Abstract
Biallelic germline mutations in the base excision repair enzyme gene MUTYH lead to multiple colorectal adenomas and carcinomas referred to as MUTYH-associated polyposis. MUTYH removes adenine misincorporated opposite the DNA oxidation product, 8-oxoguanine (OG), thereby preventing accumulation of G:C to T:A transversion mutations. The most common cancer-associated MUTYH variant proteins when expressed in bacteria exhibit reduced OG:A mismatch affinity and adenine removal activity. However, direct evaluation of OG:A mismatch repair efficiency in mammalian cells has not been assessed due to the lack of an appropriate assay. To address this, we developed a novel fluorescence-based assay of OG:A repair and measured the repair capacity of MUTYH-associated polyposis variants expressed in Mutyh−/− mouse embryonic fibroblasts (MEFs). The repair of a single site–specific synthetic lesion in a green fluorescent protein reporter leads to green fluorescent protein expression with co-expression of a red fluorescent protein serving as the transfection control. Cell lines that stably express the MUTYH-associated polyposis variants G382D and Y165C have significantly lower OG:A repair versus wild-type MEFs and MEFs expressing human wild-type MUTYH. The MUTYH allele that encodes the Q324H variant is found at a frequency above 40% in samples from different ethnic groups and has long been considered phenotypically silent but has recently been associated with increased cancer risk in several clinical studies. In vitro analysis of Q324H MUTYH expressed in insect cells showed that it has reduced enzyme activity similar to that of the known cancer variant G382D. Moreover, we find that OG:A repair in MEFs expressing Q324H was significantly lower than wild-type controls, establishing that Q324H is functionally impaired and providing further evidence that this common variant may lead to increased cancer risk.
Introduction
Oxidative DNA damage is believed to be a major underlying cause of cancer, aging and neurodegeneration (1). The modified guanine lesion, 8-oxo-7,8-dihydroguanine (OG), is a prevalent and mutagenic form of oxidative DNA damage. The base excision repair glycosylase hOGG1 removes OG present in OG:C base pairs in DNA (1). However, failure to remove OG prior to replication leads to the preferential incorporation of A opposite OG by replicative polymerases, and a subsequent round of replication results in the formation of a permanent G:C to T:A transversion mutation. Since mutagenesis due to OG depends upon the persistence of the OG:A intermediate, organisms have evolved a ‘last chance’ DNA glycosylase that removes adenine opposite OG, identified as MutY in Escherichia coli (2) and MUTYH in humans (1). Specialized repair polymerases have evolved to incorporate C opposite the OG lesion (3), facilitating subsequent full repair of the OG:C product back to a G:C base pair. When human cells are transfected with DNA containing OG lesions, OG:C bps are repaired much more rapidly than OG:A mispairs (4,5), suggesting OG:A repair may be a rate-limiting step that protects cells against OG-induced mutagenesis.
Biallelic germline mutations in MUTYH are associated with colorectal adenomas and carcinomas, referred to as MUTYH-associated polyposis (MAP (6,7)). A recent clinical follow-up study of 254 biallelic carriers found the lifetime cumulative risk of developing colorectal cancer (CRC) by age 60 was 63% and by age 70 was 86% (8), and a recent meta-analysis of clinical studies concluded there is a 28-fold increased CRC risk (9). Importantly, biallelic carriers have increased G:C to T:A transversion mutations in APC and KRAS in somatic tumor tissue (6,10), and the two most common variants, Y165C and G382D, are well-established to have reduced catalytic activity in vitro (11–14). In addition, structural and functional information on MutY enzymes further substantiate the hypothesis that compromised OG:A repair activity is the primary mechanism leading to carcinogenesis in MAP. Approximately 61% of reported MAP alleles are either Y165C or G382D, and the remaining 39% are at other positions (15). Individuals who have Y165C or G382D in one allele and an uncharacterized mutation in the second allele are of uncertain status in terms of their future risk for CRC. Thus, knowledge of the functional activity of less common variants is important for individual risk assessment in MAP. Moreover, delineating the relative ranking of functional activity of different variants is critical in cancer risk analyses.
In order to gauge their potential functional impairment, several MUTYH variants have been expressed in bacteria for in vitro glycosylase and binding studies (11,13,14,16). However, detailed analysis of the activity of MUTYH has been hampered by the low amounts and quality of the protein expressed in bacteria. More detailed kinetics and binding studies have been performed with variants of the bacterial and murine enzymes corresponding to Y165C and G382D MUTYH. These studies have revealed consistently that the Tyr-to-Cys change is more deleterious to OG:A recognition and adenine removal activity compared with the Gly-to-Asp change (12,17,18). Consistent with this, a clinical study indicates that MAP patients homozygous for Y165C have an earlier onset and more severe MAP phenotype versus G382D homozygotes (19). In addition, structural studies on bacterial MutY illustrate the roles of the native residues in recognition of the damaged OG nucleotide and are consistent with a more deleterious functional consequence for the Tyr-to-Cys substitution (20).
Five recent clinical studies have associated Q324H with increased risk of CRC and lung cancer (21–25). These studies conflict with the previous assumption that Q324H is a functionally silent polymorphism (6,7). In addition, there is conflicting evidence regarding Q324H activity in vitro (13,26), and it is unclear based on the location of Gln 324 in the enzyme sequence how this mutation would alter enzyme activity (20,27). Since the allele encoding Q324H is found in over 40% of people in some populations (28), a more thorough understanding of the intrinsic adenine glycosylase activity of this variant and its ability to mediate repair of OG:A mismatches in cells is sorely needed.
Knowledge of the intrinsic adenine glycosylase activity of a given MUTYH variant provides important information but is clearly only one aspect that may lead to functional impairment in a cellular context. Clearly, the OG:A repair activity of MUTYH variants may be further reduced in a cellular context due to reductions in expression level and stability, and altered interactions with cellular protein partners. MUTYH is known to interact with a variety of cellular proteins, including AP endonuclease and proliferating cell nuclear antigen (18), the mismatch repair protein MSH6 (29), and the DNA-damage response proteins Hus1 (30) and ATR (31). In addition, MUTYH is phosphorylated when expressed in eukaryotic cells (32). For these reasons, functional testing of MUTYH variants in living cells is expected to be particularly revealing. Recently, increased levels of OG were found in mouse embryonic fibroblasts (MEFs) deficient in endogenous mouse Mutyh and stably expressing MUTYH variants (33). Previous work has also shown that endogenous wild-type (WT) MUTYH can repair OG:A mispairs within plasmid DNA in mammalian cells (4,5), and fluorescent and luminescent reporters have been used in DNA repair assays (34,35). By combining aspects of these various approaches, we developed an assay to evaluate OG:A lesion repair using a plasmid-based fluorescent reporter in mammalian cells. A premature stop codon in the green fluorescent protein (GFP) coding region is restored to the WT codon when a site-specific synthetic OG:A mispair is repaired to OG:C. The reporter also expresses the destabilized red fluorescent protein (dsRed), which is used to identify transfection-positive cells. Thus, the green fluorescence of transfection-positive cells is a measure of OG:A repair. We found low repair in MUTYH-deficient cells and high repair in cells expressing WT MUTYH. The common cancer-associated MAP variants G382D and Y165C show significantly reduced repair versus MEFs expressing WT MUTYH. Surprisingly, the Q324H variant exhibited reduced repair levels that approached that of the two well-characterized cancer variants. In addition, analysis of the adenine glycosylase activity of Q324H expressed in insect cells showed that this variant exhibits enzyme activity similar to the G382D MUTYH variant. The low repair activity of Q324H similar to known cancer variants revealed in this study taken together with the clinical association of this variant with colorectal and lung cancer strongly suggest that Q324H should be considered a factor in cancer risk assessments.
