Abstract
The stereotypical function of kinesin superfamily motors is to transport cargo along microtubules. However, some kinesins also shape the microtubule track by regulating microtubule assembly and disassembly. Recent work has shown that the kinesin-8 family of motors are key regulators of cellular microtubule length. The studied kinesin-8s are highly processive motors that walk towards the microtubule plus-end. Once at plus-ends, they have complex effects on polymer dynamics: kinesin-8s either destabilize or stabilize microtubules, depending on the context. This review will focus on the mechanisms underlying kinesin-8-microtubule interactions and microtubule length control. We will compare and contrast kinesin-8s with the other major microtubule-regulating kinesins (kinesin-4 and kinesin-13), to survey the current understanding of the diverse ways that kinesins control microtubule dynamics.
Keywords: kinesin-8, kinesin-13, kinesin-4, microtubule dynamics, microtubule depolymerase
INTRODUCTION
The microtubule cytoskeleton, built from dimers of α/β-tubulin, takes on an astonishingly diverse range of forms and functions, and can transition between organizational states on a timescale of minutes. A plethora of microtubule-interacting proteins are involved in spatial and temporal control of microtubule dynamics. Some regulators interact with the microtubule lattice, for example the stabilizing microtubule-associated protein (MAP) tau [1] and the microtubule severing enzyme katanin [2]. Other regulators specifically control the dynamic properties of the microtubule end, for example, so called microtubule plus-end tracking proteins (+TIPs) of the EB and CLASP families [3,4]. Kinesin motors can interact with both microtubules surfaces: these motors walk along the lattice and in some cases also influence the microtubule end, functioning as motile regulators for microtubule dynamics.
Why would cells use the motile kinesins to regulate microtubule dynamics? What unique features do kinesins possess compared to other microtubule regulators? First, kinesins can send themselves to the microtubule end, without necessarily requiring additional factors. Second, because the motility of most kinesins is unidirectional, the effects of a specific motor will be restricted to one end of microtubules. Third, motility, if processive, allows kinesins to “pace out” the length of a microtubule, a capacity that may explain the length-dependent effects of kinesin-8 family in maintaining microtubule length homeostasis (Box 1). The dual function of kinesins for motility and microtubule end dynamics regulation raises interesting mechanistic questions: do kinesin motors interact with tubulin at the microtubule end in the same or different manner than the interaction that occurs when they walk along the lattice? Are walking and end dynamics regulation compatible or inherently competitive? Here we will review how kinesin motors control microtubule dynamics, focusing on recent progress in the kinesin-8 family, which are key regulators of cellular microtubule length.
Box 1. Length-dependent regulation of microtubule dynamics by kinesin-8s.
The capacity of kinesin-8 motors to regulate microtubule dynamics in a length-dependent manner depends on two key features. First, kinesin-8 motors are highly processive, which means that a significant portion of motors that land on the microtubule lattice will reach the plus-end. The heightened processivities of Kip3 and Kif18A are conferred in part by their C-terminal tail domains, which bind the microtubule lattice [20,21,60,61]. Because long microtubules contain more motor-binding sites, they will accumulate greater concentrations of kinesin-8 motors at their plus-ends. Second, kinesin-8 motors pause at the plus-end and impact their dynamics. For Kip3, which destabilizes the plus-ends of GMPCPP microtubules, these two properties enable the motor to depolymerize microtubules in a length-dependent fashion [15,22]: the depolymerization rate of short microtubules is slower than that for long microtubules.
Because GMPCPP-stabilized microtubules are considered to mimic the GTP-cap (Box 2) of growing dynamic microtubules, the ability of Kip3 to depolymerize GMPCPP-stabilized microtubules may correspond to an ability to remove the GTP-cap from dynamic microtubules and promote catastrophe. Indeed, Kip3 enhances the catastrophe rate in a length-dependent manner on reconstituted dynamic microtubules in vitro [63] and on artificially induced long kinetochore microtubules in vivo [88]. It is worth noting that the length-dependent effect is only obvious on long microtubules in these studies. The effect is subtle on microtubules shorter than two microns, the length of most yeast astral microtubules and kinetochore microtubules. Thus, for budding yeast Kip3, it is not yet clear if length-dependent destabilization, as opposed to its general microtubule destabilizing effect, is the dominant regulatory mechanism in vivo. In fission yeast, there is good evidence that the kinesin-8s promote length-dependent catastrophe but there are several factors that complicate the interpretation [89]. First, the fission yeast kinesin-8s Klp5/6 do not have detectable depolymerase activity [33]. Second, the length dependent catastrophes are observed for microtubule bundles rather than individual microtubules [89]. Third, the speed of the fission yeast kinesin-8s on yeast microtubules is slower than microtubule growth rates in cytoplasm [32], raising the possibility that additional factors slowing down of microtubule growth at the cell tip could also influence these findings. To date, length-dependent catastrophe has not been described in human cells, but this mechanism has been proposed to drive the medial positioning of chromosomes along the spindle axis [31].
MOLECULAR ARCHITECTURE OF THE KINESIN-MICROTUBULE INTERFACE
The kinesin superfamily of proteins share high sequence homology in their motor domains, which contain an ATP-binding catalytic core and microtubule-interacting surfaces. The motor domain forms an arrow-shaped structure that is built from a central β-sheet, flanked on each side by three α-helices. The arrow base lies flat on the microtubule lattice, with its tip pointed towards the plus-end. The phosphate-binding P-loop, switch-I, and switch-II regions form the nucleotide binding and hydrolysis motifs. The switch-II cluster (helices α4 - loop L12 – helices α5) follows switch II (loop L11) and composes the major motor-microtubule interaction surfaces. The switch-II cluster slides along the central β sheet in a manner that is regulated by the nucleotide–binding state, coordinating nucleotide status and microtubule binding affinity [5,6].
Features of microtubule-depolymerizing kinesins
Kinesin motors that directly regulate microtubule dynamics are indicated in Table 1. These motors share the common motor domain fold of the walking kinesins, but also contain unique structural elements. Kinesin-13, the first identified and best-understood microtubule depolymerase, contains a family-specific, extended loop 2 (L2) within its motor domain. L2 forms a rigid finger-like structure with a conserved “KVD” peptide motif located at the tip of the L2 finger. Structural modeling suggests that the kinesin-13 L2 binds the interdimer interface of tubulin, stabilizing the peeled conformation of protofilaments typical of depolymerizing microtubules. Consistent with this hypothesis, the KVD motif is critical for the depolymerase activity of kinesin-13 [7,8]. Overall, the microtubule interaction surface of the kinesin-13 motor domain has a convex conformation that appears to be more complementary to the curved tubulin conformation at microtubule plus-ends than to the straight tubulin conformation along the lattice [7,8]. That kinesin-13s stabilize the bent conformation of tubulin was first suggested by its ability to induce the formation of tubulin rings from Taxol-stabilized microtubules [9].
Table 1.
