Abstract
We have isolated the plasma membrane H+−ATPase in a phosphorylated form from spinach (Spinacia oleracea L.) leaf tissue incubated with fusicoccin, a fungal toxin that induces irreversible binding of 14–3–3 protein to the C terminus of the H+-ATPase, thus activating H+ pumping. We have identified threonine-948, the second residue from the C-terminal end of the H+-ATPase, as the phosphorylated amino acid. Turnover of the phosphate group of phosphothreonine-948 was inhibited by 14–3–3 binding, suggesting that this residue may form part of a binding motif for 14–3–3. This is the first identification to our knowledge of an in vivo phosphorylation site in the plant plasma membrane H+-ATPase.
The plant plasma membrane H+-ATPase provides the driving force for secondary active transport, and controls intra- and extracellular pH, as well as cell turgor. Considering that the H+-ATPase plays a major role in the control of many cell processes, it is not surprising that it is regulated by a number of factors, including hormones, light, and fungal toxins (for review, see Michelet and Boutry, 1995).
The only mechanism for regulation so far demonstrated is that ATP hydrolytic activity and H+ pumping may be controlled via an autoinhibitory domain located in the C-terminal region of the H+-ATPase (Palmgren et al., 1990, 1991). Removal of this domain by proteolysis or deletion at the gene level results in an activated form of the enzyme with a higher Vmax, a lower Km for ATP, a more alkaline pH optimum, and a more efficient coupling between ATP hydrolysis and H+ pumping (Palmgren et al., 1990, 1991; Regenberg et al., 1995). Very similar results were obtained after incubation of intact tissue with the fungal toxin fusicoccin (Johansson et al., 1993; Rasi-Caldogno et al., 1993; Lanfermeijer and Prins, 1994), a treatment suggested to cause a displacement of the C-terminal autoinhibitory domain.
Fusicoccin was initially suggested to activate the plasma membrane H+-ATPase by direct interaction with the enzyme (Marrè, 1979). Later, fusicoccin was demonstrated to bind to a “receptor” belonging to a family of proteins designated 14–3–3 proteins (Korthout and de Boer, 1994; Marra et al., 1994; Oecking et al., 1994). 14–3–3 proteins constitute a highly conserved family of eukaryotic proteins with multiple regulatory functions (for review, see Aitken, 1996). Recently, it was shown that 14–3–3 proteins bind directly to the C-terminal region of the H+-ATPase and that fusicoccin stabilizes the H+-ATPase/14–3–3 complex formed, rendering the association irreversible (Jahn et al., 1997; Oecking et al., 1997; Baunsgaard et al., 1998). This explains earlier observations that plasma membranes isolated from fusicoccin-treated material contained several times more 14–3–3 than plasma membranes isolated from the corresponding controls (Korthout and de Boer, 1994; Oecking et al., 1994). The strong interaction induced by fusicoccin also allowed the H+-ATPase/14–3–3 complex to be solubilized and purified from isolated plasma membranes (Jahn et al., 1997; Oecking et al., 1997). Binding of 14–3–3 proteins to the C-terminal region of the H+-ATPase was also shown to occur in the absence of fusicoccin, and it was suggested that 14–3–3 proteins are natural ligands of the H+-ATPase, regulating H+ pumping by displacing the autoinhibitory domain of the enzyme (Jahn et al., 1997; Oecking et al., 1997; Baunsgaard et al., 1998).
Binding of 14–3–3 proteins to their target proteins has been shown to involve a phosphorylated motif in the target (Muslin et al., 1996). Two optimal motifs have been identified, RSXpSXP, based on the binding motif in the animal Ser/Thr protein kinase Raf-1 (Muslin et al., 1996), and RXY/FXpSXP (Yaffe et al., 1997), identified by using phosphoserine-oriented peptide libraries to probe mammalian and yeast 14–3–3 proteins. There is, however, no amino acid sequence in the C-terminal region of the H+-ATPase with obvious similarity to any of the two optimal binding motifs, suggesting that 14–3–3 binding to this region involves a somewhat different motif. In the present study we identify a Thr at the C-terminal end of the H+-ATPase, the phosphorylation of which is affected by fusicoccin-dependent binding of 14–3–3 to this region.
