Abstract
The nucleotide-binding domain leucine-rich repeat proteins (NLRs) represent the major class of intracellular innate immune receptors in plants and animals. Understanding their functions is a major challenge in immunology. This review highlights recent efforts toward elucidating NLR functions in human and plants. We compare unconventional aspects of NLR proteins across the two kingdoms. We review recent advances describing P-loop independent activation, nuclear-cytoplasmic trafficking, oligomerization and multimerization requirements for signaling, and for expanded functions beyond pathogen recognition by several NLR proteins.
Introduction
Plants and animals are exposed to a wide array of infectious agents. In both cases, the host–pathogen interaction involves recognition of non-self by the host immune system, amplification of immune responses, and eventual attenuation or elimination of the pathogen. Host–pathogen interactions are dynamic liaisons in which the two partners influence each other’s evolution.
Plants lack an adaptive immune system and rely solely on their innate immune system to counteract infection. Perception of non-self molecules in plants occurs in two different layers. In the first, microbe-associated molecular patterns (MAMPs) conserved within a class of microbe are recognized by polymorphic plasma membrane-spanning pattern recognition receptors (PRRs), typically characterized by an extracellular domain with LRR or lysine motifs, and MAMP-triggered immunity (MTI) ensues [1,2]. Pathogens evolved effector molecules to counteract host surveillance and suppress MTI. In response, plants evolved a second layer of defense that utilizes effector-specific intracellular NLR receptors that are activated to trigger effector-triggered immunity (ETI).
Similarly, animals employ a limited repertoire of PRRs to recognize MAMPs or endogenous molecules that result from pathogen invasion, referred to as damage-associated molecular patterns (DAMPs), resulting in inflammatory signaling [3]. Animal PRRs include Toll-like receptors (TLRs), C-type lectin receptors (CLRs), retinoic acid-inducible gene (RIG)-I-like receptors (RLRs), and, the topic of this review, NLRs [4].
Plant NLRs either directly recognize specific effectors, or indirectly through the action of an effector on a host target (modification of self). The repertoire of plant NLRs is large (~160 in Arabidopsis [5•]) and highly polymorphic. The smaller number of animal NLR genes, ~20 in mammals [6], recognizes MAMPs or modified-self in the form of DAMPs. As in plants, animal NLRs likely recognize bacterial ligands through direct and specific interaction [7••]. Despite structural and, at least superficially, functional similarity, plant and animal NLRs are proposed to have evolved independently with convergent function [8]. Despite the lack of NLRs in the Caenorhabditis elegans and drosophila genomes [6], these proteins show a remarkable expansion to over 200 genes in sea urchin [9].
NLRs as molecular switches: STANDard and unconventional routes of activation
NLR proteins belong to the signal transduction ATPases with numerous domains (STAND) subclade of the AAA-ATPase superfamily. They are characterized by a centrally located nucleotide-binding domain, a variable number of highly polymorphic C-terminal leucine-rich repeats (LRRs), and diverse N-termini (Figure 1). STAND proteins are molecular switches regulated via nucleotide-binding [10,11]. The ADP-bound form represents the resting ‘off’ state. Upon pathogen recognition, a conformational change allows ADP to be exchanged for ATP, and the active ‘on’ state initiates downstream signaling. ATP hydrolysis is the core mechanism that regulates this switch and ensures return to the inactive state (Figure 2). Direct nucleotide binding to STAND proteins has been demonstrated for only a few NLR proteins [12•,13–18]. The highly conserved Walker-A (or P-loop) motif (GxxxxGK[T/S]) is an integral part of the nucleotide-binding site and the lysine residue is crucial for the coordination of the nucleotide β-phosphate. The Walker-B (hhhDD/E), or an extended Walker-B motif (DGhDE) in the case of most animal NLRs, is also common [19]. In the conventional Walker-B, the first aspartate residue coordinates the Mg2+ cation and is required for nucleotide binding, whereas the second acidic residue is thought to prime a water molecule for nucleotide hydrolysis [20].
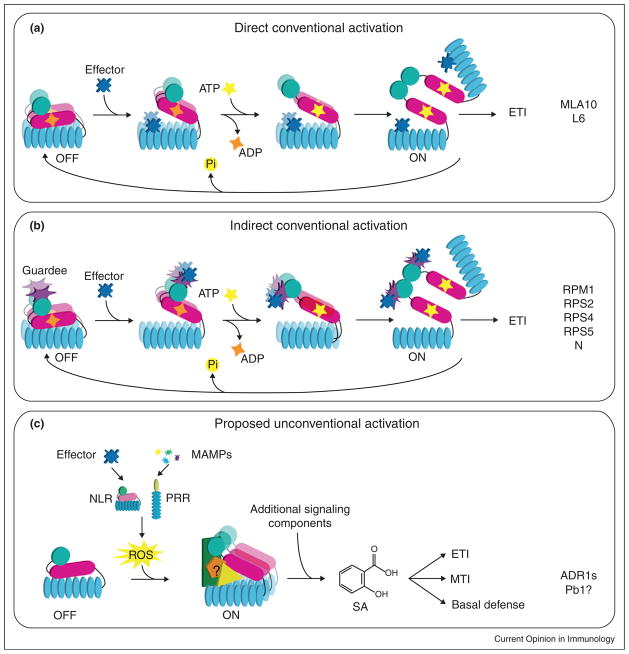
Figure 1.
Architecture of NLR proteins. Domain organization of (a) plant NLRs and (b) animal NLRs is depicted and representative members of each category are shown. (c) The conserved domains of NLR proteins are represented. N-terminal coiled-coil (CC) and Toll/interleukin-1 (TIR) and C-terminal WRKY domains are specific to plants, whereas N-terminal baculoviral inhibitory repeat (BIR) domain, caspase recruitment domain (CARD), pyrin domain (PYD), activation domain (AD), and undefined domains are specific to animals.
Figure 2.
Model for activation of plant NLR proteins. (a and b) NLRs are normally in an ‘off’ state that is achieved by the inhibitory function of the LRR domain preventing the protein activation via the NB molecular switch. The NLR can either be a dimer (MLA10) or a monomer (L6) before activation. NLR proteins can recognize the effector protein directly through the LRR domain (a), or through modifications of the host target of an effector that typically interacts with the NLR N-terminal domain (b). Recognition of the pathogen effector triggers the release of the inhibitory LRR domain and this conformational change allows for exchange of ADP to ATP. Binding of ATP results in a second conformational change that allows the N-terminal domains to physically interact, thus activating the NLR. Based on the dimerization of NOD1 through the CARD domains, this mechanism is likely to be conserved in animal NLRs. (c) When a P-loop is not required for NLR function, we propose that the NLR is activated by an oxidative burst resulting from either NLR-mediated or PRR-mediated effector-dependent or MAMP-dependent recognition. Unconventional NLRs could function as scaffolds for interactions with unknown partners. Heterotypic or homotypic interactions with accessory proteins or other NLRs could result in the activation of downstream signaling which leads to ETI, MTI, and basal defense.