Kinesins that directly regulate microtubule dynamics
| Family | Gene | Effect | Refs |
|---|---|---|---|
| Kinesin-4 | XKLP1 | Inhibits MT growth and shrinkage | [37–39] |
|
| |||
| Kinesin-7 | CENP-E | Promotes elongation of stabilized MTs | [62] |
|
| |||
| Kinesin-8 | KIP3 | Depolymerizes stabilized MTs | [14,15] |
| Promotes MT catastrophe and inhibits MT growth | [63] | ||
|
| |||
| KIF18A | Inhibits MT growth | [25] | |
|
| |||
| KLP5/6 | Promotes MT nucleation | [32] | |
|
| |||
| Kinesin-10 | NOD | Promotes tubulin polymerization | [64] |
|
| |||
| Kinesin-13 | XKCM1 | Depolymerizes stabilized MTs | [47] |
| Inhibits MT growth and promotes MT catastrophe | |||
|
| |||
| MCAK | Depolymerizes stabilized MTs | [65] | |
| Promotes MT catastrophe | [63] | ||
|
| |||
| Kinesin-14 | KAR3 | Depolymerizes stabilized MTs | [44] |
The ATPase cycle of kinesin-13s also appears to be tailored to their functions as microtubule depolymerases. ADP-release, one of the rate-limiting steps in the ATP hydrolysis cycle of kinesin-13s, appears to be maximally stimulated by microtubule ends in comparison to the stimulation from microtubule lattice [10]. This contrasts with motile kinesin-1 where ADP release is maximally stimulated by the lattice [11]. Therefore, kinesin-13 only remains at the ADP-bound state along the lattice, unable to undergo ATP hydrolysis cycles, step along the lattice, or make directional movement. Lacking motility, kinesin-13s find microtubule ends either by one dimensional diffusion along the lattice [12], or by binding to proteins at the microtubule end such as EB1 [13]. Thus kinesin-13s have made an evolutionary trade-off: losing the capacity for directional motility in the process of having acquired robust depolymerase activity.
The multifunctional kinesin-8s
However, motility and depolymerase activity are not inherently conflicting. Kinesin-8s are the most processive motile kinesins known, yet at least some family members possess depolymerase activity [14,15]. Although the molecular mechanism that enables the co-existence of both activities remains unclear, kinesin-8s do share certain structural features with both conventional motile kinesins and the kinesin-13s. Similar to other plus-end directed motile kinesins, kinesin-8s have an N-terminal motor domain, rather than the internal motor domain found in kinesin-13s. However, like kinesin-13s, kinesin-8s contain an extended loop L2 that appears to make an extra contact with α-tubulin at the minus end along the motor-microtubule interface (Figure 1) [16]. The amino acid sequence and length of this loop shares little similarity with L2 from kinesin-13s. Although it is possible that the L2 sequences of kinesin-13s and kinesin-8s have structural similarity, this remains untested because the L2 sequence of kinesin-8 is not visible in the crystal structure [16].
Figure 1. Interacting surfaces between kinesin motor domain and tubulin dimers.

Structure of the kinesin-8/Kif18A motor domain – microtubule complex. Overview of the asymmetric unit of the 13 Å helical reconstruction shows the Kif18A motor domain bound to the α/β tubulin dimer. The kinesin-8-α tubulin contact mediated by loop L2 is highlighted in sky blue dotted circle. The motor-tubulin interface was depicted by a blue curve. This figure was adapted from [16].
Because the motor domain of the human kinesin-8 Kif18A can induce the formation of tubulin rings from Taxol-stabilized microtubules [16], it is appealing to speculate that kinesin-8s, similar to kinesin-13s, destabilize microtubules by bending protofilament at plus-ends. However, Kif18A induces rings significantly less efficiently than MCAK. Moreover, as discussed below, there is also controversy over the biochemical properties of Kif18A: different groups have suggested that it either weakly depolymerizes microtubules or lacks depolymerase activity entirely. More structural information coupled with mutagenesis and domain swaps may shed light on whether the specific sequences within Loop 2 or the overall rigidity of its structure play a key role in determining the biochemical properties of kinesin-8s.
Do kinesins interact differently with the microtubule lattice and the microtubule end?
Tubulin subunits on the microtubule end have exposed surfaces that are buried when tubulin is incorporated into the microtubule lattice. The plus-end also differs from the lattice because of its “cap” composed of GTP-liganded tubulin (Box 2). The microtubule plus-end is therefore chemically and structurally different from the lattice and it is therefore possible that kinesin motors could interact differently with microtubule plus-ends compared to the lattice. Indeed, exciting recent work has demonstrated that a member of the EB-family of non-motor microtubule plus-end binding proteins makes specific contacts on microtubules that likely are important for sensing the nucleotide state of tubulin [17]. The idea that some kinesins might also sense features of the microtubule end is supported by the fact that both kinesin-13s and kinesin-8s can bind tubulin dimers. Binding to tubulin dimers is a common feature shared by many microtubule plus end-tracking proteins (+TIPs), such as CLIP-170, CLASP, and XMAP215 [18,19]. Furthermore, additional studies have also shown that kinesin-8 motors pause at microtubule plus-ends for extended periods (~60–75 seconds) of time both in vitro and at kinetochore-microtubule plus-ends. The mechanism underlying this prolonged dwell time at plus-ends is uncharacterized, but is known to require the microtubule-binding activity of its C-terminal tail domain [20,21]. Interestingly, paused motors at microtubule plus-ends appear to be “bumped off” by incoming motors, leading to the proposal of a novel mechanism of microtubule depolymerization mediated by cooperative interactions among motors (Box 3) [22]. It was also recently reported that individual Kip3 motors frequently step from one protofilament to the next during processive walking [23]. This raises the question of whether Kip3 really pauses at plus-ends or it actually steps from one protofilament to another. A more interesting question is whether torsional force generated during inter-filament stepping could destabilize lateral interactions between protofilaments at plus-ends and contribute to microtubule depolymerization.
Box 2. GTP-cap [86].
A “GTP-cap” model is believed to underlie microtubule dynamic instability. When a microtubule grows, GTP-liganded tubulins are incorporated into the lattice. Once in the lattice, GTP is hydrolyzed to GDP. Because of a time lag between tubulin incorporation and GTP hydrolysis, the tip of growing microtubules is composed of GTP-liganded tubulin, which is called “GTP-cap” whereas the lattice of microtubules is composed of GDP-liganded tubulin. GTP-tubulin was suggested to have a straight conformation in the lattice, which stabilizes microtubules and favors growing. Once the GTP-cap is physically removed or chemically removed by GTP hydrolysis, the GDP- tubulins are exposed. GDP-bound tubulins adopt a bent conformation, which destabilizes microtubule plus-ends and initiate shrinkage. The GTP-cap can be regained during the shrinkage phase and this is accompanied by the regrowth of microtubules. The transition from growing to shrinking is called catastrophe, while the transition from shrinking to growing is called rescue. Catastrophe and rescue frequencies, together with growth and shrinkage rates, are the four parameters commonly used to describe microtubule dynamics.
The GTP-cap was suggested to have sub-structures by recent studies. Microtubules were found to have tapered ends, rather than blunt ends during the growth phase. This suggests the length of GTP-cap is not uniform among the protofilaments [87]. Another study suggested it takes multiple steps to trigger a catastrophe. Structural flaws could be accumulated in the GTP-cap during the growth before a catastrophe is triggered [63]. These studies suggested a variety of mechanisms that microtubule regulators can adopt to affect the assembly dynamics.
Box 3. “bump-off” model.
Work from the Howard Laboratory showed the ~minute-long dwell time of Kip3 molecules at the microtubule plus-end is markedly decreased as the flux of motors coming to the plus-end is increased. This has led to an attractive model where incoming motors push other motors off the plus-end, triggering the removal of tubulin dimers from the end [22]. Although appealing, there are important aspects of this model that remain to be tested: the bump-off effect is well documented on chemically stabilized microtubules, but has not been examined on dynamic microtubules. It is also difficult to completely exclude the possibility that multiple motors alter the architecture of the microtubule plus-end and thus diminish the dwell time for end-bound motors indirectly. Much also remains to be learned about the nature of motor-motor interactions at the plus end: is the interaction specific or a “slap in the back” from motors crowding towards the plus-end? Would the force from the latter type of interaction be sufficient to dislodge a motor from the plus-end?
HOW DOES KINESIN-8 CONTROL MICROTUBULE LENGTH?