MATERIALS AND METHODS
Plant Material
Spinach (Spinacia oleracea L.) was grown in a greenhouse with supplementary light (23 W/m2, 350–800 nm, G/86/2 HPLR 400 W, Philips, Eindhoven, The Netherlands). Expanding leaves of 4- to 5-week-old plants were used.
In Vivo Phosphorylation
Spinach leaves (5–25 g) were cut into small pieces and infiltrated under a vacuum with 6 volumes of 0.33 m Suc, 10 mm Mes/1,3-bis(Tris[hydroxymethyl]methylamino)propane, pH 6.0, 0.1 mm EGTA, 0.1 mm EDTA, and 1 to 7 mCi 32P-orthophosphate. After 1 h of incubation at room temperature, 5 μm fusicoccin was added and incubation proceeded for another 30 min. Alternatively, fusicoccin was already present in the infiltration medium and 32P-orthophosphate was added 15 min later; total incubation was also for 90 min. Controls did not receive fusicoccin. After removing the incubation buffer, plasma membranes were isolated as described previously (Larsson et al., 1994).
Trypsin Treatment
Trypsin treatment of plasma membrane vesicles was essentially as described by Palmgren et al. (1990); 66 μg of plasma membrane protein in 66 μL of 10 mm Mops/ 1,3-bis(Tris[hydroxymethyl]methylamino)propane, pH 7.0, 20% (v/v) glycerol, 5 mm EDTA, 1 mm DTT (buffer A) containing 6 mm ATP was mixed with an equal volume of Brij 58 (10 mg/mL in buffer A) to turn all plasma membrane vesicles cytoplasmic-side-out (Johansson et al., 1995). Then, 2 μL of buffer A containing 1 μg of trypsin was added. After incubation at 20°C for 5 min, the reaction was stopped by the addition of 2 μL of 100 mm Pefablock (catalog no. 1429868, Boehringer Mannheim) and put on ice.
SDS-PAGE and Western Blotting
Samples were solubilized at room temperature in standard sample buffer, and polypeptides were separated by SDS-PAGE according to the method of Laemmli (1970). Gels were either stained with Coomassie brilliant blue R 250, or polypeptides were electrophoretically transferred to an Immobilon PVDF transfer membrane (Millipore) for immunostaining. A monoclonal antibody, 46E5B11F6, raised against a fusion protein containing the N-terminal region of the maize H+-ATPase and an antiserum raised against a barley 14–3–3 protein (Brandt et al., 1992) were used. Immunoblots were developed with nitroblue tetrazolium and 5-bromo-4-chloro-3-indolyl phosphate. 32P-proteins were visualized with a phosphor imager (Molecular Dynamics, Sunnyvale, CA).
Protein Digestion and Amino Acid Sequencing
The 100-kD H+-ATPase band, obtained by SDS-PAGE of in vivo-phosphorylated spinach leaf plasma membranes, was cut from the gel and digested with LysC from Achromobacter lyticus (Wako, Osaka, Japan), as described by Rosenfeld et al. (1992). Separation of eluted peptides by HPLC and amino acid sequencing were as described previously (Johansson et al., 1996). Determination of the phosphorylated amino acid sequence was by covalent sequencing of the radioactive peptide on arylactivated membranes (Millipore). After several washes with neat trifluoroacetic acid in a sequencer (model 477A, ABI) the AZT amino acid was extracted with neat trifluoroacetic acid, collected, and concentrated by evaporation. Each fraction, corresponding to one released amino acid, was spotted on TLC plates, and radioactive spots were visualized and quantified using a phosphor imager (Fuji, Tokyo, Japan).
Protein Determination
Protein was measured essentially as described by Bearden (1978), with BSA as the standard.
