Abstract
Transforming growth factor-beta (TGF-β) signaling pathways contain both tumor suppressor and tumor promoting activities. We have demonstrated that Nodal, another member of the TGF-β superfamily, and its receptors are expressed in prostate cancer cells. Nodal and TGF-β exerted similar biological effects on prostate cells; both inhibited proliferation in WPE, RWPE1 and DU145 cells, whereas neither had any effect on the proliferation of LNCaP or PC3 cells. Interestingly, Nodal and TGF-β induced migration in PC3 cells, but not in DU145 cells. TGF-β induced predominantly phosphorylation of Smad3, whereas Nodal induced phosphorylation of only Smad2. We also determined the expression and differential role of Ski, a corepressor of Smad2/3, in Nodal and TGF-β signaling in prostate cancer cells. Similar levels of Ski mRNA were found in several established prostate cell lines; however, high levels of Ski protein were only detected in prostate cancer cells and prostate cancer tissue samples. Exogenous Nodal and TGF-β had no effects on Ski mRNA levels. On the other hand, TGF-β induced a rapid degradation of Ski protein mediated by the proteasomal pathway, whereas Nodal had no effect on Ski protein. Reduced Ski levels correlated with increased basal and TGF-β-induced Smad2/3 phosphorylation. Knockdown of endogenous Ski reduced proliferation in DU145 cells and enhanced migration of PC3 cells. We conclude that high levels of Ski expression in prostate cancer cells may be responsible for repression of TGF-β and Smad3 signaling, but Ski protein levels do not influence Nodal and Smad2 signaling.
Introduction
Transforming growth factor-β (TGF-β) superfamily is composed of nearly 30 growth factors including TGF-β proteins, bone morphogenetic proteins (BMPs), activins, Nodal and its related proteins (1–3). These growth factors play an important role in cell proliferation and differentiation, development, tumorigenesis, extracellular matrix modification, apoptosis, angiogenesis and immunosuppression (1–3). There are two types of membrane serine/threonine kinase receptors that are required for the functions of TGF-β-like growth factors. TGF-β superfamily ligands bind to specific type II receptors which then associate with specific type I receptors resulting in phosphorylation and activation of type I receptors (1–3). The activated type I receptor phosphorylates the appropriate Smad proteins, which in turn interact with the co-Smad protein, Smad4, translocate to the nucleus and regulate expression of target genes (1–3). Smad2 and Smad3 respond to Nodal, TGF-β and activins, whereas Smad1, Smad5 and Smad8 mediate BMP signaling (4). TGF-β acts as tumor suppressor in the early stages of epithelial cancers by inhibiting proliferation and inducing apoptosis (5). However, in the later stages of the disease, TGF-β acts as tumor promoter and is associated with aggressive form of cancers due to its effects on angiogenesis, immune suppression and metastasis (5). Previous studies using prostate cancer-derived cell lines have shown differential effects of TGF-β and in different cell lines.
Nodal is a novel member of the TGF-β superfamily that inhibits differentiation, maintains the pluripotency of human embryonic stem cells (hESCs) and promotes the self-renewing capacity of mouse embryonic stem cells (6). Nodal also plays an important role in the induction of dorsal mesoderm, anterior patterning and formation of left–right asymmetry during early embryonic development (7). Nodal signals by binding to heterodimeric complexes between type I (ALK4) and type II receptors (ActRIIA/ActRIIB), whereas TGF-β has its own receptors (ALK5 and TβRII) (8). We have recently shown that Nodal and its signaling receptors are present in prostate cancer cells and exogenous Nodal modulates proliferation and migration of prostate cancer cells (9). These effects of Nodal are mediated by Smad2/3 signaling (9,10).
Smad signaling is subject to many levels of positive and negative regulation that target both the receptors and the intracellular mediators (11). Among the negative regulators of Smad2/3 function, Sloan–Kettering Institute (Ski) protein family members suppress TGF-β signaling (12). Ski was originally discovered as an oncogene of the avian Sloan–Kettering retrovirus (11), followed by identification of Ski-related novel protein N (SnoN) and its isoforms SnoN, SnoA and SnoI in several mammalian species such as monkey, dog, cow, rabbit and pig, but not in rodents (13). High levels of Ski and SnoN are associated with many types of human tumor cell lines derived from melanoma, breast cancer, and carcinoma of the esophagus, thyroid, stomach and epidermoid (14–17). Ski is an important negative regulator of TGF-β signaling through its ability to interact with and repress the activity of Smad proteins (11,18). Previous studies have shown that binding of Ski to Smad2/3 causes dissociation of the histone acetyltransferase p300 from the Smad2/3 complex and promotes association with mSin3A and histone deacetylase complex (19–21).
Although both Nodal and TGF-β have been shown to exert differential biological effects on prostate cancer cells and both share Smad2/3 signaling, differences, if any, in intracellular signaling pathways of the two cytokines remain unknown. In this study, we have compared the effects of TGF-β1 and Nodal on proliferation and migration of prostate cancer cells and have determined the expression and role of Ski in Smad2 and Smad3 signaling.
Materials and methods
Chemicals and reagents
Recombinant human Nodal and TGF-β1 were purchased from R&D systems (Minneapolis, MN). The antibodies against phospho-Smad2, phospho-Smad3 and Smad2/3 were purchased from Cell Signaling Technology (Danvers, MA). Anti-Ski, Smad2, Smad3 antibodies and protein A/G agarose were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-β-Actin antibody was purchased from Sigma–Aldrich (St Louis, MO). The anti-rabbit and anti-mouse immunoglobulins coupled with horseradish peroxidase (IgG-HRP) were obtained from Promega (Madison, WI) and donkey anti-goat IgG-HRP was obtained from Santa Cruz Biotechnology. Proteasome inhibitor (MG123) was purchased from Sigma–Aldrich. Smad3 inhibitor (SIS3) was obtained from Calbiochem (San Diego, CA). Prostate cancer tissue microarray (PR956) was obtained from US Biomax (Rockville, MD).
Cell culture and cell treatments
Normal prostate epithelial cells (PrECs) were obtained from Lonza (Walkersville, MD). Prostate epithelial stem cell line (WPE), immortalized normal PrECs (PZ-HPV7), immortalized prostate luminal epithelial cell line (RWPE1), k-ras transformed RWPE1 cell line (RWPE2) and prostate cancer cell lines (LNCaP, DU145 and PC3) were obtained from American Type Culture Collection (ATCC; Rockville, MD). LNCaP is an androgen-dependent cell line isolated from a lymph node lesion. DU145 and PC3 are androgen-independent cell lines derived from brain and bone metastatic sites, respectively. PC3M cells, derived from a PC3 xenograft, were obtained from Dr Girish Shah (University of Louisiana). The prostate cells were cultured using established procedures and treated with Nodal and TGF-β1 as described previously (9,22).
RNA isolation, reverse transcription and polymerase chain reaction
Total RNA was isolated from prostate cells using TRIzol (Invitrogen, Carlsbad, CA) and the resulting RNA samples were quantified by optical density reading at 260nm as described previously (9,23).
Polymerase chain reaction (PCR) was performed to detect mRNA levels of Ski and L-19. The PCR reaction mixture was composed of 0.1mM dNTPs, 0.5U Taq DNA polymerase, 10× PCR buffer with 3mM MgCl2 and 25 pM of the specific primers in a total volume of 15 µl. The Ski primers were 5′-TGTCTGCCGCAGATTCTCAACT-3′ (forward) and 5′-GGA TGCCCATGACTTTGAGGA-3′ (reverse). The L-19 primers were 5′-GAAATCGCCAATGCCAACTC-3′ (forward) and 5′-TCTTAGACCTGCG- AGCCTCA-3′ (reverse). L-19 (a ribosomal protein) was used as a template control. RNA samples processed without reverse transcription (RT) and PCR amplified by the L-19 and Ski primers were used as negative controls. Amplification was performed at 94°C for 20 s, 60°C for 30 s and 72°C for 45 s for 30 cycles for Ski and L-19. For all PCRs, an initial step was at 95°C for 2min and a final extension was at 72°C for 10min. The PCR products were separated on 1.5–2% agarose gels and stained with ethidium bromide.
