Abstract
DNA methylation is a major control program that modulates gene expression in a plethora of organisms. Gene silencing through methylation occurs through the activity of DNA methyltransferases, enzymes that transfer a methyl group from S-adenosyl-l-methionine to the carbon 5 position of cytosine. DNA methylation patterns are established by the de novo DNA methyltransferases (DNMTs) DNMT3A and DNMT3B and are subsequently maintained by DNMT1. Aging and age-related diseases include defined changes in 5-methylcytosine content and are generally characterized by genome-wide hypomethylation and promoter-specific hypermethylation. These changes in the epigenetic landscape represent potential disease biomarkers and are thought to contribute to age-related pathologies, such as cancer, osteoarthritis, and neurodegeneration. Some diseases, such as a hereditary form of sensory neuropathy accompanied by dementia, are directly caused by methylomic changes. Epigenetic modifications, however, are reversible and are therefore a prime target for therapeutic intervention. Numerous drugs that specifically target DNMTs are being tested in ongoing clinical trials for a variety of cancers, and data from finished trials demonstrate that some, such as 5-azacytidine, may even be superior to standard care. DNMTs, demethylases, and associated partners are dynamically shaping the methylome and demonstrate great promise with regard to rejuvenation.
Introduction
Epigenetic regulation of gene expression occurs through the addition and removal of chemical tags to DNA-associated proteins as well as to the DNA itself. DNA methylation targets the latter and involves the addition of a methyl group to the carbon 5 (C5) position of cytosine and typically leads to gene silencing.1 Genomic 5-methylcytosine (5mC) patterns are established during early development by the de novo DNA methyltransferases (DNMTs) DNMT3A and DNMT3B and are subsequently maintained by DNMT1.2 Global demethylation and remodeling of these patterns occur during two life cycle phases in mammals, one during gametogenesis and one during preimplantation development.1
DNA methylation is an evolutionarily conserved form of transcriptional repression that exists in a spectrum of genomes—both prokaryotic and eukaryotic.3,4 This epigenetic tag was originally discovered in bacterial restriction-modification systems, with some restriction endonucleases being specific to methylated DNA. This has led to the theory that prokaryotic DNA methylation evolved as a form of defense against foreign DNA molecules.3 In eukaryotes, transposable elements and repeats are densely methylated in fungi, plants, and mammals, suggesting transposon defense as another ancestral role of DNA methylation.4
Methylation of cytosine residues is thought to prevent gene transcription by directly interfering with the recognition sequence of transcription factors or with the transcriptional machinery. Gene silencing via methylation can also be mediated by the assistance of methyl-CpG binding domain proteins (MBDs), enzymes that establish silent chromatin through the recruitment of histone deacetylases.5 Action on cytosine by DNMTs occurs almost exclusively within CG dinucleotides (CpGs), and approximately 60%–90% of all mammalian CpGs are methylated. An exception is made for unmethylated CpG islands, clusters of CpG-enriched DNA that often reside within gene promoter regions.6 In general, only regions of the genome that possess CpGs are directly susceptible to methylation. Mammalian telomere repeats, for example, lack the CpG site and are thought to be exempt from direct action by DNMTs. Methylation of adjacent subtelomeric regions, however, controls both telomere recombination and telomere length in mouse embryonic stem cells (ESCs).7
The importance of DNMTs is made apparent by the phenotypes of DNMT-deficient mice: Ablation of Dnmt1, Dnmt3a, or Dnmt3b results in early embryonic lethality,2,8 and selective deletion of neuronal Dnmt3a creates mice with a shortened life span and neuromuscular defects.9 In addition, DNA methylation has been implicated in a wide array of biological processes, including genomic imprinting,10 X chromosome inactivation,11,12 autoimmunity,13 carcinogenesis, and aging.14
Aging is strongly correlated with changes in DNA methylation. DNA methylation and epigenetic alterations have been directly linked to longevity in a wide array of organisms, ranging in complexity from yeast to humans.15 The general trends, supported by an ever-increasing body of both in vitro and in vivo work, are the establishment of global hypomethylation (non-CpG islands) and regions of hypermethylation (primarily CpG islands) with age.16
Aging is traditionally thought to be caused by numerous complex and interacting factors. These include oxidative DNA damage, depletion of self-renewing stem cells, mitochondrial and nuclear genome mutations, shortening of telomeres, and other processes.17 The question of whether alterations in the methylome also play a causal role in aging has yet to be elucidated. This review addresses this issue and investigates the role of DNA methylation and its mechanisms in aging, age-related disease, and rejuvenation.
Basic Mechanisms
DNA methylation
The methylation reaction is catalyzed by DNMTs, enzymes that transfer a methyl group from S-adenosyl-l-methionine (SAM) to the C5 of a cytosine. This reaction features SAM as the electrophile methyl donor and C5 as a weak nucleophile unable to interact with SAM on its own. However, a nucleophile from a DNMT can attack the carbon-6 of cytosine, covalently binding the enzyme to the DNA. This activates the nucleophilic character of C5, facilitating the transfer of a methyl group from SAM. The enzyme nucleophile is consequently eliminated and deprotonation at C5 separates the nucleotide–DNMT complex.18
Silencing pathways
There are multiple routes to gene silencing via methylation, the most direct of which involves interfering with transcription factors or basal transcriptional machinery that interact with cytosines in the major groove of double helices. Because the majority of mammalian transcription factors possess DNA recognition elements containing CpG-rich motifs as well as GC-rich binding sites, DNA methylation can obstruct or eliminate their ability to act on many important regulatory sites. Alternatively, transcriptional machinery can be directly excluded from methylated promoter DNA by altering nucleosome stability or position.5
The effects of DNA methylation on genes are also mediated by members of the MBD protein family. To date, five major mammalian MBD proteins have been identified: methyl-CpG binding protein 2 (MeCP2), MBD1, MBD2, MBD3, and MBD4. These proteins, excluding MBD3, display a higher affinity for methylated DNA over unmethylated DNA.19 Although each MBD has been reported to enhance transcriptional repression of methylated CpGs,6 the mechanism by which MeCP2 does this is the best characterized: MeCP2 binds directly to co-repressor complexes, such as the Sin3, c-Ski, and N-CoR complexes, which then interact with histone deacetylases, recruiting them to methylated sites to establish silent chromatin.20,21
Demethylation and hydroxymethylation
Multiple mechanisms for active DNA demethylation have been proposed, including base excision repair, enzymatic removal of the methyl group, and oxidative demethylation of 5mC.22 The most compelling demethylating scheme involves the ten eleven translocation (TET) family, enzymes that iteratively oxidize 5mC to 5-hydroxymethylcytosine (5hmC), to 5-formylcytosine, and finally to 5-carboxylcytosine (5caC). 5caC has been theorized to be decarboxylated by an unknown decarboxylase, providing an elegant means of regenerating normal cytosine.23
Alternatively, 5caC nucleotides may be directly removed by thymine-DNA glycosylase (TDG). Multiple lines of evidence support this hypothesis: (1) Ablation of TDG produces abnormal DNA methylation patterns in mouse embryonic fibroblasts24; (2) ESC lysates lose glycosylase activity against 5caC in the absence of TDG; (3) overexpressing TDG in ESCs decreases the genomic content of 5caC and, conversely, lack of TDG results in genomic accumulation of 5caC.25
Hydroxymethylation by TET1 may also passively demethylate DNA by disrupting the ability of DNMTs and MBD proteins to access the previously nonhydroxylated 5mC. Whether the mechanism is passive or active, it is well established that TET1 binds to transcription start sites of CpG-rich promoters26 and promotes DNA demethylation.27 Recently, it was proposed that the demethylating effects of hydroxymethylation may work to oppose spontaneous hypermethylation. Given that promoter hypermethylation is often observed in both aging and cancer,28 we speculate that TET1 might be a novel protective factor in both senescence and age-related disease.
Regulation of DNMTs
Regulation of DNA methylation appears to be quite extensive because DNMT1 is targeted by multiple transcription factors and can be methylated, phosphorylated, acetylated, and ubiquitinated.29 The maintenance methyltransferase can also be modified by covalent attachment of small ubiquitin-like modifier (SUMO) proteins, a process referred to as SUMOylation.30 The effects of these modifications are site specific and dependent on the enzymatic players involved. For example, phosphorylation of Ser143 by AKT1 reportedly stabilizes DNMT1,31 whereas phosphorylation of Ser146 by the casein kinase 1δ/ε was found to decrease DNMT1's DNA binding affinity.32
DNMT activity is also regulated by polycomb group (PcG) proteins, which play vital roles in chromatin remodeling and in the maintenance of epigenetic memory.33 The PcG protein and histone methyltransferase EZH2 was shown to associate directly with DNMTs and to be necessary for methylating the DNA of EZH2-target promoters.34 A large overlap of genes bound by the PcG protein SUZ12 and TET1 in ESCs has also been found,28 suggesting that PcG proteins may play roles in both DNA methylation and demethylation.