RESULTS AND DISCUSSION
Fusicoccin-Dependent 32P-Labeling of the H+-ATPase
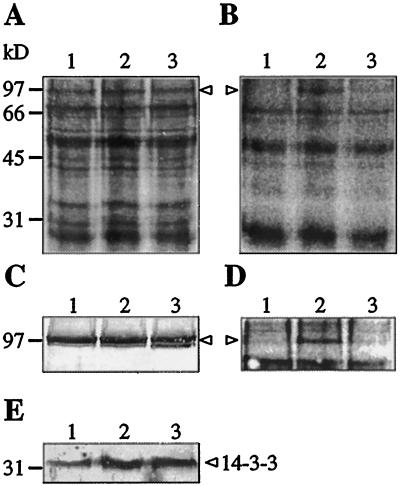
To investigate whether binding of 14–3–3 to the plasma membrane H+-ATPase may involve a phosphorylated amino acid residue, pieces of spinach leaves were infiltrated with 32P-orthophosphate and with fusicoccin to promote H+-ATPase/14–3–3 complex formation (Jahn et al., 1997; Oecking et al., 1997). This was followed by plasma membrane isolation, separation of polypeptides, and visualization of phosphorylated polypeptides. A 100-kD band coinciding with the position of the H+-ATPase was labeled, provided that the plant material was preincubated for 1 h with 32P-orthophosphate to equilibrate the ATP pool with 32P before addition of fusicoccin (Fig. 1, B and D, lanes 2). No 32P-labeling of the H+-ATPase was observed in the absence of fusicoccin (lanes 1), or if fusicoccin was added 15 min before addition of 32P-orthophosphate (lanes 3). Fusicoccin treatment led to an increase in binding of 14–3–3 to the plasma membrane (Fig. 1E), in agreement with earlier results (Korthout and de Boer, 1994; Oecking et al., 1994; Jahn et al., 1997).
Figure 1.
In vivo phosphorylation of spinach leaf plasma membrane polypeptides. Polypeptides were stained with Coomassie brilliant blue R (A), and a phosphor imager was used to visualize 32P-polypeptides in the dried gel (B). Immunostaining of the H+-ATPase (C) shows that the 100-kD H+-ATPase band coincides with the 32P-polypeptide, as visualized with a phosphor imager (D). E, Immunostaining of 14–3–3 protein. Pieces of spinach leaves were infiltrated with medium containing 32P-orthophosphate in the absence (lanes 1) or presence (lanes 2 and 3) of fusicoccin and incubated for 90 min. In lanes 2, leaves were incubated for 1 h with 32P-orthophosphate before addition of fusicoccin, whereas in lanes 3, leaves were incubated with fusicoccin for 15 min before addition of 32P-orthophosphate. Plasma membranes were isolated and polypeptides were separated by SDS-PAGE (20 μg of protein per lane). Polypeptides were either stained with Coomassie brilliant blue R, or detected by immunoblotting using antibodies raised against a maize H+-ATPase or a barley 14–3–3 protein. Arrowheads mark the position of the 100-kD H+-ATPase. Numbers at left refer to molecular mass standards.
In vivo activation of spinach leaf H+-ATPase by fusicoccin is a rapid process and reaches a plateau within minutes (Johansson et al., 1993). This means that formation of stable H+-ATPase/14–3–3 complexes (Jahn et al., 1997; Oecking et al., 1997) is completed within minutes, locking the enzyme in an activated and phosphorylated (Fig. 1, lanes 2) form. In the absence of fusicoccin, no phosphorylation of the H+-ATPase was observed (Fig. 1, B and D, lanes 1). This may indicate that the level of phosphorylation, as well as the level of H+-ATPase/14–3–3 complex formation, is normally very low, and that phosphorylation may be observed only when the equilibrium is shifted toward complex formation by the action of fusicoccin. Alternatively, the in vivo level of H+-ATPase/14–3–3 complex formation is high, but the binding is readily reversible and the complexes are disrupted and the phosphate lost due to endogenous protein phosphatase activity during homogenization of the tissue.
