Abstract
Purpose
Little information is available on genetic and epigenetic changes in duodenal adenocarcinomas. The purpose was to identify possible subsets of duodenal adenocarcinomas based on microsatellite instability (MSI), DNA methylation, mutations in the KRAS and BRAF genes, clinicopathologic features, and prognosis.
Experimental Design
Demographics, tumor characteristics and survival were available for 99 duodenal adenocarcinoma patients. Testing for KRAS and BRAF mutations, MSI, MLH1 methylation and CpG island methylator phenotype (CIMP) status was performed. A Cox proportional hazard model was built to predict survival.
Results
CIMP+ was detected in 27 of 99 (27.3%) duodenal adenocarcinomas, and was associated with MSI (P = 0.011) and MLH1 methylation (P < 0.001), but not with KRAS mutations (P = 0.114), as compared to CIMP− tumors. No BRAF V600E mutation was detected. Among the CIMP+ tumors, 15 (55.6%) were CIMP+/MLH1-unmethylated (MLH1-U). Kaplan-Meier analysis showed tumors classified by CIMP, CIMP/MLH1 methylation status or CIMP/MSI status could predict overall survival (OS; P = 0.047, 0.002, and 0.002, respectively), while CIMP/MLH1 methylation status could also predict time-to-recurrence (TTR; P = 0.016). In multivariate analysis, CIMP/MLH1 methylation status showed a significant prognostic value regarding both OS (P < 0.001) and TTR (P = 0.023). Patients with CIMP+/MLH1-U tumors had the worst OS and TTR.
Conclusions
Our results demonstrate existence of CIMP in duodenal adenocarcinomas. The combination of CIMP+/MLH1-U appears to be independently associated with poor prognosis in patients with duodenal adenocarcinomas. This study also suggests that BRAF mutations are not involved in duodenal tumorigenesis, MSI or CIMP development.
Keywords: Duodenal Adenocarcinoma, CpG Island Methylator Phenotype, Microsatellite Instability, Prognosis
Introduction
Primary adenocarcinoma of the duodenum was initially described by Hamburger in 1746, and represents about 0.3% of all malignant neoplasms of the gastrointestinal tract (1). During recent years, duodenal cancer incidence rates have increased more markedly than those for other sub-sites of small intestine (2). Integrated genetic and epigenetic analysis of duodenal adenocarcinoma is needed to better understand the pathways involved in its carcinogenic process, establish markers of resistance to traditional therapies, and contribute to the development of targeted therapies.
Much of our understanding of intestinal malignancies has developed from studies of colorectal cancers (CRCs). Two mechanisms of tumorigenesis in CRC have recently drawn a great deal of attention: microsatellite instability (MSI) and CpG island methylator phenotype (CIMP). MSI, the abnormal shortening or lengthening of DNA by 1 to 6 repeating base pair units, develops from defects in the mismatch repair (MMR) genes MLH1, MSH2, MSH6 and PMS2 (3). Patients with Lynch Syndrome have an inherited defect in MMR genes and have an increased risk around 4%, over 100 times the risk in the general population, demonstrating the importance of these genes in tumorigenesis(4). The MMR gene MLH1 has also been found to be methylated in CRCs resulting in MSI (5). Methylation of MLH1 is a more frequent mechanism of silencing of MMR genes in sporadic CRCs than mutations and is often associated with CIMP tumorigenesis (6). CIMP is defined as aberrant methylation of cytosine residues at CpG islands in the promoter regions of multiple cancer-specific genes (7). About half of CIMP positive cancers also show MSI via epigenetic inactivation of MLH1. This subset of cancers (CIMP+, MLH1 methylated) may be associated with a positive family or personal history of cancer (8). MLH1 methylation has also been detected in some CIMP negative (CIMP−) tumors (7). Although CIMP has previously been identified in duodenal cancers in a small subset of patients, little is known about the epigenetic alterations in these tumors (9).
KRAS mutations occur in about 30–40% of CRCs and have been proposed as a possible cause of aberrant methylation (10, 11). Recent studies indicated that CRCs with KRAS mutations might be associated with a unique DNA methylation profile and appeared to be independent of MSI status (12, 13). BRAF V600E mutation is present at a frequency of 5-22% in CRCs and has been correlated with CIMP and MSI (10, 14). Despite its strong association with CIMP in CRCs, the hypothesis that BRAF mutation may cause aberrant CpG island methylation remains controversial (15). The incidence of KRAS and BRAF mutations and their association with CIMP and MSI in duodenal cancers are, as yet, unknown.
In this study, we examined both genetic and epigenetic alterations associated with survival and recurrence in duodenal adenocarcinomas. To our knowledge, the current analysis included the largest number of patients with duodenal adenocarcinomas in any single study to date.
Materials and Methods
Study Population
This retrospective cohort study included patients with pathologically confirmed duodenal adenocarcinoma who had surgical resections. Patients were identified from the Johns Hopkins Hospital Oncology Clinical Information System from January 1997 to December 2009 and 155 duodenal adenocarcinomas patients who underwent surgical resection at our institution were identified. Patients who underwent preoperative chemotherapy/radiotherapy, lacked follow-up information or had missing archival primary tumors or corresponding matched normal samples were excluded. Formalin-fixed, paraffin-embedded (FFPE) tissue blocks of primary tumors and corresponding matched normal samples were collected from 107 patients. Tissue sections from the blocks were then reviewed by an expert gastrointestinal pathologist. After excluding ampullary tumors and low tumor cellularity sections, the remaining 99 cases formed the final study cohort (Table 1). Ascertainment of survival was performed by using the Johns Hopkins electronic health records, the Cancer Registry and mortality was confirmed also within the Social Security Death Index. The Johns Hopkins Hospital Institutional Review Board approved this research protocol.
Table 1.