Methylation and Aging
DNA hypomethylation and hypermethylation
The past few decades of research have uncovered a powerful link between DNA methylation and aging. A simple and elegant study conducted by Fraga et al. exemplifies this link quite nicely. By analyzing global and locus-specific differences in DNA methylation of monozygotic twins, the authors found that younger twins had indistinguishable methylomes, whereas older twins had remarkably different methylomes.35 A related study quantifying the methylation status of 27,578 CpG loci in both monozygotic twin and nontwin samples revealed 88 sites in or near 80 genes whose 5mC content changed significantly with age. Methylation of CpGs in the EDARADD, TOM1L1, and NPTX2 genes was especially correlated with age and, by building a regression model using two cytosines from these loci, the authors were able to predict the age of an individual with an average error of 5.2 years.36
All three of the aforementioned genes have been implicated in human pathologies. For instance, EDARADD mutations can slow wound healing37 and cause loss of teeth, hair, and sweat glands.38 NPTX2 is upregulated in Parkinson disease (PD)39 and in pancreatic cancer,40 whereas TOM1L1 expression is decreased in esophageal squamous cell carcinoma.41
These studies reveal an epigenetic drift with aging and suggest that DNA methylation is associated with longevity. While more speculative, these studies also hint that DNA methylation may play a role in regulating life span. A large amount of in vitro work strongly supports these suppositions. In cultured human embryonic lung fibroblasts, global hypomethylation occurs during both stress-induced and and replicative senescence.42 A separate study found that passage number and genomic 5mC content were inversely correlated for hamster, human, and mice diploid fibroblasts.43 In addition, treatment of human diploid fibroblasts with 5-azacytidine (5-azaC), a compound that inhibits methylation of cytosine in DNA, significantly reduces the doubling potential of cells in culture.44
A related paper reported that treatment of human umbilical cord blood-derived multipotent stem cells with 5-azaC or specific small interfering RNAs (siRNAs) against DNMT1 and DNMT3B induces cellular senescence and alters DNA methylation of various CpG islands.45 These data implicate actions by DNMTs in the maintenance of stem cell multipotency and self-renewal—a topic greatly relevant to aging and regeneration. Indeed, mouse hematopoietic stem cells with reduced Dnmt1 activity cannot suppress myeloerythroid regulators, disallowing them from differentiating into lymphoid progeny. This demonstrates that constitutive methylation is imperative for the renewal of hematopoietic stem cells.46 Demethylation is also thought to play a prominent role in hematopoietic differentiation.47 Of note, it is possible to generate pluripotent stem cells from adult somatic cells through the induction of defined factors. These cells exhibit pluiripotency, but they have subtle differences in their methylomes compared to ESCs. This suggests that proper epigenetic reprogramming may be required for the development of viable stem cell therapies.48
A plethora of in vivo studies have also divulged the relevance of DNA methylation in aging. Romanov and Vanyushin found that the thymus and heart tissue from cows experienced a decline in 5mC content with age.49 Genomic hypomethylation in various organs has also been observed in rats50 and mice.51 A study making use of pyrosequencing probed 217 nonpathologic human tissues from 10 anatomic sites, and analyzed DNA methylation of 1,413 autosomal CpG loci associated with 773 genes in both young and old subjects. The authors found strikingly significant CpG island-dependent connections between methylation and aging. Loci outside of CpG islands lost methylation with age whereas loci in CpG islands gained methylation with age.52 In normal human prostate tissue procured from 45 organ donors, hypermethylation was observed as a function of age for CpG islands in RARβ2, RASSF1A, GSTP1, NKX2-5, and ESR1 genes— all genes in which mutations are believed to be a risk factor for cancer.53
Bellizzi et al. analyzed the peripheral blood DNA from 318 humans of middle and advanced age, finding a significant correlation between global hypomethylation, frailty, and loss of function (as determined by a geriatrician). A 7-year follow-up disclosed that a significant decrease in 5mC content was associated with a worsening in health status.54 Bollati et al. assessed DNA methylation in two of the most characterized transposable elements, Alu and LINE-1. This study used blood DNA from elderly subjects (718 persons between 55 and 92 years old) that had been repeatedly evaluated over the course of 8 years. The authors found a negative correlation between Alu and LINE-1 methylation and age. By comparing blood samples from the same individual spread across 8 years, average Alu methylation was also reported to decrease over time.55 Given that transposable elements are thought to play prominent roles in age-related genomic instability,56 demethylation of said elements over time may contribute to human senescence.
Werner syndrome (WS) is a disorder characterized by growth retardation and premature aging. The WS gene WRN is mutated in patients with this syndrome and plays roles in telomere maintenance, DNA repair, and transcription.57 WRN has been shown to be aberrantly methylated in disease, such as advanced oral squamous cell carcinoma58; however, no studies have comprehensively investigated the methylome of WS patients. It would be interesting to know if these patients display atypical DNMT expression as well as atypical DNA methylation patterns compared to healthy controls.
Caloric restriction
Caloric restriction (CR), the decrease in nutrient intake above the level of starvation and below what an organism would consume ad libitum, is one of the most robust and consistent means of increasing life span across a spectrum of organisms.59,60 Both caloric intake and DNMT3A play a role in neuronal aging, with DNMT3A being essential for the formation of memory and synaptic and neuronal plasticity.61 A recent paper found that Dnmt3a-immunoreactivity increases with age in the CA3 and CA1-2 hippocampal regions of mice and that reducing caloric intake by 50% attenuates this increase.62 An age-related increase in 5mC occurs in these regions as well as the hippocampal dentate gyrus and is also attenuated by CR.63 Similarly, 5hmC content increases with age in all three of these regions, and CR opposes this age-related increase in the CA3 region, further implicating methylomics in hippocampal aging. It has yet to be ascertained, however, whether or not these changes in the methylomic landscape are responsible for mediating any health outputs of CR.64
DNMT modulation and longevity
DNA methylation in Drosophila melanogaster is carried out by the sole methyltransferase gene dDnmt2. Lin et al. ubiquitously expressed the UAS-dDnmt2 transgene driven by daughterless-GAL4 in Drosophila. Flies with this overexpressed gene (two- to four-fold increase as confirmed by RT-PCR) enjoyed a boost of up to 58% in mean life span compared to controls. In contrast, creation of flies with approximately 50% less dDnmt2 resulted in a 27% reduction in mean life span. Feeding Drosophila the free radical generator paraquat revealed that dDnmt2-overexpressing flies were more resistant to oxidative stress. Multiple small heat shock protein–encoding (sHsp) genes (Hsp22, Hsp23, and Hsp26) were also upregulated approximately three-fold in flies overexpressing dDnmt2. Conversely, sHsp-encoding genes were downregulated two- to three-fold in flies with diminished levels of dDnmt2.65 This study demonstrates that, in Drosophila, dDnmt2 is a regulator of life span and stress resistance. Further studies are warranted to determine if these changes in dDnmt2 expression correspond to changes in 5mC content.
In social insects such as honeybees, there exist different castes. Despite having identical genomes, female larvae diverge into two classes of bees—infertile worker bees and fertile queens. The queens are behaviorally dominant, physically larger, and substantially longer-lived compared to their worker sisters. The difference in development is thought to lie in differential feeding. Larvae fed a diet of royal jelly emerge as queen bees. This diet is thought to influence the methylome and, concordant with this, queens have significantly different 5mC content compared with workers.66 Interestingly, treating larvae with siRNAs directed against Dnmt3a engendered adult bees with queen characteristics, including fully developed ovaries. Knockdown of Dnmt3a was accompanied by decreased levels of 5mC content.67 While longevity has to our knowledge not been studied in this context, these studies suggest that differing levels of Dnmt expression and 5mC content may also account for the life span difference between the two castes.
Apart from a few in vivo studies in mice, fruit flies, and potentially honeybees, very little evidence attests to the ability of DNMTs to regulate organismal life span. Additional studies attempting to modulate longevity by modifying 5mC content and associated partners in model organisms are imperative to determine whether or not DNA methylation is simply correlated with aging or if it is indeed a modulator of aging.
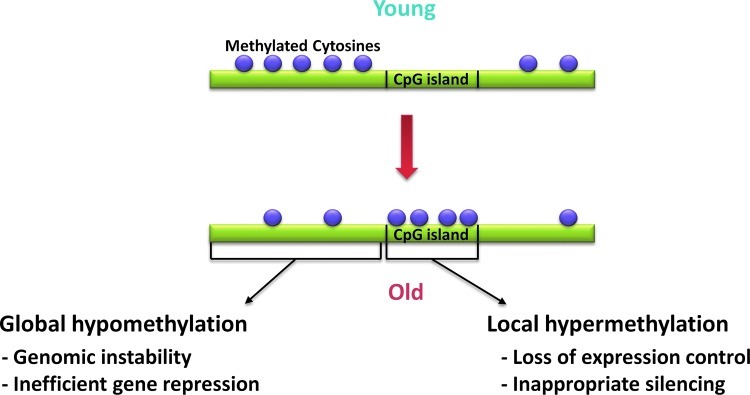
In sum, there is little doubt that DNA methylation levels and aging are strongly linked. The general trend seems to be the establishment of global hypomethylation and regions of CpG-island hypermethylation with age. Speculatively, this overall decrease in 5mC content could lead to less efficient gene regulation, and the CpG island hypermethylation could cause inappropriate silencing of specific genes. Such methylomic changes may render the genome unstable and contribute to aging phenotypes (Fig. 1). Additional studies are required to illuminate the finer details of epigenetic aging and the precise role DNA methylation plays in senescence and longevity.
FIG. 1.
Schematic representation of the changes in the methylome during aging. Purple circles are indicative of methylated CpG dinucleotides. Aging is often marked by the establishment of global hypomethylation and regions of CpG island hypermethylation. These changes in the epigenetic landscape may contribute to aging by adversely affecting genomic stability and gene regulation. (Color image is available at www.liebertpub.com/rej).
Methylation and Age-Related Disease
Aberrant methylation in cancer
Cancer is an age-related pathology that is also marked by global hypomethylation and promoter hypermethylation. Hypermethylation has been reported with age for numerous CpG islands, with affected genes including tumor-suppressor, growth/differentiation, metastasis/angiogenesis-related, DNA-repair/detoxification genes, and many others.68 RASSF1A, for example, is a tumor suppressor gene that mediates cell cycle control in healthy cells. Depletion of RASSF1A and hypermethylation of the RASSF1A promoter are common epigenetic alterations in renal cell carcinoma. RASSF1A becomes progressively methylated with age and increased adiposity— two known risk factors for renal cancer.69 Methylation of RASSF1A also correlates highly with breast cancer risk, atypical cytology, and benign breast disease requiring biopsy.70
Early studies of epigenetic regulation of ESR1 found that its gene expression is either absent or heavily diminished in colorectal tumors. This gene suppression was accompanied by significant hypermethylation compared to nontumor controls. Exogenously expressing the ESR1 gene in cultured colon carcinoma cells significantly suppressed growth, suggesting that ESR1 likely functions as a tumor suppressor. ESR1, which becomes hypermethylated with age in human colonic mucosa,71 exhibits further age-dependent methylation in atherosclerosis72 and prostate cancer.73
Liver cancer has also been linked to DNA methylation: Feeding rats a methyl-deficient diet is sufficient to induce hepatocarcinogenesis.74 This dietary methyl-deficiency is accompanied by decreased levels of SAM and genomic hypomethylation. Although refeeding of the complete diet restores liver SAM levels, it does little to correct the observed hypomethylation.75 Conversely, a methyl-balanced diet upregulates tumor suppressor genes such as RIZ1.76
Mutations in DNMT3A have been reported in approximately 20% of patients with acute myeloid leukemia (AML)77 and in 8% of patients with myelodysplastic syndromes (MDS),78 with DNMT3A mutations being associated with a less optimistic prognosis for both diseases. In this context, Gao et al. created a conditional mouse model with mutant K-ras–induced lung neoplasms. In this experimental system, mice lacked Dnmt3a solely in their tumors. The authors found that this conditional deficiency reduces life span and significantly promotes tumor growth and progression.79 The KRAS oncogene was chosen because hypermethylation of its promoter region is one of the most frequent genetic aberrations observed in lung cancer and can even be used as a screening marker for early tumor detection.80 These results indicate that DNMT3A, through silencing of cancer-promoting genes, may fulfill the role of a tumor suppressor.