The importance of nucleotide binding for plant NLR activation is reflected by the auto-activation phenotypes caused by several missense mutations in the moderately conserved, plant-specific MHD motif in the NB domain [19]. The precise effects of such mutations in nucleotide binding are largely unknown. Animal NLRs do not contain an MHD motif, but a functionally conserved histidine is part of the corresponding WH domain, and this motif is required to coordinate the β-phosphate of the nucleotide in the binding pocket [21]. Biochemical analysis of an MHD mutation (D555V) in the flax M NLR protein revealed that the auto-activation phenotype is due to increased ATP binding [12•], suggesting either: (i) a conformational change in the NLR protein that favors ATP binding versus ADP binding; or (ii) a reduced ATPase activity that prevents the protein from switching back into an inactive state.
A more complex activation/deactivation mechanism characterizes mammalian Nod1 and Nod2 NLRs, which recognize bacterial peptidoglycan (PGN). In these cases indirect elicitor recognition likely induces oligomerization, an essential step for subsequent signaling. Nod1 and Nod2 employ different mechanisms of activation. While an initial ATP hydrolysis step mediated by the first acidic residue of the extended Walker-B DE motif is necessary for both Nod1 and Nod2 activation, Nod2, but not Nod1, requires an additional ATP hydrolysis event mediated by the second acidic residue for deactivation of the signaling platform [22•].
Natural NLR variants provide evidence for unconventional activation mechanisms. A genetic study in rice showed that the panicle blast-resistance gene Pb1 encodes an unconventional NLR protein [23••] that contains two N-terminal CC domains, a degenerate NB domain that completely lacks the P-loop motif, and a C-terminal LRR domain. This unique structure indicates either that Pb1 lost its nucleotide-binding activity, and thus that activation does not conform to the traditional molecular switch, or that nucleotide-binding is achieved differently, potentially mediated by an insertion of 30 amino acids found within the Walker-B motif. The small Pb1 NLR family from rice shares the P-loop deletion and the insertion in the Walker-B, and these features seem conserved in maize.
An alternative model for NLR activation for at least some phenotypes was substantiated for the Arabidopsis ADR1 (activated disease resistance 1) family of CC-NLRs [24••]. Three members of this family (ADR1, ADR1-L1, and ADR1-L2) function additively as ‘helper NB-LRRs’ to transduce signals subsequent to specific NLR receptor activation during effector-triggered immunity, and they are required for basal defense against virulent pathogens. The ADR1 proteins are also required for accumulation of the phytohormone salicylic acid (SA), an essential signaling molecule for plant immune responses, following challenge with MAMPs encoded by a disarmed bacterial pathogen. Remarkably, in the case of ADR1-L2, none of these three phenotypes requires an intact P-loop motif, as a triple missense allele in three invariant residues (GKT > AAA) required for ATP binding retains function [24••].
Interestingly, a number of full-length and truncated NLR genes in Arabidopsis thaliana and A. lyrata could encode proteins that either possess a degenerate P-loop motif or completely lack the P-loop motif, analogous to rice Pb1 (Table 1). We propose that these proteins can act as scaffolds for interactions with as yet unknown immune function partners, as proposed for ADR1 and Pb1 (Figure 2). Such scaffolds could function as signalosomes required for downstream output responses, potentially as regulatory partners for more conventional nucleotide-binding NLRs. This is reminiscent of some human NLRs that can regulate signal transduction pathways either instead of, or in addition to, their function as microbial sensor [25•].
Table 1.
Unconventional P-loop motifs in full-length and truncated NLRs in Arabidopsis thaliana and A. lyrata.a
| Species | Protein ID # | P-loop motif | Architecture |
|---|---|---|---|
| Arabidopsis thaliana | At1g17615 | AxxxxGRS | TIR-NB |
| At1g72890 | GxxxxGRS | ||
| At1g72910 | AxxxxGRS | ||
| At1g72920 | AxxxxGRS | ||
| At1g72940 | AxxxxGRS | ||
| At1g72950 | AxxxxGRS | ||
| Arabidopsis thaliana | At1g72900 | GxxxxCRS | TIR-NB |
| Arabidopsis thaliana | At1g57650 | No consensus | NB-LRR |
| Arabidopsis lyrata | 860345 | SxxxxGGS | TIR-NB-LRR |
| Arabidopsis lyrata | 865712 | GxxxxGVA | TIR-NB-LRR |
| Arabidopsis lyrata | 882156 | GxxxxGRG | TIR-NB-LRR |
| Arabidopsis lyrata | 916973 | GxxxxGKA | TIR-NB-LRR |
| Arabidopsis lyrata | 875509 | No consensus | TIR-NB-LRR |
| 877022 | |||
| 886387 | |||
| 916966 | |||
| Arabidopsis lyrata | 880060 | AxxxxGRS | TIR-NB |
| 877388 | GxxxxGRS | ||
| 877391 | AxxxxGRS | ||
| 877392 | AxxxxGRS | ||
| 877393 | AxxxxGRS | ||
| Arabidopsis lyrata | 471970 | No consensus | TIR-NB |
| Arabidopsis lyrata | 888983 | No consensus | CC-NB-LRR |
| Arabidopsis lyrata | 856364 | No consensus | CC-NB |
| Arabidopsis lyrata | 874617 | GxxxxGVA | NB-LRR |
| Arabidopsis lyrata | 497514 | No consensus | NB-LRR |
| 856232 | |||
| 883426 | |||
| Arabidopsis lyrata | 856467 | No consensus | NB |
| 863489 | |||
| 875983 | |||
| 883439 |
Together we STAND, divided we fall
To prevent inappropriate activation and unnecessary damage, NLRs are under exquisite control. NLRs exist in an inactive state that relies on negative regulation exerted by the LRR on the NB domain [21,26–28]. Several reports indicate that pathogen recognition releases this auto-inhibition leading to activation of downstream signaling [27,29–32]. Global reduction of NLR stability via mutation of the RAR1 co-chaperone, reduces basal defense against virulent pathogens [33], suggesting that NLRs collectively cycle slowly between active and inactive states at a basal level, and that this sets the constitutive or primed basal defense output [34].
In animals, activation of NLRs can result in multimerization and the formation of a molecular scaffold that recruits additional components required for signaling [7••]. The crystal structure of the NLR-related CED-4 apoptosome reveals a homo-octomeric funnel-shaped structure [35], similarly electron microscopy data suggest the existence of the Apaf-1 apoptosome as a disc-shaped homoheptamer [36].
The NLR proteins NLRP3, NLRP1, and NLRC4 undergo homomerization and heteromerization with additional proteins to form a scaffold for the recruitment and activation of procaspase-1 [37,38]. Moreover, heteromerization of NLRC4 with either NAIP2 or NAIP5 is required for inflammasome specificity in response to direct recognition of bacterial ligands (PrgJ and flagellin respectively) [7••,39••].
