Abstract
Genetic transformation of Belgian endive (Cichorium intybus) and carrot (Daucus carota) by Agrobacterium rhizogenes resulted in a transformed phenotype, including annual flowering. Back-crossing of transformed (R1) endive plants produced a line that retained annual flowering in the absence of the other traits associated with A. rhizogenes transformation. Annualism was correlated with the segregation of a truncated transferred DNA (T-DNA) insertion. During vegetative growth, carbohydrate reserves accumulated normally in these annuals, and they were properly mobilized prior to anthesis. The effects of individual root-inducing left-hand T-DNA genes on flowering were tested in carrot, in which rolC (root locus) was the primary promoter of annualism and rolD caused extreme dwarfism. We discuss the possible adaptive significance of this attenuation of the phenotypic effects of root-inducing left-hand T-DNA.
Agrobacterium rhizogenes carries T-DNA that induces the formation of roots after its integration into the plant genome. This Ri T-DNA alters the phenotype of the transformed roots and the plants that regenerate from them (Tepfer, 1982, 1984). In species that regenerate naturally from their roots, transmission of Ri T-DNA into seed would seem probable. Several species, including morning glory and tobacco, carry sequences homologous to Ri T-DNA (Tepfer, 1982; White et al., 1983; Furner et al., 1986). Genes encoded by this vestigial Ri T-DNA are expressed in tobacco (Ichikawa et al., 1990; Meyer et al., 1995). It was suggested that this apparent natural genetic transformation could be of adaptive significance to the plant (Tepfer, 1983; Meyer et al., 1995).
In this report we describe more fully how transformation by the Ri TL-DNA could lead to beneficial or deleterious phenotypes of potential adaptive significance. We previously showed in Belgian endive (Cichorium intybus) that roots induced by A. rhizogenes regenerated into plants having an altered phenotype that segregated in meiosis. The T1 generation showed varying degrees of phenotypic alterations associated with the segregation of two insertions, one complete and one truncated (Sun et al., 1991b). We examined the relationship between the truncated insertion and the switch from biennial to annual flowering. We consider the possibility that methylation may also be important in the attenuation of the foreign genes and propose that the conversion to annualism could be beneficial in nature. The effects of individual genes from the Ri TL-DNA on phenotype were examined in carrot (Daucus carota), in which the change to annualism was produced for the most part by rolC, and rolD was associated with a disadvantageous phenotype.
MATERIALS AND METHODS
Plant Cultivation
Seeds of transgenic and wild-type endive (Cichorium intybus) were sown in the greenhouse in two parallel rows in sand-filled tanks 6.0 m long × 0.5 m deep × 0.6 m wide. They were watered with a complete mineral nutrient solution designed for endive growth (Améziane et al., 1995). Regenerated plantlets of transgenic and wild-type carrot (Daucus carota) were grown for 1 month in vitro and were then transferred to individual pots in the greenhouse and watered with an appropriate nutrient solution (Lesaint and Coïc, 1983). All plants in the greenhouse were exposed to the long-day (16 h) photoperiod necessary for inducing flowering in endive and carrot.
Endive plants were harvested at the end of the vegetative phase, 5 months after sowing, and their transformed status was verified by PCR. Shoots were discarded and the tuberized roots were placed in a dark, mist-filled chamber at 18°C and 90% RH for 21 d, where they were bathed in a recirculated nutrient solution (Lesaint and Coïc, 1983). This “forcing” procedure produced the edible, etiolated shoot known as the chicon.
Endive and Carrot Transformation
Carrot was transformed with wild-type Ri T-DNA through regeneration from roots induced by Agrobacterium rhizogenes strain A4RSII, as previously described (Tepfer, 1984). Root induction and regeneration were also used for Agrobacterium tumefaciens containing EcoRI fragment 15 (rolA, rolB, and rolC) in a binary vector. Individual genes from the Ri TL-DNA were introduced into carrot using A. tumefaciens strains carrying binary vectors pMRK or Bin19 (Bevan, 1984; Beach and Gresshoff, 1986; Vilaine et al., 1987) with or without fragments of the Ri TL-DNA and representing ORFs 8, 10, 11, 12, 14, or 15 with their native promoters. These were previously described for ORFs 10, 11, and 15 (Vilaine et al., 1987) and ORF 12 (Michael et al., 1990) or, in the case of ORF 8, were produced by standard cloning procedures (Table I).
Table I.
Plasmids used in carrot transformation
| Plasmid | Fragment | Sequence Coordinates | ORFs | Loci |
|---|---|---|---|---|
| pBin H21 | HindIII (21) | 6360 –9815 | 8, 9 | aux1-like |
| pMRK E15 | EcoRI (15) | 9075 –13445 | 10, 11, 12 | rol A, rol B, rol C |
| pMRK 10 | EcoRI-NruI | 9075 –10968 | 10 | rol A |
| pMRK 11 | HindIII (30) | 9814 –11587 | 11 | rol B |
| pLBR 11 | HindIII-EcoRI | 11587 –13445 | 12 | rol C |
| pMRK 14 | EcoRI (37a) | 15385 –17059 | 14 | (no name) |
| pMRK 15 | DraI | 16168 –18135 | 15 | rol D |
DNA fragments (Jouanin, 1984) containing genes and regulatory sequences from the Ri TL-DNA (Slightom et al., 1986) were cloned into binary plasmidsa pMRK and pBin 19.
pBin H21 was constructed for this work; see Michael et al. (1990) for pLBR 11 and Vilaine et al. (1987) for other plasmids.
