Abstract
The AP-1 transcription factor Batf3 is required for homeostatic development of CD8α+ classical dendritic cells that prime CD8 T-cell responses against intracellular pathogens. Here, we identify an alternative, Batf3-independent pathway for their development operating during infection with intracellular pathogens mediated by the cytokines IL-12 and IFN-γ. This alternative pathway results from molecular compensation for Batf3 provided by the related AP-1 factors Batf, which also functions in T and B cells, and Batf2 induced by cytokines in response to infection. Reciprocally, physiologic compensation between Batf and Batf3 also occurs in T cells for expression of IL-10 and CTLA-4. Compensation among BATF factors is based on the shared capacity of their leucine zipper domains to interact with non-AP-1 factors such as Irf4 and Irf8 to mediate cooperative gene activation. Conceivably, manipulating this alternative pathway of dendritic cell development could be of value in augmenting immune responses to vaccines.
Batf1 and Batf3 are activator protein 1 (AP-1)2 transcription factors3,4 with immune-specific functions5–8. Batf is required for development of T helper cells producing IL-17 (TH17) and follicular helper T (TFH) cells5, and class-switch recombination (CSR) in B cells6,9. Batf3 is required for development of CD8α+ classical dendritic cells (cDCs) and related CD103+ DCs8 that cross-present antigens to CD8 T cells7 and produce IL-12 in response to pathogens10.
We recently recognized a heterozygous phenotype for Batf3 of 50% fewer CX3CR1−CD8α+ cDCs in Batf3+/− mice11, implying levels of Batf3 are limiting for CD8α+ DC development under homeostatic conditions. While analyzing Toxoplasma gondii infection in Batf3−/− mice, we observed evidence suggesting Batf3-independent CD8α+ cDC development10 based on apparent priming of pathogen specific CD8 T cells in Batf3−/− mice treated with IL-12. Here, we report a Batf3-independent pathway of CD8α+ cDC development that functions physiologically during infection by intracellular pathogens and describe its molecular basis, which involves compensatory BATF factors.
Pathogens and IL-12 restore CD8α+ cDCs in Batf3−/− mice
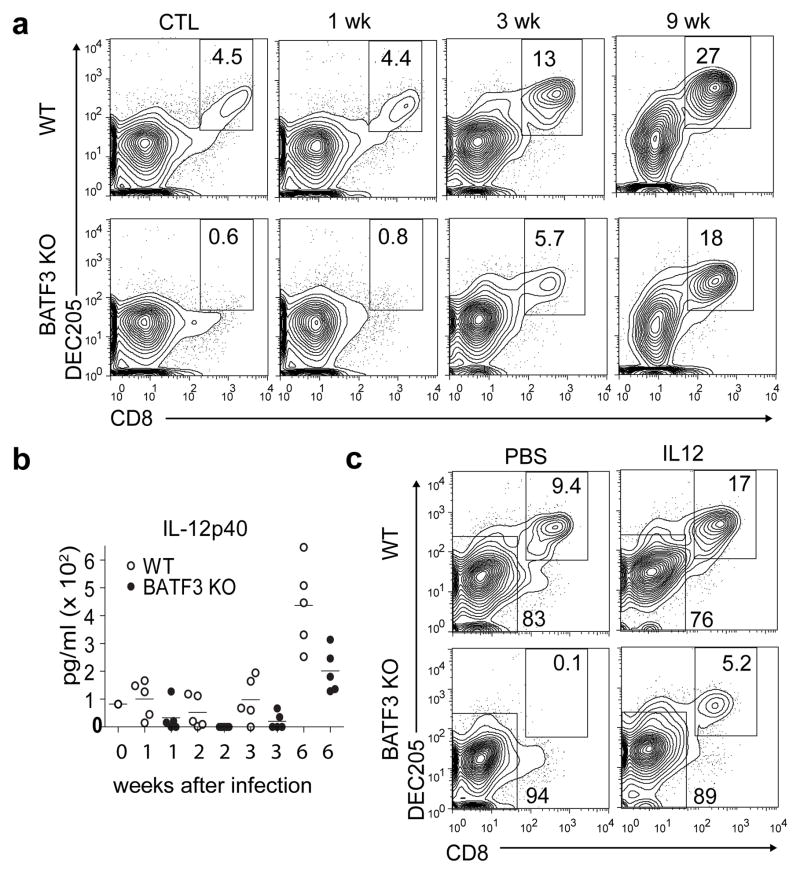
IL-12 administration to Batf3−/− mice before infection with type II Prugniaud (Pru) T. gondii reversed their susceptibility by inducing IFN-γ production not only from NK cells but also from CD8 T cells10, suggesting potentially restored cross-priming. To test this idea, we infected Batf3−/− mice with the attenuated12 RHΔku80Δrop5 strain of T. gondii and examined CD8α+ cDCs (Supplementary Fig. 1a). Surprisingly, CD8α+ cDCs reappeared in spleens of Batf3−/− mice by day 10 after infection. Infection by L. monocytogenes also restored CD8α+ cDCs in Batf3−/− mice (Supplementary Fig. 1b). Aerosolized infection with Mycobacterium tuberculosis (Mtb) caused a progressive restoration of CD8α+ cDCs in Batf3−/− mice and expanded CD8α+ cDCs in WT mice (Fig. 1a). Mtb infection of Batf3−/− mice restored the missing lung-resident CD103+ cDCs (DEC205+ CD24+ CD4− Sirp-α− CD11b−)8,13 (Supplementary Fig. 1c). Batf3−/− mice showed no difference in survival to Mtb infection compared to WT mice (Supplemental Fig. 1d), suggesting the initial lack of CD8α+ cDCs and peripheral CD103+ cDCs was not sufficient for lethality with Mtb. Since IL-12 is important in control of Mtb14, we measured serum IL-12 in Mtb-infected WT and Batf3−/− mice (Fig. 1b). IL-12 was reduced in Batf3−/− mice in the first three weeks, but increased after 6 weeks to approximately 50% of Mtb-infected WT mice.
Figure 1. Intracellular pathogens or IL-12 restore lymphoid CD8α+ cDCs and tissue-resident CD103+ cDCs in Batf3−/− mice.
a, Wild type (WT) and Batf3−/− (BATF3 KO) 129SvEv mice were uninfected (CTL) or infected with Mtb, and spleens harvested and analyzed by FACS at the indicated time. Histograms for indicated markers are gated as autofluorescent−MHCIIhighCD11c+ cells. Numbers are percent of cells in the gate. b, Serum IL-12 was measured from individual mice (a) at the indicated time. c, Wild type (WT) and Batf3−/− (BATF3 KO) 129SvEv mice were treated with vehicle (PBS) or IL-12 (IL12) and analyzed by FACS after 3 days as in (a).