Materials and methods
Mammalian cell lines and cell culture
Mutyh −/− and WT MEFs were generated from littermates by crossing Mutyh +/− mice (36) and spontaneously immortalized as described previously (37). Presence of the targeted insert in the Mutyh gene was independently verified by PCR, and reverse transcription (RT)–PCR (as detailed below) verified no Mutyh messenger RNA (mRNA). The cells were grown at 37°C with 5% CO2 with high-glucose Dulbecco’s modified Eagle’s medium, 10% fetal bovine serum, 1% non-essential amino acids, and 1% GlutMAX (Invitrogen).
Construction of fluorescent reporter
All reagents were obtained from New England Biolabs unless otherwise noted. The green fluorescent protein (GFP) region from enhanced GFP-c1 vector (Clontech) was cloned into the pIRES2-dsRed-express vector (Clontech) and codon 34 of enhanced GFP was modified from GAG to TAA (stop codon) by QuikChange site-directed mutagenesis (Stratagene) referred to as pGFP-off because it codes for a truncated version of GFP lacking the fluorescent chromophore, thus it expresses dsRed only. The pGFP-off vector is used as a negative control in the OG:A repair assays. The corresponding construct with codon 34 modified to GAA is referred to as pGFP-on since it constitutively expresses the full-length GFP along with dsRed. The pGFP-on vector was used as a positive control to represent the maximal GFP signal that can be observed in each experiment. These plasmids do not replicate in the MEFs used due to the lack of SV40 large T antigen in these cell lines. Single-stranded DNA was produced from pGFP-off based on standard protocols using M13K07 phage. The OG-containing 35 nucleotide strand (5′-TCCGGCGAGGGC/OG/AAGGCGATGCCACCTACGGCAA-3′) was annealed to the single-stranded template DNA and second strand synthesis utilized T7 DNA polymerase as detailed in the Supplementary Information, available at Carcinogenesis Online. Importantly, the pGFP-off construct contains the premature GFP stop codon, thus it cannot be a source of extraneous GFP expression. Transient transfection into MEFs was completed using Lipofectamine LTX (Invitrogen).
Stable transfection of MUTYH
The Tet-On Advanced Inducible Gene Expression System (Clontech) was used per manufacturer’s protocols. The pTRE-tight vector contains the human MUTYH type 2 (beta 3) 521 amino acid isoform (NCBI ref NM_001048174.1), although for clarity we refer to amino acid position based on the 535 amino acid human MUTYH isoform (GenBank: U63329.1), consistent with previous papers. The Y165C, G382D and Q324H variants correspond to Y179C, G396D and Q338H, respectively, using the Leiden Open Variation Database (LOVD) convention (15).
Competitive RT–PCR
To accurately measure transcription of MUTYH in stably transfected cell lines, competitive RT–PCR with Qiagen One-Step RT–PCR kit (Germantown, MD) was used with a synthetic truncated standard of MUTYH transcribed in vitro using T7 RNA polymerase as described in the Supplementary Material and Methods, available at Carcinogenesis Online.
Flow cytometry
Cell harvesting, flow cytometry (Becton Dickinson FACScan) and data analysis (FlowJo Software; Ashland, OR) were completed using standard methods, with further details included in the Supplementary Information, available at Carcinogenesis Online. Each data point in the cellular repair assay represents at least 30000 cells analyzed in at least three separate experiments. Lesion repair is defined as the percentage of GFP-positive cells in the dsRed (transfection) positive cell population, normalized within each experiment to the percentage of GFP in the parallel undamaged pGFP-on (dsRed+/GFP+) vector, which is considered the maximum possible GFP signal. Differences in the ratio of GFP-positive cells to GFP-negative cells between cell lines was tested for significance using Fisher’s exact test with a two-tailed P value. The standard deviation of OG:A repair in each cell line was calculated from the differences in OG:A repair between multiple experiments.
Construction and expression of Y165C, G382D and Q324H MUTYH in Sf9 insect cells
All proteins were expressed as maltose-binding protein fusions. Experimental procedures for expression vector construct, expression in Sf9 cells and enzyme purification are similar to that reported previously for the WT MUTYH (32). Bacmids for Y165C and G382D MUTYH were prepared using bacterial expression plasmids as reported previously (11). A tobacco etch virus recognition sequence consisting of seven amino acids (ENLYFQG) was incorporated into the Q324H MUTYH-containing expression vector between the MBP-tag and the mutyh gene to enable cleavage of the tag in future experiments. Both WT and Q324H MUTYH expression levels and activity in these constructs were compared with those of WT MUTYH reported previously (32). In these experiments, the MBP-tag was retained in Q324H MUTYH to allow for direct comparison among all variants under similar conditions.
Adenine glycosylase assays
The following oligonucleotide duplex was used for the adenine glycosylase assays:
d(5′-CGATCATGGAGCCAC(OG)AGCTCCCGTTACAG-3′
d(3′-GCTAGTACCTCG GTG (A)TCGAGGGCAATGTC-5′)
Adenine glycosylase activity assays of WT and variant MUTYH, including active-site titrations, were performed as described previously (32) and in Supplementary Material and Methods, available at Carcinogenesis Online.
Results
Fluorescent reporter design and production
To assess the ability of variants of MUTYH to repair OG:A mispairs in mammalian cells, we developed a scheme to measure the repair using a fluorescent reporter as shown in Figure 1A. A chemically synthesized OG nucleotide is placed in the sense strand opposite 2′-deoxyadenosine within the coding region of a GFP reporter construct. Without repair, the template strand codes for the stop codon TAA, leading to a 33 amino acid–truncated protein product that does not contain the chromophore core at amino acids 65–67 (38) and thus does not fluoresce. Excision of adenine and subsequent repair to cytosine causes the complementary mRNA codon to change from UAA to GAA, and translation of this change restores the WT amino acid, allowing expression of full-length WT GFP. The status of this A to C repair is amplified by the strong cytomegalovirus promoter upstream of the coding region. Transfection status of individual cells was quantified by the constitutive expression of dsRed translated from an internal ribosome entry site. The fluorescence of individual cells was detected by flow cytometry after 3 days and the resulting histograms of red versus green fluorescence were analyzed to calculate OG:A lesion repair. One concern was that nonsense-mediated mRNA destruction would alter red fluorescence, since the dsRed coding region is downstream from this codon. However, the same level of red fluorescence was measured both with and without the GFP stop codon in the corresponding pGFP-off and pGFP-on vectors, and these constructs were used as GFP-negative and GFP-positive controls in all experiments. Although endonuclease, polymerase and ligase activities are required to complete repair on the transcribed strand, this scheme hypothetically does not require the repair of the oxidized guanine on the non-transcribed strand.