Fresh, home-grown carrots that had received no cold treatment were surface sterilized by immersion for 20 min in a Bayrochlor solution (three tablets per liter; Bayrol, Germany). Carrots were then sliced into approximately 5-mm segments that were placed on water/agar medium, and 1 μL of an overnight bacterial culture (sterile water in controls) was deposited on the surface. Wound calli obtained with tissue inoculated with A. tumefaciens were transferred to medium (Monnier, 1976) containing hormones and antibiotics (0.36 μm 2,4-D, 0.72 μm kinetin, 200 mg/L cefotaxim, and 100 mg/L kanamycin). Four to five independently transformed cell lines were produced for each bacterial genotype. Embryogenesis was induced by subculture in the same medium without 2,4-D and agitation at 100 rpm/min in the dark at 25°C to 27°C. Plantlets were subcultured at 22°C to 24°C (light period) and 18°C to 20°C (dark period) for 10 h of light each day. Short-day, noninductive conditions for flowering were maintained throughout regeneration.
Analysis of Carbohydrates and Nitrogen-Containing Compounds
Tuberized roots were separated from the shoots, freeze-dried, and pulverized. Soluble carbohydrates, amino acids, and nitrate were extracted from 100 mg of freeze-dried powder by agitating for 2 h in 5 mL of ethanol (80%) at 4°C. After the sample was centrifuged for 10 min at 15,000g, the ethanol fraction was removed and the pellet was similarly extracted with 5 mL of deionized water. The ethanol and water fractions were combined and stored at −80°C. Samples were dried under a vacuum and resuspended in water before analysis. Total amino acids were determined using ninhidrin (Rosen, 1957), Arg was measured using guanidine (Rosenberg et al., 1956; Le Nard and Fiala, 1990), and nitrate was assayed by ion-exchange chromatography (model 4000i, HPLC ASA4 column, Dionex, Sunnyvale, CA). Total nitrogen was determined by ammonia distillation after Kjeldahl digestion.
The same extract was neutralized using cation- and anion-exchange resins and carbohydrates were quantified by isocratic HPLC (Spectra-Physics, Mountain View, CA) at 75°C using a 300- × 7.8-mm Aminex HPX 42 C column (Bio-Rad) with water as the eluant and a flow rate of 0.6 mL min−1. Eluted carbohydrates were detected by differential refractometry (Shimadzu Corp., Kyoto, Japan). Carbohydrates were also separated using ascending TLC on precoated 20- × 20-cm silica gel plates (F 1500, Schleicher & Schuell). Chromatograms were developed twice with butan-1-ol, propan-2-ol, and water (3:12:4, v/v) as the solvent (Kanaya et al., 1978). Carbohydrates were visualized by spraying with a ketose-specific, urea phosphoric acid stain (Wise et al., 1955).
DNA Extraction and Analysis
DNA was extracted using established methods (Dellaporta et al., 1983). Transformed genotypes were verified by PCR using primers for rolA (5′-GAATTAGCCGGACTAAACGT and 5′-TTGTTTGGATGCCCTAATT). Probes were labeled for hybridization by random priming (Pharmacia) and molecular hybridization was performed as previously described (Southern, 1975).
RESULTS
Physiological and Molecular Analysis of Endive Lines Transformed by A. rhizogenes
An R0 endive plant transformed by A. rhizogenes strain A4RSII was crossed with a normal plant. In the R1 progeny, two transformed phenotypes were identified. Transformed lines 1 and 2 exhibited attenuated phenotypes similar to the T phenotype previously described in tobacco (Tepfer, 1984). These attenuated lines had normal aerial parts and tuberized roots similar to control plants, but lateral roots were more numerous (Fig. 1). The third line was designated T′, since it was more altered, resembling the R0 parent, with stunted growth, wrinkled leaves, reduced development of tuberized roots, and overproduction of lateral roots (Fig. 1). Irrespective of other aspects of their phenotype, R1 transformed plants shifted from biennial to annual flowering and flowered without vernalization after a vegetative period, including a long-day stimulus. Flowering was earlier, however, in the more severely altered line 3 than in lines 1 and 2 (Fig. 1).
Figure 1.
Attenuation of the transformed phenotype in three transformed lines (R1 generation). Lines 1 and 2 are of the attenuated T phenotype and line 3 shows the more altered (T′) phenotype. Transformed lines flowered as annuals instead of as biennials, without vernalization.
Two R1 plants from each line of the T phenotype (designated 1a, 1b, 2a, and 2b) and four plants from the T′ line 3 (designated 3a–3d) were selfed. In the R2 progeny we assessed general morphology, time of flowering, and accumulation of inulin, the primary carbohydrate reserve in tuberized endive roots. After exposure to long-day photoperiods, all of the plants derived from the three transformed lines flowered as annuals without vernalization. The T′ line developed floral stems earlier than the T lines. As expected, the control behaved as a biennial and flowered without vernalizaton when treated with GA3 (results not shown).