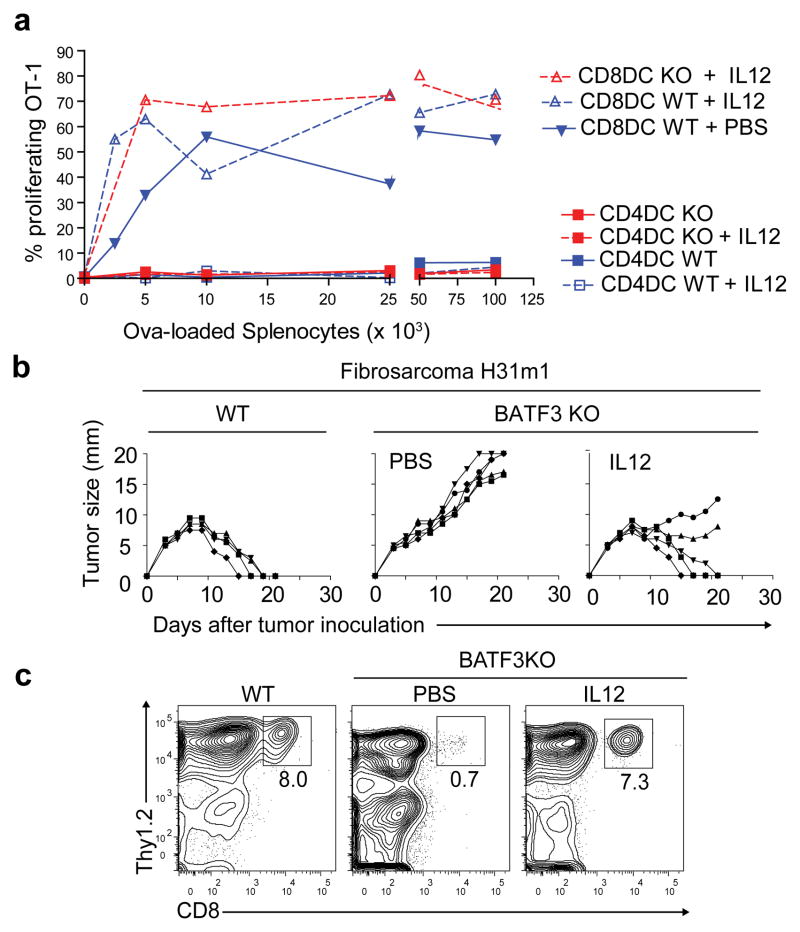
We found that IL-12 administration restored CD8α+ cDCs in all backgrounds of Batf3−/− mice (Fig. 1c, Supplementary Fig. 2a). Restored CD8α+ cDCs were functional for cross-presentation7,15,16 (Fig. 2a, Supplementary Fig. 2b). Purified splenic CD8α+ and CD4+ cDCs from WT or Batf3−/− mice, treated with vehicle or IL-12, were tested for cross-presentation. As a control, OT-I T cells proliferated in response to WT CD8α+ DCs, but not CD4+ DCs, co-cultured with ovalbumin-loaded cells with or without IL-12 treatment (Fig. 2a). Importantly, OT-I T cells proliferated similarly in response to IL-12-induced Batf3−/− CD8α+ DCs, but not Batf3−/− CD4+ DCs, as to WT CD8α+ cDCs. The CD8α+ cDCs restored by IL-12 in Batf3−/− mice were more similar in gene expression to WT CD8α+ DCs than to CD4+ cDCs (Supplementary Fig. 2c). 1855 genes differed more than 4-fold in expression between IL-12-induced CD8α+ cDCs and CD4+ cDCs within Batf3−/− mice. However, in comparing IL-12-induced CD8α+ cDCs in Batf3−/− mice with the rare CD8α+ cDCs from Batf3−/− C57BL/6 mice, or with CD8α+ cDCs in WT mice, there were only 34 and 206 genes respectively differing more than 4-fold in expression.
Figure 2. IL-12-induced CD8α+ cDCs in Batf3−/− mice can cross-present and mediate tumor rejection.
a, From mice in Fig. 1c, DCs were purified by sorting as CD3−DX5−MHCII+CD11c+Sirp-α−CD24+DEC205+ DCs (CD8DC) and CD3−DX5−MHCII+CD11c+Sirp-α+CD24−DEC205− DCs (CD4DC) and assayed for cross-presentation7. OT-I proliferation in response to cDCs mixed with the indicated number of MHC class I-deficient ovalbumin (Ova)-loaded splenocytes is shown. b, Wild type (WT) or Batf3−/− (BATF3 KO) mice treated with vehicle (PBS) or with IL-12 (IL12) were inoculated with 1×106 H31m1 fibrosarcomas. Tumor size in individual mice is shown. c, Mice in (b) were analyzed by FACS 11 days after H31m1 inoculation for CD8 T cell infiltration into tumors7.
The IL-12-induced restoration of CD8α+ cDCs in Batf3−/− mice was dependent on IFN-γ in vivo (Supplementary Fig. 2d). With administration of control antibody, IL-12 induced a 3-fold increase in CD8α+ cDCs in WT mice and restored CD8α+ cDCs in Batf3−/− mice. Both effects were blocked by anti-IFN-γ antibody. IL-12 induced IFN-γ production from NK cells but not T cells (Supplementary Fig. 2e), and IL-12 treatment was able to restore CD8α+ cDCs in Rag2−/−Batf3−/− mice (Supplementary Fig. 2f), indicating that T cells and B cells are not required for this effect.
Batf3−/− mice fail to reject highly immunogenic H31m1 and D42m1 fibrosarcomas7. However, Batf3−/− mice pre-treated with IL-12 either fully rejected or showed reduced growth of these fibrosacromas (Fig. 2b, Supplementary Fig. 2g). Rejection was not simply due to NK cell activation, since IL-12 treatment failed to alter tumor growth in Rag2−/− mice. Strikingly, IL-12-treated Batf3−/− mice re-established priming of CD8 T cells capable of infiltrating tumors similar to WT mice (Fig. 2c). Mice with the inactivating IRF8 R249C mutation17–19 failed to restore CD8α+ cDCs upon IL-12 administration (Supplementary Fig. 3a), indicating that restoration requires functional IRF8 protein. Further, these mice were highly susceptible to infection by aerosolized Mtb20 (Supplementary Fig. 3b), suggesting that Batf3−/− mice may resist Mtb infection by restoration of CD8α+ cDCs. Thus, physiological Batf3-independent development of CD8α+ cDCs is induced by pathogens, mediated by IL-12 and IFN-γ.
Batf, Batf2 and Batf3 cross-compensate in DCs and T cells
We asked if Batf could replace Batf3 for in vitro cDC development7,21 (Fig. 3a). CD103+ Sirp-α− cDCs do not develop in Flt3L-treated Batf3−/− bone marrow (BM) cultures. Retroviral expression of Batf3 into Batf3−/− BM progenitors restored CD103+ cDC development. Notably, Batf fully and cell-intrinsically restored CD103+ Sirp-α− cDC development, while c-Fos was inactive (Supplementary Fig. 3c). CD103+ cDCs restored by Batf and Batf3 in vitro were functional, showing features of mature CD103+ cDCs, including loss of Sirp-α and CD11b, upregulation of CD24, and selective production of IL-12 in response to T. gondii antigen (Supplementary Fig. 3c-d). Reciprocally, Batf3 but not c-Fos restored cell-intrinsic IL-17a production by Batf−/− T cells and CSR in Batf−/− B cells (Supplementary Fig. 3e–f). Thus, Batf and Batf3 can molecularly cross-compensate for several distinct lineage-specific functions, activities not shared by c-Fos.
Figure 3. Batf compensates for CD8α+ cDC development in Batf3−/− mice.
a, Wild type (WT), or Batf3−/− (BATF3KO) BM cells were infected with GFP-RV33 (Empty) or retrovirus expressing the indicated cDNA and cultured with Flt3L7. Histograms for the indicated markers are for B220−CD11c+ cells on day 10. Numbers are the percent of cells in the gate. b, Inguinal lymph nodes from WT, Batf3−/− (BATF3 KO) or Batf−/−Batf3−/− mice (BATF1/3 DKO) on 129SvEv or C57BL/6 backgrounds were analyzed by FACS. Shown are histograms for DEC205 and CD8α. c, CD4 T cells of the indicated genotype were differentiated twice under TH2 conditions5 and analyzed by FACS for intracellular IL-10.
We also identified in vivo compensation between Batf and Batf3 in DCs. On the 129SvEv and BALB/c backgrounds, Batf3−/− mice completely lack CD8α+ cDCs (Fig. 3b, Supplementary Fig. 2a). In contrast, C57BL/6 Batf3−/− mice retain a 3–7% population of CD8α+ cDCs19,22 in spleen and unexpectedly retain a completely normal population of CD8α+ cDCs in skin-draining inguinal lymph nodes (ILNs) (Fig. 3b). These CD8α+ cDCs are functional, since C57BL/6 Batf3−/− mice showed robust priming of CD8 T cells against HSV infection in the footpad, while 129SvEv Batf3−/− mice, lacking CD8α+ cDCs in ILNs, did not (Supplementary Fig. 3g). However, C57BL/6 Batf−/−Batf3−/− (BATF1/3DKO) mice lack CD8α+ DCs in ILNs and fail to prime T cells against HSV. Thus, retention of functional CD8α+ cDCs in ILNs of C57BL/6 Batf3−/− mice is Batf-dependent.
Batf and Batf3 also compensate in expression of genes by T cells. IL-4 and IL-10 production were not substantially affected in either Batf−/− or Batf3−/− TH2 cells (Fig. 3c, Supplementary Fig. 3i–k). However, IL-10 is reduced at least 8-fold in BATF1/3DKO TH2 cells. Also, the inhibitory receptor CTLA-4 is partially reduced in Batf−/− TH2 cells but reduced 3-fold further in BATF1/3DKO TH2 cells. In contrast, IFN-γ production by TH1 cells is unaffected by loss of Batf, Batf3 or both.