The function and dynamic properties of many microtubule-dependent systems are defined by the lengths of individual microtubules contained within them. During cell division, the lengths of astral microtubules govern the probability that their plus-ends will be captured by the cortex and thereby participate in spindle positioning. The lengths of kinetochore-fiber microtubules on the other hand dictate the location of chromosomes in the mitotic spindle. Cellular “rulers” that measure size and length are poorly characterized, generally, and the mechanisms that sense and regulate microtubule length are no exception. Kinesin-8 motors have emerged as important regulators of microtubule length and microtubule-dependent cellular processes, particularly during mitosis. Table 2 summarizes kinesin-8s’ cellular functions that have been characterized in various organisms. Many of these functions can be attributed, or partially attributed, to their plus end-directed motility and plus end-specific destabilizing activity [14,15,24,25]. Interestingly, a combination of these two activities led to a novel length-dependent mechanism that was proposed to maintain the cellular microtubule length homeostasis (Box 1) [15,22].
Table 2.
Cellular and physiological functions of Kinesin-8s
| Species | Gene | Function | Reference |
|---|---|---|---|
| Saccharomyces cerevisiae | KIP3 | Spindle positioning | [68,69] |
| Kinetochore clustering | [20,70] | ||
| Spindle size control | [26,71,72] | ||
|
| |||
| Ashbya gossypii | KIP3 | Suppressing nuclear oscillations | [73] |
|
| |||
| Aspergillus nidulans | KIPB | Spindle positioning | [29] |
| Mitotic progression | |||
|
| |||
| Schizosaccharomyces pombe | KLP5/6 | Kinetochore alignment | [74,75] |
| Meiosis progression | [30] | ||
| Silencing spindle assembly checkpoint | [76] | ||
|
| |||
| Drosophila melanogaster | KLP67A | Kinetochore congression | [77,78] |
| Spindle size control | [27] | ||
| Centrosome separation | [28] | ||
| Spindle elongation | [79] | ||
| Central spindle assembly | [28,80] | ||
|
| |||
| Homo sapiens | KIF18A | Kinetochore congression | [24,31] |
| Spindle size control | |||
| Male fertility (in mouse) | [81] | ||
| Overexpressed in breast, lung, and colorectal cancer | [82–84] | ||
| Human height association | [85] | ||
|
| |||
| KIF18B | Suppress astral microtubule overgrowth | [58,59] | |
Family-conserved functions are in bold
Enhancing Kinesin-8 mediated length control by flipping between stabilizing and destabilizing states
Kinesin-8s not only destabilize microtubules but also, in certain contexts, possess the ability to stabilize them. The importance of the microtubule destabilizing activity of kinesin-8s is underscored by the fact that loss of kinesin-8s generally leads to long cellular microtubules [24,26–30]. However, time-lapse imaging of cellular microtubule dynamics revealed unexpected complexity. Kip3 in budding yeast destabilizes growing astral microtubules by promoting catastrophe, whereas it stabilizes shrinking astral microtubules by slowing shrinkage and promoting rescue [14]. The stabilizing effect of Kip3 on shrinking microtubules requires its microtubule and tubulin-binding tail domain [20]. These dual effects on microtubule dynamics by kinesin-8s are not unique to the budding yeast motor, because live cell imaging in human cells demonstrated that Kif18A also has stabilizing effects on astral and kinetochore microtubules [25,31]. In this case, a capping mechanism for Kif18A was proposed where Kif18A would dampen both microtubule growth and shrinkage [25].
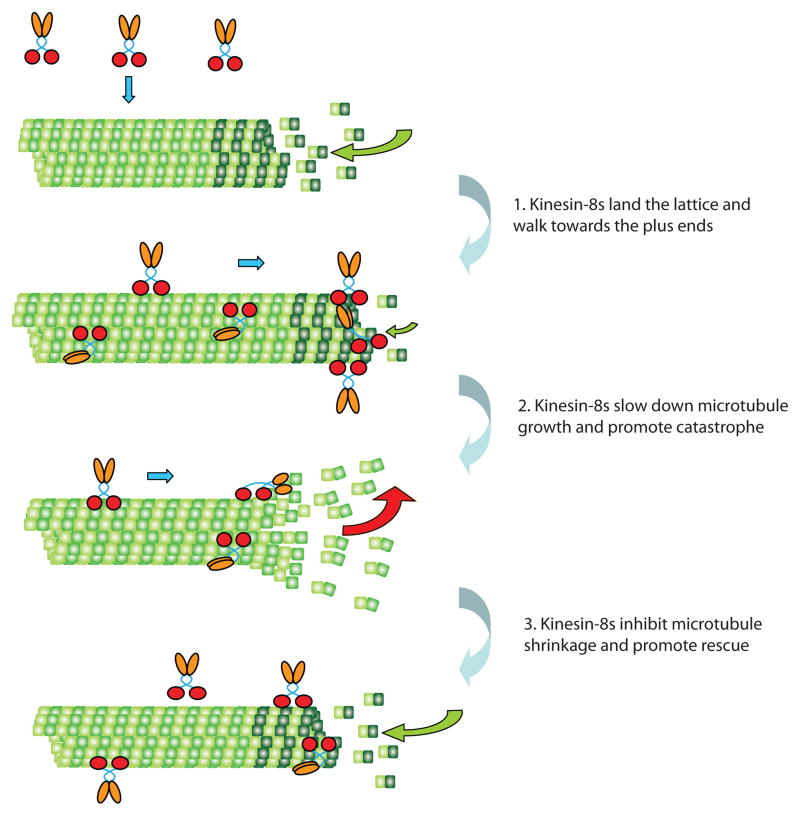
How are the opposing effects of kinesin-8s on microtubule dynamics integrated into a coherent regulatory effect? A concentration-threshold model has been proposed to explain the effects of Kip3 on cellular microtubule dynamics that may also be relevant to Kif18A and other kinesin-8s (Figure 2) [20]. When microtubules grow, Kip3 lands on the lattice, and walks towards the plus-end. Because the velocity of Kip3 is faster than the growth rate of microtubules, and because of its high processivity [14,15], Kip3 accumulates at the plus-end and promotes catastrophes. The large amount of Kip3 on growing microtubules enables cooperative interactions that favor catastrophe. After catastrophe, the high concentration of Kip3 at the plus-end is lost but a lower level of Kip3 reaches the end of the shrinking microtubule because of the ongoing flux along the lattice. At lower concentrations of Kip3, the stabilizing effects dominate, decreasing the shrinkage rate and promoting rescue. The stabilization of short shrinking microtubules combined with the destabilization of long growing microtubules is expected to synergize to narrow the length variation of microtubules in cells. A similar model has been proposed to explain Kif18As’ role in promoting metaphase chromosome congression [31].
Figure 2. A model for regulation of microtubule dynamics by kinesin-8.
1. Kinesin-8s land on the lattice of a growing microtubule and walk processively towards the plus-end in a manner facilitated by its C-terminal microtubule-binding domain. 2. Kinesin-8s accumulate at the plus-end, slowing down microtubule growth and promoting catastrophe through a depolymerizing or capping mechanism. 3. During the shrinking phase, kinesin-8s at low concentrations inhibit microtubule shrinkage and promote rescue. The figure was adapted from [20].
DIVERSE MECHANISMS FOR KINESIN-MEDIATED MICROTUBULE DYNAMICS REGULATION
Are all kinesin-8s microtubule depolymerases?
