Abstract
Skeletal muscles in the limb and body trunk are composed of heterogeneous myofibers expressing different isoforms of myosin heavy chain (Myh), including type I (slow, Myh7), IIA (intermediate, Myh2), IIX (fast, Myh1) and IIB (very fast, Myh4). While the contraction force and speed of a muscle are known to be determined by the relative abundance of myofibers expressing each Myh isoform, it is unclear how specific combinations of myofiber types are formed and regulated at the cellular and molecular level. We report here that myostatin (Mstn) positively regulates slow but negatively regulates fast Myh isoforms. Mstn was expressed at higher levels in the fast muscle myoblasts and myofibers than in the slow muscle counterparts. Interestingly, Mstn knockout led to a shift of Myh towards faster isoforms, suggesting an inhibitory role of Mstn in fast Myh expression. Consistently, when induced to differentiate, Mstn null myoblasts formed myotubes preferentially expressing fast Myh. Conversely, treatment of myoblasts with a recombinant Mstn protein upregulated Myh7 but downregulated Myh4 gene expression in newly formed myotubes. Importantly, both Mstn antibody and soluble activin type 2B receptor inhibited slow Myh7 and promoted fast Myh4 expression, indicating that myostatin acts through canonical activin receptor to regulate the expression of Myh genes. These results demonstrate a role of myostatin in the specification of myofiber types during myogenic differentiation.
Keywords: Myostatin (Mstn), satellite cells, myofiber, myosin heavy chain, TGFβ
INTRODUCTION
Myostatin (Mstn), also known as growth and differentiation factor 8 (GDF8), is a member of the transforming growth factor β (TGFβ) family of secreted growth factors [1]. Mstn is translated as a secreted protein in an inactive form, which is then activated through two separate cleavages removing the signaling peptide and inhibitory domain [2]. This active form of Mstn can bind to a number of regulatory proteins within the blood/serum [3,4]. Similar to other TGFβ family ligands, the activated Mstn can bind to activin receptors (ActR) to initiate a signal transduction cascade in the target cell [5,6].
Mstn is a highly conserved negative regulator of skeletal muscle mass. Mstn is expressed in the myotome compartment of somites and later in developing skeletal muscles throughout embryogenesis [1]. When functional Mstn is absent during fetal development, the resultant adult muscle mass is significantly increased due to both hypertrophy and hyperplasia of skeletal muscle myofibers [1,7]. In adult tissues, Mstn gene is predominantly expressed in the skeletal muscles with varying levels depending on muscle types [8].To achieve specific physiological functions, each limb muscle contains a unique combination of fast and slow myofibers, classified based on the expression of myosin heavy chain (Myh) isoforms: type I (slow oxidative, Myh7), IIA (fast oxidative, Myh2), IIX (fast glycolytic, Myh1) and IIB (very fast glycolytic, Myh4) [9]. Interestingly, fast muscles (containing predominantly type II fast myofibers) express higher levels of Mstn than slow muscles, and such differential expression is shown to be regulated by miR-27 [10,11]. In addition, loss of Mstn led to increased proportions of fast myofibers in both Mstn−/−mice and the double muscled calves [12,13]. Thus, Mstn regulates both muscle size and myofiber type composition.
Mstn has also been shown to regulate muscle progenitor cell (satellite cell) function and myogenesis. Mstn−/− muscle contains more satellite cells per unit myofiber domain and the satellite cells are more resistance to activation, suggesting that Mstn maintains satellite cells in a quiescent state [14,15]. In addition, Mstn also inhibits the proliferation of satellite cells derived from several animal species [16,17,18,19]. By contrast, Mstn seems to promote the differentiation of myoblasts in fish [20]. It has been unknown, however, whether Mstn regulates the differentiation of myoblasts into fast versus slow muscles.