Immunoprecipitation
PC3 cells were treated with Nodal (200ng/ml) or TGF-β1 (5ng/ml) for 30min and were lysed in lysis buffer (Cell Signaling Technology, Beverly, MA). Total cell lysates containing 500 µg protein were used for immunoprecipitation. Supernatants were precleared by centrifugation after 1h of incubation at 4°C with 30 µl of immobilized protein-A/G Sepharose beads. The resulting supernatants were incubated with either 5 µg of anti-Smad2 or Smad3 antibodies overnight at 4°C. Immunocomplexes were collected by centrifugation after incubation with protein-A/G Sepharose beads and were analyzed by western blot analysis with anti-Ski antibody.
Western blot analysis
Total cellular proteins were prepared and analyzed by western blot as described previously (23,24). Briefly, cell lysates were mixed with Laemmli buffer. Individual samples (30–35 µg proteins) were subjected to sodium dodecyl sulfate–polyacrylamide gel electrophoresis in 8 or 10% gels and transferred to polyvinylidene difluoride membranes (Millipore). After blocking with 5% fat-free milk in Tris-Buffered Saline Tween (TBST) for 1h at room temperature, the membranes were incubated with appropriate dilutions of specific primary antibodies (1:1000 dilution for anti-p-Smad2, anti-p-Smad3 and Smad2/3 antibodies; 1:500 for anti-Ski antibody; 1:20 000 for anti-β-actin antibody) overnight at 4°C. After washing, the blots were incubated with anti-rabbit or anti-mouse IgG-HRPs for 1h. The blots were developed in enhanced chemiluminescence mixture (Thermo Fisher Scientific , Rockford, IL) and the density of specific protein bands were determined by QuantityOne image analysis software.
Immunofluorescence of Ski
Prostate cells were cultured in four-chamber slides at the density of 1×104 cells per well. The cells were preincubated in serum or supplement-free media for 2h, followed by treatment with TGF-β1 (5ng/ml) for different time periods. Cells were fixed with 4% paraformaldehyde in PBS (phosphate buffered saline) for 20min. After washing with PBS, the cells were permeabilized in 0.2% Triton X-100 in PBS for 10min and washed with PBS. Prostate tissue sections were deparaffinized in xylene and rehydrated. Endogenous peroxide was blocked by incubating the sections in 0.3% hydrogen peroxide for 5min. The cells or tissue sections were incubated in blocking solution (5% normal goat serum in PBS) for 1h. The cells or tissue sections were then incubated with anti-Ski antibody (1:300) overnight at 4°C. After washing, the cells or tissue sections were incubated with secondary antibody, Alexa Fluor 488-conjugated anti-rabbit antibody (1:1000) (Molecular Probes), for 1h. Both were washed with PBS and incubated with 4′,6-diamidino-2-phenylindole for 10min to detect the nuclei. The tissue sections were then washed with PBS and counterstained with hematoxylin. Coverslips were mounted with Vector Vectashield mounting medium (Vector Laboratories, Burlingame, CA) for the cells, and the tissue sections were mounted using xylene mounting medium (Fisher Scientific). Images were captured using 40× magnification with an Axiovision camera using Axiovision software.
Transfection with Ski small interfering RNA
To knockdown endogenous Ski expression, prostate cells were cultured in 6-well plates at the density of 2×105 cells per well in 2ml antibiotic-free normal growth medium supplemented with 5% fetal bovine serum (FBS). The cells were cultured at 37°C until the cells were 60–80% confluent. Control and Ski small interfering RNAs (siRNAs) were transfected into DU145 and PC3 cells according to the manufacturer’s instructions (Santa Cruz Biotechnology). Briefly, the cells were washed once with 2ml of siRNA transfection medium and the siRNA duplex was overlaid onto the washed cells, incubated for 6h, followed by addition of 10% FBS to the cells without removing the transfection mixture and the cells were incubated for 24h. The medium was changed with fresh medium and the cells were incubated for an additional 24h. The cells were harvested at specific times for determination of the changes in Ski expression and/or for proliferation and wound healing assays.
Cell proliferation assay
The effects of Nodal and TGF-β on cell proliferation after Ski knockdown were determined using 3H-thymidine incorporation assay as described previously (9,23).
Wound healing assay
Confluent cell monolayers of parental and Ski siRNA transfected PC3 cells were wounded with a pipette tip. Phase contrast pictures were taken using inverted microscope at the time of wounding (0h) and 48h later. The wound closure was estimated as the percentage of the closure area to the initial wounded area. Each treatment group was assayed in replicates of three and each experiment was repeated at least three times.
Statistical analysis
All experiments were repeated at least three times using a different cell preparation. The results are presented as means ± SEM of three independent experiments and images from a single representative experiment are presented. Analysis of variance and Duncan’s modified multiple range tests were employed to assess the significance of differences among various treatment groups.
Results
Effects of Nodal and TGF-β on proliferation and migration in prostate cell lines
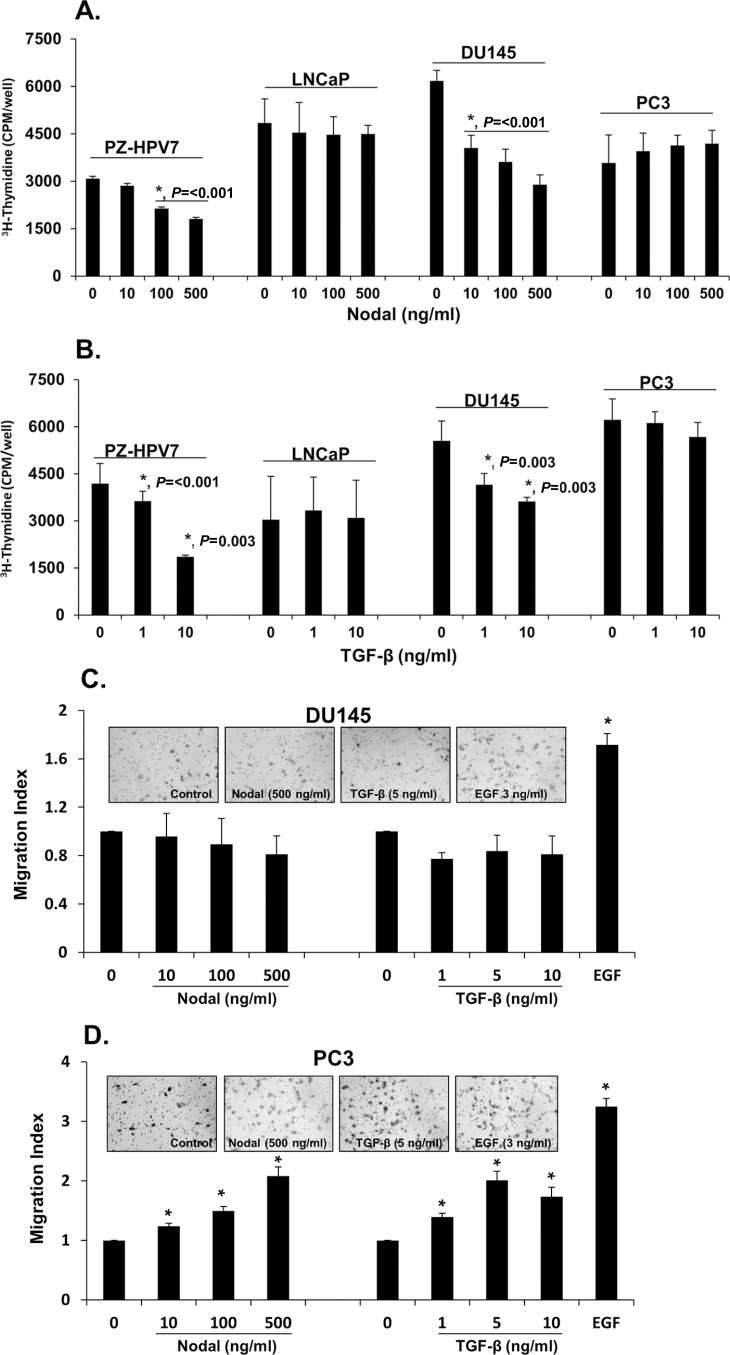
TGF-β exerts differential biological effects in different prostate cancer-derived cell lines (5). We have demonstrated that Nodal, another novel member of the TGF-β superfamily, and its receptors are expressed in prostate cancer cells and Nodal exerts differential effects on proliferation and migration in different prostate cell lines (9). Therefore, we determined the comparative effects of Nodal and TGF-β on proliferation and migration under identical experimental conditions in selected prostate cell lines. As shown in Figure 1A and 1B, both Nodal and TGF-β inhibited proliferation in a normal prostate cell line (PZ-PHV7) and in DU145 prostate cancer cells. However, both Nodal and TGF-β had no effect on proliferation of PC3 and LNCaP cells. Interestingly, both Nodal and TGF-β induced cell migration in PC3 cells, but not in DU145 cells (Figure 1C and 1D). On the other hand, epidermal growth factor (EGF) used as a positive control induced cell migration in both DU145 and PC3 cells.