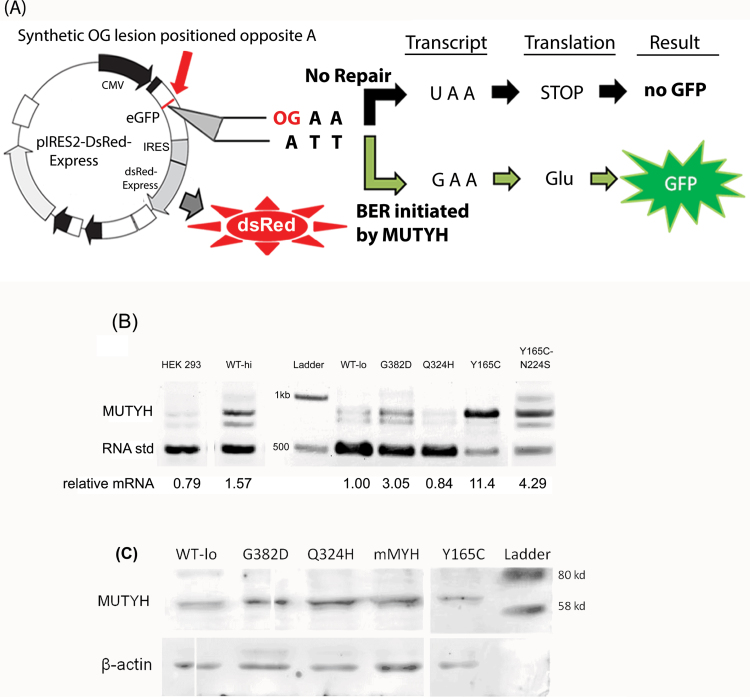
Fig. 1.
Mammalian cell-based OG:A repair assay for MUTYH variants. (A) Design of green fluorescent protein reporter of OG:A lesion repair. By default, the adenine in the template strand ATT (codon 34) is transcribed into a stop codon and the GFP protein is not produced. The OG lesion placed opposite this adenine makes the base pair a substrate for MUTYH, which catalyzes the removal of the adenine base. Upon full repair of the OG:A lesion to OG:C, the template strand codes for the WT glutamate and translation of the active GFP protein occurs. The dsRed expression is constitutively active and is a reporter for transfected cells. (B) Messenger RNA expression of MUTYH and variants in MEF cell lines. Competitive RT–PCR using a quantified RNA standard as detailed in Materials and Methods. The relative density of the upper band represents the relative cellular MUTYH mRNA abundance. The lower band is due to the addition of a quantified RNA standard to the RT–PCR, encoding a truncated form of MUTYH. The resulting cDNAs after reverse transcription compete for the same flanking MUTYH primers. The upper band is normalized versus the lower band to determine mRNA levels. The intermediate band is due to cross-annealing during PCR. WT-hi and WT-lo are the human WT high and low expression MEF cell lines, respectively. (C) Western blot of MUTYH and beta-actin in WT and Mutyh −/− MEF cell lines stably expressing MUTYH variants. Whole cell lysates of MEF cell lines loaded onto a 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis and western blot performed using anti-MUTYH (Abnova 4D10) and anti-beta-actin horse radish peroxidase (SCBT C4). Lane 1 WT-lo represents the cell line that stably expresses the human WT MUTYH, whereas lanes 2, 3 and 5 are stably expressing G382D, Q324H and Y165C MUTYH, respectively. Lane 4–labeled mMYH is endogenous mouse Mutyh from WT MEFs. This image is of a single exposure of one membrane, cut to remove extraneous samples and a diagonal tear. The full-length blot is presented in Supplementary Figure 1, available at Carcinogenesis Online.
Stable expression of MUTYH variants
To quantitatively assess the relative activity of MUTYH variants, we stably transfected the 521 amino acid nuclear isoform of human MUTYH into MEFs lacking endogenous mouse Mutyh. Doxycycline levels did not modulate MUTYH expression in stable transfected colonies as measured by mRNA levels determined via quantitative RT–PCR. Most cell lines had high levels of expression relative to endogenous MUTYH mRNA levels in HEK 293 cells. To obtain different levels of expression, we screened hygromycin-resistant colonies to isolate both low- and high-expression cell lines as shown in Figure 1B. Given that this system was designed to modulate expression levels, we verified mRNA expression did not change before and after key GFP reporter experiments. Western blot analysis of these cells lines shows similar levels of MUTYH protein as shown in Figure 1C. Interestingly, strong mRNA overexpression of the Y165C cell line results in similar protein levels as the other cell lines as measured by western blotting, suggesting reduced stability of this isoform.
GFP expression depends on the expression of active MUTYH
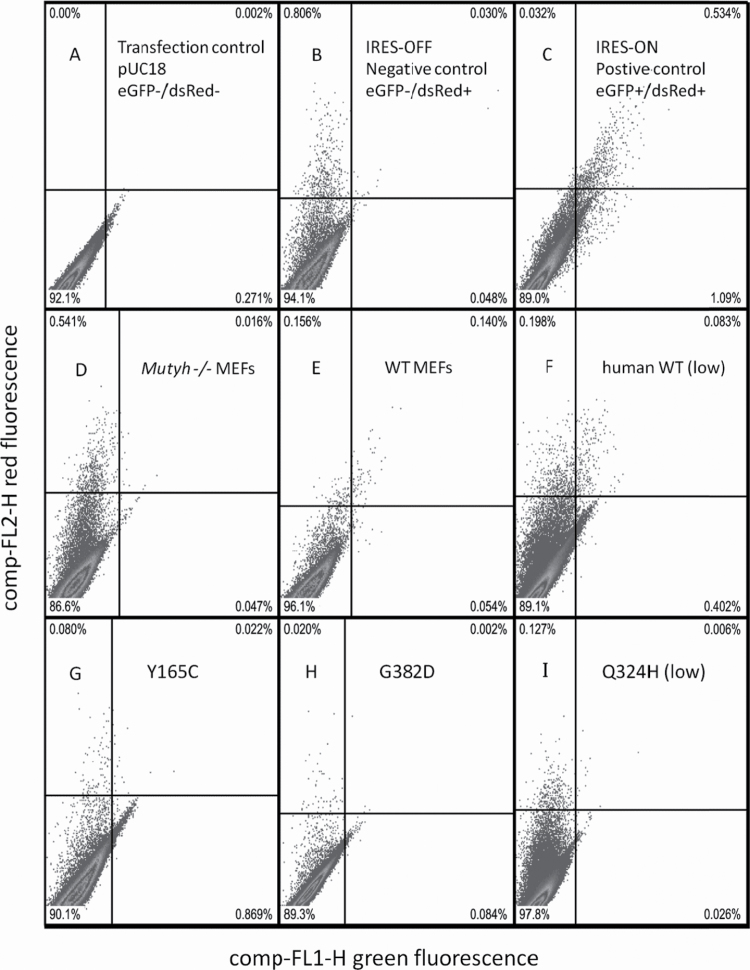
To verify that GFP fluorescence of the OG:A-containing reporter construct was a measure of enzyme-mediated cellular OG:A repair, we transiently transfected this construct into Mutyh-null and the parental WT MEFs. Red and green fluorescence intensity was quantified using flow cytometry after a 66-h incubation to allow cellular repair of the construct. Representative histograms from a set of experiments are shown in Figure 2. All data are included in Supplementary Table 1, available at Carcinogenesis Online, and summarized in Figure 3A. These data show that despite low transfection efficiency in MEFs, transfected cells are easily distinguished by simple quadrant analysis versus negative and positive controls. In addition, consistent statistically significant results were obtained in repeated trials. We categorized dsRed-positive cells transfected with the OG:A-containing construct as either GFP-positive or GFP-negative based on quadrants determined by the pGFP-off and pGFP-on control conditions. We found 51% of the dsRed (transfection)-positive cells were GFP-positive in unmodified WT MEFs, whereas Mutyh deficient MEFs had 8% GFP-positive cells (P < 0.0001, Fisher exact test). This is evidence that the A opposite the synthetic OG lesion has been replaced with C more often in cells expressing endogenous mouse Mutyh. In addition to the quadrant analysis shown, the flow cytometry data were also analyzed by quantification of median green fluorescence of dsRed-positive cells and grouping events by irregular polygons; all methods yielded similar results.
