Abstract
The quinone pharmacophore is present in many drug classes but is particularly common among antitumor drugs. Many quinones serve essentially as pro-drugs and exert their activities after reduction. Reduction of quinones may generate semiquinones or hydroquinones with subsequent generation of reactive oxygen radicals and oxidative stress, quinones can be designed so they lose a leaving group when reduced to the hydroquinone generating a reactive electrophile or the hydroquinone form of the molecule may have greater pharmacological activity than the parent quinone against a particular target. Enzyme systems that reduce quinones therefore become critically important in the pharmacological activity of this class of drugs. There are a number of enzyme systems that can catalyze reduction of quinones including cytochrome P450 reductase, cytochrome b5 reductase, NAD(P)H:quinone oxidoreductase 1 (NQO1), NAD(P)H:quinone oxidoreductase 2 (NQO2), carbonyl reductases, and thioredoxin reductase. In this context, one of the most extensively studied reductases has been NAD(P)H:quinone oxidoreductase 1 (NQO1). In this review we will focus on the role of NQO1 in the bioactivation of clinically important quinones mitomycin C, β-lapachone and 17AAG as well as the influence of the NQO1*2 polymorphism on the sensitivity and resistance to these agents.
NQO1
NAD(P)H:quinone oxidoreductase 1 (NQO1, EC 1.6.99.2) is a flavoenzyme that catalyzes the two-electron reduction of quinones to their hydroquinone forms [1]. NQO1 functions as a homodimer with one FAD bound per monomer. This enzyme utilizes reduced pyridine nucleotide cofactors NADH or NADPH to catalyze the direct two-electron reduction of a broad range of quinones [2]. The crystal structure of human NQO1 was resolved in 2000 [3] and this work demonstrated that the cofactor and the substrate share the same binding site confirming the ping-pong mechanism of catalysis [4]. NQO1 is localized primarily in the cytosol but lower levels have been detected in the nucleus [5]. In human tissues NQO1 is expressed at high levels in many epithelial cells as well as vascular endothelium and adipocytes [6, 7]. Humans, unlike most other mammals, do not express NQO1 in normal liver hepatocytes [6, 8] but NQO1 expression is seen in pre-neoplastic lesions and liver cancers [9, 10]. NQO1 is expressed at high levels in most human solid tumors including tumors from colon, breast, pancreas and lung [6, 11].
There are two characterized polymorphisms in NQO1, NQO1*2 and NQO1*3, with well-defined phenotypes and frequencies. The NQO1*2 polymorphism is characterized by a C to T change at position 609 of the human cDNA which results in a proline to serine substitution at amino acid 187 of NQO1 [12]. The resulting mutant NQO1 protein is catalytically inactive due to the inability to correctly bind the FAD cofactor [13]. The mutant NQO1*2 protein has also been shown to bind to the Hsp70 binding domain of the E3 ubiquitin ligase STUB1/CHIP which catalyzes the ubiquitination of the NQO1*2 protein resulting in proteasomal degradation [14, 15]. Individuals genotyped as homozygous for the NQO1*2 polymorphism are NQO1 null, while individuals genotyped as heterozygous have reduced levels of NQO1 activity and protein [16]. The allele frequency of the NQO1*2 polymorphism is much lower in Caucasians compared to Asian populations [17]. In some Asian populations the percentage of individuals homozygous for the NQO1*2 polymorphism can be as high as 40% [18, 19]. The NQO1*3 polymorphism has been characterized as a C465T substitution resulting in an arginine to tryptophan amino acid change in the protein [20, 21]. The variant NQO1*3 protein has similar stability to the wildtype NQO1*1 protein and is catalytically active but major differences in the two proteins in the rate of metabolism of quinone substrates have been observed [20]. The allele frequency of the NQO1*3 polymorphism ranges from >0.01 in Inuit population to 0.05 in Caucasians [17].
The high levels of expression of NQO1 in solid tumors in combination with the ability to reduced many quinone-containing antitumor drugs has drawn attention to NQO1 as a potential molecular target in cancer treatment.
Bioreductive activation of quinones by NQO1
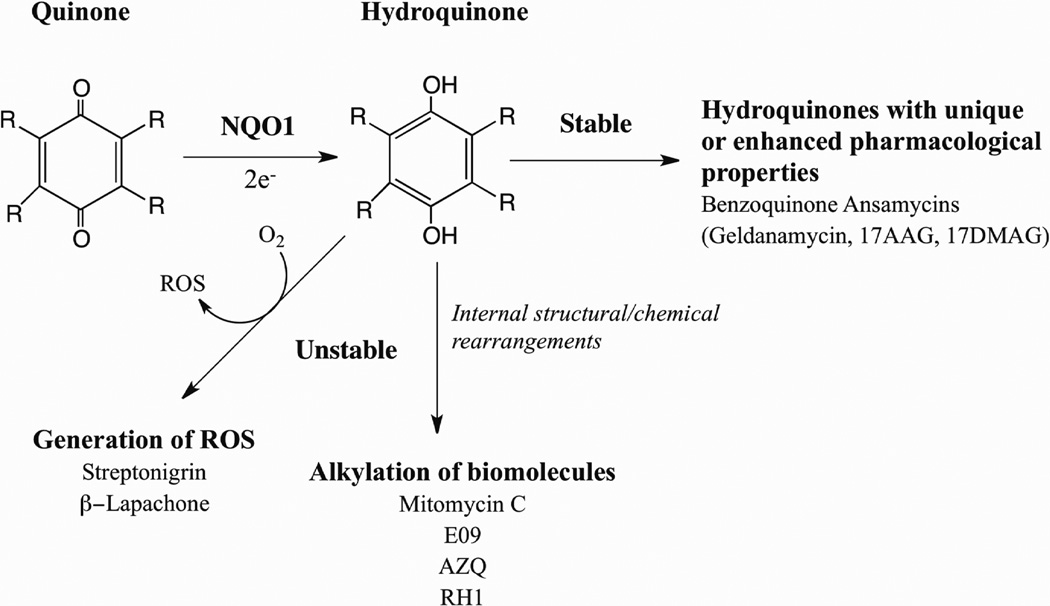
The direct two-electron reduction of quinones to hydroquinone by NQO1 is historically considered a detoxification mechanism because this reaction by-passes the formation of the highly reactive semiquinone. However, in reality whether the formation of the hydroquinone is a detoxification reaction, or alternatively, an activation reaction will depend upon the chemical reactivities of the quinone and hydroquinone. There are many examples of naturally occurring and synthetic quinones that following reduction to their corresponding hydroquinones induce toxicity. The ability of NQO1 to generate cytotoxic hydroquinones has been utilized as a strategy to combat antiproliferative diseases such as cancer. As shown in Figure 2 a hydroquinone generated following reduction by NQO1 can exert toxicity through a number of mechanisms depending upon its chemical reactivity. Unstable hydroquinones can undergo chemical rearrangements leading to alkylation of essential biomolecules such as DNA or undergo redox reactions leading to the formation of highly reactive oxygen species. Alternatively, if the hydroquinone is chemically stable it may possess unique or enhanced pharmacological properties not observed with the parent quinone. As shown in Figure 2 NQO1 has been implicated in the bioactivation of many antitumor quinones. In this review we will discuss the role of NQO1 is the bioactivation of three clinically significant quinones mitomycin C, β-lapachone and 17AAG.
Figure 2.
Pathways for bioreductive activation of antitumor quinones by NQO1 (ROS, reactive oxygen species).
Bioreductive activation of mitomycin C by NQO1
Mitomycin C (MMC) is a quinone containing antibiotic isolated from Streptomyces caespitosus. MMC has been used clinically for greater than 30 years for the treatment of solid tumors including stomach, pancreas, breast and lung. The mechanism of action of MMC is believed to be intracellular bioreductive activation leading to DNA interstrand crosslinking.
Studies using cultured tumor cell lines in combination with the NQO1 inhibitor dicumarol suggested a positive correlation between NQO1 (DT-diaphorase) catalytic activity and MMC sensitivity [22–41]. Under hypoxia, however, dicumarol potentiated MMC-induced DNA crosslinking and cytotoxicity suggesting that under hypoxic conditions MMC may be activated more efficiently by other bioreductive enzymes [22, 42–45].
