Abstract
Here we demonstrate two convenient methods to extend and narrow the useful dynamic range of a model electrochemical DNA sensor. We did so by combining DNA probes of different target affinities but with similar specificity on the same electrode. We were able to achieve an extended dynamic response spanning 3 orders of magnitude in target concentration. Using a different strategy we have also narrowed the useful dynamic range of an E-DNA sensor to only an 8-fold range of target concentrations.
Keywords: electrochemical biosensors, dynamic range, engineering, depletant, pseudo-Hill coefficients
The use of electrode-immobilized biomolecules, such as proteins and nucleic acids, represents a common feature among many emerging biotechnologies. For example, the specificity, affinity and versatility of biomolecular recognition has been exploited for the development of a wide range of electrochemical biosensors that show promise for the detection of many clinically and industrially important analytes[1,2]. Such “bioelectronic interfaces” similarly form the basis of biofuel cells[3] and molecular logic gates[4], technologies that have attracted significant recent efforts. Interest in the applications of surface-electrode-bound biomolecular systems is thus rapidly growing.
Despite their often impressive performances, technologies based on biomolecular recognition suffer from the inherent limitation of single-site binding: its fixed dose-response curve characteristics. That is, single-site binding almost invariably produces a fixed, hyperbolic relationship between target concentration and receptor binding (the Langmuir isotherm) for which the useful range (here defined as the range between 10% and 90% site occupancy) spans an 81-fold concentration range.[5] This fixed dynamic range reduces the utility of electrochemical biosensors in applications, such as viral load monitoring, in which the concentration of the target molecule can vary over many orders of magnitude. It likewise limits the usefulness of biosensors in applications requiring high sensitivity (a steep relationship between target concentration and output signal), such as in the monitoring of drugs with narrow therapeutic windows. Thus, the possibility to arbitrarily extend or narrow this fixed dynamic range would prove advantageous in several biosensing applications. Similarly, the ability to extend the dynamic range of biorecognition would likely improve the efficiency of biofuel cells,[3] and the ability to narrow the dynamic range would reduce noise in molecular logic gates,[4] further illustrating the limitations associated with the fixed dynamic range of most biomolecular recognition.
Recently we have shown that some of the mechanisms employed by nature to alter the otherwise fixed dynamic range of single-site binding can also be used to broaden and narrow the dose-response curves of solution-phase, optical biosensors.[6] For example, by combining together biosensors of identical specificity but differing in affinity we have expanded the useful 81-fold range of a molecular beacon, a model solution-phase optical biosensor, by more than 10,000-fold.[6] In parallel we have also adapted the sequestration mechanism, often employed by nature to generate “ultrasensitive” genetic networks, to narrow the dynamic range of the same biosensor down to 5-fold, thus greatly increasing the sensitivity of this category of biosensors.[6]
Following the above work we demonstrate here the application of these approaches to modifying the dynamic range of reagentless, electrochemical biosensors. Specifically, we have used these approaches to arbitrarily narrow and broaden the useful dynamic ranges of electrochemical “E-DNA” sensors,[7] a class of structure-switching DNA probes that enable the single-step detection of specific oligonucleotides directly in complex media, such as blood serum and environmental samples (Figure 1).[7b, 8]
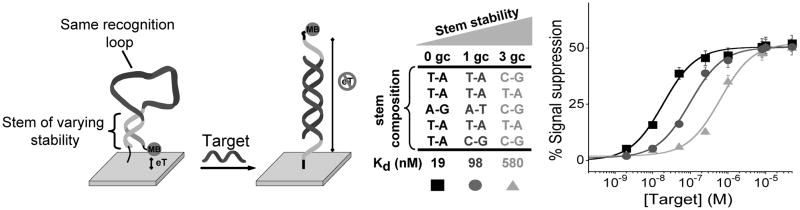
Figure 1.