Clinicopathological and molecular characteristics of patients and tumors by CpG island methylator phenotype (CIMP) status
| Characteristic | Total | CIMP− | CIMP+ | P * |
|---|---|---|---|---|
| No. of patients | 99 | 72 (72.7%) | 27 (27.3%) | |
| Sex | 1.000 | |||
| Male | 55 (55.6%) | 40 (55.6%) | 15 (55.6%) | |
| Female | 44 (44.4%) | 32 (44.4%) | 12 (44.4%) | |
| Age at surgery | 0.247 | |||
| < 70 | 64 (64.6%) | 49 (68.1%) | 15 (55.6%) | |
| ≥70 | 35 (35.4%) | 23 (31.9%) | 12 (44.4%) | |
| Stage | 0.067† | |||
| I (T1–2 N0 M0) | 8 (8.1%) | 3 (4.2%) | 5 (18.5%) | |
| II (T3–4 N0 M0) | 21 (21.2%) | 18 (25.0%) | 3 (11.1%) | |
| III (anyT N1-2 M0) | 64 (64.6%) | 47 (65.3%) | 17 (63.0%) | |
| IV (anyT anyN M1) | 6 (6.1%) | 4 (5.6%) | 2 (7.4%) | |
| Tumor differentiation | 1.000 | |||
| Well/moderate | 55 (55.6%) | 40 (55.6%) | 15 (55.6%) | |
| Poor | 44 (44.4%) | 32 (44.4%) | 12 (44.4%) | |
| Extent of resection | 1.000† | |||
| R0 | 87 (87.9%) | 63 (87.5%) | 24 (88.9%) | |
| R1/R2 | 12 (12.1%) | 9 (12.5) | 3 (11.1%) | |
| Chemotherapy/radiotherapy | 0.448 | |||
| No | 38 (38.4%) | 26 (36.1%) | 12 (44.4%) | |
| Yes | 61 (61.6%) | 46 (63.9%) | 15 (55.6%) | |
| MSI status | 0.011 | |||
| MSS | 79 (79.8%) | 62 (86.1%) | 17 (63.0%) | |
| MSI | 20 (20.2%) | 10 (13.9%) | 10 (37.0%) | |
| KRAS | 0.114 | |||
| Wild-type | 67 (67.7%) | 52 (72.2%) | 15 (55.6%) | |
| Mutated | 32 (32.3%) | 20 (27.8%) | 12 (44.4%) | |
| MLH1 methylation | < 0.001† | |||
| U | 85 (85.9%) | 70 (97.2%) | 15 (55.6%) | |
| M | 14 (14.1%) | 2 (2.8%) | 12 (44.4%) |
NOTE:
CIMP− versus CIMP+, χ2 test unless indicated otherwise;
Fisher’s exact test.
Abbreviations: CIMP, CpG island methylator phenotype; MSS, microsatellite stable; MSI, microsatellite instability; U, unmethylated; M, methylated.
Analyses of KRAS and BRAF Mutations, and Microsatellite Instability
Genomic DNA was extracted from FFPE tissues. Polymerase chain reaction (PCR) and sequencing targeted for KRAS codons 12 and 13, BRAF codon 600 were performed (16).
MSI status was determined using D2S123, D5S346, D17S250, BAT25, and BAT26 (17). Microsatellite sizes were compared with those of normal adjacent tissue, and tumors with 2 or more of the markers exhibiting instability were classified as high MSI (MSI-H). Tumors with only one marker exhibiting instability or no markers with instability were classified as low MSI (MSI-L) or microsatellite stable (MSS), respectively.
Bisulfite Modification and Methylation Analysis
Purified DNA (2 μg) was bisulfite treated and purified using the EZ DNA methylation kit (Zymo Research, Orange, CA) according to the manufacturer’s instructions.
A 5-gene signature was used to assess the CIMP methylation status of the primary tumor tissue: CACNA1G, IGF2, NEUROG1, RUNX3, and SOCS1 (6). Methylation was quantified by MethyLight, a methylation-specific, probe-based, real-time PCR technique (6, 18). Alu was used as a normalization control reaction. All CIMP probes utilized a 5′ FAM fluorophore, a 3′ IBFQ quencher, and an internal ZEN quencher (Integrated DNA Technologies, Coraville, IA). DNA methylation was reported as the percent of methylated reference (PMR) = 100 × ((methylated reaction/Alu)sample/(methylated reaction/Alu)M.SssI-reference) (6). We classified each marker as methylated when PMR ≥4. The PMR cut-off levels were set at plus two standard deviations of the average methylation levels observed in normal duodenal mucosa controls. Samples were considered CIMP+ if at least 3 out of the five studied genes were methylated (6).
Conventional methylation-specific PCR (MSP) (19) was carried out to validate the CIMP status by using a panel of 20 cancer-specific genes/loci (6, 7, 9, 20–27), as well as the five genes used for MethyLight. Gene and locus names and primers are shown in Supplementary Table S1. Methylation index was calculated as total number of genes and loci methylated/total number of genes and loci analyzed (28).
Immunohistochemistry
Immunohistochemistry (IHC) analysis for MLH1 expression was performed. In brief, FFPE tissues were sectioned at 6 μm and stained with antibody to MLH1 (BD PharMingen, San Diego, California). Tumor cells with absent nuclear staining were interpreted to have an absence of protein expression. Intact nuclear staining of adjacent non-neoplastic epithelium served as an internal positive control.
Statistical Methods
Differences in categorical variables between study groups were analyzed using χ2 test for homogeneity of Fisher’s exact test. To compare continuous variables, the Student’s t-test was used when variances were equal. The Mann-Whitney U test was used when variances were unequal. Correlation between MethyLight and MSP results were analyzed by Fisher’s exact test. All hypotheses tests were two-sided, and results were considered statistically significant for P values < 0.05.
The main outcome of this study was overall survival (OS), which was defined as the time from surgery to death resulting from any cause. In addition, time-to-recurrence (TTR) was defined as the time from surgery to recurrence, where patients without evidence of recurrence were censored for TTR at last follow-up. Survival was estimated by using the Kaplan-Meier method and log-rank statistics computed to test for differences between survival curves for various prognostic factors. Cox proportional hazard models were used to calculate hazard ratio (HR) with corresponding 95% confidence interval (CI) of recurrence or death according to molecular features (ie, CIMP/MLH1 methylation, CIMP, MLH1 methylation or MSI status), adjusted for age, sex, stage, tumor differentiation, chemotherapy and/or radiotherapy, and KRAS mutation status. All calculations were performed using SPSS 16.0 software (SPSS Inc, Chicago, IL).