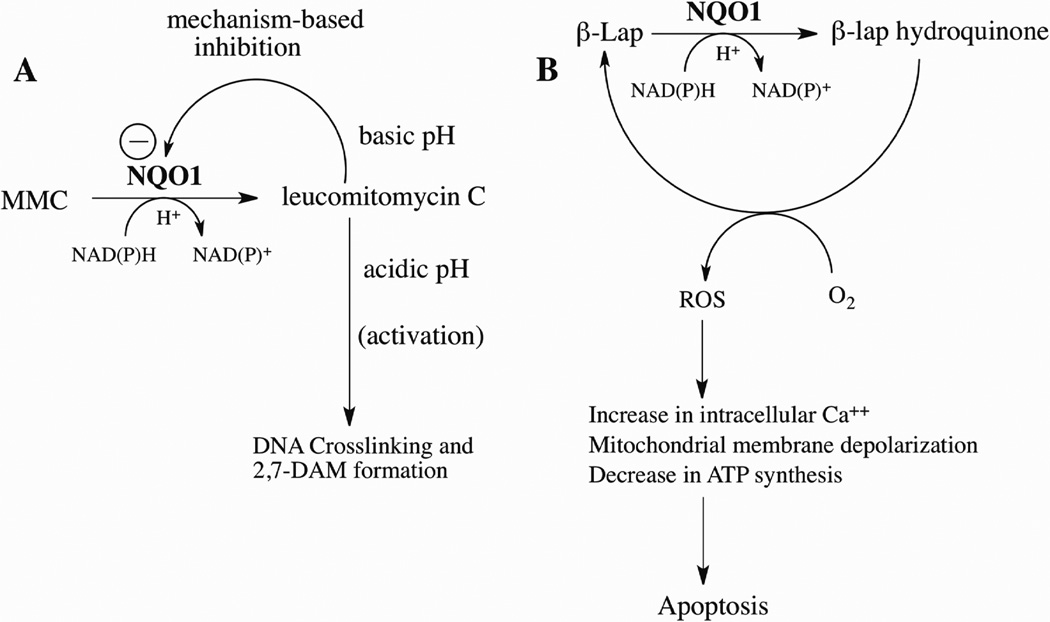
Experiments using purified rat and human NQO1 confirmed that these enzymes could bioactivate MMC, however, the metabolism of MMC by NQO1 was discovered to be pH-dependent [24, 46]. When reactions were performed under acidic conditions MMC underwent bioactivation by NQO1 to a reactive species capable of crosslinking DNA as well as metabolites including 2,7-diaminomitosene [46]. However, as the pH of the reaction was increased MMC-induced DNA crosslinking and metabolite formation decreased substantially. Biochemical studies with purified NQO1 and MMC revealed that MMC was a pH-dependent mechanism-based inhibitor of NQO1 [47]. Under basic pH conditions NQO1 underwent alkylation by leucomitomycin C (MMC hydroquinone) in or near the active site of NQO1 resulting in the inhibition of catalytic activity. As the pH of the reaction was decreased inactivation of NQO1 by leucomitomycin became less efficient resulting in the release from the active site of leucomitomycin and subsequent alkylation of biomolecules such as DNA (Figure 3A). Studies in cultured cells confirmed that under acidic conditions MMC induced greater levels of DNA crosslinking and more pronounced growth inhibition [30, 40, 48, 49].
Figure 3.
The role of NQO1 in the bioactivation of mitomycin C and β-lapachone. (A) Reduction of MMC by NQO1 generates leucomitomycin C (MMC hydroquinone) which under basic conditions alkylates NQO1 in the active site preventing further metabolism. Under acidic conditions, the leucomitomycin C escapes the active site in NQO1 and can alkylate important biomolecules such as DNA or form the major metabolite 2,7-diaminomitosene (2,7-DAM); (B) Reduction of β-lapachone by NQO1 forms an unstable hydroquinone which interacts with molecular oxygen to generate reactive oxygen species leading to apoptosis.
A role for NQO1 in MMC activation is supported by cell culture and xenograft experiments using isogenic cell lines engineered to overexpress NQO1. These studies demonstrated a positive correlation between NQO1 expression and sensitivity to MMC [35, 50–52]. Dietary induction of NQO1 by dimethylfumarate increased the sensitivity of human xenograft colon tumors to MMC [53]. In experiments where cultured cells were exposed to sub-lethal concentrations of MMC for extended periods of time resistant clones developed that were shown to have lower levels of NQO1 activity [31, 35, 54–61]. Genetic mutations in the NQO1 gene resulting in lower NQO1 protein levels and decreased catalytic activity were also seen in HCT116 human colon carcinoma cells made resistant to MMC by continuous low-dose exposure [20, 27].
Studies that have examined the relationship between NQO1 protein expression/activity and MMC sensitivity in more complex systems such as mouse xenograft models of human cancers or in human subjects with cancers are limited. Sensitivity to MMC was shown to correlate with NQO1 activity in mouse xenograft model using human NSCLC [62] while no relationship between MMC sensitivity and the NQO1*2 allele (see above) was observed in a series of human tumor xenografts [63]. In patients with disseminated peritoneal cancer receiving intraperitoneal hyperthermic chemotherapy with MMC lower levels of NQO1 activity in dissected tumor tissues was associated with reduced survival in a subset of patients [64]. The widespread use of intravesical MMC therapy for the treatment of superficial bladder cancers has generated interest in a search for biomarkers of MMC sensitivity. The expression of NQO1 and NADPH cytochrome P450 reductase in a series of bladder tumors were shown to correlate with MMC sensitivity [65]. In another study, however, no correlation was observed between immunohistochemical staining for NQO1 or NADPH cytochrome P450 reductase in resected bladder tumors and clinical response to MMC [66]. In addition, genotyping of human bladder tumors for the NQO1*2 polymorphism was also found to be a poor predictor of a clinical response to MMC [67].
A major dose limiting toxicity observed with the clinical use of MMC is bone marrow depression[68]. NQO1 levels or the NQO1*2 polymorphism have not been previously associated with an increased risk of developing complications due to MMC therapy. In experiments using NQO1 knockout mice treated with MMC it was observed that mice deficient in NQO1 were resistant to MMC-induced bone marrow toxicity [69].
Biochemical and cell based experiments clearly demonstrate that NQO1 can bioactivate MMC and is generally a good predictor of MMC sensitivity. Given the multitude of factors that could influence the antitumor response to MMC including intracellular pH and O2 concentrations, competing bioreductive enzymes, as well as DNA repair enzymes responsible for the repair of cytotoxic MMC-DNA interstrand crosslinks it is not surprising that NQO1 genotype or NQO1 protein levels by themselves may not be suitable candidates to predict clinical response to MMC therapy.
Bioactivation of β-lapachone by NQO1
β-Lapachone is a naturally occurring ortho napthoquinone isolated from the bark of the lapacho tree (Tabebuia avellanedae). β-lapachone was shown to have anti-bacterial and anti-fungal and anti-trypanosomal properties primarily due to the ability of β-lapachone to rapidly induce the formation of superoxide and hydrogen peroxide with the simultaneous oxidation of reduced pyridine nucleotides [70]. Early experiments demonstrated that β-lapachone could inhibit the repair of mammalian DNA through a mechanism involving inhibition of topoisomerase I [71–74]. β-lapachone has been shown to induce apoptosis in human leukemia and prostate cancer cells and over-expression of BCL2 could protect cells against β-lapachone induced apoptosis [75].
Studies in human breast and prostate cancer cell lines demonstrated that dicumarol could protect against β-lapachone-induced growth inhibition in NQO1-rich cells but had limited effect on NQO1-null cells implicating a role for NQO1 in β-lapachone toxicity [76, 77]. In addition, in these studies the overexpression of NQO1 increased the sensitivity of breast and prostate cancer cells to β-lapachone [76, 77]. Lysates prepared from breast cancer cells overexpressing NQO1 catalyzed the oxidation of NADH in the presence of β-lapachone. In these studies greater than 50 molar equivalents of NADH were oxidized per molar equivalent of β-lapachone suggesting that β-lapachone underwent NQO1-dependent redox cycling [76]. NQO1 catalyzes the redox cycling of β-lapachone through the generation of an unstable hydroquinone, which under aerobic conditions, is rapidly oxidized back to the parent quinone. Redox cycling of β-lapachone is characterized by the oxidation of large amounts of reduced pyridine nucleotides and the formation of reactive oxygen species including superoxide and hydrogen peroxide [78]. The reductive activation of β-lapachone by NQO1 (Figure 3B) has been shown to result in a rapid increase in intracellular calcium leading to mitochondrial membrane depolarization, loss of ATP, DNA fragmentation and apoptosis [79, 80].
In experiments where β-lapachone was used in combination with irradiation sensitivity to β-lapachone could be increased if cancer cells were irradiated in the presence of β-lapachone or if β-lapachone was added up to 24hr after irradiation suggesting that β-lapachone had a direct effect on DNA repair as well as effects that were independent of DNA repair [81, 82]. The synergy observed between irradiation and β-lapachone when the drug is administered hours after irradiation could be explained by the upregulation of NQO1 [81, 82]. X-ray irradiation has been shown to induce the expression of NQO1 in human cancer cells [83] and it has been proposed that at longer time points after irradiation the upregulation of NQO1 increased β-lapachone-dependent redox cycling leading to increased cytotoxicity. A similar mechanism has been proposed for the synergy observed between hyperthermia and β-lapachone where hyperthermia was shown to increase the expression of NQO1 in tumor cells resulting in greater sensitivity to β-lapachone [84–86].
The ability of NQO1 to efficiently catalyze the redox cycling of β-lapachone has been exploited to target human tumors with high levels of NQO1. Non-small cell lung cancers (NSCLC) have been shown to have high levels of NQO1 expression [7, 11, 62] and correspondingly NSCLC cells were shown to be very sensitive to β-lapachone. In these studies the sensitivity to β-lapachone was directly related to NQO1 protein levels [87]. NQO1 has been selected as a target for the activation of β-lapachone in pancreatic cancers since pancreatic cancers have significantly higher levels of NQO1 when compared to normal pancreatic tissue [88]. In human pancreatic cancer cells the cytotoxicity of β-lapachone was shown to be significantly decreased by dicumarol pretreatment and in experiments using siRNA to knockdown NQO1 protein levels [89]. In a more detailed study using the MiaPaCa-2 cell line in combination with shRNA targeted against NQO1 stable cell lines were created expressing varying levels of NQO1 [80]. In studies using these isogenic cell lines it was clear that there was a correlation between NQO1 activity and bioactivation of β-lapachone and it was concluded that a threshold level of NQO1 catalytic activity of 90U was required to efficiently activate β-lapachone in MiaPaCa-2 cells [80].