Hydroxymethylation and cancer
AML is commonly marked by chromosomal abnormalities implicating the Mixed Lineage Leukemia (MLL) gene at 11q23. In a case of pediatric AML containing the chromosomal translocation t(10;11)(q22;q23), the 5mC hydroxylase TET1 was found to be the fusion partner of MLL. This observation was replicated in a separate leukemia patient, suggesting that TET1 may be the recurrent chromosomal target in AML patients harboring this translocation.81 Mutations or deletions in the related TET2 have been found in patients with AML, MDS, or myeloproliferative disorders. Of 320 bone marrow/blood samples analyzed from patients with myeloid disease, a somatic mutation rate of 15% was found for TET2.82 Concordant with this, 5hmC levels are severely decreased in cancerous breast, colon, and prostate compared to normal tissues.83 This decrease in 5hmC content could contribute to CpG island hypermethylation in cancer by allowing DNMTs to recognize additional cytosines.
Recent studies have also linked isocitrate dehyrogenases (IDHs) to both cancer and TET1. Cytoplasmic IDH1 and mitochondrial IDH2 are two genes that play vital roles in cellular respiration and are recurrently mutated in cancer. Mutant IDH1 and IDH2 demonstrate a neomorphic ability to convert α-ketoglutarate (α-KG) into 2-hydroxyglutarate, an oncometabolite capable of inhibiting TET hydroxymethylases. Because α-KG is thought to be required for the conversion of 5mC to 5hmC, this inhibition may work by decreasing the availability of the substrate α-KG.84 Given that elevated levels of α-KG are considered to be a risk factor for the formation of malignant tumors,85 it would be interesting to study the roles IDH1, IDH2, and α-KG play in mediating aberrant DNA methylation in cancers.
Aberrant methylation in noncancerous diseases
While data implicating DNA methylation in noncancerous diseases is more recent and tentative, numerous age-related pathologies have been linked to aberrant 5mC content. Alzheimer disease (AD), a neurological disorder that primarily affects the elderly, is exemplary of such changes. By using specific PCR assays, Silva et al. found that patients with AD had a higher methylation frequency of hTERT compared to elderly controls.86 Repetitive LINE-1 elements were also reported to be significantly hypermethylated in AD patients compared to healthy controls.87 AD patients have been further characterized by a decrease in brain SAM levels,88 and, concomitant with this, temporal neocortex neuronal nuclei were found to be hypomethylated in a patient with AD compared to his non-AD monozygotic twin.89
5hmC, which exists at high levels within the brain, has also been implicated in AD, inferring a potential involvement of demethylation with neurodegeneration.90 In accordance with this possibility, a single nucleotide polymorphism in the gene encoding TET1 (rs5030882) was reported to be associated with late-onset AD.91 Two DNMT3B promoter polymorphisms (rs2424913 and rs1569686) were also studied as candidate risk factors for late-onset AD, though no significant effect on disease age of onset was found for either one.92
Not all cases of abnormal methylation in disease are hypermethylation, however. For example, promoter-specific demethylation has been found in patients with PD. The major hallmark of PD is the presence of Lewy bodies, spherical masses composed of aggregated α-synuclein. In rare cases of PD, the neurodegenerative disease is caused by mutations in the SNCA gene, with duplication and triplication of the SCNA gene resulting in α-synuclein accumulation. Patients with triplet SCNA demonstrate a more severe phenotype and greater aggregation, suggesting that SCNA regulates disease severity. SCNA has two CpG islands, one located in the first exon and one located in the first intron (CpG-2). Recently, it was shown that CpG-2 is specifically demethylated in the presence of dopamine and this demethylation is associated with increased α-synuclein accumulation. CpG-2 was also found to be significantly demethylated in the substantia nigra in postmortem PD brains compared to healthy controls.93
Mutations in human DNMT1 were recently shown to cause both peripheral and central neurodegeneration. This neurodegeneration is one form of hereditary autonomic and sensory neuropathy that is accompanied by both hearing loss and dementia. The mutations causal for this disease also caused global hypomethylation, site-specific hypermethylation, premature degradation of mutant protein, and reduced methyltransferase activity.94
In osteoarthritis, there is a characteristic loss of articular cartilage that occurs as a function of age. The detailed mechanisms underlying this are not known, but it has been theorized that a decrease in essential growth factors may contribute to this wear and tear. Indeed, an approximately four-fold decline in the growth factor OP-1, a regulator of chondrocyte activity, has been observed in the cartilage of individuals between 35 and 75 years of age. Using tissue obtained from older adults, Loeser et al. isolated adult chondrocytes and found that OP-1 methylation and age are positively correlated with one another. In addition, inhibiting DNA methylation in cultured chondrocyte cells with 5-azaC increased OP-1 gene expression and OP-1 protein production.95
Further examples of implicated ailments include type 2 diabetes,96 renal disease,97 and undoubtedly many others. A complete listing of all genes reported to be abnormally methylated in age-related disease (both cancerous and noncancerous) is beyond the scope of this review; however, these examples serve to illustrate a point. Disease pathogenesis involves concomitant changes in DNMT and demethylase activity as well as 5mC and 5hmC content. Aberrant methylation is clearly a hallmark of disease and, at least in some cases, may even contribute to disease phenotypes. Akin to aging, whether or not aberrant methylation is causal for disease or simply a product of disease is enigmatic. Some studies suggest a more causal role, while others suggest differently. For example, oxidative stress can cause DNA lesions and induce genomic hypomethylation by interfering with the ability of DNMTs to bind cytosine.98 This raises the possibility that age-related hypomethylation may be the result of free radical–induced DNA damage. Future research attempting to alleviate or worsen disease symptoms through modulation of DNMTs or 5mC content will help address this debate.
Therapeutic Interventions
DNA methylation, with its purported roles in gene regulation, aging, and disease, is an appealing target for therapeutics. Interventions for DNA methylation currently exist and consist almost exclusively of DNMT inhibitors. Such classes of intervention include nucleoside analogues, nonnucleoside analogs, nutritional modifiers (methyl donors and bioactive components), antisense oligonucleotides, and siRNAs.
Nucleoside analogs
Nucleoside analog inhibitors, comprising of either ribonucleosides or deoxyribonucleosides, are incorporated into DNA, where they take the place of cytosine. These dissembling bases then bind and sequester DNMTs, effectively depleting DNMT activity. Some of the best-characterized DNMT-specific analog inhibitors include zebularine, 5-azaC, and decitabine.99 The latter two were approved by the Food and Drug Administration (FDA) in 2004 and 2006, respectively, and are in ongoing trials for a variety of cancers.100
Inspired by promising results from Phase I–II testing, a Phase III clinical study investigated the efficacy of 5-azaC in treating patients with MDS. Patients who received subcutaneous injections of 5-azaC fared significantly better compared to patients who received standard care alone. Observed improvements include delayed progression time to AML, increased survival rates, and enhanced hematologic responses.101 Decitabine has also been found to be clinically effective in patients with MDS. Results from a Phase III study demonstrate that patients receiving dosages of decitabine displayed hematologic improvement and a delayed onset of both death and AML. In addition to being well tolerated and having a manageable toxicity profile,101,102 both of these drugs demethylate and reactivate tumor suppressor genes in vitro.103,104
Although the use of nucleoside analog inhibitors has been very useful for patients with hematological cancers, only modest effects have been observed in patients with solid tumors. In non-MDS studies, however, these DNMT inhibitors were tested in late-stage patients. Due to the likely development of multidrug resistance and greater difficulty of treating advanced disease, the actual effectiveness of these drugs may have been masked.100
Nonnucleoside analogs
Nonnucleoside analog inhibitors do not require incorporation into DNA and include compounds such as procaine, procainamide, and hydralazine. Unlike the DNMT inhibitors 5-azaC and decitabine, the mechanisms of action of these nonnucleoside analogs are not well understood, and their ability to impact DNA methylation has yet to be established concretely.
Procaine is a FDA-approved anesthetic that was shown to demethylate DNA dose-dependently and exert growth-inhibitory effects in the MCF-7 breast cancer cell line. The drug also demethylated hypermethylated CpG islands and restored previously silenced expression of the RARβ2 tumor suppressor gene.105 Procainamide and hydralazine were also shown to demethylate and reactivate tumor suppressor genes in cultured cells.106 In addition, both drugs have been reported to inhibit DNMT activity in vitro.107,108
In a Phase I study involving patients with untreated cervical cancer, hydralazine was given to groups of patients at one of four doses. Hydralazine, which was well tolerated, demethylated and reactivated tumor suppressor genes without affecting global 5mC content.109 A Phase III study treated advanced cervical cancer patients with hydralazine and the histone deacetylase inhibitor valproate added to cisplatin topotecan. Drugs were administered a week before chemotherapy in either group and continually applied until signs of disease progression were observed. Patients treated with these compounds enjoyed significantly greater progression-free survival compared to those that received cisplatin topotecan and placebo alone.110
Methyl donors
Methyl donors are thought to play important roles in the maintenance of DNA methylation and are interconnected through one-carbon metabolism, a series of biological reactions that involve the transfer of a methyl group. Methyl donors (e.g., methionine and choline) as well as the crucial coenzymes in one-carbon metabolism (e.g., B-vitamins like folate) can all affect 5mC content by altering the concentrations of SAM or its demethylated product S-adenosylhomocysteine, a known inhibitor of DNMTs.111
Each of these methyl donors seems to be unique in its ability to impact organismal health. A 14-month choline-devoid diet, for example, induces large hepatocellular carcinomas and hepatic hypomethylation in rats.112 Mice fed a methionine-deficient diet, however, enjoy a longer life span and higher resistance to oxidative liver cell injury induced by acetaminophen.113 The restriction of methionine was reported to also boost longevity in Drosophila114 and rats115 and was shown to inhibit colon carcinogenesis.116 In addition, methionine restriction decreased oxidative damage to mitochondrial DNA and proteins as well as decreased the production of mitochondrial reaction oxygen species. These antideleterious properties were concomitant with decreases in genomic 5mC content.117 Whether these methyl donor–specific effects are significantly mediated through changes in DNA methylation or through other factors is currently unknown.