Fig. 1.
Effects of Nodal and TGF-β on cell proliferation and migration of prostate cell lines. (A and B). The effects of Nodal and TGF-β on DNA synthesis in PZ-HPV7, LNCaP, DU145 and PC3 cells as determined by 3H-thymidine incorporation assay. The cells were serum-starved for 24h and treated with different concentrations of rhNodal and TGF-β for 18h in the presence of 5% FBS. The cells were then pulse labeled for 4h with 1 µCi/ml 3H-thymidine and DNA incorporated radioactivity was determined by liquid scintillation counting. Each bar represents mean ± SEM (n = 3). *Significantly different compared with untreated controls. (C and D) Nodal and TGF-β dose-dependently induced migration in PC3 cells but not in DU145 cells in a transwell migration assay. Representative images of DU145 and PC3 cell lines after different treatments. Cells were visualized under 10× objectives. EGF (3ng/ml) was used as a positive control. Each bar represents mean ± SEM (n = 3). *Significantly different (P < 0.05) compared with untreated controls.
Distinct role of Nodal and TGF-β-induced Smad signaling in prostate cell lines
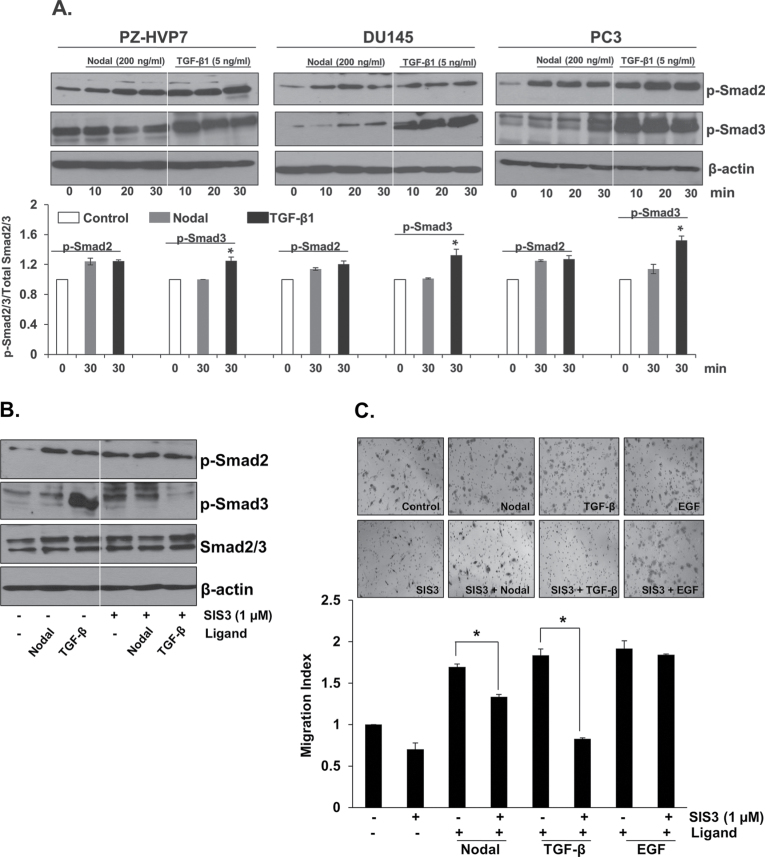
Nodal and TGF-β signaling is initiated by binding of the ligand to type II receptors that form heterodimers with type I receptors leading to the phosphorylation of Smad2 and Smad3 proteins (8,25); therefore, we investigated whether Nodal and TGF-β effects are mediated by similar signaling components. We studied the effects of exogenous Nodal and TGF-β on phosphorylation of Smad2 and Smad3 in PZ-HVP7, DU145 and PC3 cells. Western blot analysis showed that Smad2 was phosphorylated in a time-dependent manner in PZ-HVP7, DU145 and PC3 cells in response to Nodal treatment; however, Nodal had only a minor, if any, effect on Smad3 phosphorylation (Figure 2A). Interestingly, exogenous TGF-β induced both Smad2 and Smad3 phosphorylation. Phosphorylation of Smad3 was much higher than that of Smad2 in response to TGF-β treatment (Figure 2A). These findings suggest that Nodal primarily induces Smad2 signaling, whereas TGF-β can induce both Smad2 and Smad3 phosphorylation.
Fig. 2.
Activation of Nodal and TGF-β signaling in prostate cell lines. (A) Western blot analyses of phosphorylated Smad2 and Smad3 and β-actin in PZ-HPV7, DU145 and PC3 cells at different time periods after treatment with Nodal (200ng/ml) or TGF-β1 (5ng/ml). Western blots using anti-β-actin antibody were used as internal controls. Quantitative analysis of p-Smad2 and p-Smad3 in PZ-HPV7, DU145 and PC3 cells treated with Nodal and TGF-β were relative to that of the untreated control (designated as one) after normalization to the signal obtained with Smad2/3. Each bar represents mean ± SEM (n = 3). *Significantly different (P < 0.05) compared with untreated controls. (B) Western blot analyses of phosphorylated Smad2 and Smad3, total Smad2/3 and β-actin in PC3 cells after pretreatment with Smad3 inhibitor (SIS3, 1 µM). (C) Pretreatment with SIS3 (1 µM) for 30min completely blocked the migration of PC3 cells induced by TGF-β (5ng/ml) but not in Nodal (500ng/ml) and EGF (3ng/ml) treated cells. The data were presented as mean ± SEM (n = 3). *Significantly different (P < 0.05) compared with untreated controls.
Previous studies have shown that a specific inhibitor of Smad3 (SIS3) completely diminished the constitutive phosphorylation of Smad3, Smad3 binding to DNA and the interaction of Smad3 with Smad4 (26). As shown in Figure 2B, pretreatment with Smad3 inhibitor (SIS3) suppressed TGF-β-induced Smad3 phosphorylation. On the other hand, this inhibitor had no effect on the phosphorylation of Smad2 in the presence or absence of TGF-β. Additionally, pretreatment with SIS3 completely blocked the stimulatory effects of TGF-β on migration of PC3 cells (Figure 2C) but caused only a partial blockage of Nodal effects. The inhibitor did not influence EGF-induced migration of PC3 cells (Figure 2C). These results indicate that TGF-β effects in prostate cancer cells are mediated primarily by Smad3, whereas the effects of Nodal are mediated primarily by Smad2.