As discussed above, cells lacking kinesin-8s have long microtubules and yeast Kip3 directly depolymerizes microtubules in vitro. However, whether a generalized mechanism can explain how kinesin-8s cause microtubules to catastrophe is unclear. One study has reported that human Kif18A can depolymerize GMPCPP-stabilized microtubules, albeit inefficiently [24]. However, other studies have shown that Kif18A suppresses the growth of microtubule plus-ends and promotes microtubule pausing in a concentration-dependent manner [21,25]. This activity is consistent with the ability of Kif18A to suppress kinetochore oscillations in cells [31]. How might such a pausing activity lead to catastrophes in cells? In principle, prolonged pausing of a microtubule plus-end would deplete GTP-tubulin from the GTP cap, leading to a catastrophe. Notably, fission yeast Kinesin-8s Klp5/6 do not have a detectable depolymerase activity [32,33]. Biochemical evidence of their ability to suppress microtubule plus-end dynamics has not been reported.
What is the mechanistic basis for the differing activities of Kip3 and the other kinesin-8 family members? One possibility is that during the evolution, Kip3 acquired new functions and new ways of interacting with the microtubule not present in the kinesin-8s in other organisms. Alternatively, Kip3 and other kinesin-8s could interact with microtubule ends in essentially the same way, but with differences in affinity. Formin biochemistry is particularly illustrative of this possibility. These molecules bind the barbed ends of actin filaments where they promote filament elongation. Although it is now well-appreciated that all formins remain attached to the barbed end while simultaneously promoting barbed end assembly [34], the fission yeast formin Cdc12 initially appeared to be a barbed end capping protein. This effect could be relieved, however, by the addition of profilin, a natural formin binding partner. In the presence of profilin, the block of Cdc12 for the barbed end assembly is released, triggering a regime where Cdc12 becomes a processive barbed end polymerase, just like all other formins [35]. The mechanism for the profilin effect is not fully defined, but is likely to be due in part to profilin increasing the off-rate of Cdc12 from the barbed end of the actin filament (D. Kovar, personal communication). It will be interesting to determine whether the differing biochemical properties of Kip3 and Kif18A represent qualitative or quantitative differences in how they interact with microtubule ends and influence their dynamics.
Another kinesin-mediated mechanism for microtubule length control
Kinesin-4 is another example of a processive plus-end-directed kinesin important for microtubule length control. Kinesin-4 is recruited to anti-parallel overlapping microtubules by the non-motor MAP PRC1. PRC1 is a member of the Ase1/Map65 family of proteins that crosslink anti-parallel microtubules. In metazoans, both PRC1 and kinesin-4s are required to maintain the anti-parallel arrays of microtubules in the anaphase spindle midzone [36]. A mini-midzone has been reconstituted from dynamic microtubules, PRC1 and kinesin-4 (Xklp1 in Xenopus). Once recruited by PRC1, Xklp1 walks processively back and forth between anti-parallel microtubules, giving the appearance of “patrolling” the zone of microtubule overlap [37]. Xklp1 limits the length of the overlap zone by dampening both microtubule growth and shrinkage in a manner similar to Kif18A. Unlike Kip3, Xklp1 has little or no effect on the catastrophe frequency [38]. Xklp1 therefore stabilizes the overlap between antiparallel microtubules, but prevents overgrowth of the overlap zone. The in vitro results are consistent with the observation that human kinesin-4 Kif4 suppresses microtubule growth [39] and restricts spindle midzone size in cells [40]. Together, these findings suggest an appealing self-rectifying mechanism to control microtubule length in the mid-zone: when microtubule overlap is reduced by midzone sliding motors, there is reduced kinesin-4 and thus more microtubule growth, enabling regrowth to its normal size.
Although both kinesin-4s and kinesin-8s inhibit microtubule growth and promote microtubule pausing [25,38,39], the underlying mechanism could be different. Kinesin-8 strongly accumulates on growing microtubule ends [14,21], whereas kinesin-4/Xklp1 evenly decorates microtubules [38]. Cryoelectron microscopy experiments suggest that Xklp1 induces disruptions of the microtubule lattice and generates frayed microtubule ends. These observations suggest Xklp1 could affect the dynamic properties of microtubule ends through allosteric effects on the microtubule lattice [38]. In principle this would be quite different from what is thought to be the end-specific regulation of microtubule dynamics by kinesin-8s and kinesin-13s. However, Xklp1 shares the ability to bind to tubulin dimers with kinesin-8 and kinesin-13s [38], suggesting a counterargument that kinesin-4s may in fact interact with microtubule ends in a manner that is not wholly dissimilar to that of kinesin-8s.
A minus-end-directed motor that promotes plus-end depolymerization
Kar3, the budding yeast kinesin-14, is an interesting example of a minus-end-directed motor that regulates plus-end dynamics. Kar3 crosslinks and promotes the shortening of microtubule plus-ends during karyogamy, the process of nuclear fusion that occurs during mating [41,42]. Unlike other kinesin-14s, which usually form homodimers, Kar3 forms heterodimers with non-catalytic partners, Cik1 or Vik1 [43]. In vitro, the Kar3-Cik1 heterodimer promotes microtubule depolymerization from the plus-end [44]. By comparison with MCAK or Kip3, this depolymerizing activity is relatively weak, being revealed only in the presence of a low concentration of Taxol. The biochemical properties of Kar3-Cik1 suggest a model where Cik1 anchors the heterodimer to the plus-end, where the Kar3 motor could promote plus-end disassembly by generating a compressive force between tubulin subunits at the plus-ends. Interestingly, it has recently been reported that catastrophe is triggered when microtubules hit an artificial barrier coated with cytoplasmic dynein, another microtubule minus-end-directed motor. Dynein pulls microtubule against the barrier, which potentially causes plus-end deformation and triggers destabilization [45]. Thus, this type of mechanism appears to be general.
OPTIMALITY IN THE EVOLUTION OF MOTORS THAT GOVERN CELLULAR MICROTUBULE DYNAMICS
A minimum of 11 kinesin families were predicted to be present in the last common eukaryotic ancestor. These include all the major microtubule-regulating kinesin families discussed above: kinesin-4, -8, -13, and -14 [46]. During evolution, certain kinesins have been lost or gained whereas the functional requirements for kinesins in microtubule regulation seem to remain. For example, budding yeasts do not have kinesin-13, the most robust microtubule depolymerases. Since simple eukaryotes such as Giardia encode kinesin-13 proteins, it is likely that kinesin-13s were lost in fungi. Yeast kinesin-14/Kar3, which is unique among kinesin-14s in the ability to destabilize microtubule ends, may have acquired the ability to depolymerize microtubules as a compensatory mechanism.
Gene duplication events in higher eukaryotes has markedly increased the number of kinesins, enabling specialization of function and potentially the acquisition of novel activities. The functions of these motors are intimately related to their biophysical properties and association with other proteins that impact their distribution within the cell. As shown for the kinesin-13s, even members of the same subfamily can have striking differences in localization and biochemical activity. While comparably active as GMPCPP microtubule depolymerases in vitro [47,48], MCAK/Kif2C targets the centromere of mitotic chromosomes [49,50] and tracks with microtubule plus-ends [51] whereas Kif2A localizes primarily to spindle poles [52] and fails to tip-track [51]. In contrast, Kif24 is concentrated at the mother centriole and interestingly, exhibits a reduced capacity to disassemble Taxol or GMPCPP-stabilized microtubules [53]; (S. Gayek and P. Ohi, unpublished observations).