Plant fresh weight and morphology were recorded 5 months after sowing, at the end of the vegetative period. In lines 1 and 2, fresh weights and the root-to-shoot ratio were similar to the control (Table II). In the T′ line (no. 3) root fresh weights were reduced by a factor of 0.46, shoot fresh weight was reduced by a factor of 0.3, and the shoot-to-root ratio was 1.6 compared with 2.5 in the controls (Table II). All R2 plants in T lines 1 and 2 had normal leaves and tuberized roots like the control, with no increase in lateral roots. Furthermore, the shape of the transformed tuberized roots more closely resembled the controls in the R2 than in the R1 generation (Fig. 2). Progeny in line 3 were heterogeneous: the phenotype T′ was recovered in 70%, 45%, 75%, and 100% of the plants derived from, respectively, lines 3a, 3b, 3c, and 3d. The remainder were of the T phenotype.
Table II.
Effects of transformation on root and shoot fresh weight, root system architecture, leaf morphology, and flowering time
| Genotype | Fresh
Wt
|
Phenotypea
|
||||
|---|---|---|---|---|---|---|
| Root | Shoot | Root/Shoot | Lateral roots | Wrinkled leaves | Flowering time | |
| g/organ | months | |||||
| Wild type | 135 (21) | 340 (45) | 2.5 | − | − | 13 –15b |
| 1 | 96 (29) | 219 (61) | 2.3 | + | − | 7 –9 |
| 2 | 136 (19) | 313 (35) | 2.3 | + | − | 6 –8 |
| 3 | 63 (21) | 102 (31) | 1.6 | ++ | + | 5 –7 |
Numbers in parentheses are ses (P = 0.05, n = 6).
−, Absence of trait; +, presence of trait.
The wild-type plants flowered only after vernalization.
Figure 2.
Production of edible shoots in control and transformed endive lines 1 to 3 (R2 generation). In the control shoot, chicon formation required 2 weeks of cold treatment but not in the transformed lines. The size and shape of the tuberized roots in lines 1 and 2 more closely resembled the control in this R2 generation (Fig. 1, R1 generation).
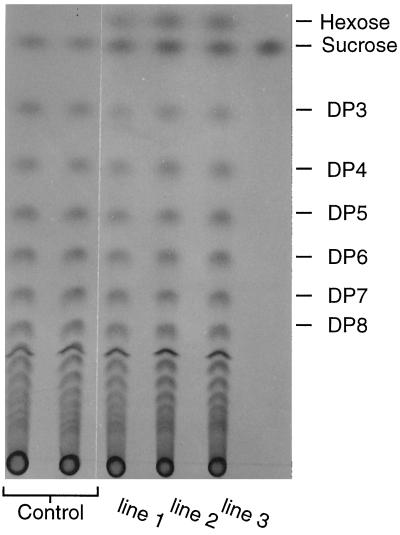
The amount of inulin ranged between 71% and 75% of root dry weight in transformed and wild-type plants (Table III), and no qualitative changes were detected in transgenic tuberized roots; the transformed roots were capable of synthesizing inulin oligomers, as well as highly polymerized molecules (Fig. 3). At the end of the vegetative phase, free Fru content increased 8- to 10-fold in the roots of transformed plants (Table III), indicating high inulin hydrolysis activity at the end of the vegetative period. As a consequence of this activity, hexose was detected in the three transformed lines but not in the controls (Fig. 3).
Table III.
Total N, total N in amino acids, Arg, inulin, and Fru in tuberized roots of transformed endive harvested at the end of the vegetative phase
| Genotype | Total N | Total N in
|
|||
|---|---|---|---|---|---|
| Amino acids | Arg | Inulin | Fru | ||
| mg 100 mg−1 dry wt | μg 100 mg−1 dry wt | mg g−1 dry wt | μmol g−1 dry wt | ||
| Wild type | 1.2 (0.2) | 720 (83) | 430.00 | 750 (25) | 8.3 (1.2) |
| 1 | 1.7 (0.3) | 517 (61) | 2.48 | 711 (32) | 79.6 (11.5) |
| 2 | 1.4 (0.1) | 293 (21) | 3.51 | 722 (52) | 76.4 (12.3) |
| 3 | 0.9 (0.1) | 82 (9) | 3.66 | 720 (71) | 68.9 (13.9) |
Numbers in parentheses are ses (P = 0.05, n = 6).
Figure 3.
Hexose, Suc, and inulin in various degrees of polymerization (DP) in endive tuberized roots. An equivalent of 40 mg of dry matter was loaded in each lane. Hexose was detected in the transformed lines only, as expected for annuals ready to flower.
Total nitrogen was lower in the roots of the T′ plants (line 3) than in the controls and in the T lines (lines 1 and 2). Amino acids made up 60% of the total nitrogen pool in control plants (Table III), whereas in transformed plants they were reduced to 30% in line 1, 21% in line 2, and 10% in line 3. Arg was severely reduced in all transformed lines, where it represented 0.6% (line 1), 0.8% (line 2), and 0.89% (line 3) of the Arg in the controls (Table III).
Control and transformed roots were harvested at the end of vegetative growth and were then forced for 3 weeks in the dark (see Methods) to produce chicons. Forcing immediately after harvesting without cold treatment produced chicons in transformed lines 1 and 2. In line 3, only roots derived from plants showing the T phenotype produced chicons, since plants of the T′ phenotype did not produce storage roots suitable for forcing (Fig. 2). Control roots did not develop chicons unless they were exposed to 0°C to 2°C for 2 weeks.
