Abstract
Despite increasing understanding of its pathophysiology, the aetiology of systemic mast cell activation disease (MCAD) remains largely unknown. Research has shown that somatic mutations in kinases are necessary for the establishment of a clonal mast cell population, in particular mutations in the tyrosine kinase Kit and in enzymes and receptors with crucial involvement in the regulation of mast cell activity. However, other, as yet undetermined, abnormalities are necessary for the manifestation of clinical disease. The present article reviews molecular genetic research into the identification of disease-associated genes and their mutational alterations. The authors also present novel data on familial systemic MCAD and review the associated literature. Finally, the importance of understanding the molecular basis of inherited mutations in terms of diagnostics and therapy is emphasized.
Keywords: heritability, mast cell activation disease, mast cell activation syndrome, mastocytosis, molecular genetics
Introduction
The term systemic mast cell activation disease (MCAD; synonym mast cell disease) refers to disorders characterized by an enhanced release of mast cell mediators sometimes accompanied by accumulation of such dysfunctional mast cells which may or may not be readily histologically detectable (ref. 1 and further references therein). According to the current classification1–3 three variants of systemic MCAD are distinguished: systemic mastocytosis (SM), mast cell activation syndrome (MCAS) and mast cell leukaemia (MCL). The first of these, SM, is characterized by specific pathological mutations and immunohistochemical findings, which are known as the World Health Organization criteria.4 A diagnosis of MCAS1–3,5,6 is assigned to patients who present with multiple mast cell mediator-induced symptoms but who do not fulfil the requirements for a diagnosis of SM, and in whom relevant differential diagnoses have been excluded. The last, MCL, is an aggressive mast cell neoplasm which is defined by increased numbers of mast cells in bone marrow smears (≥ 20%) and the presence of circulating mast cells (reviewed in ref. 4).
Few data are available concerning the prevalence of systemic MCAD. Both SM and MCL are rare disorders.4,7 For SM, data from the French mastocytosis network Association Française pour les Initiatives de Recherche sur le Mastocyte et Les Mastocytoses,8 the Spanish mastocytosis network Red Española de Mastocitosis,9,10 the Italian Mastocytosis Registry,11 and the German Competence Network on Mastocytosis (own unpublished results), suggest a prevalence of at least one in 364 000 in Europe. However, given that these data represent only a proportion of all cases, the true prevalence will be higher. Data from a clinical population suggest that the prevalence of MCL is two orders of magnitude lower than that of SM.12 In contrast, MCAS seems to be a more common disorder. Evidence has been presented that MCAS may be an underlying cause of various clinical presentations, e.g. in subsets of patients with fibromyalgia13 and irritable bowel syndrome.14 Hence, the prevalence of MCAS is likely to lie within the single-digit percentage range.
The relationship between systemic MCAD and cutaneous mastocytosis (CM, synonyms: paediatric or childhood-onset mastocytosis) remains unclear. Early studies suggested that CM and systemic MCAD were separate disease entities, because the majority of CM patients were found to lack mutations of the tyrosine kinase KIT gene.15,16 However, subsequent studies have demonstrated that the frequency of clonal KIT mutations is similar in patients with CM, SM and MCAS, and that they are present in up to 86% of patients from each diagnostic group.17–20 In addition, several studies have reported the evolution of CM into SM, suggesting that the two disorders are not distinct, but may instead be part of a continuous spectrum of mast cell-related dysfunction.18,21–24 However, unless otherwise stated, the present review is confined to discussion of the systemic MCAD variants in the definitions of the current consensus, i.e. SM, MCAS and MCL.
Genetic alterations in patients with systemic MCAD
The establishment of an aberrant clonal mast cell population is dependent upon mutations in genes that encode proteins with a crucial involvement in the regulation of mast cell activity. Those studied best are the mutations occurring in the tyrosine kinase Kit.18,25–27 In the majority of patients with systemic MCAD, these mutations are limited to derivatives of haematopoietic stem cells.28,29 This indicates that these mutations did not arise during early embryonic development, and were therefore somatic rather than germline. The mutations underlying systemic MCAD drive aberrant mediator production/release with or without readily histologically detectable mast cell accumulation. Mast cell accumulation is due predominantly to a decrease in mast cell apoptosis (refs 30,31 and further references therein). On a limited scale, it is also due to an increase in proliferation.32,33 The mutations underlying MCL appear to drive malignant mast cell proliferation in addition to aberrant mediator production and release.4,34,35
A general view on genetic alterations in tyrosine kinase Kit
The type-III transmembrane receptor tyrosine kinase Kit plays a crucial role in the development of mast cells, as well as of haematopoietic progenitor cells, melanocytes, primordial germ cells and the interstitial cells of Cajal (for review, see ref. 36). In humans, the KIT gene (previously termed c-kit), which encodes the tyrosine kinase Kit, is located in the pericentromeric region of the long arm of chromosome 4 (4q12; http://www.ncbi.nlm.nih.gov/sites/entrez?db=gene&cmd=retrieve&dopt=default&list_uids=3815#ubor2_RefSeq). The human KIT gene spans approximately 89 kb and contains 21 exons, which are transcribed/translated into a protein with a molecular mass of 145 000 and a length of 976 amino acids (http://atlasgeneticsoncology.org/Genes/KITID127.html). Under physiological conditions, binding of the endogenous Kit ligand, i.e. the stem-cell factor, to the extracellular domain of the receptor results in receptor dimerization, activation of the intracellular tyrosine kinase domain through autophosphorylation of specific tyrosine residues, and receptor activation (for review, see ref. 37). In normal mature mast cells, activation of Kit signalling through stem-cell factor leads to increased cell proliferation and survival, changes in mast cell migration and adhesion, mast cell degranulation and mediator release.
Various somatic heterozygous Kit alterations (point mutations and deletions/insertions) have been detected in patients with systemic MCAD (Table 1). In any given patient, one Kit alteration or a combination of two or more Kit alterations can occur. For several of these genetic alterations of KIT (Table 1, bold typed) it has already been demonstrated that they cause changes in the downstream Kit signalling pathways in mast cells. These convert Kit into a constitutively active, dysregulated, stem-cell-factor-independent tyrosine kinase. Independence of stem-cell factor binding may be explained, at least in part, by kinase activity within unusual mutation-induced subcellular locations, such as the Golgi compartment or the endoplasmic reticulum.38,39 Abnormalities in the growth, proliferation and survival of mutated mast cells are mediated by activation of the phosphoinositide 3-kinase/protein kinase B/mammalian Target of Rapamycin pathway. Research suggests that enhancement of mast cell degranulation is mediated by tyrosine phosphorylation of non-T-cell activation linker.40 Within this context, it is interesting to note that within any given patient, mutations that are functionally activating in mast cells may be functionally inactivating in other Kit-expressing cells, e.g. in the Cajal cells of the gastrointestinal tract.41 This may be a result of differences in the signalling cascades. This could explain why a minority of patients with systemic MCAD present with constipation rather than diarrhoea.1
Table 1.