That 14–3–3 binding to the H+-ATPase protects the phosphorylated amino acid residue from dephosphorylation was demonstrated by incubating the leaf tissue with fusicoccin before addition of 32P-orthophosphate (i.e. H+-ATPase/14–3–3 complex formation preceded 32P-labeling of the ATP pool). In that experiment, no 32P-labeling of the H+-ATPase was observed, suggesting that the fusicoccin-induced binding of 14–3–3 somehow shielded the phosphorylated amino acid residue from protein phosphatase/kinase activity (Fig. 1, B and D, lanes 3). Thus, taken together, our results suggest that binding of 14–3–3 to the H+-ATPase protects a phosphorylated amino acid residue in the enzyme from being turned over.
Localization of the Phosphorylation Site
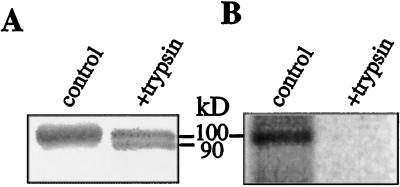
Mild trypsin treatment removes a 7- to 10-kD fragment from the C-terminal end of the H+-ATPase, thereby activating the enzyme (Palmgren et al., 1991). Hence, trypsin treatment provides a simple means for testing whether the phosphorylation observed in Figure 1 was due to an amino acid residue located in the C-terminal region of the H+-ATPase.
Plasma membranes labeled with 32P as in Figure 1 (lanes 2) were treated with trypsin in the presence of Brij 58, a detergent producing 100% sealed, cytoplasmic-side-out vesicles (Johansson et al., 1995), thus making the C-terminal region of all H+-ATPase molecules accessible for trypsin cleavage. Mild trypsin treatment resulted in a partial removal of the native 100-kD H+-ATPase band and the appearance of a 90-kD band (Fig. 2A). This 90-kD band, representing the H+-ATPase lacking the C-terminal region, was not phosphorylated (Fig. 2B), indicating that the phosphorylated amino acid residue was located in the C-terminal region; the remaining 100-kD H+-ATPase species was not labeled. The latter result is in agreement with the recent finding (Jahn et al., 1997; Oecking et al., 1997) that trypsin treatment causing removal of the C-terminal region from about 50% of the H+-ATPase molecules was sufficient for the removal of all bound 14–3–3, indicating that the C termini of H+-ATPase molecules binding 14–3–3 are more susceptible to proteolysis.
Figure 2.
Trypsin treatment of isolated plasma membrane vesicles removes all 32P label from the H+-ATPase. A, Immunostaining of the H+-ATPase; B, 32P-polypeptides in A, as visualized with a phosphor imager. Plasma membranes containing [32P]H+-ATPase were treated with trypsin in the presence of Brij 58, thus making the C-terminal region of all H+-ATPase molecules accessible for proteolysis. Polypeptides were separated by SDS-PAGE and transferred to a membrane for immunostaining. The positions of the native 100-kD H+-ATPase and the 90-kD species resulting from proteolytic removal of the C-terminal region of the H+-ATPase are indicated.