β-lapachone (ARQ 501) has entered Phase 1 and 2 clinical trials for the treatment of solid tumors. Despite the large volume of data implicating NQO1 in the bioactivation and cytotoxicity of β-lapachone, including many studies with human cell lines that are deficient in NQO1 due to the NQO1*2*2 genotype and were very resistant to β-lapachone, there are no published papers or abstracts from these clinical trials that have reported if sensitivity to β-lapachone is related to NQO1 activity and whether resistance to β-lapachone is observed in patients because they carry the NQO1*2*2 genotype.
Bioactivation of benzoquinone ansamycins by NQO1
The benzoquinone ansamycins (BQAs) including geldanamycin, 17-AAG and 17-DMAG are a group of quinone containing polyketide antibiotics. Geldanamycin (GA) was isolated from Streptomyces hygroscopicus [90, 91] and GA was originally found to have antitumor properties due to its ability to inhibit RNA and DNA replication [92–94]. Later it was shown that GA could inhibit the expression of the oncogene cMyc [92] and inhibit the activity of vSrc [95, 96]. Studies using GA affinity chromatography led to the discovery that heat shock protein 90 (Hsp90) was the target of GA [96]. BQAs bind to the ATP binding pocket in Hsp90 and inhibit the ATPase activity of the enzyme preventing the correct folding of newly synthesized proteins. Hsp90 has been shown to play a role in the maturation of many oncogenic proteins such as Raf-1 [97], HER2 [98, 99], BCR-ABL [100], KIT [101, 102] as well as steroid hormone receptors [103]. Hsp90 has become an attractive antitumor target since the inhibition of the chaperone function of Hsp90 can alter many key oncogenic pathways simultaneously. Preclinical studies with GA resulted in hepatotoxicity and less toxic analogs of GA including 17-AAG (17-N-allylamino-17-demethoxygeldanamycin) and 17-DMAG (17-(dimethylaminoethylamino)-17-demethoxygeldanamycin) were developed that focused on substitutions on 17-position on the ansamycin ring [104]. 17-AAG and 17-DMAG have shown antitumor activity against a wide spectrum of human cancers in laboratory studies and are currently in clinical trials.
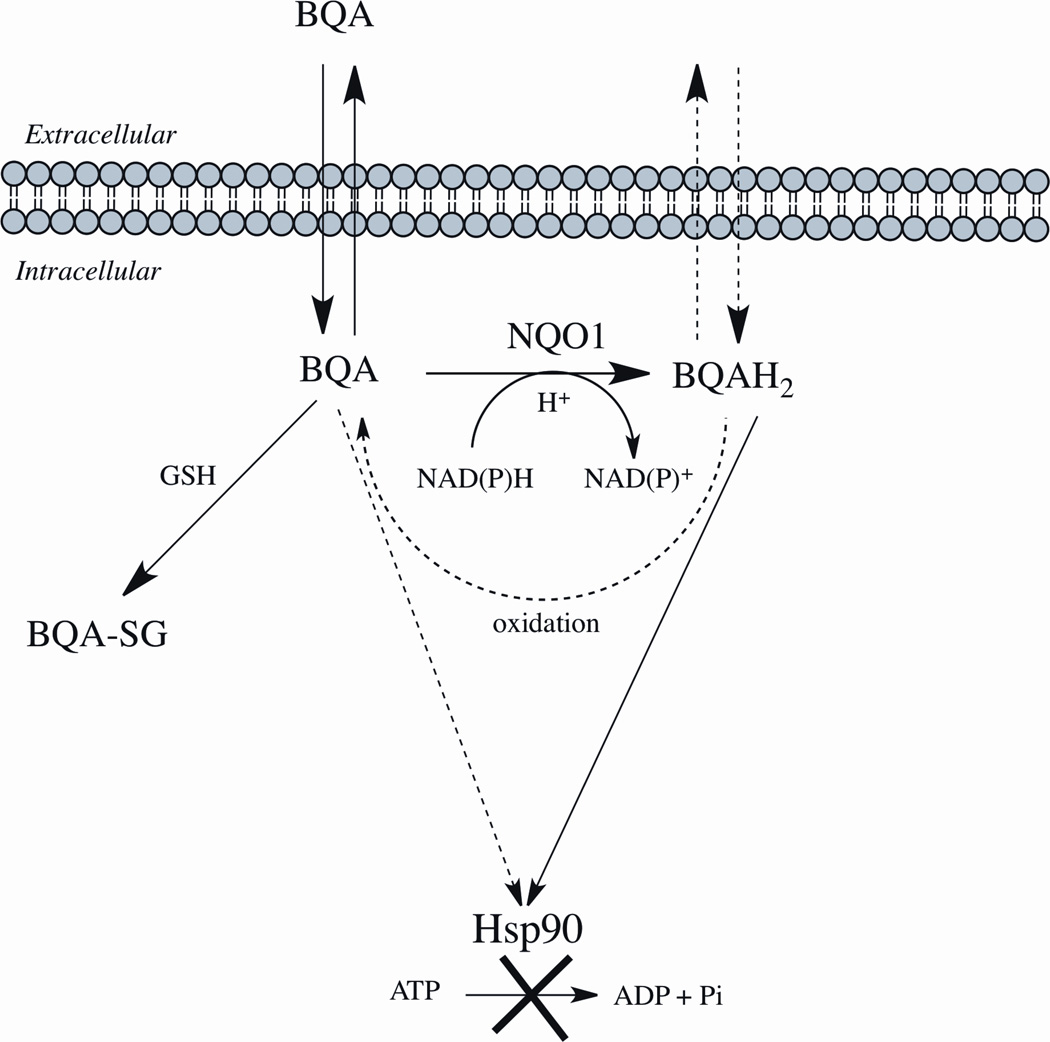
A correlation between NQO1 catalytic activity and sensitivity of human cancer cell lines and xenografts to GA and 17-AAG was first reported by Kelland et al. [105]. Later it was shown in cell-free systems that purified NQO1 could reduce a series of BQA including 17-AAG to their corresponding hydroquinones [106–108]. These studies also demonstrated that the hydroquinones formed following reduction by NQO1 were relatively stable and could be isolated for analysis but were susceptible to metal-catalyzed oxidation [109]. Polarographic studies showed that the addition of SOD significantly inhibited the oxidation of 17-AAG hydroquinone generated by NQO1 implicating superoxide in the oxidation of 17-AAG hydroquinone [110]. In cell-free studies it was also shown that the hydroquinones formed from BQAs were resistant to direct conjugation by GSH at the 19-position on the ansamycin ring [107]. The ability of cells to generate the hydroquinone would prevent drug inactivation due to GSH conjugation (Figure 4). The most important feature of the hydroquinone, however, became apparent from cell-free studies that showed that the hydroquinone of 17-AAG was a more potent inhibitor of purified Hsp90 when compared to 17-AAG (quinone) [106]. Computational-based molecular simulation studies with BQAs confirmed these data and showed that the hydroquinones bound with greater affinity to the ATPase active site in Hsp90 when compared with their corresponding quinones [106, 108, 111]. The greater interaction energies observed with hydroquinone ansamycins can be explained by the stronger and greater number of hydrogen-bonding interactions with key amino acid residues in the ATP-binding domain of Hsp90.
Figure 4.
The Role of NQO1 in potentiating the antitumor activity of benzoquinone ansamycins. Solid lines represent major pathways. Dashed lines represent minor pathways. BQA, benzoquinone ansamycin quinone; BQAH2, benzoquinone ansamycin hydroquinone; BQA-GS, benzoquinone ansamycin quinone-glutathione conjugate; Hsp90, heatshock 90 protein.
Studies in NQO1 null and NQO1-overexpressing breast and pancreatic cancer isogenic cell lines confirmed earlier observations that NQO1 expression increases sensitivity to 17-AAG [106, 110]. HPLC analysis confirmed greater 17-AAG hydroquinone generation in cell lines expressing NQO1 and hydroquinone formation could be inhibited by pretreatment with the NQO1 mechanism-based inhibitor ES936 (5-methoxy-1,2-dimethyl-3-[(4-nitrophenoxy)methyl]indole-4,7-dione) [106, 110]. Intracellular reduction of 17-AAG to the hydroquinone resulted in substantially higher intracellular concentrations of 17-AAG (quinone and hydroquinone) suggesting that the hydroquinone may not diffuse readily across cell membranes (Figure 4). Correspondingly, biomarkers of Hsp90 inhibition were more pronounced in cells expressing NQO1 compared to NQO1 null cells [106, 110]. Moreover, human glioblastoma and melanoma cell lines made resistant to 17-AAG by continuous low dose exposure had markedly decreased NQO1 activity and protein [112]. Interestingly, genetic analysis revealed that the resistant glioblastomas had acquired mutations in NQO1 and now expressed the NQO1 null (NQO1*2*2) genotype [112]. Taken together these data strongly implicate a role for NQO1 and reduction to the hydroquinone in the cytotoxicity of BQAs.