Expression of Ski in prostate cell lines and primary prostate tissues
Several studies have shown that Ski is a negative regulator of TGF-β signaling pathway through its ability to interact with and repress the activity of Smad2/3 proteins (11,18,27–31). Since Nodal and TGF-β receptors are coupled with Smad2 and Smad3 signaling, we investigated the expression of Ski and its potential regulation of Nodal and/or TGF-β signaling in prostate cancer cells.
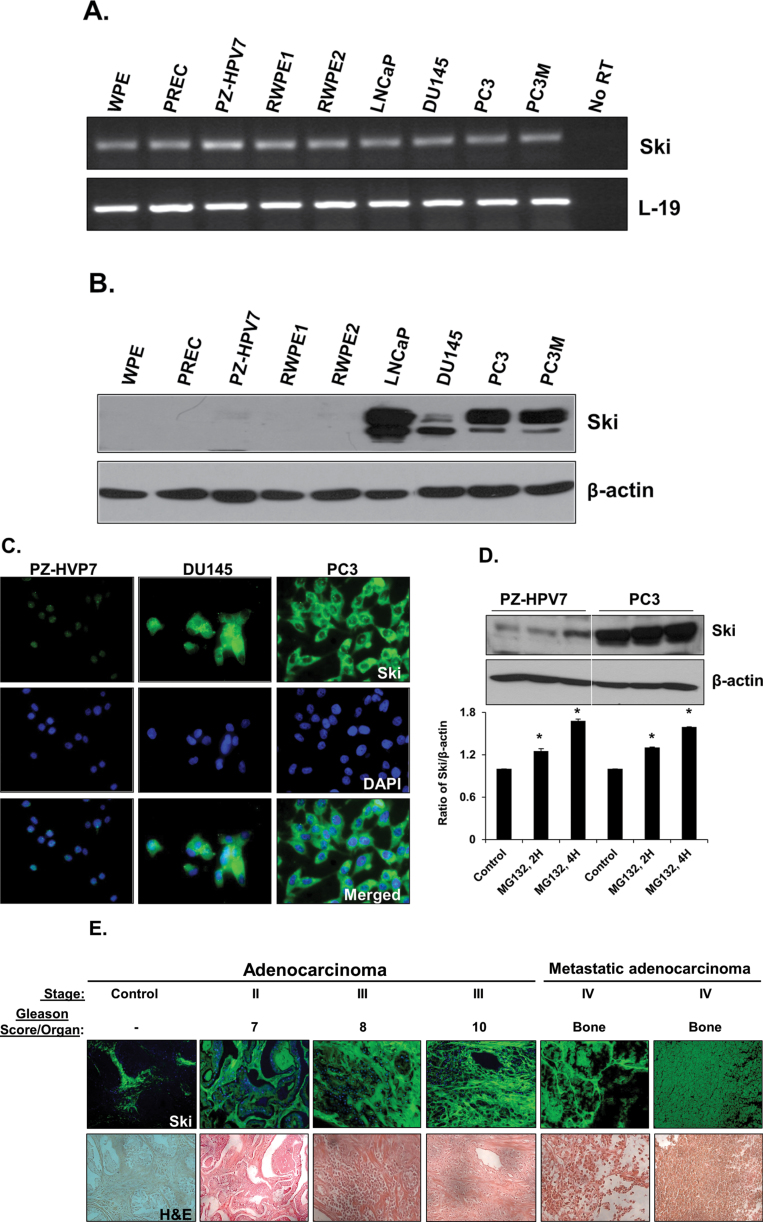
Total RNAs and proteins were extracted from prostate stem cells (WPE), normal PrECs, immortalized normal epithelial cells (PZ-HPV7 and RWPE1), k-ras transformed RWPE1 (RWPE2) cells and prostate cancer cell lines (LNCaP, DU145, PC3 and PC3M). As shown in Figure 3A, RT–PCR detected Ski mRNA in all cell lines. The expression levels were not significantly different in various cell lines. The identity of the RT–PCR product with Ski was confirmed by DNA sequencing. To examine the presence of Ski protein in these prostate cell lines, total cellular proteins were analyzed by western blots using specific anti-Ski antibody. Ski protein was highly expressed in all prostate cancer cell lines (LNCaP, DU145, PC3 and PC3M) (Figure 3B); however, it was either very low or undetectable in prostate stem cells (WPE) and normal prostate cells (PrEC, PZ-HPV7, RWPE1 and RWPE2) (Figure 4B). Treatment with proteasome inhibitor (MG132) increased Ski protein levels in PZ-HVP7 and PC3 cells (Figure 3D), indicating that posttranslational degradation of Ski by ubiquitin–proteasome pathway is responsible for low Ski protein levels in normal prostate cells.
Fig. 3.
Basal expression of Ski in prostate cell lines. (A) Total RNAs were isolated and semi-quantitative RT–PCR was performed to determine the mRNA levels of Ski in prostate cell lines. L-19 was used as an internal control. No RT samples derived from the same RNAs were also included. (B) Western blot analysis of Ski protein levels in prostate cell lines. Total cellular proteins were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis and blotted using anti-Ski antibody. (C) Ski (green) expression in PZ-HVP7, DU145 and PC3 prostate carcinoma cells. Nuclei were counterstained with 4′,6-diamidino-2-phenylindole (blue). Ski was predominately localized in cytoplasm of the cells; the basal expression of Ski was high in prostate cancer cell lines but low in PZ-HPV7 cells. (D) Western blot analyses of Ski in PZ-HVP7 and PC3 cells treated with MG132 (25 µM). Anti-β-actin antibody was served as internal controls. Quantitative analysis of Ski in PZ-HPV7 and PC3 cells with or without treatment with MG132 (25 µM) (lower panel) were relative to untreated control (designated as one) after normalization to the signal obtained with β-actin. Each bar represents mean ± SEM (n = 3). *Significantly different (P < 0.05) compared with untreated controls. (E) Tissue sections were deparaffinized, rehydrated and blocked for endogenous peroxidase. Sections were blocked using normal goat serum in PBS and incubated with Ski antibody overnight. Sections were washed and incubated with Alexa Flour 488 anti-rabbit for 30min, washed again, and incubated with 4′,6-diamidino-2-phenylindole. Sections were then washed and counterstained with hematoxylin and mounted using xylene mounting medium. Immunofluorescence and H&E staining were analyzed by Zeiss microscope using 40× magnification. Representative images of the control and cancer tissues are shown.