Homodimerization is also inferred to be an essential step for plant NLR activation from co-immunoprecipitation of differentially tagged molecules [28,40,41]. Whether this is biologically relevant for function remains elusive. The N-termini of the barley CC-NLR MLA10 [42••] and of the flax TIR-NLR L6 [43••] were crystallized and structural analysis combined with mutagenesis confirmed homotypic dimer association for the respective N-terminal domains. Interestingly, loss-of-function mutations in other plant TIR-NLRs correspond to residues located on the L6 dimer interface, suggesting that homodimerization is also essential in these cases [43••].
Over-expression of the TIR domains of a number of plant NLRs can result in cell death, and this is interpreted to reflect ectopic activation [43••,44–46]. Over-expression of the CC domain of N-required gene 1 (NRG1), ADR1s [47•], or MLA10 [42••] all result in similar ectopic cell death phenotypes. NRG1 and ADR1 are both members of a rare class of divergent CCR-NLRs (previously CCRPW8) [47•] that lack the conserved EDVID motif thought to regulate intra-molecular interactions between the CC domain and the NB-LRR [48]. CCR-NLRs are ancient and conserved among flowering plants. The common evolution history of NRG1 and ADR1, and their ‘helper functions’ [24••,49] suggest that NRG1 might also act in a P-loop independent fashion. Transient over-expression of their respective CCR domains is sufficient to induce HR responses. However, this is not a unique feature of CCR proteins, since over-expression of the canonical MLA10 CC domain is also sufficient to trigger cell death [42••].
Unconventional functions of NLRs
As more reports on NLR protein function emerge, common features of activation appear less evident. Divergence in the mechanism of NLR is also reflected by novel functions beyond pathogen recognition. Functions for human NLRs in processes unrelated to pathogen detection have been reviewed [25•].
Interestingly, the NLR homolog Kaposi’s sarcoma-associated herpesvirus Orf63 [50•] shares homology with the NB and the LRR domains of human NLRP1, but lacks either the CARD or the PYD domain. Orf63 protein interacts with and subverts NLRP1 and NLRP3 inflammasomes [50•], perhaps because it lacks an N-terminal signaling domain and inhibits NLR multimerization.
This observation is reminiscent of bacterial TIR domains. TIR-containing proteins TlpA, TcpB, TcpC, and Btp1 from a range of pathogenic bacteria interfere with host TLR signaling and immune responses [51–53]. Interestingly, the phylogeny of the bacterial TIR domains is not congruent with the respective bacterial genome phylogeny, indicating that the bacterial TIR domains have spread via horizontal gene transfer [54].
NLR activation in plants typically results in a localized form of cell death around the infection site, known as hypersensitive response (HR). Arabidopsis lesion simulating disease 1 (lsd1) and accelerated cell death 11 (acd11) loss-of-function mutants are characterized by the constitutive activation of immune responses in the absence of an invader [55,56]. In lsd1 and acd11, programmed cell death is initiated but cannot be contained. Both lsd1 and acd11 phenotypes are associated with uncontrolled accumulation of SA, as mutations in the SA biosynthetic enzyme SID2 in these backgrounds suppresses cell death [57,58]. The CCR-NLR ADR1-L2 and the TIR-NLR LAZ5 function as positive regulators of lsd1 and acd11 cell death respectively [24••,59••], suggesting that the ectopic cell death phenotypes of acd11 and lsd1 are a result of the inappropriate activation of NLR proteins.
Set the control
Aberrant responses following inappropriate activation of the NLR receptors are common to animals and plants. Many gain-of-function mutations in human NLRs result in autoinflammatory diseases [25•]. Similarly, several aberrant phenotypes have been shown for a number of constitutively active plant NLRs [60]. In Arabidopsis, the uni-1D mutant results from a gain-of-function mutation in the LRR domain of a gene encoding a CC-NLR. uni-1D up-regulates SA-dependent Pathogenesis-Related (PR) genes and is characterized by SA-independent morphological defects through the accumulation of cytokinin, indicating that activation of this NLR protein activation can engage the cytokinin pathway [61].
Gain-of-function NLR mutations have also been linked to chilling responses. A mutation in the NB domain of Arabidopsis RPP4 (chs2) results in sensitivity to low temperature and autoimmunity [62]. However, no obvious phenotype can be observed at normal temperatures. Similarly chs3 exhibits temperature-sensitive phenotypes at chilling temperature as a result of activated defense responses, in this case mediated by the gain-of-function mutation in the CHS3 gene encoding an unusual TIR-NLR that carries a C-terminal zinc-binding LIM domain [63]. Additional evidence of inappropriate activation of NLR proteins in plants is represented by hybrid necrosis. Many inter-species or intra-species crosses result in drastic phenotypes such as dwarfism, necrosis, and sometimes lethality. The molecular events that trigger hybrid necrosis in tomato and Arabidopsis were recently shown to involve epistatic interaction between NLRs [64], suggesting that the incompatibility is due to a mis-regulated activation of the immune responses. All these studies suggest that exquisite control must be exerted on innate immune receptor activation in order to prevent aberrant and detrimental responses.
NLRs: insiders or outsiders?
Previous studies revealed the role of NLRs as cytoplasmic (in animals) or membrane-bound sensors (in plants) for MAMPs or pathogen effectors. A classic example is Arabidopsis CC-NLR Resistance to Pseudomonas syringae pv. maculicula 1 (RPM1) which resides on the inner plasma-membrane before, and upon activation [65••]. Although Nod1 and Nod2 were first described as cytosolic receptors, their distribution at the plasma membrane is crucial for the recruitment of RIP2, the activation of NF-kB, and for the recently described Nod-dependent autophagy to limit intracellular bacterial entry [22•,66,67•].
Emerging evidence denotes the importance of nucleo-cytoplasm trafficking of some immune receptors for proper immune responses. A classic example in animals is the master co-activator CIITA, a key regulator of major histocompatibility complex (MHC) class II gene expression that functions as a scaffold for transcription factors that target MHC class II gene promoters [68]. Similar to CIITA, NLRC5 was recently described as a transcriptional regulator of MHC class I that shuttles between cytosol and nucleus to mediate MHC class I immune responses [69••].
In plants, nuclear localization seems crucial for a number of NLRs. A putative function as transcriptional regulator for NLRs was suggested for Arabidopsis resistance to Ralstonia solanacearum 1 (RRS1), an atypical immune receptor that contains the TIR-NB-LRR domains and a WRKY motif of plant transcription factors. Although transcriptional activity has not yet been described for this protein, its nuclear localization [70] suggests that RRS1 could be a transcriptional regulator of the plant immune responses.