IPI504 (Retaspimycin) is the hydroquinone of 17-AAG and was developed as a more water-soluble alternative to 17-AAG. Interestingly, during the same time period it was demonstrated by our own work that the hydroquinone of 17AAG was the active Hsp90 inhibitor and was markedly more potent that parent quinone [106]. While studies using human tumor isogenic cell lines from colon, breast and pancreas which express a range of NQO1 protein levels showed a positive correlation between NQO1 activity and sensitivity to 17-AAG, a study using the 17-AAG prodrug IPI504 did not reach the same conclusion [113]. In experiments where cell lines were treated with IPI504 for 3 consecutive days no significant correlation was observed between NQO1 activity and growth inhibition induced by IPI504. These conclusions were puzzling since the most sensitive cell lines had the highest levels of NQO1 activity and cell lines genotyped as homozygous for the NQO1*2 polymorphism (MDA468 and MDA231) were the most resistant to IPI504 [113]. The lack of sensitivity to IPI504 in cells homozygous for NQO1*2 polymorphism is consistent with data obtained with 17-AAG [105, 106, 110, 114].
Despite studies which show resistance to BQAs in cell lines homozygous for the NQO1*2 polymorphism, there are no published studies at this time that have examined whether clinical response to BQAs is associated with the NQO1*2 genotype. A single study has examined the effect of the NQO1*2 allele on 17-AAG distribution and toxicity in a small number of patients in a phase 1 clinical trial. In this study no correlation was observed between the NQO1*2 allele and 17-AAG distribution and toxicity [115]. However, it was not clear if any of the participants in this study were homozygous for the NQO1*2 polymorphism.
NQO1 in the clinic
The use of NQO1 as a predictive biomarker for sensitivity to quinone antitumor drugs is compelling. NQO1 activity and protein expression can be easily measured in biopsied tumor samples but the results of a single biopsy may not be an accurate predictor of NQO1 activity over the course of therapy. NQO1 protein expression can be rapidly induced by a host of dietary components, xenobiotics and environmental factors. Therefore, the predictive value of immunohistochemical staining for NQO1 is limited since NQO1 levels may vary considerably over time. An alternative test for predicting NQO1 expression in tumors would involve genotyping patients for the NQO1*2 polymorphism. This simple genetic test may be a more practical measure of the influence of NQO1 on quinone drug activation since this test would identify NQO1-null individuals. Complicating the association between the NQO1*2 polymorphism and response to chemotherapy are studies in women with breast cancer that have demonstrated an association of the NQO1*2 polymorphism with decreased survival in patents receiving anthracycline therapy [116, 117], however, in these studies the role of NQO1 in survival is believed to be linked to its role in p53 stabilization and or modulation of TNF-alpha and not through drug activation [116].
Given the substantial amount of preclinical data clearly implicating NQO1 in the bioactivation of quinones such as β-lapachone and 17-AAG it would be predicted that NQO1 null individuals would respond less favorably or may have increased toxicities due to their inability to efficiently activate these drugs in tumor cells. However, to date there are no published papers that have examined whether the NQO1*2 polymorphism correlates with clinical outcomes for patients in clinical trials with β-lapachone and 17-AAG. Hopefully in the future as β-lapachone and the 17-AAG progress through clinical trials with larger numbers of patients the question of whether NQO1 plays a clinically significant role in the antitumor activity of these drugs will be addressed.
Summary
NQO1 is important in the bioreductive activation of a number of different types of antitumor quinones. The role of NQO1 in the bioreductive activation of mitomycin C remains controversial but pre-clinical data strongly suggests a role for NQO1 in the activation of β-lapachone and the benzoquinone ansamycin class of Hsp90 inhibitors. Currently, there is little clinical data to reinforce the biological relevance of NQO1 in either sensitivity or resistance to antitumor quinones but the availability of simple assays for the determination of a relatively common null polymorphism in NQO1 should facilitate clinical testing of this hypothesis in the future.
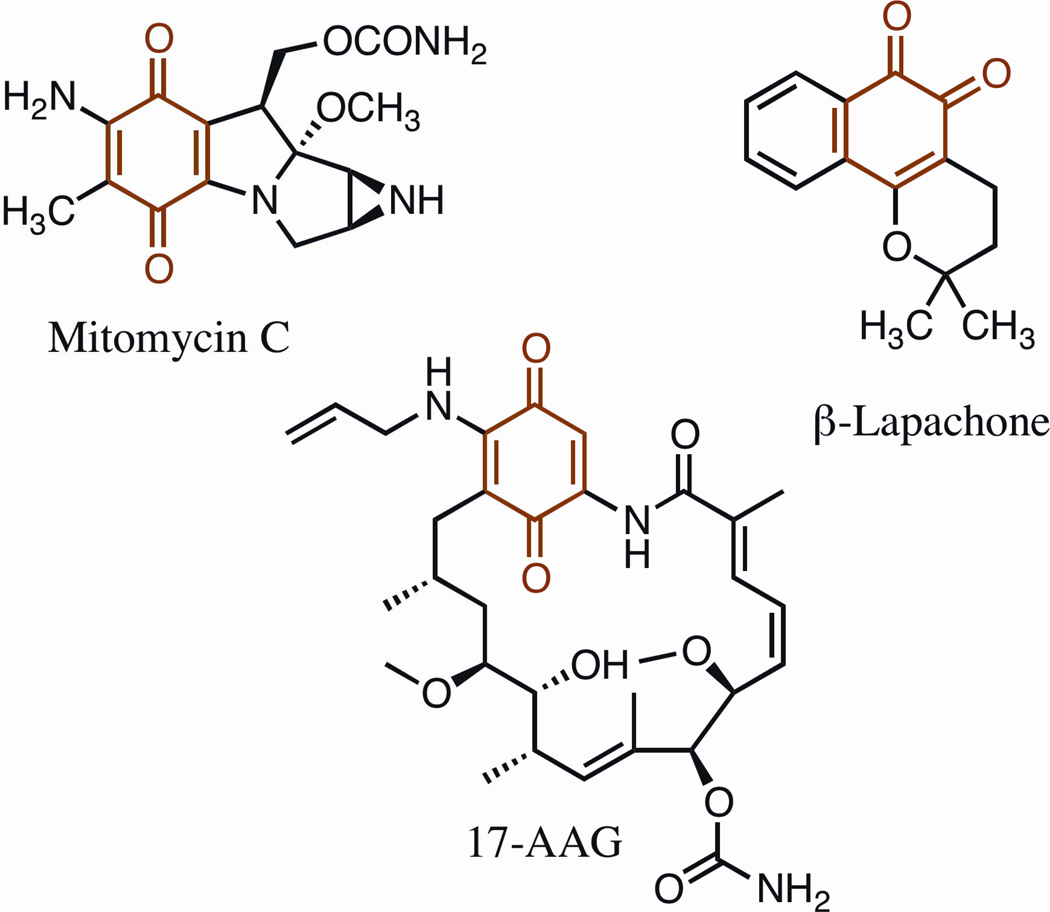
Figure 1.