Distribution of alterations in the amino acids sequence of the tyrosine kinase Kit (deduced from the nucleotide sequence of the 21 exons of the gene KIT) in mast cells from patients with systemic mast cell activation disease (mast cell activation syndrome and systemic mastocytosis)
| Signal transduction exons 12–21, aa 592–976 | |||||||
|---|---|---|---|---|---|---|---|
| Ligand(SCF)-binding domain exons 1–5 aa 1–308 | Dimerization domain exons 6–7 aa 309–410 | Proteolytic cleavage site exons 8–9 aa 411–513 | Membrane-spanning region exon 10 aa 514–549 | Auto-inhibition exon 11 aa 550–591 | Kinase domain1 Kinase insert sequence, | Kinase domain 2 | C-terminus |
| W8R1 | D327N2 | D419H/G2 | Del 510–5131,2,5 | E554K2 | Del 592–6261 | F782S1 | Frequently splicing errors1,2 |
| C12S1 | E338K1,,2 | 419InsFF3 | Del 5211 | V559I/A/G6,10 | G658E2 | N787D1 | |
| L18P2 | Q346L1 | Del 4193 | F522C6 | V560G2,6,11 | Y672S2 | H790R1 | |
| P31T2 | M351E/I1,2 | Del 417– 419InsY3 | V530I7 | D572A3 | 683 Ins R2 | H802Y2 | |
| 41 Stop codon1 | F355L1 | A533D6,8 | S688L2 | 814 Stop codon2 | |||
| E53K1,2 | E359V1 | C443Y3 | M541L1,2,3,8,9 | S709A1 | A814V/T13 | ||
| Del59–4371 | 359 Stop codon1 | S464L2 | Del 7151,2,12 | R815K6 | |||
| E73R1 | 475 Stop codon1,2 | E720K2 | Ins V815-I8166 | ||||
| T74R1 | M724I2 | D816V/Y/F/H/ | |||||
| K116N2 | S476I3 | A736V1 | I3,6,11 | ||||
| V214L2 | K484R2 | D751Y1 | I817V6 | ||||
| 252Ins Q1,2 | ITD501–5023 | D760V2 | D820G14 | ||||
| K259E1 | ITD502–5033 | E761K2 | S821F1 | ||||
| H265Q1 | ITD505–5083 | 764 Stop codon2 | N822I/K15,16 | ||||
| E270K1,2 | K509I3,4 | A829T1 | |||||
| L276S1 | 830 Stop codon2 | ||||||
| G286R2 | A837V1 | ||||||
| E839K6 | |||||||
| L862V1,2 | |||||||
Ref. 26;
Ref. 25;
Ref. 18;
Ref. 68;
Ref. 43;
Ref. 27;
Ref. 74;
Ref. 17;
Ref. 19;
Ref. 64;
Ref. 75;
Ref. 45;
Ref. 76;
Ref. 77;
Ref. 20;
Ref. 78.
Three isoforms resulting from alternative splicing are indicated by italic type. Mutations which have been demonstrated to induce an increased mast cell activity are indicated by bold type. aa, amino acid; SCF, stem cell factor.
The described Kit isoforms
Six Kit isoforms exist as a result of the presence of three alternative splicing sites. Each isoform is of differing pathophysiological relevance. An additional glutamine residue at amino acid position 252 is present because of the insertion of an alternative splicing consensus sequence at the 3′ end (− 5 nucleotides) of intron 4, at the beginning of the coding sequence of exon 5.25 The two isoforms, i.e. 252Q(+) and 252Q(−), are present in similar numbers in patients with MCAD and healthy volunteers (Figure 1),25,26 which suggests that they have no crucial involvement in the induction of an unregulated increase in mast cell activity.
Figure 1.
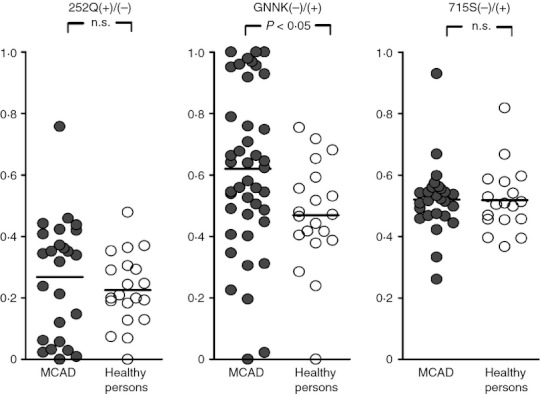
Distribution of the following ratios: (i) of Q252(+)/Q252(−) expression intensity (i.e. the amount of PCR amplification product with and without the additional glutamine residue [Q]); (ii) of the GNNK(–) isoform compared to the GNNK(+) isoform [i.e. the amount of PCR amplification product without and with the tetrapeptide sequence glycine-asparagine-asparagine-lysine (GNNK)]; and (iii) of S715(−)/S715(+) expression intensity (i.e. the amount of PCR amplification product with and without the serine residue [S]). Comparison of the distribution of the ratios between patients with systemic mast cell activation disease (MCAD, filled symbols) and healthy probands (healthy persons, open symbols). Ordinate: Ratio of the expression intensities. The black bars represent the respective mean. P < 0·05, Fisher′s exact test; n.s., not significant.
Two further isoforms result from alternative 5' splice donor sites,42 and these are characterized by the presence or absence of the tetrapeptide sequence glycine-asparagine-asparagine-lysine (GNNK) in the extracellular part of the juxtamembrane region. In vitro, these isoforms differ substantially in terms of their functional activities, with the GNNK(−) isoform showing tumorigenic potency.43,44 Hence, a predominance of the GNNK(−) isoform may contribute to or reflect a pathological increase in the activation of affected mast cells. Studies comparing MCAD patients with healthy controls have reported significantly stronger expression of the GNNK(−) isoform compared with the GNNK(+) isoform in mast cells from patients with MCAD (Figure 1).26 An expression intensity ratio for GNNK(−)/(+) of ≥ 80% was associated almost exclusively with high symptom intensity. Although the reason for the predominant expression of the GNNK(−) isoform in these patients is unknown, its presence appears to support a diagnosis of MCAD. However, the absence of this predominant expression does not exclude the diagnosis.
The third alternative splicing site in KIT results in the presence or absence of a serine residue at position 715 in the kinase insert region of human Kit. This results from an alternative 3' splice acceptor site usage.45 The two isoforms S715(−) and S715(+) are co-expressed in similar quantities in patients with MCAD and in healthy controls (Figure 1).25,26 This suggests that these isoforms have no substantial involvement in the induction of a pathological increase in mast cell activity.