An additional striking example is Suppressor of npr1-1, Constitutive 1 (SNC1), an Arabidopsis TIR-NLR protein with unknown pathogen detection function. A mutation in the snc1 mutant leads to ectopic auto-activation and to enhanced disease resistance [71]. Intriguingly, SNC1 might function analogously to transcriptional regulators CIITA and NLRC5. Recent genetic and biochemical evidence suggest that Topless-related 1 (TPR1) functions as a transcriptional co-repressor of negative regulators of immune responses [72•]. This function requires the physical association between the nuclear pools of TPR1 and SNC1. Thus SNC1 likely activates defense responses by modulating the transcriptional repression activity of TPR1 on targets that would otherwise be detrimental to proper immune responses.
Nuclear-cytoplasmic partitioning is an effector-independent feature of MLA10, RPS4, N, and SNC1 and is required for the induction of defense responses [73]. Additionally, Rx was shown to localize both to the cytoplasm and the nucleus [74•] and to physically associate with a member of the Ran GTPase Activating Protein family (RanGAP2) that controls nucleo-cytoplasmic trafficking of macromolecules through the nuclear pore [75]. Effector-dependent activation of Rx occurs in the cytoplasm and nucleotide binding is an essential step for nuclear import. The LRR domain prevents Rx from being imported into the nucleus, indicating that intra-molecular interactions regulate nuclear accessibility [74•]. RanGAP2 was shown to function as a cytoplasmic retention factor for Rx in order to fine-tune defense signaling. Enforced nuclear accumulation of Rx prevents a normal HR, and thus suppresses resistance to PVX [76•]. These data suggest that effector-dependent, P-loop-dependent activation of Rx occurs in the cytoplasm and triggers an intra-molecular conformational change that releases the inhibiting function of the LRR domain. It is tempting to speculate that this change might destabilize the interaction between RanGAP2 and the Rx CC domain, thus allow Rx trafficking into the nucleus.
Although nuclear-cytoplasmic partitioning is crucial for several immune receptors (Figure 3), the molecular events subsequent to nuclear import remain elusive. We can only speculate that NLRs shuttling into the nucleus might function as activators or repressors of defense-related gene expression. To investigate this possibility, the isolation of the nuclear complexes that associate to these plant NLRs will be essential for the characterization of putative target genes.
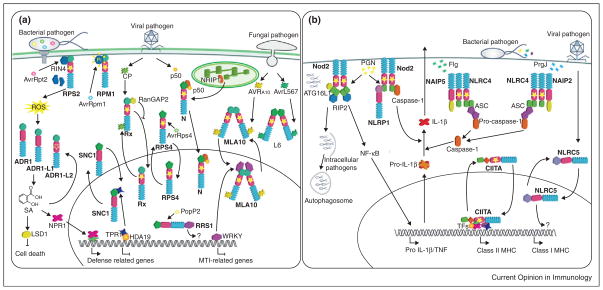
Figure 3.
NLR functions in plant and animal cells. (a) In plants, exposure to pathogen effectors results in activation of NLR proteins. NLRs can either be localized at the plasma membrane, the cytosol, or shuttle between the cytosol and the nucleus. Recognition of effectors can either be direct (L6, MLA10, Rx) or mediated by host proteins (RPM1, RPS2, N). RPM1 and RPS2 recognize the bacterial effectors AvrRpm1 and AvrRpt2 respectively through either phosphorylation or cleavage of the host protein RIN4. N recognizes the 50 kDa helicase (p50) domain of Tobacco Mosaic Virus, which recruits the chloroplastic protein NRIP1 to form an immune receptor complex through N TIR domain. Activation of MLA10 and L6 following recognition of AVRA10 and AvrL567through their respective LRR domains results in homodimerization through their N-terminal domains. Rx is activated by recognition of the Potato Virus X coat protein (CP) and activation results in the inhibition of the cytoplasmic retention protein RanGAP2. SNC1 shuttles from the cytoplasm to the nucleus upon activation from a yet unknown signal. In the nucleus SNC1 recruits the transcriptional co-repressor TPR1, which normally associates to histone deacetylase 19 (HDA19) to repress transcription of defense-related genes. RRS1 is a TIR-NLR that carries a WRKY domain at its C-terminus and localizes to the nucleus. PopP2 is an effector from Ralstonia solanacearum that associates to RRS1 in the nucleus. Transcriptional activity for the WRKY domain of RRS1 has not yet been reported. ADR1 NLRs are activated downstream of an oxidative burst derived from either effector-mediated activation of unrelated NLRs or MAMP recognition. ADR1 proteins are regulators of SA accumulation and SA levels are controlled by LSD1, a negative regulator of cell death. SA is essential for Non-expressor or PR genes 1 (NPR1) function to transcriptionally regulate defense-related genes. (b) In animals MAMPs are perceived by cytoplasmic NLRs, however Nod2 is functional at the plasma membrane where it perceives PGN. Nod2 responds to PGN and its activation triggers the recruitment of the adaptor protein RIP2, resulting in downstream signaling and the expression of pro-inflammatory cytokines. Moreover, Nod2 recruits the autophagy protein ATG16L to the plasma membrane at the bacterial entry site in a mechanism that does not require RIP2. Additionally, Nod2 acts in synergy with NLRP1 in PNG sensing and subsequent caspase-1 activation, thus cleavage of IL-1β zymogen. The NLRC4 inflammasome is activated in response to the bacterial proteins flagellin (flg) and PrgJ. The bacterial ligands trigger oligomerization of NLRC4 with the bacterial sensor NAIP NLRs. The NAIP2-NLRC4 complex confers specificity for PrgJ recognition, whereas NAIP5-NLRC4 to flg. In both cases activation of the downstream signaling results in caspase-1 activation and cytokines secretion via recruitment of the adaptor protein ASC (Apoptosis-associated Speck-like Protein Containing a CARD). CIITA and NLRC5 shuttle between the cytoplasm and the nucleus and function as transcriptional regulators of genes encoding the major histocompatibility complex I and II. CIITA requires its GTPase activity to access the nucleus, thus to recruit transcription factors (TFs) and histone modifying enzymes. Filled stars indicate a requirement of an intact P-loop for the NLR function, an empty star indicates that an intact P-loop is dispensable for the function, as suggested by mutagenesis and/or biochemical analysis.
Conclusions
Since the first NLRs were cloned in the mid 1990s [6,77], we have witnessed a remarkable growth in understanding of their functions in both animal and plant immune systems. However, the more we know, the less clear are generalizable analogies among this fascinating class of proteins. In the past two years, several studies on varied sites of pre-activation and post-activation localization, expanded functions beyond pathogen recognition, and dispensable P-loop activity for several NLRs do not support a simple unifying model to describe how these proteins are activated and how they function. This is likely due to an evolutionarily flexible juxtaposition of highly variable self-association interfaces with a hair trigger conformational switch. This flexibility proved to be useful in a variety of intracellular contexts and in coordination with defense signaling machines.