Chemical structures of mitomycin C, β-lapachone and 17-AAG The quinone moiety is highlighted in red. 17-AAG, 17-N-allylamino-17-demethoxygeldanamycin.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Vasiliou V, Ross D, Nebert DW. Update of the NAD(P)H:quinone oxidoreductase (NQO) gene family. Hum Genomics. 2006;2:329–335. doi: 10.1186/1479-7364-2-5-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lind C, Cadenas E, Hochstein P, Ernster L. DT-diaphorase: purification, properties, and function. Methods Enzymol. 1990;186:287–301. doi: 10.1016/0076-6879(90)86122-c. [DOI] [PubMed] [Google Scholar]
- 3.Faig M, Bianchet MA, Talalay P, Chen S, Winski S, Ross D, et al. Structures of recombinant human and mouse NAD(P)H:quinone oxidoreductases: species comparison and structural changes with substrate binding and release. Proc Natl Acad Sci U S A. 2000;97:3177–3182. doi: 10.1073/pnas.050585797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bianchet MA, Faig M, Amzel LM. Structure and mechanism of NADP.H:quinone acceptor oxidoreductases (NQO) Methods Enzymol. 2004;382:144–174. doi: 10.1016/S0076-6879(04)82009-3. [DOI] [PubMed] [Google Scholar]
- 5.Winski SL, Koutalos Y, Bentley DL, Ross D. Subcellular localization of NAD(P)H:quinone oxidoreductase 1 in human cancer cells. Cancer Res. 2002;62:1420–1424. [PubMed] [Google Scholar]
- 6.Siegel D, Ross D. Immunodetection of NAD(P)H:quinone oxidoreductase 1 (NQO1) in human tissues. Free Radic Biol Med. 2000;29:246–253. doi: 10.1016/s0891-5849(00)00310-5. [DOI] [PubMed] [Google Scholar]
- 7.Siegel D, Franklin WA, Ross D. Immunohistochemical detection of NAD(P)H:quinone oxidoreductase in human lung and lung tumors. Clin Cancer Res. 1998;4:2065–2070. [PubMed] [Google Scholar]
- 8.Strassburg A, Strassburg CP, Manns MP, Tukey RH. Differential gene expression of NAD(P)H:quinone oxidoreductase and NRH:quinone oxidoreductase in human hepatocellular and biliary tissue. Mol Pharmacol. 2002;61:320–325. doi: 10.1124/mol.61.2.320. [DOI] [PubMed] [Google Scholar]
- 9.Schor NA, Morris HP. The activity of the D-T diaphorase in experimental hepatomas. Cancer Biochem Biophys. 1977;2:5–9. [PubMed] [Google Scholar]
- 10.Cresteil T, Jaiswal AK. High levels of expression of the NAD(P)H:quinone oxidoreductase (NQO1) gene in tumor cells compared to normal cells of the same origin. Biochem Pharmacol. 1991;42:1021–1027. doi: 10.1016/0006-2952(91)90284-c. [DOI] [PubMed] [Google Scholar]
- 11.Schlager JJ, Powis G. Cytosolic NAD(P)H:(quinone-acceptor)oxidoreductase in human normal and tumor tissue: effects of cigarette smoking and alcohol. Int J Cancer. 1990;45:403–409. doi: 10.1002/ijc.2910450304. [DOI] [PubMed] [Google Scholar]
- 12.Traver RD, Horikoshi T, Danenberg KD, Stadlbauer TH, Danenberg PV, Ross D, et al. NAD(P)H:quinone oxidoreductase gene expression in human colon carcinoma cells: characterization of a mutation which modulates DT-diaphorase activity and mitomycin sensitivity. Cancer Res. 1992;52:797–802. [PubMed] [Google Scholar]
- 13.Chen S, Wu K, Zhang D, Sherman M, Knox R, Yang CS. Molecular characterization of binding of substrates and inhibitors to DT-diaphorase: combined approach involving site-directed mutagenesis, inhibitor-binding analysis, and computer modeling. Mol Pharmacol. 1999;56:272–278. doi: 10.1124/mol.56.2.272. [DOI] [PubMed] [Google Scholar]
- 14.Tsvetkov P, Adamovich Y, Elliott E, Shaul Y. E3 ligase STUB1/CHIP regulates NAD(P)H:quinone oxidoreductase 1 (NQO1) accumulation in aged brain, a process impaired in certain Alzheimer disease patients. J Biol Chem. 2011;286:8839–8845. doi: 10.1074/jbc.M110.193276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Siegel D, Anwar A, Winski SL, Kepa JK, Zolman KL, Ross D. Rapid polyubiquitination and proteasomal degradation of a mutant form of NAD(P)H:quinone oxidoreductase 1. Mol Pharmacol. 2001;59:263–268. doi: 10.1124/mol.59.2.263. [DOI] [PubMed] [Google Scholar]
- 16.Siegel D, McGuinness SM, Winski SL, Ross D. Genotype-phenotype relationships in studies of a polymorphism in NAD(P)H:quinone oxidoreductase 1. Pharmacogenetics. 1999;9:113–121. doi: 10.1097/00008571-199902000-00015. [DOI] [PubMed] [Google Scholar]
- 17.Gaedigk A, Tyndale RF, Jurima-Romet M, Sellers EM, Grant DM, Leeder JS. NAD(P)H:quinone oxidoreductase: polymorphisms and allele frequencies in Caucasian, Chinese and Canadian Native Indian and Inuit populations. Pharmacogenetics. 1998;8:305–313. doi: 10.1097/00008571-199808000-00004. [DOI] [PubMed] [Google Scholar]
- 18.Kelsey KT, Ross D, Traver RD, Christiani DC, Zuo ZF, Spitz MR, et al. Ethnic variation in the prevalence of a common NAD(P)H quinone oxidoreductase polymorphism and its implications for anti-cancer chemotherapy. Br J Cancer. 1997;76:852–854. doi: 10.1038/bjc.1997.474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ross D, Siegel D. NAD(P)H:quinone oxidoreductase 1 (NQO1, DT-diaphorase), functions and pharmacogenetics. Methods Enzymol. 2004;382:115–144. doi: 10.1016/S0076-6879(04)82008-1. [DOI] [PubMed] [Google Scholar]
- 20.Pan SS, Forrest GL, Akman SA, Hu LT. NAD(P)H:quinone oxidoreductase expression and mitomycin C resistance developed by human colon cancer HCT 116 cells. Cancer Res. 1995;55:330–335. [PubMed] [Google Scholar]
- 21.Hu LT, Stamberg J, Pan S. The NAD(P)H:quinone oxidoreductase locus in human colon carcinoma HCT 116 cells resistant to mitomycin C. Cancer Res. 1996;56:5253–5259. [PubMed] [Google Scholar]
- 22.Begleiter A, Robotham E, Leith MK. Role of NAD(P)H:(quinone acceptor) oxidoreductase (DT-diaphorase) in activation of mitomycin C under hypoxia. Mol Pharmacol. 1992;41:677–682. [PubMed] [Google Scholar]
- 23.Keyes SR, Fracasso PM, Heimbrook DC, Rockwell S, Sligar SG, Sartorelli AC. Role of NADPH:cytochrome c reductase and DT-diaphorase in the biotransformation of mitomycin C1. Cancer Res. 1984;44:5638–5643. [PubMed] [Google Scholar]
- 24.Siegel D, Gibson NW, Preusch PC, Ross D. Metabolism of mitomycin C by DT-diaphorase: role in mitomycin C-induced DNA damage and cytotoxicity in human colon carcinoma cells. Cancer Res. 1990;50:7483–7489. [PubMed] [Google Scholar]
- 25.Marshall RS, Paterson MC, Rauth AM. Studies on the mechanism of resistance to mitomycin C and porfiromycin in a human cell strain derived from a cancer-prone individual. Biochem Pharmacol. 1991;41:1351–1360. doi: 10.1016/0006-2952(91)90108-h. [DOI] [PubMed] [Google Scholar]
- 26.Marshall RS, Paterson MC, Rauth AM. DT-diaphorase activity and mitomycin C sensitivity in non-transformed cell strains derived from members of a cancer-prone family. Carcinogenesis. 1991;12:1175–1180. doi: 10.1093/carcin/12.7.1175. [DOI] [PubMed] [Google Scholar]
- 27.Pan SS, Akman SA, Forrest GL, Hipsher C, Johnson R. The role of NAD(P)H:quinone oxidoreductase in mitomycin C- and porfiromycin-resistant HCT 116 human colon-cancer cells. Cancer Chemother Pharmacol. 1992;31:23–31. doi: 10.1007/BF00695990. [DOI] [PubMed] [Google Scholar]
- 28.Lee JH, Naito M, Nakajima M, Tsuruo T. Isolation and characterization of a mitomycin C-resistant variant of human colon carcinoma HT-29 cells. Cancer Chemother Pharmacol. 1993;33:215–220. doi: 10.1007/BF00686219. [DOI] [PubMed] [Google Scholar]
- 29.Nishiyama M, Saeki S, Aogi K, Hirabayashi N, Toge T. Relevance of DT-diaphorase activity to mitomycin C (MMC) efficacy on human cancer cells: differences in in vitro and in vivo systems. Int J Cancer. 1993;53:1013–1016. doi: 10.1002/ijc.2910530626. [DOI] [PubMed] [Google Scholar]
- 30.Begleiter A, Leith MK. Role of NAD(P)H:(quinone acceptor) oxidoreductase (DT-diaphorase) in activation of mitomycin C under acidic conditions. Mol Pharmacol. 1993;44:210–215. [PubMed] [Google Scholar]
- 31.Bando T, Kasahara K, Shibata K, Nakatsumi Y, Fujimura M, Matsuda T. Role of dt-diaphorase as a determinant of sensitivity to mitomycin analogs in nonsmall cell lung-cancer cell-lines. Int J Oncol. 1994;5:819–825. doi: 10.3892/ijo.5.4.819. [DOI] [PubMed] [Google Scholar]
