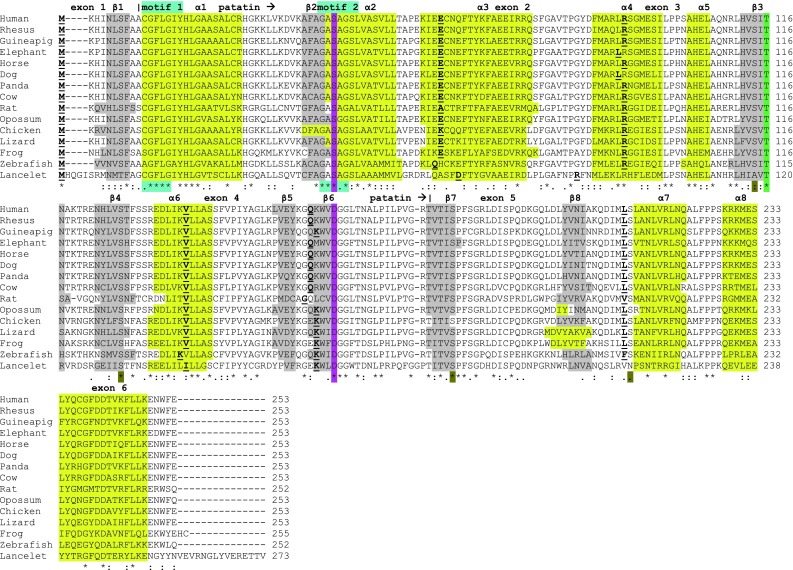
Amino acid sequence alignments for PNPLA4 sequences. See Table 1 for sources of PNPLA4 sequences; * identical residues; 1 or 2 conservative substitutions; 1 or 2 non-conservative substitutions; patatin refers to predicted motif residues (6–173); motif 1 (residues 11–15) refers to putative active site region; motif 2 refers to active site region; active site catalytic dyad residues Ser43 and Asp163; predicted helix (designated as α1, α2 etc.); predicted sheet (designated as β1, β2, etc.); conserved Thr116 and serine residues; and bold underlined font shows predicted exon junctions
Site-directed mutagenesis studies for human PNPLA4 (Gao and Simon 2005; Gao et al. 2009) and potato patatin (Hirschberg et al. 2001) have enabled the identification of key catalytic residues among those aligned for the vertebrate PNPLA4 sequences examined (Fig. 1). These included an active site motif (Gly-Xaa-Ser-Yaa-Gly designated as motif 2) (human PNPLA4 residues 41–45); active site residues Ser43 and Asp163 which serve as the catalytic dyad during catalysis; and a putative oxy-anion hole with a consensus sequence for this motif (Cys-Gly-Phe-Leu-Gly for residues 11–15 designated as motif 1). These residues are conserved among all of the vertebrate PNPLA4 sequences examined (with the exception of a Ala10 → Ser10 substitution for opossum PNPLA4), in addition to Thr116 (except for Ser116 in marmoset PNPLA4 [sequence not shown]), which is a site subject to site-specific phosphorylation (Daub et al. 2008). High theoretical isoelectric points (pI) were observed for each of the vertebrate PNPLA4 subunits examined (pI values range from 8.2–9.3), as compared with the other PNPLA-like subunits examined which exhibited lower predicted pI values, with the exception of the vertebrate PNPLA8 subunits (pI values of 9.2–9.3) (Table 1; Supplementary Table 1).
Predicted secondary and tertiary structures for vertebrate PNPLA4 subunits
Analyses of predicted secondary structures for PNPLA4 sequences revealed similar α-helix and β-sheet structures for all of the vertebrate subunits examined, particularly near key residues or functional domains (Fig. 1). Predicted secondary (Fig. 1) and tertiary structures (Fig. 2) were very similar to those reported for potato patatin (Rydel et al. 2003), which have been retained for all of the vertebrate PNPLA4 sequences examined. The predicted PNPLA4 tertiary structure (Fig. 2) is based on a partial sequence for this enzyme (residues 6–173) revealing the relative positioning and predicted structures for each of 5α-helices and 5β-sheets. These included the N-terminus α-helix (designated as α1), which may serve as a membrane anchor for PNPLA4 (no predicted trans-membrane properties were, however, observed for the α1 helix); an oxy-anion hole proposed for the motif previously reported (Cys-Gly-Phe-Leu-Gly for residues 11–15 designated as motif 1) located near the active site cleft (Fig. 2) which is similar to the oxy-anion hole reported for potato patatin (Rydel et al. 2003) and human PNPLA8 (encoding cytosolic phospholipase A2) (Dessen et al. 1999); a second α-helix (α2) and β-sheet (β2) which contain the active site motif Gly-Xaa-Ser-Yaa-Gly (residues 41–45 for human PNPLA4 designated as motif 2); and a β-sheet (β5) which contains Asp163, the second member of the active site dyad of catalytic residues. These structures are proximally located within a putative active site cleft supported by the predicted three-dimensional structure for this enzyme, however, any firm conclusions must await further studies. Several conserved serine residues were also observed for the vertebrate PNPLA4 sequences which may correspond to residues previously proposed for performing structural roles in potato patatin phospholipase A (Hirschberg et al. 2001; Rydel et al. 2003).