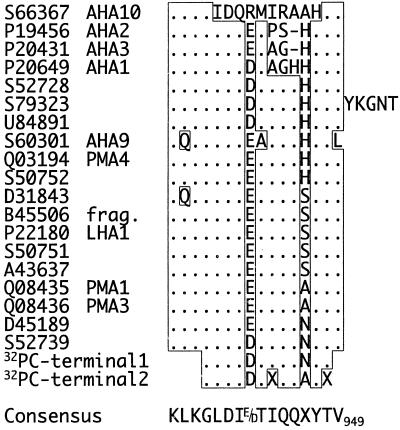
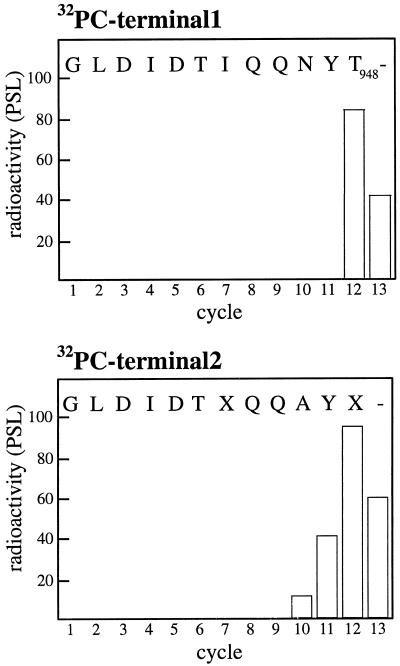
To determine the position of the phosphorylation site in the C terminus, the labeled H+-ATPase was excised from the gel and digested with the proteinase LysC, cleaving polypeptides C-terminal to Lys residues. The resulting peptides were separated by reversed-phase HPLC and the radioactivity in each fraction determined. Only two radiolabeled peaks, with the sequences GLDIDTIQQNYT and GLDIDTXQQAYX, were found (data not shown). These amino acid sequences are homologous to the relatively conserved C-terminal end of the plant plasma membrane H+-ATPase (Fig. 3), except that the C-terminal Val residue was missing in both sequences (this residue probably fell under the detection limit). The sequences obtained differ in at least one position and probably represent two different H+-ATPase isoforms in spinach leaves. To identify the phosphorylated amino acid residue, the radioactivity released during each sequencing cycle was measured. Most of the radioactivity applied was released with the 12th amino acid (Fig. 4), i.e. the second Thr in the sequence corresponding to Thr-948 in the Arabidopsis isoform AHA 1 (compare with Fig. 3). Thr-948 is a highly conserved residue, present in all H+-ATPases found in the database (Fig. 3).
Figure 3.
Alignment of the amino acid sequences obtained for the two 32P]H+-ATPase peptides with C-terminal H+-ATPase sequences. The amino acid sequences obtained for the two 32P-peptides (32PC-terminal1 and 32PC-terminal2) are compared with C-terminal sequences (the last 15–21 amino acids, depending on isoform) of plant plasma membrane H+-ATPases found in the database. GeneWorks (IntelliGenetics, Oxford Molecular Group, Oxford, UK) was used for the multiple alignment. Sequence identity is indicated with boxes and dots. Dashes indicate gaps introduced to maximize sequence identity. Numbering in the consensus sequence is according to the Arabidopsis isoform AHA1.
Figure 4.
Identification of the in vivo-phosphorylated amino acid residue. The graphs show the radioactivity released for each cycle during sequencing of the two 32P-peptides (32PC-terminal1 and 32PC-terminal2 in Fig. 3) and identifies Thr-948 (numbering is according to the Arabidopsis isoform AHA1, Fig. 3) as the phosphorylated amino acid residue. PSL, Photostimulated luminescence.
Role of Phosphothreonine-948
Assuming that phosphothreonine-948 is part of a binding motif for 14–3–3 protein, synthetic peptides corresponding to this domain were tested for their ability to disrupt the H+-ATPase/14–3–3 complex and deactivate the H+-ATPase. Thus, plasma membranes isolated from fusicoccin-treated leaves were incubated with synthetic peptides corresponding to the last 16 amino acids in the C terminus of the H+-ATPase (KLKGLDIDT942IQQAYT948V; numbering is according to the Arabidopsis isoform AHA1, Fig. 3); one peptide was not phosphorylated, a second was phosphorylated on Thr-948, and a third was phosphorylated on Thr-942. However, none of the peptides (in concentrations up to 100 μm, and incubation times up to 30 min) had any effect on H+-ATPase activity nor on the amount of 14–3–3 bound to the plasma membrane (data not shown). This is in agreement with earlier results, where incubation with phosphorylated Raf-1 peptide did not remove the bound 14–3–3 after incubation with fusicoccin (Jahn et al., 1997), which demonstrates the irreversible nature of fusicoccin-induced H+-ATPase/14–3–3 complex formation.