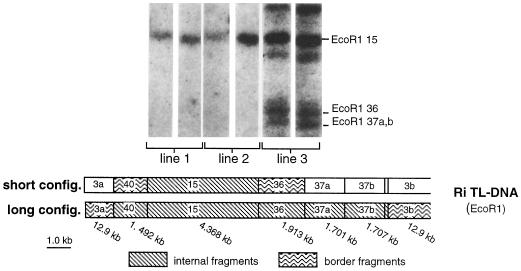
The transformed genotype was examined in detail in plants of the T phenotype from lines 1, 2, and 3 and in the T′ phenotype from line 3. Transformation was verified by PCR in the three lines corresponding to phenotypes T and T′ using primers that amplify a 0.5-kb segment of the rolC gene (not shown). Plant DNA was also digested by EcoRI, which cuts seven times in the Ri TL-DNA, and hybridization was carried out with a probe (pLJ1) that covers the entire TL-DNA (Jouanin, 1984). Hybridization patterns differed in the T and T′ samples. As expected from previous work (Sun et al., 1991b), plants showing the T′ phenotype carried the complete 21-kb insert consisting of EcoRI fragments 40, 15, 36, 37a, and 37b, extending to the left into fragment 3a and to the right into fragment 3b (Fig. 4). Plants of the T phenotype had a truncated TL-DNA, including EcoRI fragment 15, with junctions in EcoRI fragment 40 on the left and 36 on the right (Fig. 4). The R0 parent (phenotype T′) was the same as in plants from line 3 exhibiting the T′ phenotype (Sun et al., 1991b).
Figure 4.
Southern hybridization after digestion of transformed plant DNA with EcoRI and hybridization using a probe covering the TL-DNA from A. rhizogenes strain A4. Fragments internal to the TL-DNA are identified on the right. No hybridization was detected in control samples (not shown). The positions of restriction fragments are given below the figure. config., Configuration.
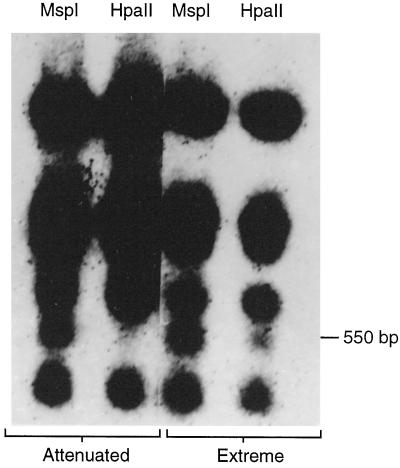
The foreign DNA was examined for methylation in two R1 siblings bearing the complete insertion (as verified by Southern blotting) but exhibiting T or T′ levels of phenotypic change (Sun et al., 1991b). Total DNA was digested by the isoschizoenzymes MspI and HpaII, and hybridization was carried out with a probe covering EcoRI fragment 15, including rolA, rolB, and rolC. The pattern of hybridization was similar with both enzymes in the plant of the T′ phenotype, whereas DNA from the T phenotype showed two different hybridization patterns with MspI and HpaII (Fig. 5). Among the five fragments detected with MspI in the T phenotype, one at 550 bp was not seen with HpaII, indicating that at least one restriction site in EcoRI 15 was methylated in plants of the T phenotype.
Figure 5.
Southern hybridization with DNA from two transformed R1 siblings bearing complete T-DNA insertions using isoschizomers MspI and HpaII. The probe was EcoRI 15 from the Ri TL-DNA. In the extreme (T′) phenotype the pattern of hybridization was similar with both enzymes, whereas in the attenuated phenotype the fragment at 550 bp was not seen with HpaII, indicating that at least one site was methylated in plants of the T phenotype.
Effects of Ri TL-DNA Genes in Carrot
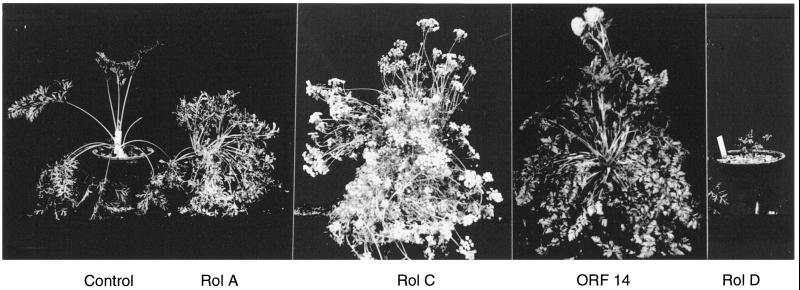
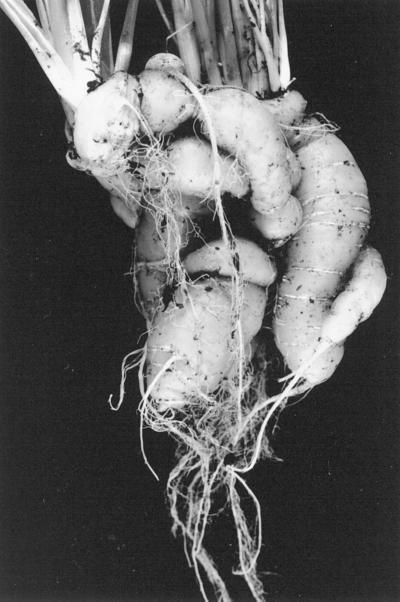
Carrot was chosen for ease of transformation with single genes in a binary vector. Carrot plants transformed by A. rhizogenes strain A4RSII exhibited the previously reported phenotype (Tepfer, 1984) consisting of root plagiotropism, accentuated branching, and inability to form a tuberized root. Plants transformed by A. tumefaciens carrying a binary vector containing rol genes (rolABC or rolA, rolB, or rolC) were modified to varying degrees. Wrinkled leaves were observed in only a few A4RSII and rolABC plants, whereas wild-type, vector control, and rolB plants generally developed one shoot meristem per plant. A4RSII, rolABC, rolA, rolC, and ORF 14 plants produced two to three meristems. The few rolD plants that were recovered were severely dwarfed, with wrinkled leaves and curved petioles (Fig. 6). Plants transformed by rolC were annuals, but they nevertheless developed storage roots that were branched and intertwined (Fig. 7).