The effects of environmentally induced demethylation, such as injurious γ-radiation, can be mitigated by supplementation with methyl donors. Mice fed either a normal control diet or a methyl-supplemental diet for 2 weeks were exposed to γ-radiation. Control mice displayed global hypomethylation as well as a significant decrease in SAM levels and reduced DNMT activity postradiation. In methyl-supplemented mice, SAM levels and DNMT activity both increased and 5mC content was stably maintained. These data show that the dietary intake of methyl donors can play a pivotal role in maintaining DNA methylation in response to demethylating environmental influences.118
Bioactive components
Although their epigenetic roles are poorly understood, many nutrients have also been reported to influence DNA methylation. (−)-Epigallocatechin-3-gallate (EGCG), the major polyphenol of green tea, is known for its alleged anticarcinogenic, antidiabetic, and general health-conferring properties.119 Interestingly, EGCG treatment reduces 5mC content of cancer cell lines in a dose-dependent manner. This demethylation is reportedly accompanied by a decrease in DNMT protein levels and the reactivation of the p16, p21, RARβ, and hMLH1 tumor suppressor genes.119,120
Similar results have been obtained by treating cells with genistein, the major isoflavone present in soybean. In KYSE 510 cells, genistein repressed cell growth and dose-dependently inhibited DNMT activity. The isoflavone also demethylated and reactivated expression of the RARβ, p16, and MGMT genes.121 This is intriguing, because dietary genistein was shown to protect rats from chemically induced mammary tumors in a dose-dependent manner. This conferred protection was highly significant, with rats that received the lowest or highest dosages of genistein developing 20% or 50% less tumors, respectively.122
Further work is required to determine if any of the health benefits conferred by ECGC, genistein, or other bioactive components work through changes in DNA methylation.
Further interventions
Further avenues leading to methylome-modification include antisense DNMT oligodeoxynucleotides and siRNA. In vitro studies have demonstrated that the DNMT inhibitor and oligodeoxynucleotide MG98 is capable of reactivating tumor suppressor genes.123 A Phase I study treated advanced solid tumor patients with MG98 and found that DNMT1 expression was suppressed in 26 out of 32 patients analyzed. While one patient achieved protracted disease stabilization and another had a partial response, this124 and a Phase II study failed to find a consistent response to MG98 in most patients. In addition, the latter study failed to observe any conclusive pattern of reduced DNMT activity.125 The clinical efficacy of MG98 as well as its ability to suppress DNMT activity are controversial and require further study.
In cell culture models, siRNA directed against DNMT1 and DNMT3B led to partial demethylation from numerous methylated loci, including the RASSF1A, HIN-1, and TWIST promoter regions. Double knockdown against both of these DNMTs produced a 7- to 15-fold increase in expression for all three of these genes.126 Use of siRNAs against DNMT1 and DNMT3B in stem cells decreased CpG island methylation, increased expression of the p21 and p16 genes, and induced cellular senescence.45 Outside of in vitro work, no clinical data currently exists attesting to the potential of siRNA knockdown in methylome therapeutics.
Exosomes, which are endogenous nanovesicles that ferry proteins and RNAs, can be targeted to deliver siRNA to mice in a tissue- and cell type-specific manner. This delivery method resulted in efficient knockdown of neuronal mRNA and protein expression of the AD-relevant BACE1.127 RNA interference can produce toxic and off-target effects, but the use of library selection screens to develop highly specific siRNAs effective at low concentrations has the potential to rectify these problems.128 Multiple clinical trials (Phases I–III) are currently in progress using RNA interference to treat diseases such as age-related macular degeneration, diabetic macular edema, and solid tumors.129
A few of these 5mC-modifying therapeutics show clinical promise with regard to treating human disease (Table 1), however their mechanisms of action are, for the most part, poorly understood. Given that age-related diseases are marked by changes in DNA methylation that are both sequence- and tissue-specific, greater specificity may be required to make these treatments more useful. Recent advances in bioinformatics and biomedical research allow for high-throughput survey of DNA–protein interactions as well as methylome analysis. These techniques allow scientists to quickly gain biological insights from vast amounts of data130 and demonstrate promise for the development of new drugs with greater specificity and utility.
Table 1.
Clinically Relevant Drugs Reported to Influence DNA Methylaiton
| Drug | Therapeutic Class | Clinical Data | Reference(s) |
|---|---|---|---|
| Azacitidine Decitabine | Nucleoside analogs | Delayed progression time to AML and enhanced hematologic activity and survival in patients with MDS | 101, 102 |
| Hydralazine | Nonnucleoside analogs | Demethylated and reactivated tumor suppressor genes and, when used with valproate, improved progression-free survival in patients with cervical cancer | 109, 110 |
| MG98 | Antisense oligonucleotides | Suppressed DNMT1 expression and exhibited some clinical activity in patients with advanced solid malignances | 124 |
AML, Acute myeloid leukemia; DNMT, DNA methyltransferase; MDS, myelodysplastic syndromes.
Concluding Remarks
Multiple targets have been proposed to be promising in the effort to extend human health span. Such targets include telomeres,131 free radicals,132 DNA repair machinery,133 mitochondrial enzymes,134 and many others. This review, drawing upon all of the data mentioned in the previous discussion, asserts that DNA methylation is also a promising target for age-related ailments and rejuvenation.
Despite decades of research, one fundamental question remains to be answered: Do changing DNA methylation levels contribute to aging and age-related disease or are they simply a correlative product of other factors? It is well established that aberrant 5mC content, particularly global hypomethylation and CpG island hypermethylation, are hallmarks of both aged and cancerous methylomes. Although they are not as thoroughly characterized, atypical methylomic patterns have also been reported in patients with AD, PD, osteoarthritis, and other age-related pathologies. All of these studies clearly demonstrate a correlative link between DNA methylation, aging, and age-related disease.
Other studies suggest that DNA methylation and associated partners (e.g., DNMTs) are regulators of life span and health span. The most exciting of these demonstrates that overexpression of dDnmt2 significantly increases mean and maximum life span in Drosophila. Conversely, reducing dDnmt2 expression substantially accelerates fruit fly aging.65 In mice, neuronal deletion of Dnmt3a reduces life span9 and deletion of Dnmt3a in lung tumors decreases murine survival.79 Feeding rats a methyl-deficient diet induces hepatocarcinogenesis,74 and treatment of MDS patients with 5-azaC or decitabine delays progression time to AML and boosts survival rates.101,102 Mutations in TET2 are relatively common in patients with myeloid disease,82 and cellular senescence can be induced through inhibition of DNMTs.44,45 Taken together, these data demonstrate great promise for 5mC-targeting drugs in rejuvenation.
Despite these data, the question of whether changes in methylation are downstream effects of aging and pathology or actually one of the causative factors has not been conclusively answered yet. It is evident that a broad range of age-related diseases show aberrant methylation and many potential treatments based on rejuvenating the methylome remain unexplored or undeveloped. Future research is imperative to explore this potential properly and will require a greater understanding of putative mechanisms surrounding DNMTs and their associated partners. In sum, further research into methylomic aging, disease, drug development, and regulatory mechanisms is required to determine the true role of DNA methylation in aging, rejuvenation, and age-related disease.
Acknowledgments
The authors thank Daniel Kimbel and Bryan Vukorepa for collaborative interactions and helpful discussions. J.P.M. is grateful to the BBSRC, the Wellcome Trust, the Ellison Medical Foundation, and a Marie Curie International Reintegration Grant within EC-FP7 for supporting work in his laboratory.
Author Disclosure Statement
The authors declare that they have no conflict of interest.