CONCLUDING REMARKS
Phosphorylation of an amino acid residue in the C-terminal region of the H+-ATPase has been suggested as a possible mechanism to regulate H+-ATPase activity (Palmgren et al., 1991). In vivo phosphorylation of the oat root plasma membrane H+-ATPase, as well as a pH- and Ca2+-dependent in vitro phosphorylation of Ser and Thr residues of the H+-ATPase in isolated plasma membrane vesicles, was shown by Schaller and Sussman (1988), but no effect on activity was found. More recently, Vera-Estrella et al. (1994) and Xing et al. (1996) have described an elicitor-induced, in vivo dephosphorylation of the plasma membrane H+-ATPase of cultured tomato cells, a dephosphorylation that caused activation of the enzyme. Furthermore, Sekler et al. (1994) have demonstrated in vivo phosphorylation of the H+-ATPase of the acidophilic alga Dunaliella acidophila. In none of these cases was the phosphorylated amino acid identified. However, for D. acidophila, the phosphorylation site was clearly localized to the C-terminal part of the enzyme.
In this work we present the first identification to our knowledge of an in vivo phosphorylation site in the plant plasma membrane H+-ATPase. We show that after incubation of spinach leaf tissue with fusicoccin, the plasma membrane H+-ATPase may be isolated in a phosphorylated form (Fig. 1, B and D, lanes 2). Furthermore, we show that the phosphorylated amino acid is located in the C-terminal region of the H+-ATPase (Figs. 2 and 3) and is identical to Thr-948 (Fig. 4), the second amino acid from the C-terminal end in most isoforms of the H+-ATPase (Fig. 3). Because phosphothreonine-948 is protected from turnover by 14–3–3 binding, it may be part of a binding motif for 14–3–3 protein. Alternatively, phosphothreonine-948 may be shielded from protein phosphatase/kinase activity due to a conformational change induced by 14–3–3 binding. The putative 14–3–3-binding motif in which phosphothreonine-948 resides is far from a perfect match to any of the recently identified optimal binding motifs for 14–3–3 proteins (Muslin et al., 1996; Yaffe et al., 1997). However, as proposed by Yaffe et al. (1997), “The nature of 14–3–3 interaction is determined by the extent to which the binding motif matches the optimal consensus sequence”, and “determines if the role of 14–3–3 is to act as a sequestering molecule, a chaperone, or an adaptor.” For the H+-ATPase/14–3–3 interaction, where the 14–3–3 protein acts as a regulatory protein, a relatively weak and readily reversible binding should be expected. A weak interaction is supported by experimental data showing that the H+-ATPase when isolated is devoid of 14–3–3 unless the tissue has been incubated with fusicoccin to induce a stronger interaction (Jahn et al., 1997; Oecking et al., 1997). Thus, the irreversible binding of 14–3–3 induced by fusicoccin is lethal; the hosts of the fungus Fusicoccum amygdali, peach and almond trees, wilt and die because the irreversible activation of the H+-ATPase in stomatal cells results in a permanent opening of the stomata (for review, see de Boer, 1997).
ACKNOWLEDGMENTS
We wish to thank Adine Karlsson for excellent technical assistance and Dr. Louise Race (Department of Plant Cell Biology, Lund University, Sweden) for critically reading the manuscript. We thank Dr. Wolfgang Michalke (Institut für Biologie III, Albert-Ludwigs-Universität, Freiburg, Germany) for kindly providing the monoclonal antibody to the H+-ATPase and Dr. David B. Collinge (Department of Plant Biology, Royal Veterinary and Agricultural University, Frederiksberg C, Denmark) for a generous gift of the antiserum to the 14–3–3 protein.