Figure 6.
Effects of Ri TL-DNA genes on growth and flowering in carrots. Control plants developed one shoot meristem, whereas rolA, rolC, and ORF 14 plants produced two to three shoot meristems. rolC plants shifted from biennial to annual flowering without vernalization. One of 17 ORF 14 plants flowered, but only two flowers were produced on this plant, compared with the numerous flowers on the rolC plant. rolD plants were severely dwarfed.
Figure 7.
Effects of rolC on root architecture in carrots. Branched and intertwined tuberized root of the rolC plant shown in Figure 6.
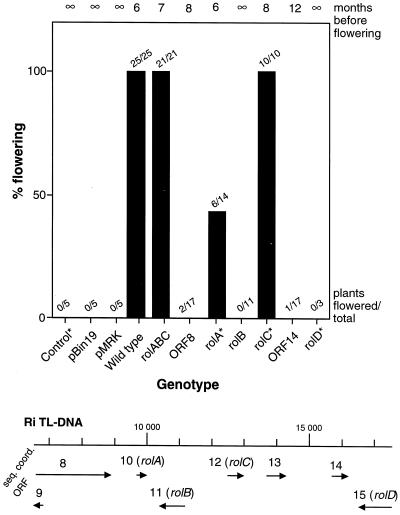
A4RSII, rolABC, and rolC plants flowered without vernalization as annuals at the end of vegetative growth (Table II; Fig. 8). Six of 14 rolA plants were also annuals. Others, including the vector control, wild-type, and rolB plants, were biennials, with the exception of 2 ORF 8 plants (out of 17) and 1 ORF 14 plant (out of 14), which flowered after 8 and 12 months, respectively, of vegetative growth without vernalization (Figs. 6 and 8). The transformed status of selected carrot transgenics was verified by Southern hybridization (results not shown).
Figure 8.
Quantification of the effects of Ri TL-DNA genes on flowering in carrots. Flowering frequency without vernalization (percentage and number of plants flowered/total) and time of flowering were recorded for nontransformed controls, two vector controls (Bin19 and MRK), transformants expressing wild-type Ri T-DNA (A4RSII), and plants transformed by individual genes with their endogenous promoters: ORF 8 (an aux1 homolog), rolABC, rolA, rolB, rolC, or rolD and ORF 14. Map positions of genes tested are given below the graph. ∞, Flowering did not occur without vernalization; *, phenotypes illustrated in Figure 6. seq. coord., Sequence coordinates.
DISCUSSION
Metabolic Changes in Endive Roots
Inulin accumulation in tuberized endive roots was not changed by genetic transformation by A. rhizogenes, but at the end of the vegetative phase free Fru represented 8 to 10 times the levels found in the controls. Similar Fru buildup due to inulin hydrolysis occurs in wild-type plants vernalized for 2 to 4 months in the field or in roots harvested and stored at 0°C for at least 5 weeks. Therefore, transformation by wild-type Ri T-DNA induced in endive roots the shift from sink to source without vernalization.
Nitrogen is stored in endive roots either as protein (40% of total nitrogen) or as amino acids, among which Arg predominates (60% of total amino acids). Nitrate and ammonium are present in negligible amounts (Limami et al., 1993). Transformation was associated with decreases in amino acid titers (principally Arg) that were directly proportional to the change in phenotype. Effects on total nitrogen were less dramatic, suggesting that transformation altered the balance between amino acid and non-amino acid nitrogen stocks. The shoot-to-root ratio (Table II) decreased with severe phenotypic change. A similar stimulation of root-system development occurs in endive plants lacking nitrogen (Améziane et al., 1995). In tobacco phenotypic changes due to the Ri T-DNA were correlated with reduced polyamine accumulation (Martin-Tanguy et al., 1990) in direct proportion to the severity of the transformed phenotype. Polyamine synthesis depends on Arg, which was reduced in transformed endive as a function of severity of phenotype. Lack of Arg could be important in establishing the transformed phenotype through interference with polyamine synthesis, although in tobacco this is probably not the case (J. Martin-Tanguy, personal communication).