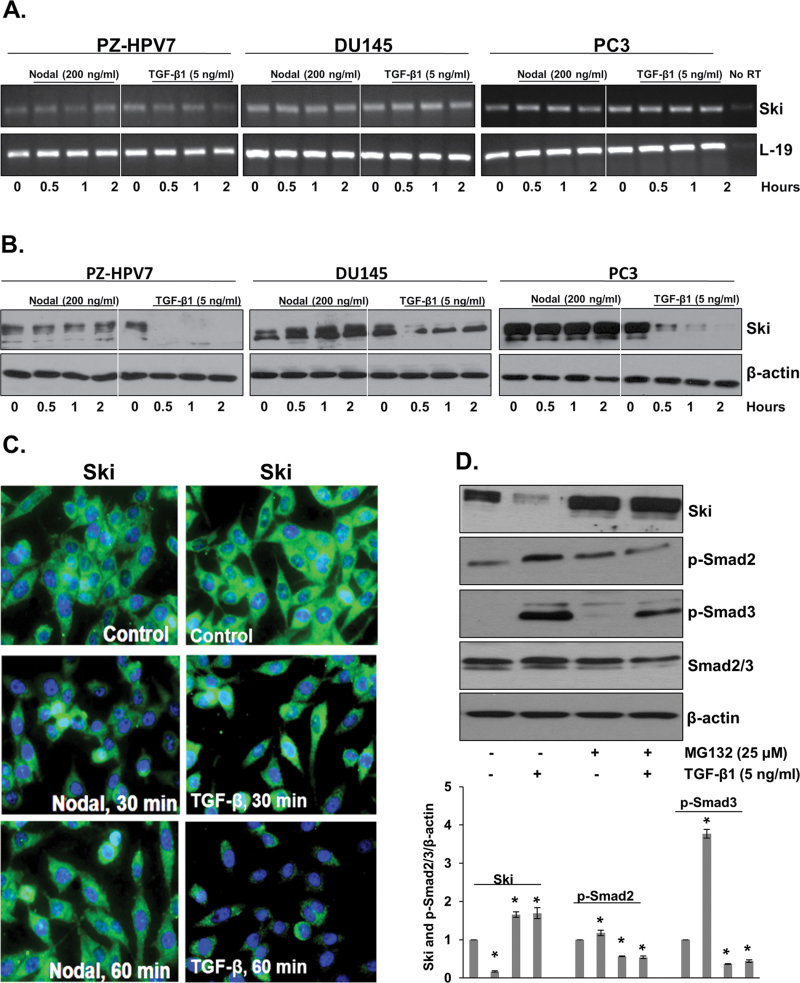
Fig. 4.
The effects of Nodal and TGF-β on Ski in prostate cell lines. (A) Semi-quantitative RT–PCR was performed to determine the mRNA levels of Ski in PZ-HVP7, DU145 and PC3 cells at different time periods after treatment with Nodal (200ng/ml) and TGF-β1 (5ng/ml). L-19 was used as an internal control. No RT samples derived from the same RNAs were also included. (B) Western blot analyses of Ski protein in PZ-HVP7, DU145 and PC3 cells at different time periods after treatment with Nodal (200ng/ml) and TGF-β1 (5ng/ml). Western blots using anti-β-actin antibody served as internal controls. (C) Prostate cells were analyzed by immunofluorescence with anti-Ski antibody after treatment with Nodal (200ng/ml) and TGF-β1 (5ng/ml) for 30 and 60min. Immunofluorescence showed that treatment with TGF-β decreases the levels of Ski protein in PC3 cells. (D) Western blot analyses of Ski, p-Smad2 and p-Smad3 in PC3 cells (upper panel) treated with TGF-β1 (5ng/ml) in the presence or absence of MG132 (25 µM). Western blots using anti-β-actin antibody were used as internal controls. Quantitative analysis of Ski, p-Smad2 and p-Smad3 in PC3 cells with TGF-β1 (5ng/ml) with or without MG132 (lower panel). For further details, see legend to Figure 4C. Each bar represents mean ± SEM (n = 3). * Significantly different (P<0.05) compared with untreated controls.
To determine the intracellular localization of Ski in PZ-HPV7, DU145 and PC3 cells, immunofluorescence was performed with specific anti-Ski antibody. As shown in Figure 3C, Ski was predominately localized in the cytoplasm of the cells. Next, we determined whether Ski was expressed in human prostate tissues, and whether its levels, cellular localization and/or activity correlated with prostate tumor progression. Prostate tissue microarrays containing normal prostate and prostate adenocarcinomas tissues at different stages and Gleason scores and metastatic cancers were analyzed for presence of Ski protein by immunofluorescence. As shown in Figure 3E, Ski protein was absent in normal prostate tissues; however, it was highly expressed in adenocarcinomas and metastatic cancer tissues.
The effects of Nodal and TGF-β on Ski in prostate cell lines
Next, we determined the effects of Nodal and TGF-β on Ski protein in normal prostate cells (PZ-HVP7) and in prostate cancer cells (DU145 and PC3). Cells were cultured in the presence or absence of Nodal (200ng/ml) or TGF-β (5ng/ml) for specific time periods and the expression of Ski was determined by RT–PCR, western blotting and immunofluorescence. As shown in Figure 4A, exogenous Nodal and TGF-β did not influence the levels of Ski mRNA in any of the cell lines. On the other hand, TGF-β treatment led to a significant decrease in the levels of Ski protein in all three cell lines. Interestingly, Nodal had no effect on Ski protein levels (Figure 4B). Immunofluorescence confirmed that treatment with TGF-β decreased the levels of Ski protein in PC3 cells, but not in Nodal effects (Figure 4C).
Several studies have shown that rapid decrease in Ski protein levels following TGF-β treatment is the result of Smad3 targeting of Ski to the proteasome for degradation. To address this, DU145 (data not shown) and PC3 cells were treated with TGF-β in the presence or absence of MG132, an inhibitor of proteasome activity. As shown in Figure 4E, proteasome inhibitor blocked TGF-β-induced reduction in Ski protein indicating that TGF-β-induced degradation of Ski is mediated by the proteasome pathway. Treatment with MG132 resulted in decreased basal and TGF-β-induced phosphorylation of both Smad2 and Smad3 (Figure 4D). Taken together, these findings indicate that TGF-β-initiated degradation of Ski is mediated by the proteasome pathway in prostate cancer cells and this degradation is required for increased Smad2 and Smad3 phosphorylation in response to TGF-β.
Differential roles of Ski in TGF-β and Nodal signaling
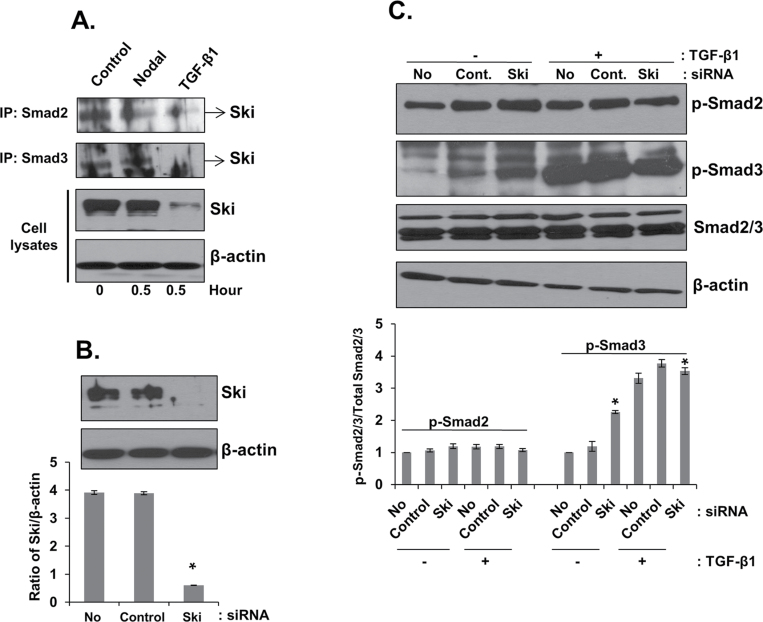
To determine whether differential effects of Nodal and TGF-β on Ski protein in prostate cancer cells result in differential regulation of Smad2 and Smad3 signaling, we investigated the interaction of Ski with Smad2 and Smad3 in Nodal and TGF-β-treated PC3 cells. Total cellular proteins were immune-precipitated with anti-Smad2 or anti-Smad3 antibodies followed by western blotting for Ski protein. As shown in Figure 5A, treatment with Nodal resulted in dissociation of Smad2 protein from Ski without affecting Smad3 or total Ski protein levels. On the other hand, TGF-β treatment resulted in degradation of Ski protein leading to dissociation of both Smad2 and Smad3 from the Ski protein.