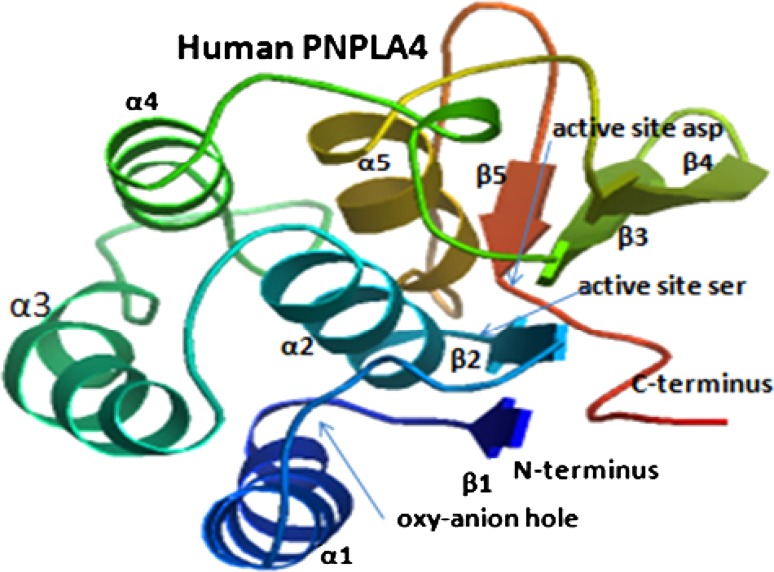
Fig. 2.
Predicted tertiary structure for human PNPLA4. The predicted structure for human PNPLA4 is based on the reported structure for potato patatin (Rydel et al. 2003) and obtained using the SWISS MODEL web site http://swissmodel.expasy.org/workspace/. The rainbow color code describes the 3D structures from the N- (blue) to C-termini (red color); predicted α-helices, β-sheets, active site residues (Ser43 and Asp163) and active site ‘motifs’ (1 and 2) are shown
Predicted gene locations, exonic structures and expression for vertebrate PNPLA4 genes
Table 1 summarizes the predicted locations for vertebrate PNPLA4 genes based on BLAT interrogations of several vertebrate genomes using the sequence for human PNPLA4 (Gao and Simon 2005; Gao et al. 2009) and the predicted sequences for other vertebrate PNPLA4 enzymes and the UC Santa Cruz Web Browser (Kent et al. 2003). Eutherian mammalian PNPLA4 genes were located on the X-chromosome in each case, however, the marsupial PNPLA4 gene (opossum; Monodelphis domestica) was located on an autosome (chromosome 7), suggesting that the X-chromosome location for PNPLA4 is restricted to eutherian mammalian genomes. Table 1 also provides data for other vertebrate PNPLA4 genes, including the previously reported chicken PNPLA4 sequence (Saarela et al. 2008), and those predicted for lizard (Anolis carolensis), frog (Xenopus tropicalis), zebrafish (Danio rerio) and lancelet (Branchiostomafloridae) genomes, which have distinct locations to those reported here for the other vertebrate PNPLA-like genes. Figure 1 summarizes the predicted exonic start sites for several vertebrate PNPLA4 genes with each having six coding exons in identical or similar positions. In contrast, lancelet PNPLA4 contained 5 coding exons, with exon 5 corresponding to exons 5 and 6 for the vertebrate PNPLA4 genes.