- 32.Shibata K, Kasahara K, Bando T, Nakatsumi Y, Fujimura M, Tsuruo T, et al. Establishment and characterization of non-small cell lung cancer cell lines resistant to mitomycin C under aerobic conditions. Jpn J Cancer Res. 1995;86:460–469. doi: 10.1111/j.1349-7006.1995.tb03079.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yoshida T, Tsuda H. Gene targeting of DT-diaphorase in mouse embryonic stem cells: establishment of null mutant and its mitomycin C-resistance. Biochem Biophys Res Commun. 1995;214:701–708. doi: 10.1006/bbrc.1995.2342. [DOI] [PubMed] [Google Scholar]
- 34.Fitzsimmons SA, Workman P, Grever M, Paull K, Camalier R, Lewis AD. Reductase enzyme expression across the National Cancer Institute Tumor cell line panel: correlation with sensitivity to mitomycin C and EO9. J Natl Cancer Inst. 1996;88:259–269. doi: 10.1093/jnci/88.5.259. [DOI] [PubMed] [Google Scholar]
- 35.Mikami K, Naito M, Tomida A, Yamada M, Sirakusa T, Tsuruo T. DT-diaphorase as a critical determinant of sensitivity to mitomycin C in human colon and gastric carcinoma cell lines. Cancer Res. 1996;56:2823–2826. [PubMed] [Google Scholar]
- 36.Begleiter A, Leith MK, Curphey TJ. Induction of DT-diaphorase by 1,2-dithiole-3-thione and increase of antitumour activity of bioreductive agents. Br J Cancer Suppl. 1996;27:S9–S14. [PMC free article] [PubMed] [Google Scholar]
- 37.Wang X, Doherty GP, Leith MK, Curphey TJ, Begleiter A. Enhanced cytotoxicity of mitomycin C in human tumour cells with inducers of DT-diaphorase. Br J Cancer. 1999;80:1223–1230. doi: 10.1038/sj.bjc.6690489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Suzuki K, Yamamoto W, Park JS, Hanaoka H, Okamoto R, Kirihara Y, et al. Regulatory network of mitomycin C action in human colon cancer cells. Jpn J Cancer Res. 1999;90:571–577. doi: 10.1111/j.1349-7006.1999.tb00785.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seow HA, Penketh PG, Belcourt MF, Tomasz M, Rockwell S, Sartorelli AC. Nuclear overexpression of NAD(P)H:quinone oxidoreductase 1 in Chinese hamster ovary cells increases the cytotoxicity of mitomycin C under aerobic and hypoxic conditions. J Biol Chem. 2004;279:31606–31612. doi: 10.1074/jbc.M404910200. [DOI] [PubMed] [Google Scholar]
- 40.Nishiyama M, Suzuki K, Kumazaki T, Yamamoto W, Toge T, Okamura T, et al. Molecular targeting of mitomycin C chemotherapy. Int J Cancer. 1997;72:649–656. doi: 10.1002/(sici)1097-0215(19970807)72:4<649::aid-ijc17>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 41.Lin YL, Ho IC, Su PF, Lee TC. Arsenite pretreatment enhances the cytotoxicity of mitomycin C in human cancer cell lines via increased NAD(P)H quinone oxidoreductase 1 expression. Toxicol Appl Pharmacol. 2006;214:309–317. doi: 10.1016/j.taap.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 42.Keyes SR, Heimbrook DC, Fracasso PM, Rockwell S, Sligar SG, Sartorelli AC. Chemotherapeutic attack of hypoxic tumor cells by the bioreductive alkylating agent mitomycin C. Adv Enzyme Regul. 1985;23:291–307. doi: 10.1016/0065-2571(85)90053-6. [DOI] [PubMed] [Google Scholar]
- 43.Keyes SR, Rockwell S, Sartorelli AC. Enhancement of mitomycin C cytotoxicity to hypoxic tumor cells by dicoumarol in vivo and in vitro. Cancer Res. 1985;45:213–216. [PubMed] [Google Scholar]
- 44.Dulhanty AM, Whitmore GF. Chinese hamster ovary cell lines resistant to mitomycin C under aerobic but not hypoxic conditions are deficient in DT-diaphorase. Cancer Res. 1991;51:1860–1865. [PubMed] [Google Scholar]
- 45.Bizanek R, Chowdary D, Arai H, Kasai M, Hughes CS, Sartorelli AC, et al. Adducts of mitomycin C and DNA in EMT6 mouse mammary tumor cells: effects of hypoxia and dicumarol on adduct patterns. Cancer Res. 1993;53:5127–5134. [PubMed] [Google Scholar]
- 46.Siegel D, Beall H, Senekowitsch C, Kasai M, Arai H, Gibson NW, et al. Bioreductive activation of mitomycin C by DT-diaphorase. Biochemistry. 1992;31:7879–7885. doi: 10.1021/bi00149a019. [DOI] [PubMed] [Google Scholar]
- 47.Siegel D, Beall H, Kasai M, Arai H, Gibson NW, Ross D. pH-dependent inactivation of DT-diaphorase by mitomycin C and porfiromycin. Mol Pharmacol. 1993;44:1128–1134. [PubMed] [Google Scholar]
- 48.Pan SS, Yu F, Hipsher C. Enzymatic and pH modulation of mitomycin C-induced DNA damage in mitomycin C-resistant HCT 116 human colon cancer cells. Mol Pharmacol. 1993;43:870–877. [PubMed] [Google Scholar]
- 49.Kennedy KA, McGurl JD, Leondaridis L, Alabaster O. pH dependence of mitomycin C-induced cross-linking activity in EMT6 tumor cells. Cancer Res. 1985;45:3541–3547. [PubMed] [Google Scholar]
- 50.Sharp SY, Kelland LR, Valenti MR, Brunton LA, Hobbs S, Workman P. Establishment of an isogenic human colon tumor model for NQO1 gene expression: application to investigate the role of DT-diaphorase in bioreductive drug activation in vitro and in vivo. Mol Pharmacol. 2000;58:1146–1155. doi: 10.1124/mol.58.5.1146. [DOI] [PubMed] [Google Scholar]
- 51.Winski SL, Hargreaves RH, Butler J, Ross D. A new screening system for NAD(P)H:quinone oxidoreductase (NQO1)-directed antitumor quinones: identification of a new aziridinylbenzoquinone, RH1, as a NQO1-directed antitumor agent. Clin Cancer Res. 1998;4:3083–3088. [PubMed] [Google Scholar]
- 52.Cowen RL, Patterson AV, Telfer BA, Airley RE, Hobbs S, Phillips RM, et al. Viral delivery of P450 reductase recapitulates the ability of constitutive overexpression of reductase enzymes to potentiate the activity of mitomycin C in human breast cancer xenografts. Mol Cancer Ther. 2003;2:901–909. [PubMed] [Google Scholar]
- 53.Begleiter A, Leith MK, Thliveris JA, Digby T. Dietary induction of NQO1 increases the antitumour activity of mitomycin C in human colon tumours in vivo. Br J Cancer. 2004;91:1624–1631. doi: 10.1038/sj.bjc.6602171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Eickelmann P, Schulz WA, Rohde D, Schmitz-Drager B, Sies H. Loss of heterozygosity at the NAD(P)H: quinone oxidoreductase locus associated with increased resistance against mitomycin C in a human bladder carcinoma cell line. Biol Chem Hoppe Seyler. 1994;375:439–445. doi: 10.1515/bchm3.1994.375.7.439. [DOI] [PubMed] [Google Scholar]
- 55.Perry RR, Kang Y, Greaves B. Biochemical characterization of a mitomycin C resistant colon cancer cell line variant. Biochem Pharmacol. 1993;46:1999–2005. doi: 10.1016/0006-2952(93)90642-a. [DOI] [PubMed] [Google Scholar]
- 56.Xu BH, Gupta V, Singh SV. Characterization of a human bladder cancer cell line selected for resistance to mitomycin C. Int J Cancer. 1994;58:686–692. doi: 10.1002/ijc.2910580512. [DOI] [PubMed] [Google Scholar]
- 57.Xu BH, Gupta V, Singh SV. Mechanism of differential sensitivity of human bladder cancer cells to mitomycin C and its analogue. Br J Cancer. 1994;69:242–246. doi: 10.1038/bjc.1994.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ward TH, Haran MS, Whittaker D, Watson AJ, Howard TD, Butler J. Cross-resistance studies on two K562 sublines resistant to diaziridinylbenzoquinones. Biochem Pharmacol. 1995;50:459–464. doi: 10.1016/0006-2952(95)00155-s. [DOI] [PubMed] [Google Scholar]
- 59.Singh SV, Xu BH, Gupta V, Emerson EO, Zaren HA, Jani JP. Characterization of a human bladder cancer cell line selected for resistance to BMY 25067, a novel analogue of mitomycin C. Cancer Lett. 1995;95:49–56. doi: 10.1016/0304-3835(95)03864-s. [DOI] [PubMed] [Google Scholar]
- 60.Lambert PA, Kang Y, Greaves B, Perry RR. The importance of DT-diaphorase in mitomycin C resistance in human colon cancer cell lines. J Surg Res. 1998;80:177–181. doi: 10.1006/jsre.1998.5481. [DOI] [PubMed] [Google Scholar]
- 61.Baumann RP, Hodnick WF, Seow HA, Belcourt MF, Rockwell S, Sherman DH, et al. Reversal of mitomycin C resistance by overexpression of bioreductive enzymes in Chinese hamster ovary cells. Cancer Res. 2001;61:7770–7776. [PubMed] [Google Scholar]