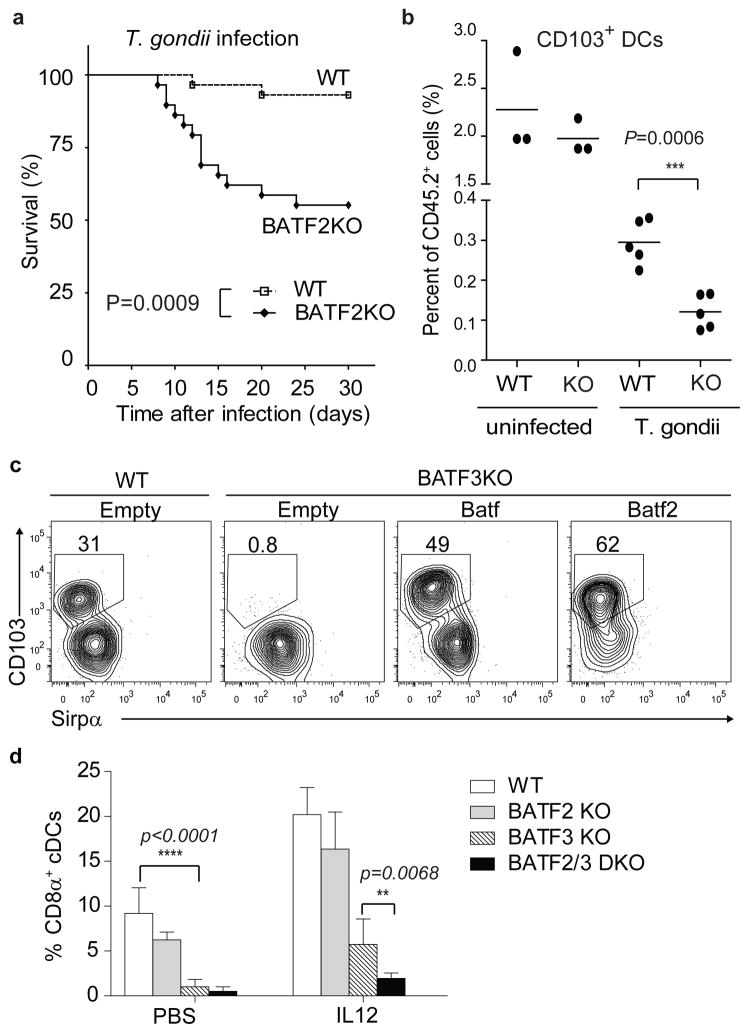
We asked if IL-12-induced restoration of CD8α+ cDCs in Batf3−/− mice was due to compensation by Batf (Supplementary Fig. 3h). Restoration of splenic CD8α+ cDCs in IL-12-treated BATF1/3DKO mice was reduced to 5% from 11% in IL-12-treated Batf3−/− mice. While Batf appears responsible for roughly half of the IL-12-induced restoration of CD8α+ cDCs in Batf3−/− mice, some residual compensation remained in BATF1/3DKO mice. We asked if a third factor might compensate for Batf and Batf3. Batf2 (SARI)23 is closely related to Batf and Batf3 and is induced by LPS and IFN-γ in macrophages and CD103+ DC populations (Supplementary Fig. 4a–c). We found that Batf2 was induced by IFN-γ in WT and Batf3−/− DCs derived from Flt3L-cultured BM (Supplementary Fig. 4d–e) and in vivo by IL-12 in DCs in Batf3−/− mice (Supplementary Fig. 4f). Induction of Batf2 by IFN-γ in cDCs made it a potential candidate to mediate IFN-γ-dependent compensation for Batf3.
We made Batf2−/− mice by targeting exons 1 and 2, eliminating Batf2 protein expression (Supplementary Fig. 5). Batf2−/− mice had normal development of NK, T and B cells, pDCs, neutrophils, resting cDCs, and peritoneal, liver and lung macrophages (Supplementary Fig. 6a–c). However, Batf2−/− mice displayed significantly decreased survival after infection by T. gondii (Pru) (Fig. 4a), although parasite burden and serum cytokines were similar to WT mice (Supplementary Fig. 7a–b). Notably, Batf2−/− mice showed significantly decreased numbers of lung-resident CD103+CD11b− DCs and CD103− CD11b− macrophages after infection (Fig. 4b, Supplementary Fig. 7c). These changes were specific, since there were no differences in other myeloid subsets (Supplementary Fig. 7d-e) Thus, Batf2 plays a role in maintaining numbers of Batf3-dependent CD103+ DCs in the lung following infection with T. gondii.
Figure 4. Batf2 compensates for Batf3 in CD8α+ and CD103+ cDC development during T. gondii infection.
a, Wild-type (WT) and Batf2−/− (BATF2KO) mice were infected with T. gondii and monitored for survival. n=29 for WT (dashed line) and Batf2−/− (solid line) mice. b, Shown are percentages of lung CD103+ DCs of total CD45.2+ cells for uninfected and infected (T. gondii) mice on day 10. n=5 from one of three experiments. c, WT or Batf3−/− (BATF3KO) BM cells were infected with the indicated retrovirus, cultured with Flt3L and analyzed by FACS on day 10. d, Groups of 5 mice each of the indicated genotypes were treated with vehicle (PBS) or IL-12 (IL12) and analyzed by FACS after 3 days. Shown are percentages of CD8α+ cDCs as a total of splenic cDCs.
We asked if Batf2 could compensate for DC defects in Batf3−/− mice and for T and B cell defects in Batf−/− mice (Fig. 4c, Supplementary Fig. 8a-b). Like Batf, retroviral expression of Batf2 restored development of CD103+Sirp-α− DCs in Flt3L-treated Batf3−/− BM. Unlike Batf and Batf3, expression of Batf2 did not restore TH17 development Batf−/− T cells and only weakly restored CSR in Batf−/− B cells. Thus, Batf2 selectively compensates for Batf and Batf3 in cDCs but not in T or B cells. We next examined in vivo IL-12-induced restoration of CD8α+ cDCs in Batf2−/−, Batf3−/− and Batf2−/−Batf3−/− (BATF2/3DKO) mice (Fig. 4d, Supplementary Fig. 8c). IL-12 treatment restored CD8α+ cDCs from <1% to >5% of total cDCs in Batf3−/− mice, but this was significantly reduced to ~2% in BATF2/3DKO mice. This suggests that Batf2 is responsible for roughly half of IL-12-induced CD8α+ cDC restoration in Batf3−/− mice. We found similar results with in vitro CD103+ cDC development. While GM-CSF restored only CD103 and not DEC205 expression in Flt3L-treated Batf3−/− BM cultures19, adding IFN-γ with GM-CSF restored DEC205+CD103+CD11b− DCs (Supplementary Fig. 8d-e). We used this system to analyze compensation between Batf, Batf2, and Batf3 (Supplementary Fig. 8d–e). Relative to WT BM, CD103+DEC205+CD11b− cDCs were partially reduced in Batf−/−, Batf2−/− and Batf3−/− singly-deficient BM, but were reduced to an even greater extent in both BATF1/3DKO and BATF2/3DKO BM relative to all single-deficient BM cultures. Collectively, these results suggest that Batf and Batf2 both act in the cytokine-dependent rescue of CD8α+ cDC development in Batf3−/− mice.
The BATF LZ domain interacts with IRF4 and IRF8
To understand BATF cross-compensation, we analyzed chimeric proteins containing fused domains of Batf, Batf2, and c-Fos (Supplementary Fig. 9). c-Fos inactivity could result from inhibitory actions of N- and C-terminal domains missing in Batf and Batf3 (Fig. 5a). However, removing these domains from c-Fos (Δ5′c-FosΔ3′) or fusing the Batf DBD with the c-Fos LZ (BBRFFΔ3′) failed to restore CD103+ cDC development (Fig. 5a), TH17 development or CSR activity (Supplementary Fig. 10a–b). However, fusing the Batf LZ with the c-Fos DBD (Δ5′FFQBB) fully restored CD103+ cDC development in Ftl3L-treated Batf3−/− BM cultures, fully restored CSR in Batf−/− B cells and exhibited 20% of WT activity for restoring IL-17 production. Further, Batf with a C-terminal GFP (Batf-GFP) or c-Fos (BBBF) domain had >40% of WT Batf activity in all three assays (Supplementary Fig. 11). Thus, the LZ domain, not the DBD, determines BATF specificity.