References
- 1.Morgan HD. Santos F. Green K. Dean W. Reik W. Epigenetic reprogramming in mammals. Hum Mol Genet. 2005;14(Spec No 1):R47–R58. doi: 10.1093/hmg/ddi114. [DOI] [PubMed] [Google Scholar]
- 2.Okano M. Bell DW. Haber DA. Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 3.Casadesús J. Low D. Epigenetic gene regulation in the bacterial world. Microbiol Mol Biol Rev. 2006;70:830–856. doi: 10.1128/MMBR.00016-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zemach A. McDaniel IE. Silva P. Zilberman D. Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science. 2010;328:916–919. doi: 10.1126/science.1186366. [DOI] [PubMed] [Google Scholar]
- 5.Bird AP. Wolffe AP. Methylation-induced repression—belts, braces, and chromatin. Cell. 1999;99:451–454. doi: 10.1016/s0092-8674(00)81532-9. [DOI] [PubMed] [Google Scholar]
- 6.Bogdanović O. Veenstra GJ. DNA methylation and methyl-CpG binding proteins: Developmental requirements and function. Chromosoma. 2009;118:549–565. doi: 10.1007/s00412-009-0221-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gonzalo S. Jaco I. Fraga MF. Chen T. Li E. Esteller M. Blasco MA. DNA methyltransferases control telomere length and telomere recombination in mammalian cells. Nat Cell Biol. 2006;8:416–424. doi: 10.1038/ncb1386. [DOI] [PubMed] [Google Scholar]
- 8.Li E. Bestor TH. Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- 9.Nguyen S. Meletis K. Fu D. Jhaveri S. Jaenisch R. Ablation of de novo DNA methyltransferase Dnmt3a in the nervous system leads to neuromuscular defects and shortened lifespan. Dev Dyn. 2007;236:1663–1676. doi: 10.1002/dvdy.21176. [DOI] [PubMed] [Google Scholar]
- 10.Ferguson-Smith AC. Genomic imprinting: The emergence of an epigenetic paradigm. Nat Rev Genet. 2011;12:565–575. doi: 10.1038/nrg3032. [DOI] [PubMed] [Google Scholar]
- 11.Basu R. Zhang LF. X chromosome inactivation: A silence that needs to be broken. Genesis. 2011;49:821–834. doi: 10.1002/dvg.20792. [DOI] [PubMed] [Google Scholar]
- 12.Sharp AJ. Stathaki E. Migliavacca E. Brahmachary M. Montgomery SB. Dupre Y. Antonarakis SE. DNA methylation profiles of human active and inactive X chromosomes. Genome Res. 2011;21:1592–1600. doi: 10.1101/gr.112680.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Richardson B. DNA methylation and autoimmune disease. Clin Immunol. 2003;109:72–79. doi: 10.1016/s1521-6616(03)00206-7. [DOI] [PubMed] [Google Scholar]
- 14.Issa JP. Age-related epigenetic changes and the immune system. Clin Immunol. 2003;109:103–108. doi: 10.1016/s1521-6616(03)00203-1. [DOI] [PubMed] [Google Scholar]
- 15.Sedivy JM. Banumathy G. Adams PD. Aging by epigenetics—a consequence of chromatin damage? Exp Cell Res. 2008;314:1909–1917. doi: 10.1016/j.yexcr.2008.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fraga MF. Agrelo R. Esteller M. Cross-talk between aging and cancer: The epigenetic language. Ann NY Acad Sci. 2007;1100:60–74. doi: 10.1196/annals.1395.005. [DOI] [PubMed] [Google Scholar]
- 17.Kirkwood TB. Understanding the odd science of aging. Cell. 2005;120:437–447. doi: 10.1016/j.cell.2005.01.027. [DOI] [PubMed] [Google Scholar]
- 18.Zangi R. Arrieta A. Cossío FP. Mechanism of DNA methylation: The double role of DNA as a substrate and as a cofactor. J Mol Biol. 2010;400:632–644. doi: 10.1016/j.jmb.2010.05.021. [DOI] [PubMed] [Google Scholar]
- 19.Fraga MF. Ballestar E. Montoya G. Taysavang P. Wade PA. Esteller M. The affinity of different MBD proteins for a specific methylated locus depends on their intrinsic binding properties. Nucleic Acids Res. 2003;31:1765–1774. doi: 10.1093/nar/gkg249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jones PL. Veenstra GJ. Wade PA. Vermaak D. Kass SU. Landsberger N. Strouboulis J. Wolffe AP. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat Genet. 1998;19:187–191. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- 21.Kokura K. Kaul SC. Wadhwa R. Nomura T. Khan MM. Shinagawa T. Yasukawa T. Colmenares C. Ishii S. The Ski protein family is required for MeCP2-mediated transcriptional repression. J Biol Chem. 2001;276:34115–34121. doi: 10.1074/jbc.M105747200. [DOI] [PubMed] [Google Scholar]
- 22.Wu SC. Zhang Y. Active DNA demethylation: Many roads lead to Rome. Nat Rev Mol Cell Biol. 2010;11:607–620. doi: 10.1038/nrm2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ito S. Shen L. Dai Q. Wu SC. Collins LB. Swenberg JA. He C. Zhang Y. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cortázar D. Kunz C. Selfridge J. Lettieri T. Saito Y. MacDougall E. Wirz A. Schuermann D. Jacobs AL. Siegrist F. Steinacher R. Jiricny J. Bird A. Schär P. Embryonic lethal phenotype reveals a function of TDG in maintaining epigenetic stability. Nature. 2011;470:419–423. doi: 10.1038/nature09672. [DOI] [PubMed] [Google Scholar]
- 25.He YF. Li BZ. Li Z. Liu P. Wang Y. Tang Q. Ding J. Jia Y. Chen Z. Li L. Sun Y. Li X. Dai Q. Song CX. Zhang K. He C. Xu GL. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333:1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu Y. Wu F. Tan L. Kong L. Xiong L. Deng J. Barbera AJ. Zheng L. Zhang H. Huang S. Min J. Nicholson T. Chen T. Xu G. Shi Y. Zhang K. Shi YG. Genome-wide regulation of 5hmC, 5mC, and gene expression by Tet1 hydroxylase in mouse embryonic stem cells. Mol Cell. 2011;42:451–464. doi: 10.1016/j.molcel.2011.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guo JU. Su Y. Zhong C. Ming GL. Song H. Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell. 2011;145:423–434. doi: 10.1016/j.cell.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Williams K. Christensen J. Pedersen MT. Johansen JV. Cloos PA. Rappsilber J. Helin K. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature. 2011;473:343–348. doi: 10.1038/nature10066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qin W. Leonhardt H. Pichler G. Regulation of DNA methyltransferase 1 by interactions and modifications. Nucleus. 2011;2:392–402. doi: 10.4161/nucl.2.5.17928. [DOI] [PubMed] [Google Scholar]
- 30.Lee B. Muller MT. SUMOylation enhances DNA methyltransferase 1 activity. Biochem J. 2009;421:449–461. doi: 10.1042/BJ20090142. [DOI] [PubMed] [Google Scholar]
- 31.Estève PO. Chang Y. Samaranayake M. Upadhyay AK. Horton JR. Feehery GR. Cheng X. Pradhan S. A methylation and phosphorylation switch between an adjacent lysine and serine determines human DNMT1 stability. Nat Struct Mol Biol. 2011;18:42–48. doi: 10.1038/nsmb.1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sugiyama Y. Hatano N. Sueyoshi N. Suetake I. Tajima S. Kinoshita E. Kinoshita-Kikuta E. Koike T. Kameshita I. The DNA-binding activity of mouse DNA methyltransferase 1 is regulated by phosphorylation with casein kinase 1delta/epsilon. Biochem J. 2010;427:489–497. doi: 10.1042/BJ20091856. [DOI] [PubMed] [Google Scholar]
- 33.Lund AH. van Lohuizen M. Polycomb complexes and silencing mechanisms. Curr Opin Cell Biol. 2004;16:239–246. doi: 10.1016/j.ceb.2004.03.010. [DOI] [PubMed] [Google Scholar]
- 34.Viré E. Brenner C. Deplus R. Blanchon L. Fraga M. Didelot C. Morey L. Van Eynde A. Bernard D. Vanderwinden JM. Bollen M. Esteller M. Di Croce L. de Launoit Y. Fuks F. The Polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;439:871–874. doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]
- 35.Fraga MF. Ballestar E. Paz MF. Ropero S. Setien F. Ballestar ML. Heine-Suñer D. Cigudosa JC. Urioste M. Benitez J. Boix-Chornet M. Sanchez-Aguilera A. Ling C. Carlsson E. Poulsen P. Vaag A. Stephan Z. Spector TD. Wu Y-Z. Plass C. Esteller M. Epigenetic differences arise during the lifetime of monozygotic twins. Proc Natl Acad Sci USA. 2005;102:10604–10609. doi: 10.1073/pnas.0500398102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bocklandt S. Lin W. Sehl ME. Sánchez FJ. Sinsheimer JS. Horvath S. Vilain E. Epigenetic predictor of age. PLoS One. 2011;6:e14821. doi: 10.1371/journal.pone.0014821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Langton AK. Herrick SE. Headon DJ. An extended epidermal response heals cutaneous wounds in the absence of a hair follicle stem cell contribution. J Invest Dermatol. 2008;128:1311–1318. doi: 10.1038/sj.jid.5701178. [DOI] [PubMed] [Google Scholar]
- 38.Yan M. Zhang Z. Brady JR. Schilbach S. Fairbrother WJ. Dixit VM. Identification of a novel death domain-containing adaptor molecule for ectodysplasin-A receptor that is mutated in crinkled mice. Curr Biol. 2002;12:409–413. doi: 10.1016/s0960-9822(02)00687-5. [DOI] [PubMed] [Google Scholar]