Results
Clinicopathologic Characteristics by CIMP
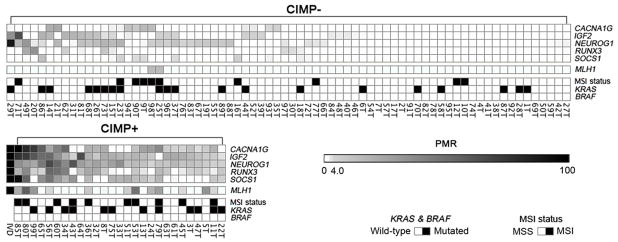
DNA extraction and CIMP testing by MethyLight were successful in all 99 patients. Twenty-seven patients (27.3%) out of the 99 patients tested were CIMP+ (Fig. 1, Table 1).
Figure 1.
Heat map demonstrating relationship of specific gene methylation, KRAS mutations, MSI status, and categorization as CIMP+ and CIMP− in duodenal adenocarcinomas. IVD, In vitro-methylated DNA (M.SssI-reference); PMR, percent of methylated reference.
To further determine whether the 5-gene signature accurately classifies patients as CIMP+ and validate the CIMP status as characterized by MethyLight, we determined the methylation of an additional panel of 20 genes/loci using conventional MSP in a group of samples. These genes were selected since they have either been previously used to identify CIMP or show frequent methylation in various cancers including duodenal cancers (6, 7, 9, 20–27). Aberrant methylation was significantly more frequent in tumors characterized as CIMP+, using the 5-gene signature, with a methylation index of 0.67 (average 13.4 genes methylated out of 20 genes examined) compared to a methylation index of 0.14 (average 2.8 genes methylated out of 20 genes examined) in tumors characterized as CIMP−, showing a marked difference (P < 0.001; Supplementary Fig. S1). In addition, comparison between the 5-gene methylation status using MethyLight technology and MSP analysis revealed significant correlations (κ = 0.583–0.813). These results suggested that the 5-gene signature was successful in identifying a CIMP+ subset of tumors.
Median age at diagnosis of duodenal cancer was 66.0 years (65.4 ± 13.4; mean ± standard deviation). Comparison of the CIMP+ and CIMP− subgroups showed that there were no differences in gender, age, tumor differentiation, extent of resection and undergoing chemotherapy/radiotherapy between the two groups (Table 1). Although not statistically significant, a trend towards association between stage and CIMP status was observed (P = 0.067). The CIMP+ group had more stage I tumors than the CIMP− group (18.5% vs. 4.2%).
MSI, CIMP and MLH1 Methylation
Among the 99 duodenal cancer patients, 20 (20.2%) displayed MSI-high; 14 (14.1%) MSI-low and 65 (65.7%) MSS status. In this study, MSI-low and MSS tumors were grouped together and henceforth are referred to as MSS. Among the twenty-seven (27.3%) patients demonstrating CIMP+, 10 (37.0%) were MSI as well (Fig. 1, Table 1). A statistically significant correlation between MSI and CIMP status was observed (P = 0.011, Table 1).
MLH1 methylation was detected in 14 (14.1%) patients and 12 (85.7%) were also CIMP+. Further associations showed that 8 (57.1%) of these were CIMP+/MSI, 4 (28.6%) were CIMP+/MSS, 2 (14.3%) were CIMP−/MSI and none showed CIMP−/MSS (Fig. 1). There were strong associations between MLH1 methylation and both MSI and CIMP+ (P < 0.001, all). Distributions of MSI, CIMP and MLH1 methylation in all patients are shown in Supplementary Fig. S2.
IHC Analysis of MLH1 in Tumors
IHC analysis of MLH1 expression was performed on selected MLH1 unmethylated (MLH1-U) and MLH1 methylated (MLH1-M) tumors. All tested MLH1-M tumors (including 4 MSS/MLH1-M tumors) had negative or low protein expression level (Fig. 2).
Figure 2.
Immunohistochemistry analysis of MLH1 expression in normal duodenal tissue and duodenal adenocarcinoma. (A), normal duodenal tissue stained with anti-MLH1 antibody showing positive nuclear staining for MLH1, particularly in the crypts. (B and C), tumors with MSS/MLH1-U stained with anti-MLH1 antibody showing positive nuclear staining for MLH1. (D–I), tumors with MLH1-M stained with anti-MLH1 antibody showing no or low MLH1 expression (D and E, two tumors with MSI/MLH1-M; F, G, H, and I, four tumors with MSS/MLH1-M).
Frequency and Associations of Tumor Mutations
Mutation analysis successfully performed in all 99 tumors and matched normal duodenal tissue specimens for KRAS and BRAF. KRAS mutations were prevalent in 32.3% (32/99) of patients and the characteristics of patients with KRAS mutations are shown in Supplementary Table S2. The most prevalent KRAS mutations were GGT>GAT (G12D) and GGT>GTT (G12V) within codon 12, and GGC>GAC (G13D) within codon 13. All mutations appear to be somatic since the same alterations were not detected in the corresponding normal tissues. Twenty-five out of 32 cases (78.1%) with KRAS mutations occurred in tumors exhibiting methylation in at least one of the six study genes (Odds Ratio 3.08, 1.17 to 8.08; P = 0.020). However, KRAS mutations were not associated with CIMP (P = 0.114, Table 1), as compared to wild-type tumors. No BRAF V600E mutation was found in any tumor or corresponding normal duodenal tissue.
Survival Analysis by CIMP and MSI
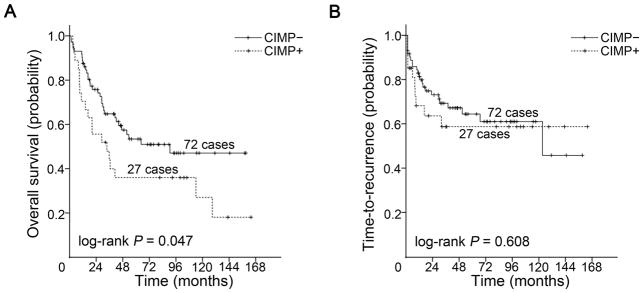
Median follow-up of patients was 36.9 months for OS analysis and 30.5 months for TTR analysis. Kaplan–Meier survival curves were generated according to clinicopathological and molecular characteristics. The median OS for the entire group was 53.7 months, with 5- and 10-year OS of 49% and 35% respectively. Age, stage, and MSI status were three important predictors of OS with older age and late stage conferring worse OS while MSI was associated with improved OS, as expected (log-rank P < 0.05, all; Supplementary Fig. S3). CIMP+ was significantly associated with shorter OS (log-rank P = 0.047; Fig. 3A). The median OS time was 33.9 months in patients with CIMP+ tumors (5- and 10-year OS of 36% and 27% respectively) compared with 90.8 months in patients with CIMP− tumors (5- and 10-year OS of 53% and 47%; Supplementary Table S3). CIMP alone was, however, not a predictor for TTR (log-rank P = 0.608; Fig. 3B). The median TTR time for the CIMP− group was 123.4 months and had not been reached for the CIMP+ group. Age, stage, differentiation, and chemotherapy and/or radiotherapy were predictors of TTR with young age, late stage, poor differentiation and undergoing chemotherapy and/or radiotherapy conferring worse TTR (log-rank P < 0.05, all; Supplementary Fig. S4).