The cellular functions of these motors reflect these important differences. At centromeres, MCAK/Kif2C destabilizes kinetochore-microtubules which both minimizes flawed kinetochore-microtubule attachments [54] and coordinates the oscillatory behavior of sister kinetochores [55]. At spindle poles, Kif2A destabilizes microtubule minus-ends thereby “reeling in” microtubules to drive polewards microtubule flux [56]. The unique ability of MCAK/Kif2C, but not other kinesin-13s, to track with microtubule plus-ends via EB1-binding [13] specifically enables this motor to increase microtubule dynamicity by causing microtubule plus-ends to more rapidly transit between growth and shortening [57]. Since kinesin-13s are nonmotile and reach microtubule ends by diffusion along the microtubule lattice [12], positioning MCAK/Kif2C near microtubule ends would facilitate its action particularly if its microtubule substrate was long in length or short-lived. Plus-end tracking may thus be important in remodeling interphase microtubule arrays and may also account for the observation that MCAK is more critical for spindle assembly than Kif2A [48]. Intriguingly, Kif24 selectively destabilizes centriolar microtubules [53] which is consistent with its modest ability to depolymerize stabilized microtubules in vitro. The ability of Kif24 to discriminate between microtubule substrates, as well as the mechanism by which the motor localizes to the mother centriole, is a fascinating area of future work.
Emerging work suggests that kinesin-8s will also exhibit functional diversity. While the canonical kinesin-8s accumulate at kinetochore-microtubule plus-ends and act to align chromosomes at the midzone of the mitotic spindle, Kif18B localizes to all spindle microtubule plus-ends [58,59]. This distinction can in part be explained by the different functions of the Kip3/Kif18A and Kif18B C-terminal tail domains. Whereas the tail domain of Kif18A serves as a processivity module [21,60,61], the C-terminus of Kif18B interacts with EB1 [58,59]. This change in domain structure has important implications for the motile and biophysical properties of Kif18B. In particular, it will be interesting to see if Kif18B, like Kif18A and Kip3, is highly processive, a property that up to now has been stereotypical of the kinesin-8 family. Whether Kif18B acts to regulate microtubule dynamics is also unclear. Microtubules in cancer cells depleted of Kif18B are abnormally long [58,59], but Tanenbaum et al. have proposed that the primary function of Kif18B is to deliver MCAK to microtubule plus-ends. This is interesting albeit surprising since most kinesin-8 motor domains characterized thus far have some capacity to regulate microtubule dynamics.
Concluding remarks
Kinesin superfamily members have evolved a diverse repertoire of biochemical activities that allow them to sculpt the microtubule cytoskeleton in addition to driving intracellular transport. Kinesin motors are thus well-suited to power cellular processes that occur on timescales of seconds to minutes and underlie the more global organization of the microtubule cytoskeleton. In this review, we have attempted to summarize why the microtubule regulating kinesins are particularly instrumental. These motors have biophysical properties that allow them to measure and influence microtubule length, enabling them to control the size and dynamics of diverse cellular structures. Motors are also fast-acting. Their action at microtubule tips is principally limited by their lattice on-rate, walking speed, and processivity. By comparison, chance microtubule plus-end interactions with cellular structures that influence their dynamics, such as kinetochores and cortical attachment sites, are slow. The prevailing model of how Kif18A drives chromosome congression is a useful example of a self-assembled system that relies on the unique biophysical properties of kinesin-8 motors, kinetochore-microtubule length and lifetime.
An outstanding question is whether dynamics-regulating motors interact with the microtubule in different ways than a purely motile motor or whether dynamics regulation is enabled by more subtle changes in binding affinity or kinetic parameters. This issue is important because it impacts the degree to which motors must undergo specialization for their cellular functions. The extreme example is kinesin-13/MCAK, which has lost motility in favor of gaining structural elements that enable robust depolymerase activity. Microtubule plus-end regulation by kinesin-8s could involve both qualitative and quantitative differences of how the motors interact with plus-ends compared to purely motile kinesins. Indeed, cryoelectron microscopy data suggest that the kinesin-8 specific loop 2 makes unique contacts at the tubulin dimer interface. But the high processivity of these motors raises the possibility that the strength of contacts might also contribute. We have speculated that such quantitative differences might explain why Kip3 has robust depolymerase activity whereas other family members do not. It is worth noting that most of the dynamics regulating motors we have discussed destabilize microtubules. A hypothetical explanation for this trend is that the strain exerted by the dimeric motor on adjacent tubulin subunits distorts the protofilament and favors destabilization. This would imply that every kinesin has some potential to affect microtubule dynamics; whether they do so would then be a question of affinity and time spent at the microtubule end.
Looking forward, we anticipate that additional structural data, integrated with single molecule imaging, mutational analysis, and in vivo cell biological approaches will resolve how kinesin-8 motors, as well as other kinesin superfamily members, affect microtubule plus-end dynamics. These approaches will also enable us to relate the activities of various microtubule regulating kinesins to each other. In addition, biochemical reconstitution and mathematical modeling is enabling complex microtubule structures to be built from the ground up. This is enabling us to see how simple regulatory building blocks, such as dynamics-regulating kinesins, facilitate emergent properties of the microtubule cytoskeleton.
Acknowledgments
We thank H. Arellano-Santoyo for helpful comments on the manuscript. D. Pellman was supported by Howard Hughes Medical Institute and a National Institute of Health grant (GM61345). R. Ohi was supported by National Institute of Health Grant GM086610.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Maccioni RB, Cambiazo V. Role of microtubule-associated proteins in the control of microtubule assembly. Physiol Rev. 1995;75:835–864. doi: 10.1152/physrev.1995.75.4.835. [DOI] [PubMed] [Google Scholar]
- 2.Roll-Mecak A, McNally FJ. Microtubule-severing enzymes. Curr Opin Cell Biol. 2010;22:96–103. doi: 10.1016/j.ceb.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gouveia SM, Akhmanova A. Cell and molecular biology of microtubule plus end tracking proteins: end binding proteins and their partners. Int Rev Cell Mol Biol. 2010;285:1–74. doi: 10.1016/B978-0-12-381047-2.00001-3. [DOI] [PubMed] [Google Scholar]