Many questions remain unanswered to drive research unfold the mechanisms that characterize NLR proteins: What are the molecular dynamics that regulate NLR activation? What are the downstream events that follow NLR activation? How is enzymatic activity dispensable for some NLR functions? What is the diversity of sites and modes of NLR activation in plant and animal cells? And what does this tell us about the cellular machinery with which they interact?
Acknowledgments
We thank Dr. Tatiana Mucyn for critical reading of the manuscript. J.L.D. is a Howard Hughes Medical Institute-Gordon and Betty Moore Foundation Plant Science Investigator. This work was funded by the National Institutes of Health (Grants RO1GM066025 and R01GM057171 with an American Recovery & Reinvestment Act supplement to J.L.D.) and the National Science Foundation (Arabidopsis 2010 Program Grant IOS-0929410 to J.L.D.); V.B. was supported by the Human Frontier Science Program (Grant LT00905/2006-L).
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Jones JD, Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 2.Segonzac C, Zipfel C. Activation of plant pattern-recognition receptors by bacteria. Curr Opin Microbiol. 2011;14:54–61. doi: 10.1016/j.mib.2010.12.005. [DOI] [PubMed] [Google Scholar]
- 3.Chen GY, Nunez G. Sterile inflammation: sensing and reacting to damage. Nat Rev Immunol. 2010;10:826–837. doi: 10.1038/nri2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davis BK, Wen H, Ting JP. The inflammasome NLRs in immunity, inflammation, and associated diseases. Annu Rev Immunol. 2011;29:707–735. doi: 10.1146/annurev-immunol-031210-101405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5•.Guo YL, Fitz J, Schneeberger K, Ossowski S, Cao J, Weigel D. Genome-wide comparison of NB-LRR encoding genes in Arabidopsis. Plant Physiol. 2011;157:757–769. doi: 10.1104/pp.111.181990. The authors compared the repertoire of NLR proteins of Arabidopsis thaliana with its outcrossing relative A. lyrata and analyzed inter-specific and intra-specific diversity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ting JP, Davis BK. CATERPILLER: a novel gene family important in immunity, cell death, and diseases. Annu Rev Immunol. 2005;23:387–414. doi: 10.1146/annurev.immunol.23.021704.115616. [DOI] [PubMed] [Google Scholar]
- 7••.Zhao Y, Yang J, Shi J, Gong YN, Lu Q, Xu H, Liu L, Shao F. The NLRC4 inflammasome receptors for bacterial flagellin and type III secretion apparatus. Nature. 2011;477:596–600. doi: 10.1038/nature10510. This study presents biochemical evidence of direct and specific recognition of bacterial ligands by animal NLRs. Recognition of flagellin by NAIP5 and recognition of type III secretion system rod proteins by NAIP2 trigger oligomerization of NLRC4 and inflammasome activation. [DOI] [PubMed] [Google Scholar]
- 8.Ausubel FM. Are innate immune signaling pathways in plants and animals conserved? Nat Immunol. 2005;6:973–979. doi: 10.1038/ni1253. [DOI] [PubMed] [Google Scholar]
- 9.Hibino T, Loza-Coll M, Messier C, Majeske AJ, Cohen AH, Terwilliger DP, Buckley KM, Brockton V, Nair SV, Berney K, et al. The immune gene repertoire encoded in the purple sea urchin genome. Dev Biol. 2006;300:349–365. doi: 10.1016/j.ydbio.2006.08.065. [DOI] [PubMed] [Google Scholar]
- 10.Marquenet E, Richet E. How integration of positive and negative regulatory signals by a STAND signaling protein depends on ATP hydrolysis. Mol Cell. 2007;28:187–199. doi: 10.1016/j.molcel.2007.08.014. [DOI] [PubMed] [Google Scholar]
- 11.Takken FL, Tameling WI. To nibble at plant resistance proteins. Science. 2009;324:744–746. doi: 10.1126/science.1171666. [DOI] [PubMed] [Google Scholar]
- 12•.Williams SJ, Sornaraj P, de Courcy-Ireland E, Menz RI, Kobe B, Ellis JG, Dodds PN, Anderson PA. An autoactive mutant of the M flax rust resistance protein has a preference for binding ATP, whereas wild-type M protein binds ADP. Mol Plant Microbe Interact. 2011;24:897–906. doi: 10.1094/MPMI-03-11-0052. This is the first report to describe that an autoactive NLR allele mutated in the MHD motif has preference for binding ATP. [DOI] [PubMed] [Google Scholar]
- 13.Tameling WI, Elzinga SD, Darmin PS, Vossen JH, Takken FL, Haring MA, Cornelissen BJ. The tomato R gene products I-2 and MI-1 are functional ATP binding proteins with ATPase activity. Plant Cell. 2002;14:2929–2939. doi: 10.1105/tpc.005793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duncan JA, Bergstralh DT, Wang Y, Willingham SB, Ye Z, Zimmermann AG, Ting JP. Cryopyrin/NALP3 binds ATP/dATP, is an ATPase, and requires ATP binding to mediate inflammatory signaling. Proc Natl Acad Sci U S A. 2007;104:8041–8046. doi: 10.1073/pnas.0611496104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harton JA, Cressman DE, Chin KC, Der CJ, Ting JP. GTP binding by class II transactivator: role in nuclear import. Science. 1999;285:1402–1405. doi: 10.1126/science.285.5432.1402. [DOI] [PubMed] [Google Scholar]
- 16.Kim HE, Du F, Fang M, Wang X. Formation of apoptosome is initiated by cytochrome c-induced dATP hydrolysis and subsequent nucleotide exchange on Apaf-1. Proc Natl Acad Sci U S A. 2005;102:17545–17550. doi: 10.1073/pnas.0507900102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ye Z, Lich JD, Moore CB, Duncan JA, Williams KL, Ting JP. ATP binding by monarch-1/NLRP12 is critical for its inhibitory function. Mol Cell Biol. 2008;28:1841–1850. doi: 10.1128/MCB.01468-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ueda H, Yamaguchi Y, Sano H. Direct interaction between the tobacco mosaic virus helicase domain and the ATP-bound resistance protein. N factor during the hypersensitive response in tobacco plants. Plant Mol Biol. 2006;61:31–45. doi: 10.1007/s11103-005-5817-8. [DOI] [PubMed] [Google Scholar]
- 19.Takken FL, Albrecht M, Tameling WI. Resistance proteins: molecular switches of plant defence. Curr Opin Plant Biol. 2006;9:383–390. doi: 10.1016/j.pbi.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 20.Iyer LM, Leipe DD, Koonin EV, Aravind L. Evolutionary history and higher order classification of AAA+ ATPases. J Struct Biol. 2004;146:11–31. doi: 10.1016/j.jsb.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 21.Riedl SJ, Li W, Chao Y, Schwarzenbacher R, Shi Y. Structure of the apoptotic protease-activating factor 1 bound to ADP. Nature. 2005;434:926–933. doi: 10.1038/nature03465. [DOI] [PubMed] [Google Scholar]