Figure 3.
Kaplan-Meier survival estimates of overall survival and time-to-recurrence in patients with CIMP+ and CIMP− duodenal adenocarcinomas. (A) overall survival, (B) time-to-recurrence.
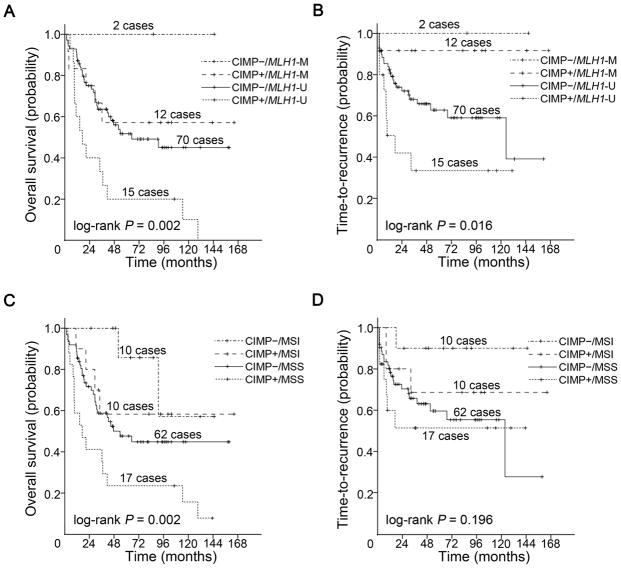
Tumors were further classified by CIMP and MLH1 methylation status into: CIMP−/MLH1-U (n = 70), CIMP−/MLH1-M (n = 2), CIMP+/MLH1-U (n = 15) and CIMP+/MLH1-M (n = 12) groups. There were significant differences both in OS (log-rank P = 0.002; Fig. 4A) and TTR (log-rank P = 0.016; Fig. 4B) in the groups classified by CIMP/MLH1 methylation status. CIMP+/MLH1-U group had the shortest OS and TTR whereas CIMP−/MLH1-M group had the longest OS and TTR. CIMP−/MLH1-M group consisted of two patients with a remarkable recurrence-free follow-up of 85.8 and 144.9 months at the conclusion of the study, respectively.
Figure 4.
Kaplan-Meier survival estimates of overall survival and time-to-recurrence in patients with duodenal adenocarcinomas. Overall survival in (A) groups classified by CIMP/MLH1 methylation status, (C) groups classified by CIMP/MSI status. Time-to-recurrence in (B) groups classified by CIMP/MLH1 methylation status, (D) groups classified by CIMP/MSI status. The P values shown have been pooled over strata.
Tumors were also categorized by CIMP and MSI status into: CIMP−/MSS (n = 62), CIMP−/MSI (n = 10), CIMP+/MSS (n = 17) and CIMP+/MSI (n = 10) groups. In the groups classified by CIMP/MSI status, there was significant difference in OS (log-rank P = 0.002; Fig. 4C), but not in TTR (log-rank P = 0.196; Fig. 4D) with CIMP+/MSS group having the worst OS.
Multivariate Analysis of Outcome Predictors
ACox proportional hazards model for multivariate analysis including CIMP/MLH1 methylation status, age, sex, stage, tumor differentiation, chemotherapy and/or radiotherapy, and KRAS mutation status in relation to OS and TTR was performed (Table 2). Only CIMP/MLH1 methylation status (P < 0.001), age (P = 0.002), and stage (P < 0.001) remained statistically significant as predictors of OS. CIMP/MLH1 methylation status (P = 0.023), stage (P = 0.020), along with tumor differentiation (P = 0.034) were also associated with risk of recurrence and independently predicted TTR.
Table 2.