- 62.Malkinson AM, Siegel D, Forrest GL, Gazdar AF, Oie HK, Chan DC, et al. Elevated DT-diaphorase activity and messenger RNA content in human non-small cell lung carcinoma: relationship to the response of lung tumor xenografts to mitomycin Cl. Cancer Res. 1992;52:4752–4757. [PubMed] [Google Scholar]
- 63.Phillips RM, Burger AM, Fiebig HH, Double JA. Genotyping of NAD(P)H:quinone oxidoreductase (NQO1) in a panel of human tumor xenografts: relationship between genotype status, NQO1 activity and the response of xenografts to Mitomycin C chemotherapy in vivo(1) Biochem Pharmacol. 2001;62:1371–1377. doi: 10.1016/s0006-2952(01)00769-9. [DOI] [PubMed] [Google Scholar]
- 64.Fleming RA, Drees J, Loggie BW, Russell GB, Geisinger KR, Morris RT, et al. Clinical significance of a NAD(P)H: quinone oxidoreductase 1 polymorphism in patients with disseminated peritoneal cancer receiving intraperitoneal hyperthermic chemotherapy with mitomycin C. Pharmacogenetics. 2002;12:31–37. doi: 10.1097/00008571-200201000-00005. [DOI] [PubMed] [Google Scholar]
- 65.Gan Y, Mo Y, Kalns JE, Lu J, Danenberg K, Danenberg P, et al. Expression of DT-diaphorase and cytochrome P450 reductase correlates with mitomycin C activity in human bladder tumors. Clin Cancer Res. 2001;7:1313–1319. [PubMed] [Google Scholar]
- 66.Basu S, Brown JE, Flannigan GM, Gill JH, Loadman PM, Martin SW, et al. Immunohistochemical analysis of NAD(P)H:quinone oxidoreductase and NADPH cytochrome P450 reductase in human superficial bladder tumours: relationship between tumour enzymology and clinical outcome following intravesical mitomycin C therapy. Int J Cancer. 2004;109:703–709. doi: 10.1002/ijc.20005. [DOI] [PubMed] [Google Scholar]
- 67.Basu S, Brown JE, Flannigan GM, Gill JH, Loadman PM, Martin SW, et al. NAD(P)H:Quinone oxidoreductase-1 C609T polymorphism analysis in human superficial bladder cancers: relationship of genotype status to NQO1 phenotype and clinical response to Mitomycin C. Int J Oncol. 2004;25:921–927. [PubMed] [Google Scholar]
- 68.Buzdar AU, Tashima CK, Blumenschein GR, Hortobagyi GN, Yap HY, Krutchik AN, et al. Mitomycin-C and megestrol acetate in treatment of breast cancer refractory to hormonal and combination chemotherapy. Cancer. 1978;41:392–395. doi: 10.1002/1097-0142(197802)41:2<392::aid-cncr2820410202>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 69.Adikesavan AK, Barrios R, Jaiswal AK. In vivo role of NAD(P)H:quinone oxidoreductase 1 in metabolic activation of mitomycin C and bone marrow cytotoxicity. Cancer Res. 2007;67:7966–7971. doi: 10.1158/0008-5472.CAN-06-4480. [DOI] [PubMed] [Google Scholar]
- 70.Boveris A, Docampo R, Turrens JF, Stoppani AO. Effect of beta-lapachone on superoxide anion and hydrogen peroxide production in Trypanosoma cruzi. Biochem J. 1978;175:431–439. doi: 10.1042/bj1750431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Boorstein RJ, Pardee AB. Coordinate inhibition of DNA synthesis and thymidylate synthase activity following DNA damage and repair. Biochem Biophys Res Commun. 1983;117:30–36. doi: 10.1016/0006-291x(83)91536-x. [DOI] [PubMed] [Google Scholar]
- 72.Boorstein RJ, Pardee AB. Beta-lapachone greatly enhances MMS lethality to human fibroblasts. Biochem Biophys Res Commun. 1984;118:828–834. doi: 10.1016/0006-291x(84)91469-4. [DOI] [PubMed] [Google Scholar]
- 73.Boothman DA, Trask DK, Pardee AB. Inhibition of potentially lethal DNA damage repair in human tumor cells by beta-lapachone, an activator of topoisomerase I. Cancer Res. 1989;49:605–612. [PubMed] [Google Scholar]
- 74.Li CJ, Averboukh L, Pardee AB. beta-Lapachone, a novel DNA topoisomerase I inhibitor with a mode of action different from camptothecin. J Biol Chem. 1993;268:22463–22468. [PubMed] [Google Scholar]
- 75.Planchon SM, Wuerzberger-Davis SM, Pink JJ, Robertson KA, Bornmann WG, Boothman DA. Bcl-2 protects against beta-lapachone-mediated caspase 3 activation and apoptosis in human myeloid leukemia (HL-60) cells. Oncol Rep. 1999;6:485–492. doi: 10.3892/or.6.3.485. [DOI] [PubMed] [Google Scholar]
- 76.Pink JJ, Planchon SM, Tagliarino C, Varnes ME, Siegel D, Boothman DA. NAD(P)H:Quinone oxidoreductase activity is the principal determinant of beta-lapachone cytotoxicity. J Biol Chem. 2000;275:5416–5424. doi: 10.1074/jbc.275.8.5416. [DOI] [PubMed] [Google Scholar]
- 77.Planchon SM, Pink JJ, Tagliarino C, Bornmann WG, Varnes ME, Boothman DA. beta-Lapachone-induced apoptosis in human prostate cancer cells: involvement of NQO1/xip3. Exp Cell Res. 2001;267:95–106. doi: 10.1006/excr.2001.5234. [DOI] [PubMed] [Google Scholar]
- 78.Docampo R, Cruz FS, Boveris A, Muniz RP, Esquivel DM. beta-Lapachone enhancement of lipid peroxidation and superoxide anion and hydrogen peroxide formation by sarcoma 180 ascites tumor cells. Biochem Pharmacol. 1979;28:723–728. doi: 10.1016/0006-2952(79)90348-4. [DOI] [PubMed] [Google Scholar]
- 79.Tagliarino C, Pink JJ, Dubyak GR, Nieminen AL, Boothman DA. Calcium is a key signaling molecule in beta-lapachone-mediated cell death. J Biol Chem. 2001;276:19150–19159. doi: 10.1074/jbc.M100730200. [DOI] [PubMed] [Google Scholar]
- 80.Li LS, Bey EA, Dong Y, Meng J, Patra B, Yan J, et al. Modulating endogenous NQO1 levels identifies key regulatory mechanisms of action of beta-lapachone for pancreatic cancer therapy. Clin Cancer Res. 2011;17:275–285. doi: 10.1158/1078-0432.CCR-10-1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Choi EK, Terai K, Ji IM, Kook YH, Park KH, Oh ET, et al. Upregulation of NAD(P)H:quinone oxidoreductase by radiation potentiates the effect of bioreductive beta-lapachone on cancer cells. Neoplasia. 2007;9:634–642. doi: 10.1593/neo.07397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Suzuki M, Amano M, Choi J, Park HJ, Williams BW, Ono K, et al. Synergistic effects of radiation and beta-lapachone in DU-145 human prostate cancer cells in vitro. Radiat Res. 2006;165:525–531. doi: 10.1667/RR3554.1. [DOI] [PubMed] [Google Scholar]
- 83.Boothman DA, Meyers M, Fukunaga N, Lee SW. Isolation of x-ray-inducible transcripts from radioresistant human melanoma cells. Proc Natl Acad Sci U S A. 1993;90:7200–7204. doi: 10.1073/pnas.90.15.7200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Song CW, Chae JJ, Choi EK, Hwang TS, Kim C, Lim BU, et al. Anti-cancer effect of bio-reductive drug beta-lapachon is enhanced by activating NQO1 with heat shock. Int J Hyperthermia. 2008;24:161–169. doi: 10.1080/02656730701781895. [DOI] [PubMed] [Google Scholar]
- 85.Dong GZ, Youn H, Park MT, Oh ET, Park KH, Song CW, et al. Heat shock increases expression of NAD(P)H:quinone oxidoreductase (NQO1), mediator of beta-lapachone cytotoxicity, by increasing NQO1 gene activity and via Hsp70-mediated stabilisation of NQO1 protein. Int J Hyperthermia. 2009;25:477–487. doi: 10.1080/02656730903049836. [DOI] [PubMed] [Google Scholar]
- 86.Park HJ, Choi EK, Choi J, Ahn KJ, Kim EJ, Ji IM, et al. Heat-induced up-regulation of NAD(P)H:quinone oxidoreductase potentiates anticancer effects of beta-lapachone. Clin Cancer Res. 2005;11:8866–8871. doi: 10.1158/1078-0432.CCR-05-0818. [DOI] [PubMed] [Google Scholar]
- 87.Bey EA, Bentle MS, Reinicke KE, Dong Y, Yang CR, Girard L, et al. An NQO1- and PARP-1-mediated cell death pathway induced in non-small-cell lung cancer cells by beta-lapachone. Proc Natl Acad Sci U S A. 2007;104:11832–11837. doi: 10.1073/pnas.0702176104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Awadallah NS, Dehn D, Shah RJ, Russell Nash S, Chen YK, Ross D, et al. NQO1 expression in pancreatic cancer and its potential use as a biomarker. Appl Immunohistochem Mol Morphol. 2008;16:24–31. doi: 10.1097/PAI.0b013e31802e91d0. [DOI] [PubMed] [Google Scholar]
- 89.Ough M, Lewis A, Bey EA, Gao J, Ritchie JM, Bornmann W, et al. Efficacy of beta-lapachone in pancreatic cancer treatment: exploiting the novel, therapeutic target NQO1. Cancer Biol Ther. 2005;4:95–102. doi: 10.4161/cbt.4.1.1382. [DOI] [PubMed] [Google Scholar]
