Abstract
The organic extract of a marine sponge Petrosia alfiani selectively inhibited iron chelator-induced hypoxia-inducible factor-1 (HIF-1) activation in a human breast tumor T47D cell-based reporter assay. Bioassay-guided fractionation yielded seven xestoquinones (1 – 7) including three new compounds 14-hydroxymethylxestoquinone (1), 15-hydroxymethylxestoquinone (2), and 14,15-dihydroxestoquinone (3). Compounds 1 – 7 were evaluated for their effects on HIF-1 signaling, mitochondrial respiration, and tumor cell proliferation/viability. The known metabolites adociaquinones A (5) and B (6), that possess a 3,4-dihydro-2H-1,4-thiazine-1,1-dioxide moiety, potently and selectively inhibited iron chelator-induced HIF-1 activation in T47D cells, each with an IC50 value of 0.2 μM. Mechanistic studies revealed that adociaquinones promote oxygen consumption without affecting mitochondrial membrane potential. Compound 1 both enhances respiration and decreases mitochondrial membrane potential, suggesting that it acts as a protonophore that uncouples mitochondrial respiration.
Mitochondria are primary consumers of cellular oxygen and act as key regulators of cellular signaling pathways.1 Under hypoxic (decreased oxygen tension) conditions, elevated reactive oxygen species (ROS) production at mitochondrial electron transport chain (ETC) complex III is believed to mediate the activation of hypoxia-inducible factor-1 (HIF-1) by inhibiting HIF-1α hydroxylases.1 The transcription factor HIF-1 promotes tumor cell survival and represents an important molecular target for anticancer drug discovery.2 Mitochondrial respiratory chain inhibitors such as rotenone (complex I) and myxothiazol (complex III) suppress HIF-1 activation by blocking hypoxia-induced increases in ROS production.3,4 We reported that mitochondrial uncouplers including FCCP ([[4-(trifluoromethoxy)phenyl]hydrazono]-propanedinitrile) and mammea-type isoprenylated dihydroxycoumarins from Mammea americana potently inhibited HIF-1 activation in human breast tumor T47D cells.5,6 However, the underlying mechanism(s) for uncoupling agents to suppress HIF-1 activation remains unclear.
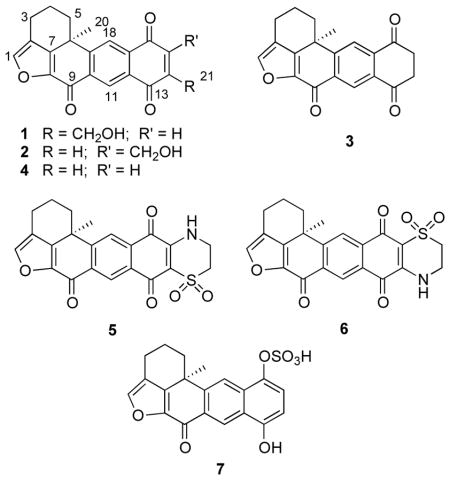
In our continuing search for natural product-derived small-molecule HIF-1 inhibitors, a lipid extract of the sponge Petrosia (Petrosia) alfiani de Voogd & van Soest (Petrosiidae) inhibited chemical hypoxia (1,10-phenanthroline, 10 μM)-induced HIF-1 activation by 63% at 5 μg mL−1 in a T47D cell-based reporter assay. Bioassay-guided isolation of the active extract and subsequent structure elucidation of the purified compounds afforded three new xestoquinones 14-hydroxymethylxestoquinone (1), 15-hydroxymethylxestoquinone (2), 14,15-dihydroxestoquinone (3), and four previously reported analogues xestoquinone (4),7,8 adociaquinones A (5) and B (6),9,10 and xestoquinol sulfate (7).11
Xestoquinone (4) and its analogues are polycyclic quinone-type metabolites that have primarily been associated with Petrosiidae sponges of the genus Xestospongia.7–11 The xestoquinone family has been shown to exhibit a wide spectrum of effects, including 12–14 antifungal,15 cytotoxic,15 and antimalarial16 activities. At the molecular level, xestoquinones have been shown to exert inhibitory effects against myosin Ca2+ ATPase,17 Cdc25B, MKP-1, and MKP-3 phosphatases,18,19 pp60v-src protein tyrosine kinase,20 and topoisomerase II.21 This report describes the isolation, identification and characterization of bioactive constituents of P. alfiani and examines the potential importance of certain structural features of xestoquinones (1 – 7) on mitochondrial respiration and HIF-mediated signaling in T47D cells.