Fig. 2.
Representative flow cytometry histograms of red versus green fluorescence in MEF cell lines used to quantify levels of WT and variant MUTYH-mediated OG:A repair. Top row (panels A, B, C): Controls run in parallel with all OG:A repair trials to determine boundaries of quadrant analysis. MEFs transfected with a non-fluorescent construct pUC18 (A) are used to determine the horizontal quadrant line, thus events above this line are categorized as dsRed (transfection)-positive. MEFs transfected with (B) pGFP-off (GFP-/dsRed+) and (C) pGFP-on (GFP+/dsRed+) serve as negative and positive control conditions. The cells transfected with pGFP-off and pGFP-on are used to define the upper left quadrant and upper right quadrants, respectively. Middle row (panels D–F): Representative single trial histogram of Mutyh−/− MEFs, WT MEFs and Mutyh−/− MEFs expressing WT MUTYH, each transiently transfected with the OG:A-containing reporter construct. Cell lines in each panel: Mutyh −/− MEFs (D), WT MEFs (E), Mutyh −/− MEFs expressing human WT MUTYH at low level (F) as shown in Figure 1B. Bottom row (panels G–I): Representative single trial histograms of Mutyh −/− MEFs expressing MUTYH variants transiently transfected with the OG:A-containing reporter construct. Cell lines in each panel: Mutyh −/− MEFs expressing Y165C (G), G382D (H) and Q324H (I) at low levels as shown in Figure 1B. Using this quadrant analysis, the percent repair in a given cell line and trial is the number of dsRed+/GFP+ cells in the upper right quadrant divided by the number of dsRed+ (transfection-positive) cells above the horizontal quadrant line. Note quadrant dividing lines vary between the displayed trials because they are determined by within-experiment controls (as shown in panels A–C). The total number of cells from several experiments (Supplementary Table 1, available at Carcinogenesis Online) were used to determine levels of OG:A repair summarized in Figure 3.
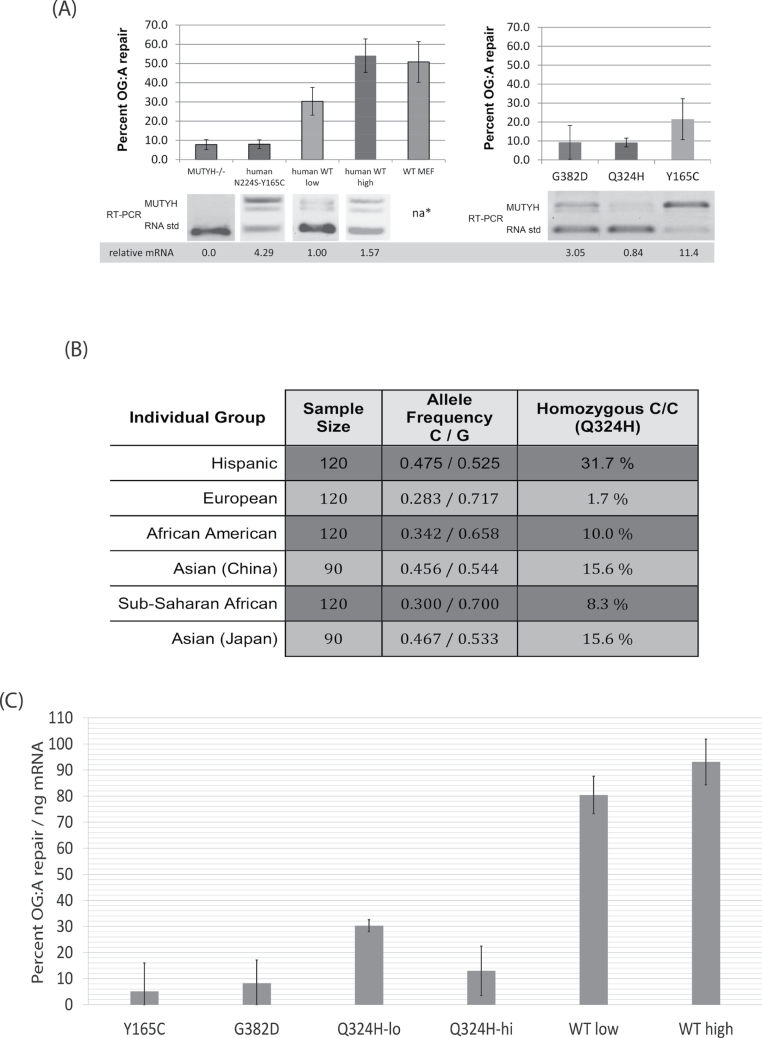
Fig. 3.
(A) Summary of flow cytometry results with the OG:A-containing fluorescent reporter and competitive RT–PCR of MUTYH expression in the corresponding MEF cell lines. Error bars represent the standard deviation from at least three separate experiments. na* endogenous mouse Mutyh not detected by RT–PCR due to differences in the gene sequence between species; western blot (Figure 1C) shows protein expression of WT MEFs is similar to the stable cell lines. GFP repair of the WT-lo cell line is significantly different from all other cell lines (P < 0.05). Also see Supplementary Table 1, available at Carcinogenesis Online) (B) Evidence that the SNP corresponding to the Q324H variant is common in diverse human groups. Matched subset of NCBI data for SNP rs3219489, format adapted from dbSNP (28) (http://www.ncbi.nlm.nih.gov/projects/SNP/snp_ref.cgi?rs=3219489) collected by the Environmental Genome Project (54). The minor C allele encodes histidine (CAC), leading to the Q324H protein variant, whereas the major G allele encodes glutamine (CAG). Right: The frequency of homozygous carriers of Q324H in these samples. The minor allele frequency of all NCBI recorded samples is 31.9%. (C) Repair of OG:A (as measured by GFP fluorescence) normalized to MUTYH mRNA levels (as measured by competitive RT–PCR). Error bars are the square root of the sum of the variances from the RT–PCR and GFP fluorescence data, thus account for variation from both these data sources.
Analysis of stably transfected Mutyh −/− MEF cell lines expressing the WT human 521 amino acid nuclear isoform of MUTYH found that a low mRNA expression cell line (‘WT-lo’) had 30% GFP-positive cells, whereas a high mRNA expression cell line (‘WT-hi’) had 57% GFP-positive cells, evidence that MUTYH expression level affects the number of GFP-positive cells (P < 0.0001), as shown in Figure 3A and detailed in Supplementary Table 1, available at Carcinogenesis Online.
Amino acid 224 is in a highly conserved loop lining the adenine binding pocket (39) and N224S is a rare variant associated with CRC (40), thus the N224S-Y165C double mutant should be catalytically inactive. The percentage of GFP-positive cells stably transfected with the N224S-Y165C double mutant was not significantly different from Mutyh-null cells (P = 0.87), despite strong mRNA overexpression, as shown in Figure 3A. Taken together, the data from WT, Mutyh-null and Mutyh-null cells expressing active and inactive forms of MUTYH provide evidence that the enzymatic activity of MUTYH mediates increased GFP expression and thus this assay can be used to directly measure the repair of a single OG:A mispair in mammalian cells.