- 22•.Zurek B, Proell M, Wagner RN, Schwarzenbacher R, Kufer TA. Mutational analysis of human NOD1 and NOD2 NACHT domains reveals different modes of activation. Innate Immun. 2011 doi: 10.1177/1753425910394002. By mutational analysis of the ATPase domain the authors show that Nod1 and Nod2 differ in their mode of activation/deactivation. Moreover they confirm a plasma membrane localization for functional Nod1 and Nod2. [DOI] [PubMed] [Google Scholar]
- 23••.Hayashi N, Inoue H, Kato T, Funao T, Shirota M, Shimizu T, Kanamori H, Yamane H, Hayano-Saito Y, Matsumoto T, et al. Durable panicle blast-resistance gene Pb1 encodes an atypical CC-NBS-LRR protein and was generated by acquiring a promoter through local genome duplication. Plant J. 2010;64:498–510. doi: 10.1111/j.1365-313X.2010.04348.x. This study describes the mapping and cloning of the blast-resistance gene Pb1 in rice, a NLR protein that does not possess a P-loop motif. [DOI] [PubMed] [Google Scholar]
- 24••.Bonardi V, Tang S, Stallmann A, Roberts M, Cherkis K, Dangl JL. Expanded functions for a family of plant intracellular immune receptors beyond specific recognition of pathogen effectors. Proc Natl Acad Sci U S A. 2011;108:16463–16468. doi: 10.1073/pnas.1113726108. In this work the ADR1 proteins are described as unconventional NLRs that function as effector-dependent and effector-independent regulators of SA accumulation. ADR1-L2 is the first NLR described for which mutations in its P-loop motif do not affect the function. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25•.Kufer TA, Sansonetti PJ. NLR functions beyond pathogen recognition. Nat Immunol. 2011;12:121–128. doi: 10.1038/ni.1985. This review deserves a special recomendation as it offers a comprehensive and up-to-date description of animal NLRs that share unconventional functions. [DOI] [PubMed] [Google Scholar]
- 26.Tanabe T, Chamaillard M, Ogura Y, Zhu L, Qiu S, Masumoto J, Ghosh P, Moran A, Predergast MM, Tromp G, et al. Regulatory regions and critical residues of NOD2 involved in muramyl dipeptide recognition. EMBO J. 2004;23:1587–1597. doi: 10.1038/sj.emboj.7600175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bendahmane A, Farnham G, Moffett P, Baulcombe DC. Constitutive gain-of-function mutants in a nucleotide binding site-leucine rich repeat protein encoded at the Rx locus of potato. Plant J. 2002;32:195–204. doi: 10.1046/j.1365-313x.2002.01413.x. [DOI] [PubMed] [Google Scholar]
- 28.Ade J, DeYoung BJ, Golstein C, Innes RW. Indirect activation of a plant nucleotide binding site-leucine-rich repeat protein by a bacterial protease. Proc Natl Acad Sci U S A. 2007;104:2531–2536. doi: 10.1073/pnas.0608779104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jia Y, McAdams SA, Bryan GT, Hershey HP, Valent B. Direct interaction of resistance gene and avirulence gene products confers rice blast resistance. EMBO J. 2000;19:4004–4014. doi: 10.1093/emboj/19.15.4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ellis JG, Lawrence GJ, Luck JE, Dodds PN. Identification of regions in alleles of the flax rust resistance gene L that determine differences in gene-for-gene specificity. Plant Cell. 1999;11:495–506. doi: 10.1105/tpc.11.3.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shen QH, Zhou F, Bieri S, Haizel T, Shirasu K, Schulze-Lefert P. Recognition specificity and RAR1/SGT1 dependence in barley Mla disease resistance genes to the powdery mildew fungus. Plant Cell. 2003;15:732–744. doi: 10.1105/tpc.009258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hsu LC, Ali SR, McGillivray S, Tseng PH, Mariathasan S, Humke EW, Eckmann L, Powell JJ, Nizet V, Dixit VM, Karin M. A NOD2-NALP1 complex mediates caspase-1-dependent IL-1beta secretion in response to Bacillus anthracis infection and muramyl dipeptide. Proc Natl Acad Sci U S A. 2008;105:7803–7808. doi: 10.1073/pnas.0802726105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Holt BF, 3rd, Belkhadir Y, Dangl JL. Antagonistic control of disease resistance protein stability in the plant immune system. Science. 2005;309:929–932. doi: 10.1126/science.1109977. [DOI] [PubMed] [Google Scholar]
- 34.Cheng YT, Li Y, Huang S, Huang Y, Dong X, Zhang Y, Li X. Stability of plant immune-receptor resistance proteins is controlled by SKP1-CULLIN1-F-box (SCF)-mediated protein degradation. Proc Natl Acad Sci U S A. 2011;108:14694–14699. doi: 10.1073/pnas.1105685108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qi S, Pang Y, Hu Q, Liu Q, Li H, Zhou Y, He T, Liang Q, Liu Y, Yuan X, et al. Crystal structure of the Caenorhabditis elegans apoptosome reveals an octameric assembly of CED-4. Cell. 2010;141:446–457. doi: 10.1016/j.cell.2010.03.017. [DOI] [PubMed] [Google Scholar]
- 36.Acehan D, Jiang X, Morgan DG, Heuser JE, Wang X, Akey CW. Three-dimensional structure of the apoptosome: implications for assembly, procaspase-9 binding, and activation. Mol Cell. 2002;9:423–432. doi: 10.1016/s1097-2765(02)00442-2. [DOI] [PubMed] [Google Scholar]
- 37.Franchi L, Eigenbrod T, Munoz-Planillo R, Nunez G. The inflammasome: a caspase-1-activation platform that regulates immune responses and disease pathogenesis. Nat Immunol. 2009;10:241–247. doi: 10.1038/ni.1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tschopp J, Schroder K. NLRP3 inflammasome activation: the convergence of multiple signalling pathways on ROS production? Nat Rev Immunol. 2010;10:210–215. doi: 10.1038/nri2725. [DOI] [PubMed] [Google Scholar]
- 39••.Kofoed EM, Vance RE. Innate immune recognition of bacterial ligands by NAIPs determines inflammasome specificity. Nature. 2011;477:592–595. doi: 10.1038/nature10394. This paper shows that the ‘helper NLRs’ NAIP2 and NAIP5 confer specificity to the NLRC4 inflammasome. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mestre P, Baulcombe DC. Elicitor-mediated oligomerization of the tobacco N disease resistance protein. Plant Cell. 2006;18:491–501. doi: 10.1105/tpc.105.037234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gutierrez JR, Balmuth AL, Ntoukakis V, Mucyn TS, Gimenez-Ibanez S, Jones AM, Rathjen JP. Prf immune complexes of tomato are oligomeric and contain multiple Pto-like kinases that diversify effector recognition. Plant J. 2010;61:507–518. doi: 10.1111/j.1365-313X.2009.04078.x. [DOI] [PubMed] [Google Scholar]