RESULTS AND DISCUSSION
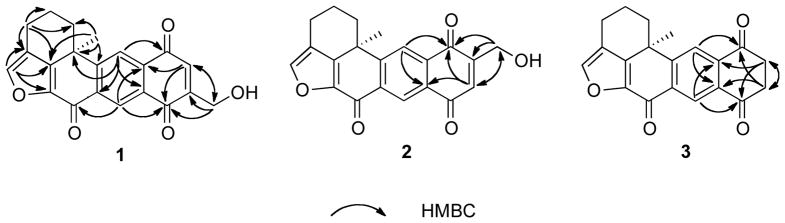
Compound 1 was obtained as a yellow powder and its molecular formula was assigned as C21H16O5 based on its HRESIMS data, indicating 14 degrees of unsaturation. The IR spectrum suggested the presence of hydroxy and carbonyl functional groups at 3433 (br) and 1672 cm−1, respectively. The 13C NMR and DEPT spectra demonstrated 21 carbons, accounting for one methyl, four methylenes, four methines, and 12 quaternary carbons (Table 1). The 1H and 13C NMR data for 1 (Table 1) were similar to the published values for xestoquinone (4),7 except for one major difference being the presence of resonances for an extra hydroxymethyl methylene [δH 4.73 (2H, s), δC 60.0] in 1. The C-14 position of the hydroxymethyl methylene was established by observation of key HMBC correlations from H-11 [δH 8.97 (1H, s)] and H-15 [δH 7.07 (1H, s)] to C-13 (δC 184.2) and C-17 (δC 133.5), and from the hydroxymethyl singlet at δH 4.73 to the resonances for C-13, C-14, and C-15 (Figure 1). Compound 1 exhibited a similar positive specific rotation value ([α]24D +24.3) to that of xestoquinone.8 Because the structures of 1 and 4 bear only a single stereogenic center, the absolute configuration of C-6 in 1 was inferred to be S, as in 4. Thereby, 1 was deduced to be a 14-hydroxymethyl-substituted analogue of xestoquinone and was assigned the name 14-hydroxymethylxestoquinone.
Table 1.
1H and 13C NMR Data of 1 – 3 (400 and 100 MHz, CDCl3, δ ppm)
| No. |
1
|
2
|
3
|
|||
|---|---|---|---|---|---|---|
| δC | δH (J in Hz) | δC | δH (J in Hz) | δC | δH (J in Hz) | |
|
|
|
|
||||
| 1 | 145.1, CH | 7.54, s | 145.1, CH | 7.53, s | 145.1, CH | 7.53, s |
| 2 | 121.7, C | 121.6, C | 121.6, C | |||
| 3 | 18.6, CH2 | 2.87, dd (7.6, 16.8) 2.65, dd (8.8, 16.8) |
18.6, CH2 | 2.87, dd (7.6, 16.6) 2.65, dd (9.3, 16.6) |
18.5, CH2 | 2.87, dd (7.8, 17.0) 2.64, dd (8.7, 17.0) |
| 4 | 17.0, CH2 | 2.27, m 2.18, m |
17.0, CH2 | 2.28, m 2.18, m |
17.0, CH2 | 2.27, m 2.18, m |
| 5 | 31.3, CH2 | 2.56, m 1.74, td (3.8, 12.6) |
31.4, CH2 | 2.56, dt (3.6, 12.9) 1.75, td (4.3, 12.9) |
31.4, CH2 | 2.56, dt (3.5, 12.9) 1.74, td (4.4, 12.9) |
| 6 | 37.5, C | 37.5, C | 37.4, C | |||
| 7 | 147.6, C | 147.4, C | 147.4, C | |||
| 8 | 144.2, C | 144.2, C | 144.2, C | |||
| 9 | 170.5, C | 170.4, C | 170.4, C | |||
| 10 | 137.9, C | 138.2, C | 138.5, C | |||
| 11 | 127.0, CH | 8.97, s | 127.1, CH | 9.01, s | 127.6, CH | 9.00, s |
| 12 | 130.7, C | 130.7, C | 133.9, C | |||
| 13 | 184.2, C | 183.9, C | 194.7, C | |||
| 14 | 150.4, C | 134.3, CH | 7.10, s | 37.5, CH2 | 3.13, m | |
| 15 | 133.5, CH | 7.07, s | 149.6, C | 37.8, CH2 | 3.13, m | |
| 16 | 184.7, C | 185.2, C | 196.0, C | |||
| 17 | 133.5, C | 133.6, C | 136.6, C | |||
| 18 | 123.2, CH | 8.19, s | 123.2, CH | 8.22, s | 123.5, CH | 8.19, s |
| 19 | 156.5, C | 156.2, C | 156.5, C | |||
| 20 | 32.7, CH3 | 1.52, s | 32.7, CH3 | 1.53, s | 32.8, CH3 | 1.51, s |
| 21 | 60.0, CH2 | 4.73, s | 60.0, CH2 | 4.72, s | ||
Figure 1.
Selected HMBC correlations observed in 1 – 3.
The molecular formula of 2 was established as C21H16O5 by HRESIMS. The 1H NMR and 13C NMR spectra of 2 were nearly identical to those of 1 (Table 1), suggesting that both compounds were isomers. The key HMBC correlations from H-14 [δH 7.10 (1H, s)] and H-18 [δH 8.22 (1H, s)] to C-12 (δC 130.7) and C-16 (δC 185.2), and from the hydroxymethyl singlet [δH 4.72 (3H, s)] to the resonances for C-14, C-15, and C-16 (Figure 1) indicated the hydroxymethyl branch in 2 was attached to the C-15 carbon. As in the structures of 1 and 4, the positive optical rotation ([α]24D +32.7) of 2 indicated that C-6 in 2 possessed a similar S-configuration and was a 15-hydroxymethyl-substituted regioisomer of 1, and was given the trivial name 15-hydroxymethylxestoquinone.