Univariate and multivariate Cox proportional hazard analysis of overall survival (OS) and time-to-recurrence (TTR)
| Characteristic | Total n | OS
|
TTR
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| Univariate
|
Multivariate
|
Univariate
|
Multivariate
|
||||||
| HR (95% CI) | P value | HR (95% CI) | P value | HR (95% CI) | P value | HR (95% CI) | P value | ||
| CIMP/MLH1 methylation | 0.011 | < 0.001 | 0.062 | 0.023 | |||||
| CIMP−/MLH1-U | 70 | 1.00 (Referent) | 1.00 (Referent) | 1.00 (Referent) | 1.00 (Referent) | ||||
| CIMP+/MLH1-U | 15 | 2.76 (1.46, 5.19) | 0.002 | 4.73 (2.34, 9.54) | < 0.001 | 2.26 (1.04, 4.88) | 0.039 | 3.33 (1.39, 8.00) | 0.007 |
| CIMP+/MLH1-M | 12 | 0.80 (0.31, 2.07) | 0.649 | 0.43 (0.15, 1.24) | 0.117 | 0.21 (0.03, 1.57) | 0.212 | 0.26 (0.03, 2.10) | 0.204 |
| CIMP−/MLH1-M | 2 | 0 (0, –) | 0.971 | 0 (0, –) | 0.973 | 0 (0, –) | 0.977 | 0 (0, ) | 0.978 |
| Age | |||||||||
| <70 | 64 | 1.00 (Referent) | 1.00 (Referent) | 1.00 (Referent) | 1.00 (Referent) | ||||
| ≥70 | 35 | 1.73 (0.99, 3.00) | 0.053 | 2.65 (1.42, 4.92) | 0.002 | 0.27 (0.09, 0.75) | 0.013 | 0.47 (0.15, 1.41) | 0.177 |
| Sex | |||||||||
| Male | 55 | 1.00 (Referent) | 1.00 (Referent) | 1.00 (Referent) | 1.00 (Referent) | ||||
| Female | 44 | 0.96 (0.55, 1.67) | 0.887 | 1.45 (0.80, 2.65) | 0.221 | 0.50 (0.24, 1.03) | 0.061 | 0.90 (0.42, 1.94) | 0.790 |
| Stage | |||||||||
| Stage I and II | 29 | 1.00 (Referent) | 1.00 (Referent) | 1.00 (Referent) | 1.00 (Referent) | ||||
| Stage III and IV | 70 | 2.72 (1.35, 5.45) | 0.005 | 5.27 (2.25, 12.38) | < 0.001 | 4.88 (1.71, 13.97) | 0.003 | 4.28 (1.26, 14.50) | 0.020 |
| Differentiation | |||||||||
| Well/moderately | 55 | 1.00 (Referent) | 1.00 (Referent) | 1.00 (Referent) | 1.00 (Referent) | ||||
| Poorly | 44 | 1.65 (0.95, 2.87) | 0.078 | 1.42 (0.78, 2.56) | 0.252 | 3.01 (1.48, 6.12) | 0.002 | 2.27 (1.06,4.84) | 0.034 |
| Chemotherapy/radiotherapy | |||||||||
| No | 38 | 1.00 (Referent) | 1.00 (Referent) | 1.00 (Referent) | 1.00 (Referent) | ||||
| Yes | 61 | 1.42 (0.79, 2.57) | 0.246 | 1.10 (0.55, 2.20) | 0.778 | 3.67 (1.49, 9.03) | 0.005 | 1.87 (0.71, 4.92) | 0.203 |
| KRAS mutations | |||||||||
| Absent | 67 | 1.00 (Referent) | 1.00 (Referent) | 1.00 (Referent) | 1.00 (Referent) | ||||
| Present | 32 | 1.09 (0.61, 1.95) | 0.772 | 0.64 (0.34, 1.20) | 0.160 | 1.02 (0.50, 2.09) | 0.960 | 0.51 (0.22, 1.17) | 0.111 |
Abbreviations: OS, overall survival; TTR, time-to-recurrence; HR, hazard ratio; CI, confidence interval; CIMP, CpG island methylator phenotype; MLH1-M, MLH1-methylated; MLH1-U, MLH1-unmethylated; MSS, microsatellite stable; MSI, microsatellite instability
The influence of CIMP, MLH1 methylation, or MSI on OS and TTR, independent of the clinicopathological and molecular variables were separately assessed (Supplementary Table S4). In multivariate analyses, CIMP by itself only showed a trend toward correlation with both OS (P = 0.081) and TTR (P = 0.176). MLH1 methylation status was independently associated with OS (P = 0.021), but not TTR (P = 0.070). MSI independently correlated with both OS (P = 0.003) and TTR (P = 0.018).
Discussion
The CIMP was first characterized in human CRC by our group as cancer-specific CpG island hypermethylation of a subset of genes in a subset of tumors (7). Weisenberger et al. confirmed and further characterized CRC CIMP using MethyLight technology (6). Since then CIMP has been demonstrated in multiple other malignancies including gastric (29), pancreatic (20), lung (21), oral (22), breast (30), and small intestinal cancers (31), as well as neuroblastoma (32), malignant melanoma (23), and glioma (33). In present study, we analyzed a large cohort of patients with duodenal adenocarcinomas and showed that CIMP+ existed in 27.3% of the tumors.
There is no consensus regarding the best markers for defining CIMP in duodenal cancer. Fang et al. compared the CIMP-associated loci from breast cancer, colon cancer, and glioma, and found that the CIMP signature was shared by multiple human malignancies (30). By using CIMP-associated loci in CRC, previous studies have successfully identified CIMP tumors in duodenal cancers (9, 31). In this study, CIMP was defined by a panel of five markers proposed and validated by Weisenberger and colleagues (6). This 5-gene signature used has been shown to be highly accurate and the most cost-effective screening method for CIMP status in CRC (6). The question therefore arises as to whether this panel of markers would also be applicable in duodenal cancers. Ideally, it would be helpful to utilize a whole epigenome approach to define CIMP in cancers. However, this is not feasible given the rarity of duodenal adenocarcinomas and lack of appropriate fresh tissue samples to perform this analysis. In order to confirm if the 5-gene signature truly differentiates a CIMP+ group, we screened a panel of 20 commonly used markers for CIMP. Importantly, duodenal tumors that were identified as CIMP+ by the 5-gene signature were concordant with those positive on the large-scale screen. Our results demonstrated that this 5-gene signature correlated with CIMP and accurately define CIMP in duodenal adenocarcinomas.
It has been established that KRAS and BRAF mutations have a number of downstream effectors that can activate or repress genes, and which may then contribute to patterning the epigenome (34). In our data KRAS gene mutation was associated with tumors that had at least one gene methylated, and this is in accordance with the evidence of induction of the ras oncogenic pathway may result in DNA methylation (35). Yet it appears that KRAS mutations alone do not dictate duodenal cancer CIMP status since KRAS mutations were not associated with CIMP as compared to wild-type tumors.
Moreover in CRC, CIMP status has been associated with mutations of the BRAF gene and has been felt to be mutually exclusive to KRAS mutations (6). In fact, we did not detect any mutations in codon 600 of the BRAF gene. This is in keeping with Blaker and colleagues who described only one mutation (a 3 bp (GAT) deletion at codon 603/604) in a panel of 21 adenocarcinomas of the small intestine (36). It is possible that other mutations exist outside codon 600 of the BRAF gene and would have been detected if we had screened the whole of the BRAF gene, however most BRAF mutations in human cancers are within codon 600 (10, 37). These data lead us to conclude that BRAF mutations are not critically involved in duodenal tumorigenesis, MSI or CIMP development.