- 39.Moran LB. Hickey L. Michael GJ. Derkacs M. Christian LM. Kalaitzakis ME. Pearce RK. Graeber MB. Neuronal pentraxin II is highly upregulated in Parkinson's disease and a novel component of Lewy bodies. Acta Neuropathol. 2008;115:471–478. doi: 10.1007/s00401-007-0309-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Park JK. Ryu JK. Lee KH. Lee JK. Yoon WJ. Lee SH. Yoo JW. Woo SM. Lee GY. Lee CH. Kim YT. Yoon YB. Quantitative analysis of NPTX2 hypermethylation is a promising molecular diagnostic marker for pancreatic cancer. Pancreas. 2007;35:e9–e15. doi: 10.1097/MPA.0b013e318153fa42. [DOI] [PubMed] [Google Scholar]
- 41.Qi Y. Li X. Zhao L. Seykora JT. Decreased Srcasm expression in esophageal squamous cell carcinoma in a Chinese population. Anticancer Res. 2010;30:3535–3539. [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang W. Ji W. Yang J. Yang L. Chen W. Zhuang Z. Comparison of global DNA methylation profiles in replicative versus premature senescence. Life Sci. 2008;83:475–480. doi: 10.1016/j.lfs.2008.07.015. [DOI] [PubMed] [Google Scholar]
- 43.Wilson VL. Jones PA. DNA methylation decreases in aging but not in immortal cells. Science. 1983;220:1055–1057. doi: 10.1126/science.6844925. [DOI] [PubMed] [Google Scholar]
- 44.Holliday R. Strong effects of 5-azacytidine on the in vitro lifespan of human diploid fibroblasts. Exp Cell Res. 1986;166:543–552. doi: 10.1016/0014-4827(86)90499-4. [DOI] [PubMed] [Google Scholar]
- 45.So AY. Jung JW. Lee S. Kim HS. Kang KS. DNA methyltransferase controls stem cell aging by regulating BMI1 and EZH2 through microRNAs. PLoS One. 2011;6:e19503. doi: 10.1371/journal.pone.0019503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bröske AM. Vockentanz L. Kharazi S. Huska MR. Mancini E. Scheller M. Kuhl C. Enns A. Prinz M. Jaenisch R. Nerlov C. Leutz A. Andrade-Navarro MA. Jacobsen SE. Rosenbauer F. DNA methylation protects hematopoietic stem cell multipotency from myeloerythroid restriction. Nat Genet. 2009;41:1207–1215. doi: 10.1038/ng.463. [DOI] [PubMed] [Google Scholar]
- 47.Calvanese V. Fernández AF. Urdinguio RG. Suárez-Alvarez B. Mangas C. Pérez-García V. Bueno C. Montes R. Ramos-Mejía V. Martínez-Camblor P. Ferrero C. Assenov Y. Bock C. Menendez P. Carrera AC. Lopez-Larrea C. Fraga MF. A promoter DNA demethylation landscape of human hematopoietic differentiation. Nucleic Acids Res. 2011;40:116–131. doi: 10.1093/nar/gkr685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Okita K. Ichisaka T. Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313–317. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- 49.Romanov GA. Vanyushin BF. Methylation of reiterated sequences in mammalian DNAs. Effects of the tissue type, age, malignancy and hormonal induction. Biochim Biophys Acta. 1981;653:204–218. doi: 10.1016/0005-2787(81)90156-8. [DOI] [PubMed] [Google Scholar]
- 50.Vanyushin BF. Nemirovsky LE. Klimenko VV. Vasiliev VK. Belozersky AN. The 5-methylcytosine in DNA of rats. Tissue and age specificity and the changes induced by hydrocortisone and other agents. Gerontologia. 1973;19:138–152. [PubMed] [Google Scholar]
- 51.Singhal RP. Mays-Hoopes LL. Eichhorn GL. DNA methylation in aging of mice. Mech Ageing Dev. 1987;41:199–210. doi: 10.1016/0047-6374(87)90040-6. [DOI] [PubMed] [Google Scholar]
- 52.Christensen BC. Houseman EA. Marsit CJ. Zheng S. Wrensch MR. Wiemels JL. Nelson HH. Karagas MR. Padbury JF. Bueno R. Sugarbaker DJ. Yeh R-F. Wiencke JK. Kelsey KT. Aging and environmental exposures alter tissue-specific DNA methylation dependent upon CpG island context. PLoS Genet. 2009;5:e1000602. doi: 10.1371/journal.pgen.1000602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kwabi-Addo B. Chung W. Shen L. Ittmann M. Wheeler T. Jelinek J. Issa JP. Age-related DNA methylation changes in normal human prostate tissues. Clin Cancer Res. 2007;13:3796–3802. doi: 10.1158/1078-0432.CCR-07-0085. [DOI] [PubMed] [Google Scholar]
- 54.Bellizzi D. D'Aquila P. Montesanto A. Corsonello A. Mari V. Mazzei B. Lattanzio F. Passarino G. Global DNA methylation in old subjects is correlated with frailty. Age (Dordr) 2012;34:169–179. doi: 10.1007/s11357-011-9216-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bollati V. Schwartz J. Wright R. Litonjua L. Tarantini L. Su H. Sparrow D. Vokonas P. Baccarelli A. Decline in genomic DNA methylation through aging in a cohort of elderly subjects. Mech Ageing Dev. 2009;130:234–239. doi: 10.1016/j.mad.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nikitin AG. Shmookler Reis RJ. Role of transposable elements in age-related genomic instability. Genet Res. 1997;69:183–195. doi: 10.1017/s0016672397002772. [DOI] [PubMed] [Google Scholar]
- 57.Lutomska A. Lebedev A. Scharffetter-Kochanek K. Iben S. The transcriptional response to distinct growth factors is impaired in Werner syndrome cells. Exp Gerontol. 2008;43:820–826. doi: 10.1016/j.exger.2008.06.007. [DOI] [PubMed] [Google Scholar]
- 58.Supić G. Kozomara R. Branković-Magić M. Jović N. Magić Z. Gene hypermethylation in tumor tissue of advanced oral squamous cell carcinoma patients. Oral Oncol. 2009;45:1051–1057. doi: 10.1016/j.oraloncology.2009.07.007. [DOI] [PubMed] [Google Scholar]
- 59.de Magalhães JP. Wuttke D. Wood SH. Plank M. Vora C. Genome-environment interactions that modulate aging: Powerful targets for drug discovery. Pharmacol Rev. 2012;64:88–101. doi: 10.1124/pr.110.004499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Joy TK. Arik AJ. Corby-Harris V. Johnson AA. Riehle MA. The impact of larval and adult dietary restriction on lifespan, reproduction and growth in the mosquito Aedes aegypti. Exp Gerontol. 2010;45:685–690. doi: 10.1016/j.exger.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Miller CA. Sweatt JD. Covalent modification of DNA regulates memory formation. Neuron. 2007;53:857–869. doi: 10.1016/j.neuron.2007.02.022. [DOI] [PubMed] [Google Scholar]
- 62.Chouliaras L. van den Hove DL. Kenis G. Dela Cruz J. Lemmens MA. van Os J. Steinbusch HW. Schmitz C. Rutten BP. Caloric restriction attenuates age-related changes of DNA methyltransferase 3a in mouse hippocampus. Brain Behav Immun. 2011;25:616–623. doi: 10.1016/j.bbi.2010.11.016. [DOI] [PubMed] [Google Scholar]
- 63.Chouliaras L. van den Hove DL. Kenis G. Keitel S. Hof PR. van Os J. Steinbusch HW. Schmitz C. Rutten BP. Prevention of age-related changes in hippocampal levels of 5-methylcytidine by caloric restriction. Neurobiol Aging. 2012;33:1672–1681. doi: 10.1016/j.neurobiolaging.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chouliaras L. van den Hove DL. Kenis G. Keitel S. Hof PR. van Os J. Steinbusch HW. Schmitz C. Rutten BP. Age-related increase in levels of 5-hydroxymethylcytosine in mouse hippocampus is prevented by caloric restriction. Curr Alzheimer Res. 2012;9:536–544. doi: 10.2174/156720512800618035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lin MJ. Tang LY. Reddy MN. Shen CK. DNA methyltransferase gene dDnmt2 and longevity of Drosophila. J Biol Chem. 2005;280:861–864. doi: 10.1074/jbc.C400477200. [DOI] [PubMed] [Google Scholar]
- 66.Lyko F. Foret S. Kucharski R. Wolf S. Falckenhayn C. Maleszka R. The honey bee epigenomes: differential methylation of brain DNA in queens and workers. PLoS Biol. 2010;8:e1000506. doi: 10.1371/journal.pbio.1000506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kucharski R. Maleszka J. Foret S. Maleszka R. Nutritional control of reproductive status in honeybees via DNA methylation. Science. 2008;319:1827–1830. doi: 10.1126/science.1153069. [DOI] [PubMed] [Google Scholar]
- 68.Ahuja N. Issa JP. Aging, methylation and cancer. Histol Histopathol. 2000;15:835–842. doi: 10.14670/HH-15.835. [DOI] [PubMed] [Google Scholar]
- 69.Peters I. Vaske B. Albrecht K. Kuczyk MA. Jonas U. Serth J. Adiposity and age are statistically related to enhanced RASSF1A tumor suppressor gene promoter methylation in normal autopsy kidney tissue. Cancer Epidemiol Biomarkers Prev. 2007;16:2526–2532. doi: 10.1158/1055-9965.EPI-07-0203. [DOI] [PubMed] [Google Scholar]
- 70.Euhus DM. Bu D. Milchgrub S. Xie XJ. Bian A. Leitch AM. Lewis CM. DNA methylation in benign breast epithelium in relation to age and breast cancer risk. Cancer Epidemiol Biomarkers Prev. 2008;17:1051–1059. doi: 10.1158/1055-9965.EPI-07-2582. [DOI] [PubMed] [Google Scholar]
- 71.Issa JP. Ottaviano YL. Celano P. Hamilton SR. Davidson NE. Baylin SB. Methylation of the oestrogen receptor CpG island links ageing and neoplasia in human colon. Nat Genet. 1994;7:536–540. doi: 10.1038/ng0894-536. [DOI] [PubMed] [Google Scholar]
- 72.Post WS. Goldschmidt-Clermont PJ. Wilhide CC. Heldman AW. Sussman MS. Ouyang P. Milliken EE. Issa JP. Methylation of the estrogen receptor gene is associated with aging and atherosclerosis in the cardiovascular system. Cardiovasc Res. 1999;43:985–991. doi: 10.1016/s0008-6363(99)00153-4. [DOI] [PubMed] [Google Scholar]