Compound 3 was isolated as an optically active ([α]24D +5.1) yellow solid and deduced to have the molecular formula C22H28O4 by HRESIMS. Comparison of the 1H and 13C NMR spectra of 3 (Table 1) with those of 47 indicated that both compounds possessed the identical pentacyclic architecture except that, in 3, the 14,15-double bond was saturated (Figure 1). In the 1H NMR spectrum of 3, the two aromatic protons that were assigned to H-14 and H-15 in 4 were replaced by a resonance at δH 3.13 that integrated for four methylene protons (Table 1). The C-13 and C-16 carbonyl carbon resonances in the 13C NMR spectrum of 3 were accordingly shifted downfield to δC 194.7 and δC 196.0, respectively. Further, based on key HMBC correlations from the methylene resonance at δH 3.13 to the resonances for C-12, C-13, C-16, and C-17 (Figure 1), the structure of 3 was deduced to be the 14,15-saturated analogue of 4 (trivially named as 14,15-dihydroxestoquinone).
Spectroscopic dereplication efforts identified the other isolated compounds as known metabolites xestoquinone (4),7,8 adociaquinones A (5) and B (6),9,10 and xestoquinol sulfate (7).11
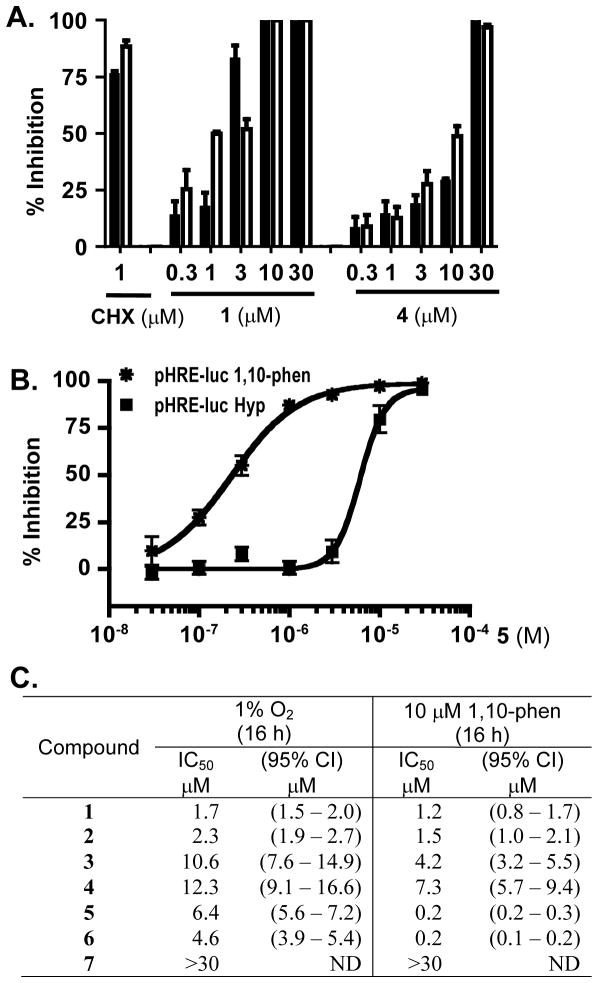
Concentration-response studies were performed to determine the effects of 1 – 7 on HIF-1 activation in a T47D cell-based reporter assay.22 Data for the representative compounds 1, 4, and 5 are shown in Figure 2A (1 and 4) and B (5), and the IC50 values for 1 – 7 summarized in Figure 2C. The hydroxymethylated metabolites 1 and 2 inhibited both hypoxia-induced and iron chelator (chemical hypoxia)-induced HIF-1 activation with comparable low micromolar IC50 values (1.2 – 2.3 μM, Figure 2A and C). Xestoquinone (4) and its 14,15-saturated analogue 3 exhibited reduced potency (IC50 values 4.2 – 12.3 μM, Figure 2A and C). Compounds 5 and 6 selectively suppressed iron chelator-induced HIF-1 activation (IC50 values 0.2 and 0.2 μM, respectively, Figure 2B and C) relative to their inhibitory effects on hypoxia-induced HIF-1 activation (IC50 values 6.4 and 4.6 μM, respectively, Figure 2B and C). The 3,4-dihydro-2H-1,4-thiazine-1,1-dioxide moiety in 5 and 6 appears to be an important pharmacophore for this enhanced selectivity towards chemical hypoxia-induced HIF-1 activation. At the concentrations tested (0.3 – 30 μM in half-log increments), xestoquinol sulfate (7) did not affect HIF-1 activation (IC50 > 30 μM, Figure 2C). Similar results were observed in separate experiments (Supporting Information, Figure S1). These observations suggest that the 1,4-quinone moieties in 1 – 6 are critical for xestoquinones to suppress HIF-1 activation. The lack of inhibitory activity of xestoquinol sulfate (7) may also be attributed to its more polar charged nature that may render it unable to penetrate lipophilic plasma and mitochondrial membranes.
Figure 2.
Compounds 1 – 7 inhibit HIF-1 activation. (A) Concentration-dependent inhibition of HIF-1 activation by 1 and 4 in a T47D cell-based reporter assay for HIF-1 activity [the inducing conditions are hypoxia (1% O2, black bar) or chemical hypoxia (1,10-phenanthroline, 10 μM, open bar)]. The protein synthesis inhibitor cyclohexamide (CHX, 1 μM) was included as a positive control. Luciferase activities are presented as “% Inhibition” of the induced control in the absence of test compounds. Data shown are averages + standard deviation of one experiment performed in triplicate. (B) Concentration-response results of 5. Experimental procedures and data presentation are the same as those described in (A). (C) IC50 values of 1 – 7 suppressing HIF-1 activation in a T47D cell-based reporter assay. The IC50 and 95% confidence interval (CI) values were determined from one experiment performed in triplicate.