Figure 5. BATF leucine zipper interactions with non-AP-1 factors mediate lineage-specific actions.
a, Structures of chimeric proteins are shown below a diagram of c-Fos. DNA binding domain (DB), hinge (H), leucine zipper (LZ), amino- (5′) and carboxy-terminus (3′). Flt3L-treated WT or Batf3−/− (BATF3 KO) BM infected with the indicated retrovirus were analyzed after 10 days. b, 293FT cells expressing both Batf and Irf4 (upper panel) or Batf and Irf4 as indicated (lower panel) were analyzed by EMSA with the indicated probes and antibodies c, 293FT cells expressing Irf4 (+) and the indicated Batf chimera were analyzed by EMSA with the AICE1 probe. d, B cells were analyzed by EMSA with the indicated probe and competitor oligonucleotides (comp). e, Batf3−/− (BATF3 KO) BM infected with the indicated Batf retroviruses s encoding Batf were analyzed as in (a). f, 293FT cells expressing the indicated Batf mutants were analyzed by EMSA with the AICE1 probe.
Removing the Batf2 C-terminal domain (Batf2 DM) allowed for full restoration of CD103+ cDC development, but led to only partial restoration of CSR activity (~15%), and no restoration of TH17 development (Supplementary Fig. 10c–h). Fusing the Batf DBD with the Batf2 LZ (B1HB2 DM) allowed for approximately 50% of WT Batf activity for CSR and TH17 development in Batf−/− B and T cells. Fusing the Batf2 DBD with Batf LZ (B2QB1) allowed for 50% of WT Batf activity in CD103+ cDC development, but provided less than 5% WT Batf activity in TH17 development. Thus, the Batf2 DBD restricts activity in T and B cells, but its LZ functions for all BATF lineage-specific activities.
A recent study24 proposed that Irf4 and Batf may interact. Correspondence between the phenotypes of Batf−/− with Irf4−/− mice, lacking TH17 development and CSR, and Batf3−/− with Irf8−/− mice, lacking CD8α+ cDC development (Supplementary Fig. 12)5–7,25–28, suggested that Batf and Batf3 may cooperate with both Irf4 and Irf8. Further, ChIP-Seq analysis of Batf and Irf4 revealed coincident binding to composite AP-1 and IRF motifs (Li et al., In Press; Glasmacher et al, In Press). By analogy with the Ets-IRF composite elements (EICE)29, the AP-1-IRF composite elements are designated as AICE (Glasmacher et al., In Press; Li et al., In Press). Recalling the c-Fos LZ interaction with NF-AT that mediates cooperative binding to Il2 regulatory regions30, we therefore asked if the Batf LZ interacted with non-AP-1 factors, including Irf4.
Electrophoretic mobility shift assays (EMSA) demonstrated interactions between BATF and both Irf4 and Irf8 (Fig. 5, Supplementary Figs. 13, 14). The Batf/Jun complex that formed on an AP-1 consensus probe1,2 was unchanged by addition of Irf4 or Irf8. Its abundance was increased by additional JunB (Supplementary Fig. 13a). However, using an AICE from the CTLA-4 locus, a slower mobility complex formed with addition of either Irf4 or Irf8, which required the IRF consensus element (Fig. 5b, Supplementary Fig. 13b). This Batf/Jun/Irf4 complex required both Batf and IRF protein, and Irf4 was unable to bind the AICE1 probe without Batf (Fig. 5b). In contrast, Irf4 was able to bind to and Ets/IRF consensus elements31 (EICE) in the presence of PU.1 independently of Batf (Fig. 5b). The Batf/Jun/Irf4 complex had similar mobility to the Fos/Jun complex in activated cells, but could be distinguished by antibody supershifts (Supplementary Fig. 13c). Spatial constraints on the Batf/Irf4 interaction are suggested by lack of Batf/Irf4 complex formation on the AICE2 probe with reversed orientation of the AP-1 and IRF elements (Supplementary 13a, c).
The ability of chimeric Batf proteins to interact with Irf4 correlated with their functional activity (Fig. 5c). The active Δ5′FFQBB interacted with Irf4, but the inactive Δ5′c-FosΔ3′ and BBRFFΔ3′ did not (Fig. 5c), although all three bound an AP-1 consensus (Supplementary Fig. 13d). A Batf/IRF complex also formed in B and T cells, requiring both the IRF and AP-1 consensus elements (Fig. 5d, Supplementary Fig. 14c), and involved contribution by endogenous Irf4 and Irf8 (Supplementary Fig. 14a, b, d). The Batf/IRF complex was completely absent in Batf−/− B cells, but was partially retained in single-deficient Irf4−/− or Irf8−/− B cell extracts (Supplementary Fig. 14a-b). Supershifts using anti-Irf4 and anti-Irf8 antibodies in Irf4−/− or Irf8−/− B cell nuclear extracts showed that the endogenous Batf/IRF complex is composed of a mixture of Irf4 and Irf8 (Supplementary Fig. 14b). Similarly, supershifts of primary TH17 cells demonstrate an interaction between Batf and both Irf4 and Irf8 (Supplementary Fig. 14d).
We identified Batf LZ mutations which abrogated both function and Irf4 interactions (Fig. 5e–f). Positions b, c and f of the c-Fos LZ face away from the parallel coiled-coil of the Jun LZ and mediate interactions with NF-AT30 (Supplementary Fig. 9d). Several Batf mutations in these positions had no effect on activity (Supplementary Table 1, Supplementary Fig. 9c). However, four residues together controlled nearly all BATF-specific activity (Fig. 5e, Supplementary 15). Batf L56A, Batf K63D and Batf E77K showed no reduction in CD103+ cDC development, and retained >60% of WT Batf activity in CSR. In contrast, Batf H55Q activity was reduced by nearly 70% (Fig. 5e). Double mutants Batf K63D E77K and Batf H55Q L56A were reduced by 50% and 75%, but quadruple mutant Batf H55Q L56A K63D E77K had less than 10% of WT Batf activity in CD103+ cDC development (Fig. 5e). This loss of activity was specific since two other quadruple mutants, Batf E59Q D60Q K63D E77K and Batf K63D R69Q K70D E77K, maintained >50% of WT Batf activity (Supplementary Fig. 15c–d). A similar pattern was seen for CSR in Batf−/− B cells (Supplementary Fig. 15b). The activity of these mutants correlated with Irf4 interaction (Fig. 5f). WT Batf and functional mutant K63D E77K, and both functional quadruple mutants formed a complex with Irf4, whereas the less active Batf H55Q and Batf H55Q L56A, and the completely inactive Batf H55Q L56A K63D E77K did not, although all were stably expressed and could bind an AP-1 site (Supplementary 13e–f).
Discussion
We uncovered a cytokine-driven pathway for expansion of functional CD8α+ cDCs occurring physiologically during infection by intracellular pathogens. This pathway relies on functional compensation for Batf3 provided by Batf and Batf2 through a shared specificity defined by the BATF LZ domain to support CD8α+ cDC and TH17 development, and CSR in B cells. This compensatory pathway of CD8α+ cDCs development may provide a basis for augmenting therapeutic immune responses. The basis of this shared specificity is the LZ interaction with non-AP-1 factors, including Irf4 and Irf8, extending the repertoire of AP-1 interactions beyond the recognized association of Fos with NF-AT30. The ability of both Batf and Batf3 to support both Irf4- and Irf8-dependent lineage activities implies that the specificity for gene activation is determined largely by the IRF factors. An important next step will be to determine how regulatory elements discriminate between these distinct complexes.