The mutation KitD816V
In the mast cells of the majority (> 90%) of patients with SM, mutations in the activation loop of Kit are present (most frequently D816V; reviewed in ref. 27). According to recent studies, the pathological immunohistochemical alterations that constitute the World Health Organization criteria (formation of mast cell clusters; spindle-shaped morphology of mast cells; expression of CD25 in mast cells; marked increase in blood tryptase concentration) seem to be both causally related and specific to the occurrence of a mutation at amino acid position 816 of tyrosine kinase Kit in the affected mast cells.4,46,47 During SM progression, the Kit mutant D816V may disappear (ref. 48 and own unpublished observation).
The D816V Kit mutation is present to a varying degree in patients with myeloid and lymphoproliferative mastocytosis-associated clonal haematological non-mast cell lineage diseases.49,50 The occurrence of this mutation in these diseases points to a common bi-committed neoplastic progenitor clone. In several studies, the KIT mutation at codon 816 in mast cells was also detected on CD34+ haematopoietic cells, eosinophils, monocytic neutrophil-lineage bone marrow precursor cells, and lymphocytes in around 30% of patients with SM.28,51
Genetic polymorphisms associated with Kit mutations
KitD816V-positive SM has been associated with a polymorphism in the IL13 promoter gene (Table 2).52 This polymorphism leads to a high transcription rate. Hence, the known proliferation-promoting influence of interleukin-13 (IL-13) on mast cells53 should be enhanced. In accordance with this hypothesis, this polymorphism has been correlated with an elevated serum tryptase level, which is a marker of increased mast cell activation in SM.4
Table 2.
Genes and genetic alterations related to systemic mast cell activation disease
| Gene | Function | Genetic alteration | Cells investigated | Summary of findings | Reference |
|---|---|---|---|---|---|
| KIT | Tyrosine kinase Kit | Multiple alterations | Mast cells and multiple other haematopoetic lineages | Clearly associated with pathological mast cell activation | Present paper |
| JAK2 | Janus kinase 2 | V617F | BMMC | Associated with pathological mast cell activation | 79,80 |
| PDGFRα | Tyrosine kinase platelet-derived growth factor receptor α | Fusion of FIP1L1 gene to PDGFRα gene in exon 12 | PBMC; BMMC; multiple other haematopoetic lineages including mast cells | Associated with pathological mast cell activation | 81–84 |
| NRAS | Protein with intrinsic GTPase activity | G12D | BMMC | Aggressive systemic mastocytosis | 85 |
| G13D | |||||
| RASGRP4 | Ras guanyl nucleotide-releasing protein | Failure to remove intron 5 | Bone marrow mast cells | Systemic mastocytosis | 35 |
| CBL | E3-ligase | Multiple alterations | Retroviral expression of Cbl mutants in transplanted bone marrow in mice | Cytokine-independent mast cell activation | 86 |
| HRH4 | Histamine H4 receptor | Truncated splice variants | Cord blood-derived mast cells; eosinophils; basophils; dendritic cells | Inhibition of full-length histamine-H4-receptor function | 87 |
| IL13 | Interleukin 13 | −1112C/T | PBMC | Association with KitD816V-positive systemic mastocytosis | 52 |
| IL4 | Interleukin 4 | Q576R | PBMC | Association with KitD816V-positive systemic mastocytosis | 54 |
| TET2 | Candidate tumour suppressor gene | Multiple | BMMC; cord blood-derived mast cells and eosinophils | Association with KitD816V-positive systemic mastocytosis | 88,89 |
| TNF | Tumour necrosis factor α | 238G/A | No information provided | Association with KitD816V-positive systemic mastocytosis | 90 |
| VEGFA | Vascular endothelial growth factor | −1154A/G | No information provided | Association with KitD816V-positive systemic mastocytosis | 91 |
BMMC, bone marrow mononuclear cells; PBMC, peripheral blood mononuclear cells/leucocytes.
Daley et al.54 reported that the polymorphism Q576R in the cytoplasmic domain of the α-subunit of the IL-4 receptor (Table 2) was found more frequently in SM patients whose disease was limited to skin, and who exhibited lower levels of surrogate disease markers. These data suggest that this polymorphism may mitigate disease expression, which may be a result of an increase in the efficacy of IL-4 at the mutated receptor. In accordance with this hypothesis, the addition of IL-4 to mast cell cultures has been shown to induce apoptosis.55
Finally, associations between SM and polymorphisms in TET2 (a candidate tumour-suppressor gene), the TNFα gene, and the VEGFA promoter gene have been reported. No association with SM was found for IL8 (Table 2).
However, none of the findings from the above mentioned candidate gene studies have been consistently replicated in independent studies, and so these findings cannot be regarded as proven.
Genome-wide gene expression profiling of human mast cells carrying KitD816V
D'Ambrosio et al.56 obtained gene expression profiles from bone marrow mononuclear cells of eight patients with SM and the KitD816V mutation, and compared them with those of five healthy controls. Analysis of global median differential expression defined 130 genes with a known functional product that were differentially and significantly expressed in SM patients compared with controls. This suggests that SM is accompanied by an altered gene expression profile in the bone marrow. The most prominent of these differentially expressed genes was tryptase, which showed a 44·6-fold increase compared with controls. Genes involved in cell proliferation, neoplastic transformation, and apoptosis were also up-regulated. However, these findings require confirmation in experiments on purified mast cells from patients with SM and MCAS.
Cytogenetic findings associated with the KitD816V mutation
In a third of SM patients, genetic instability of the haematopoietic cells is reflected in chromosome abnormalities, as detected during cytogenetic analysis of bone marrow cells.57–59 These abnormalities include deletions of chromosomes 5, 7, 11 and 20. No correlation between clinical features and cytogenetic findings has been reported.
Genetic alterations beyond Kit
Research has demonstrated induction of SM and MCAS secondary to mutations in the genes encoding the kinases JAK2 and PDGFRα; the Src-kinases; and proteins of cellular signal transduction (RASGRP4, CBL-encoded E3 ligase, histamine H4 receptor; Table 2, references therein).
Heritability of systemic MCAD
Although numerous reports of familial occurrence and concordant twin pairs have strongly suggested a genetic basis for CM,16,17,60–67 few data are available concerning the genetic contribution to systemic MCAD. To date, systemic MCAD has been assumed to be largely sporadic in nature15, and only three familial cases of systemic MCAD (two SM and one MCAS) have been reported. In one SM family, the functionally activating point mutation KitK509I was identified in both affected patients (mother and daughter).68 In a second SM family, a functionally activating mutation in Kit exon 8, which resulted in the deletion of codon 419, was detected in the three affected patients (mother, daughter, granddaughter) but not in the healthy sister.69 One MCAS family has also been reported in which the father and his two children (daughter and son) presented with varying clinical features.70 However, the underlying genetic alterations were not investigated.
