Abstract
Introduction:
Enterococcus faecalis has long been implicated in persistent root canal infections and therapy-resistant endodontic infections. It has also been associated with bacteremia, that is, infective endocarditis arising from certain invasive dental procedures. E. faecalis endocarditis antigen (efaA) has been identified as one of the principal virulence factors associated with infective endocarditis.
Aim:
To detect the presence of putative E. faecalis virulence factor, efaA in root canals of therapy-resistant endodontic infections using polymerase chain reaction (PCR) amplification.
Materials and Methods:
Samples were obtained from 32 patients (20–70 years) undergoing endodontic retreatment, which were incubated in prereduced thioglycollate broth and subcultured onto ethyl violet azide broth (EVA; selective medium for E. faecalis). Deoxyribonucleic acid (DNA) was extracted from the samples and analyzed for the endocarditis virulence factor efaA using PCR.
Results:
Among the positive E. faecalis samples, efaA gene was identified in 11 out of 15 samples.
Conclusion:
Our findings indicate that efaA, a potent E. faecalis virulence gene can be found in E. faecalis strains detected in root canals of therapy-resistant endodontic infections similar to reports for ‘medical’ strains.
Keywords: Enterococcus faecalis endocarditis antigen, infective endocarditis, polymerase chain reaction
INTRODUCTION
Enterococci are Gram-positive facultative anaerobic bacteria which can cause a variety of diseases in humans, including septicaemia, endocarditis, urinary tract infections, wound infections, and meningitis.[1] In the past few years, Enterococcus faecalis has been mentioned with increased frequency with regard to teeth with asymptomatic persistent endodontic infections, predominantly in therapy-resistant endodontic infections.[2] E. faecalis have been able to form biofilms in root canals, and this ability can be important for bacterial resistance and persistence after endodontic procedures.[3]
Virulence factors like collagen-binding protein (ace), enterococcal surface proteins (esp), and hemolysin activator (cylA) play a significant role in enterococcal adhesion to collagen and extracellular matrix.[4] E. faecalis endocarditis antigen (efaA) was first identified from the antiserum of a patient with E. faecalis endocarditis.[5] The amino acid sequence of the associated protein efaA revealed 55–60% homology to a group of streptococci proteins known as adhesins.[5] Hemolysin, gelatinase, ace, and esp also function as putative virulence factors of endocarditis-causing E. faecalis.[6]
E. faecalis strains derived from different sources such as endocarditis, urinary tract infections, and even endodontic infections have been shown to possess distinct patterns of ‘virulence factors’. However, the significance of efaA in persistent endodontic infections has not been investigated.
Over the past few years, microbiological techniques such as polymerase chain reaction (PCR) have improved the sensitivity of microbial detection compared to culturing and have therefore enabled the identification of enterococci with greater precision. Microbial identification rely on the fact that certain genes contain information about microbial identity.[7]
Hence the aim of our study was to detect the presence of putative E. faecalis virulence factor efaA in root canals of therapy-resistant endodontic infections using the molecular-based PCR method.
MATERIALS AND METHODS
All clinical procedures performed in this study were approved by the Institutional Review Board of the University and were described in advance to patients, who provided written informed consent. Samples were obtained from 32 patients (12 women and 20 men between 20–70 yrs) undergoing endodontic retreatment for teeth determined clinically and radiologically to have nonhealing periradicular lesions. The teeth were single rooted (18 maxillary anterior teeth and 14 mandibular anterior teeth) and contained a root filling with associated periradicular lesions. Of the teeth included in the study, four had no restorations, 18 had temporary or defective permanent restorations, and 10 had adequate permanent restorations. Symptoms were reported for 10 teeth and four had lesions with the sinus tract. All the root-filled teeth had endodontic therapy completed more than one year previously. The terminus of the root canal fillings ranged from 0 to 5 mm short of the radiographic apex. The tooth was isolated with rubber dam and disinfected with 30% hydrogen peroxide and 5% tincture of iodine for two minutes. Disinfectants were neutralized with 5% sodium thiosulfate. Old restoration and caries were removed. A standard access opening was created with a sterile #2 round bur at high speed. Pulp space was accessed and root fillings were mechanically removed with Gates-Glidden burs and hand files with no solvents. A sterile #15 file was placed and working length determined. The canal was flushed with saline and the #15 file was used with a gentle filing motion to agitate root canal contents for one minute. The entire contents of the canal were absorbed into sterile paper points and transferred to prereduced thioglycollate broth. The broth tubes were stored aerobically at 37°C;2.0 ml of overnight-grown culture from thioglycollate broth was transferred to ethyl violet azide (EVA) broth [Figure 1] which is a selective medium for E. faecalis. The positive samples were inoculated on trypticase soy agar plates [Figure 1] for observation of colony morphology. Further confirmation of the growth of E. faecalis was obtained by biochemical characterization using bile esculin, pyruvate, and mannitol tests [Figure 1].