OG:A repair of MAP variants
To measure the OG:A lesion repair capacity of MAP variants in mammalian cells, the GFP assay was performed on Mutyh −/− MEFs that were stably transfected with vectors that encode Y165C or G382D MUTYH (Figure 3A). Cells overexpressing the Y165C variant had 22% OG:A repair, lower than WT-lo (30%, P < 0.04) and WT-hi cells (57%, P < 0.0001) despite Y165C mRNA levels that were approximately 11-fold and 7-fold higher than these cell lines, respectively. Western blot quantification suggests that this Y165C cell line has protein levels similar to WT-lo (Figure 1C), suggesting reduced protein stability of this variant. However, this cell line did have increased OG:A repair above the MUTYH-deficient background (22% versus 8%, P < 0.0001), evidence that Y165C does retain enzymatic activity in living cells. The G382D cell line had approximately a 3-fold higher mRNA expression of MUTYH versus WT-lo; despite overexpression in this cell line, significantly lower OG:A repair (9% versus 30%, P < 0.0001) was observed. Notably, this G382D cell line had mRNA expression 3.7-fold lower than the Y165C cell line, yet protein levels by western blotting were generally higher for G382D over Y165C.
OG:A repair in MEFs expressing MUTYH variant Q324H
The single-nucleotide polymorphism (SNP) rs3219489 corresponds to the Q324H amino acid variant, and a matched subset of the NCBI data of this SNP (28) is shown in Figure 3B, evidence that this polymorphism is widespread in diverse human populations. Indeed, in three of six groups over 15% of the samples are homozygous for Q324H and in three of six groups the allele frequency is above 45%. This underscores the importance of evaluating the OG:A repair capacity in cells mediated by Q324H. To directly measure OG:A repair of this common variant, cell lines that stably express Q324H in the Mutyh-null MEF background were developed. Colonies were screened to find a cell line expressing Q324H MUTYH at levels similar to the WT-lo cell line and similar to endogenous MUTYH mRNA levels in the human cell line HEK 293 as shown in Figure 1B. The MUTYH mRNA expression of this Q324H cell line was not significantly different from WT-lo in seven separate total RNA extractions. However, in the GFP assay, this Q324H cell line had significantly lower levels of OG:A lesion repair versus WT-lo (9% versus 30%, P < 0.0001). Aliquots of the OG:A reporter construct were divided between these two cell lines and run in parallel, ruling out the possibility that differences in the number of GFP-positive cells was due to differences in the reporter construct itself.
To further investigate the OG:A repair activity of the Q324H variant, two additional cell lines were analyzed that overexpress this variant as measured by competitive RT–PCR. Overexpression of Q324H led to 39% OG:A repair; significantly lower repair than WT-hi (P < 0.006), despite the fact that this Q324H cell line had approximately 5-fold higher mRNA expression. However, when compared with the cell line overexpressing Y165C at similar levels, the Q324H cell lines mediate significantly higher levels of OG:A repair (39% versus 22%, P < 0.0001). This analysis indicates that the Q324H variant has an intermediate capacity to repair OG:A lesions between that of Y165C and WT MUTYH.
OG:A repair normalized to mRNA levels
Given that we have a set of cell lines stably transfected with WT and variant MUTYH cDNAs with different mRNA expression levels and that the competitive RT–PCR assay had very high reliability, the OG:A repair capacity was normalized using mRNA expression levels as shown in Figure 3C. The validity of this normalization is supported by the fact that the repair efficiency values of the two WT MUTYH cell lines are similar. Moreover, the WT MUTYH cell lines exhibit the highest repair efficiency. The low repair observed in the Y165C cell line despite strong overexpression of the Y165C mRNA provided the lowest repair efficiency, and the values with G382D variant cell line was not significantly different (P = 0.62). The low-expression Q324H cell line had significantly higher repair efficiency versus G382D and Y165C (P < 0.01), but the high-expression Q324H cell line did not. The reason that higher levels of Q324H are less efficient at OG:A repair in this assay is unclear; however, it suggests that the assay is able to distinguish the distinctly different behavior of specific MUTYH variants in a cellular context.
Adenine glycosylase activity of MUTYH variants
We previously reported that expression of a maltose-binding fusion protein of MUTYH in insect cells provides enzyme that has considerably higher active enzyme concentration than protein expressed in bacteria (32). This is presumably due to the superior folding and processing during overexpression in eukaryotic cells. In order to directly compare the intrinsic adenine glycosylase activity of the variants to the cellular OG:A repair, the three variants analyzed above were overexpressed using a baculovirus expression vector in Sf9 cells as we reported previously (32). WT MUTYH exhibits biphasic kinetic behavior under conditions of [E] < [OG:A-DNA], which provides a means to determine the active-site concentration of the enzyme, which varies between different preparations. Discrepancies between previous reports on activities of MUTYH variants may be due to lack of consideration of differences in active concentration, and thus active-site correction is important when evaluating enzymatic properties of MUTYH variants. In comparing several enzyme preparations, both G382D and Q324H MUTYH exhibited similar ranges of active fraction compared with WT MUTYH, whereas Y165C MUTYH consistently gave lower levels of active protein (~10% of WT). Rate constants for adenine removal for WT and variant MUTYH (Table 1) were measured under single-turnover conditions as reported previously (32). Under buffer conditions of physiological salt (150mM NaCl), both G382D and Q324H have adenine excision rates that are 60% of that measured for WT MUTYH, whereas no activity was observed with Y165C MUTYH. At lower salt (100mM NaCl), adenine removal activity for Y165C MUTYH was detected, but reactions did not go to completion. Of note, at this lower salt, both Q324H and G382D MUTYH exhibit adenine glycosylase activity similar to the WT enzyme (Table 1). We have previously found that the activity of the bacterial and murine enzyme variants corresponding to Y165C and G382D are sensitive to buffer salt concentration and the presence of other proteins (such as AP endonuclease), whereas the WT forms are relatively insensitive to these altered conditions. We attribute the sensitivity of the variant proteins to compromised interactions with the DNA substrate that are important in the adenine removal activity. These results indicate that in terms of adenine glycosylase activity, Q324H resembles G382D MUTYH.
Table 1.
Single-turnover rate constants for Q324H, G382D, Y165C and WT MUTYHa
| Enzyme form | kst (min–1)a | |
|---|---|---|
| OG:A100mM NaCl | OG:A150mM NaCl | |
| WT MUTYH | 0.6±0.1 | 0.5±0.1 |
| Q324H MUTYH | 0.5±0.1 | 0.3±0.1 |
| G382D MUTYH | 0.5±0.1 | 0.3±0.1 |
| Y165C MUTYH | ~0.1 | NCb |
aRate constants were determined with an OG:A containing 30-bp DNA duplex at 37°C with either 100 or 150mM NaCl-containing assay buffer. Rate constants kst were determined under single-turnover conditions ([DNA] < [Enzyme]). Values reported have been averaged over at least three separate experiments.
bNC: No Cleavage. Strand scission (at the A nucleotide) above baseline levels was not detected under these conditions.