Several important questions remain to be answered. Mstn is highly expressed in fast than in slow muscles, yet its mutation paradoxically leads to increased proportion of fast myofibers. It is unclear whether the observed myofiber type switching in Mstn−/− muscles is due to a secondary effect of muscle hypertrophy. Does Mstn regulate myofiber composition differentially in fast and slow muscles? Does Mstn regulate myofiber type formation developmentally during myogenesis or postnatally during muscle hypertrophy? Given that Mstn blockage-induced postnatal muscle hypertrophy does not seem to require progenitor cell activity [21], we hypothesize that Mstn regulates myofiber type specification early during myogenic differentiation. We provide novel evidence that Mstn is differentially expressed in myoblasts reside in the slow and fast muscles to regulate their differentiation potential to become slow and fast myotubes, respectively.
MATERIALS AND METHODS
Animals
All procedures involving the use of animals were approved by Purdue University’s Animal Care and Use Committee and performed in accordance with NIH guidelines. Mice were housed in the animal facility with free access to water and standard rodent chow. Mstn mutant mice were generated by Dr. Se-Jin Lee (The Johns Hopkins University, Baltimore, MD) [1]. Heterozygous mice were bred to generate Mstn null and wildtype littermates used in this study. PCR genotyping was done using protocols as previously described [22].
Primary myoblast isolation and culture
Primary myoblasts were isolated from hind limb skeletal muscles of 2-month old wild type mice. Muscles were minced and digested in type I collagenase and dispase B mixture (Roche Applied Science). Cells were then filtered from debris, centrifuged, and cultured in growth media (F-10 Ham’s medium supplemented with 20% fetal bovine serum, 4 ng/mL basic fibroblast growth factor, and 1% penicillin-streptomycin) on collagen-coated dishes, as described [23].
Quantitative realtime polymerase chain reaction (qPCR)
RNA was extracted and purified from muscle tissue or cell cultures with Trizol (Invitrogen) and contaminating DNA was removed with DNase I. Random hexamer primers were used to convert RNA into cDNA. Genomic DNA of cell transplantation samples were extracted and purified with phenol: chloroform. QPCR was performed using a light cycler 480 (Roche) machine and reagents for 40 cycles, and the fold changes for all the samples were calculated by 2−ΔΔct methods. 18s was used as housekeeping gene for mRNA qPCR. Primers used were listed in [23], except for Mstn: F 5′-CAC TCT ACA AAG TAC GAG TCT CTC T-3′ and R 5′-CTA GAG TTG ACT GAA AGT TGA CCT-3′.
Cryosectioning
EDL and SOL muscle samples (fresh) were embedded in optimal cutting temperature (O.C.T) compound (Sakura Finetek) and quickly frozen in dry ice cooled isopentane. Muscles were cut into 10μm thickness by a Leica CM 1850 cryostat and the sections were placed on superfrost plus glass slides (Electron Microscopy Sciences).
Immunostaining and image capture
Tissue sections were blocked for 30 minutes with blocking buffer (5% horse serum, 2% bovine serum albumin, 0.2% triton X-100 and 0.1% sodium azide in PBS). Tissues were then incubated with primary antibodies NOQ (Abcam) and MF20 (Developmental Studies Hybridoma Bank, Iowa, USA) diluted in blocking buffer for 1 hour at room temperature, followed incubating with secondary antibodies and Hoechst (diluted in PBS) for 30 minutes at room temperature, and mounted with Dako fluorescent mounting media (Glostrup, Denmark). Fluorescent images were captured with a Coolsnap HQ CCD camera (Photometrics, USA) driven by IP Lab software (Scanalytics Inc, USA) using Leica DMI 6000B fluorescent microscope (Mannheim, Germany) with a 20X objective (NA = 0.70).
Mstn antibody and Activin receptor type IIB treatment
When primary myoblasts in growth medium were 90% confluent, they were switched to differentiation medium (5% horse serum in DMEM) with 0 ng/mL, 200 ng/mL and 2000 ng/mL Mstn antibody [24] or soluble activin receptor type IIB [25]. Differentiation medium was changed every day for 6 days. For each treatment, 3 different batches of cells were used, and each batch of cells was duplicated.