- 90.DeBoer C, Meulman PA, Wnuk RJ, Peterson DH. Geldanamycin, a new antibiotic. J Antibiot (Tokyo) 1970;23:442–447. doi: 10.7164/antibiotics.23.442. [DOI] [PubMed] [Google Scholar]
- 91.BeBoer C, Dietz A. The description and antibiotic production of Streptomyces hygroscopicus var. Geldanus. J Antibiot (Tokyo) 1976;29:1182–1188. doi: 10.7164/antibiotics.29.1182. [DOI] [PubMed] [Google Scholar]
- 92.Yamaki H, Iguchi-Ariga SM, Ariga H. Inhibition of c-myc gene expression in murine lymphoblastoma cells by geldanamycin and herbimycin, antibiotics of benzoquinoid ansamycin group. J Antibiot (Tokyo) 1989;42:604–610. doi: 10.7164/antibiotics.42.604. [DOI] [PubMed] [Google Scholar]
- 93.Li LH, Clark TD, Cowie CH, Rinehart KL., Jr Effects of geldanamycin and its derivatives on RNA-directed DNA polymerase and infectivity of Rauscher leukemia virus. Cancer Treat Rep. 1977;61:815–824. [PubMed] [Google Scholar]
- 94.Yamaki H, Suzuki H, Choi EC, Tanaka N. Inhibition of DNA synthesis in murine tumor cells by geldanamycin, an antibiotic of the benzoquinoid ansamycin group. J Antibiot (Tokyo) 1982;35:886–892. doi: 10.7164/antibiotics.35.886. [DOI] [PubMed] [Google Scholar]
- 95.Uehara Y, Hori M, Takeuchi T, Umezawa H. Phenotypic change from transformed to normal induced by benzoquinonoid ansamycins accompanies inactivation of p60src in rat kidney cells infected with Rous sarcoma virus. Mol Cell Biol. 1986;6:2198–2206. doi: 10.1128/mcb.6.6.2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Whitesell L, Mimnaugh EG, De Costa B, Myers CE, Neckers LM. Inhibition of heat shock protein HSP90-pp60v-src heteroprotein complex formation by benzoquinone ansamycins: essential role for stress proteins in oncogenic transformation. Proc Natl Acad Sci U S A. 1994;91:8324–8328. doi: 10.1073/pnas.91.18.8324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Schulte TW, Neckers LM. The benzoquinone ansamycin 17-allylamino-17-demethoxygeldanamycin binds to HSP90 and shares important biologic activities with geldanamycin. Cancer Chemother Pharmacol. 1998;42:273–279. doi: 10.1007/s002800050817. [DOI] [PubMed] [Google Scholar]
- 98.Basso AD, Solit DB, Munster PN, Rosen N. Ansamycin antibiotics inhibit Akt activation and cyclin D expression in breast cancer cells that overexpress HER2. Oncogene. 2002;21:1159–1166. doi: 10.1038/sj.onc.1205184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Munster PN, Marchion DC, Basso AD, Rosen N. Degradation of HER2 by ansamycins induces growth arrest and apoptosis in cells with HER2 overexpression via a HER3, phosphatidylinositol 3'-kinase-AKT-dependent pathway. Cancer Res. 2002;62:3132–3137. [PubMed] [Google Scholar]
- 100.Nimmanapalli R, O'Bryan E, Bhalla K. Geldanamycin and its analogue 17-allylamino-17-demethoxygeldanamycin lowers Bcr-Abl levels and induces apoptosis and differentiation of Bcr-Abl-positive human leukemic blasts. Cancer Res. 2001;61:1799–1804. [PubMed] [Google Scholar]
- 101.Fumo G, Akin C, Metcalfe DD, Neckers L. 17-Allylamino-17-demethoxygeldanamycin (17-AAG) is effective in down-regulating mutated, constitutively activated KIT protein in human mast cells. Blood. 2004;103:1078–1084. doi: 10.1182/blood-2003-07-2477. [DOI] [PubMed] [Google Scholar]
- 102.Yu W, Rao Q, Wang M, Tian Z, Lin D, Liu X, et al. The Hsp90 inhibitor 17-allylamide-17-demethoxygeldanamycin induces apoptosis and differentiation of Kasumi-1 harboring the Asn822Lys KIT mutation and down-regulates KIT protein level. Leuk Res. 2006;30:575–582. doi: 10.1016/j.leukres.2005.08.028. [DOI] [PubMed] [Google Scholar]
- 103.Bagatell R, Khan O, Paine-Murrieta G, Taylor CW, Akinaga S, Whitesell L. Destabilization of steroid receptors by heat shock protein 90-binding drugs: a ligand-independent approach to hormonal therapy of breast cancer. Clin Cancer Res. 2001;7:2076–2084. [PubMed] [Google Scholar]
- 104.Supko JG, Hickman RL, Grever MR, Malspeis L. Preclinical pharmacologic evaluation of geldanamycin as an antitumor agent. Cancer Chemother Pharmacol. 1995;36:305–315. doi: 10.1007/BF00689048. [DOI] [PubMed] [Google Scholar]
- 105.Kelland LR, Sharp SY, Rogers PM, Myers TG, Workman P. DT-Diaphorase expression and tumor cell sensitivity to 17-allylamino, 17-demethoxygeldanamycin, an inhibitor of heat shock protein 90. J Natl Cancer Inst. 1999;91:1940–1949. doi: 10.1093/jnci/91.22.1940. [DOI] [PubMed] [Google Scholar]
- 106.Guo W, Reigan P, Siegel D, Zirrolli J, Gustafson D, Ross D. Formation of 17-allylamino-demethoxygeldanamycin (17-AAG) hydroquinone by NAD(P)H:quinone oxidoreductase 1: role of 17-AAG hydroquinone in heat shock protein 90 inhibition. Cancer Res. 2005;65:10006–10015. doi: 10.1158/0008-5472.CAN-05-2029. [DOI] [PubMed] [Google Scholar]
- 107.Guo W, Reigan P, Siegel D, Ross D. Enzymatic reduction and glutathione conjugation of benzoquinone ansamycin heat shock protein 90 inhibitors: relevance for toxicity and mechanism of action. Drug Metab Dispos. 2008;36:2050–2057. doi: 10.1124/dmd.108.022004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Guo W, Reigan P, Siegel D, Zirrolli J, Gustafson D, Ross D. The bioreduction of a series of benzoquinone ansamycins by NAD(P)H:quinone oxidoreductase 1 to more potent heat shock protein 90 inhibitors, the hydroquinone ansamycins. Mol Pharmacol. 2006;70:1194–1203. doi: 10.1124/mol.106.025643. [DOI] [PubMed] [Google Scholar]
- 109.Guo W, Siegel D, Ross D. Stability of the Hsp90 inhibitor 17AAG hydroquinone and prevention of metal-catalyzed oxidation. J Pharm Sci. 2008;97:5147–5157. doi: 10.1002/jps.21394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Siegel D, Shieh B, Yan C, Kepa JK, Ross D. Role for NAD(P)H:quinone oxidoreductase 1 and manganese-dependent superoxide dismutase in 17-(allylamino)-17-demethoxygeldanamycin-induced heat shock protein 90 inhibition in pancreatic cancer cells. J Pharmacol Exp Ther. 2011;336:874–880. doi: 10.1124/jpet.110.176438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Reigan P, Siegel D, Guo W, Ross D. A mechanistic and structural analysis of the inhibition of the 90-kDa heat shock protein by the benzoquinone and hydroquinone ansamycins. Mol Pharmacol. 2011;79:823–832. doi: 10.1124/mol.110.070086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Gaspar N, Sharp SY, Pacey S, Jones C, Walton M, Vassal G, et al. Acquired resistance to 17-allylamino-17-demethoxygeldanamycin (17-AAG, tanespimycin) in glioblastoma cells. Cancer Res. 2009;69:1966–1975. doi: 10.1158/0008-5472.CAN-08-3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Douglas M, Lim AR, Porter JR, West K, Pink MM, Ge J, et al. The antiproliferative activity of the heat shock protein 90 inhibitor IPI-504 is not dependent on NAD(P)H:quinone oxidoreductase 1 activity in vivo. Mol Cancer Ther. 2009;8:3369–3378. doi: 10.1158/1535-7163.MCT-09-0568. [DOI] [PubMed] [Google Scholar]
- 114.Zajac M, Gomez G, Benitez J, Martinez-Delgado B. Molecular signature of response and potential pathways related to resistance to the HSP90 inhibitor, 17AAG, in breast cancer. BMC Med Genomics. 2010;3:44. doi: 10.1186/1755-8794-3-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Goetz MP, Toft D, Reid J, Ames M, Stensgard B, Safgren S, et al. Phase I trial of 17-allylamino-17-demethoxygeldanamycin in patients with advanced cancer. J Clin Oncol. 2005;23:1078–1087. doi: 10.1200/JCO.2005.09.119. [DOI] [PubMed] [Google Scholar]
- 116.Fagerholm R, Hofstetter B, Tommiska J, Aaltonen K, Vrtel R, Syrjakoski K, et al. NAD(P)H:quinone oxidoreductase 1 NQO1*2 genotype (P187S) is a strong prognostic and predictive factor in breast cancer. Nat Genet. 2008;40:844–853. doi: 10.1038/ng.155. [DOI] [PubMed] [Google Scholar]
- 117.Jamieson D, Cresti N, Bray J, Sludden J, Griffin MJ, Hawsawi NM, et al. Two minor NQO1 and NQO2 alleles predict poor response of breast cancer patients to adjuvant doxorubicin and cyclophosphamide therapy. Pharmacogenet Genomics. 2011;21:808–819. doi: 10.1097/FPC.0b013e32834b6918. [DOI] [PubMed] [Google Scholar]