(Left) E-DNA sensors consist of a stem-loop DNA modified with a redox reporter (here methylene blue) and attached to an interrogating gold electrode via an introduced thiol group.[7b] This probe undergoes a large-scale conformational switch upon hybridization with a DNA complementary to the loop, leading to large change in Faradaic current from the redox reporter. The affinity of such “switch-based” probes can be rationally tuned by many orders of magnitude, without affecting their specificity, by simply altering the stability of their nonbinding, non-signalling state (e.g., by varying the stability of the E-DNA probe’s stem with the change of the GC base pairs content).[9] (Right) Here we have employed a set of three E-DNA probes sharing a common recognition element but spanning almost three orders of magnitude of target affinity. Error bars in this figure and in the following figures represent the average and standard deviations of measurements performed on at least three independently sensors.
E-DNA sensors are comprised of a redox-reporter-modified stem-loop DNA-probe (receptor) attached to an interrogating electrode.[7b] In the absence of target, the formation of the stem holds the redox reporter into proximity with the electrode, supporting efficient electron transfer. Upon hybridization with a complementary oligonucleotide target, the terminus of the probe is pushed away from the electrode, which, in turn, hinders the efficiency with which electrons are transferred to the electrode and reduces the observed Faradaic current (Figure 1). The first strategy we have employed to narrow or extend the dynamic range of this sensor requires the availability of probes directed against the same target molecule but differing in affinity.[6] For the E-DNA sensor this can be achieved by using a set of stem-loop probes that share a common recognition loop, and thus target the same DNA sequence, but differ in the stability of their double-stranded stems. Doing so we can arbitrarily vary the target-probe dissociation constant –here over three orders of magnitude– without affecting the target-recognizing loop, and thus without changing the probe’s sequence specificity (Figure 1). [6, 9] While the affinity of E-DNA sensors is easily tuned via changes in their stem stability, reaching this objective can be more challenging for recognition elements displaying more complex structures. A number of rational and semi-rational strategies have, however, been reported by which to engineer (and tune) similar switching mechanisms into aptamers, aptazymes. and even proteins.[10] Loh and co-workers, for example, have recently demonstrated a generic strategy to design novel protein-based switches, termed “alternate frame folding”, in which duplication of a portion of a protein’s sequence is used to stabilize an alternative, nonbinding, circularly permuted conformation.[10d] Proteins and nucleic acids can also be engineered to undergo folding-induced conformational changes via the introduction of destabilizing mutations (typically remote from the target binding site so as to ensure that specificity is retained) that push the folding equilibrium toward the nonbinding, unfolded state, thus coupling binding to a conformational change (folding) and simultaneously coupling binding affinity to folding stability.[10]
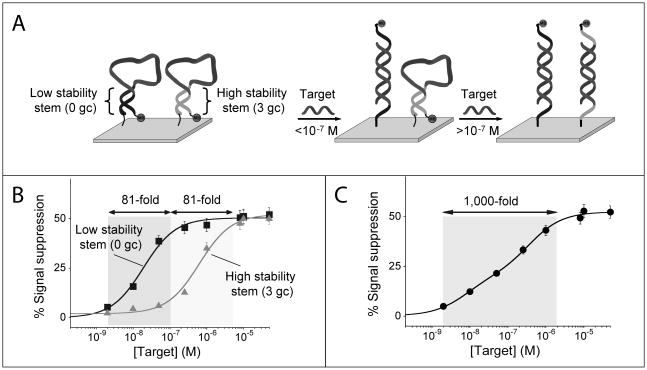
As noted above, traditional E-DNA sensors exhibit a useful dynamic range of 81-fold (Figure 1), again defined as the change in concentration required to transition from receptor occupancy of 10% to occupancy of 90%. We can extend this useful dynamic range by co-immobilizing two E-DNA probes differing in affinity for their (common) target DNA onto a single electrode. (Of note, the E-DNA probes we have employed are equally modified with the same methylene blue redox reporter and thus they both signal at the same redox potential and with the same relative signal change at saturating target concentrations, Figure 1). To achieve optimal log-linear behavior in the modified sensor, the affinities of the two probes should differ by approximately 30-fold.[6] For example, by combining on the same electrode surface an equimolar concentration of the low-stability 0GC stem-loop probe (Figure 1, bottom), which exhibits a dissociation constant of 19 nM, with the more stable 3GC stem-loop probe, exhibiting a dissociation constant of 580 nM, we expand the normally 81-fold dynamic range of this approach to approximately 1,000 fold (spanning from 2 nM to 2,000 nM) and achieve excellent linearity on a log[concentration] plot (R2 = 0.978; Figure 2).