Familial clustering of systemic MCAD
Demonstration of familial clustering would be an important step towards defining the genetic contribution to the risk of systemic MCAD. However, at the time of writing, no systematic investigation of the heritability of systemic MCAD has been reported. Therefore, the present authors analysed data from 120 Caucasian index patients (Figure 2) with MCAS (n = 102) or SM (n = 18), who presented to our research group between May 2005 and November 2010, to determine familial aggregation. A sample of 258 probands was used as a control group representative of the German population. These individuals had been randomly recruited from the German inhabitants of the city of Bonn. Analysis of our data revealed that irrespective of systemic MCAD variant and gender, around 75% of our index patients had at least one first-degree relative with systemic MCAD (Figure 2). Within any given family, the disease variant, the severity of the mediator-related symptoms, and the age at clinical onset differed between individuals. Similar findings were reported in abstracts by Burks et al. 70 and Bursztein et al. 2009.70,71 The prevalence of systemic MCAD among the first-degree relatives in our sample was 33% (Figure 2), which differed significantly (P < 0·0001; Fisher′s exact test) from the prevalence in the control group (around 14%; Figure 2). The age distribution was similar in both groups. In both patients and controls, a female preponderance was observed (Figure 2). Our data suggest that systemic MCAD pedigrees include more systemic MCAD cases than would be expected by chance. This finding is consistent with the hypothesis that systemic MCAD has a heritable component, at least in a considerable number of patients.
Figure 2.
Characteristics of the presently investigated population in terms of the familial aggregation of systemic mast cell activation disease (MCAD). In the index patients, the diagnosis was assigned according to the criteria for MCAD.1 Due to the anonymous nature of the survey of the control population, clinical diagnoses were assigned in the control group on the basis of a self-report checklist and the exclusion of relevant differential diagnoses (for the checklist, see ref. 92). Asterisks: number of affected persons/total number of persons in the respective group (percentage). A sample of 258 probands randomly recruited from the German inhabitants of the city of Bonn was used as a control group representative of the German population.
At the time of writing, the molecular processes that result in a familial aggregation of MCAD remain speculative. The detection of differing systemic MCAD-associated somatic mutations within given families (refs 18,72; own unpublished data) is compatible with the hypothesis that disease arises secondary to a dysfunction of mutated, and as yet unidentified, operator or regulator genes. This dysfunction could result in mutations in KIT and other mast-cell-regulatory genes. Within this context, it is interesting to note that most of the insertions and deletions in KIT in patients with MCAS involve intron–exon junctions, suggesting that distortion of alternative splicing may contribute to the generation of these novel transcripts.26
Conclusions and perspective
Despite increasing understanding of its pathophysiology, the aetiology of MCAD remains largely unknown. The establishment of an aberrant clonal mast cell population is dependent upon mutations in kinases (particularly in the tyrosine kinase Kit) and in enzymes and receptors crucially involved in the regulation of mast cell activity. Other genetic alterations, some of which are as yet undetermined, appear to be related to systemic MCAD phenotype and prognosis.
Recent findings suggest that the various systemic MCAD variants and clinical phenotypes do not represent distinct disease entities, but rather varying presentations of a common generic root process of mast cell dysfunction.2,7,25,26 Interestingly, in contrast to adult-onset systemic MCAD, more than 50% of paediatric cases of cutaneous mastocytosis appear to enter long-term remission spontaneously,23,65 though whether such remissions are permanent or relapse in adulthood as systemic MCAD is unknown. The reason for this difference in prognosis – if both are clonal diseases with common roots – awaits clarification. Improved understanding of the biochemical processes leading to this resolution may open new avenues in the treatment of adult systemic MCAD.
To optimize research into the genetic basis of MCAD, a number of approaches may be suitable. First, next-generation sequencing could facilitate identification of the putative mutated disease-related operator and regulator genes in familial cases. Second, studies of animal models displaying disturbances secondary to primary mast cell dysfunction will implicate further genes of relevance to systemic MCAD. In dogs, mast cell tumours are the second most common form of neoplastic disease (7–21% of all cutaneous neoplasms). In up to 40% of all canine mast cell tumours, KIT mutations are present. Well-documented breed predispositions indicate an underlying heritable component (for review, see ref. 73). Third, genome-wide association studies could allow the unbiased and systematic identification of systemic MCAD risk genes. Since their introduction, genome-wide association studies have identified risk genes for several multifactorial diseases (http://www.genome.gov/gwastudies/). However, these studies showed that most risk genes have only moderate effect sizes, and that analysis of large samples is necessary. Given the high prevalence of the MCAS variant, the recruitment of such samples should be possible at least for MCAS.
Taken together, improved knowledge of the genetic aetiology of MCAD may facilitate the development of new therapeutic strategies, which possibly may also imply curative in addition to symptomatic treatment options.
Acknowledgments
We thank Dr Christine Schmäl for her revision of the manuscript. This study was supported by grants from the B. Braun-Stiftung, Germany and the Förderclub Mastzellforschung e.V., Germany.
Glossary
- CM
cutaneous mastocytosis
- GNNK
glycine-asparagine-asparagine-lysine
- IL-4
interleukin 4
- MCAD
mast cell activation disease
- MCAS
mast cell activation syndrome
- MCL
mast cell leukaemia
- SM
systemic mastocytosis
Disclosures
The authors declare that they have no competing interests.
Authors' contribution
All authors contributed equally to conception, design and drafting of the manuscript. All authors read and approved the final manuscript.