It was reported that CIMP+ CRCs have a distinct clinicopathological and molecular features, such as associations with proximal tumor location, female sex, poor differentiation and mucinous tumor histology, MSI as a consequence of hypermethylation of the MLH1 gene, and high BRAF mutation rate (6). In our study, we were unable to find any association of CIMP with gender or tumor differentiation, but not surprisingly, we found a strong correlation between CIMP and both MLH1 methylation and MSI in duodenal cancers. Interestingly, we also found that only half of CIMP+ tumors showed MLH1 methylation and a small number of MLH1 methylated tumors were CIMP−. IHC analysis of MLH1 expression in those MLH1-M tumors also validated the methylation. These observations are consistent with a stochastic process of cancer methylation and a gradually increasing probability of MLH1 methylation (38). On a basis of a limited number of cases, we found that CIMP+ duodenal cancers had a relatively earlier stage when compared with CIMP− tumors though this was not statistically significant. The result indicates that CIMP development is an early event in some cases of duodenal cancer.
Several studies have investigated the relationship between CIMP status and survival in various malignancies. However, these results are inconsistent (30, 39–46). The association of better clinical outcome with CIMP+ tumors has been reported in CRC, gliomas and breast cancer. Poor prognosis with CIMP+ tumors has also been reported across CRC (44), esophageal cancer (45), gastric cancer (46), myelodysplastic syndromes (43), neuroblastomas (41), and leukemia (42). The discrepancy of these observations might be due to different methylation markers of CIMP panels, methodologies for methylation detection, patient populations, distribution of tumor stages and differentiations, terms of follow-up, other factors associated with prognosis being included (such as chemotherapy and/or radiotherapy).
The prognostic significance of CIMP status in duodenal cancer has not previously been described. We identified a patient population that was CIMP+ and found that this inversely correlated with survival. Stratification of CIMP by MSI status was predictive of OS but not TTR. However, stratification of CIMP by MLH1 methylation status further enhances the ability to predict OS as well as predict TTR in CIMP+ patients on multivariate analysis. CIMP+/MLH1-U patients had a poorer outcome than individuals with CIMP−/MLH1-U tumors and indeed compared with all other individuals in the study. Interestingly, there were only two patients with CIMP− but MLH1 methylated tumors, both of them did extremely well with long term follow-up. This is particularly surprising since one of the tumors was poorly differentiated, the other was moderately differentiated, and both were advanced stage (stage III). Most importantly, in 27 CIMP+ tumors, there were 17 MSS and 10 MSI tumors. Even though multivariate analysis showed that MSI was an independent predictor of OS and TTR, stratification of CIMP+ tumors by MSI could not clearly define two subtypes. On the contrary, MLH1 methylation status segregated the CIMP+ tumors into 15 MLH1-U tumors and 12 MLH1-M tumors. Both subtypes behave differently with significantly different OS and TTR, which indicate that CIMP+ tumors may follow two different pathways. The significant survival difference between the groups classified by CIMP/MLH1 methylation status might imply a clue to the complexity of CIMP development. It suggests that not only oncogenic pathways but also epigenetic pathways themselves jointly affect CIMP in a pattern of reciprocal causation.
An important limitation of our study is the lack of statistical power because of a low number of patients in some subgroups might obscure more subtle relations. The analysis in a larger cohort of duodenal adenocarcinomas is needed to validate our findings.
In conclusion, our data suggests that CIMP does exist in duodenal adenocarcinomas and it may assist in the prognostic classification of these patients. Stratification of CIMP by MLH1 methylation status enhances the ability to predict OS as well as predict TTR. Patients with CIMP+ duodenal adenocarcinomas, especially those with CIMP+ tumors in absence of MLH1 methylation, may need more intensive surveillance and subtype-specific adjuvant therapy strategies. We did not detect any BRAF V600E mutation, which suggests that BRAF mutations are not critically involved in duodenal tumorigenesis, MSI or CIMP development. These results give new insight into the genetic and epigenetic pathways of duodenal adenocarcinoma and demonstrate the need for further understanding of these unique tumors.
Supplementary Material
Translational relevance.
CpG island methylator phenotype (CIMP) has been found in multiple malignancies including duodenal adenocarcinoma but has not been further characterized due to the rarity of this disease. Using a large cohort of duodenal adenocarcinomas, we prove that CIMP exists in duodenal adenocarcinomas and is associated with MSI and MLH1 methylation. No BRAF V600E mutation has been detected in this study, indicating that BRAF mutations are not critically involved in duodenal tumorigenesis, MSI or CIMP development. CIMP+ is a prognostic marker for poor overall survival in patients with duodenal adenocarcinomas. CIMP+ in the absence of MLH1 methylation is a marker for poor overall survival and time-to-recurrence in duodenal cancers. Our findings highlight the usefulness of CIMP classification for prognosis prediction. Patients with CIMP+ duodenal adenocarcinomas especially those with CIMP+/MLH1-U tumors may need more intensive surveillance and novel subtype-specific adjuvant therapy strategies after surgery.
Acknowledgments
We would like to thank Kathy Bender and Rickey Moore for administrative support. We would also like to thank Sharon Metzger-Gaud, Theresa Sanlorenzo-Caswell, and the Johns Hopkins Cancer Registry for assistance with the primary cancer databases.
Grant Support
This work was supported by National Institutes of Health grant CA140599 and CA127141, and National Natural Science Foundation of China grant 81000898.
Footnotes
Disclosure of Potential Conflicts of Interest
S.B.B has commercial grant funding and serves on the advisory board for MDx Health Inc. and BioNumerik Pharmaceuticals Inc.