Figure 2.
Employing a pair of signalling probes differing in affinity we can broaden the dynamic range of E-DNA sensors. (A) We did so by co-immobilizing (1:1 ratio) on a single electrode surface a relatively low affinity E-DNA probe (e.g., probe 3GC, Kd = 580 nM) with a higher affinity E-DNA probe (e.g., probe 0GC, Kd = 19 nM). (B) The useful dynamic range (defined as the fold-concentration change upon transition from 10% occupancy to 90% occupancy) of these individual probes spans an 81-fold range of target concentrations over two distinct concentration regimes. (C) With this strategy the resulting dose-response curve is extended and spans a 1,000-fold range of target concentrations.
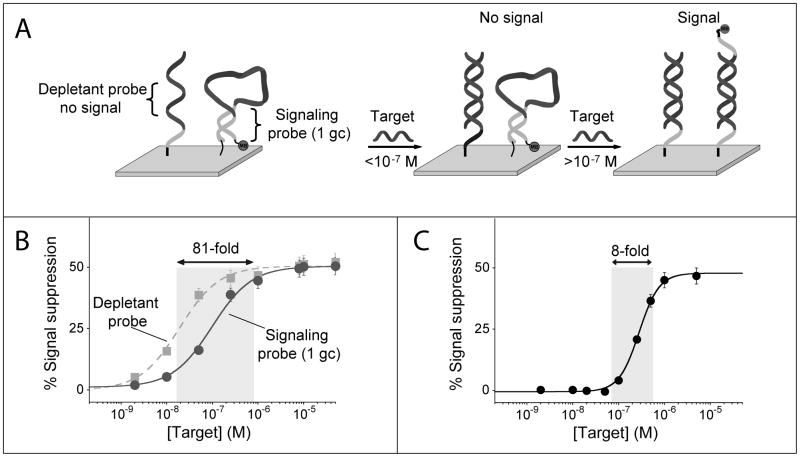
The availability of probes retaining a common specificity profile but differing in affinity also provides a means of narrowing the useful dynamic range of E-DNA sensors, thus enhancing their sensitivity (the steepness of the input/output curve) and improving their ability to measure small changes in concentration. Specifically, we adapted here the sequestration mechanism used by nature to improve the sensitivity of many regulatory cascades through the competition between a high-affinity, but not signaling, recognition element (the depletant) and a low-affinity signaling receptor.[11] To demonstrate this we co-immobilized two E-DNA probes, the stem-loop sequence 1GC and an equivalent, fully linear probe lacking a complementary stem, both of which are complementary to the same 13-base target sequence. Because the linear probe does not undergo a binding-induced conformational change its affinity for the DNA target is significantly greater than that of the stem loop 1GC probe. In this application the higher affinity linear probe lacks any redox reporter (methylene blue) and thus the hybridization of the target to this probe does not produce any measurable signal change. This linear probe therefore acts as the depletant, “silently” sequestering the target until the threshold concentration is surpassed.[11c] The lower-affinity signalling probe (1GC) is only activated (and thus only signals the presence of the target) when the depletant is saturated and this threshold is surpassed.
Using this approach we convert the hyperbolic dose-response curve of a traditional E-DNA sensor into an ultrasensitive response with a dynamic range spanning only an 8-fold range of target concentration, an order of magnitude narrower than the dynamic range of a traditional E-DNA sensor (Figure 3).
Figure 3.
Using the sequestration mechanism we can dramatically narrow the useful dynamic range of an E-DNA sensor, thus greatly improving its sensitivity (i.e., its ability to measure small changes in target concentration). (A) We do so by co-immobilizing on a single electrode surface a low affinity, signaling E-DNA probe with a higher affinity probe (depletant) which, lacking the redox reporter, does not signal upon binding its target. At low concentrations the target preferentially binds the depletant, which removes (sequesters) target from the sample without generating a signal. When the total target amount surpasses that of the depletant (the sink is saturated), a threshold response is achieved in which further addition of target dramatically raises the relative concentration of free target. This gives rise to a much steeper dose-response curve than this would occur in the absence of a depletant. (C) Using this approach we have narrowed the 81-fold useful dynamic range of an unmodified E-DNA sensor to a mere 8-fold, thus increasing its sensitivity by an order of magnitude.