References
- 1.Molderings GJ, Brettner S, Homann J, Afrin LB. Mast cell activation disease: a concise practical guide for diagnostic workup and therapeutic options. J Hematol Oncol. 2011;4:10. doi: 10.1186/1756-8722-4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akin C, Valent P, Metcalfe DD. Mast cell activation syndrome: proposed diagnostic criteria. J Allergy Clin Immunol. 2010;126:1099–104. doi: 10.1016/j.jaci.2010.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Valent P, Akin C, Arock M, et al. Definitions, Criteria and Global Classification of Mast Cell Disorders with Special Reference to Mast Cell Activation Syndromes: a Consensus Proposal. Int Arch Allergy Immunol. 2012;157:215–25. doi: 10.1159/000328760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Valent P, Akin C, Escribano L, et al. Standards and standardization in mastocytosis: consensus statements on diagnostics, treatment recommendations and response criteria. Eur J Clin Invest. 2007;37:435–53. doi: 10.1111/j.1365-2362.2007.01807.x. [DOI] [PubMed] [Google Scholar]
- 5.Alvarez-Twose I, González de Olano D, Sánchez-Muñoz L, et al. Clinical, biological, and molecular characteristics of clonal mast cell disorders presenting with systemic mast cell activation symptoms. J Allergy Clin Immunol. 2010;125:1269–78. doi: 10.1016/j.jaci.2010.02.019. [DOI] [PubMed] [Google Scholar]
- 6.Hamilton MJ, Hornick JL, Akin C, Castells MC, Greenberger NJ. Mast cell activation syndrome: a newly recognized disorder with systemic clinical manifestations. J Allergy Clin Immunol. 2011;128:147–52. doi: 10.1016/j.jaci.2011.04.037. [DOI] [PubMed] [Google Scholar]
- 7.Hermine O, Lortholary O, Leventhal PS, et al. Case–control cohort study of patients' perceptions of disability in mastocytosis. PLoS One. 2008;3:e2266. doi: 10.1371/journal.pone.0002266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lanternier F, Cohen-Akenine A, Palmerini F, et al. Phenotypic and genotypic characteristics of mastocytosis according to the age of onset. PLoS One. 2008;3:e1906. doi: 10.1371/journal.pone.0001906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Escribano L, Alvarez-Twose I, Sánchez-Muñoz L, et al. Prognosis in adult indolent systemic mastocytosis: a long-term study of the Spanish Network on Mastocytosis in a series of 145 patients. J Allergy Clin Immunol. 2009;124:514–21. doi: 10.1016/j.jaci.2009.05.003. [DOI] [PubMed] [Google Scholar]
- 10.Alvarez-Twose I, González-de-Olano D, Sánchez-Muñoz L, et al. Validation of the REMA Score for Predicting Mast Cell Clonality and Systemic Mastocytosis in Patients with Systemic Mast Cell Activation Symptoms. Int Arch Allergy Immunol. 2011;157:275–80. doi: 10.1159/000329856. [DOI] [PubMed] [Google Scholar]
- 11.Merante S, Magliacane D, Neri I, et al. The New Italian Mastocytosis Registry. 53rd ASH Annual Meeting and Exposition 2010; Poster 3805, [ http://ash.confex.com/ash/2010/webprogram/Paper28567.html]
- 12.Lim KH, Tefferi A, Lasho TL, et al. Systemic mastocytosis in 342 consecutive adults: survival studies and prognostic factors. Blood. 2009;113:5727–36. doi: 10.1182/blood-2009-02-205237. [DOI] [PubMed] [Google Scholar]
- 13.Lucas HJ, Brauch CM, Settas L, Theoharides TC. Fibromyalgia – new concepts of pathogenesis and treatment. Int J Immunopathol Pharmacol. 2006;19:5–9. [PubMed] [Google Scholar]
- 14.Frieling T, Meis K, Kolck UW, et al. Evidence for mast cell activation in patients with therapy-resistant irritable bowel syndrome. Z Gastroenterol. 2011;49:191–4. doi: 10.1055/s-0029-1245707. [DOI] [PubMed] [Google Scholar]
- 15.Longley BJ, Metcalfe DD, Tharp M, Wang X, Tyrrell L, Lu SZ, Heitjan D, Ma Y. Activating and dominant inactivating c-kit catalytic domain mutations in distinct clinical forms of human mastocytosis. Proc Natl Acad Sci U S A. 1999;96:1609–14. doi: 10.1073/pnas.96.4.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rosbotham JL, Malik NM, Syrris P, et al. Lack of c-kit mutation in familial urticaria pigmentosa. Br J Dermatol. 1999;140:849–52. doi: 10.1046/j.1365-2133.1999.02814.x. [DOI] [PubMed] [Google Scholar]
- 17.Tang X, Boxer M, Drummond A, Ogston P, Hodgins M, Burden AD. A germline mutation in KIT in familial diffuse cutaneous mastocytosis. J Med Genet. 2004;41:e88. doi: 10.1136/jmg.2003.015156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bodemer C, Hermine O, Palmérini F, et al. Pediatric mastocytosis is a clonal disease associated with D(816)V and other activating c-KIT mutations. J Invest Dermatol. 2010;130:804–15. doi: 10.1038/jid.2009.281. [DOI] [PubMed] [Google Scholar]
- 19.Foster R, Byrnes E, Meldrum C, et al. Association of paediatric mastocytosis with a polymorphism resulting in an amino acid substitution (M541L) in the transmembrane domain of c-KIT. Br J Dermatol. 2008;159:1160–9. doi: 10.1111/j.1365-2133.2008.08827.x. [DOI] [PubMed] [Google Scholar]
- 20.Wasag B, Niedoszytko M, Piskorz A, et al. Novel activating KIT-N822I mutation in familial cutaneous mastocytosis. Exp Hematol. 2011;39:859–65. doi: 10.1016/j.exphem.2011.05.009. [DOI] [PubMed] [Google Scholar]
- 21.Noack F, Escribano L, Sotlar K, Nunez R, Schuetze K, Valent P, Horny HP. Evolution of urticaria pigmentosa into indolent systemic mastocytosis: abnormal immunophenotype of mast cells without evidence of c-Kit mutation ASP-816-VAL. Leuk Lymphoma. 2003;44:313–9. doi: 10.1080/1042819021000037967. [DOI] [PubMed] [Google Scholar]
- 22.Vogel NM, Lichtin AE, Hsieh FH. Longitudinal disease progression in mastocytosis syndromes: a retrospective chart review. J Allergy Clin Immunol. 2006;117(Suppl):S125. [Google Scholar]
- 23.Uzzaman A, Maric I, Noel P, Kettelhut BV, Metcalfe DD, Carter MC. Pediatric-onset mastocytosis: a long term clinical follow-up and correlation with bone marrow histopathology. Pediatr Blood Cancer. 2009;53:629–34. doi: 10.1002/pbc.22125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bhamidimarri K, Sidhu L, Xie J, Pulakhandam U, Gumstae V. Gastrointestinal manifestations in cutaneous mastocytosis evolving into systemic mastocytosis. Am J Gastroenterol. 2010;105(Suppl. 1):S348. [Google Scholar]