Figure 3 examined the predicted location of the human PNPLA4 gene on the human X-chromosome as well as comparative sequence identities for vertebrate PNPLA4 sequences. The absence of a mouse PNPLA4 gene was readily apparent from this study. Moreover, a major decrease in sequence identities for vertebrate PNPLA4 genes with the human PNPLA4 gene was observed for the more distantly related species examined, especially for the intronic sequences and for exons 5 and 6 of chicken, frog and zebrafish PNPLA4 genes. It is suggested that this may reflect a higher level of conservation for the ‘patatin’ encoding regions for the vertebrate PNPLA4 sequences, which are encoded by exons 1–4 of the vertebrate PNPLA4 genes examined (Fig. 1).
Fig. 3.
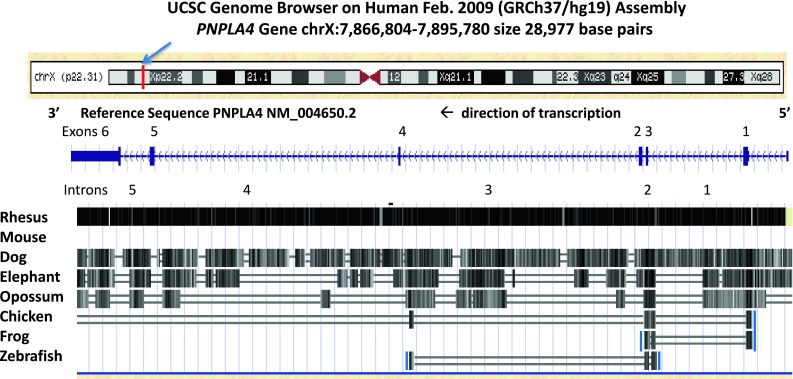
Comparative sequences for vertebrate PNPLA4 genes derived from the UCSC Genome Browser (Kent et al. 2003) using the Comparative Genomics track to examine alignments and evolutionary conservation of PNPLA4 gene sequences; a diagram of human chromosome X and the positioning for the human PNPLA4 gene (in red) was taken from the UCSC Genome Browser; genomic sequences aligned for this study included primate (human and rhesus), non-primate eutherian mammal (mouse, dog and elephant), a marsupial (opossum), bird (chicken), amphibian (frog) and fish (zebrafish); conservation measures were based on conserved sequences across all of these species in the alignments which included the 5′-untranslated, exons (exons 1–6), introns (introns 1–5) and 3′ untranslated regions for the PNPLA4 gene; regions shadedfromblackto grey showing decreasing levels of sequence identity; exons 1–4 showed highest levels of gene sequence conservation
Supplementary Table 3 examined the comparative sizes for several vertebrate PNPLA4 genes and intronic sequences (introns 1–5 for vertebrate PNPLA4 genes and introns 1–4 for the lancelet PNPLA4 gene examined). The rat PNPLA4 gene was much smaller than other PNPLA4 genes examined, being >10 times smaller than the human gene, which is reflected in the smaller sizes observed for introns 1, 3, 4 and 5. Moreover, a mouse PNPLA4 gene was not detected in this and previous studies and further investigations are required to demonstrate whether this gene is absent from the mouse genome or has escaped detection at this stage. The guinea pig (Cavia porcellus) PNPLA4 gene, however, resembled other mammalian PNPLA4 genes in the comparative sizes of introns, which suggested that the small size for the rat PNPLA4 gene was not a common feature for other rodent PNPLA4 genes. Comparisons of intron sizes for vertebrate and invertebrate PNPLA4 genes also showed that intron 2 was much smaller for all mammalian (also chicken and lizard) PNPLA4 genes examined than other introns, although intron 2 sequences for frog (Xenopus tropicalis), zebrafish (Danio rerio) and lancelet (Branchiostoma floridae) PNPLA4 genes were much larger than for the mammalian PNPLA4 genes.