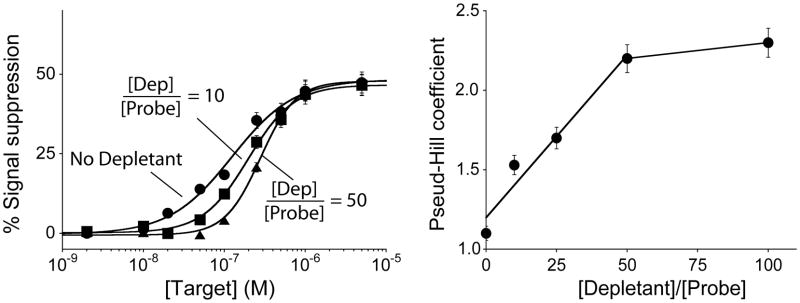
The sensitivity achieved via the sequestration mechanism depends on the relative amounts of depletant and signaling probe,[11c] and thus on the relative density of the two on the sensor’s surface. To demonstrate this we have altered the ratio of probe and depletant on our sensors by altering the depletant/probe concentration ratio employed during sensor fabrication.[12] To compare sensors fabricated using differing depletant/probe ratios we fitted their input-output curves to the Hill equation, which, although physically meaningful only when used to describe the ultrasensitivity associated with allosteric cooperativity,[13] provides a convenient means of quantifying the steepness of a binding curve. As expected, we observe a “pseudo”-Hill coefficient near unity (1.1±0.1) for sensors lacking the depletant. Upon the addition of the depletant probe, this coefficient increases monotonically with increasing depletant/probe ratios until it plateaus at 2.3 for ratios above 50 (Figure 4, right). The highest pseudo-Hill coefficient we have achieved compresses the 81-fold useful dynamic range of an unmodified E-DNA sensor to only 8-fold, significantly increasing the steepness of the dose-response curve of the sensor and, in turn, improving its ability to detect smaller relative changes in target concentration.
Figure 4.
The sensitivity (i.e., steepness of the dose-response curve) achieved using the sequestration mechanism depends on the ratio of depletant to probe employed during sensor fabrication. To show this we have fitted our data to obtain pseudo-Hill coefficients, which, although our system is not classically cooperative, are analogous to the Hill coefficient commonly used to describe cooperative enzymatic systems.[13] We find that the pseudo-Hill coefficient increases monotonically with this ratio until plateauing at values above 50.
The above arguments notwithstanding, we must note that this strategy is not without limitations. Specifically, the sequestration approach only works for fixed, small sample volumes (here we have employed 3 μL samples) in order to avoid the “premature” saturation of the fixed number of depletant molecules on the electrode surface. Moreover, as discussed before, the probe/depletant ratio on surface is a key factor which must be carefully controlled. We did so by assuming that the density ratios on surface are linearly correlated with the concentration ratios deployed in solution during deposition. This (seemingly reasonable) assumption seems confirmed by the linear dependence of the absolute current signals (which are correlated to surface density[14]) versus [probe]/[depletant] ratio (Figure SI1). However, we note that this correlation could be more complicated for less defined recognition elements which can induce a non-linear immobilization of probe and depletant (Figure SI2). Finally, the approach proposed is limited to [depletant]/[probe] ratio below 100: over this limit, the density of the probe, and therefore its signal, becomes so low that it is not possible to observe a measurable current (Figure SI3). In order to circumvent these limitations, we also propose the use of an alternative strategy where a fixed concentration of depletant is exogenously added to the mixture solution (Figure 5) thus overcoming possible problems due to uncontrolled density ratios. Moreover, because the depletant is now free to diffuse in solution, its affinity for the target is greatly increased compared to the surface-bound probe. This enables to use the same recognition element for both the depletant and the signalling probe thus making the approach also suitable to the use of more complex biorecognition elements whose affinity cannot be easily tuned.