- 25.Molderings GJ, Kolck UW, Scheurlen C, Brüss M, Homann J, von Kügelgen I. Multiple novel alterations in Kit tyrosine kinase in patients with gastrointestinally pronounced systemic mast cell activation disorder. Scand J Gastroenterol. 2007;42:1045–53. doi: 10.1080/00365520701245744. [DOI] [PubMed] [Google Scholar]
- 26.Molderings GJ, Meis K, Kolck UW, Homann J, Frieling T. Comparative analysis of mutation of tyrosine kinase kit in mast cells from patients with systemic mast cell activation syndrome and healthy subjects. Immunogenetics. 2010;62:721–7. doi: 10.1007/s00251-010-0474-8. [DOI] [PubMed] [Google Scholar]
- 27.Orfao A, Garcia-Montero AC, Sanchez L, Escribano L REMA. Recent advances in the understanding of mastocytosis: the role of KIT mutations. Br J Haematol. 2007;138:12–30. doi: 10.1111/j.1365-2141.2007.06619.x. [DOI] [PubMed] [Google Scholar]
- 28.Garcia-Montero AC, Jara-Acevedo M, Teodosio C, et al. KIT mutation in mast cells and other bone marrow hematopoietic cell lineages in systemic mast cell disorders: a prospective study of the Spanish Network on Mastocytosis (REMA) in a series of 113 patients. Blood. 2006;108:2366–72. doi: 10.1182/blood-2006-04-015545. [DOI] [PubMed] [Google Scholar]
- 29.Kristensen T, Broesby-Olsen S, Vestergaard H, Bindslev-Jensen C, Møller MB on behalf of the Mastocytosis Centre Odense University Hospital (MastOUH) Circulating KIT D816V mutation-positive non-mast cells in peripheral blood are characteristic of indolent systemic mastocytosis. Eur J Haematol. 2012;89:42–6. doi: 10.1111/j.1600-0609.2012.01789.x. [DOI] [PubMed] [Google Scholar]
- 30.Kohno M, Yamasaki S, Tybolewicz VLJ, Saito T. Rapid and large amount of autocrine IL-3 production is responsible for mast cell survival by IgE in the absence of antigen. Blood. 2005;105:2059–65. doi: 10.1182/blood-2004-07-2639. [DOI] [PubMed] [Google Scholar]
- 31.Aichberger KJ, Gleixner KV, Mirkina I, et al. Identification of proapoptotic Bim as a tumor suppressor in neoplastic mast cells: role of KIT D816V and effects of various targeted drugs. Blood. 2009;114:5342–51. doi: 10.1182/blood-2008-08-175190. [DOI] [PubMed] [Google Scholar]
- 32.Rottem M, Okada T, Goff JP, Metcalfe DD. Mast cells cultured from the peripheral blood of normal donors and patients with mastocytosis originate from a CD34+/FcεRI-cell population. Blood. 1994;84:2489–96. [PubMed] [Google Scholar]
- 33.Jordan JH, Jäger E, Sperr WR, et al. Numbers of colony-forming progenitors in patients with systemic mastocytosis: potential diagnostic implications and comparison with myeloproliferative disorders. Eur J Clin Invest. 2003;33:611–8. doi: 10.1046/j.1365-2362.2003.01172.x. [DOI] [PubMed] [Google Scholar]
- 34.Ning ZQ, Li J, Arceci RJ. Signal transducer and activator of transcription 3 activation is required for Asp816 mutant c-kit-mediated cytokine-independent survival and proliferation in human leukemia cells. Blood. 2001;97:3559–67. doi: 10.1182/blood.v97.11.3559. [DOI] [PubMed] [Google Scholar]
- 35.Yang Y, Li L, Wong GW, Krilis SA, Madhusudhan MS, Sali A, Stevens RL. RasGRP4, a new mast cell-restricted Ras guanine nucleotide-releasing protein with calcium- and diacylglycerol-binding motifs. Identification of defective variants of this signaling protein in asthma, mastocytosis, and mast cell leukemia patients and demonstration of the importance of RasGRP4 in mast cell development and function. J Biol Chem. 2002;277:25756–74. doi: 10.1074/jbc.M202575200. [DOI] [PubMed] [Google Scholar]
- 36.Roskoski R. Structure and regulation of Kit protein-tyrosine kinase – the stem cell factor receptor. Bichem Biophys Res Commun. 2005;338:1307–15. doi: 10.1016/j.bbrc.2005.09.150. [DOI] [PubMed] [Google Scholar]
- 37.Roskoski R. Signaling by Kit protein-tyrosine kinase – the stem cell factor receptor. Biochem Biophys Res Commun. 2005;337:1–13. doi: 10.1016/j.bbrc.2005.08.055. [DOI] [PubMed] [Google Scholar]
- 38.Bougherara H, Subra F, Crépin R, Tauc P, Auclair C, Poul MA. The aberrant localization of oncogenic kit tyrosine kinase receptor mutants is reversed on specific inhibitory treatment. Mol Cancer Res. 2009;7:1525–33. doi: 10.1158/1541-7786.MCR-09-0138. [DOI] [PubMed] [Google Scholar]
- 39.Toffalini F, Demoulin JB. New insights into the mechanisms of hematopoietic cell transformation by activated receptor tyrosine kinases. Blood. 2010;116:2429–37. doi: 10.1182/blood-2010-04-279752. [DOI] [PubMed] [Google Scholar]
- 40.Gilfillan AM, Rivera J. The tyrosine kinase network regulating mast cell activation. Immunol Rev. 2009;228:149–69. doi: 10.1111/j.1600-065X.2008.00742.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Breuer C, Oh J, Molderings GJ, Schemann M, Kuch B, Mayatepek E, Adam R. Therapy-refractory gastrointestinal motility disorder in a child with c-kit mutations. World J Gastroenterol. 2010;16:4363–6. doi: 10.3748/wjg.v16.i34.4363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hayashi S, Kunisada T, Ogawa M, Yamaguchi K, Nishikawa S. Exon skipping by mutation of an authentic splice site of c-kit gene in W/W mouse. Nucleic Acids Res. 1991;19:1267–71. doi: 10.1093/nar/19.6.1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Caruana G, Cambareri AC, Ashman LK. Isoforms of c-KIT differ in activation of signalling pathways and transformation of NIH3T3 fibroblasts. Oncogen. 1999;18:5573–81. doi: 10.1038/sj.onc.1202939. [DOI] [PubMed] [Google Scholar]
- 44.Voytyuk O, Lennartsson J, Mogi A, Caruana G, Courtneidge S, Ashman LK, Rönnstrand L. Src family kinases are involved in the differential signaling from two splice forms of c-Kit. J Biol Chem. 2003;278:9159–66. doi: 10.1074/jbc.M211726200. [DOI] [PubMed] [Google Scholar]
- 45.Crosier PS, Ricciardi ST, Hall LR, Vitas MR, Clark SC, Crosier KE. Expression of isoforms of the human receptor tyrosine kinase c-kit in leukemic cell lines and acute myeloid leukemia. Blood. 1993;82:1151–8. [PubMed] [Google Scholar]