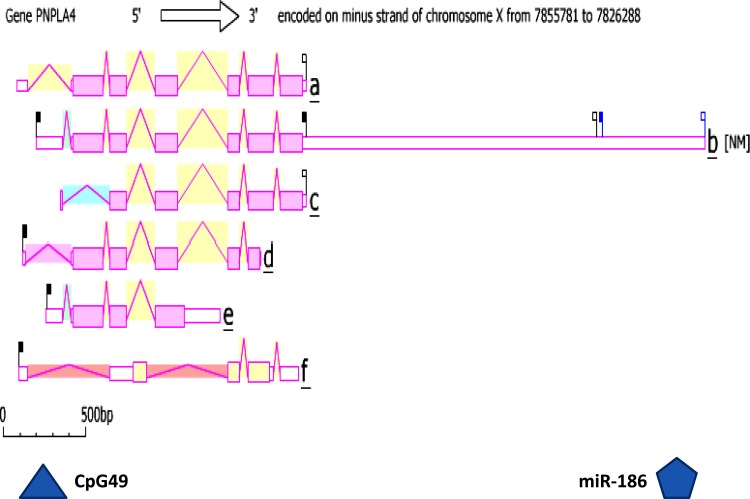
Figure 4 illustrates the comparative predicted structures of pre-messenger RNA human PNPLA4 gene transcripts (http://www.ncbi.nlm.nih.gov/IEB/Research/Acembly/) (Thierry-Mieg and Thierry-Mieg 2006). There were 6 introns present for the pre-messenger mRNA PNPLA4a and PNPLA4b transcripts, with the latter containing a CpG49 island in the 5′-noncoding segment corresponding to the promoter for this gene. In addition, the PNPLA4b transcript contained an extended 3′-noncoding segment with a predicted miRNA-186 binding site. These predicted gene regulation sites may contribute to the high level of gene expression (×1.5 times the expression of the average gene) and wide tissue expression observed for PNPLA4. Elango and Yi (2011) have previously reported that larger CpG islands are associated with gene promoters of housekeeping genes showing a broad range of gene expression and containing more RNA polymerase II binding sites than other promoters. Moreover, miRNAs are post-transcriptional regulators that bind to complementary sequences on target messenger RNA transcripts (mRNAs), usually resulting in translational repression or target degradation and gene silencing (Bartel 2009). Consequently, the presence of CpG49 and miRNA-186 within the PNPLA4 gene may contribute significantly to the broad tissue expression observed for PNPLA4 transcripts. Figure 5 presents ‘heat maps’ showing the comparative gene expression for various human tissues obtained from GNF Expression Atlas Data using U133A and GNF1H PNPLA4 chips (Su et al. 2004) with higher levels being observed in bronchial epithelial cells and heart as well as significant expression in the other tissues examined.
Fig. 4.
Gene structure and major isoforms for human PNPLA4. From AceView website (Thierry-Mieg and Thierry-Mieg 2006) http://www.ncbi.nlm.nih.gov/IEB/Research/Acembly/ mature isoform variants (designated as ‘a’, ‘b’ etc.) are shown for each PNPLA4 transcript; capped 5′- and 3′- ends for the predicted mRNA sequences are identified; a predicted CpG49 island, a miRNA binding site (miR-186) and a scale of base pairs of nucleotide sequences are shown
Fig. 5.

Human tissue gene expression ‘heat maps’ for PNPLA4 taken from the human genome browser (Kent et al. 2003) (http://genome.ucsc.edu); GNF Expression Atlas 2 data using expression chips for human PNPLA4 (http://biogps.gnf.org) (Su et al. 2004); comparative gene expression levels among human tissues: red (high) and black (intermediate) expression levels
Phylogeny of vertebrate PNPLA4 and other PNPLA-like lipases
A phylogenetic tree has been previously described from alignments of vertebrate ATGL-like amino acid sequences (PNPLA1, ATGL, PNPLA3, PNPLA4 and PNPLA5) with the predicted fruit fly (Drosophila melanogaster) ATGL sequence serving to ‘root’ the tree (Holmes 2012). Clustering was reported for five major groups of vertebrate ATGL-like sequences: PNPLA1; ATGL (or PNPLA2); PNPLA3; PNPLA4; and PNPLA5. Clustering into sub-groupings was also described, including PNPLA3 and PNPLA5, with ATGL; and PNPLA4 with PNPLA1. These results were consistent with the presence of ATGL-like and PNPLA4-like genes within primitive vertebrate genomes examined, and were suggestive of an initial gene duplication event for ATGL generating both of these genes, during the evolutionary appearance of vertebrates. This is consistent with PNPLA4 being an ancient gene, appearing in some primitive vertebrate genomes and being present throughout vertebrate evolution over a period of evolution of >500 million years, which is reported for the timing of the appearance of vertebrates during evolution (Donoghue and Benton 2007).