Footnotes
This work was supported by the Swedish Foundation for Strategic Research, the Swedish Natural Science Research Council, the Swedish Council for Forestry and Agricultural Research, and the European Union Biotechnology Program.
LITERATURE CITED
- Aitken A. 14–3–3 and its possible role in co-ordinating multiple signalling pathways. Trends Cell Biol. 1996;6:341–347. doi: 10.1016/0962-8924(96)10029-5. [DOI] [PubMed] [Google Scholar]
- Baunsgaard L, Fuglesang AT, Jahn T, Korthout HAAJ, de Boer AH, Palmgren MG. The 14–3–3 proteins associate with the plant plasma membrane H+-ATPase to generate a fusicoccin binding complex and a fusicoccin responsive system. Plant J. 1998;13:661–671. doi: 10.1046/j.1365-313x.1998.00083.x. [DOI] [PubMed] [Google Scholar]
- Bearden JC., Jr Quantitation of submicrogram quantities of protein by an improved protein-dye binding assay. Biochim Biophys Acta. 1978;533:525–529. doi: 10.1016/0005-2795(78)90398-7. [DOI] [PubMed] [Google Scholar]
- Brandt J, Thordal-Christensen H, Vad H, Gregersen PL, Collinge DB. A pathogen-induced gene of barley encodes a protein showing high similarity to a protein kinase regulator. Plant J. 1992;2:815–820. [PubMed] [Google Scholar]
- de Boer AH. Fusicoccin: a key to multiple 14–3–3 locks? Trends Plant Sci. 1997;2:60–66. [Google Scholar]
- Jahn T, Fuglesang AT, Olsson A, Brüntrup IM, Collinge DB, Volkmann D, Sommarin M, Palmgren MG, Larsson C. The 14–3–3 protein interacts directly with the C-terminal region of the plant plasma membrane H+-ATPase. Plant Cell. 1997;9:1805–1814. doi: 10.1105/tpc.9.10.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson F, Olbe M, Sommarin M, Larsson C. Brij 58, a polyoxyethylene acyl ether, creates membrane vesicles of uniform sidedness: a new tool to obtain inside-out (cytoplasmic side-out) plasma membrane vesicles. Plant J. 1995;7:165–173. doi: 10.1046/j.1365-313x.1995.07010165.x. [DOI] [PubMed] [Google Scholar]
- Johansson F, Sommarin M, Larsson C. Fusicoccin activates the plasma membrane H+-ATPase by a mechanism involving the C-terminal inhibitory domain. Plant Cell. 1993;5:321–327. doi: 10.1105/tpc.5.3.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson I, Larsson C, Ek B, Kjellbom P. The major integral proteins of spinach leaf plasma membranes are putative aquaporins and are phosphorylated in response to Ca2+ and apoplastic water potential. Plant Cell. 1996;8:1181–1191. doi: 10.1105/tpc.8.7.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korthout HAAJ, de Boer AH. A fusicoccin binding protein belongs to the family of 14–3–3 brain protein homologs. Plant Cell. 1994;6:1682–1692. doi: 10.1105/tpc.6.11.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lanfermeijer FC, Prins HBA. Modulation of H+-ATPase activity by fusicoccin in plasma membrane vesicles from oat (Avena sativa L.) roots. A comparison of modulation by fusicoccin, trypsin and lysophosphatidylcholine. Plant Physiol. 1994;104:1277–1285. doi: 10.1104/pp.104.4.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson C, Sommarin M, Widell S. Isolation of highly purified plasma membranes and the separation of inside-out and right-side-out vesicles. Methods Enzymol. 1994;228:451–469. [Google Scholar]
- Marra M, Fullone MR, Fogliano V, Masi S, Mattei M, Pen J, Aducci P. The 30-kD protein present in purified fusicoccin receptor preparations is a 14–3–3-like protein. Plant Physiol. 1994;106:1497–1501. doi: 10.1104/pp.106.4.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrè E. Fusicoccin: a tool in plant physiology. Annu Rev Plant Physiol. 1979;30:273–288. [Google Scholar]