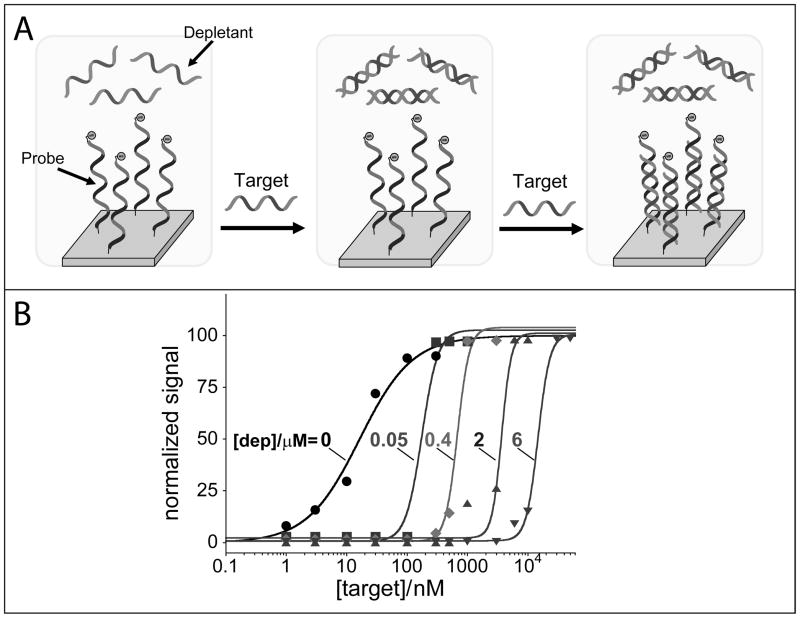
Figure 5.
To overcome the limitations inherent to the surface attached depletants (which are easily saturated), we also show that the depletant probe can be simply added in solution at a fixed concentration. Here we use an unlabeled non-signalling probe (with the exact same sequence of the signalling redox-labelled probe) that sequesters the target DNA till a threshold level (fixed by the depletant concentration in solution) over which further increase in target concentration results in a steep dose-response curve. Because the depletant is free in solution, it rapidly reacts with the target (and with higher affinity) before this later can diffuse to the electrode surface and “activate” the signalling probe. (B) By using different concentrations of depletant in the reaction mix (0, 0.05, 0.4, 2, 6 μM) we can not only achieve steeper transitions than those observed with the depletant co-immobilized with the probe but we can also easily tune the threshold level at which we observe the sharp digital-like response of the sensor.
The unattached “non-signalling” depletant probe sequesters the target DNA until a threshold level (fixed by the depletant concentration) over which further increase in target concentration results in a steep dose-response curve (Figure 5). To improve the convenience of this approach, the specific amount of depletant was added by non-covalently absorbing it on the electrode surface. As soon as the sample is applied on the electrode surface, the depletant diffuses in solution, which maintains the single-step convenience of the reagentless sensor. With this strategy we have built an array of electrodes, each of which containing various concentration of depletant, and thus various target threshold with pseudo-Hill coefficient values between 3 and 4 and a dynamic range spanning only 2–3-fold of target concentration (Figure 5),
Here we have demonstrated convenient methods to extend and narrow the useful dynamic range of a model electrochemical DNA sensor. We did so by combining DNA probes of different target affinities but with similar specificity on the same electrode.[6] Employing a pair of signaling probes with dissociation constants differing by approximately an order of magnitude we produced a pseudo-log linear response spanning 3 orders of magnitude in target concentration. And, by employing a pair of probes in which the higher affinity probe is non-signaling we have narrowed the useful dynamic range of an E-DNA sensor to only an 8-fold range of target concentrations, significantly improving its sensitivity. Moreover, because the relevant probes are all strongly chemiadsorbed onto their interrogating electrodes, the modified sensors remain reagentless, reusable, highly selective electrochemical devices readily amenable to lab-on-a-chip applications and point-of-care use.[7b] To overcome possible limitations in the application of the strategy employed to narrow the sensor’s dynamic range, we have also demonstrated an alternative “sequestration” approach where the depletant is added in solution. A great advantage of this strategy is that it doesn’t require variants of the receptor with different affinities: the depletant displays a higher affinity than the probe itself since it is free to diffuse in solution.