Flowering and Growth
Flowering occurs as a function of environmental stimuli and metabolic states. Biennialism is considered to be an adaptation to northern climates. Annualism is an undesirable trait in biennial crops such as endive, carrot, and sugar beet, in which the genetics of bolting have been studied in detail (Sadeghian et al., 1993). In sugar beet the B allele confers flowering (bolting) under constant high temperatures (Munerati, 1931), in the absence of the effects on the long-day requirement (Margara, 1960). The gene is frequently found in wild beets in southern climates but it is not found in northern ecotypes (Van Dijk et al., 1997). In certain Arabidopsis ecotypes a dominant gene is associated with retarded flowering, which is accelerated by vernalization (Koornneef et al., 1991; Lee et al., 1993; Clarke and Dean, 1994).
Endive and carrot are strict biennials, requiring 7 to 9 weeks of vernalization followed by a long-day photoperiod for at least 1 month before flowering. The conversion to annualism by the Ri T-DNA in carrot and endive (Tepfer, 1984; Sun et al., 1991b; Kamada et al., 1992) is in contrast to its effects in tobacco, in which flowering is retarded in proportion to inhibition of the accumulation of polyamines and their hydroxycinnamic acid conjugates (Martin-Tanguy et al., 1990). Single genes from the Ri TL-DNA have contrasting effects on phenotype. When controlled by the 35S cauliflower mosaic virus promoter, rolC strongly stimulated flowering in belladonna (Kurioka et al., 1992) and weakly stimulated flowering in tobacco (Martin-Tanguy et al., 1993). rolA retarded flowering in tobacco with its native promoter and completely inhibited flowering with the 35S promoter (Martin-Tanguy et al., 1996), but flowering was restored by grafting onto root stock that had been induced to flower. Therefore, our finding that rolC with its native promoter caused annual flowering in carrot is consistent with previous results (Kamada et al., 1992), but the potential of rolA to stimulate flowering at a low level in carrot is not consistent with its effects in tobacco.
Flowering was also stimulated in plants transformed by ORFs 8 and 14 but at low levels and after a delay. ORF 8 is homologous to iaaM and aux1 (Levesque et al., 1988), which is responsible for the first step in auxin synthesis in Pseudomonas savastanoi and in T-DNA from A. tumefaciens. ORF 14 bears weak sequence similarity to rolC (Levesque et al., 1988). These low-level stimulations suggest that a stronger promoter might confer efficient annual flowering in biennial plants. They also reinforce the conclusion (Levesque et al., 1988) that the Ri TL-DNA contains redundant information.
As expected from results with other species, rolC caused increased branching in carrot shoots and branched and twisted carrot storage organs. The dwarf phenotype produced by rolD is in contrast to its effect in day-neutral tobacco, in which the primary effect was to accelerate and stimulate flowering (Mauro et al., 1996).
In tobacco methylation of a region 3′ to the rolA-coding sequence was correlated with the inactivation of the gene and attenuation of its inhibition of flowering (Sun et al., 1991a). In endive we also found differences in the pattern of methylation in the region covering rolA, rolB, and rolC as a function of the T and T′ phenotypes, again indicating the potential of methylation to attenuate the effects of Ri TL-DNA genes. Attenuation through methylation could contribute to the functional integration of the T-DNA into the plant genome.
Natural Transformation and Adaptation
Natural genetic transformation by exogenous DNA is documented in bacteria (Romanowski et al., 1993; Lorenz and Wackernagel, 1994), and DNA transfer from bacteria to higher organisms occurs in the laboratory (Tepfer, 1984; Heinemann and Sprague, 1989). The occurrence and significance of DNA transfer from bacteria to eukaryotes has been discussed (Tepfer, 1983, 1984; Heinemann and Sprague, 1989; Meyer et al., 1995), but its significance in nature is difficult to assess. Among the probable cases of “horizontal” gene transfer, interactions between the soil bacterium A. rhizogenes and higher plants provide arguments for the importance of gene transfer from bacteria to plants.
Given the mobility of seeds through vectors such as wind, animals, and water, it seems reasonable that a biennial species or ecotype could find itself growing in a southern latitude where annualism is advantageous. Natural genetic transformation by A. rhizogenes might provide genes that allow flowering in the absence of a cold treatment. The wild-type Ri TL-DNA, which causes root formation and is compatible with plant regeneration, should be able to enter the germ line in plants that regenerate naturally from their roots. Root-inducing T-DNA is not disadvantageous in scented geranium (Pellegrineschi et al., 1994), but in other species such as endive or tobacco it can interfere with essential plant functions. To compensate, the expression of this information becomes attenuated, e.g. through increasing ploidy level and methylation (Martin-Tanguy et al., 1996), or, as we show here, through segregation of truncated T-DNA insertions.
ACKNOWLEDGMENTS
The authors are grateful to C. Dufosse for amino acid and inulin analysis and for growing and selfing transgenic plants and to F. Vilaine and A. Michael for providing bacterial strains and plasmids.