These phylogenetic studies were also extended to include other PNPLA-like genes and proteins, namely PNPLA6, PNPLA7 and PNPLA8 sequences (Holmes 2012). The results were indicative of at least three major PNPLA-like sequence groups, including the ATGL-like sequences (PNPLA1, ATGL (PNPLA2), PNPLA3, PNPLA4 and PNPLA5 (Group 1); the PNPLA6 and PNPLA7 sequences (Group 2); and the PNPLA8 sequences (Group 3). Group 1 sequences were further divided according to the designation of ATGL-like gene families, which clustered with the sea squirt ATGL-like sequence, and were suggestive of an ancestral relationship between early vertebrate ATGL and PNPLA4 genes, with other members of PNPLA-like group 1 sequences, which appeared later during vertebrate evolution: PNPLA1 and PNPLA3/PNPLA5. This report (Holmes 2012) also suggested that vertebrate PNPLA6 and PNPLA7 sequences shared a common evolutionary origin distinct to the ATGL-like and PNPLA8 sequences, which were ‘rooted’ with the sea squirt (Ciona intestinalis) PNPLA7 sequence, whereas the vertebrate PNPLA8 sequences were also distinct and separately ‘rooted’ with the sea squirt (Ciona intestinalis) PNPLA8 sequence.
Overall, it is likely that the three PNPLA-like groups of genes and proteins have arisen from distinct ancestral genes during vertebrate evolution, namely ATGL (the proposed ancestral gene for group 1 PNPLA-like genes [PNPLA1; ATGL; PNPLA3/PNPLA5; and PNPLA4]; PNPLA6/PNPLA7 for group 2 PNPLA-like genes; and PNPLA8 for group 3 PNPLA-like genes (Holmes 2012).
Summary
The results of this study support previous studies (Wilson et al. 2006; Kienesberger et al. 2009; Saarela et al. 2008; Holmes 2012) for at least eight vertebrate PNPLA-like genes and encoded lipases, including five ATGL-like genes, namely PNPLA4 (encoding PNPLA4) and PNPLA1, ATGL (encoding adipose triglyceride lipase), PNPLA3 and PNPLA5 genes; two PNPLA6-like genes, PNPLA6 (encoding neuropathy target esterase) and PNPLA7; and PNPLA8 (encoding cytosolic phospholipase A2). Vertebrate PNPLA4 sequences shared key conserved sequences reported for human PNPLA4 (Gao and Simon 2005; Wilson et al. 2006; Gao et al. 2009), including active site residues, an oxy-anion ‘hole’ sequence, a phosphorylated Thr site and several conserved serine residues. Gene expression data showed that the human PNPLA4 gene is broadly expressed at higher levels than those for the average gene, for which a CpG island localized in the PNPLA4 promoter and a miRNA binding site localized in the extended 3′noncoding region of PNPLA4b mRNA isoform may contribute to these high expression levels. A recent phylogenetic study (Holmes 2012) has suggested that PNPLA4 is an ancient gene in vertebrate evolution derived from a duplication of an ancestral ATGL-like gene within a primitive vertebrate genome.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
The advice of Dr Laura Cox of the Texas Biomedical Research Institute is gratefully acknowledged.