Our work is not the first to rationally extend the useful dynamic range of an electrochemical biosensor. Our approach, however, appears rather easier to implement than other, previously reported approaches to this end. This includes approaches based on the use of multiple sensors combined with chemometric strategy[15] or on the use of diffusion barrier membranes.[16] In addition, the use of sets of recognition elements differing only in affinity, and not specificity, represents an advantage over other approaches, such as those utilizing combinations of enzymes differing in both affinity and specificity,[17] in that it leads to a fixed specificity profile across the sensor’s entire dynamic range.
In contrast to broadening the useful dynamic range of electrochemical biosensors, a goal that has seen significant prior literature exploration, we are not aware of any prior literature regarding the narrowing of their dynamic range. The steep dose response curves we achieved open the door to a number of sensing applications requiring high sensitivity and a low signal-to-noise ratio at certain specific target concentration. Of note, compared to a sensor that responds gradually to target inputs, an ultrasensitive electrochemical sensor would be far more useful to generate electrochemical logic gates, ideas that have attracted significant recent interest.[18]
The approaches demonstrated here are general, and can be applied to extend or narrow the dynamic range of other electrochemical biosensors provided that the affinities of the biomolecular recognition elements upon which they are based can be appropriately tuned. This is the case of, for example, structure-switching ribozymes and aptamers whose affinity has been rationally modulated through quantitative and predictive model to meet certain performance requirements.[19] Despite being a more challenging task, rational and semi-rational engineering strategies are also available to tune the affinity of proteins or more complex recognition elements.[10] Indeed, several examples have been reported, which suggest that our approach to affinity tuning may be broadly applicable.[10]
The ability to broaden or narrow the dynamic range of biomolecular recognition could also be of utility in biotechnologies beyond biosensing. The fixed dynamic range of single site binding, for example, limits the utility of biomolecular recognition in biofuel cells, for which wider dynamic range equates to better power efficiencies.[3] It also limits the performance of bio-electronic “logic gates” used in biocomputing, as a steeper, nearly all-or-none “digital” response could significantly reduce the noise floor in such systems.[18a, 4]
Experimental Section
Experimental details in supporting information
Supplementary Material
Footnotes
This work was supported by the Italian Ministry of University and Research (MIUR) through the project FIRB “Futuro in Ricerca” and by NIH through grant AI076899.
Supporting information for this article is available on the WWW under http://www.angewandte.org or from the author.
Contributor Information
Di Kang, Department of Chemistry and Biochemistry, Center for Bioengineering, University of California, Santa Barbara, Santa Barbara, CA 93106 (USA).
Dr. Alexis Vallée-Bélisle, Department of Chemistry and Biochemistry, Center for Bioengineering, University of California, Santa Barbara, Santa Barbara, CA 93106 (USA)
Alessandro Porchetta, Dipartimento di Scienze e Tecnologie Chimiche, University of Rome Tor Vergata, Via della Ricerca Scientifica, Rome 00133, (Italy), Consorzio Interuniversitario Biostrutture e Biosistemi “INBB”, Rome 00136, (Italy).
Prof. Dr. Kevin W. Plaxco, Department of Chemistry and Biochemistry, Center for Bioengineering, University of California, Santa Barbara, Santa Barbara, CA 93106 (USA). Interdepartmental Program in Biomolecular Science and Engineering, University of California, Santa Barbara, Santa Barbara, CA 93106 (USA)
Dr. Francesco Ricci, Email: francesco.ricci@uniroma2.it, Department of Chemistry and Biochemistry, Center for Bioengineering, University of California, Santa Barbara, Santa Barbara, CA 93106 (USA). Dipartimento di Scienze e Tecnologie Chimiche, University of Rome Tor Vergata, Via della Ricerca Scientifica, Rome 00133, (Italy), Consorzio Interuniversitario Biostrutture e Biosistemi “INBB”, Rome 00136, (Italy).