Luciferase Reporter Assay
The fast fiber-specific luciferase reporter constructs (Myh4) and pRL-TK plasmid expressing Renilla luciferase were obtained from David Gerrad (Purdue University, IN, USA) [26]. Primary myoblasts were co-transfected with pGL-Myh4 and pRL-TK plasmids using the NeonTM Transfection System (Invitrogen Inc) according to the manufacturer’s recommended protocol. 1 × 106 cells were transfected with 2 μg of plasmid DNA at a condition of 1500 V, 10 ms and 3 pulses. The transfected cells were then plated into 48- well plate and then induced to fuse into myotubes for 5 days. The reporter assays were performed with Dual-Luciferase Reporter Assay System (Promega, Madison, WI), according to the manufacturer’s recommendations. The samples were read at Tecan Genios Pro (Tecan Group Ltd.) plate reader. Luciferase activity was calculated as the ratio between Firefly and Renilla Luciferase.
ImageJ analysis
The signal density of identically threshold adjusted fluorescent images was calculated with ImageJ system. Color image was converted into grayscale by changing it into 8-bit. Rolling ball radius under Substract Background was needed to be changed into 50 to remove some of the background coloration from the image. All the images were measured using the same standards.
Statistical analysis
The data were presented with mean ± standard error of the mean (SEM). P-values were calculated using two-tailed student’s t-test. The ones with P-value less than 0.05 were considered as statistic significant.
RESULTS AND DISCUSSION
Mstn is differentially expressed in the fast and slow muscles at the whole muscle, myofiber and primary myoblast levels
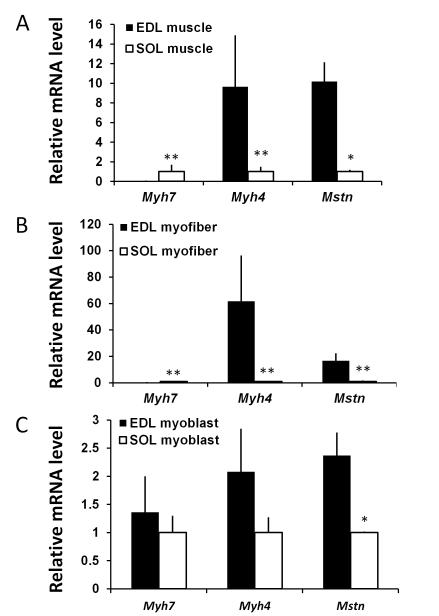
The extensor digitorum longus (EDL) and soleus (SOL) muscles have been widely used as representative fast and slow muscles, respectively [27,28,29,30,31]. They both are hind limb muscles and very small in size (each contains about 1000 myofibers) in mouse, thus allowing for unbiased analysis of myofiber composition and gene expression at the whole muscle level [23]. In addition, single myofibers can be enzymatically released from these muscles, free of connective tissue and other interstitial cells, thus providing great opportunities to examine muscle cell specific gene expression. As expected, the expression of Type IIB fast myosin heavy chain (Myh4) was about 10 times higher, but Type I slow myosin heavy chain (Myh7) was about 100 times lower, in the EDL than SOL muscles (Fig. 1A). Strikingly, Mstn was expressed ~10 times higher in EDL than SOL muscles (Fig. 1A). This result is consistent with previous reports that Mstn expression level is correlated to the abundance of Type IIB myofibers [32].
Figure 1. Mstn is differently expressed in the fast (EDL) and slow (SOL) muscles at the whole muscle, myofibers and myoblast levels.
(A) Relative mRNA abundance of Mstn, Myh7 and Myh4 in EDL and SOL whole muscles. (B) Relative mRNA abundance of Mstn, Myh7 and Myh4 in EDL and SOL myofibers free of connective tissues and non-muscle cells. (C) Relative mRNA abundance of Mstn, Myh7 and Myh4 in myoblasts derived from the EDL and SOL muscles. The expression levels in the SOL muscle/myofiber/myoblast are normalized to 1 for each gene examined. N = 3 replicates using samples from 3 pairs of mice, *p <0.05 and **p < 0.01 by Student t-Test.