- 4.Schuyler SC, Pellman D. Microtubule “plus-end-tracking proteins”: The end is just the beginning. Cell. 2001;105:421–424. doi: 10.1016/s0092-8674(01)00364-6. [DOI] [PubMed] [Google Scholar]
- 5.Marx A, Hoenger A, Mandelkow E. Structures of kinesin motor proteins. Cell Motil Cytoskeleton. 2009;66:958–966. doi: 10.1002/cm.20392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sack S, Kull FJ, Mandelkow E. Motor proteins of the kinesin family. Structures, variations, and nucleotide binding sites. Eur J Biochem. 1999;262:1–11. doi: 10.1046/j.1432-1327.1999.00341.x. [DOI] [PubMed] [Google Scholar]
- 7.Ogawa T, Nitta R, Okada Y, Hirokawa N. A common mechanism for microtubule destabilizers-M type kinesins stabilize curling of the protofilament using the class-specific neck and loops. Cell. 2004;116:591–602. doi: 10.1016/s0092-8674(04)00129-1. [DOI] [PubMed] [Google Scholar]
- 8.Shipley K, Hekmat-Nejad M, Turner J, Moores C, Anderson R, Milligan R, Sakowicz R, Fletterick R. Structure of a kinesin microtubule depolymerization machine. EMBO J. 2004;23:1422–1432. doi: 10.1038/sj.emboj.7600165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moores CA, Yu M, Guo J, Beraud C, Sakowicz R, Milligan RA. A mechanism for microtubule depolymerization by KinI kinesins. Mol Cell. 2002;9:903–909. doi: 10.1016/s1097-2765(02)00503-8. [DOI] [PubMed] [Google Scholar]
- 10.Friel CT, Howard J. The kinesin-13 MCAK has an unconventional ATPase cycle adapted for microtubule depolymerization. EMBO J. 2011;30:3928–3939. doi: 10.1038/emboj.2011.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hackney DD. The rate-limiting step in microtubule-stimulated ATP hydrolysis by dimeric kinesin head domains occurs while bound to the microtubule. J Biol Chem. 1994;269:16508–16511. [PubMed] [Google Scholar]
- 12.Helenius J, Brouhard G, Kalaidzidis Y, Diez S, Howard J. The depolymerizing kinesin MCAK uses lattice diffusion to rapidly target microtubule ends. Nature. 2006;441:115–119. doi: 10.1038/nature04736. [DOI] [PubMed] [Google Scholar]
- 13.Honnappa S, Gouveia SM, Weisbrich A, Damberger FF, Bhavesh NS, Jawhari H, Grigoriev I, van Rijssel FJ, Buey RM, Lawera A, et al. An EB1-binding motif acts as a microtubule tip localization signal. Cell. 2009;138:366–376. doi: 10.1016/j.cell.2009.04.065. [DOI] [PubMed] [Google Scholar]
- 14.Gupta ML, Jr, Carvalho P, Roof DM, Pellman D. Plus end-specific depolymerase activity of Kip3, a kinesin-8 protein, explains its role in positioning the yeast mitotic spindle. Nat Cell Biol. 2006;8:913–923. doi: 10.1038/ncb1457. [DOI] [PubMed] [Google Scholar]
- 15.Varga V, Helenius J, Tanaka K, Hyman AA, Tanaka TU, Howard J. Yeast kinesin-8 depolymerizes microtubules in a length-dependent manner. Nat Cell Biol. 2006;8:957–962. doi: 10.1038/ncb1462. [DOI] [PubMed] [Google Scholar]
- 16.Peters C, Brejc K, Belmont L, Bodey AJ, Lee Y, Yu M, Guo J, Sakowicz R, Hartman J, Moores CA. Insight into the molecular mechanism of the multitasking kinesin-8 motor. EMBO J. 2010;29:3437–3447. doi: 10.1038/emboj.2010.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maurer SP, Fourniol FJ, Bohner G, Moores CA, Surrey T. EBs recognize a nucleotide-dependent structural cap at growing microtubule ends. Cell. 2012;149:371–382. doi: 10.1016/j.cell.2012.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Slep KC, Vale RD. Structural basis of microtubule plus end tracking by XMAP215, CLIP-170, and EB1. Mol Cell. 2007;27:976–991. doi: 10.1016/j.molcel.2007.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Al-Bassam J, Kim H, Brouhard G, van Oijen A, Harrison SC, Chang F. CLASP promotes microtubule rescue by recruiting tubulin dimers to the microtubule. Dev Cell. 2010;19:245–258. doi: 10.1016/j.devcel.2010.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Su X, Qiu W, Gupta ML, Jr, Pereira-Leal JB, Reck-Peterson SL, Pellman D. Mechanisms underlying the dual-mode regulation of microtubule dynamics by Kip3/kinesin-8. Mol Cell. 2011;43:751–763. doi: 10.1016/j.molcel.2011.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stumpff J, Du Y, English CA, Maliga Z, Wagenbach M, Asbury CL, Wordeman L, Ohi R. A tethering mechanism controls the processivity and kinetochore-microtubule plus-end enrichment of the kinesin-8 Kif18A. Mol Cell. 2011;43:764–775. doi: 10.1016/j.molcel.2011.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Varga V, Leduc C, Bormuth V, Diez S, Howard J. Kinesin-8 motors act cooperatively to mediate length-dependent microtubule depolymerization. Cell. 2009;138:1174–1183. doi: 10.1016/j.cell.2009.07.032. [DOI] [PubMed] [Google Scholar]
- 23.Bormuth V, Nitzsche B, Ruhnow F, Mitra A, Storch M, Rammner B, Howard J, Diez S. The Highly Processive Kinesin-8, Kip3, Switches Microtubule Protofilaments with a Bias toward the Left. Biophys J. 2012;103:L4–6. doi: 10.1016/j.bpj.2012.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mayr MI, Hummer S, Bormann J, Gruner T, Adio S, Woehlke G, Mayer TU. The human kinesin Kif18A is a motile microtubule depolymerase essential for chromosome congression. Curr Biol. 2007;17:488–498. doi: 10.1016/j.cub.2007.02.036. [DOI] [PubMed] [Google Scholar]
- 25.Du Y, English CA, Ohi R. The kinesin-8 Kif18A dampens microtubule plus-end dynamics. Curr Biol. 2010;20:374–380. doi: 10.1016/j.cub.2009.12.049. [DOI] [PubMed] [Google Scholar]
- 26.Straight AF, Sedat JW, Murray AW. Time-lapse microscopy reveals unique roles for kinesins during anaphase in budding yeast. J Cell Biol. 1998;143:687–694. doi: 10.1083/jcb.143.3.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goshima G, Wollman R, Stuurman N, Scholey JM, Vale RD. Length control of the metaphase spindle. Curr Biol. 2005;15:1979–1988. doi: 10.1016/j.cub.2005.09.054. [DOI] [PubMed] [Google Scholar]
- 28.Gandhi R, Bonaccorsi S, Wentworth D, Doxsey S, Gatti M, Pereira A. The Drosophila kinesin-like protein KLP67A is essential for mitotic and male meiotic spindle assembly. Mol Biol Cell. 2004;15:121–131. doi: 10.1091/mbc.E03-05-0342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rischitor PE, Konzack S, Fischer R. The Kip3-like kinesin KipB moves along microtubules and determines spindle position during synchronized mitoses in Aspergillus nidulans hyphae. Eukaryot Cell. 2004;3:632–645. doi: 10.1128/EC.3.3.632-645.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.West RR, Malmstrom T, Troxell CL, McIntosh JR. Two related kinesins, klp5+ and klp6+, foster microtubule disassembly and are required for meiosis in fission yeast. Mol Biol Cell. 2001;12:3919–3932. doi: 10.1091/mbc.12.12.3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stumpff J, von Dassow G, Wagenbach M, Asbury C, Wordeman L. The kinesin-8 motor Kif18A suppresses kinetochore movements to control mitotic chromosome alignment. Dev Cell. 2008;14:252–262. doi: 10.1016/j.devcel.2007.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Erent M, Drummond DR, Cross RA. S. pombe kinesins-8 promote both nucleation and catastrophe of microtubules. PLoS One. 2012;7:e30738. doi: 10.1371/journal.pone.0030738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grissom PM, Fiedler T, Grishchuk EL, Nicastro D, West RR, McIntosh JR. Kinesin-8 from fission yeast: a heterodimeric, plus-end-directed motor that can couple microtubule depolymerization to cargo movement. Mol Biol Cell. 2009;20:963–972. doi: 10.1091/mbc.E08-09-0979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goode BL, Eck MJ. Mechanism and function of formins in the control of actin assembly. Annu Rev Biochem. 2007;76:593–627. doi: 10.1146/annurev.biochem.75.103004.142647. [DOI] [PubMed] [Google Scholar]