References
- 1.a) Wang J. Chem Rev. 2008;108:814–825. doi: 10.1021/cr068123a. [DOI] [PubMed] [Google Scholar]; b) Wang J. Biosens Bioelectron. 2006;21:1887–1892. doi: 10.1016/j.bios.2005.10.027. [DOI] [PubMed] [Google Scholar]
- 2.a) Drummond TG, Hill MG, Barton JK. Nat Biotechnol. 2003;21:1192–1199. doi: 10.1038/nbt873. [DOI] [PubMed] [Google Scholar]; b) Palecek E, Fojta M. Anal Chem. 2001;73:74a–83a. doi: 10.1021/ac0123936. [DOI] [PubMed] [Google Scholar]
- 3.Bullen RA, Arnot TC, Lakeman JB, Walsh FC. Biosens Bioelectron. 2006;21:2015–2045. doi: 10.1016/j.bios.2006.01.030. [DOI] [PubMed] [Google Scholar]
- 4.a) Privman V, Pedrosa V, Melnikov D, Pita M, Simonian A, Katz E. Biosens Bioelectron. 2009;25:695–701. doi: 10.1016/j.bios.2009.08.014. [DOI] [PubMed] [Google Scholar]; b) Wang J, Katz E. Isr J Chem. 2011;51:141–150. [Google Scholar]
- 5.(a) Koshland DE. The molecular basis for enzyme regulation. Vol. 1. Academic Press; New York: 1970. [Google Scholar]; b) Goldbeter A, Koshland DE. Proc Natl Acad Sci USA. 1981;78:6840–6844. doi: 10.1073/pnas.78.11.6840. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Koshland DE, Goldbeter A, Stock JB. Science. 1982;217:220–225. doi: 10.1126/science.7089556. [DOI] [PubMed] [Google Scholar]; d) Ferrell JE. Trends Biochem Sci. 1996;21:460–466. doi: 10.1016/s0968-0004(96)20026-x. [DOI] [PubMed] [Google Scholar]
- 6.Vallee-Belisle A, Ricci F, Plaxco KW. J Am Chem Soc. 2011 doi: 10.1021/ja209850j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.a) Fan CH, Plaxco KW, Heeger AJ. Proc Natl Acad Sci USA. 2003;100:9134–9137. doi: 10.1073/pnas.1633515100. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Ricci F, Plaxco KW. Microchim Acta. 2008;163:149–155. [Google Scholar]
- 8.Lubin AA, Plaxco KW. Accounts Chem Res. 2010;43:496–505. doi: 10.1021/ar900165x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vallee-Belisle A, Ricci F, Plaxco KW. Proc Natl Acad Sci USA. 2009;106:13802–13807. doi: 10.1073/pnas.0904005106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.a) Vallee-Belisle A, Plaxco KW. Curr Opin Struct Biol. 2010;20:518–526. doi: 10.1016/j.sbi.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Golynskiy MV, Koay MS, Vinkenborg JL, Merkx M. ChemBioChem. 2011;12:353–361. doi: 10.1002/cbic.201000642. [DOI] [PubMed] [Google Scholar]; c) Guntas G, Mansell TJ, Kim JR, Ostermeier M. Proc Natl Acad Sci USA. 2005;102:11224–11229. doi: 10.1073/pnas.0502673102. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Stratton MM, Loh SN. Protein Sci. 2011;20:19–29. doi: 10.1002/pro.541. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Strickland D, Yao XL, Gawlak G, Rosen MK, Gardner KH, Sosnick TR. Nat Methods. 2010;7:623–626. doi: 10.1038/nmeth.1473. [DOI] [PMC free article] [PubMed] [Google Scholar]; f) Palmer AE, Giacomello M, Kortemme T, Hires SA, Lev-Ram V, Baker D, Tsien RY. Chem Biol. 2006;13:521–530. doi: 10.1016/j.chembiol.2006.03.007. [DOI] [PubMed] [Google Scholar]; g) Kohn JE, Plaxco KW. Proc Natl Acad Sci USA. 2005;102:10841–10845. doi: 10.1073/pnas.0503055102. [DOI] [PMC free article] [PubMed] [Google Scholar]; h) Marvin JS, Hellinga HW. Nat Struct Biol. 2001;8:795–798. doi: 10.1038/nsb0901-795. [DOI] [PubMed] [Google Scholar]