- 73.Li LC. Shiina H. Deguchi M. Zhao H. Okino ST. Kane CJ. Carroll PR. Igawa M. Dahiya R. Age-dependent methylation of ESR1 gene in prostate cancer. Biochem Biophys Res Commun. 2004;321:455–461. doi: 10.1016/j.bbrc.2004.06.164. [DOI] [PubMed] [Google Scholar]
- 74.Mikol YB. Hoover KL. Creasia D. Poirier LA. Hepatocarcinogenesis in rats fed methyl-deficient, amino acid-defined diets. Carcinogenesis. 1983;4:1619–1629. doi: 10.1093/carcin/4.12.1619. [DOI] [PubMed] [Google Scholar]
- 75.Pogribny IP. Ross SA. Wise C. Pogribna M. Jones EA. Tryndyak VP. James SJ. Dragan YP. Poirier LA. Irreversible global DNA hypomethylation as a key step in hepatocarcinogenesis induced by dietary methyl deficiency. Mutat Res. 2006;593:80–87. doi: 10.1016/j.mrfmmm.2005.06.028. [DOI] [PubMed] [Google Scholar]
- 76.Zhou W. Alonso S. Takai D. Lu SC. Yamamoto F. Perucho M. Huang S. Requirement of RIZ1 for cancer prevention by methyl-balanced diet. PLoS One. 2008;3:e3390. doi: 10.1371/journal.pone.0003390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shah MY. Licht JD. DNMT3A mutations in acute myeloid leukemia. Nat Genet. 2011;43:289–290. doi: 10.1038/ng0411-289. [DOI] [PubMed] [Google Scholar]
- 78.Walter MJ. Ding L. Shen D. Shao J. Grillot M. McLellan M. Fulton R. Schmidt H. Kalicki-Veizer J. O'Laughlin M. Kandoth C. Baty J. Westervelt P. DiPersio JF. Mardis ER. Wilson RK. Ley TJ. Graubert TA. Recurrent DNMT3A mutations in patients with myelodysplastic syndromes. Leukemia. 2011;25:1153–1158. doi: 10.1038/leu.2011.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gao Q. Steine EJ. Barrasa MI. Hockemeyer D. Pawlak M. Fu D. Reddy S. Bell GW. Jaenisch R. Deletion of the de novo DNA methyltransferase Dnmt3a promotes lung tumor progression. Proc Natl Acad Sci USA. 2011;108:18061–18066. doi: 10.1073/pnas.1114946108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Belinsky SA. Gene-promoter hypermethylation as a biomarker in lung cancer. Nat Rev Cancer. 2004;4:707–717. doi: 10.1038/nrc1432. [DOI] [PubMed] [Google Scholar]
- 81.Lorsbach RB. Moore J. Mathew S. Raimondi SC. Mukatira ST. Downing JR. TET1, a member of a novel protein family, is fused to MLL in acute myeloid leukemia containing the t(10;11)(q22;q23) Leukemia. 2003;17:637–641. doi: 10.1038/sj.leu.2402834. [DOI] [PubMed] [Google Scholar]
- 82.Delhommeau F. Dupont S. Della Valle V. James C. Trannoy S. Massé A. Kosmider O. Le Couedic JP. Robert F. Alberdi A. Lécluse Y. Plo I. Dreyfus FJ. Marzac C. Casadevall N. Lacombe C. Romana SP. Dessen P. Soulier J. Viguié F. Fontenay M. Vainchenker W. Bernard OA. Mutations in TET2 in myeloid cancers. N Engl J Med. 2009;360:2289–2301. doi: 10.1056/NEJMoa0810069. [DOI] [PubMed] [Google Scholar]
- 83.Haffner MC. Chaux A. Meeker AK. Esopi DM. Gerber J. Pellakuru LG. Toubaji A. Argani P. Iacobuzio-Donahue C. Nelson WG. Netto GJ. De Marzo AM. Yegnasubramanian S. Global 5-hydroxymethylcytosine content is significantly reduced in tissue stem/progenitor cell compartments and in human cancers. Oncotarget. 2011;2:627–637. doi: 10.18632/oncotarget.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dang L. White DW. Gross S. Bennett BD. Bittinger MA. Driggers EM. Fantin VR. Jang HG. Jin S. Keenan MC. Marks KM. Prins RM. Ward PS. Yen KE. Liau LM. Rabinowitz JD. Cantley LC. Thompson CB. Vander Heiden MG. Su SM. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462:739–744. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Prensner JR. Chinnaiyan AM. Metabolism unhinged: IDH mutations in cancer. Nat Med. 2011;17:291–293. doi: 10.1038/nm0311-291. [DOI] [PubMed] [Google Scholar]
- 86.Silva PN. Gigek CO. Leal MF. Bertolucci PH. de Labio RW. Payão SL. Smith Me A. Promoter methylation analysis of SIRT3, SMARCA5, HTERT and CDH1 genes in aging and Alzheimer's disease. J Alzheimers Dis. 2008;13:173–176. doi: 10.3233/jad-2008-13207. [DOI] [PubMed] [Google Scholar]
- 87.Bollati V. Galimberti D. Pergoli L. Dalla Valle E. Barretta F. Cortini F. Scarpini E. Bertazzi PA. Baccarelli A. DNA methylation in repetitive elements and Alzheimer disease. Brain Behav Immun. 2011;25:1078–1083. doi: 10.1016/j.bbi.2011.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Morrison LD. Smith DD. Kish SJ. Brain S-adenosylmethionine levels are severely decreased in Alzheimer's disease. J Neurochem. 1996;67:1328–1331. doi: 10.1046/j.1471-4159.1996.67031328.x. [DOI] [PubMed] [Google Scholar]
- 89.Mastroeni D. McKee A. Grover A. Rogers J. Coleman PD. Epigenetic differences in cortical neurons from a pair of monozygotic twins discordant for Alzheimer's disease. PLoS One. 2009;4:e6617. doi: 10.1371/journal.pone.0006617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.van den Hove DL. Chouliaras L. Rutten BP. The role of 5-hydroxymethylcytosine in aging and Alzheimer's disease: Current status and prospects for future studies. Curr Alzheimer Res. 2012;9:545–549. doi: 10.2174/156720512800618008. [DOI] [PubMed] [Google Scholar]
- 91.Morgan AR. Hamilton G. Turic D. Jehu L. Harold D. Abraham R. Hollingworth P. Moskvina V. Brayne C. Rubinsztein DC. Lynch A. Lawlor B. Gill M. O'Donovan M. Powell J. Lovestone S. Williams J. Owen MJ. Association analysis of 528 intra-genic SNPs in a region of chromosome 10 linked to late onset Alzheimer's disease. Am J Med Genet B Neuropsychiatr Genet. 2008;147B:727–731. doi: 10.1002/ajmg.b.30670. [DOI] [PubMed] [Google Scholar]
- 92.Coppedè F. Zitarosa MT. Migheli F. Lo Gerfo A. Bagnoli S. Dardano A. Nacmias B. Mancuso M. Monzani F. Siciliano G. Sorbi S. Migliore L. DNMT3B promoter polymorphisms and risk of late onset Alzheimer's disease. Curr Alzheimer Res. 2012;9:550–554. doi: 10.2174/156720512800618062. [DOI] [PubMed] [Google Scholar]
- 93.Matsumoto L. Takuma H. Tamaoka A. Kurisaki H. Date H. Tsuji S. Iwata A. CpG demethylation enhances alpha-synuclein expression and affects the pathogenesis of Parkinson's disease. PLoS One. 2010;5:e15522. doi: 10.1371/journal.pone.0015522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Klein CJ. Botuyan MV. Wu Y. Ward CJ. Nicholson GA. Hammans S. Hojo K. Yamanishi H. Karpf AR. Wallace DC. Simon M. Lander C. Boardman LA. Cunningham JM. Smith GE. Litchy WJ. Boes B. Atkinson EJ. Middha S. B Dyck PJ. Parisi JE. Mer G. Smith DI. Dyck PJ. Mutations in DNMT1 cause hereditary sensory neuropathy with dementia and hearing loss. Nat Genet. 2011;43:595–600. doi: 10.1038/ng.830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Loeser RF. Im HJ. Richardson B. Lu Q. Chubinskaya S. Methylation of the OP-1 promoter: Potential role in the age-related decline in OP-1 expression in cartilage. Osteoarthritis Cartilage. 2009;17:513–517. doi: 10.1016/j.joca.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Liu ZH. Chen LL. Deng XL. Song HJ. Liao YF. Zeng TS. Zheng J. Li HQ. Methylation status of CpG Sites in the MCP-1 promoter is correlated to serum MCP-1 in type 2 diabetes. J Endocrinol Invest. 2012;6:585–589. doi: 10.3275/7981. [DOI] [PubMed] [Google Scholar]
- 97.Sapienza C. Lee J. Powell J. Erinle O. Yafai F. Reichert J. Siraj ES. Madaio M. DNA methylation profiling identifies epigenetic differences between diabetes patients with ESRD and diabetes patients without nephropathy. Epigenetics. 2011;6:20–28. doi: 10.4161/epi.6.1.13362. [DOI] [PubMed] [Google Scholar]
- 98.Franco R. Schoneveld O. Georgakilas AG. Panayiotidis MI. Oxidative stress, DNA methylation and carcinogenesis. Cancer Lett. 2008;266:6–11. doi: 10.1016/j.canlet.2008.02.026. [DOI] [PubMed] [Google Scholar]
- 99.Egger G. Liang G. Aparicio A. Jones PA. Epigenetics in human disease and prospects for epigenetic therapy. Nature. 2004;429:457–463. doi: 10.1038/nature02625. [DOI] [PubMed] [Google Scholar]
- 100.Mack GS. To selectivity and beyond. Nat Biotechnol. 2010;28:1259–1266. doi: 10.1038/nbt.1724. [DOI] [PubMed] [Google Scholar]
- 101.Sullivan M. Hahn K. Kolesar JM. Azacitidine: A novel agent for myelodysplastic syndromes. Am J Health Syst Pharm. 2005;62:1567–1573. doi: 10.2146/ajhp040385. [DOI] [PubMed] [Google Scholar]
- 102.Kantarjian H. Issa JP. Rosenfeld CS. Bennett JM. Albitar M. DiPersio J. Klimek V. Slack J. de Castro C. Ravandi F. Helmer R., 3rd Shen L. Nimer SD. Leavitt R. Raza A. Saba H. Decitabine improves patient outcomes in myelodysplastic syndromes: Results of a phase III randomized study. Cancer. 2006;106:1794–1803. doi: 10.1002/cncr.21792. [DOI] [PubMed] [Google Scholar]
- 103.Scott SA. Dong WF. Ichinohasama R. Hirsch C. Sheridan D. Sanche SE. Geyer CR. Decoteau JF. 5-Aza-2′-deoxycytidine (decitabine) can relieve p21WAF1 repression in human acute myeloid leukemia by a mechanism involving release of histone deacetylase 1 (HDAC1) without requiring p21WAF1 promoter demethylation. Leuk Res. 2006;30:69–76. doi: 10.1016/j.leukres.2005.05.010. [DOI] [PubMed] [Google Scholar]