- 46.Mayerhofer M, Gleixner KV, Hoelbl A, et al. Unique effects of KIT D816V in BaF3 cells: induction of cluster formation, histamine synthesis, and early mast cell differentiation antigens. J Immunol. 2008;180:5466–76. doi: 10.4049/jimmunol.180.8.5466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang Y, Létard S, Borge L. Pediatric mastocytosis-associated KIT extracellular domain mutations exhibit different functional and signaling properties compared with KIT-phosphotransferase domain mutations. Blood. 2010;116:1114–23. doi: 10.1182/blood-2009-06-226027. [DOI] [PubMed] [Google Scholar]
- 48.Gotlib J, Berubé C, Growney JD, et al. Activity of the tyrosine kinase inhibitor PKC412 in a patient with mast cell leukemia with the D816V KIT mutation. Blood. 2005;106:2865–70. doi: 10.1182/blood-2005-04-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fritsche-Polanz R, Fritz M, Huber A, Sotlar K, Sperr WR, Mannhalter C, Födinger M, Valent P. High frequency of concomitant mastocytosis in patients with acute myeloid leukemia exhibiting the transforming KIT mutation D816V. Mol Oncol. 2010;4:335–46. doi: 10.1016/j.molonc.2010.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sotlar K, Colak S, Bache A, Berezowska S, Krokowski M, Bültmann B, Valent P, Horny HP. Variable presence of KIT(D816V) in clonal haematological non-mast cell lineage diseases associated with systemic mastocytosis (SM-AHNMD) J Pathol. 2010;220:586–95. doi: 10.1002/path.2677. [DOI] [PubMed] [Google Scholar]
- 51.Akin C, Kirshenbaum AS, Semere T, Worobec AS, Scott LM, Metcalfe DD. Analysis of the surface expression of c- kit Asp816Val activating mutation in T cells, B cells, and myelomonocytic cells in patients with mastocytosis. Exp Hematol. 2000;28:140–7. doi: 10.1016/s0301-472x(99)00145-9. [DOI] [PubMed] [Google Scholar]
- 52.Nedoszytko B, Niedoszytko M, Lange M, et al. Interleukin-13 promoter gene polymorphism –1112C/T is associated with the systemic form of mastocytosis. Allergy. 2009;64:287–94. doi: 10.1111/j.1398-9995.2008.01827.x. [DOI] [PubMed] [Google Scholar]
- 53.Kaur D, Hollins F, Woodman L, Yang W, Monk P, May R, Bradding P, Brightling CE. Mast cells express IL-13Rα1: IL-13 promotes human lung mast cell proliferation and FcεRI expression. Allergy. 2006;61:1047–53. doi: 10.1111/j.1398-9995.2006.01139.x. [DOI] [PubMed] [Google Scholar]
- 54.Daley T, Metcalfe DD, Akin C. Association of the Q576R polymorphism in the interleukin-4 receptor α chain with indolent mastocytosis limited to the skin. Blood. 2001;98:880–2. doi: 10.1182/blood.v98.3.880. [DOI] [PubMed] [Google Scholar]
- 55.Bailey DP, Kashyap M, Mirmonsef P, Bouton LA, Domen J, Zhu J, Dessypris EN, Ryan JJ. Interleukin-4 elicits apoptosis of developing mast cells via a Stat6-dependent mitochondrial pathway. Exp Hematol. 2004;32:52–9. doi: 10.1016/j.exphem.2003.10.011. [DOI] [PubMed] [Google Scholar]
- 56.D'Ambrosio C, Akin C, Wu Y, Magnusson MK, Metcalfe DD. Gene expression analysis in mastocytosis reveals a highly consistent profile with candidate molecular markers. J Allergy Clin Immunol. 2003;112:1162–70. doi: 10.1016/j.jaci.2003.07.008. [DOI] [PubMed] [Google Scholar]
- 57.Swolin B, Rödjer S, Roupe G. Cytogenetic Studies in Patients with Mastocytosis. Cancer Genet Cytogenet. 2000;120:131–5. doi: 10.1016/s0165-4608(99)00256-3. [DOI] [PubMed] [Google Scholar]
- 58.Swolin B, Rödjer S, Ogärd I, Roupe G. Trisomies 8 and 9 not detected with fish in patients with mastocytosis. Am J Hematol. 2002;70:324–5. doi: 10.1002/ajh.10147. [DOI] [PubMed] [Google Scholar]
- 59.Jost E, Michaux L, van den Abeele M, et al. Complex karyotype and absence of mutation in the c-Kit receptor in aggressive mastocytosis presenting with pelvic osteolysis, eosinophilia and brain damage. Ann Hematol. 2001;80:302–7. doi: 10.1007/s002770000271. [DOI] [PubMed] [Google Scholar]
- 60.Offidani A, Cellini A, Simonetti O, Bossi G. Urticaria pigmentosa in monozygotic twins. Arch Dermatol. 1994;130:935–6. [PubMed] [Google Scholar]
- 61.Pec J, Palencarova E, Malisova S, Dobrota D, Hajtman A, Pec M, Lepej J. Urticaria pigmentosa in identical male twins. Acta Derm Venereol. 1995;75:244. doi: 10.2340/0001555575244. [DOI] [PubMed] [Google Scholar]
- 62.Nishida T, Hirota S, Taniguchi M, et al. Familial gastrointestinal stromal tumours with germline mutation of the KIT gene. Nat Genet. 1998;19:323–4. doi: 10.1038/1209. [DOI] [PubMed] [Google Scholar]
- 63.Trevisan G, Pauluzzi P, Gatti A, Semeraro A. Familial mastocytosis associated with neurosensory deafness. J Eur Acad Dermatol Venereol. 2000;14:119–22. doi: 10.1046/j.1468-3083.2000.00018.x. [DOI] [PubMed] [Google Scholar]
- 64.Beghini A, Tibiletti MG, Roversi G, Chiaravalli AM, Serio G, Capella C, Larizza L. Germline mutation in the juxtamembrane domain of the kit gene in a family with gastrointestinal stromal tumors and urticaria pigmentosa. Cancer. 2001;92:657–62. doi: 10.1002/1097-0142(20010801)92:3<657::aid-cncr1367>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 65.Ben-Amitai D, Metzker A, Cohen HA. Pediatric cutaneous mastocytosis: a review of 180 patients. Isr Med Assoc J. 2005;7:320–2. [PubMed] [Google Scholar]
- 66.Yanagihori H, Oyama N, Nakamura K, Kaneko F. C-kit mutations in patients with childhood-onset mastocytosis and genotype–phenotype correlation. J Mol Diagn. 2005;7:252–7. doi: 10.1016/S1525-1578(10)60552-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.de la Sotta PD, Romero WA, Kramer D, Cárdenas C, González S. Cutaneous mastocytosis in twins: multiple mastocytomas and urticaria pigmentosa in two pairs of monozygotic twins. Pediatr Dermatol. 2011;28:585–7. doi: 10.1111/j.1525-1470.2011.01351.x. [DOI] [PubMed] [Google Scholar]
- 68.Zhang LY, Smith ML, Schultheis B, Fitzgibbon J, Lister TA, Melo JV, Cross NCP, Cavenagh JD. A novel K5091 mutation of KIT identified in familial mastocytosis – in vitro and in vivo responsiveness to imatinib therapy. Leuk Res. 2006;30:373–8. doi: 10.1016/j.leukres.2005.08.015. [DOI] [PubMed] [Google Scholar]