- 35.Kovar DR, Kuhn JR, Tichy AL, Pollard TD. The fission yeast cytokinesis formin Cdc12p is a barbed end actin filament capping protein gated by profilin. J Cell Biol. 2003;161:875–887. doi: 10.1083/jcb.200211078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kurasawa Y, Earnshaw WC, Mochizuki Y, Dohmae N, Todokoro K. Essential roles of KIF4 and its binding partner PRC1 in organized central spindle midzone formation. EMBO J. 2004;23:3237–3248. doi: 10.1038/sj.emboj.7600347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bieling P, Telley IA, Surrey T. A minimal midzone protein module controls formation and length of antiparallel microtubule overlaps. Cell. 2010;142:420–432. doi: 10.1016/j.cell.2010.06.033. [DOI] [PubMed] [Google Scholar]
- 38.Bringmann H, Skiniotis G, Spilker A, Kandels-Lewis S, Vernos I, Surrey T. A kinesin-like motor inhibits microtubule dynamic instability. Science. 2004;303:1519–1522. doi: 10.1126/science.1094838. [DOI] [PubMed] [Google Scholar]
- 39.Stumpff J, Wagenbach M, Franck A, Asbury CL, Wordeman L. Kif18A and Chromokinesins Confine Centromere Movements via Microtubule Growth Suppression and Spatial Control of Kinetochore Tension. Dev Cell. 2012;22:1017–1029. doi: 10.1016/j.devcel.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hu CK, Coughlin M, Field CM, Mitchison TJ. KIF4 regulates midzone length during cytokinesis. Curr Biol. 2011;21:815–824. doi: 10.1016/j.cub.2011.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Molk JN, Salmon ED, Bloom K. Nuclear congression is driven by cytoplasmic microtubule plus end interactions in S. cerevisiae. J Cell Biol. 2006;172:27–39. doi: 10.1083/jcb.200510032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maddox PS, Stemple JK, Satterwhite L, Salmon ED, Bloom K. The minus end-directed motor Kar3 is required for coupling dynamic microtubule plus ends to the cortical shmoo tip in budding yeast. Curr Biol. 2003;13:1423–1428. doi: 10.1016/s0960-9822(03)00547-5. [DOI] [PubMed] [Google Scholar]
- 43.Manning BD, Barrett JG, Wallace JA, Granok H, Snyder M. Differential regulation of the Kar3p kinesin-related protein by two associated proteins, Cik1p and Vik1p. J Cell Biol. 1999;144:1219–1233. doi: 10.1083/jcb.144.6.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sproul LR, Anderson DJ, Mackey AT, Saunders WS, Gilbert SP. Cik1 targets the minus-end kinesin depolymerase kar3 to microtubule plus ends. Curr Biol. 2005;15:1420–1427. doi: 10.1016/j.cub.2005.06.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Laan L, Pavin N, Husson J, Romet-Lemonne G, van Duijn M, Lopez MP, Vale RD, Julicher F, Reck-Peterson SL, Dogterom M. Cortical Dynein Controls Microtubule Dynamics to Generate Pulling Forces that Position Microtubule Asters. Cell. 2012;148:502–514. doi: 10.1016/j.cell.2012.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wickstead B, Gull K, Richards TA. Patterns of kinesin evolution reveal a complex ancestral eukaryote with a multifunctional cytoskeleton. BMC Evol Biol. 2010;10:110. doi: 10.1186/1471-2148-10-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Desai A, Verma S, Mitchison TJ, Walczak CE. Kin I kinesins are microtubule-destabilizing enzymes. Cell. 1999;96:69–78. doi: 10.1016/s0092-8674(00)80960-5. [DOI] [PubMed] [Google Scholar]
- 48.Ohi R, Burbank K, Liu Q, Mitchison TJ. Nonredundant functions of Kinesin-13s during meiotic spindle assembly. Curr Biol. 2007;17:953–959. doi: 10.1016/j.cub.2007.04.057. [DOI] [PubMed] [Google Scholar]
- 49.Wordeman L, Mitchison TJ. Identification and partial characterization of mitotic centromere-associated kinesin, a kinesin-related protein that associates with centromeres during mitosis. J Cell Biol. 1995;128:95–104. doi: 10.1083/jcb.128.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Walczak CE, Mitchison TJ, Desai A. XKCM1: a Xenopus kinesin-related protein that regulates microtubule dynamics during mitotic spindle assembly. Cell. 1996;84:37–47. doi: 10.1016/s0092-8674(00)80991-5. [DOI] [PubMed] [Google Scholar]
- 51.Moore AT, Rankin KE, von Dassow G, Peris L, Wagenbach M, Ovechkina Y, Andrieux A, Job D, Wordeman L. MCAK associates with the tips of polymerizing microtubules. J Cell Biol. 2005;169:391–397. doi: 10.1083/jcb.200411089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ganem NJ, Compton DA. The KinI kinesin Kif2a is required for bipolar spindle assembly through a functional relationship with MCAK. J Cell Biol. 2004;166:473–478. doi: 10.1083/jcb.200404012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kobayashi T, Tsang WY, Li J, Lane W, Dynlacht BD. Centriolar kinesin Kif24 interacts with CP110 to remodel microtubules and regulate ciliogenesis. Cell. 2011;145:914–925. doi: 10.1016/j.cell.2011.04.028. [DOI] [PubMed] [Google Scholar]
- 54.Huang H, Feng J, Famulski J, Rattner JB, Liu ST, Kao GD, Muschel R, Chan GK, Yen TJ. Tripin/hSgo2 recruits MCAK to the inner centromere to correct defective kinetochore attachments. J Cell Biol. 2007;177:413–424. doi: 10.1083/jcb.200701122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wordeman L, Wagenbach M, von Dassow G. MCAK facilitates chromosome movement by promoting kinetochore microtubule turnover. J Cell Biol. 2007;179:869–879. doi: 10.1083/jcb.200707120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ganem NJ, Upton K, Compton DA. Efficient mitosis in human cells lacking poleward microtubule flux. Curr Biol. 2005;15:1827–1832. doi: 10.1016/j.cub.2005.08.065. [DOI] [PubMed] [Google Scholar]
- 57.Montenegro Gouveia S, Leslie K, Kapitein LC, Buey RM, Grigoriev I, Wagenbach M, Smal I, Meijering E, Hoogenraad CC, Wordeman L, et al. In vitro reconstitution of the functional interplay between MCAK and EB3 at microtubule plus ends. Curr Biol. 2010;20:1717–1722. doi: 10.1016/j.cub.2010.08.020. [DOI] [PubMed] [Google Scholar]
- 58.Tanenbaum ME, Macurek L, van der Vaart B, Galli M, Akhmanova A, Medema RH. A complex of Kif18b and MCAK promotes microtubule depolymerization and is negatively regulated by Aurora kinases. Curr Biol. 2011;21:1356–1365. doi: 10.1016/j.cub.2011.07.017. [DOI] [PubMed] [Google Scholar]
- 59.Stout JR, Yount AL, Powers JA, Leblanc C, Ems-McClung SC, Walczak CE. Kif18B interacts with EB1 and controls astral microtubule length during mitosis. Mol Biol Cell. 2011;22:3070–3080. doi: 10.1091/mbc.E11-04-0363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Weaver LN, Ems-McClung SC, Stout JR, LeBlanc C, Shaw SL, Gardner MK, Walczak CE. Kif18A uses a microtubule binding site in the tail for plus-end localization and spindle length regulation. Curr Biol. 2011;21:1500–1506. doi: 10.1016/j.cub.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mayr MI, Storch M, Howard J, Mayer TU. A Non-Motor Microtubule Binding Site Is Essential for the High Processivity and Mitotic Function of Kinesin-8 Kif18A. PLoS One. 2011;6:e27471. doi: 10.1371/journal.pone.0027471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sardar HS, Luczak VG, Lopez MM, Lister BC, Gilbert SP. Mitotic kinesin CENP-E promotes microtubule plus-end elongation. Curr Biol. 2010;20:1648–1653. doi: 10.1016/j.cub.2010.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gardner MK, Zanic M, Gell C, Bormuth V, Howard J. Depolymerizing Kinesins Kip3 and MCAK Shape Cellular Microtubule Architecture by Differential Control of Catastrophe. Cell. 2011;147:1092–1103. doi: 10.1016/j.cell.2011.10.037. [DOI] [PubMed] [Google Scholar]