Abbreviations:
- ORF
open reading frame
- Ri TL-DNA
root-inducing, left-hand transferred DNA
- T
low-level transformed phenotype
- T′
high-level transformed phenotype
- T-DNA
transferred DNA
LITERATURE CITED
- Améziane R, Limami MA, Morot-Gaudry JF. Effect of growth nitrate concentration on carbon partitioning and sink strength in chicory. J Exp Bot 46. 1995;46:1423–1428. [Google Scholar]
- Beach K, Gresshoff P (1986) In vitro culture of legume root tissue transformed by Agrobacterium rhizogenes. In D Somers, B Gengenbach, D Biesoboer, W Hackett, C Green, eds, VIth International Congress of Plant Tissue and Cell Culture, University of Minnesota, Minneapolis, pp 155
- Bevan M. Binary Agrobacterium vectors for plant transformation. Nucleic Acids Res. 1984;12:8711–8721. doi: 10.1093/nar/12.22.8711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke JH, Dean C. Mapping FRI, a locus controlling flowering time and vernalization response in Arabidopsis thaliana. Mol Gen Genet. 1994;242:81–89. doi: 10.1007/BF00277351. [DOI] [PubMed] [Google Scholar]
- Dellaporta S, Wood J, Hicks J. A plant DNA minipreparation: version II. Plant Mol Biol Rep. 1983;1:19–21. [Google Scholar]
- Furner I, Huffman G, Amasino R, Garfinkel D, Gordon M, Nester E. An Agrobacterium transformation in the evolution of the genus Nicotiana. Nature. 1986;319:422–427. [Google Scholar]
- Heinemann J, Sprague GFJ. Bacterial conjugative plasmids mobilize DNA transfer between bacteria and yeast. Nature. 1989;340:205–209. doi: 10.1038/340205a0. [DOI] [PubMed] [Google Scholar]
- Ichikawa T, Ozeki Y, Syono K. Evidence for the expression of the rol genes of Nicotiana glauca in genetic tumors of N. galuca × N. langsdorffii. Mol Gen Genet. 1990;220:177–180. doi: 10.1007/BF00260478. [DOI] [PubMed] [Google Scholar]
- Jouanin L. Restriction map of an agropine-type Ri plasmid and its homologies with Ti plasmids. Plasmid. 1984;12:91–102. doi: 10.1016/0147-619x(84)90055-6. [DOI] [PubMed] [Google Scholar]
- Kamada H, Saitou T, Harada H. No requirement of vernalization for flower formation in Ri-transformed Cichorium plants. Plant Tissue Culture Lett. 1992;9:206–208. [Google Scholar]
- Kanaya KI, Chiba S, Shimomura T. Thin-layer chromatography of linear oligosaccharides. Agric Biol Chem. 1978;42:1947–1948. [Google Scholar]
- Koornneef M, Hanhart CJ, Veen VD. A genetic and physiological analysis of late flowering mutants in Arabidopsis thaliana. Mol Gen Genet. 1991;229:57–66. doi: 10.1007/BF00264213. [DOI] [PubMed] [Google Scholar]
- Kurioka Y, Suzuki Y, Kamada H, Harada H. Promotion of flowering and morphological alterations in Atropa belladonna transformed with a CaMV 35S-rolC chimeric gene of the Ri plasmid. Plant Cell Rep. 1992;12:1–6. doi: 10.1007/BF00232412. [DOI] [PubMed] [Google Scholar]
- Le Nard M, Fiala V. Post harvest variation of free arginine in basal plate tissues of tulip bulbs; relation to bulb physiological evolution. Acta Hortic. 1990;266:293–298. [Google Scholar]
- Lee I, Bleecker A, Amasino R. Analysis of naturally occurring late flowering in Arabidopsis thaliana. Mol Gen Genet. 1993;237:171–176. doi: 10.1007/BF00282798. [DOI] [PubMed] [Google Scholar]
- Lesaint C, Coïc Y (1983) Cultures hydroponiques. La maison rustique. Flammarion, Paris
- Levesque H, Delepelaire P, Rouzé P, Slightom J, Tepfer D. Common evolutionary origin of the central portions of the Ri TL-DNA of Agrobacterium rhizogenes and the Ti T-DNAs of Agrobacterium tumefaciens. Plant Mol Biol. 1988;11:731–744. doi: 10.1007/BF00019514. [DOI] [PubMed] [Google Scholar]
- Limami A, Roux L, Laville J, Roux Y. Nitrogen compound dynamics in the chicory (Cichorium intybus L.) tuberised tap root during the growing season and cold-storage period. J Plant Physiol. 1993;141:263–268. [Google Scholar]
- Lorenz MG, Wackernagel W. Bacterial gene transfer by natural genetic transformation in the environment. Microbiol Rev. 1994;58:563–602. doi: 10.1128/mr.58.3.563-602.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margara J. Ann Amelior Plant. 1960;10:361–471. [Google Scholar]
- Martin-Tanguy J, Corbineau F, Burtin D, Ben-Hayyim G, Tepfer D. Genetic transformation with a derivative of rolC from Agrobacterium rhizogenes and treatment with α-aminoisobutyric acid produce similar phenotypes and reduce ethylene production and the accumulation of water-insoluble polyamine-hydroxycinnamic acid conjugates in tobacco flowers. Plant Sci. 1993;93:63–76. [Google Scholar]