References
- 1.Alwmark A, Andersson A, Lasson A. Primary carcinoma of the duodenum. Ann Surg. 1980;191:13–8. doi: 10.1097/00000658-198001000-00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Qubaiah O, Devesa SS, Platz CE, Huycke MM, Dores GM. Small intestinal cancer: a population-based study of incidence and survival patterns in the United States, 1992 to 2006. Cancer Epidemiol Biomarkers Prev. 2010;19:1908–18. doi: 10.1158/1055-9965.EPI-10-0328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vasen HF, Watson P, Mecklin JP, Lynch HT. New clinical criteria for hereditary nonpolyposis colorectal cancer (HNPCC, Lynch syndrome) proposed by the International Collaborative group on HNPCC. Gastroenterology. 1999;116:1453–6. doi: 10.1016/s0016-5085(99)70510-x. [DOI] [PubMed] [Google Scholar]
- 4.Koornstra JJ, Kleibeuker JH, Vasen HF. Small-bowel cancer in Lynch syndrome: is it time for surveillance? Lancet Oncol. 2008;9:901–5. doi: 10.1016/S1470-2045(08)70232-8. [DOI] [PubMed] [Google Scholar]
- 5.Herman JG, Umar A, Polyak K, Graff JR, Ahuja N, Issa JP, et al. Incidence and functional consequences of hMLH1 promoter hypermethylation in colorectal carcinoma. Proc Natl Acad Sci U S A. 1998;95:6870–5. doi: 10.1073/pnas.95.12.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weisenberger DJ, Siegmund KD, Campan M, Young J, Long TI, Faasse MA, et al. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet. 2006;38:787–93. doi: 10.1038/ng1834. [DOI] [PubMed] [Google Scholar]
- 7.Toyota M, Ahuja N, Ohe-Toyota M, Herman JG, Baylin SB, Issa JP. CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci U S A. 1999;96:8681–6. doi: 10.1073/pnas.96.15.8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frazier ML, Xi L, Zong J, Viscofsky N, Rashid A, Wu EF, et al. Association of the CpG island methylator phenotype with family history of cancer in patients with colorectal cancer. Cancer Res. 2003;63:4805–8. [PubMed] [Google Scholar]
- 9.Kim SG, Chan AO, Wu TT, Issa JP, Hamilton SR, Rashid A. Epigenetic and genetic alterations in duodenal carcinomas are distinct from biliary and ampullary carcinomas. Gastroenterology. 2003;124:1300–10. doi: 10.1016/s0016-5085(03)00278-6. [DOI] [PubMed] [Google Scholar]
- 10.Garnett MJ, Marais R. Guilty as charged: B-RAF is a human oncogene. Cancer Cell. 2004;6:313–9. doi: 10.1016/j.ccr.2004.09.022. [DOI] [PubMed] [Google Scholar]
- 11.Guan RJ, Fu Y, Holt PR, Pardee AB. Association of K-ras mutations with p16 methylation in human colon cancer. Gastroenterology. 1999;116:1063–71. doi: 10.1016/s0016-5085(99)70009-0. [DOI] [PubMed] [Google Scholar]
- 12.Shen L, Toyota M, Kondo Y, Lin E, Zhang L, Guo Y, et al. Integrated genetic and epigenetic analysis identifies three different subclasses of colon cancer. Proc Natl Acad Sci U S A. 2007;104:18654–9. doi: 10.1073/pnas.0704652104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ogino S, Kawasaki T, Kirkner GJ, Loda M, Fuchs CS. CpG island methylator phenotype-low (CIMP-low) in colorectal cancer: possible associations with male sex and KRAS mutations. J Mol Diagn. 2006;8:582–8. doi: 10.2353/jmoldx.2006.060082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oliveira C, Westra JL, Arango D, Ollikainen M, Domingo E, Ferreira A, et al. Distinct patterns of KRAS mutations in colorectal carcinomas according to germline mismatch repair defects and hMLH1 methylation status. Hum Mol Genet. 2004;13:2303–11. doi: 10.1093/hmg/ddh238. [DOI] [PubMed] [Google Scholar]
- 15.Hinoue T, Weisenberger DJ, Pan F, Campan M, Kim M, Young J, et al. Analysis of the association between CIMP and BRAF in colorectal cancer by DNA methylation profiling. PLoS One. 2009;4:e8357. doi: 10.1371/journal.pone.0008357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yachida S, Mudali S, Martin SA, Montgomery EA, Iacobuzio-Donahue CA. β-catenin nuclear labeling is a common feature of sessile serrated adenomas and correlates with early neoplastic progression after BRAF activation. Am J Surg Pathol. 2009;33:1823–32. doi: 10.1097/PAS.0b013e3181b6da19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dietmaier W, Wallinger S, Bocker T, Kullmann F, Fishel R, Ruschoff J. Diagnostic microsatellite instability: definition and correlation with mismatch repair protein expression. Cancer Res. 1997;57:4749–56. [PubMed] [Google Scholar]
- 18.Eads CA, Danenberg KD, Kawakami K, Saltz LB, Blake C, Shibata D, et al. MethyLight: a high-throughput assay to measure DNA methylation. Nucleic Acids Res. 2000;28:E32. doi: 10.1093/nar/28.8.e32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci U S A. 1996;93:9821–6. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ueki T, Toyota M, Sohn T, Yeo CJ, Issa JP, Hruban RH, et al. Hypermethylation of multiple genes in pancreatic adenocarcinoma. Cancer Res. 2000;60:1835–9. [PubMed] [Google Scholar]
- 21.Suzuki M, Shigematsu H, Iizasa T, Hiroshima K, Nakatani Y, Minna JD, et al. Exclusive mutation in epidermal growth factor receptor gene, HER-2, and KRAS, and synchronous methylation of nonsmall cell lung cancer. Cancer. 2006;106:2200–7. doi: 10.1002/cncr.21853. [DOI] [PubMed] [Google Scholar]