References
- Alfoldi J, Di Palma F, Grabherr M, Williams C, Kong L, Mauceli E, et al. The genome of the green anole lizard and a comparative analysis with birds and mammals. Nature. 2011;477:587–591. doi: 10.1038/nature10390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul F, Vyas V, Cornfield A, Goodin S, Ravikumar TS, Rubin EH, Gupta E. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenna V, Sugawara R, Koike T, Lopez R, Gibson TJ, Higgins DJ, Thompson JD, et al. Multiple sequence alignment with the clustal series of programs. Nucleic Acids Res. 2003;31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chimpanzee Sequencing and Analysis Consortium Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- Cygler M, Schrag JD. Structure as basis for understanding interfacial properties of lipases. Methods Enzymol. 1997;284:3–27. doi: 10.1016/S0076-6879(97)84003-7. [DOI] [PubMed] [Google Scholar]
- Daub H, Olsen JV, Barlein M, Gnad F, Opermann FS, Korner R, et al. Kinase-selective enrichment enables phophoproteomics of the kinome across the cell cycle. Mol Cell. 2008;31:438–448. doi: 10.1016/j.molcel.2008.07.007. [DOI] [PubMed] [Google Scholar]
- Dehal P, Satou Y, Campbell RK, Chapman J, Degnan B, De Tomaso A, et al. The draft genome of Ciona intestinalis: insights into chordate and vertebrate origins. Science. 2002;298:2157–2167. doi: 10.1126/science.1080049. [DOI] [PubMed] [Google Scholar]
- Dessen A, Tang A, Schmidt H, Stahl H, Clark JD, Seehra J, Somers WS. Crystal structure of human phopholipase A(2) reveals a novel topology and catalytic mechanism. Cell. 1999;97:349–360. doi: 10.1016/S0092-8674(00)80744-8. [DOI] [PubMed] [Google Scholar]
- Donoghue PCJ, Benton MJ. Rocks and clocks: calibrating the tree of life using fossils and molecules. Trends Genet. 2007;22:424–431. doi: 10.1016/j.tree.2007.05.005. [DOI] [PubMed] [Google Scholar]
- Dunham I, Shimizu N, Roe BA, Chissoe S, Hunt AR, Collins JE, et al. The DNA sequence of human chromosome 22. Nature. 1999;402:489–495. doi: 10.1038/990031. [DOI] [PubMed] [Google Scholar]
- Elango N, Yi SV. Functional relevance of CpG island length for regulation of gene expression. Genetics. 2011;187:1077–1083. doi: 10.1534/genetics.110.126094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Simon M. Identification of a novel keratinocyte retinyl ester hydrolase as a transacylase and lipase. J Invest Dermatol. 2005;124:1259–1266. doi: 10.1111/j.0022-202X.2005.23761.x. [DOI] [PubMed] [Google Scholar]
- Gao JG, Shin A, Gruber R, Schmuth M, Simon M. GS2 as a retinol transacylase and as a catalytic dyad independent regulator of retinylester accretion. Mol Genet Metabol. 2009;96:253–260. doi: 10.1016/j.ymgme.2008.12.007. [DOI] [PubMed] [Google Scholar]
- Gasteiger R, Hoogland C, Gattiker A, Duvaud S, Wilkins MR et al (2005) Protein identification and analysis tools on the ExPASy Server. In: Walker JM (ed) The proteomics protocols handbook, Humana Press, USA
- Gibbs RA, Rogers J, Katze MG, Bumgarner R, Weinstock GM, Mardis ER, et al. Evolutionary and biomedical insights from the rhesus macaque genome. Science. 2007;316:222–234. doi: 10.1126/science.1139247. [DOI] [PubMed] [Google Scholar]
- Grimwood J, Gordon LA, Olsen A, Terry A, Schmutz J, Lamerdin J, et al. The DNA sequence of human chromosome 19. Nature. 2004;428:528–535. doi: 10.1038/nature02399. [DOI] [PubMed] [Google Scholar]
- Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modelling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- Haemmerle G, Moustafa T, Woelkart G, Büttner S, Schmidt A, van de Weijer T, et al. ATGL-mediated fat catabolism regulates cardiac mitochondrial function via PPAR-α and PGC-1. Nature Med. 2011;17:1076–1085. doi: 10.1038/nm.2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellsten U, Harland RM, Gilchrist MJ, Hendrix D, Jurka J, Kapitonov V, et al. The genome of the western clawed frog Xenopus tropicalis. Science. 2010;328:633–636. doi: 10.1126/science.1183670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschberg HJHB, Simons J-WFA, Dekker N, Egmond MR. Cloning, expression, purification and characterization of patatin, a novel phospholipase A. Eur J Biochem. 2001;268:5037–5044. doi: 10.1046/j.0014-2956.2001.02411.x. [DOI] [PubMed] [Google Scholar]