- 11.a) Buchler NE, Louis M. J Mol Biol. 2008;384:1106–1119. doi: 10.1016/j.jmb.2008.09.079. [DOI] [PubMed] [Google Scholar]; b) Buchler NE, Cross FR. Mol Syst Biol. 2009;5:272. doi: 10.1038/msb.2009.30. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Ricci F, Vallee-Belisle A, Plaxco KW. PLoS Comput Biol. 2011;7(10):e1002171. doi: 10.1371/journal.pcbi.1002171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.a) Xiao Y, Lai RY, Plaxco KW. Nat Protoc. 2007;2:2875–2880. doi: 10.1038/nprot.2007.413. [DOI] [PubMed] [Google Scholar]; b) Rowe AA, White RJ, Bonham AJ, Plaxco KW. J Vis Exp. 2011:e2922. doi: 10.3791/2922. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Kang D, Zuo XL, Yang RQ, Xia F, Plaxco KW, White RJ. Anal Chem. 2009;81:9109–9113. doi: 10.1021/ac901811n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hill AV. J Physiol. 1910;40:iv–vii. doi: 10.1113/jphysiol.1910.sp001366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ricci F, Lai RY, Heeger AJ, Plaxco KW, Sumner JJ. Langmuir. 2007;23:6827. doi: 10.1021/la700328r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chow E, Wong ELS, Pascoe O, Hibbert DB, Gooding JJ. Anal Bioanal Chem. 2007;387:1489–1498. doi: 10.1007/s00216-006-1022-0. [DOI] [PubMed] [Google Scholar]
- 16.Mullen WH, Keedy FH, Churchouse SJ, Vadgama PM. Anal Chim Acta. 1986;183:59–66. [Google Scholar]
- 17.Yamazaki T, Kojima K, Sode K. Anal Chem. 2000;72:4689–4693. doi: 10.1021/ac000151k. [DOI] [PubMed] [Google Scholar]
- 18.a) Stojanovic MN, Stefanovic D. Nat Biotechnol. 2003;21:1069–1074. doi: 10.1038/nbt862. [DOI] [PubMed] [Google Scholar]; b) Saghatelian A, Volcker NH, Guckian KM, Lin VSY, Ghadiri MR. J Am Chem Soc. 2003;125:346–347. doi: 10.1021/ja029009m. [DOI] [PubMed] [Google Scholar]; c) Adleman LM. Science. 1994;266:1021–1024. doi: 10.1126/science.7973651. [DOI] [PubMed] [Google Scholar]; d) Xia F, Zuo XL, Yang RQ, White RJ, Xiao Y, Kang D, Gong XO, Lubin AA, Vallee-Belisle A, Yuen JD, Hsu BYB, Plaxco KW. J Am Chem Soc. 2010;132:8557–8559. doi: 10.1021/ja101379k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.a) Hamaguchi N, Ellington A, Stanton M. Anal Biochem. 2001;294:126–131. doi: 10.1006/abio.2001.5169. [DOI] [PubMed] [Google Scholar]; b) Chen X, Ellington AD. PLoS Comput Biol. 2009;5(12):e1000620. doi: 10.1371/journal.pcbi.1000620. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Beisel CL, Smolke CD. PLoS Comput Biol. 2009;5(4):e1000363. doi: 10.1371/journal.pcbi.1000363. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Lau PS, Coombes BK, Li YF. Angew Chem Int Ed. 2010;49:7938–7942. doi: 10.1002/anie.201002621. [DOI] [PubMed] [Google Scholar]; e) Sefah K, Phillips JA, Xiong XL, Meng L, Van Simaeys D, Chen H, Martin J, Tan WH. Analyst. 2009;134:1765–1775. doi: 10.1039/b905609m. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.