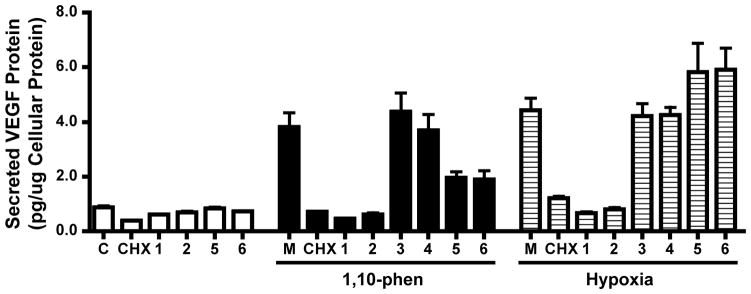
To investigate if the HIF-1 inhibition observed in the reporter assay translates into the suppression of HIF-1 target gene expression, the effects of 1 – 6 (10 μM) on expression of the HIF-1 target gene vascular endothelial growth factor (VEGF) were examined at the protein level by ELISA in T47D cells. Exposure to hypoxia and treatment with 1,10-phenanthroline each increased secreted VEGF protein levels in T47D cells, relative to the normoxic control (Figure 3). Compounds 1 and 2 blocked the induction of secreted VEGF protein by both hypoxia and chemical hypoxia (Figure 3). At the concentration of 10 μM, the less potent HIF-1 inhibitors 3 and 4 did not affect the increase in VEGF protein levels following either hypoxic exposure or 1,10-phenanthroline treatment (Figure 3). In contrast, 5 and 6 suppressed the induction of secreted VEGF protein by 1,10-phenanthroline while moderately increasing the levels of secreted VEGF protein under hypoxic conditions (Figure 3). However, the inhibitory effects exerted by 5 and 6 on chemical hypoxia-stimulated VEGF production were less pronounced than those observed in the presence of 1 and 2, although 5 and 6 were more potent in the T47D cell-based reporter assay (Figure 2C). Further studies are required to address this difference in potency. A similar trend was observed for cellular VEGF protein levels (Supporting Information, Figure S2).
Figure 3.
Effects of 1 – 6 on hypoxia- and chemical hypoxia (1,10-phenanthroline)-induced secreted VEGF protein production in T47D cells. T47D cells were exposed to 1,10-phenanthroline (10 μM, 16 h) or hypoxia (1% O2, 16 h) in the presence of 1 – 6 (10 μM). The level of secreted VEGF protein in the conditioned medium was determined by ELISA and normalized to the quantity of cellular protein. Cyclohexamide (CHX, 10 μM) was included as a positive control. “C” indicates media control under normoxic conditions, and “M” represents media under induced conditions (hypoxia or 1,10-phenanthroline as appropriate). Data shown are average + standard deviation from one experiment performed in triplicate.
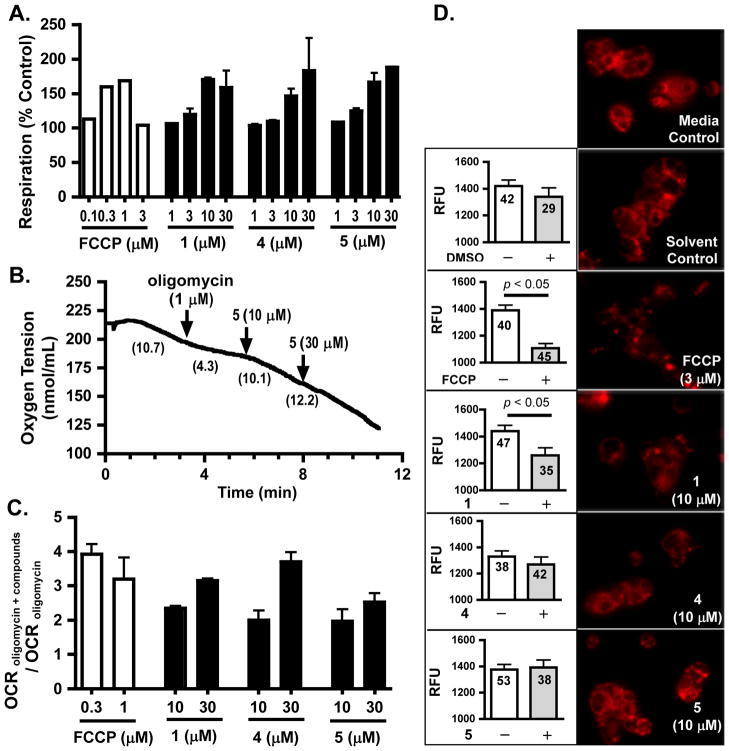
In previous studies, the plant-derived 1,4-quinones known as royleanones, displayed protonophoric activity and uncoupled mitochondrial respiration in vitro.23 Mitochondrial uncouplers have been shown to disrupt HIF-1 signaling in cell lines.5,6,24 To determine if xestoquinones disrupt mitochondrial respiration, a T47D cell-based respiration assay25 was performed with three representative xestoquinone derivatives 1, 4, and 5. The commonly used mitochondrial uncoupler FCCP was used as a positive control. At the concentrations of 10 and 30 μM, compounds 1, 4, and 5 all significantly enhanced the rate of cellular oxygen consumption by 47% – 89% in comparison to the untreated control, results similar to those observed with FCCP addition (Figure 4A). To determine if the respiratory responses were a consequence of increased ATP turnover, the effects of 1, 4, and 5 on respiration were further examined in the presence of oligomycin, an inhibitor of F0F1ATPase (Figure 4B and C). By inhibiting proton influx into the matrix through F0F1ATPase, oligomycin increases the proton gradient across the inner mitochondrial membrane and thus triggers a decrease in oxygen consumption rate. The observation that 1, 4, 5, and the positive control FCCP all stimulated state 4 respiration is consistent with the hypothesis that, mechanistically, xestoquinones exert pharmacological effects by functioning as protonophores (Figure 4B and C). In human hepatocarcinoma Hep3B cells, compounds 1 – 6 and FCCP also stimulated oligomycin-suppressed respiration (Supporting Information, Figure S3). However, it is possible that the increased respiration results from direct stimulation of mitochondrial substrate oxidation reactions rather than uncoupling. To assess the effects of xestoquinones and related quinones on mitochondrial membrane potential, the fluorescent cationic dye tetramethyl rhodamine methyl ester (TMRM) was used as an indicator. The TMRM dye accumulates in the matrix and its corresponding fluorescence intensity correlates with the magnitude of mitochondrial inner membrane potential. The positive control FCCP decreased membrane potential and resulted in reduced fluorescence (Figure 4D, FCCP). Among the compounds (1, 4, and 5) examined, 1 reduced mitochondrial membrane potential (Figure 4D, 1), while 4 and 5 did not exhibit a pronounced effect (Figure 4D, 4 and 5). Thus, xestoquinones such as 1 appear to act as protonophoric uncouplers of oxidative phosphorylation and interfere with mitochondria-mediated HIF-1 signaling, while other related compounds (e.g., 4 and 5) may function through mechanism(s) that remain to be determined.