Fig. 5.
Roles of Ski in Nodal and TGF-β signaling in prostate cancer cell lines. (A) PC3 cells were treated with Nodal (200ng/ml) and TGF-β1 (5ng/ml) for 0.5h. Proteins were immunoprecipitated from cell lysates using anti-Smad2/3 antibodies followed by western blot analysis with anti-Ski antibody. Total cell lysates were also collected to determine the expression of Ski and β-actin (internal control). (B) Knockdown of endogenous Ski protein in PC3 cells by specific Ski siRNA. Equal amount of cell lysates were analyzed by western blot using anti-Ski antibody (upper panel). The levels of β-actin in cell lysates were used as internal controls. Quantitative analysis of PC3 cells with or without Ski siRNA or control siRNA after normalization to the signal obtained with β-actin (lower panel). * Significantly different (P < 0.05) from cell transfected with control siRNA. (C) Western blot analysis of phosphorylated Smad2 and Smad3, total Smad2/3 and β-actin in PC3 cells transfected with control or Ski siRNA. The cells were incubated in the presence or absence of TGF-β1 (5ng/ml) for 10min. Western blots using anti-Smad2/3 and β-actin antibodies as internal controls. Quantitative analysis of p-Smad2 and p-Smad3 in PC3 cells with or without treatment with TGF-β1 (5ng/ml) (lower panel) were relative to untreated control (designated as one) after normalization to the signal obtained with Smad2/3. Each bar represents mean ± SEM (n = 3). *Significantly different (P < 0.05) compared with untreated controls.
Knockdown of endogenous Ski enhances TGF-β signaling in prostate cancer cells
To determine whether knockdown of endogenous Ski protein will lead to enhanced TGF-β signaling, we performed transient transfection in DU145 and PC3 cells using siRNA specific for human Ski. The protein levels of Ski were significantly reduced (P < 0.05) in both DU145 (data not shown) and PC3 cells (Figure 5B). As shown in Figure 5C, knockdown of endogenous Ski alone was sufficient to increase Smad3 (1.4-fold; P < 0.05) phosphorylation in PC3 cells compared with that in cells transfected with control siRNA. Exogenous TGF-β further increased the phosphorylation of Smad3 in PC3 cells transfected with both control and Ski siRNA (Figure 5C; P < 0.05). Knockdown of Ski did not have any significant effect on phosphorylation of Smad2 in PC3 cells. These results indicate that Ski plays a direct role in the regulation of Smad3 phosphorylation and that TGF-β primarily employs Smad3 for intracellular signaling in prostate cancer cells.
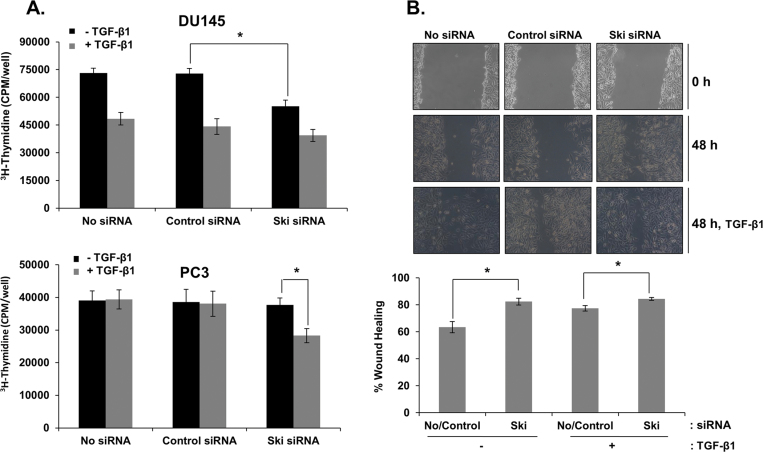
Next, we determined whether knockdown of Ski will enhance TGF-β effects on proliferation and migration of prostate cancer cells. For these experiments, we utilized DU145 cells, in which TGF-β inhibits proliferation, and PC3 cells, in which TGF-β induces migratory and invasive behavior. Knockdown of endogenous Ski expression significantly reduced basal cell proliferation (P < 0.05) in DU145 cells, which was further reduced after treatment with TGF-β (Figure 6A). On the other hand, although knockdown of endogenous Ski protein did not influence basal cell proliferation in PC3 cells, it was sufficient to make these cells responsive to growth inhibitory effects of TGF-β (Figure 6A; P < 0.05).
Fig. 6.
Effects of TGF-β on proliferation and migration in DU145 and PC3 cells with or without Ski knockdown. (A) After transfection with control or Ski siRNA, the cells were cultured in serum-free media for 24h and treated with TGF-β (5ng/ml) for 18h in the presence of 5% FBS. The cells were then pulse-labeled for 4h with 1 µCi/ml 3H-thymidine and radioactivity was determined by liquid scintillation counting. Each bar represents mean ± SEM (n = 3). *Significantly different (P < 0.05) compared with controls. (B) Confluent cell monolayers of PC3 cells transfected with Ski or control siRNA were wounded with a pipette tip. The cells were treated with TGF-β (5ng/ml) for 48h. Phase contrast pictures were taken under an inverted microscope at the time of wounding (0h) and 48h later. Percent wound closure was calculated by measuring the wound closure distance, and this value is expressing as a percentage of the initial wound length. Each bar represents mean ± SEM (n = 3). *Significantly different (P < 0.05) from cells transfected with control siRNA.
We also examined whether reducing Ski expression affects the motility of PC3 cells in a wound healing assay. Knockdown of endogenous Ski alone in PC3 cells was sufficient to cause a significant increase (P < 0.05) in migration of these cells (Figure 6B). Exogenous TGF-β (5ng/ml) did not further enhance these effects of Ski knockdown in PC3 cells. These results suggest that increased Ski protein levels in prostate cancer cells are partially responsible for reduced TGF-β and Smad signaling in these cells.
Discussion
In this study, we report that TGF-β superfamily members, TGF-β1 and Nodal exert similar effects on proliferation and migration of several normal and prostate cancer cell lines. However, the two cytokines exert their effects by inducing the phosphorylation of different Smad proteins; TGF-β1 effects are mediated primarily by Smad3, whereas Nodal effects are exerted exclusively by Smad2 phosphorylation. We also show that the levels of Smad-regulating Ski protein are high in prostate cancer cell lines and prostate cancer patient tissues and that its downregulation is required for the expression of basal and TGF-β1-dependent phosphorylation of Smad3 and TGF-β1 effects on proliferation and migration in prostate cancer cells. On the other hand, Ski protein does not appear to regulate Smad2 function and Nodal signaling in prostate cancer cells.
TGF-β inhibits proliferation of PrECs and prostate cancer cells in earlier stages of the disease; in the later stages, the cancer cells develop resistance to growth inhibitory effects of TGF-β but become responsive to its effects on invasive and metastatic behavior (32). Several previous studies have addressed the role of TGF-β produced by the epithelial cells or by stromal cells in the prostate and have investigated the development of resistance to inhibitory effects of TGF-β on proliferation of prostate cancer cells (33–36). Our recent demonstration of the expression of Nodal and its receptors in prostate cancer cells and differential effects of Nodal on proliferation and migration of prostate cancer cells prompted us to compare the biological effects of these two TGF-β superfamily members in prostate cells. Interestingly, Nodal and TGF-β exerted similar biological effects on cell proliferation and migration that are specific to different prostate cell lines indicating that two cytokines may be able to substitute each other in prostate cancer progression. Hence, any future therapeutic strategies aimed at TGF-β family will need to consider the overlapping roles of TGF-β1 and Nodal during different stages of prostate cancer.