Discussion
We have developed a fluorescence-based assay to directly measure the repair of a single site–specific OG:A lesion in mammalian cells and have applied this assay to measure DNA repair mediated by WT and cancer-associated variants of MUTYH. In addition, we compared these cellular OG:A repair levels to the adenine glycosylase activity of overexpressed protein isolated from insect cells. Using similar MEF cell lines, Molatore et al. (33) found that the two most common MUTYH variants Y165C and G382D are associated with levels of genomic OG significantly higher than cells expressing WT MUTYH. MUTYH activity presumably alters cellular OG levels because the repair of OG:A mispairs to OG:C enables OG glycosylases to then initiate removal of the OG. The correlation of MUTYH expression to GFP signal in MUTYH-deficient and proficient cells lines supports the validity of this assay to measure cellular repair of OG:A mispairs. In addition, the reduced GFP signal in cell lines expressing well-characterized MAP variants (Y165C, G382D) further validates this approach. Importantly, pGFP-off and pGFP-on vector constructs, differing only at a single base pair from the OG-containing construct, were used as positive and negative controls in all assays, and the constitutive expression of red fluorescence controlled for transfection efficiency.
This GFP assay depends on the repair of a non-replicating episomal plasmid. Hayashi, et al. (5) presented data suggesting that replication strongly enhances OG:A repair. Since transcription can only occur in the nucleus and these were serum-stimulated cells that are actively dividing, cellular repair cofactors such as proliferating cell nuclear antigen (PCNA) are present and may be important for MUTYH activity or to complete the repair process; however, it appears that MUTYH activity does not require the DNA substrate itself to replicate. Overexpression of the WT human MUTYH cDNA in Mutyh-null MEFs does not increase OG:A repair above that mediated by endogenous mouse Mutyh, suggesting that the activity of downstream repair enzymes in the base excision repair pathway may be limiting factors when MUTYH is overexpressed. Background repair of the fluorescent reporter is very low in Mutyh-null MEFs, evidence that only MUTYH can repair OG:A mispairs in non-replicating DNA. Previous studies suggest that mismatch repair enzymes influence OG damage repair (41,42), but in this system (possibly due to the non-replicating substrate), MUTYH-independent repair appears minor or non-existent.
Expression of the Y165C and G382D variants in MEFs was associated with an approximate 10-fold reduction in OG:A repair versus WT MUTYH expression when results are normalized to mRNA expression (Figure 3C). The RT–PCR quantification of mRNA levels was very reliable and was used as a baseline measure of expression since amino acid changes can affect protein stability and protein expression levels. Strong overexpression of Y165C (as measured by RT–PCR) did mediate significantly more OG:A repair versus the parental MUTYH-deficient MEFs (22% versus 9%, respectively). This is interesting considering that in vitro kinetic analysis of the insect cell expressed Y165C MUTYH was significantly reduced compared with the WT MUTYH (Table 1). This is evidence that cellular overexpression can compensate for low intrinsic enzyme activity. In addition, western blotting results suggest that the Y165C isoform has reduced protein stability, since protein levels are similar to WT cell lines despite the strong mRNA overexpression. In contrast, an approximate 3-fold increase in mRNA expression of G382D results in only 9% repair, not significantly different from the background signal in Mutyh-null MEFs. The adenine glycosylase activity of G382D MUTYH (Table 1) was only modestly reduced consistent with extensive previous in vitro work that has shown that G382D is less catalytically impaired than Y165C (6,12,14,16,18). Thus, there is a striking difference in the activity of these MUTYH variants in kinetic analysis of purified proteins versus the GFP-based mammalian cell assay, which suggests that G382D is relatively more impaired within cells. Even when the apparent differences in GFP repair are normalized to the differences in mRNA expression, these two variants are not significantly different in their activity in mammalian cells. The functional deficiency of G382D MUTYH may be magnified in a cellular context due to added demand of locating the OG:A mismatch in a cellular context and competition for DNA with other cellular proteins. Indeed, physiological (150mM) salt versus low salt buffer conditions uncover this deficiency (as shown in Table 1), which is equally present in the Q324H variant as well. This demonstrates that although careful biochemical analysis provides important mechanistic insights, this GFP assay integrates numerous factors that can affect the rate of OG:A mismatch repair in cells, including intrinsic enzyme activity, substrate binding affinity, protein stability and expression, post-translational modifications and protein–protein interactions. Thus, this assay provides a needed quantitative measure of MUTYH activity in living cells.
In this study, expression of the human Q324H variant led to significantly lower OG:A repair versus WT human MUTYH when mRNA expression was normalized. Importantly, these mRNA levels were similar to endogenous MUTYH levels in HEK 293 cells. To verify this reduced activity, two other MEF cell lines overexpressing Q324H were analyzed and also had reduced OG:A repair versus human WT MUTYH overexpression. This reduction was more pronounced when repair was normalized to mRNA expression. The fact that the assay detected reduced OG:A repair by G382D and Y165C variants, which are associated with a 28-fold increased CRC risk (9) and increased G:C to T:A transversion mutations in APC and KRAS in somatic tumor tissue in CRC patients (6,10) suggests that the reduced repair of the Q324H variant observed in this assay could be clinically relevant. This is further supported by the adenine glycosylase activity assays that show the Q324H variant has reduced activity at physiological salt concentrations.
Q324 is within the interdomain connector region between the catalytic N-terminal and the OG-interacting C-terminal domains. Although it appears to not be directly involved in catalysis or DNA binding, this flexible connector region may be critical for proper alignment of the two domains needed to couple OG:A recognition and adenine excision. Since the interdomain connector is not present in bacterial MutY homologs and has been shown to interact with the 9-1-1 clamp (27,30), amino acid substitutions in this region may also alter important protein–protein interactions. Human AP endonuclease-1 also interacts with MUTYH near the interdomain connector region (43) and AP endonuclease-1 enhances WT and G382D catalytic turnover in vitro (18). The Q324H variant may be altering the conformation of this region, affecting these protein–protein interactions and thus reducing the rate of cellular OG:A repair.
The nucleotide change leading to the Q324H variant is a G to T change that is designated as NCBI SNP rs3219489. The NCBI SNP database (28) shows a sample of Hispanics in the USA and people from China and Japan have minor allele frequencies between 45% and 47%, evidence that this allele is very common. Early clinical studies of MUTYH-associated polyposis show that Q324H is found in control groups and CRC patients at a similar frequency (44,45). This data coupled with biochemical data showing that Q324H had WT activity (26) probably contributed to the absence of analysis or reporting on Q324H status. Two older studies that did present data on Q324H are informative of the possible increased risk. Croitoru et al. (46) found 12 out of 1238 (1%) CRC patients were biallelic for known cancer-associated MUTYH variants, but additionally showed that out of the 29 people categorized as monoallelic for Y165C or G382D, 8 had Q324H in the second allele, including 4 people with colorectal adenomas; this corresponds to a 67% increase in affected population. Fleischmann et al. (47) found 2 of 358 CRC patients with biallelic MUTYH mutations but noted 2 additional Q324H/G382D heterozygotes, doubling the affected population. More recently, Casper et al. (48) found two monoallelic carriers of MAP variants with the second allele encoding Q324H in an unselected pool of 352 CRC patients, and these authors suggested that Q324H carriers may have a higher risk of CRC.
In a larger case–control study, Picelli et al. (21) assayed 1785 CRC patients and 1722 controls at a site in Sweden and found homozygous carriers of Q324H at increased risk for rectal cancer [odds ratio (OR) = 1.52; confidence interval (CI) = 1.06–2.17], but did not account for any other known pathogenic MUTYH variants. Similarly, Miyaishi et al. (24) found that homozygous carriers of Q324H had an increased risk of lung cancer in a Japanese population (n = 108, OR = 3.03, CI = 1.31–7.00, P = 0.010). Tao et al. (22) found that the presence of four SNP isoforms together, including one coding for Q324H, was associated with increased CRC risk in the distal colon (n=685, OR = 1.43; 95% CI = 1.005–2.029; P = 0.046); however, the Q324H SNP alone was not, evidence that other genetic factors influenced risk in this study. Kasahara et al. (23) found that Q324H carriers had an increased risk of CRC in a comparison of 68 CRC patients versus 121 controls (OR = 3.95, CI = 1.28–12.20, P = 0.017) in a Japanese population. Finally, in a sample of African-Americans, Q324H was found at a significantly higher frequency in 123 CRC patients with single or multiple adenomas versus a control population of African-Americans (P < 0.001).