Online Methods
Generation of Batf2−/− mice
The targeting construct was assembled by Gateway recombination cloning system (Invitrogen). To construct pENTR loxFRT rNEO, a floxed PGK–neor (phosphoglycerate kinase promoter - neomycin phosphotransferase) gene cassette (1982 bp) was excised from the pLNTK targeting vector34 using SalI and XhoI. After incubation with Easy-A cloning enzyme (Stratagene) and dNTPs to generate 3′-A overhangs, the PGK–neor cassette was ligated into the multiple cloning site of pGEM-T Easy (Promega). The resulting 2022 bp PGK–neor gene cassette was released using NotI and ligated into the 2554 bp backbone of NotI digested pENTR lox-Puro35. Flippase Recognition Target (FRT) sites were sequentially inserted at the SacI and HindIII sites using DNA fragments generated by following annealing oligonucleotides: SacII-FRT-A (GAAGTTCCTATTCCGAAGTTCCTATTCTCTAGAAAGTATAGGAACTTCCGC) and SacII-FRT-B (GGAAGTTCCTATACTTTCTAGAGAATAGGAACTTCGGAATAGGAACTTCGC), or HindIII-FRT-A (AGCTTGAAGTTCCTATTCCGAAGTTCCTATTCTCTAGAAAGTATAGGAACTTC) and HindIII-FRT-B (AGCTGAAGTTCCTATACTTTCTAGAGAATAGGAACTTCGGAATAGGAACTTCA). To construct pENTR-Batf2-5HA, the 5′ homology arm was generated by PCR from genomic DNA using the following oligonucleotides, which contain attB4 and attB1r site: GGGGACAACTTTGTATAGAAAAGTTGGCAGGCTGAAGCAGGGACAC, and GGGGACTGCTTTTTTGTACAAACTTGCCCTAGCACACCCAGTTTCAGTTTC. The attB4-attB1r PCR fragment was ligated into pDONR(P4-P1R) plasmid (Invitrogen) by BP recombination reaction. To construct pENTR-Batf2-3HA, the 3′ homology arm was generated by PCR from genomic DNA using the following oligonucleotides which contain attB2 and attB3site: GGGGACAGCTTTCTTGTACAAAGTGGGCAGTCCCAGCATGACCCAGCCCCAGCATGA CCCAGCATCTGC, and GGGGACAACTTTGTATAATAAAGTTGCCGTCTCACTCAGTTCTGTGTGTG. The attB2-attB3 PCR fragment was ligated into pDONR (P2-P3) plasmid (Invitrogen) by BP recombination reaction, following by the LR recombination reaction to generate final targeting construct by using pENTR-Batf2-5HA, pENTR-Batf2-3HA, pENTR-loxFRT rNEO, and pDEST DTA-MLS. The linearized vector was electroporated into EDJ22 embryonic stem cells, 129 SvEv background, and targeted clones were identified by Southern blot analysis with 5′ and 3′ probes. Blastocyst injections were performed, and male chimeras were bred to female 129S6/SvEv mice. Probes for Southern Blot analysis were amplified from genomic DNA using the following primers: 5′-GTTGGTGTGAGGTGATGAGGTCCC -3′ and 5′-CTTGACTTCCTAGACCAGGGGC-3′ for the 5′ probe; 5′-GGACCATATACTCTTACTGTTGAAACC-3′ and 5′-GGGCTGGGGACACACATG-3′ for the 3′ probe. Southern Blot analysis and genotyping PCR were performed to confirm germline transmission and the genotype of the progeny. Primers used for genotyping PCR are shown as follows: KO screen forward, 5′-GAAACTGAAACTGGGTGTGCTAGGG-3′; KO screen reverse, 5′ CGCCTTCTTGACGAGTTCTTCTGAG-3′; WT screen reverse, 5′-GCCTTCCTTGTCTCTCTCCATAGCG-3′.
Generation of Batf2-specific rabbit polyclonal antibody
Murine full-length Batf2 cDNA was cloned into pET-28a (+) expression vector (Novagen) to generate recombinant Batf2 protein with His tagged. Recombinant Batf2 was then expressed using Escherichia coli BL21 (Invitrogen). Purified recombinant Batf2 was used to immunize New Zealand White rabbits (Harlan), and rabbit anti-mouse Batf2 sera were collected and tested by ELISA and Western Blot analysis.
Pathogen infections
Listeria monocytogenes and T. gondii infections were carried out as previously described36,37. The type II avirulent Prugniaud (Pru) strain of T. gondii expressing a firefly luciferase and GFP transgene (provided by J. Boothroyd, Stanford University, Palo Alto, CA) was used for infections of Batf2−/− mice for the experiments shown in Fig. 4 and Supplementary Fig. 7. Briefly, 800 tachyzoites were injected intraperitoneally (i.p.) into mice. For Herpes simplex virus 1 (HSV-1) infections, mice were infected with the KOS strain subcutaneously (s.c.) with 1.5 × 105 PFU/mouse in the foot pad. Viral supernatant and HSV peptide were provided by M. Colonna. For Mycobacterium tuberculosis (Mtb) infections, Erdman strain was grown to a density of 8×106 CFU/ml, and mice were exposed to aerosol infection for 40 minutes using a Glas-Col aerosol exposure system (AES). After 24 hours, two mice per group were sacrificed, and lungs were harvested to determine infection efficiency, which was about 100 CFU/lung. Lungs and spleens were collected at 1, 3 and 9 weeks post infection. The left lobe of the lung and one third of the spleen were used for histological examination upon formalin fixation. The right lobe of the lung and two thirds of the spleen were divided into CFU analysis and FACS analysis. For CFU analysis, organs were homogenized and plated at various dilutions on 7H-10 culture plates, and after three weeks, colonies were counted. For FACS analysis, organs were prepared as described38. For lungs, Ficoll gradient purification was performed prior to surface staining. All samples were fixed upon staining for 20 minutes in 4% formalin. Serum was collected by retro-orbital bleeding at week 1, 2, 3 and 6 and decontaminated by double filtering through a 0.2mm filter.
Bioluminescence Imaging
Imaging was done as previously described39. In brief, mice were injected intraperitoneally with D-luciferin (Biosynth AG, Switzerland) at 150 mg/kg and allowed to remain active for 5 minutes before being anesthetized with 2% isoflurane for 5 minutes. Animals were than imaged with a Xenogen IVIS200 machine (Caliper Life Sciences). Data were analyzed with the Living Image software (Caliper Life Sciences).
In vitro T cell restimulation after HSV infection
One week after infection, spleens were harvested and restimulated with the HSV peptide HSV-gB2 (498-505) (Anaspec). Briefly, 2×106 splenocytes were restimulated for 5 hours in the presence of Brefeldin A at 1 μg/mL and analyzed by FACS for intracellular IFN-γ and TNF-α production as described later.
In vitro cross-presentation
DC cross-presentation of antigen to CD8+ OT-I T cells was assessed as previously described40. Briefly, Spleens from naïve, vehicle or IL12-treated WT or Batf3−/− mice were digested with collagenase B (Roche) and DNase I (Sigma-Aldrich). CD11c+ DCs were obtained by negative selection using B220, Thy1.2, and DX5 microbeads followed by positive selection with CD11c microbeads by MACS purification (Miltenyi Biotec). CD8α+ and CD4+ cDCs were cell sorted on a FACSAria II flow cytometer (post-sort purity >96%). Splenocytes from Kb−/−Db−/−β2m−/− mice were prepared in serum-free medium, loaded with 10 mg/ml ovalbumin (EMD) by osmotic shock, and irradiated (13.5 Gy) as described previously40. OT-I T cells were purified from OT-I/Rag2−/− mice by CD11c and DX5 negative selection, followed by positive selection with CD8α microbeads (purity >96%). T cells were fluorescently labeled by incubation with 1 μM CFSE (Sigma-Aldrich) for 9 min at 25°C at a density of 2 × 107 cells/ml. For the cross presentation assay, 5 × 104 –105 purified DCs were incubated with 5 × 104 –105 CFSE-labeled OT-I T cells in the presence of increasing numbers of irradiated, ovalbumin-loaded Kb−/−Db−/−β2m−/− splenocytes (5 × 103 to 1 × 105). After 3 days, OT-I T cell proliferation was analyzed by CSFE dilution gating on CD3+ CD8+ CD45.1+ cells.
Tumor transplantation
MCA-induced fibrosarcomas were derived from 129/SvEv strain Rag2−/− or WT mice as described previously41. Tumor cells were propagated in vitro and 106 tumor cells (129SvEv fibrosarcoma tumor line H31m1 or D42m1) were injected s.c. in a volume of 150 μl endotoxin-free PBS into the shaved flanks of naïve or IL12 conditioned (as described above) wild type (WT) Batf3−/− and Rag2 −/− recipient mice as described previously40. Injected cells were >90% viable as assessed by trypan blue exclusion. Tumor size was measured on the indicated days and is presented as the mean of two perpendicular diameters.
Tumor harvest
WT and naïve or IL12 treated Batf3−/− mice were inoculated with the H31m1 fibrosarcoma tumor line. 11 days after, tumors were removed, minced and treated with 1μmg/ml type A collagenase (Sigma) in HBSS (Hyclone) for 1h at 37°C. Cell suspension was analyzed by FACS for CD8 T cell recruitment at the tumor site gating on CD45+, Thy1.2+ and CD8α+ cells.