- 22.Shaw RJ, Hall GL, Lowe D, Bowers NL, Liloglou T, Field JK, et al. CpG island methylation phenotype (CIMP) in oral cancer: associated with a marked inflammatory response and less aggressive tumour biology. Oral Oncol. 2007;43:878–86. doi: 10.1016/j.oraloncology.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 23.Tanemura A, Terando AM, Sim MS, van Hoesel AQ, de Maat MF, Morton DL, et al. CpG island methylator phenotype predicts progression of malignant melanoma. Clin Cancer Res. 2009;15:1801–7. doi: 10.1158/1078-0432.CCR-08-1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Williams CS, Zhang B, Smith JJ, Jayagopal A, Barrett CW, Pino C, et al. BVES regulates EMT in human corneal and colon cancer cells and is silenced via promoter methylation in human colorectal carcinoma. J Clin Invest. 2011;121:4056–69. doi: 10.1172/JCI44228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schuebel KE, Chen W, Cope L, Glockner SC, Suzuki H, Yi JM, et al. Comparing the DNA hypermethylome with gene mutations in human colorectal cancer. PLoS Genet. 2007;3:1709–23. doi: 10.1371/journal.pgen.0030157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yi JM, Dhir M, Van Neste L, Downing SR, Jeschke J, Glockner SC, et al. Genomic and epigenomic integration identifies a prognostic signature in colon cancer. Clin Cancer Res. 2011;17:1535–45. doi: 10.1158/1078-0432.CCR-10-2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brandes JC, van Engeland M, Wouters KA, Weijenberg MP, Herman JG. CHFR promoter hypermethylation in colon cancer correlates with the microsatellite instability phenotype. Carcinogenesis. 2005;26:1152–6. doi: 10.1093/carcin/bgi058. [DOI] [PubMed] [Google Scholar]
- 28.Marsit CJ, Houseman EA, Christensen BC, Eddy K, Bueno R, Sugarbaker DJ, et al. Examination of a CpG island methylator phenotype and implications of methylation profiles in solid tumors. Cancer Res. 2006;66:10621–9. doi: 10.1158/0008-5472.CAN-06-1687. [DOI] [PubMed] [Google Scholar]
- 29.Toyota M, Ahuja N, Suzuki H, Itoh F, Ohe-Toyota M, Imai K, et al. Aberrant methylation in gastric cancer associated with the CpG island methylator phenotype. Cancer Res. 1999;59:5438–42. [PubMed] [Google Scholar]
- 30.Fang F, Turcan S, Rimner A, Kaufman A, Giri D, Morris LG, et al. Breast cancer methylomes establish an epigenomic foundation for metastasis. Sci Transl Med. 2011;3:75ra25. doi: 10.1126/scitranslmed.3001875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Warth A, Kloor M, Schirmacher P, Blaker H. Genetics and epigenetics of small bowel adenocarcinoma: the interactions of CIN, MSI, and CIMP. Mod Pathol. 2011;24:564–70. doi: 10.1038/modpathol.2010.223. [DOI] [PubMed] [Google Scholar]
- 32.Abe M, Watanabe N, McDonell N, Takato T, Ohira M, Nakagawara A, et al. Identification of genes targeted by CpG island methylator phenotype in neuroblastomas, and their possible integrative involvement in poor prognosis. Oncology. 2008;74:50–60. doi: 10.1159/000139124. [DOI] [PubMed] [Google Scholar]
- 33.Noushmehr H, Weisenberger DJ, Diefes K, Phillips HS, Pujara K, Berman BP, et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17:510–22. doi: 10.1016/j.ccr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nagasaka T, Sasamoto H, Notohara K, Cullings HM, Takeda M, Kimura K, et al. Colorectal cancer with mutation in BRAF, KRAS, and wild-type with respect to both oncogenes showing different patterns of DNA methylation. J Clin Oncol. 2004;22:4584–94. doi: 10.1200/JCO.2004.02.154. [DOI] [PubMed] [Google Scholar]
- 35.Gazin C, Wajapeyee N, Gobeil S, Virbasius CM, Green MR. An elaborate pathway required for Ras-mediated epigenetic silencing. Nature. 2007;449:1073–7. doi: 10.1038/nature06251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blaker H, Helmchen B, Bonisch A, Aulmann S, Penzel R, Otto HF, et al. Mutational activation of the RAS-RAF-MAPK and the Wnt pathway in small intestinal adenocarcinomas. Scand J Gastroenterol. 2004;39:748–53. doi: 10.1080/00365520410005847. [DOI] [PubMed] [Google Scholar]
- 37.Wan PT, Garnett MJ, Roe SM, Lee S, Niculescu-Duvaz D, Good VM, et al. Mechanism of activation of the RAF-ERK signaling pathway by oncogenic mutations of B-RAF. Cell. 2004;116:855–67. doi: 10.1016/s0092-8674(04)00215-6. [DOI] [PubMed] [Google Scholar]
- 38.Hawkins N, Norrie M, Cheong K, Mokany E, Ku SL, Meagher A, et al. CpG island methylation in sporadic colorectal cancers and its relationship to microsatellite instability. Gastroenterology. 2002;122:1376–87. doi: 10.1053/gast.2002.32997. [DOI] [PubMed] [Google Scholar]
- 39.Dahlin AM, Palmqvist R, Henriksson ML, Jacobsson M, Eklof V, Rutegard J, et al. The role of the CpG island methylator phenotype in colorectal cancer prognosis depends on microsatellite instability screening status. Clin Cancer Res. 2010;16:1845–55. doi: 10.1158/1078-0432.CCR-09-2594. [DOI] [PubMed] [Google Scholar]
- 40.An C, Choi IS, Yao JC, Worah S, Xie K, Mansfield PF, et al. Prognostic significance of CpG island methylator phenotype and microsatellite instability in gastric carcinoma. Clin Cancer Res. 2005;11:656–63. [PubMed] [Google Scholar]
- 41.Abe M, Ohira M, Kaneda A, Yagi Y, Yamamoto S, Kitano Y, et al. CpG island methylator phenotype is a strong determinant of poor prognosis in neuroblastomas. Cancer Res. 2005;65:828–34. [PubMed] [Google Scholar]
- 42.Roman-Gomez J, Jimenez-Velasco A, Agirre X, Prosper F, Heiniger A, Torres A. Lack of CpG island methylator phenotype defines a clinical subtype of T-cell acute lymphoblastic leukemia associated with good prognosis. J Clin Oncol. 2005;23:7043–9. doi: 10.1200/JCO.2005.01.4944. [DOI] [PubMed] [Google Scholar]
- 43.Shen L, Kantarjian H, Guo Y, Lin E, Shan J, Huang X, et al. DNA methylation predicts survival and response to therapy in patients with myelodysplastic syndromes. J Clin Oncol. 2010;28:605–13. doi: 10.1200/JCO.2009.23.4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barault L, Charon-Barra C, Jooste V, de la Vega MF, Martin L, Roignot P, et al. Hypermethylator phenotype in sporadic colon cancer: study on a population-based series of 582 cases. Cancer Res. 2008;68:8541–6. doi: 10.1158/0008-5472.CAN-08-1171. [DOI] [PubMed] [Google Scholar]
- 45.Ling Y, Huang G, Fan L, Wei L, Zhu J, Liu Y, et al. CpG island methylator phenotype of cell-cycle regulators associated with TNM stage and poor prognosis in patients with oesophageal squamous cell carcinoma. J Clin Pathol. 2011;64:246–51. doi: 10.1136/jcp.2010.082875. [DOI] [PubMed] [Google Scholar]
- 46.Park SY, Kook MC, Kim YW, Cho NY, Jung N, Kwon HJ, et al. CpG island hypermethylator phenotype in gastric carcinoma and its clinicopathological features. Virchows Arch. 2010;457:415–22. doi: 10.1007/s00428-010-0962-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.