Figure 4.
Compounds 1, 4, and 5 disrupt cellular respiration. (A) Concentration–response results of 1, 4, and 5, and the standard uncoupler FCCP on oxygen consumption of intact T47D cells. Compounds were tested at the specified concentrations. Oxygen consumption rates are expressed as a percentage of the untreated controls. Data shown for 1, 4, and 5 are average + standard deviation for the concentrations of 10 and 30 μM from three independent experiments, and average + deviation from the average for the lower concentrations (n = 2). Data for FCCP are from one representative experiment and were similar to those previously reported.5 The compounds were added in half-log increments to achieve the desired concentrations. (B) Compound 5 stimulated state 4 respiration. Oligomycin and 5 were added to T47D cells in a sequential manner at the specified concentrations. The rates of oxygen consumption (in nmol O2/min) are indicated after each addition. Data shown are representative of two independent experiments. (C) Compounds 1, 4, 5, and FCCP stimulate oligomycin-inhibited respiration. The rates of oxygen consumption (OCR) in T47D cells in the presence of oligomycin and test compounds (OCR oligomycin + compounds) were determined and normalized to those obtained in the presence of oligomycin (OCR oligomycin). Data shown are average + deviation from average from two independent experiments. (D) Effects of 1, 4, 5, and FCCP on mitochondrial membrane potential. T47D cells pre-incubated with TMRM were exposed to compounds at the specified concentrations. “Solvent Control” indicates cells exposed to 0.26% DMSO. Representative images are shown in the right panel. Fluorescence intensity (RFU) was obtained by quantifying images acquired before (“−”) and after compound addition (“+”) with Adobe Photoshop. The number inside each column represents the number of cells analyzed. Data were compared by unpaired t-test using AxioVision software.
The effects of 1 – 7 on cell proliferation/viability were examined in human breast tumor T47D and MDA-MB-231 cell lines. Compounds 1 – 6 exhibited more pronounced cytostatic/cytotoxic effects in T47D cells in comparison to MDA-MB-231 cells following 48 h treatment (IC50 values, Table 2). Similar results obtained in separate experiments are shown in Supporting Information (Figure S4). The inactive HIF-1 inhibitor xestoquinol sulfate (7) did not suppress cell proliferation/viability (< 5% inhibition) in either cell line. The 1,4-quinone moiety in 1 – 6 appears to be an important structural element required for the cytostatic/cytotoxic activity of these sponge metabolites.
Table 2.
IC50 values of 1 – 7 on the proliferation/viability of human breast tumor T47D and MDA-MB-231 cells in a 48 h exposure concentration-response study. The IC50 and 95% CI values were determined from one experiment performed in triplicate.