- 104.Uenogawa K. Hatta Y. Arima N. Hayakawa S. Sawada U. Aizawa S. Yamamoto T. Takeuchi J. Azacitidine induces demethylation of p16INK4a and inhibits growth in adult T-cell leukemia/lymphoma. Int J Mol Med. 2011;28:835–839. doi: 10.3892/ijmm.2011.756. [DOI] [PubMed] [Google Scholar]
- 105.Villar-Garea A. Fraga MF. Espada J. Esteller M. Procaine is a DNA-demethylating agent with growth-inhibitory effects in human cancer cells. Cancer Res. 2003;63:4984–4989. [PubMed] [Google Scholar]
- 106.Segura-Pacheco B. Trejo-Becerril C. Perez-Cardenas E. Taja-Chayeb L. Mariscal I. Chavez A. Acuña C. Salazar AM. Lizano M. Dueñas-Gonzalez A. Reactivation of tumor suppressor genes by the cardiovascular drugs hydralazine and procainamide and their potential use in cancer therapy. Clin Cancer Res. 2003;9:1596–1603. [PubMed] [Google Scholar]
- 107.Lee BH. Yegnasubramanian S. Lin X. Nelson WG. Procainamide is a specific inhibitor of DNA methyltransferase 1. J Biol Chem. 2005;280:40749–40756. doi: 10.1074/jbc.M505593200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Deng C. Lu Q. Zhang Z. Rao T. Attwood J. Yung R. Richardson B. Hydralazine may induce autoimmunity by inhibiting extracellular signal-regulated kinase pathway signaling. Arthritis Rheum. 2003;48:746–756. doi: 10.1002/art.10833. [DOI] [PubMed] [Google Scholar]
- 109.Zambrano P. Segura-Pacheco B. Perez-Cardenas E. Cetina L. Revilla-Vazquez A. Taja-Chayeb L. Chavez-Blanco A. Angeles E. Cabrera G. Sandoval K. Trejo-Becerril C. Chanona-Vilchis J. Duenas-González A. A phase I study of hydralazine to demethylate and reactivate the expression of tumor suppressor genes. BMC Cancer. 2005;5:44. doi: 10.1186/1471-2407-5-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Coronel J. Cetina L. Pacheco I. Trejo-Becerril C. González-Fierro A. de la Cruz-Hernandez E. Perez-Cardenas E. Taja-Chayeb L. Arias-Bofill D. Candelaria M. Vidal S. Dueñas-González A. A double-blind, placebo-controlled, randomized phase III trial of chemotherapy plus epigenetic therapy with hydralazine valproate for advanced cervical cancer. Preliminary results. Med Oncol. 2011;(Suppl 1):S540–546. doi: 10.1007/s12032-010-9700-3. [DOI] [PubMed] [Google Scholar]
- 111.Park LK. Friso S. Choi SW. Nutritional influences on epigenetics and age-related disease. Proc Nutr Soc. 2012;71:75–83. doi: 10.1017/S0029665111003302. [DOI] [PubMed] [Google Scholar]
- 112.Locker J. Reddy TV. Lombardi B. DNA methylation and hepatocarcinogenesis in rats fed a choline-devoid diet. Carcinogenesis. 1986;7:1309–1312. doi: 10.1093/carcin/7.8.1309. [DOI] [PubMed] [Google Scholar]
- 113.Miller RA. Buehner G. Chang Y. Harper JM. Sigler R. Smith-Wheelock M. Methionine-deficient diet extends mouse lifespan, slows immune and lens aging, alters glucose, T4, IGF-I and insulin levels, and increases hepatocyte MIF levels and stress resistance. Aging Cell. 2005;4:119–125. doi: 10.1111/j.1474-9726.2005.00152.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Troen AM. French EE. Roberts JF. Selhub J. Ordovas JM. Parnell LD. Lai CQ. Lifespan modification by glucose and methionine in Drosophila melanogaster fed a chemically defined diet. Age (Dordr) 2007;29:29–39. doi: 10.1007/s11357-006-9018-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Richie JP. Leutzinger Y. Parthasarathy S. Malloy V. Orentreich N. Zimmerman JA. Methionine restriction increases blood glutathione and longevity in F344 rats. FASEB J. 1994;8:1302–1307. doi: 10.1096/fasebj.8.15.8001743. [DOI] [PubMed] [Google Scholar]
- 116.Komninou D. Leutzinger Y. Reddy BS. Richie JP. Methionine restriction inhibits colon carcinogenesis. Nutr Cancer. 2006;54:202–208. doi: 10.1207/s15327914nc5402_6. [DOI] [PubMed] [Google Scholar]
- 117.Sanchez-Roman I. Gomez A. Gomez J. Suarez H. Sanchez C. Naudi A. Ayala V. Portero-Otin M. Lopez-Torres M. Pamplona R. Barja G. Forty percent methionine restriction lowers DNA methylation, complex I ROS generation, and oxidative damage to mtDNA and mitochondrial proteins in rat heart. J Bioenerg Biomembr. 2011;43:699–708. doi: 10.1007/s10863-011-9389-9. [DOI] [PubMed] [Google Scholar]
- 118.Batra V. Sridhar S. Devasagayam TP. Enhanced one-carbon flux towards DNA methylation: Effect of dietary methyl supplements against gamma-radiation-induced epigenetic modifications. Chem Biol Interact. 2010;183:425–433. doi: 10.1016/j.cbi.2009.11.010. [DOI] [PubMed] [Google Scholar]
- 119.Nandakumar V. Vaid M. Katiyar SK. (−)-Epigallocatechin-3-gallate reactivates silenced tumor suppressor genes, Cip1/p21 and p16INK4a, by reducing DNA methylation and increasing histones acetylation in human skin cancer cells. Carcinogenesis. 2011;32:537–544. doi: 10.1093/carcin/bgq285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Fang MZ. Wang Y. Ai N. Hou Z. Sun Y. Lu H. Welsh W. Yang CS. Tea polyphenol (−)-epigallocatechin-3-gallate inhibits DNA methyltransferase and reactivates methylation-silenced genes in cancer cell lines. Cancer Res. 2003;63:7563–7570. [PubMed] [Google Scholar]
- 121.Fang MZ. Chen D. Sun Y. Jin Z. Christman JK. Yang CS. Reversal of hypermethylation and reactivation of p16INK4a, RARbeta, and MGMT genes by genistein and other isoflavones from soy. Clin Cancer Res. 2005;11(19 Pt 1):7033–7041. doi: 10.1158/1078-0432.CCR-05-0406. [DOI] [PubMed] [Google Scholar]
- 122.Fritz WA. Coward L. Wang J. Lamartiniere CA. Dietary genistein: Perinatal mammary cancer prevention, bioavailability and toxicity testing in the rat. Carcinogenesis. 1998;19:2151–2158. doi: 10.1093/carcin/19.12.2151. [DOI] [PubMed] [Google Scholar]
- 123.Amato RJ. Inhibition of DNA methylation by antisense oligonucleotide MG98 as cancer therapy. Clin Genitourin Cancer. 2007;5:422–426. doi: 10.3816/CGC.2007.n.029. [DOI] [PubMed] [Google Scholar]
- 124.Plummer R. Vidal L. Griffin M. Lesley M. de Bono J. Coulthard S. Sludden J. Siu LL. Chen EX. Oza AM. Reid GK. McLeod AR. Besterman JM. Lee C. Judson I. Calvert H. Boddy AV. Phase I study of MG98, an oligonucleotide antisense inhibitor of human DNA methyltransferase 1, given as a 7-day infusion in patients with advanced solid tumors. Clin Cancer Res. 2009;15:3177–3183. doi: 10.1158/1078-0432.CCR-08-2859. [DOI] [PubMed] [Google Scholar]
- 125.Winquist E. Knox J. Ayoub JP. Wood L. Wainman N. Reid GK. Pearce L. Shah A. Eisenhauer E. Phase II trial of DNA methyltransferase 1 inhibition with the antisense oligonucleotide MG98 in patients with metastatic renal carcinoma: A National Cancer Institute of Canada Clinical Trials Group investigational new drug study. Invest New Drugs. 2006;24:159–167. doi: 10.1007/s10637-006-5938-1. [DOI] [PubMed] [Google Scholar]
- 126.Leu YW. Rahmatpanah F. Shi H. Wei SH. Liu JC. Yan PS. Huang TH. Double RNA interference of DNMT3b and DNMT1 enhances DNA demethylation and gene reactivation. Cancer Res. 2003;63:6110–6115. [PubMed] [Google Scholar]
- 127.Alvarez-Erviti L. Seow Y. Yin H. Betts C. Lakhal S. Wood MJ. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat Biotechnol. 2011;29:341–345. doi: 10.1038/nbt.1807. [DOI] [PubMed] [Google Scholar]
- 128.Castanotto D. Sensor and sensitivity: A screen for elite shRNAs. Mol Ther. 2011;19:823–825. doi: 10.1038/mt.2011.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Castanotto D. Rossi JJ. The promises and pitfalls of RNA-interference-based therapeutics. Nature. 2009;457:426–433. doi: 10.1038/nature07758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.de Magalhães JP. Finch CE. Janssens G. Next-generation sequencing in aging research: Emerging applications, problems, pitfalls and possible solutions. Ageing Res Rev. 2010;9:315–323. doi: 10.1016/j.arr.2009.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Cox LS. Mason PA. Prospects for rejuvenation of aged tissue by telomerase reactivation. Rejuvenation Res. 2010;13:749–754. doi: 10.1089/rej.2010.1140. [DOI] [PubMed] [Google Scholar]
- 132.Obrenovich ME. Nair NG. Beyaz A. Aliev G. Reddy VP. The role of polyphenolic antioxidants in health, disease, and aging. Rejuvenation Res. 2010;13:631–643. doi: 10.1089/rej.2010.1043. [DOI] [PubMed] [Google Scholar]
- 133.Best BP. Nuclear DNA damage as a direct cause of aging. Rejuvenation Res. 2009;12:199–208. doi: 10.1089/rej.2009.0847. [DOI] [PubMed] [Google Scholar]
- 134.Dubec SJ. Aurora R. Zassenhaus HP. Mitochondrial DNA mutations may contribute to aging via cell death caused by peptides that induce cytochrome c release. Rejuvenation Res. 2008;11:611–619. doi: 10.1089/rej.2007.0617. [DOI] [PMC free article] [PubMed] [Google Scholar]