We speculate that the high allele frequency and reduced activity of Q324H leads to an increase in cancer risk by two mechanisms. First, it may be present in monoallelic carriers of known cancer-associated MAP variants. Thus, it may be relevant to the ongoing controversy regarding the possible increased risk of CRC for monoallelic carriers of cancer-associated MUTYH variants (see (9,49) and references therein). These clinical studies have not considered Q324H as a possible modifier of risk; however, many people carry a second allele encoding Q324H. Thus, a previously identified monoallelic carrier of Y165C who is carrying Q324H in the other allele is probably at greater risk of CRC. Then, the high frequency of individuals biallelic for Q324H leads to an increased risk of cancer in the general population, but given the subtle functional deficiency, these cases may not be easily distinguishable in their clinical presentation and age of onset versus sporadic CRC cases. Together these two factors probably expand what is considered to be the population of individuals at higher risk of CRC due to germline MUTYH mutations.
Analysis of WT and variant MUTYH activity at different mRNA expression levels in this study suggests that cellular OG:A repair is altered by both the intrinsic activity of the protein isoform and cellular expression. This is striking, given that there is a wide variation in MUTYH expression in humans (50,51) and that reduced MUTYH expression is associated with poor survival in gastric cancers (52). The fact that MAP is typically not diagnosed until 40 years of age (7) underlines the subtle phenotype of biallelic MUTYH deficiency and is evidence that the accumulation of mutations due to oxidative DNA damage is a slow process. These G:C to T:A transversion mutations are also common in sporadic cancers (53). Taken together, this suggests that MAP may simply represent a severe MUTYH deficiency and individual differences in MUTYH activity may influence sporadic cancer risk and clinical outcomes.
Supplementary material
Supplementary Table 1, Figure 1, and supplementary material and methods can be found at http://carcin.oxford.org/
Funding
National Cancer Institute of the National Institutes of Health (CA67985); California Cancer Research Coordinating Committee to S.S.D; National Institute of Environmental Health Sciences (T32 ES007058-33) to A.G.R.
Supplementary Material
Acknowledgement
We would like to thank Adaeze Okeh, Oliver Munyaradzi, Rizpah Bellard and Stephen Chan for technical assistance, Cheryl Clauson for technical advice and the Privalsky laboratory (UCD) for a hygromycin vector. We also appreciate constructive comments on the manuscript provided by Prof. Jeffrey Bell and members of the David laboratory.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- CI
confidence interval
- CRC
colorectal cancer
- GFP
green fluorescent protein
- MAP
MUTYH-associated polyposis
- MEF
mouse embryonic fibroblast
- mRNA
messenger RNA
- OG
8-oxoguanine
- OR
odds ratio
- RT–PCR
reverse transcription–PCR
- SNP
single-nucleotide polymorphism
- WT
wild type
References
- 1. David S.S. et al. (2007). Base-excision repair of oxidative DNA damage. Nature 447 941–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Nghiem Y. et al. (1988). The mutY gene: a mutator locus in Escherichia coli that generates G.C—T.A transversions. Proc. Natl. Acad. Sci. USA 85 2709–2713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. van Loon B. et al. (2009). An 8-oxo-guanine repair pathway coordinated by MUTYH glycosylase and DNA polymerase lambda. Proc. Natl. Acad. Sci. USA 106 18201–18206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Le Page F. et al. (1998). Repair and mutagenic potency of 8-oxoG:A and 8-oxoG:C base pairs in mammalian cells. Nucleic Acids Res. 26 1276–1281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hayashi H. et al. (2002). Replication-associated repair of adenine:8-oxoguanine mispairs by MYH. Curr. Biol. 12 335–339 [DOI] [PubMed] [Google Scholar]
- 6. Al-Tassan N. et al. (2002). Inherited variants of MYH associated with somatic G:C–>T:A mutations in colorectal tumors. Nat. Genet. 30 227–232 [DOI] [PubMed] [Google Scholar]
- 7. Cheadle J.P. et al. (2007). MUTYH-associated polyposis–from defect in base excision repair to clinical genetic testing. DNA Repair (Amst.) 6 274–279 [DOI] [PubMed] [Google Scholar]
- 8. Nieuwenhuis M.H. et al. (2012). Evidence for accelerated colorectal adenoma–carcinoma progression in MUTYH-associated polyposis? Gut 61 734–738 [DOI] [PubMed] [Google Scholar]
- 9. Theodoratou E. et al. (2010). A large-scale meta-analysis to refine colorectal cancer risk estimates associated with MUTYH variants. Br. J. Cancer 103 1875–1884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lipton L. et al. (2003). Carcinogenesis in MYH-associated polyposis follows a distinct genetic pathway. Cancer Res. 63 7595–7599 [PubMed] [Google Scholar]
- 11. Kundu S. et al. (2009). Adenine removal activity and bacterial complementation with the human MutY homologue (MUTYH) and Y165C, G382D, P391L and Q324R variants associated with colorectal cancer. DNA Repair (Amst.) 8 1400–1410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Livingston A.L. et al. (2005). Insight into the roles of tyrosine 82 and glycine 253 in the Escherichia coli adenine glycosylase MutY. Biochemistry 44 14179–14190 [DOI] [PubMed] [Google Scholar]
- 13. Ali M. et al. (2008). Characterization of mutant MUTYH proteins associated with familial colorectal cancer. Gastroenterology 135 499–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Goto M. et al. (2010). Adenine DNA glycosylase activity of 14 human MutY homolog (MUTYH) variant proteins found in patients with colorectal polyposis and cancer. Hum. Mutat. 31 E1861–E1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Out A.A. et al. (2010). Leiden Open Variation Database of the MUTYH gene. Hum. Mutat. 31 1205–1215 [DOI] [PubMed] [Google Scholar]
- 16. D’Agostino V.G. et al. (2010). Functional analysis of MUTYH mutated proteins associated with familial adenomatous polyposis. DNA Repair (Amst.) 9 700–707 [DOI] [PubMed] [Google Scholar]
- 17. Chmiel N.H. et al. (2003). Insight into the functional consequences of inherited variants of the hMYH adenine glycosylase associated with colorectal cancer: complementation assays with hMYH variants and pre-steady-state kinetics of the corresponding mutated E.coli enzymes. J. Mol. Biol. 327 431–443 [DOI] [PubMed] [Google Scholar]
- 18. Pope M.A. et al. (2005). Insight into the functional consequences of hMYH variants associated with colorectal cancer: distinct differences in the adenine glycosylase activity and the response to AP endonucleases of Y150C and G365D murine MYH. DNA Repair (Amst.) 4 315–325 [DOI] [PubMed] [Google Scholar]
- 19. Nielsen M. et al. (2009). Analysis of MUTYH genotypes and colorectal phenotypes in patients With MUTYH-associated polyposis. Gastroenterology 136 471–476 [DOI] [PubMed] [Google Scholar]