Quantitative RT-PCR
For gene expression analysis, RNA was prepared from various cell types with either RNeasy Mini Kit (Qiagen) or RNeasy Micro kit (QIAGEN), and cDNA was synthesized with Superscript III reverse transcription (Invitrogen). Real-time PCR and a StepOnePlus Real-Time PCR system (Applied Biosystems) were used according to the manufacturer’s instructions, with the Quantitation Standard-Curve method and HotStart-IT SYBR Green qPCR Master Mix (Affymetrix/USB). PCR conditions were 10 min at 95°C, followed by 40 two-step cycles consisting of 15 s at 95°C and 1 min at 60°C. Primers used for measurement of Batf2 expression are as follows: Batf2 qRT forward, 5′-GGCAGAAGCACACCAGTAAGG-3′; Batf2 qRT reverse, 5′-GAAGGGCGTGGTTCTGTTTC-3′. Hprt was used as normalization control, and primers used are as follows: Hprt forward, 5′-TCAGTCAACGGGGGACATAAA-3′; and Hprt reverse, 5′-GGGGCTGTACTGCTTAACCAG-3′.
DC preparation
Dendritic cells from lymphoid organs and non-lymphoid organs were harvested and prepared as described 38. Briefly, spleens, MLNs, SLNs (inguinal), and liver were minced and digested in 5 ml Iscove’s modified Dulbecco’s media + 10% FCS (cIMDM) with 250 μg/ml collagenase B (Roche) and 30 U/ml DNase I (Sigma-Aldrich) for 30 min at 37°C with stirring. Cells were passed through a 70-μm strainer before red blood cells were lysed with ACK lysis buffer. Cells were counted on a Vi-CELL analyzer, and 5–10 × 106 cells were used per antibody staining reaction. Lung cell suspensions were prepared after perfusion with 10 ml Dulbeccos PBS (DPBS) via injections into the right ventricle after transection of the lower aorta. Dissected and minced lungs were digested in 5ml cIMDM with 4mg/ml collagenase D (Roche) for 1 h at 37°C with stirring. Cells from the peritoneal cavity were collected by washing the peritoneal cavity with 10 ml HBSS+2%FCS and 2mM EDTA.
Bone marrow derived DC and macrophage
Bone marrow (BM)-derived DCs were generated from BM with 100 ng/mL of recombinant Flt3L as described40. BM-derived macrophages were generated from BM with 20 ng/mL of recombinant M-CSF (Peprotech) for 7 days, and rested in cIMDM without M-CSF for 24 hours before stimulation with various conditions as described in the figure legends.
Preparation of T. gondii lysate antigen (STAg)
T gondii antigen was prepared by lysing tachyzoites of Pru strain in foreskin fibroblast cultures through two cycles of freeze-thaw in liquid nitrogen. Lysate was then filtered, aliquoted at a concentration of 1 mg/ml in PBS and stored at −80 degree. For DC stimulation, 0.05 ug/ml were used and intracellular IL12 production was analyzed by FACS.
Cytokine induced CD8α+ DCs
WT and Batf3−/− mice bone marrow was prepared as described above. GM-CSF (10ng/ml) and or IFN-γ (0.1ng/ml) was added to the cultures between day 8 and 10. Cells were analysed two days after cytokine addition.
Antibodies and flow cytometry
Staining was performed at 4°C in the presence of Fc Block (anti CD16/32 clone 2.4G2, BioXCell) in FACS buffer (DPBS + 0.5% BSA + 2 mm EDTA). The antibodies used for DC analysis were as recently described42. Cells were analyzed on BD FACSCanto II or FACSAria II and analyzed with FlowJo software (Tree star, Inc.).
Intracellular Cytokine Staining
For intracellular cytokine staining, cells were surface stained, then fixed in 2% paraformaldehyde for 15 min at 4°C, permeabilized in DPBS + 0.1% BSA + 0.5% saponin, and stained for intracellular cytokines as previously described43. Additional antibodies included anti-TNF-α (MP6-XT22) and anti-IFN-γ (XMG1.2) from BioLegend.
ELISA and CBA
IL-12p40 concentration was measured from serum samples with the Mouse IL-12p40 OptEIA ELISA set (BD Bioscience) according to the manufacturer instructions. The concentration of inflammatory cytokines was measured in the serum with the BD CBA Mouse Inflammation Kit (BD Biosciences), and data were analyzed with FCAP Array software (Soft Flow, Inc. USA).
Statistical analysis
Differences between groups in survival were analyzed by the log-rank test. Analysis of all other data was done with an unpaired, two-tailed Student’s t test with a 95% confidence interval (Prism; GraphPad Software, Inc.). P values less than 0.05 were considered significant. *0.01 < p < 0.05, **0.001 < p < 0.01, ***p < 0.001, ****p < 0.0001.
Expression microarray analysis
Total RNA was isolated from cells using the Ambion RNAqueous-Micro Kit. For Mouse Genome 430 2.0 Arrays, RNA was amplified, labeled, fragmented, and hybridized using the 3′ IVT Express Kit (Affymetrix). Data were normalized and expression values were modeled using DNA-Chip analyzer (dChip) software (www.dChip.org)44. Mouse Gene 1.0 ST Arrays, RNA was amplified with the WT Expression Kit (Ambion) and labeled, fragmented, and hybridized with the WT Terminal Labeling and Hybridization Kit (Affymetrix). Data was processed using RMA quantile normalization and expression values were modeled using ArrayStar software (DNASTAR). All original microarray data have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO.
CD4+ T cell cultures
CD4+ T cells were purified using DynaBeads FlowComp mouse CD4 kit (Invitrogen) and were activated on αCD3/αCD28 coated plates under the following culture conditions: TH1 conditions: αIL4 10μg/ml (11B11, BioXcell) 0.1μg/ml IFNγ (Peprotech), 10u/ml IL12, IL2 40u/ml; TH2 conditions: αIL12 10μg/ml (Tosh, BioXCell), αIFNγ 10μg/ml (XMG1.2, BioXCell), 10ng/ml IL4 (Peprotech), IL2 40u/ml; TH17 conditiions: IL-6 25ng/ml (Peprotech), TGFβ 2ng/ml (Peprotech), IL1β 10ng/ml (Peprotech), αIL4 10 μg/ml (11B11, BioXCell), αIFNγ 10μg/ml (XMG1.2, BioXCell), and αIL12 10μg/ml (Tosh, BioXCell). IL2 (40u/ml) was added for the TH17 cultures in Supplementary Figure 12a. Cells were diluted three fold in fresh media on day 3. On day 5 cells were activated with PMA/ionomycin for analysis of cytokines and surface markers by FACS. For some experiments, cells were restimulated on day 7 under the same conditions and analyzed 5 days later.
Retroviral analysis of BATF functional activity
For CD8α+ DC development, BM was cultured with Flt3L as described40, infected with retrovirus and 2μg/ml polybrene on day 1, and analyzed for development of cDCs (CD11c+ B220−) on day 10.
CD4+ T cells were cultured under TH17 conditions43,45 and infected with retrovirus and 6μg/ml polybrene on day 1. Cells were analyzed on day 5 for cytokine expression by intracellular staining. For some experiments, cells were restimulated on day 7 under the same conditions and analyzed 5 days later.
For analysis of class switch recombination, B220 cells purified by αB220 Macs microbeads (Miltenyi) were cultured with LPS and IL4 as described46, infected with retrovirus and 6μg/ml polybrene on day 1, and analyzed for switching to IgG1 on day 4 by FACS.
For infection of primary mouse cells, GFP-RV47 containing cDNAs for transcription factors, chimeric proteins and mutated Batf were transfected into Phoenix E cells as described previously48. Viral supernatants were collected 2 days later and concentrated by centrifugation49. Cells were infected with viral supernatants by spin infection at 1800 rpm for 45 min at room temperature.
Electromobility shift assays (EMSA)
For stable expression in 293FT cells, retroviruses (GFP-RV containing Batf, chimeric proteins or Batf mutants, and/or truncated hCD4-RV47 containing Irf4 or Irf8 cDNA) were packaged in Phoenix A cells and concentrated by centrifugation49. Infected 293FT cells were sorted for retroviral marker expression and when indicated were transiently transfected with JunB-GFP-RV using calcium phosphate. For transient expression, 293FT cells were transfected with retroviral constructs using calcium phosphate. Nuclear extracts were prepared 48hrs after transfection. For 293FT cells, nuclei were obtained after cellular lysis with Buffer A containing 0.2% NP40 and nuclear extracts in Buffer C were dialyzed against buffer D as described50. Nuclear extracts from T and B cells were prepared as described46.