- 69.Hartmann K, Wardelmann E, Ma Y, et al. Novel germline mutation of KIT associated with familial gastrointestinal stromal tumors and mastocytosis. Gastroenterology. 2005;129:1042–6. doi: 10.1053/j.gastro.2005.06.060. [DOI] [PubMed] [Google Scholar]
- 70.Burks KD, Wenzel SE, Jones SM. Familial cases of mast cell diseases. J Invest Med. 2005;53:S300. [abstract] [Google Scholar]
- 71.Bursztejn AC, Bronner M, Kanny G, Barbaud A, Morisset M, Jonveaux P. Genetic evaluation of familial mastocytosis. J Invest Dermatol. 2009;129:2528. [abstract] [Google Scholar]
- 72.Verzijl A, Heide R, Oranje AP, van Schaik RHN. c-Kit Asp-816-Val mutation analysis in patients with mastocytosis. Dermatology. 2007;214:15–20. doi: 10.1159/000096907. [DOI] [PubMed] [Google Scholar]
- 73.Welle MM, Bley CR, Howard J, Rüfenacht S. Canine mast cell tumours: a review of the pathogenesis, clinical features, pathology and treatment. Vet Dermatol. 2008;19:321–39. doi: 10.1111/j.1365-3164.2008.00694.x. [DOI] [PubMed] [Google Scholar]
- 74.O'Brien S, Tefferi A, Valent P. Chronic myelogenous leukemia and myeloproliferative disease. Hematology Am Soc Hematol Educ Program. 2004:146–62. doi: 10.1182/asheducation-2004.1.146. [DOI] [PubMed] [Google Scholar]
- 75.Buttner C, Henz BM, Welker P, Sepp NT, Grabbe J. Identification of activating c-kit mutations in adult-, but not in childhood-onset indolent mastocytosis: a possible explanation for divergent clinical behavior. J Invest Dermatol. 1998;111:1227–31. doi: 10.1046/j.1523-1747.1998.00414.x. [DOI] [PubMed] [Google Scholar]
- 76.Sotlar K, Horny HP, Simonitsch I, et al. CD25 indicates the neoplastic phenotype of mast cells: a novel immunohistochemical marker for the diagnosis of systemic mastocytosis (SM) in routinely processed bone marrow biopsy specimens. Am J Surg Pathol. 2004;28:1319–25. doi: 10.1097/01.pas.0000138181.89743.7b. [DOI] [PubMed] [Google Scholar]
- 77.Pignon JM, Giraudier S, Duquesnoy P, Jouault H, Imbert M, Vainchenker W, Vernant JP, Tulliez M. A new c-kit mutation in a case of aggressive mast cell disease. Br J Haematol. 1997;96:374–6. doi: 10.1046/j.1365-2141.1997.d01-2042.x. [DOI] [PubMed] [Google Scholar]
- 78.Baek JO, Kang HK, Na SY, Lee JR, Roh JY, Lee JH, Kim HJ, Park S. N822K c-kit mutation in CD30-positive cutaneous pleomorphic mastocytosis after germ cell tumour of the ovary. Br J Dermatol. 2012;166:1370–3. doi: 10.1111/j.1365-2133.2012.10816.x. [DOI] [PubMed] [Google Scholar]
- 79.Galderisi CD, Corless CL, Wolford J, Harrell T, Heinrich MC, Press RD. Simultaneous JAK2 V617F and KIT D816V mutations in systemic mastocytosis. Mod Pathol. 2006;19:225A–6A. [Google Scholar]
- 80.Steensma DP, Dewald GW, Lasho TL, Powell HL, McClure RF, Levine RL, Gilliland DG, Tefferi A. The JAK2 V617F activating tyrosine kinase mutation is an infrequent event in both “atypical” myeloproliferative disorders and myelodysplastic syndromes. Blood. 2005;106:1207–9. doi: 10.1182/blood-2005-03-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cools J, DeAngelo DJ, Gotlib J, et al. A tyrosine kinase created by fusion of the PDGFRA and FIP1L1 genes as a therapeutic target of imatinib in idiopathic hypereosinophilic syndrome. N Engl J Med. 2003;348:1201–14. doi: 10.1056/NEJMoa025217. [DOI] [PubMed] [Google Scholar]
- 82.Pardanani A, Ketterling RP, Brockman SR, et al. CHIC2 deletion, a surrogate for FIP1L1–PDGFRA fusion, occurs in systemic mastocytosis associated with eosinophilia and predicts response to imatinib mesylate therapy. Blood. 2003;102:3093–6. doi: 10.1182/blood-2003-05-1627. [DOI] [PubMed] [Google Scholar]
- 83.Robyn J, Lemery S, McCoy JP, et al. Multilineage involvement of the fusion gene in patients with FIP1L1/PDGFRA-positive hypereosinophilic syndrome. Br J Haematol. 2006;132:286–92. doi: 10.1111/j.1365-2141.2005.05863.x. [DOI] [PubMed] [Google Scholar]
- 84.Gotlib J, Cross NCR, Gilliland DG. Eosinophilic disorders: molecular pathogenesis, new classification, and modern therapy. Best Pract Res Clin Haematol. 2006;19:535–69. doi: 10.1016/j.beha.2005.07.013. [DOI] [PubMed] [Google Scholar]
- 85.Wilson TM, Maric I, Simakova O, et al. Clonal analysis of NRAS activating mutations in KIT-D816V systemic mastocytosis. Haematologica. 2011;96:459–63. doi: 10.3324/haematol.2010.031690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bandi SR, Brandts C, Rensinghoff M, et al. E3 ligase-defective Cbl mutants lead to a generalized mastocytosis and a myeloproliferative disease. Blood. 2009;114:4197–208. doi: 10.1182/blood-2008-12-190934. [DOI] [PubMed] [Google Scholar]
- 87.van Rijn RM, van Marle A, Chazot PL, et al. Cloning and characterization of dominant negative splice variants of the human histamine H4 receptor. Biochem J. 2008;414:121–31. doi: 10.1042/BJ20071583. [DOI] [PubMed] [Google Scholar]
- 88.Tefferi A, Levine RL, Lim KH, et al. Frequent TET2 mutations in systemic mastocytosis: clinical, KITD816V and FIP1L1–PDGFRA correlates. Leukemia. 2009;23:900–4. doi: 10.1038/leu.2009.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Schroer BC, Han YC, Hsieh FH. Identification of TET2 mutations in subjects with systemic mastocytosis and hypereosinophilic syndrome. J Allergy Clin Immunol. 2010;125:AB232. [abstract] [Google Scholar]
- 90.Nedoszytko B, Niedoszytko M, Lange M, et al. Associations of TNF-α gene polymorphisms and mastocytosis. A study of the European Competence Network in Mastocytosis (ECNM) Allergy. 2008;63(Suppl. 88):154. [abstract] [Google Scholar]
- 91.Niedoszytko M, Nedoszytko B, Lange M, et al. Vascular Endothelial Growth Factor (VEGF) promoter gene polymorphism in mastocytosis. A study of the European Competence Network on Mastocytosis (ECNM) Allergy. 2008;63(Suppl. 88):322. [abstract] [Google Scholar]
- 92.Alfter K, von Kügelgen I, Haenisch B, et al. New aspects of liver abnormalities as part of the systemic mast cell activation syndrome. Liver Int. 2009;29:181–6. doi: 10.1111/j.1478-3231.2008.01839.x. [DOI] [PubMed] [Google Scholar]