- 42••.Maekawa T, Cheng W, Spiridon LN, Toller A, Lukasik E, Saijo Y, Liu P, Shen QH, Micluta MA, Somssich IE, et al. Coiled-coil domain-dependent homodimerization of intracellular barley immune receptors defines a minimal functional module for triggering cell death. Cell Host Microbe. 2011;9:187–199. doi: 10.1016/j.chom.2011.02.008. In this paper the first crystal structure of a CC domain is presented. A mutagenic and genetic analysis indicates that the interaction of the CC domains of MLA10 monomers is essential for MLA10 function. [DOI] [PubMed] [Google Scholar]
- 43••.Bernoux M, Ve T, Williams S, Warren C, Hatters D, Valkov E, Zhang X, Ellis JG, Kobe B, Dodds PN. Structural and functional analysis of a plant resistance protein TIR domain reveals interfaces for self-association, signaling, and autoregulation. Cell Host Microbe. 2011;9:200–211. doi: 10.1016/j.chom.2011.02.009. Similar to Ref. [42••], this paper describes the crystal structure of L6 TIR domain and provides evidence that the association of the TIR domains is essential for L6 function. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Swiderski MR, Birker D, Jones JD. The TIR domain of TIR-NB-LRR resistance proteins is a signaling domain involved in cell death induction. Mol Plant Microbe Interact. 2009;22:157–165. doi: 10.1094/MPMI-22-2-0157. [DOI] [PubMed] [Google Scholar]
- 45.Krasileva KV, Dahlbeck D, Staskawicz BJ. Activation of an Arabidopsis resistance protein is specified by the in planta association of its leucine-rich repeat domain with the cognate oomycete effector. Plant Cell. 2010;22:2444–2458. doi: 10.1105/tpc.110.075358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Frost D, Way H, Howles P, Luck J, Manners J, Hardham A, Finnegan J, Ellis J. Tobacco transgenic for the flax rust resistance gene L expresses allele-specific activation of defense responses. Mol Plant Microbe Interact. 2004;17:224–232. doi: 10.1094/MPMI.2004.17.2.224. [DOI] [PubMed] [Google Scholar]
- 47•.Collier SM, Hamel LP, Moffett P. Cell death mediated by the N-terminal domains of a unique and highly conserved class of NB-LRR protein. Mol Plant Microbe Interact. 2011;24:918–931. doi: 10.1094/MPMI-03-11-0050. In this paper, the authors present a phylogenetic analysis of the CCR-NLRs and they show that over-expression of the CCR domains for NRG1 and ADR1 proteins leads to ectopic activation of defense responses. [DOI] [PubMed] [Google Scholar]
- 48.Rairdan GJ, Collier SM, Sacco MA, Baldwin TT, Boettrich T, Moffett P. The coiled-coil and nucleotide binding domains of the Potato Rx disease resistance protein function in pathogen recognition and signaling. Plant Cell. 2008;20:739–751. doi: 10.1105/tpc.107.056036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Peart JR, Mestre P, Lu R, Malcuit I, Baulcombe DC. NRG1, a CC-NB-LRR protein, together with N, a TIR-NB-LRR protein, mediates resistance against tobacco mosaic virus. Curr Biol. 2005;15:968–973. doi: 10.1016/j.cub.2005.04.053. [DOI] [PubMed] [Google Scholar]
- 50•.Gregory SM, Davis BK, West JA, Taxman DJ, Matsuzawa S, Reed JC, Ting JP, Damania B. Discovery of a viral NLR homolog that inhibits the inflammasome. Science. 2011;331:330–334. doi: 10.1126/science.1199478. For the first time a viral homolog of a human NLR protein that subverts the human immune responses was characterized. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Newman RM, Salunkhe P, Godzik A, Reed JC. Identification and characterization of a novel bacterial virulence factor that shares homology with mammalian Toll/interleukin-1 receptor family proteins. Infect Immun. 2006;74:594–601. doi: 10.1128/IAI.74.1.594-601.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cirl C, Wieser A, Yadav M, Duerr S, Schubert S, Fischer H, Stappert D, Wantia N, Rodriguez N, Wagner H, et al. Subversion of Toll-like receptor signaling by a unique family of bacterial Toll/interleukin-1 receptor domain-containing proteins. Nat Med. 2008;14:399–406. doi: 10.1038/nm1734. [DOI] [PubMed] [Google Scholar]
- 53.Salcedo SP, Marchesini MI, Lelouard H, Fugier E, Jolly G, Balor S, Muller A, Lapaque N, Demaria O, Alexopoulou L, et al. Brucella control of dendritic cell maturation is dependent on the TIR-containing protein Btp1. PLoS Pathog. 2008;4:e21. doi: 10.1371/journal.ppat.0040021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang Q, Zmasek CM, Cai X, Godzik A. TIR domain-containing adaptor SARM is a late addition to the ongoing microbe-host dialog. Dev Comp Immunol. 2011;35:461–468. doi: 10.1016/j.dci.2010.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dietrich RA, Delaney TP, Uknes SJ, Ward ER, Ryals JA, Dangl JL. Arabidopsis mutants simulating disease resistance response. Cell. 1994;77:565–577. doi: 10.1016/0092-8674(94)90218-6. [DOI] [PubMed] [Google Scholar]
- 56.Brodersen P, Petersen M, Pike HM, Olszak B, Skov S, Odum N, Jorgensen LB, Brown RE, Mundy J. Knockout of Arabidopsis accelerated-cell-death11 encoding a sphingosine transfer protein causes activation of programmed cell death and defense. Genes Dev. 2002;16:490–502. doi: 10.1101/gad.218202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Torres MA, Jones JD, Dangl JL. Pathogen-induced. NADPH oxidase-derived reactive oxygen intermediates suppress spread of cell death in Arabidopsis thaliana. Nat Genet. 2005;37:1130–1134. doi: 10.1038/ng1639. [DOI] [PubMed] [Google Scholar]
- 58.Brodersen P, Malinovsky FG, Hematy K, Newman MA, Mundy J. The role of salicylic acid in the induction of cell death in Arabidopsis acd11. Plant Physiol. 2005;138:1037–1045. doi: 10.1104/pp.105.059303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59••.Palma K, Thorgrimsen S, Malinovsky FG, Fiil BK, Nielsen HB, Brodersen P, Hofius D, Petersen M, Mundy J. Autoimmunity in Arabidopsis acd11 is mediated by epigenetic regulation of an immune receptor. PLoS Pathog. 2010;6:e1001137. doi: 10.1371/journal.ppat.1001137. In this study the authors describe a TIR-NLR protein that is required for the autoimmune phenotypes of acd11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tameling WI, Joosten MH. The diverse roles of NB-LRR proteins in plants. Physiol Mol Plant Pathol. 2007;71:126–134. [Google Scholar]