- 64.Cui W, Sproul LR, Gustafson SM, Matthies HJ, Gilbert SP, Hawley RS. Drosophila Nod protein binds preferentially to the plus ends of microtubules and promotes microtubule polymerization in vitro. Mol Biol Cell. 2005;16:5400–5409. doi: 10.1091/mbc.E05-06-0582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hunter AW, Caplow M, Coy DL, Hancock WO, Diez S, Wordeman L, Howard J. The kinesin-related protein MCAK is a microtubule depolymerase that forms an ATP-hydrolyzing complex at microtubule ends. Mol Cell. 2003;11:445–457. doi: 10.1016/s1097-2765(03)00049-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Drummond DR. Regulation of microtubule dynamics by kinesins. Semin Cell Dev Biol. 2011;22:927–934. doi: 10.1016/j.semcdb.2011.09.021. [DOI] [PubMed] [Google Scholar]
- 67.Daire V, Pous C. Kinesins and protein kinases: key players in the regulation of microtubule dynamics and organization. Arch Biochem Biophys. 2011;510:83–92. doi: 10.1016/j.abb.2011.02.012. [DOI] [PubMed] [Google Scholar]
- 68.Cottingham FR, Hoyt MA. Mitotic spindle positioning in Saccharomyces cerevisiae is accomplished by antagonistically acting microtubule motor proteins. J Cell Biol. 1997;138:1041–1053. doi: 10.1083/jcb.138.5.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.DeZwaan TM, Ellingson E, Pellman D, Roof DM. Kinesin-related KIP3 of Saccharomyces cerevisiae is required for a distinct step in nuclear migration. J Cell Biol. 1997;138:1023–1040. doi: 10.1083/jcb.138.5.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wargacki MM, Tay JC, Muller EG, Asbury CL, Davis TN. Kip3, the yeast kinesin-8, is required for clustering of kinetochores at metaphase. Cell Cycle. 2010:9. doi: 10.4161/cc.9.13.12076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Woodruff JB, Drubin DG, Barnes G. Mitotic spindle disassembly occurs via distinct subprocesses driven by the anaphase-promoting complex, Aurora B kinase, and kinesin-8. J Cell Biol. 2010;191:795–808. doi: 10.1083/jcb.201006028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Woodruff JB, Drubin DG, Barnes G. Spindle assembly requires complete disassembly of spindle remnants from the previous cell cycle. Mol Biol Cell. 2011 doi: 10.1091/mbc.E11-08-0701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Grava S, Philippsen P. Dynamics of multiple nuclei in Ashbya gossypii hyphae depend on the control of cytoplasmic microtubules length by Bik1, Kip2, Kip3, and not on a capture/shrinkage mechanism. Mol Biol Cell. 2010;21:3680–3692. doi: 10.1091/mbc.E10-06-0527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.West RR, Malmstrom T, McIntosh JR. Kinesins klp5(+) and klp6(+) are required for normal chromosome movement in mitosis. J Cell Sci. 2002;115:931–940. doi: 10.1242/jcs.115.5.931. [DOI] [PubMed] [Google Scholar]
- 75.Garcia MA, Koonrugsa N, Toda T. Two kinesin-like Kin I family proteins in fission yeast regulate the establishment of metaphase and the onset of anaphase A. Curr Biol. 2002;12:610–621. doi: 10.1016/s0960-9822(02)00761-3. [DOI] [PubMed] [Google Scholar]
- 76.Meadows JC, Shepperd LA, Vanoosthuyse V, Lancaster TC, Sochaj AM, Buttrick GJ, Hardwick KG, Millar JB. Spindle checkpoint silencing requires association of PP1 to both Spc7 and kinesin-8 motors. Dev Cell. 2011;20:739–750. doi: 10.1016/j.devcel.2011.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Savoian MS, Gatt MK, Riparbelli MG, Callaini G, Glover DM. Drosophila Klp67A is required for proper chromosome congression and segregation during meiosis I. J Cell Sci. 2004;117:3669–3677. doi: 10.1242/jcs.01213. [DOI] [PubMed] [Google Scholar]
- 78.Savoian MS, Glover DM. Drosophila Klp67A binds prophase kinetochores to subsequently regulate congression and spindle length. J Cell Sci. 2010;123:767–776. doi: 10.1242/jcs.055905. [DOI] [PubMed] [Google Scholar]
- 79.Wang H, Brust-Mascher I, Cheerambathur D, Scholey JM. Coupling between microtubule sliding, plus-end growth and spindle length revealed by kinesin-8 depletion. Cytoskeleton (Hoboken) 2010;67:715–728. doi: 10.1002/cm.20482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gatt MK, Savoian MS, Riparbelli MG, Massarelli C, Callaini G, Glover DM. Klp67A destabilises pre-anaphase microtubules but subsequently is required to stabilise the central spindle. J Cell Sci. 2005;118:2671–2682. doi: 10.1242/jcs.02410. [DOI] [PubMed] [Google Scholar]
- 81.Liu XS, Zhao XD, Wang X, Yao YX, Zhang LL, Shu RZ, Ren WH, Huang Y, Huang L, Gu MM, et al. Germinal Cell Aplasia in Kif18a Mutant Male Mice Due to Impaired Chromosome Congression and Dysregulated BubR1 and CENP-E. Genes Cancer. 2010;1:26–39. doi: 10.1177/1947601909358184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhang C, Zhu C, Chen H, Li L, Guo L, Jiang W, Lu SH. Kif18A is involved in human breast carcinogenesis. Carcinogenesis. 2010;31:1676–1684. doi: 10.1093/carcin/bgq134. [DOI] [PubMed] [Google Scholar]
- 83.Tooker BC, Newman LS, Bowler RP, Karjalainen A, Oksa P, Vainio H, Pukkala E, Brandt-Rauf PW. Proteomic detection of cancer in asbestosis patients using SELDI-TOF discovered serum protein biomarkers. Biomarkers. 2011 doi: 10.3109/1354750X.2010.543289. [DOI] [PubMed] [Google Scholar]
- 84.Nagahara M, Nishida N, Iwatsuki M, Ishimaru S, Mimori K, Tanaka F, Nakagawa T, Sato T, Sugihara K, Hoon DS, et al. Kinesin 18A expression clinical relevance to colorectal cancer progression. Int J Cancer. 2011 doi: 10.1002/ijc.25916. [DOI] [PubMed] [Google Scholar]
- 85.Kim JJ, Park YM, Baik KH, Choi HY, Yang GS, Koh I, Hwang JA, Lee J, Lee YS, Rhee H, et al. Exome sequencing and subsequent association studies identify five amino acid-altering variants influencing human height. Hum Genet. 2011 doi: 10.1007/s00439-011-1096-4. [DOI] [PubMed] [Google Scholar]
- 86.Mitchison T, Kirschner M. Dynamic instability of microtubule growth. Nature. 1984;312:237–242. doi: 10.1038/312237a0. [DOI] [PubMed] [Google Scholar]
- 87.Gardner MK, Charlebois BD, Janosi IM, Howard J, Hunt AJ, Odde DJ. Rapid microtubule self-assembly kinetics. Cell. 2011;146:582–592. doi: 10.1016/j.cell.2011.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gandhi SR, Gierlinski M, Mino A, Tanaka K, Kitamura E, Clayton L, Tanaka TU. Kinetochore-dependent microtubule rescue ensures their efficient and sustained interactions in early mitosis. Dev Cell. 2011;21:920–933. doi: 10.1016/j.devcel.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tischer C, Brunner D, Dogterom M. Force- and kinesin-8-dependent effects in the spatial regulation of fission yeast microtubule dynamics. Mol Syst Biol. 2009;5:250. doi: 10.1038/msb.2009.5. [DOI] [PMC free article] [PubMed] [Google Scholar]