Similar functions of Nodal and TGF-β in prostate cells prompted us to determine the differences in the intracellular signaling pathways employed by the two cytokines. Nodal and TGF-β receptors directly activate Smad2 and/or Smad3; however, Smad3 has been shown to be the essential mediator of most Smad-dependent TGF-β effects on gene expression, cell growth, apoptosis and tumor suppression (16). On the other hand, Smad2 only transmodulated Smad3-dependent transcription (37) suggesting that Smad2 and Smad3 have distinct roles in TGF-β signaling. We observed that TGF-β1 stimulation led to predominantly Smad3 phosphorylation whereas Nodal induced primarily Smad2 phosphorylation with little, if any, effect on Smad3 phosphorylation in PZ-HPV7, DU145 and PC3 cells. In addition, a SIS3 also completely blocked TGF-β1 effects but had only minor effects on Nodal signaling indicating that although Smad3 plays an essential role in TGF-β1 signaling, Nodal effects are exerted independent of Smad3 and presumably require only Smad2. Smad2 has shown to act as a tumor suppressor in the basal epithelial or stem cell compartment of the prostate cells (37). Since Nodal maintains the pluripotency of human embryonic stem cells, it is possible that Smad2 has a selective role in stem cell function and involvement in Nodal signaling. Our data suggest that in the presence of Nodal and its receptors in prostate cancer cells, inhibition of TGF-β receptors and Smad3 alone may not be enough to treat advanced stages of prostate cancer.
Previous studies have shown that Ski protein is overexpressed in human tumor cell lines and human tumor tissues from melanoma (38), breast (39), esophagus (40), cervical, colorectal (41), gastric (42) and pancreatic cancers (43), but is weakly expressed in normal epithelial cells; mislocalization and upregulation of Ski may contribute to malignant progression (14,44). Ski mRNA levels were ubiquitously expressed in all prostate cell lines in this study; however, higher levels of Ski protein were observed in prostate cancer cells and prostate cancer tissue samples. Gene Expression Omnibus and Oncomine Database also showed that Ski mRNA levels are ubiquitously expressed in both normal and prostate cancer cells. These differences in Ski protein levels indicate differential regulation of this protein in normal and cancer cells and suggest the involvement of posttranscriptional and posttranslational mechanisms in its regulation. Our data showing significantly increased Ski protein levels in normal prostate cell line when cultured in the presence of proteasomal inhibitor (MG132) indicating a selective inhibition of proteasomal degradation of Ski protein in prostate cancer cells. These results indicate that Ski expression and activity may vary during different stages of prostate cancer progression and may serve as a diagnostic or prognostic biomarker and therapeutic target in the advanced metastatic stage of prostate cancer.
Ski has been shown to be an important negative regulator of TGF-β and BMPs signaling via its interactions with Smad proteins (27). Previous studies have shown that Ski is effectively degraded by TGF-β signaling via Arkadia, which interacts with Ski through Smad2 and Smad3 to mediate its ubiquitination and degradation (45,46). Our results confirmed the effects of TGF-β on proteasomal degradation of Ski protein in normal PrECs and prostate cancer cells and that this degradation of Ski protein is required for basal and TGF-β-induced Smad3 phosphorylation. Therefore, it is logical to assume that degradation of Ski may be a prerequisite for TGF-β-induced biological responses during differential stages of prostate cancer. As mentioned earlier, TGF-β exerts differential effects on cell proliferation and migration in prostate cell lines. Several human cancer cell lines express high levels of Ski and are refractory to TGF-β-induced growth arrest (40,47). We show here that knockdown of endogenous Ski reduced proliferation in DU145 cells and enhanced migration in PC3 cells. These results indicate that reduced Ski protein levels in PC3 cells enhance TGF-β signaling and Ski may play a role in regulating tumor cell metastasis and invasive behavior. High levels of Ski protein in prostate cancer cells may be partially responsible for reduced TGF-β and Smad signaling in these cells. On the other hand, Nodal had no effect on Ski mRNA or protein levels suggesting that Ski protein does not influence Smad2 signaling and Nodal effects in prostate cells. Furthermore, immunoprecipitation experiments after Nodal and TGF-β treatments showed that Nodal induces selective dissociation of Smad2 protein from Ski but does not affect the interaction between Smad3 and Ski or the levels of Ski protein. Therefore, it is logical to assume that high levels of Ski protein during prostate cancer development interfere with Smad3 and TGF-β1 signaling and resistance to antiproliferatory effects of TGF-β1 in earlier stages of cancer development. On the other hand, Nodal effects on Smad2 activation and on prostate cancer cells will not be affected by high levels of Ski protein and it will be especially important during the later stages of the disease where Nodal exerts positive effects on cell migration and invasion (9).
In conclusion, our study demonstrates that Nodal and TGF-β have similar biological effects on cell proliferation and migration in prostate cells; however, these cytokines employ distinct Smad proteins to exert their effects. High levels of Ski protein were only expressed in prostate cancer cells and prostate cancer tissues. TGF-β treatment induced proteasomal degradation of Ski protein which is prerequisite for increased Smad3 phosphorylation and TGF-β signaling. On the other hand, Ski does not play a role in Smad2 function and Nodal effects on prostate cancer cells. These findings have significant implications for treatment of prostate cancers during different stages of the disease using therapeutic strategies based on inhibition of TGF-β and Smad signalings.
Funding
These studies were supported by the National Institutes of Health (grant numbers: 2G12RR003062, 5P20MD002285-02 and 2R25GM060414) and by the Department of Defense (DOD) prostate cancer research program (PCRP) grant #W8I-08-1-007.
Acknowledgements
We thank Dr Miao Zhong, Dr Peri Nagappan and Mojgan Zavareh for their help in this study.
Glossary
Abbreviations:
- BMP
bone morphogenetic protein
- EGF
epidermal growth factor
- FBS
fetal bovine serum
- hESCs
human embryonic stem cells
- PBS
phosphate buffered saline
- PrECs
prostate epithelial cells
- RT
reverse transcription
- siRNA
small interfering RNA
- SIS3
specific inhibitor of Smad3
- Ski
Sloan–Kettering Institute
- TGF-β
transforming growth factor-beta.
Footnotes
Conflict of Interest Statement: None declared.