- 61.Igari K, Endo S, Hibara K, Aida M, Sakakibara H, Kawasaki T, Tasaka M. Constitutive activation of a CC-NB-LRR protein alters morphogenesis through the cytokinin pathway in Arabidopsis. Plant J. 2008;55:14–27. doi: 10.1111/j.1365-313X.2008.03466.x. [DOI] [PubMed] [Google Scholar]
- 62.Huang X, Li J, Bao F, Zhang X, Yang S. A gain-of-function mutation in the Arabidopsis disease resistance gene RPP4 confers sensitivity to low temperature. Plant Physiol. 2010;154:796–809. doi: 10.1104/pp.110.157610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yang H, Shi Y, Liu J, Guo L, Zhang X, Yang S. A mutant CHS3 protein with TIR-NB-LRR-LIM domains modulates growth, cell death and freezing tolerance in a temperature-dependent manner in Arabidopsis. Plant J. 2010;63:283–296. doi: 10.1111/j.1365-313X.2010.04241.x. [DOI] [PubMed] [Google Scholar]
- 64.Bomblies K, Lempe J, Epple P, Warthmann N, Lanz C, Dangl JL, Weigel D. Autoimmune response as a mechanism for a Dobzhansky-Muller-type incompatibility syndrome in plants. PLoS Biol. 2007;5:e236. doi: 10.1371/journal.pbio.0050236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65••.Gao Z, Chung EH, Eitas TK, Dangl JL. Plant intracellular innate immune receptor Resistance to Pseudomonas syringae pv. maculicola 1 (RPM1) is activated at, and functions on, the plasma membrane. Proc Natl Acad Sci U S A. 2011;108:7619–7624. doi: 10.1073/pnas.1104410108. This paper establishes the plasma-membrane as the site of activation for the RPM1 NLR and uses a clever plasma-membrane tethering allele of an auto-active allele to establish that no re-localization from the membrane compartment is required for response. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lecine P, Esmiol S, Metais JY, Nicoletti C, Nourry C, McDonald C, Nunez G, Hugot JP, Borg JP, Ollendorff V. The NOD2-RICK complex signals from the plasma membrane. J Biol Chem. 2007;282:15197–15207. doi: 10.1074/jbc.M606242200. [DOI] [PubMed] [Google Scholar]
- 67•.Travassos LH, Carneiro LA, Ramjeet M, Hussey S, Kim YG, Magalhaes JG, Yuan L, Soares F, Chea E, Le Bourhis L, et al. Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry. Nat Immunol. 2010;11:55–62. doi: 10.1038/ni.1823. This study shows that Nod1 and Nod2, independently of RIP2, are essential for the autophagic responses to invasive bacteria, providing a link between microbe sensing and induction of autophagy. [DOI] [PubMed] [Google Scholar]
- 68.Ting JP, Trowsdale J. Genetic control of MHC class II expression. Cell. 2002;109(Suppl):S21–S33. doi: 10.1016/s0092-8674(02)00696-7. [DOI] [PubMed] [Google Scholar]
- 69••.Meissner TB, Li A, Biswas A, Lee KH, Liu YJ, Bayir E, Iliopoulos D, van den Elsen PJ, Kobayashi KS. NLR family member NLRC5 is a transcriptional regulator of MHC class I genes. Proc Natl Acad Sci U S A. 2010;107:13794–13799. doi: 10.1073/pnas.1008684107. In this report NLRC5 is described as a transcriptional regulator of MHC class I genes that shuttles between the cytoplasm and the nucleus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Deslandes L, Olivier J, Peeters N, Feng DX, Khounlotham M, Boucher C, Somssich I, Genin S, Marco Y. Physical interaction between RRS1-R, a protein conferring resistance to bacterial wilt, and PopP2, a type III effector targeted to the plant nucleus. Proc Natl Acad Sci U S A. 2003;100:8024–8029. doi: 10.1073/pnas.1230660100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang Y, Goritschnig S, Dong X, Li X. A gain-of-function mutation in a plant disease resistance gene leads to constitutive activation of downstream signal transduction pathways in suppressor of npr1-1, constitutive 1. Plant Cell. 2003;15:2636–2646. doi: 10.1105/tpc.015842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72•.Zhu Z, Xu F, Zhang Y, Cheng YT, Wiermer M, Li X. Arabidopsis resistance protein SNC1 activates immune responses through association with a transcriptional corepressor. Proc Natl Acad Sci U S A. 2010;107:13960–13965. doi: 10.1073/pnas.1002828107. In this study, the authors show that SNC1 associates with the transcriptional corepressor TPR1 that inhibits the transcription of negative regulators of immune responses. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Maekawa T, Kufer TA, Schulze-Lefert P. NLR functions in plant and animal immune systems: so far and yet so close. Nat Immunol. 2011;12:817–826. doi: 10.1038/ni.2083. [DOI] [PubMed] [Google Scholar]
- 74•.Slootweg E, Roosien J, Spiridon LN, Petrescu AJ, Tameling W, Joosten M, Pomp R, van Schaik C, Dees R, Borst JW, et al. Nucleocytoplasmic distribution is required for activation of resistance by the potato NB-LRR receptor Rx1 and is balanced by its functional domains. Plant Cell. 2010;22:4195–4215. doi: 10.1105/tpc.110.077537. This report shows that Rx is distributed between the cytoplasm and the nucleus. While Rx is activated in the cytoplasm, a balanced distribution is required for full Rx responses. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tameling WI, Baulcombe DC. Physical association of the NB-LRR resistance protein Rx with a Ran GTPase-activating protein is required for extreme resistance to Potato virus X. Plant Cell. 2007;19:1682–1694. doi: 10.1105/tpc.107.050880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76•.Tameling WI, Nooijen C, Ludwig N, Boter M, Slootweg E, Goverse A, Shirasu K, Joosten MH. RanGAP2 mediates nucleocytoplasmic partitioning of the NB-LRR immune receptor Rx in the Solanaceae, thereby dictating Rx function. Plant Cell. 2010;22:4176–4194. doi: 10.1105/tpc.110.077461. In agreement with ref. [74] the authors show that Rx nuclear-cytoplasmic distribution is crucial for defense signaling and that RanGAP2 mediates this partitioning by acting as a cytoplasmic retention factor for Rx. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Staskawicz BJ, Ausubel FM, Baker BJ, Ellis JG, Jones JD. Molecular genetics of plant disease resistance. Science. 1995;268:661–667. doi: 10.1126/science.7732374. [DOI] [PubMed] [Google Scholar]
- 78.Meyers BC, Kozik A, Griego A, Kuang H, Michelmore RW. Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell. 2003;15:809–834. doi: 10.1105/tpc.009308. [DOI] [PMC free article] [PubMed] [Google Scholar]