| Compound | T47D | MDA-MB-231 | ||
|---|---|---|---|---|
|
| ||||
| IC50 | (95% CI) | IC50 | (95% CI) | |
| μM | μM | μM | μM | |
| 1 | 3.6 | (2.7 – 4.8) | 6.5 | (5.6 – 7.4) |
| 2 | 4.3 | (3.5 – 5.4) | 6.3 | (5.4 – 7.3) |
| 3 | 15.9 | (15.2 – 16.7) | 28.1 | (25.9 – 30.4) |
| 4 | 13.8 | (12.0 – 15.9) | 20.6 | (18.5 – 22.8) |
| 5 | 2.6 | (2.2 – 3.1) | 6.3 | (4.2 – 9.4) |
| 6 | 2.9 | (2.5 – 3.2) | 6.1 | (4.8 – 7.7) |
| 7 | > 30 | ND | > 30 | ND |
The xestoquinone family has been reported to display a wide range of biological activities.7–21,23 Previous SAR studies indicated that the 1,4-quinone moiety in xestoquinones and adociaquinones is a common and essential pharmacophore for inhibitory activities against enzymes such as myosin Ca2+-ATPase,17 Cdc25B phosphatase,18 and protein tyrosine kinase pp60v-src.20 Our results demonstrate that a terminal quinone is required for the HIF-1 inhibitory and cytostatic/cytotoxic activities. Further mechanistic studies suggest that xestoquinone derivatives such as 1 can function as potent uncouplers of mitochondrial respiration and oxidative phosphorylation. The hydrophobicity of the polyunsaturated framework may facilitate the penetration of these sponge metabolites through lipophilic membranes. Xestoquinones may translocate protons from the intermembrane space across the mitochondrial inner membrane to the matrix. This protonophoric activity dissipates the mitochondrial proton gradient required to drive ATP synthesis. Compounds such as 4 and 5 may affect cellular respiration (e.g., stimulation of both substrate oxidation and uncoupling, or non-mitochondrial respiration) interfere with mitochondrial function via other pathway(s)/mechanism(s). In T47D cells, the adociaquinones A (5) and B (6) exhibited a highly selective inhibitory effect towards chemical hypoxia-induced HIF-1 activation. While further studies are required to determine the possible mechanism(s) responsible for this unusual pattern of stimulus-selective HIF-1 inhibitory activity, the 3,4-dihydro-2H-1,4-thiazine-1,1-dioxide moiety appears to be an important functional feature of adociaquinones that is associated with this phenomenon.
EXPERIMENTAL SECTION
General Experimental Procedures
Optical rotations were obtained on an AP IV/589-546 digital polarimeter. A Bruker Tensor 27 Genesis Series FTIR was used to obtain the IR spectra, and a Varian 50 Bio spectrophotometer was used to record the UV spectra. The NMR spectra were recorded in CDCl3 on AMX-NMR spectrometers (Bruker) operating at 400 MHz for 1H and 100 MHz for 13C, respectively. Residual solvent resonances (δ 7.26 for 1H and δ 77.16 for 13C) were used as internal references for the NMR spectra recorded running gradients. The HRESIMS were determined on a Bruker Daltonic micro TOF fitted with an Agilent 1100 series HPLC and an electrospray ionization source. TLC was performed using Merck Si 60 F254 or Si 60 RP18 F254 plates, sprayed with a 10% H2SO4 solution in EtOH, heated, and visualized under UV at 254 nm. HPLC was performed on a Waters system, equipped with a 600 controller and a 996 photodiode array detector. Two semipreparative HPLC columns [(I) Phenomenex Luna RP-18, 5 μm, 250 × 10.00 mm; (II) Phenomenex Luna 5 μm, Si gel (2), 5 μm, 250 × 10.00 mm] were employed for isolation. Various isocratic and gradient solvent systems comprised of MeCN-H2O were used to generate the best resolution of each compound. The purities of all compounds were judged on the percentage of the integrated signal at UV 220 nm. All final compounds submitted for bioassay were at least 95% pure as judged by this method.
Sponge Material
The sponge material was obtained from the U.S. National Cancer Institute’s Open Repository Program. Petrosia alfiani was collected at a depth of 25 m off the coast of Malaysia (Oct. 18, 2006), frozen at −20 °C, and ground in a meat grinder. The sponge material was identified by Dr. Belinda Alvarez de Glasby of the Museum and Art Gallery of the Northern Territory, Darwin, Australia, and a voucher specimen (collection No. 0CDN9473) was placed on file with the Department of Invertebrate Zoology, National Museum of Natural History, Smithsonian Institution, Washington, D.C.
Extraction and Isolation
Ground P. alfiani material was extracted with H2O (extract No. C028961). The residual sample was then lyophilized and extracted with 50% MeOH in CH2Cl2, 26 residual solvents were removed under vacuum, and the extract (collection number 0CDN9473) was stored at −20 °C in the NCI repository at the Frederick Cancer Research and Development Center (Frederick, MD). The extract (4.0 g sample from an NCI stock supply) was separated into nine fractions by passage over a Diaion HP-20SS gel column (40 g) eluted with gradients of MeOH in H2O (10:90, 30:70, 50:50, 70:30, 80:20, 90:10, 100:0) and 90% and 50% MeOH in CHCl3. To isolate the less active structurally related compounds, the fractions that eluted with 30% and 50% MeOH in H2O, respectively (inhibited HIF-1 by 15% and 20%, respectively; 5 μg mL−1) were combined (284 mg combined mass) and further separated by passage over a Sephadex LH-20 column eluted with MeOH to obtain 7 (113 mg, 2.8% yield). Three active fractions that eluted from the original Diaion HP-20SS column with 70%, 80%, and 90% MeOH, respectively (inhibited HIF-1 by 82%, 88%, and 76%, respectively; 5 μg mL−1) were combined (598 mg combined mass) and suspended with 50% MeOH in CH2Cl2. Following filtration, the residue was obtained as a pure compound 6 (60 mg, 1.5% yield). The supernatant (492 mg) was dried and separated by passage over a Sephadex LH-20 column eluted with 50% MeOH in CH2Cl2 to yield four fractions. The third subfraction (310 mg, inhibited HIF-1 by 84%; 5 μg mL−1) was further separated by C18 column chromatography [eluted with gradients of MeCN in H2O (20:80, 30:70, 40:60, 50:50, and 60:40) to afford five fractions]. The subfraction that eluted with 60% MeOH in H2O (199 mg) was subjected to reversed-phase HPLC (Luna 5 μm, ODS-3, 100 Å, 250 × 10.00 mm, isocratic 50% CH3CN in H2O, 4.0 mL min−1) to yield 3 (3.1 mg, 0.078% yield) and another nine fractions. Another impure subfraction (10.1 mg) was further separated by normal-phase HPLC [Luna 5 μm, Silica (2), 100 Å, 250 × 10.00 mm, isocratic 65% hexanes in EtOAc, 4.0 mL min−1] to obtain 1 (3.3 mg, 0.083% yield) and 2 (2.2 mg, 0.055% yield). The subfraction that eluted from the previous RP-18 CC with 40% MeCN in H2O were combined (31.6 mg combined mass) and further purified by reversed-phase HPLC (Luna 5 μm, ODS-3, 100 Å, 250 × 10.00 mm, isocratic 33% CH3CN in H2O, 4.0 mL min−1) to afford 5 (8.0 mg, 0.2% yield). Two other active fractions eluted from the original Diaion HP-20SS column with 100% MeOH and 50% MeOH in CHCl3, respectively (inhibited HIF-1 by 53% and 53%, respectively; 5 μg mL−1) were combined (1307 mg combined mass) and further separated by passage through a Sephadex LH-20 column eluted with 50% MeOH in CH2Cl2 and C18 column chromatography eluted with gradients of MeCN in H2O (40:60, 50:50, 60:40, 70:30, 100:0) to produce 4 (192 mg, 4.8% yield) that eluted with 60% MeCN.