- Holmes RS. Comparative studies of adipose triglyceride lipase genes and proteins: an ancient gene in vertebrate evolution. Open Access Bioinformatics. 2012;4:1–15. [Google Scholar]
- Humphray SJ, Oliver K, Hunt AR, Plumb RW, Loveland JE, Howe KL, et al. DNA sequence and analysis of human chromosome 9. Nature. 2009;429:369–374. doi: 10.1038/nature02465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Chicken Genome Sequencing Consortium (2004) Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature 432:695–716 [DOI] [PubMed]
- International Human Genome Sequencing Consortium Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2003;12:994–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kienesberger PC, Oberer M, Lass A, Zechner R. Mammalian patatin domain containing proteins: a family with diverse lipoplytic activities involved in multiple biological functions. J Lipid Res. 2009;50:S63–S68. doi: 10.1194/jlr.R800082-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopp J, Schwede T. The SWISS-MODEL repository of annotated three-dimensional protein structure homology models. Nucleic Acids Res. 2004;32:D230–D234. doi: 10.1093/nar/gkh008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee WC, Salido E, Yen PH. Isolation of a new gene GS2 (DXS1283E) from a CpG island between STS and KAL1 on Xp22.3. Genomics. 1994;22:372–376. doi: 10.1006/geno.1994.1397. [DOI] [PubMed] [Google Scholar]
- Lindblad-Toh K, Wade CM, Mikkelsen TS, Karlsson EK, Jaffe DB, Kamai M, et al. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature. 2005;438:803–819. doi: 10.1038/nature04338. [DOI] [PubMed] [Google Scholar]
- Locke DP, Hillier LW, Warren WC, Worley KC, Nazareth LV, Muzny DM et al (2011) Comparative and demographic analysis of orang-utan genomes. Nature 469:529–533 [DOI] [PMC free article] [PubMed]
- Long AC, Bomser JA, Grzybowski DM, Chandler HL. All-trans retinoic acid regulates cx43 expression, gap junction communication and differentiation in primary lens epithelial cells. Curr Eye Res. 2010;35:670–679. doi: 10.3109/02713681003770746. [DOI] [PubMed] [Google Scholar]
- Lush MJ, Li Y, Read DJ, Wills AC, Glynn P. Neuropathy target esterase and a homologous Drosophila neurodegeneration-associated mutant protein contain a novel domain conserved from bacteria to man. Biochem J. 1998;332:1–4. doi: 10.1042/bj3320001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancuso DJ, Jenkins CM, Gross RW. The genomic organisation, complete mRNA sequence, cloning, and expression of a novel membrane-associated calcium-independent phospholipase A(2) J Biol Chem. 2000;275:9937–9945. doi: 10.1074/jbc.275.14.9937. [DOI] [PubMed] [Google Scholar]
- McGuffin J, Bryson K, Jones DT. The PSIPRED protein structure prediction server. Bioinformatics. 2000;16:404–405. doi: 10.1093/bioinformatics/16.4.404. [DOI] [PubMed] [Google Scholar]
- Mikkelsen TS, Wakefield MJ, Aken B, Amemiya CT, Chang JL, Duke S, et al. Genome of the marsupial Monodelphis domestica reveals innovation in non-coding sequences. Nature. 2007;447:167–177. doi: 10.1038/nature05805. [DOI] [PubMed] [Google Scholar]
- Moller S, Croning MDR, Apweiler R. Evaluation of methods for the prediction of membrane spanning regions. Bioinformatics. 2001;17:646–653. doi: 10.1093/bioinformatics/17.7.646. [DOI] [PubMed] [Google Scholar]
- Mouse Genome Sequencing Consortium Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- Mungall AJ, Palmer SA, Sims SK, Edwards CA, Ashurst JL, Wilming L, et al. The DNA sequence and analysis of human chromosome 6. Nature. 2003;425:805–811. doi: 10.1038/nature02055. [DOI] [PubMed] [Google Scholar]
- Oki Y, Watanabe S, Endo T, Kano K. Mature adipocyte-derived dedifferentiated fat cells can trans-differentiate into osteoblasts in vitro and in vivo only by all-trans retinoic acid. Cell Struct Funct. 2008;33:211–222. doi: 10.1247/csf.08038. [DOI] [PubMed] [Google Scholar]
- Palczewski K (2011) Chemistry and biology of vision. J Biol Chem 287:1612–1619 [DOI] [PMC free article] [PubMed]
- Pino-Lagos K, Guo Y, Noelle RJ. Retinoic acid: a key player in immunity. BioFactors. 2010;36:430–436. doi: 10.1002/biof.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam NH, Butts T, Ferrier DEK, Furlong RF, Hellsten U, Kawashima T, et al. The amphioxus genome and the evolution of the chordate karyotype. Nature. 2008;453:1064–1071. doi: 10.1038/nature06967. [DOI] [PubMed] [Google Scholar]