Figure 1.
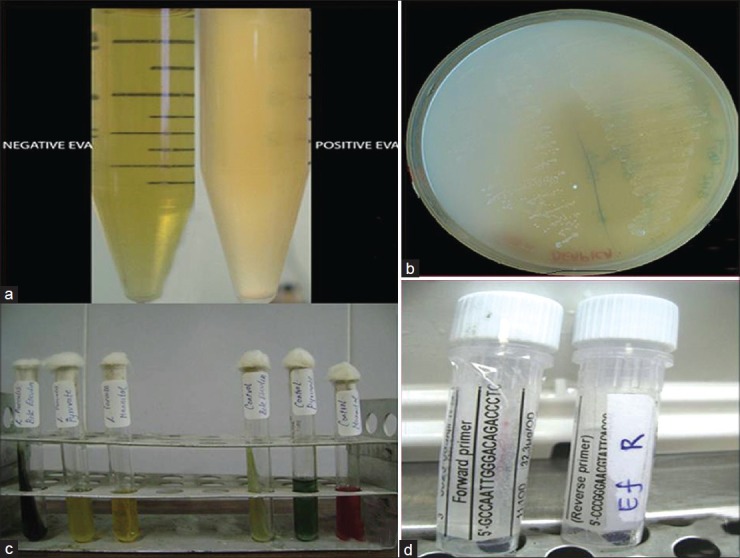
efaA gene identification: (a) ethyl violet azide (EVA) broth (b) trypticase soy agar plates (c) biochemical characterization: bile esculin, pyruvate, and mannitol tests (d) efaA primer and Taq polymerase
PCR was performed using the colonies. A single colony was used as the source for the DNA template. A positive control (ATCC 29212) and a negative control were also included. The efaA primer (Sigma-Aldrich) sequence [Figure 1] used for the PCR was 5’-CCAATTGGGACAGACCCTC-3’ (forward primer) and 5’-CGCCTTCTGTTCCTTCTTTGGC-3’ (reverse primer). PCR was carried out in a thermal cycler (Eppendorf® Mastercycler Gradient 5331, version 2.30.31.09). The reaction mix contained efaA primers, Taq DNA polymerase, reaction buffer, and dNTPs mix [mixture of equal amounts (10 mM) of dGTP, dTTP, dATP, and dCTP in a buffer (pH 7.5)]. PCR cycling parameters included an initial denaturation at 95°C followed by 30 cycles of denaturation step at 95°C for one minute, a primer-annealing step at 53°C for 10 minutes, extension at 70°C for one minute, and a final extension step at 70°C for 10 minutes. The products of PCR were analyzed using gel electrophoresis (Broviga®) technique using ethidium bromide as the DNA intercalating agent. The gel was viewed using an ultraviolet transilluminator (Vilber Lourmat, USA) and documented using a digital closed circuit (CC) camera.
RESULTS
The presence of E. faecalis was positive in 15 out of 32 samples as indicated by turbidity of the EVA broth and biochemical results. Among the 15 positive samples, efaA gene was identified in 11 out of 15 samples as demonstrated in Figure 2. The distribution of patient samples positive for efaA gene was as follows: Mean age 42.5; 8 (20) male and 3 (12) female patients; 7 positive samples involving mandibular anterior teeth and 4 involving maxillary anterior teeth.
Figure 2.