14-Hydroxymethylxestoquinone (1)
yellow amorphous solid; [α]24D +24.3 (c 0.16, CH2Cl2); UV (MeOH) λmax (log ε) 219 (4.27), 262 (4.35), 294 (4.15) nm; IR (film) νmax 3433, 2926, 2853, 1672, 1447, 1330, 1291, 1213, 1150, 1008, 886 cm−1; 1H NMR (CDCl3, 400 MHz) and 13C NMR (CDCl3, 100 MHz) data, see Table 1; HRESIMS m/z 349.1075 [M+H]+ (calcd for C21H17O5, 349.1076).
15-Hydroxymethylxestoquinone (2)
yellow amorphous solid; [α]24D +32.7 (c 0.22, CH2Cl2); UV (MeOH) λmax (log ε) 218 (4.27), 262 (4.32), 293 (4.15) nm; IR (film) νmax 3417, 2927, 2850, 1667, 1445, 1320, 1219, 1152, 1056, 1037, 936, 892 cm−1; 1H NMR (CDCl3, 400 MHz) and 13C NMR (CDCl3, 100 MHz) data, see Table 1; HRESIMS m/z 349.1072 [M+H]+ (calcd for C21H17O5, 349.1076).
14,15-Dihydroxestoquinone (3)
yellow amorphous solid; [α]24D +5.1 (c 0.04, CH2Cl2); UV (MeOH) λmax (log ε) 216 (4.08), 248 (4.19), 316 (3.87) nm; IR (film) νmax 2911, 2845, 1698, 1665, 1603, 1441, 1418, 1308, 1184, 1146, 1032, 980, 856 cm−1; 1H NMR (CDCl3, 400 MHz) and 13C NMR (CDCl3, 100 MHz) data, see Table 1; HRESIMS m/z 321.1130 [M+H]+ (calcd for C20H17O4, 321.1127).
Xestoquinone (4)
yellow amorphous solid; [α]24D +11.4 (c 1.12, CH2Cl2). Literature value [α]25D +17.2 (c 1.16, CH2Cl2).7
Cell-Based Reporter and Proliferation/Viability Assays
T47D and MDA-MB-231 cells were obtained from ATCC. Cell maintenance, experimental procedures, and data presentation for the cell-based reporter and proliferation/viability assays were the same as previously described.5 All extract, fraction, and compound samples were prepared as stock solutions in DMSO (final solvent concentration less than 0.5% in all assays).
ELISA Assay for Secreted VEGF Protein
Experimental procedures and data presentation were the same as previously described.27
Cell-Based Respiration and Mitochondrial Membrane Potential Assays
A T47D cell-based respiration assay25 was employed to determine the effects of test compounds on cellular respiration. For mechanistic studies, oligomycin A (1 μM) was added to respiring intact T47D cells, followed by test compounds or the standard uncoupler FCCP at the specified concentrations. Data presentation was the same as previously described.5
Mitochondrial membrane potential was determined with the dye TMRM as described.5 Live cell images were acquired before and 15 – 20 min after compound addition, using an Axiovert 200M epifluorescence microscope (Zeiss). Whole-cell fluorescence intensity was quantified using AxioVision software.
Statistical Analysis
Data analyses were performed with GraphPad Prism 5. Differences between data sets were considered statistically significant when p < 0.05.
Supplementary Material
Acknowledgments
The authors thank the Natural Products Branch Repository Program at the National Cancer Institute for providing marine extracts from the NCI Open Repository used in these studies, D. J. Newman and E. C. Brown (NCI, Frederick, MD) for assistance with sample logistics and collection information, S. L. McKnight (University of Texas Southwestern Medical Center at Dallas) for providing the pTK-HRE3-luc construct. This work was supported in part by the National Institutes of Health National Cancer Institute (grant CA98787). This investigation was conducted in a facility constructed with Research Facilities Improvement Grant C06 RR-14503-01 from the National Institutes of Health.