EMSA was as described51 using 3μg of nuclear extract, 1mg poly dIdC (Sigma) and 7%T 3.3%C polyacrylamide gels. For competition assays, extracts were incubated with excess unlabelled competitor DNA for 20 min before addition of labeled probe. For supershifts, extracts were incubated with antibodies (αc-Fos (4) X (Santa Cruz) (for 293FT cells) or αc-Fos 2G9C3 (Abcam) (for mouse T cells), αIrf4 (H- 140) X (Santa Cruz), αIrf8 (ICSBP) C-19) X (Santa Cruz), αJun B (C-11) X (Santa Cruz) and rabbit αBatf 43, rabbit αBatf2 (described above), and rabbit αBatf3 (KC et al, submitted) for 1hr on ice prior to addition of labeled probe and incubation at room temperature for 35minutes. The following pairs of oligonucleotides were annealed to generate probes which were labeled with 32P-dCTP using Klenow polymerase. The AP1consensus binding site is underlined. The Irf consensus binding site is in italics. Mutated bases are in lower case.
AP1: 5′GATCAGCTTCGCTTGATGAGTCAGCCGG/5′GATCCGGCTGACTCATCAAGCGAAG
AICE1 (from 3rd intron of mouse CTLA4): 5′CTTGCCTTAGAGGTTTCGGGATGACTAATACTGTA/5′TCACGTACAGTATTAGTCATCCCGAAACCTCTAAGG
AICE1 M1 (mutates the Irf consensus binding site): 5′CTTGCCTTAGAGGccaCGGGATGACTAATACTGTA/5′TCACGTACAGTATTAGTCATCCCGtggCCTCTAAGG
AICE M2 (mutates the AP-1 consensus binding site):
5′CTTGCCTTAGAGGTTTCGGGAgacCTAATACTGTA/5′TCACGTACAGTATTAGgtcTCCCGAAACCTCTAAGG
AICE2 (from 3rd intron of IL23R): 5′GATGTTTCAGGGAAAGCACTGACTCACTGGCTCTCCA/5′GGTGGAGAGCCAGTGAGTCAGTGCTTTCCCTGAAA
EICE52: 5′GAAAAAGAGAAATAAAAGGAAGTGAAACCAAG/5′GATCCTTGGTTTCACTTCCTTTTATTTCTCTTT
Eα: 5′TCGACATTTTTCTGATTGGTTAAA/5′GACTTTTAACCAATCAGAAAAATG
Plasmids
cDNAs for Batf, Batf2, Batf3, Irf8 and JunB were generated by PCR from primary cells. MigR1 containing HA tagged Irf4 was from H. Singh (Genentech). HA-Irf4 and Irf8 cDNAs were subcloned into hCD4-RV47. Mutations in Batf were generated using QuickChange mutagenesis with Batf -GFP-RV as the template. cDNAs for chimeric proteins were generated by overlap extension and PCR and cloned into GFP-RV.
Supplementary Material
Acknowledgments
Supported by the Howard Hughes Medical Institute, National Institutes of Health (AI076427-02) and Department of Defense (W81XWH-09-1-0185) (K.M.M.), the American Heart Association (12PRE8610005) (A.S.), German Research Foundation (AL 1038/1-1) (J.C.A), American Society of Hematology Scholar Award and Burroughs Welcome Fund Career Award for Medical Scientists (B.T.E.), and Cancer Research Institute predoctoral fellowship (W.L.). We thank the ImmGen consortium32, Mike White for blastocyst injections and generation of mouse chimeras, the Alvin J. Siteman Cancer Center at Washington University School of Medicine for use of the Center for Biomedical Informatics and Multiplex Gene Analysis Genechip Core Facility. The Siteman Cancer Center is supported in part by the NCI Cancer Center Support Grant P30 CA91842. IL-12 was a gift from Pfizer.
Footnotes
Author Contributions R.T., W.L., T.L.M., and M.M. performed experiments with Batf−/−, Batf2−/−, Batf3−/− and DKO mice; T.L.M. made and analyzed all Batf mutants; J.A.R. aided with EMSA; W.KC., J.C.A., and A.T.S. were involved with microarray analysis and generation of mutant mice. J.C.A. was involved in generating of Batf2−/− mice. B.T.E. was involved with L. monocytogenes analysis; N.M.K was involved with cross-presentation analysis. X.W. was involved with bioinformatic analysis; L.A.W. C.L.S. were involved with Mtb infection; E.G. and H.S. helped identify EMSA probes; P.L., W.L. and W.J.L. performed ChIP-Seq and helped identify EMSA probes; M.B. and D.S. aided with T. gondii infections; S.S.K.L., C.T.A. and R.D.S. aided with tumor models. H.S. and W.J.L. provided helpful discussions. K.M.M. directed the work and wrote the manuscript. All authors discussed the results and contributed to the manuscript.
The authors have no conflicting financial interests.
References
- 1.Dorsey MJ, et al. B-ATF: a novel human bZIP protein that associates with members of the AP-1 transcription factor family. Oncogene. 1995;11:2255–2265. [PubMed] [Google Scholar]
- 2.Wagner EF, Eferl R. Fos/AP-1 proteins in bone and the immune system. Immunol Rev. 2005;208:126–140. doi: 10.1111/j.0105-2896.2005.00332.x. [DOI] [PubMed] [Google Scholar]
- 3.Williams KL, et al. Characterization of murine BATF: a negative regulator of activator protein-1 activity in the thymus. Eur J Immunol. 2001;31:1620–1627. doi: 10.1002/1521-4141(200105)31:5<1620::aid-immu1620>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 4.Echlin DR, Tae HJ, Mitin N, Taparowsky EJ. B-ATF functions as a negative regulator of AP-1 mediated transcription and blocks cellular transformation by Ras and Fos. Oncogene. 2000;19:1752–1763. doi: 10.1038/sj.onc.1203491. [DOI] [PubMed] [Google Scholar]
- 5.Schraml BU, et al. The AP-1 transcription factor Batf controls T(H)17 differentiation. Nature. 2009;460:405–409. doi: 10.1038/nature08114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ise W, et al. The transcription factor BATF controls the global regulators of class-switch recombination in both B cells and T cells. Nat Immunol. 2011 doi: 10.1038/ni.2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hildner K, et al. Batf3 deficiency reveals a critical role for CD8alpha+ dendritic cells in cytotoxic T cell immunity. Science. 2008;322:1097–1100. doi: 10.1126/science.1164206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Edelson BT, et al. Peripheral CD103+ dendritic cells form a unified subset developmentally related to CD8alpha+ conventional dendritic cells. J Exp Med. 2010;207:823–836. doi: 10.1084/jem.20091627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Betz BC, et al. Batf coordinates multiple aspects of B and T cell function required for normal antibody responses. J Exp Med. 2010;207:933–942. doi: 10.1084/jem.20091548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mashayekhi M, et al. CD8a+ Dendritic Cells Are the Critical Source of Interleukin-12 that Controls Acute Infection by Toxoplasma gondii Tachyzoites. Immunity. 2011;35(2):249–259. doi: 10.1016/j.immuni.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bar-On L, et al. CX3CR1+ CD8alpha+ dendritic cells are a steady-state population related to plasmacytoid dendritic cells. Proc Natl Acad Sci U S A. 2010;107:14745–14750. doi: 10.1073/pnas.1001562107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Behnke MS, et al. Virulence differences in Toxoplasma mediated by amplification of a family of polymorphic pseudokinases. Proc Natl Acad Sci U S A. 2011;108:9631–9636. doi: 10.1073/pnas.1015338108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ginhoux F, et al. The origin and development of nonlymphoid tissue CD103+ DCs. J Exp Med. 2009;206:3115–3130. doi: 10.1084/jem.20091756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Modlin RL, Barnes PF. IL12 and the human immune response to mycobacteria. Res Immunol. 1995;146:526–531. doi: 10.1016/0923-2494(96)83027-6. [DOI] [PubMed] [Google Scholar]
- 15.den Haan JM, Lehar SM, Bevan MJ. CD8(+) but not CD8(−) dendritic cells cross-prime cytotoxic T cells in vivo. J Exp Med. 2000;192:1685–1696. doi: 10.1084/jem.192.12.1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wilson NS, et al. Systemic activation of dendritic cells by Toll-like receptor ligands or malaria infection impairs cross-presentation and antiviral immunity. Nat Immunol. 2006;7:165–172. doi: 10.1038/ni1300. [DOI] [PubMed] [Google Scholar]
- 17.Aliberti J, et al. Essential role for ICSBP in the in vivo development of murine CD8alpha + dendritic cells. Blood. 2003;101:305–310. doi: 10.1182/blood-2002-04-1088. [DOI] [PubMed] [Google Scholar]