Distribution of efaA gene among 15 E. faecalis isolates efaA (688 bp), P: positive marker, N: negative marker. Lane 1 and 2: 1, 2, 3, 6, 7, 8, 9 11, 12, 14, 15 are positive samples; 4, 5, 10, 13 are negative samples
DISCUSSION
E. faecalis is a micro-organism that can survive extreme challenges. Its pathogenicity ranges from life-threatening diseases in compromised individuals such as bacteremia, septicemia, endocarditis, and urinary tract infections to less severe conditions, such as infection of obturated root canals with chronic apical periodontitis.[8,9] A potential explanation for the persistence of E. faecalis in the root canal might be an expression of adherence (virulence) factors like ace and esp.[4] Virulence factors of E. faecalis enable adherence to host tissue, invasion and modulation of host inflammatory response, and secretion of various products which enhance biofilm formation.[4]
Love et al.[2] suggested the role of serum in the invasion of dentinal tubules by E. faecalis. Serum, which originates from alveolar bone and periodontal ligament, facilitates enhanced dental invasion and collagen adhesion of E. faecalis. This synergistic effect between serum and enterococci cells promotes binding and allows invasion of the dentine to occur in the presence of serum.[2] Expression of endocarditis virulence factor efaA has been shown to be induced by the growth of E. faecalis in medium containing serum.[6]
The extracellular matrix of all mammalian tissues consists of glycoproteins (e.g. collagen, laminin, fibronectin) and proteoglycans that can be exploited by micro-organisms for colonization and initiation. The ability of a bacterium to adhere to collagen has been shown to play an important role in the pathogenesis of endocarditis. As dentinal tissues share common proteins with the heart tissues, a role for ace, esp, and surface adhesins like efaA should facilitate bacterial adhesion to collagen and extracellular matrix relevant in endodontic infections.[5,9,10]
Coque conducted an in vivo testing of an E. faecalis efaA mutant (Enterococcus virulence gene) and use of efaA homologs for species identification using PCR. It was found that the use of gene probes efaA (endocardial adherence factor antigen) enabled the identification of isolates of E. faecalis.[11]
The results of our study demonstrated that although 32 contaminated root canal samples were analyzed, only 15 of them were positive for E. faecalis (46.88% prevalence). Among the 15 positive samples, efaA gene was identified in 11 samples (73.5% prevalence), and herein lies the emphasis of our study. A similar recent molecular-based study by Salah [2008] indicated that virulence determinants endocarditis antigen (efaA) genes and ace genes has been found in E. faecalis isolates of oral rinse samples of patients suffering from dental diseases.[12] Bittencourt de Marques[1] observed that the efaA gene is frequently associated with esp or eep (enhanced expression of pheromone) and to a lesser extent with virulence markers aggA (aggregation substance gene involved in adherence to mammalian cells and conjugation) or gelE (extracellular metalloendopeptidase)
Four samples were negative for the presence of efaA. It is probable that E. faecalis uses other virulence factors, such as ace and gelE more than efaA for its pathogenicity. Nevertheless, this initial study on the prevalence and relevance of the putative virulent gene efaA of E.faecalis is of great importance. Previous studies by Creti et al.[9] showed that although E.faecalis strains derived from different sources such as endocarditis, urinary tract infections possessed distinct patterns of virulence factors ace, efaA, and gelE genes were found to be the most common virulence factors.
The clinical relevance of this study is that bacteremia arising from infected root canals (E. faecalis and its virulence factors efaA) in the blood stream may present risks for systemic complications like infective endocarditis in a susceptible heart.[13] With reference to enterococcal bacteremia arising from dental procedures, procedures like tooth extractions, implant placement, periodontal surgical procedures and nonsurgical endodontic instrumentation beyond the apex, or endodontic surgery offer a higher risk of bacteremia.[13,14] Although bacteremia does not persist for more than 15 minutes, they are potentially more dangerous for patients with valvular heart disease.[13] These risks can be minimized if appropriate therapeutic measures are applied, in addition to prophylactic antibiotic coverage for high-risk patients.[13]
A limitation to this study was that the samples collected were cultured in E.faeclis-selective medium. This action could change the molecular repertoire of the organisms leading to changes in virulence that could be different than those present in biofilms in failing endodontic therapy.
Although molecular-based identification methods (PCR) are designed to detect molecular DNA, a limitation of PCR is that it cannot distinguish between DNA from viable and dead cells and it is unclear whether the results form PCR method represent authentic living flora or rather a historical record of organisms that have entered but not survived in the canal.[8,15] Therefore despite the increased detection capability of the molecular method, a combination of both molecular and culture techniques provide a comprehensive understanding of the role of E. faecalis in the root canal infection process. Nevertheless, it may be emphasized that the primary aim of this study is to characterize E. faecalis strains from infected root canals by targeting the efaA gene.
CONCLUSION
Within the limitations of this study, it may be concluded that efaA, a potent E. faecalis virulence gene can be found in E. faecalis strains detected in root canals of therapy-resistant endodontic infections similar to reports for ‘medical’ strains. Further studies should aim at obtaining blood samples from patients immediately after endodontic retreatment procedure to comprehend the role of the efaA gene in causing bacteremia.
Footnotes
Source of Support: Nil
Conflict of Interest: None declared.