- Michelet B, Boutry M. The plasma membrane H+-ATPase. A highly regulated enzyme with multiple physiological functions. Plant Physiol. 1995;108:1–6. doi: 10.1104/pp.108.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muslin AJ, Tanner JW, Allen PM, Shaw AS. Interaction of 14–3–3 with signaling proteins is mediated by the recognition of phosphoserine. Cell. 1996;84:889–897. doi: 10.1016/s0092-8674(00)81067-3. [DOI] [PubMed] [Google Scholar]
- Oecking C, Eckerskorn C, Weiler EW. The fusiccocin receptor of plants is a member of the 14–3–3 superfamily of eukaryotic regulatory proteins. FEBS Lett. 1994;352:163–166. doi: 10.1016/0014-5793(94)00949-x. [DOI] [PubMed] [Google Scholar]
- Oecking C, Piotrowski M, Hagemeier J, Hageman K. Topology and target interaction of the fusicoccin-binding 14–3–3 homologs of Commelina communis. Plant J. 1997;12:441–453. [Google Scholar]
- Palmgren MG, Larsson C, Sommarin M. Proteolytic activation of the plant plasma membrane H+-ATPase by removal of a terminal segment. J Biol Chem. 1990;265:13423–13426. [PubMed] [Google Scholar]
- Palmgren MG, Sommarin M, Serrano R, Larsson C. Identification of an autoinhibitory domain in the C-terminal region of the plant plasma membrane H+-ATPase. J Biol Chem. 1991;266:20470–20475. [PubMed] [Google Scholar]
- Rasi-Caldogno F, Pugliarello MC, Olivari C, De Michelis MI. Controlled proteolysis mimics the effect of fusicoccin on the plant plasma membrane H+-ATPase. Plant Physiol. 1993;103:391–398. doi: 10.1104/pp.103.2.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regenberg B, Villalba JM, Lanfermeijer FJ, Palmgren MG. C-terminal deletion analysis of plant plasma membrane H+-ATPase: yeast as a model system for solute transport across the plant plasma membrane. Plant Cell. 1995;7:1655–1666. doi: 10.1105/tpc.7.10.1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld J, Capdevielle J, Guillemot JC, Ferrara P. In-gel digestion of proteins for internal sequence analysis after one- or two-dimensional gel electrophoresis. Anal Biochem. 1992;203:173–179. doi: 10.1016/0003-2697(92)90061-b. [DOI] [PubMed] [Google Scholar]
- Schaller GE, Sussman MR. Phosphorylation of the plasma membrane H+-ATPase of oat roots by a calcium-stimulated protein kinase. Planta. 1988;173:509–518. doi: 10.1007/BF00958964. [DOI] [PubMed] [Google Scholar]
- Sekler I, Weiss M, Pick U. Activation of the Dunaliella acidophila plasma membrane H+-ATPase by trypsin cleavage of a fragment that contains a phosphorylation site. Plant Physiol. 1994;105:1125–1132. doi: 10.1104/pp.105.4.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vera-Estrella R, Barkla BJ, Higgins VJ, Blumwald E. Plant defense response to fungal pathogens. Activation of host-plasma membrane H+-ATPase by elicitor-induced enzyme dephosphorylation. Plant Physiol. 1994;104:209–215. doi: 10.1104/pp.104.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing T, Higgins VJ, Blumwald E. Regulation of plant defense response to fungal pathogens: two types of protein kinases in the reversible phosphorylation of the host plasma membrane H+-ATPase. Plant Cell. 1996;8:555–564. doi: 10.1105/tpc.8.3.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe MB, Rittinger K, Volinia S, Caron PR, Aitken A, Leffers H, Gamblin SJ, Smerdon SJ, Cantley LC. The structural basis for 14–3–3: phosphopeptide binding specificity. Cell. 1997;91:961–971. doi: 10.1016/s0092-8674(00)80487-0. [DOI] [PubMed] [Google Scholar]