- 18.Tailor P, Tamura T, Morse HC, Ozato K. The BXH2 mutation in IRF8 differentially impairs dendritic cell subset development in the mouse. Blood. 2008;111:1942–1945. doi: 10.1182/blood-2007-07-100750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Edelson BT, et al. Batf3-dependent CD11b(low/−) peripheral dendritic cells are GM-CSF-independent and are not required for Th cell priming after subcutaneous immunization. PLoS ONE. 2011;6:e25660. doi: 10.1371/journal.pone.0025660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marquis JF, Lacourse R, Ryan L, North RJ, Gros P. Disseminated and rapidly fatal tuberculosis in mice bearing a defective allele at IFN regulatory factor 8. J Immunol. 2009;182:3008–3015. doi: 10.4049/jimmunol.0800680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jackson JT, et al. Id2 expression delineates differential checkpoints in the genetic program of CD8alpha(+) and CD103(+) dendritic cell lineages. EMBO J. 2011;30:2690–704. doi: 10.1038/emboj.2011.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Edelson BT, et al. CD8a+ Dendritic Cells Are an Obligate Cellular Entry Point for Productive Infection by Listeria monocytogenes. Immunity. 2011;35:236–248. doi: 10.1016/j.immuni.2011.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Su ZZ, et al. Cloning and characterization of SARI (suppressor of AP-1, regulated by IFN) Proc Natl Acad Sci U S A. 2008;105:20906–20911. doi: 10.1073/pnas.0807975106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ravasi T, et al. An atlas of combinatorial transcriptional regulation in mouse and man. Cell. 2010;140:744–752. doi: 10.1016/j.cell.2010.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brustle A, et al. The development of inflammatory T(H)-17 cells requires interferon-regulatory factor 4. Nat Immunol. 2007;8:958–966. doi: 10.1038/ni1500. [DOI] [PubMed] [Google Scholar]
- 26.Klein U, et al. Transcription factor IRF4 controls plasma cell differentiation and class-switch recombination. Nat Immunol. 2006;7:773–782. doi: 10.1038/ni1357. [DOI] [PubMed] [Google Scholar]
- 27.Tamura T, Ozato K. ICSBP/IRF-8: its regulatory roles in the development of myeloid cells. J Interferon Cytokine Res. 2002;22:145–152. doi: 10.1089/107999002753452755. [DOI] [PubMed] [Google Scholar]
- 28.Sciammas R, et al. Graded expression of interferon regulatory factor-4 coordinates isotype switching with plasma cell differentiation. Immunity. 2006;25:225–236. doi: 10.1016/j.immuni.2006.07.009. [DOI] [PubMed] [Google Scholar]
- 29.Brass AL, Zhu AQ, Singh H. Assembly requirements of PU. 1-Pip (IRF-4) activator complexes: inhibiting function in vivo using fused dimers. EMBO J. 1999;18:977–991. doi: 10.1093/emboj/18.4.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen L, Glover JN, Hogan PG, Rao A, Harrison SC. Structure of the DNA-binding domains from NFAT, Fos and Jun bound specifically to DNA. Nature. 1998;392:42–48. doi: 10.1038/32100. [DOI] [PubMed] [Google Scholar]
- 31.Eisenbeis CF, Singh H, Storb U. Pip, a novel IRF family member, is a lymphoid-specific, PU. 1-dependent transcriptional activator. Genes Dev. 1995;9:1377–1387. doi: 10.1101/gad.9.11.1377. [DOI] [PubMed] [Google Scholar]
- 32.Heng TS, Painter MW. The Immunological Genome Project: networks of gene expression in immune cells. Nat Immunol. 2008;9:1091–1094. doi: 10.1038/ni1008-1091. [DOI] [PubMed] [Google Scholar]
- 33.Ranganath S, et al. GATA-3-dependent enhancer activity in IL-4 gene regulation. J Immunol. 1998;161:3822–3826. [PubMed] [Google Scholar]
- 34.Gorman JR, et al. The Ig(kappa) enhancer influences the ratio of Ig(kappa) versus Ig(lambda) B lymphocytes. Immunity. 1996;5:241–252. doi: 10.1016/s1074-7613(00)80319-2. [DOI] [PubMed] [Google Scholar]
- 35.Iiizumi S, et al. Simple one-week method to construct gene-targeting vectors: application to production of human knockout cell lines. Biotechniques. 2006;41:311–316. doi: 10.2144/000112233. [DOI] [PubMed] [Google Scholar]
- 36.Edelson BT, et al. CD8a+ Dendritic Cells Are an Obligate Cellular Entry Point for Productive Infection by Listeria monocytogenes. Immunity. 2011;35:236–248. doi: 10.1016/j.immuni.2011.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mashayekhi M, et al. CD8a+ Dendritic Cells Are the Critical Source of Interleukin-12 that Controls Acute Infection by Toxoplasma gondii Tachyzoites. Immunity. 2011;35:249–259. doi: 10.1016/j.immuni.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Edelson BT, et al. Peripheral CD103+ dendritic cells form a unified subset developmentally related to CD8alpha+ conventional dendritic cells. J Exp Med. 2010;207:823–836. doi: 10.1084/jem.20091627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saeij JP, Boyle JP, Grigg ME, Arrizabalaga G, Boothroyd JC. Bioluminescence imaging of Toxoplasma gondii infection in living mice reveals dramatic differences between strains. Infect Immun. 2005;73:695–702. doi: 10.1128/IAI.73.2.695-702.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hildner K, et al. Batf3 deficiency reveals a critical role for CD8alpha+ dendritic cells in cytotoxic T cell immunity. Science. 2008;322:1097–1100. doi: 10.1126/science.1164206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Matsushita H, et al. Cancer exome analysis reveals a T-cell-dependent mechanism of cancer immunoediting. Nature. 2012;482:400–404. doi: 10.1038/nature10755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Satpathy AT, et al. Zbtb46 expression distinguishes classical dendritic cells and their committed progenitors from other immune lineages. J Exp Med. 2012;209:1135–1152. doi: 10.1084/jem.20120030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schraml BU, et al. The AP-1 transcription factor Batf controls T(H)17 differentiation. Nature. 2009;460:405–409. doi: 10.1038/nature08114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li C, Wong WH. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci U S A. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bending D, et al. Highly purified Th17 cells from BDC2.5NOD mice convert into Th1-like cells in NOD/SCID recipient mice. J Clin Invest. 2009 doi: 10.1172/JCI37865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ise W, et al. The transcription factor BATF controls the global regulators of class-switch recombination in both B cells and T cells. Nat Immunol. 2011;12:536–543. doi: 10.1038/ni.2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ranganath S, et al. GATA-3-dependent enhancer activity in IL-4 gene regulation. J Immunol. 1998;161:3822–3826. [PubMed] [Google Scholar]
- 48.Sedy JR, et al. B and T lymphocyte attenuator regulates T cell activation through interaction with herpesvirus entry mediator. Nat Immunol. 2005;6:90–98. doi: 10.1038/ni1144. [DOI] [PubMed] [Google Scholar]
- 49.Kanbe E, Zhang DE. A simple and quick method to concentrate MSCV retrovirus. Blood Cells Mol Dis. 2004;33:64–67. doi: 10.1016/j.bcmd.2004.04.001. [DOI] [PubMed] [Google Scholar]
- 50.Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Szabo SJ, Gold JS, Murphy TL, Murphy KM. Identification of cis-acting regulatory elements controlling interleukin-4 gene expression in T cells: roles for NF-Y and NF-ATc [published erratum appears in Mol Cell Biol 1993 Sep;13(9):5928] Mol Cell Biol. 1993;13:4793–4805. doi: 10.1128/mcb.13.8.4793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Eisenbeis CF, Singh H, Storb U. Pip, a novel IRF family member, is a lymphoid-specific, PU. 1-dependent transcriptional activator. Genes Dev. 1995;9:1377–1387. doi: 10.1101/gad.9.11.1377. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.