REFERENCES
- 1.de Marques E Bittencourt, Suzart S. Occurrence of virulence-associated genes in clinical enterococcus faecalis isolated in Londrina, Brazil. J Medical Microbiology. 2004;53:1069–73. doi: 10.1099/jmm.0.45654-0. [DOI] [PubMed] [Google Scholar]
- 2.Love RM. Enterococcus faecalis--a mechanism for its role in endodontic failure. Int Endod J. 2001;34:399–405. doi: 10.1046/j.1365-2591.2001.00437.x. [DOI] [PubMed] [Google Scholar]
- 3.Arias-Moliz MT, Ferrer-Luque CM, Espigares-García M, Baca P. Enterococcus faecalis biofilms eradication by root canal irrigants. J Endod. 2009;35:711–4. doi: 10.1016/j.joen.2009.01.018. [DOI] [PubMed] [Google Scholar]
- 4.Kayaoglu G, Ørstavik D. Virulence factors of enterococcus faecalis: Relationship to endodontic disease. Critical Rev Oral Biol Med. 2004;15:308–20. doi: 10.1177/154411130401500506. [DOI] [PubMed] [Google Scholar]
- 5.Sedgley CM, Molander A, Flannagan SE, Nagel AC, Appelbe OK, Clewell DB, et al. Virulence, phenotype and genotype characteristics of endodontic enterococcus spp. Oral Microbiol Immunol. 2005;20:10–9. doi: 10.1111/j.1399-302X.2004.00180.x. [DOI] [PubMed] [Google Scholar]
- 6.Lowe AM, Lambert PA, Smith AW. Cloning of an Enterococcus faecalis endocarditis antigen: Homology with adhesins from some oral streptococci. Infect Immun. 1995;63:703–6. doi: 10.1128/iai.63.2.703-706.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baldasarri L, Creti R, Arciola CR, Montanaro L, Venditti M, Di Rosa R. Analysis of virulence factors in cases of enterococcal endocarditis. Clin Microbiol Infect. 2004;10:1006–8. doi: 10.1111/j.1469-0691.2004.00999.x. [DOI] [PubMed] [Google Scholar]
- 8.Siqueira JF, Jr, Rôças IN. Exploiting molecular methods to explore endodontic infections: Part 1-current molecular technologies for microbiological diagnosis. J Endod. 2005;31:411–23. doi: 10.1097/01.don.0000157989.44949.26. [DOI] [PubMed] [Google Scholar]
- 9.Creti R, Imperi M, Bertuccini L, Fabretti F, Orefici G, Di Rosa R, et al. Survey for virulence determinants among enterococcus faecalis isolated from different sources. J Med Microbiol. 2004;53:13–20. doi: 10.1099/jmm.0.05353-0. [DOI] [PubMed] [Google Scholar]
- 10.Stuart CH, Schwartz SA, Beeson TJ, Owatz CB, Owatz Enterococcus faecalis: Its role in root canal treatment failure and current concepts in retreatment. J Endod. 2006;32:93–8. doi: 10.1016/j.joen.2005.10.049. [DOI] [PubMed] [Google Scholar]
- 11.Coque Teresa M, Weinstock George M, Murray Barbara E. In vivo testing of an Enterococcus faecalis efaA mutant and use of a efaA homologs for species identification. Immunology and Medical Microbiology. 1998;21:323–331. doi: 10.1111/j.1574-695X.1998.tb01180.x. [DOI] [PubMed] [Google Scholar]
- 12.Randa Salah, Najla Dar-Odeh, Osama, Osam Abu Hammad, Shahabi Asem A. Prevalence of putative virulence factors and antimicrobial susceptibility of enterococcus faecalis isolates from patients with dental diseases. Biomedical Oral Health. 2008;8:1–7. doi: 10.1186/1472-6831-8-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brincat M, Savarrio L, Saunders W. Endodontics and infective endocarditis-is antimicrobial chemoprophylaxis required? Int Endod J. 2006;39:671–82. doi: 10.1111/j.1365-2591.2006.01124.x. [DOI] [PubMed] [Google Scholar]
- 14.Fujmori Masaki, Matsudo Mitsuyoshi, Sato Yasuyoshi, Otsubo Seiji, Kawashima Eiji. A case of infective endocarditis caused by odontogenic infection. J Jap Stomatot Soc. 2006;55:67–71. [Google Scholar]
- 15.Sathorn C, Parashos P, Messer HH. How useful is root culturing in predicting treatment outcome. J Endod. 2007;33:220–5. doi: 10.1016/j.joen.2006.11.006. [DOI] [PubMed] [Google Scholar]