- 20. Fromme J.C. et al. (2004). Structural basis for removal of adenine mispaired with 8-oxoguanine by MutY adenine DNA glycosylase. Nature 427 652–656 [DOI] [PubMed] [Google Scholar]
- 21. Picelli S. et al. (2010). Common variants in human CRC genes as low-risk alleles. Eur. J. Cancer 46 1041–1048 [DOI] [PubMed] [Google Scholar]
- 22. Tao H. et al. (2008). Association between genetic polymorphisms of the base excision repair gene MUTYH and increased colorectal cancer risk in a Japanese population. Cancer Sci. 99 355–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kasahara M. et al.(2008)Association of MUTYH Gln324His and APEX1 Asp148Glu with colorectal cancer and smoking in a Japanese population. J. Exp. Clin. Cancer Res. 2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miyaishi A. et al.(2009)MUTYH Gln324His gene polymorphism and genetic susceptibility for lung cancer in a Japanese population. J. Exp. Clin. Cancer Res. 2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yuan Z. et al. (2007)Q324H, A germnline variant of the MUTY homolog (MYH) gene is strongly associated with early onset adenomatous colorectal polyp formation among African Americans The 2nd biennial scientific meeting of international society for gastrointestinal hereditary tumours.Pacifico-Yokohama [Google Scholar]
- 26. Shinmura K. et al. (2000). Adenine excisional repair function of MYH protein on the adenine:8-hydroxyguanine base pair in double-stranded DNA. Nucleic Acids Res. 28 4912–4918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Luncsford P.J. et al. (2010). A structural hinge in eukaryotic MutY homologues mediates catalytic activity and Rad9-Rad1-Hus1 checkpoint complex interactions. J. Mol. Biol. 403 351–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sherry S.T. et al. (2001). dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 29 308–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gu Y. et al. (2002). Human MutY homolog, a DNA glycosylase involved in base excision repair, physically and functionally interacts with mismatch repair proteins human MutS homolog 2/human MutS homolog 6. J. Biol. Chem. 277 11135–11142 [DOI] [PubMed] [Google Scholar]
- 30. Shi G. et al. (2006). Physical and functional interactions between MutY glycosylase homologue (MYH) and checkpoint proteins Rad9-Rad1-Hus1. Biochem. J. 400 53–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hahm S.H. et al. (2011). Knock-down of human MutY homolog (hMYH) decreases phosphorylation of checkpoint kinase 1 (Chk1) induced by hydroxyurea and UV treatment. BMB Rep. 44 352–357 [DOI] [PubMed] [Google Scholar]
- 32. Kundu S. et al. (2010). Ser 524 is a phosphorylation site in MUTYH and Ser 524 mutations alter 8-oxoguanine (OG): a mismatch recognition. DNA Repair (Amst.) 9 1026–1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Molatore S. et al. (2010). MUTYH mutations associated with familial adenomatous polyposis: functional characterization by a mammalian cell-based assay. Hum. Mutat. 31 159–166 [DOI] [PubMed] [Google Scholar]
- 34. Masaoka A. et al. (2009). DNA polymerase beta and PARP activities in base excision repair in living cells. DNA Repair (Amst.) 8 1290–1299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Burger K. et al.(2010)A modified fluorimetric host cell reactivation assay to determine the repair capacity of primary keratinocytes, melanocytes and fibroblasts. BMC Biotechnol. 1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Xie Y. et al. (2004). Deficiencies in mouse Myh and Ogg1 result in tumor predisposition and G to T mutations in codon 12 of the K-ras oncogene in lung tumors. Cancer Res. 64 3096–3102 [DOI] [PubMed] [Google Scholar]
- 37. Xie Y. et al. (2008). Cells deficient in oxidative DNA damage repair genes Myh and Ogg1 are sensitive to oxidants with increased G2/M arrest and multinucleation. Carcinogenesis 29 722–728 [DOI] [PubMed] [Google Scholar]
- 38. Tsien R.Y. (1998). The green fluorescent protein. Ann. Rev. Biochem. 67 509–544 [DOI] [PubMed] [Google Scholar]
- 39. Guan Y. et al. (1998). MutY catalytic core, mutant and bound adenine structures define specificity for DNA repair enzyme superfamily. Nat. Struct. Biol. 5 1058–1064 [DOI] [PubMed] [Google Scholar]
- 40. Dallosso A.R. et al. (2008). Inherited predisposition to colorectal adenomas caused by multiple rare alleles of MUTYH but not OGG1, NUDT1, NTH1 or NEIL 1, 2 or 3. Gut 57 1252–1255 [DOI] [PubMed] [Google Scholar]
- 41. Russo M.T. et al. (2009). Role of MUTYH and MSH2 in the control of oxidative DNA damage, genetic instability, and tumorigenesis. Cancer Res. 69 4372–4379 [DOI] [PubMed] [Google Scholar]
- 42. Colussi C. et al. (2002). The mammalian mismatch repair pathway removes DNA 8-oxodGMP incorporated from the oxidized dNTP pool. Curr. Biol. 12 912–918 [DOI] [PubMed] [Google Scholar]
- 43. Parker A. et al. (2001). Human homolog of the MutY repair protein (hMYH) physically interacts with proteins involved in long patch DNA base excision repair. J. Biol. Chem. 276 5547–5555 [DOI] [PubMed] [Google Scholar]
- 44. Cheadle J.P. et al. (2003). Inherited defects in the DNA glycosylase MYH cause multiple colorectal adenoma and carcinoma Carcinogenesis 24 1281–1282 [DOI] [PubMed] [Google Scholar]
- 45. Sieber O.M. et al. (2003). Multiple colorectal adenomas, classic adenomatous polyposis, and germ-line mutations in MYH. N. Engl. J. Med. 348 791–799 [DOI] [PubMed] [Google Scholar]
- 46. Croitoru M.E. et al. (2004). Association between biallelic and monoallelic germline MYH gene mutations and colorectal cancer risk. J. Natl. Cancer Inst. 96 1631–1634 [DOI] [PubMed] [Google Scholar]
- 47. Fleischmann C. et al. (2004). Comprehensive analysis of the contribution of germline MYH variation to early-onset colorectal cancer Int. J. Cancer 109 554–558 [DOI] [PubMed] [Google Scholar]
- 48. Casper M. et al. (2011). MUTYH hotspot mutations in unselected colonoscopy patients Colorectal Dis 33 1067–1074 [DOI] [PubMed] [Google Scholar]
- 49. Win A.K. et al. (2011). Cancer risks for monoallelic MUTYH mutation carriers with a family history of colorectal cancer. Int. J. Cancer 129 2256–2262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Dixon A.L. et al. (2007). A genome-wide association study of global gene expression. Nat. Genet. 39 1202–1207 [DOI] [PubMed] [Google Scholar]
- 51. Fry R.C. et al. (2008). Genomic predictors of interindividual differences in response to DNA damaging agents. Genes Dev. 22 2621–2626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Shinmura K. et al. (2011). Reduced expression of MUTYH with suppressive activity against mutations caused by 8-hydroxyguanine is a novel predictor of a poor prognosis in human gastric cancer. J. Pathol. 225 414–423 [DOI] [PubMed] [Google Scholar]
- 53. Pleasance E.D. et al. (2010). A comprehensive catalogue of somatic mutations from a human cancer genome. Nature 463 191–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Rieder M.J. et al. (2008). The environmental genome project: reference polymorphisms for drug metabolism genes and genome-wide association studies. Drug Metab. Rev. 40 241–261 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.