References
- 1. Chang H., et al. (2002). Genetic analysis of the mammalian transforming growth factor-beta superfamily. Endocr. Rev 23 787–823 [DOI] [PubMed] [Google Scholar]
- 2. Massagué J., et al. (2000). Controlling TGF-beta signaling. Genes Dev 14 627–644 [PubMed] [Google Scholar]
- 3. Zimmerman C.M., et al. (2000). Transforming growth factor beta signaling mediators and modulators. Gene 249 17–30 [DOI] [PubMed] [Google Scholar]
- 4. Miyazawa K., et al. (2002). Two major Smad pathways in TGF-beta superfamily signalling. Genes Cells 7 1191–1204 [DOI] [PubMed] [Google Scholar]
- 5. Siegel P.M., et al. (2003). Cytostatic and apoptotic actions of TGF-beta in homeostasis and cancer. Nat. Rev. Cancer 3 807–821 [DOI] [PubMed] [Google Scholar]
- 6. Vallier L., et al. (2004). Nodal inhibits differentiation of human embryonic stem cells along the neuroectodermal default pathway. Dev. Biol 275 403–421 [DOI] [PubMed] [Google Scholar]
- 7. Schier A.F., et al. (2000). Nodal signalling in vertebrate development. Nature 403 385–389 [DOI] [PubMed] [Google Scholar]
- 8. Yeo C., et al. (2001). Nodal signals to Smads through Cripto-dependent and Cripto-independent mechanisms Mol. Cell 7 949–957 [DOI] [PubMed] [Google Scholar]
- 9. Vo B.T., et al. (2011). Expression of nodal and nodal receptors in prostate stem cells and prostate cancer cells: autocrine effects on cell proliferation and migration Prostate 71 1084–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lawrence M.G., et al. (2011). Reactivation of embryonic nodal signaling is associated with tumor progression and promotes the growth of prostate cancer cells Prostate 71 1198–1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Luo K., et al. (1999). The Ski oncoprotein interacts with the Smad proteins to repress TGFbeta signaling Genes Dev 13 2196–2206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Li Y., et al. (1986). Unique sequence, ski, in Sloan-Kettering avian retroviruses with properties of a new cell-derived oncogene J. Virol 57 1065–1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nomura N., et al. (1989). Isolation of human cDNA clones of ski and the ski-related gene, sno Nucleic Acids Res 17 5489–5500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fumagalli S., et al. (1993). Expression of the c-ski proto-oncogene in human melanoma cell lines Melanoma Res 3 23–27 [DOI] [PubMed] [Google Scholar]
- 15. Reed J.A., et al. (2005). SKI pathways inducing progression of human melanoma Cancer Metastasis Rev 24 265–272 [DOI] [PubMed] [Google Scholar]
- 16. Zhang F., et al. (2003). Ski-related novel protein N (SnoN), a negative controller of transforming growth factor-beta signaling, is a prognostic marker in estrogen receptor-positive breast carcinomas Cancer Res 63 5005–5010 [PubMed] [Google Scholar]
- 17. Imoto I., et al. (2001). SNO is a probable target for gene amplification at 3q26 in squamous-cell carcinomas of the esophagus Biochem. Biophys. Res. Commun 286 559–565 [DOI] [PubMed] [Google Scholar]
- 18. Sun Y., et al. (1999). Interaction of the Ski oncoprotein with Smad3 regulates TGF-beta signaling Mol. Cell 4 499–509 [DOI] [PubMed] [Google Scholar]
- 19. Nomura T., et al. (1999). Ski is a component of the histone deacetylase complex required for transcriptional repression by Mad and thyroid hormone receptor Genes Dev 13 412–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tokitou F., et al. (1999). Viral ski inhibits retinoblastoma protein (Rb)-mediated transcriptional repression in a dominant negative fashion J. Biol. Chem 274 4485–4488 [DOI] [PubMed] [Google Scholar]
- 21. Kokura K., et al. (2001). The Ski protein family is required for MeCP2-mediated transcriptional repression J. Biol. Chem 276 34115–34121 [DOI] [PubMed] [Google Scholar]
- 22. Zhong M., et al. (2010). Oxytocin induces the migration of prostate cancer cells: involvement of the Gi-coupled signaling pathway Mol. Cancer Res 8 1164–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Millena A.C., et al. (2004). Autocrine regulation of steroidogenic function of Leydig cells by transforming growth factor-alpha Mol. Cell. Endocrinol 224 29–39 [DOI] [PubMed] [Google Scholar]
- 24. Vo B.T., et al. Expression of nodal and nodal receptors in prostate stem cells and prostate cancer cells: autocrine effects on cell proliferation and migration Prostate 71 1084–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Miyazawa K., et al. (2002). Two major Smad pathways in TGF-beta superfamily signalling Genes Cells 7 1191–1204 [DOI] [PubMed] [Google Scholar]
- 26. Jinnin M., et al. (2006). Characterization of SIS3, a novel specific inhibitor of Smad3, and its effect on transforming growth factor-beta1-induced extracellular matrix expression Mol. Pharmacol 69 597–607 [DOI] [PubMed] [Google Scholar]
- 27. Luo K. (2004). Ski and SnoN: negative regulators of TGF-beta signaling Curr. Opin. Genet. Dev 14 65–70 [DOI] [PubMed] [Google Scholar]
- 28. Sun Y., et al. (1999). SnoN and Ski protooncoproteins are rapidly degraded in response to transforming growth factor beta signaling Proc. Natl. Acad. Sci. U.S.A 96 12442–12447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Xu W., et al. (2000). Ski acts as a co-repressor with Smad2 and Smad3 to regulate the response to type beta transforming growth factor Proc. Natl. Acad. Sci. U.S.A 97 5924–5929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Akiyoshi S., et al. (1999). c-Ski acts as a transcriptional co-repressor in transforming growth factor-beta signaling through interaction with smads J. Biol. Chem 274 35269–35277 [DOI] [PubMed] [Google Scholar]
- 31. Deheuninck J., et al. (2009). Ski and SnoN, potent negative regulators of TGF-beta signaling Cell Res 19 47–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Robson C.N., et al. (1999). Transforming growth factor-beta1 up-regulates p15, p21 and p27 and blocks cell cycling in G1 in human prostate epithelium J. Endocrinol 160 257–266 [DOI] [PubMed] [Google Scholar]
- 33. Bhowmick N.A., et al. (2005). Tumor-stroma interactions Curr. Opin. Genet. Dev 15 97–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sharif-Afshar A.R., et al. (2005). Stromal hyperplasia in male bladders upon loss of transforming growth factor-beta signaling in fibroblasts J. Urol 174(4 Pt 2)1704–7; discussion 1707 [DOI] [PubMed] [Google Scholar]
- 35. Pu H., et al. (2009). Dysfunctional transforming growth factor-beta receptor II accelerates prostate tumorigenesis in the TRAMP mouse model Cancer Res 69 7366–7374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yang F., et al. (2008). Fibroblast growth factor-2 mediates transforming growth factor-beta action in prostate cancer reactive stroma Oncogene 27 450–459 [DOI] [PubMed] [Google Scholar]
- 37. Yang J., et al. (2009). Critical role of Smad2 in tumor suppression and transforming growth factor-beta-induced apoptosis of prostate epithelial cells Cancer Res 69 2185–2190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Javelaud D., et al. Efficient TGF-beta/SMAD signaling in human melanoma cells associated with high c-SKI/SnoN expression. Mol. Cancer. (2011);10:2. doi: 10.1186/1476-4598-10-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Theohari I., et al. (2012). Differential effect of the expression of TGF-beta pathway inhibitors, Smad-7 and Ski, on invasive breast carcinomas: relation to biologic behavior APMIS 120 92–100 [DOI] [PubMed] [Google Scholar]
- 40. Villanacci V., et al. (2008). Ski/SnoN expression in the sequence metaplasia-dysplasia-adenocarcinoma of Barrett’s esophagus Hum. Pathol 39 403–409 [DOI] [PubMed] [Google Scholar]
- 41. Bravou V., et al. (2009). TGF-beta repressors SnoN and Ski are implicated in human colorectal carcinogenesis Cell. Oncol 31 41–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Takahata M., et al. (2009). SKI and MEL1 cooperate to inhibit transforming growth factor-beta signal in gastric cancer cells J. Biol. Chem 284 3334–3344 [DOI] [PubMed] [Google Scholar]
- 43. Heider T.R., et al. (2007). Ski promotes tumor growth through abrogation of transforming growth factor-beta signaling in pancreatic cancer Ann. Surg 246 61–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Berk M., et al. (1997). Mice lacking the ski proto-oncogene have defects in neurulation, craniofacial, patterning, and skeletal muscle development Genes Dev 11 2029–2039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Le Scolan E., et al. (2008). Transforming growth factor-beta suppresses the ability of Ski to inhibit tumor metastasis by inducing its degradation Cancer Res 68 3277–3285 [DOI] [PubMed] [Google Scholar]
- 46. Nagano Y., et al. (2007). Arkadia induces degradation of SnoN and c-Ski to enhance transforming growth factor-beta signaling J. Biol. Chem 282 20492–20501 [DOI] [PubMed] [Google Scholar]
- 47. Zhu Q., et al. (2007). Dual role of SnoN in mammalian tumorigenesis Mol. Cell. Biol 27 324–339 [DOI] [PMC free article] [PubMed] [Google Scholar]