- Rainier S, Bui M, Mark E, Thomas D, Tokarz D, Ming L, et al. Neuropathy target esterase gene mutations cause motor neurone disease. Am J Human Genet. 2008;82:780–785. doi: 10.1016/j.ajhg.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rat Genome Sequencing Project Consortium Genome sequence of the brown Norway rat yields insights into mammalian evolution. Nature. 2004;428:493–521. doi: 10.1038/nature02426. [DOI] [PubMed] [Google Scholar]
- Romeo M, Kozlitina A, Xing C, Pertsimlidis A, Cox D, Pennachio LA, et al. Genetic variation in PNPLA3 confers susceptibility to non-alcoholic fatty liver disease. Nature Genet. 2008;40:1461–1465. doi: 10.1038/ng.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rydel TJ, Williams JM, Krieger E, Moshiri E, Stallings WC, Brown SM, et al. The crystal structure, mutagenesis, and activity studies reveal that patatin is a lipid acyl hydrolase with a Ser-Asp catalytic dyad. Biochemistry. 2003;42:6696–6708. doi: 10.1021/bi027156r. [DOI] [PubMed] [Google Scholar]
- Saarela J, Jung G, Hermann M, Nimpf J, Schneider WJ. The patatin-like lipase family in Gallus gallus. BMC Genomics. 2008;9:281. doi: 10.1186/1471-2164-9-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer SW, Cheung J, MacDonald JR, Osborne LR, Nakabayashi K, Herbrick JA, et al. Human chromosome 7: DNA sequence and biology. Science. 2005;300:767–772. doi: 10.1126/science.1083423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenborn V, Heid IM, Vollmert C, Lingenhel A, Adams TD, Hopkins PN, et al. The ATGL gene is associated with free fatty acids, triglycerides and type 2 diabetes. Diabetes. 2006;55:1270–1275. doi: 10.2337/db05-1498. [DOI] [PubMed] [Google Scholar]
- Sprague J, Bayraktaroglu L, Bradford Y, Conlin T, Dunn N, Fashena D, et al. The zebrafish information network: the zebrafish model organism database. Nucleic Acids Res. 2005;34:D581–D585. doi: 10.1093/nar/gkj086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su AI, Wiltshire T, Batalov S, Lapp H, Ching K, et al. A gene atlas of the human and mouse protein encoding transcriptomes. Proc Natl Acad Sci USA. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka H, Takeya R, Sumimoto H. A novel intracellular membrane-bound calcium-independent phospholipase A(2) Biochem Biophys Res Commun. 2000;27:320–326. doi: 10.1006/bbrc.2000.2776. [DOI] [PubMed] [Google Scholar]
- The Bovine Genome Sequencing and Analysis Consortium et al (2009) The genome sequence of taurine cattle: a window to ruminant biology and evolution. Science 24:522–528 [DOI] [PMC free article] [PubMed]
- Thierry-Mieg D, Thierry-Mieg J. AceView: a comprehensive cDNA-supported gene and transcripts annotation. Genome Biol. 2006;7:S12. doi: 10.1186/gb-2006-7-s1-s12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade CM, Giulotto E, Sigurdsson S, Zoli M, Gnerre S, Imsland F, et al. Genome sequence, comparative analysis and population genetics of the domestic horse. Science. 2009;326:865–867. doi: 10.1126/science.1178158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson PA, Gardner SD, Lambie NM, Commans SA, Crowther DJ. Characterization of the human papatin-like phospholipase family. J Lipid Res. 2006;47:1940–11949. doi: 10.1194/jlr.M600185-JLR200. [DOI] [PubMed] [Google Scholar]
- Ye X, Tao Q, Wang Y, Cheng Y, Lotan R. Mechanisms underlying the induction of the putative human tumor suppressor GPRC5A by retinoic acid. Cancer Biol Ther. 2009;8:951–962. doi: 10.4161/cbt.8.10.8244. [DOI] [PubMed] [Google Scholar]
- Zaccheo O, Dinsdale D, Meacock PA, Glynn P. Neuropathy target esterase and its yeast homologue degrade phosphatidylcholine to glycerophosphocholine in living cells. J Biol Chem. 2004;279:24024–24033. doi: 10.1074/jbc.M400830200. [DOI] [PubMed] [Google Scholar]
- Zimmermann R, Strauss JG, Haemmerle G, Schoiswohl G, Birner-Gruenberger R, Riederer M, et al. Fat mobilization in adipose tissue is promoted by adipose triglyceride lipase. Science. 2004;306:1383–1386. doi: 10.1126/science.1100747. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.