Footnotes
Supporting Information. NMR spectra for 1 – 3, compound purities, and effects of compound treatment on cellular VEGF protein levels in T47D cells. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Hamanaka RB, Chandel NS. Curr Opin Cell Biol. 2009;21:894–899. doi: 10.1016/j.ceb.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Semenza GL. Oncogene. 2010;29:625–634. doi: 10.1038/onc.2009.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chandel NS, Maltepe E, Goldwasser E, Mathieu CE, Simon MC, Schumacker PT. Proc Natl Acad Sci USA. 1998;95:11715–11720. doi: 10.1073/pnas.95.20.11715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chandel NS, McClintock DS, Feliciano CE, Wood TM, Melendez JA, Rodriguez AM, Schumacker PT. J Biol Chem. 2000;275:25130–25138. doi: 10.1074/jbc.M001914200. [DOI] [PubMed] [Google Scholar]
- 5.Du L, Mahdi F, Jekabsons MB, Nagle DG, Zhou YD. J Nat Prod. 2010;73:1868–1872. doi: 10.1021/np100501n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Du L, Mahdi F, Jekabsons MB, Nagle DG, Zhou YD. J Nat Prod. 2011;74:240–248. doi: 10.1021/np100762s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nakamura H, Kobayashi J, Kobayashi M, Ohizumi Y, Hirata Y. Chem Lett. 1985:713–716. [Google Scholar]
- 8.Harada N, Sugioka T, Uda H, Kuriki T. J Org Chem. 1990;55:3158–3163. [Google Scholar]
- 9.Schmitz FJ, Bloor SJ. J Org Chem. 1988;53:3922–3925. [Google Scholar]
- 10.Harada N, Sugioka T, Soutome T, Hiyoshi N, da Hisashi U, Kuriki T. Tetrahedron Asym. 1995;6:375–376. [Google Scholar]
- 11.Kobayashi J, Hirase T, Shigemori H, Ishibashi M. J Nat Prod. 1992;55:994–998. [Google Scholar]
- 12.Kobayashi M, Nakamura H, Kobayashi J, Ohizumi Y. J Pharmacol Exp Ther. 1991;257:82–89. [PubMed] [Google Scholar]
- 13.Kobayashi M, Muroyama A, Nakamura H, Kobayashi J, Ohizumi Y. J Pharmacol Exp Ther. 1991;257:90–94. [PubMed] [Google Scholar]
- 14.Ito M, Hirata Y, Nakamura H, Ohizumi Y. J Pharmacol Exp Ther. 1999;291:976–981. [PubMed] [Google Scholar]
- 15.Nakamura M, Kakuda T, Qi J, Hirata M, Shintani T, Yoshioka Y, Okamoto T, Oba Y, Nakamura H, Ojika M. Biosci Biotechnol Biochem. 2005;69:1794–1752. doi: 10.1271/bbb.69.1749. [DOI] [PubMed] [Google Scholar]
- 16.Laurent D, Jullian V, Parenty A, Knibiehler M, Dorin D, Schmitt S, Lozach O, Lebouvier N, Frostin M, Alby F, Manurel S, Doerig C, Meijer L, Sauvain M. Bioorg Med Chem. 2006;14:4477–4482. doi: 10.1016/j.bmc.2006.02.026. [DOI] [PubMed] [Google Scholar]
- 17.Nakamura M, Kakuda T, Oba Y, Ojika M, Nakamura H. Bioorg Med Chem. 2003;11:3077–3082. doi: 10.1016/s0968-0896(03)00276-1. [DOI] [PubMed] [Google Scholar]
- 18.Cao S, Foster C, Brisson M, Lazo JS, Kingston DG. Bioorg Med Chem. 2005;13:999–1003. doi: 10.1016/j.bmc.2004.11.039. [DOI] [PubMed] [Google Scholar]
- 19.Cao S, Murphy BT, Foster C, Lazo JS, Kingston DGI. Bioorg Med Chem. 2009;17:2276–2281. doi: 10.1016/j.bmc.2008.10.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alvi KA, Rodríguez J, Diaz MC, Moretti R, Wilhelm RS, Lee RH, Slate DL, Crews P. J Org Chem. 1993;58:4871–4880. [Google Scholar]
- 21.Concepción GP, Foderaro TA, Eldredge GS, Lobkovsky E, Clardy J, Barrows LR, Ireland CM. J Med Chem. 1995;38:4503–4507. doi: 10.1021/jm00022a016. [DOI] [PubMed] [Google Scholar]
- 22.Hodges TW, Hossain CF, Kim YP, Zhou YD, Nagle DG. J Nat Prod. 2004;67:767–771. doi: 10.1021/np030514m. [DOI] [PubMed] [Google Scholar]
- 23.Spiridonov NA, Arkhipov VV, Foigel AG, Shipulina LD, Fomkina MG. Phytother Res. 2003;17:1228–1230. doi: 10.1002/ptr.1403. [DOI] [PubMed] [Google Scholar]
- 24.Thomas R, Kim MH. Mol Cell Biochem. 2007;296:35–44. doi: 10.1007/s11010-006-9295-3. [DOI] [PubMed] [Google Scholar]
- 25.Liu Y, Veena CK, Morgan JB, Mohammed KA, Jekabsons MB, Nagle DG, Zhou YD. J Biol Chem. 2009;284:5859–5868. doi: 10.1074/jbc.M806744200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McCloud TC. Molecules. 2010;15:4526–4563. doi: 10.3390/molecules15074526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou YD, Kim YP, Mohammed KA, Jones DK, Muhammad I, Dunbar DC, Nagle DG. J Nat Prod. 2005;68:947–950. doi: 10.1021/np050029m. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.