- Martin-Tanguy J, Sun L-Y, Burtin D, Vernoy R, Rossin N, Tepfer D. Attenuation of the phenotype caused by the root-inducing, left-hand, transferred DNA and its rolA gene. Plant Physiol. 1996;111:259–267. doi: 10.1104/pp.111.1.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Tanguy J, Tepfer D, Paynot M, Burtin D, Heisler L, Martin C. Inverse relationship between polyamine levels and the degree of phenotypic alteration induced by the Ri TL-DNA from Agrobacterium rhizogenes. Plant Physiol. 1990;92:912–918. doi: 10.1104/pp.92.4.912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauro M, Trovato M, Paolis A, Gallelli A, Costantino P, Altamura M. The plant oncogene rolD stimulates flowering in transgenic tobacco plants. Dev Biol. 1996;180:693–700. doi: 10.1006/dbio.1996.0338. [DOI] [PubMed] [Google Scholar]
- Meyer AD, Takanari I, Meins F., Jr Horizontal gene transfer: regulated expression of a tobacco homologue of the Agrobacterium rhizogenes rolC gene. Mol Gen Genet. 1995;249:265–273. doi: 10.1007/BF00290526. [DOI] [PubMed] [Google Scholar]
- Michael A, Burtin D, Martin-Tanguy J, Tepfer D (1990) Effects of the rolC locus from the Ri TL-DNA of Agrobacterium rhizogenes on development and polyamine metabolism in tobacco. In H Helsot, J Davis, J Florint, L Bobichon, G Durand, L Pénasse, eds, GIM 90 Proceedings. Société Française de Microbiologie, Paris, pp 863–868
- Monnier M. Culture in vitro de l'embryon immature de Capsella bursa-pastoris. Rev Cytol Biol Veg. 1976;39:1–120. [Google Scholar]
- Munerati O. L'eredità dela tendenza alla annualità nella commune barbabietola coltivata. Z Pflanzenzucht. 1931;17:84–89. [Google Scholar]
- Pellegrineschi A, Damon JP, Valtorta N, Paillard N, Tepfer D. Improvement of ornamental characters and fragrance production in lemon-scented geranium through genetic transformation by Agrobacterium rhizogenes. Biotechnology. 1994;12:64–68. [Google Scholar]
- Romanowski G, Lorenz MG, Wackernagel W. Plasmid DNA in a groundwater aquifer microcosm-adsorption, DNAase resistance and natural genetic transformation of Bacillus subtilis. Mol Ecol. 1993;2:171–181. doi: 10.1111/j.1365-294x.1993.tb00106.x. [DOI] [PubMed] [Google Scholar]
- Rosen H. A modified ninhydrin colorimetric analysis for amino acids. Arch Biochem Biophys. 1957;67:10–15. doi: 10.1016/0003-9861(57)90241-2. [DOI] [PubMed] [Google Scholar]
- Rosenberg H, Heron A, Morisson F. Estimation of arginine. Biochem J. 1956;63:163–159. doi: 10.1042/bj0630153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadeghian S, Becker H, Johansson E. Inheritance of bolting in three sugar beet crosses after different periods of vernalization. Plant Breeding. 1993;110:328–333. [Google Scholar]
- Slightom J, Durand-Tardif M, Jouanin L, Tepfer D. Nucleotide sequence analysis of TL-DNA of Agrobacterium rhizogenes agropine type plasmid. J Biol Chem. 1986;261:108–121. [PubMed] [Google Scholar]
- Southern E. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Sun L-Y, Monneuse M-O, Martin-Tanguy J, Tepfer D. Changes in flowering and accumulation of polyamines and hydroxycinnamic acid-polyamine conjugates in tobacco plants transformed by the rolA locus from the Ri TL-DNA of Agrobacterium rhizogenes. Plant Sci. 1991a;80:145–146. [Google Scholar]
- Sun L-Y, Touraud G, Charbonnier C, Tepfer D. Modification of phenotype in Belgian endive (Cichorium intybus) through genetic transformation by Agrobacterium rhizogenes: conversion from biennial to annual flowering. Transgenic Res. 1991b;1:14–22. [Google Scholar]
- Tepfer D (1982) La transformation génétique de plantes supérieures par Agrobacterium rhizogenes. In 2e Colloque sur les Recherches Fruitières. Centre Technique Interprofessionnel des Fruits et Légumes, Bordeaux, France, pp 47–59
- Tepfer D (1983) The biology of genetic transformation of higher plants by Agrobacterium rhizogenes. In A Puhler, ed, Molecular Genetics of the Bacteria Plant Interaction. Springer Verlag, Berlin, pp 248–258
- Tepfer D. Transformation of several species of higher plants by Agrobacterium rhizogenes: sexual transmission of the transformed genotype and phenotype. Cell. 1984;37:959–967. doi: 10.1016/0092-8674(84)90430-6. [DOI] [PubMed] [Google Scholar]
- Van Dijk H, Boutry P, McCombie H, Vernet P. Flowering time in wild beet (Beta vulgaris ssp. maritima) along a latitudinal cline. Acta Oecol. 1997;18:47–60. [Google Scholar]
- Vilaine F, Charbonnier C, Casse-Delbart F. Further insight concerning the TL-region of the Ri plasmid of Agrobacterium rhizogenes strain A4: transfer of a 1.9 kb fragment is sufficient to induce transformed roots on tobacco leaf fragments. Mol Gen Genet. 1987;210:111–115. [Google Scholar]
- White F, Garfinkel D, Huffman G, Gordon M, Nester E. Sequences homologous to Agrobacterium rhizogenes T-DNA in the genomes of uninfected plants. Nature. 1983;301:348–350. [Google Scholar]
- Wise C, Dimler R, Davis H. Determination of easily hydrolysable fructose units in dextran preparations. Anal Chem. 1955;27:33–36. [Google Scholar]