To confirm that the higher level of Mstn expression in the EDL muscle is myofiber specific (not due to non-muscle cells in the whole muscle), we extracted individual myofibers from EDL and SOL muscles. Consistently, Mstn mRNA and Myh4 levels were 17 and 62 times higher, but Myh7 was 25 times lower, in EDL myofibers than in SOL myofibers (Fig. 1B). We further asked if Mstn is differentially expressed in satellite cells associated with EDL and SOL muscles of adult mice by examining satellite cell-derived primary myoblasts established from these two muscles. Interestingly, Mstn mRNA level was 2.4 times higher in the EDL myoblasts than SOL myoblasts (Fig. 1C). By contrast, Myh4 and Myh7 expression were not significantly different in the EDL and SOL derived myoblasts during proliferation prior to differentiation (Fig. 1C). Thus, Mstn expression is higher in the fast muscles at the whole muscle, myofiber and myoblast levels. The novel finding that Mstn is differentially expressed in fast and slow muscle derived myogenic progenitor cells (myoblasts) prior to their differentiation into myotubes indicates that Mstn has important roles in myofiber type determination during myogenesis.
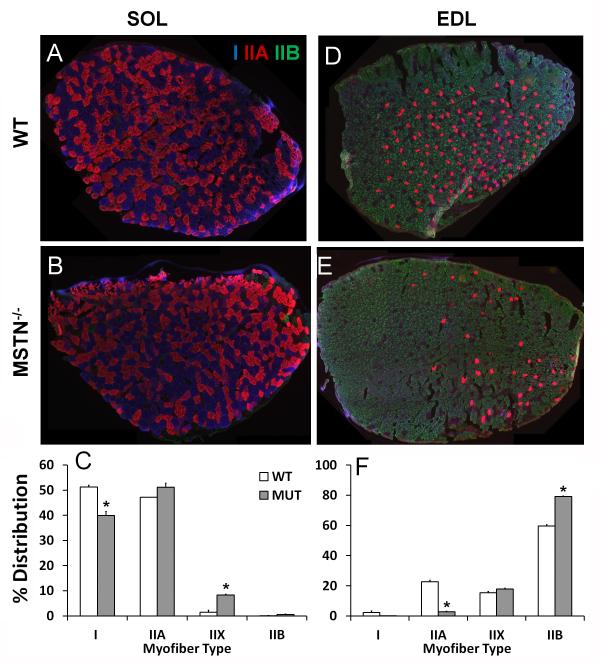
Mstn mutation reduces slow myofibers and leads to myofiber type switching towards faster myosin heavy chain isoforms in mature fast and slow muscles
To further explore the roles of Mstn in muscle myofiber type determination, we compared myofiber composition of Mstn knockout mice and their wild type littermates. SOL and EDL muscles were isolated from young adults (2-3 months of age), cryosectioned and labeled with monoclonal antibodies specific for Type I (Myh7), IIA (Myh2) and IIB (Myh4) myosin heavy chain (Fig. 2A, B, D, E). In the slow SOL muscles, loss of Mstn reduced the percentage of Type I myofibers from 51% in the wildtype to 40% in the Mstn−/− (Fig. 2C). Meanwhile, the percentages of Type IIX and IIB myofibers were both increased in the Mstn−/− than in wildtype SOL (Fig. 2C; Type IIX myofibers: 1% in the wildtype vs 8% in the mutant). In the fast EDL muscles, the abundance of Type IIB myofibers increased from 60% in the wildtype to 80% in the Mstn−/−, accompanied by reduced abundance of Type I and IIA myofibers (Fig. 5F; Type IIA myofibers: 23% in the wildtype vs 3% in the Mstn−/−). As myofiber type distribution is known to exhibit regional clustering even within a muscle [33,34], our analysis of myofiber type across the entire cross sectional area of both fast and slow muscles provides unbiased assessment of myofiber type distribution. Our results demonstrate that loss of Mstn leads to the myofiber type transition from slow to fast in mature resting muscles.
Figure 2. Myofiber composition in the fast and slow muscles of Mstn knockout and wildtype mice.
(A B, D, E) Representative staining of EDL and SOL muscle sections collected from 2-month-old Mstn knockout (Mstn−/−) mice and wildtype littermates. Blue, red and green signals are for Myh7, Myh2 and Myh4 immunofluorescence that mark Type I, IIA, and IIB myofibers, respectively. The unlabeled myofibers are presumably Type IIX (Myh1). (C, F) Distribution (%) of Type I, IIA, IIX and IIB myofibers in SOL (C) and EDL (F) muscles, respectively. N = 2 pair of mice, *p <0.05 by Student t-Test. WT: Wildtype, MUT: Mstn mutant.
Reduced formation of slow myosin heavy chain-expressing myotubes in Mstn mutant myoblasts
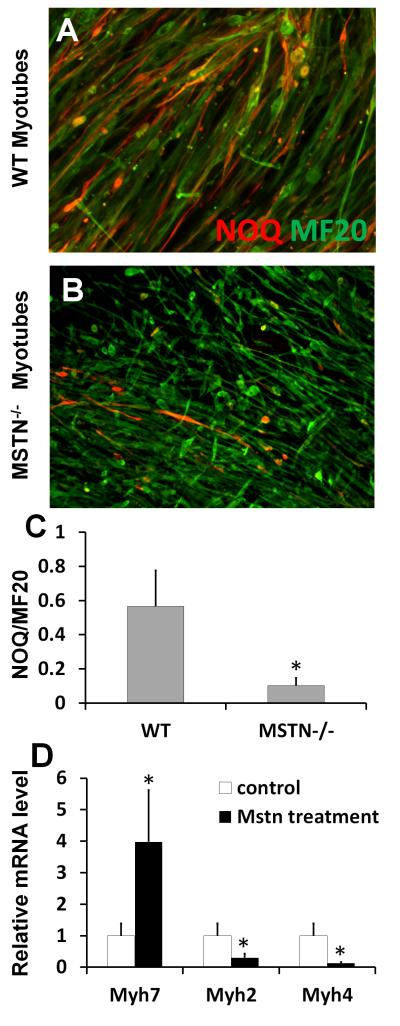
The observed increases in fast and decreases in slow myofiber types in steady state Mstn−/− muscles can be established during development, or during postnatal muscle hypertrophy, which is known to be associated with myofiber type switching towards fast Myh [35]. To distinguish these different possibilities, we investigated if Mstn plays a role in the determination of myofiber types during myogenesis by examaining the expression of Myh isoforms in cultures of satellite cell-derived myoblasts of Mstn−/− mice and wildtype littermates. Primary myoblasts derived from Mstn−/− mice and wildtype littermates were induced to differentiate by serum withdrawal. At day 6 after induction, nascent myotubes were formed in both wildtype and Mstn−/− myoblast cultures (Fig. 3A-B). Immunolabeling of slow myosin heavy chain with NOQ antibody and total myosin heavy chain with MF20 antibody indicates that the abundance of NOQ-expressing slow myotubes are robustly reduced in the Mstn−/− myoblast cultures (Fig. 3C). Interestingly, the nascent myotubes formed by Mstn−/− myoblasts are much smaller than those formed by the wildtype myoblasts (Fig. 3A-B). The smaller size of nascent myotubes allows sufficient muscle hyperplasia without increasing the overall muscle mass/body size during development, as observed in Mstn−/− mouse embryos [36].
Figure 3. Mstn enhances slow myosin and inhibits fast myosin expression during myogenic differentiation.
(A-B) Primary myoblasts collected from the hindlimb muscles of 2-month-old Mstn−/− and their littermate wildtype mice were induced to differentiate for 6 days and stained for NOQ (red) and MF20 (green) antibodies that specifically label slow and pan- myosin heavy chain. (C) Ratio of NOQ/MF20 staining signals quantified using Image-J software. (D) Relative mRNA expression levels of Myh7, Myh2 and Myh4 genes in nascent myotubes differentiated from wildtype myoblasts treated with vehicle control or recombinant myostatin (600 ng/ml) for 5 days. Error bars represent SEM (N = 3). *p<0.05 (student’s T test).
The reduced abundance of nascent slow myotubes in Mstn−/− myoblast cultures indicates that Mstn promotes slow myosin heavy chain expression during myogenesis. To directly test this possibility, we treated wildtype primary myoblasts with Mstn recombination protein (600 ng/ml) for 5 days in differentiation medium. We then extracted RNA from the control (vehicle) and Mstn treated cultures and conducted quantitative realtime PCR analysis to examine the expression levels of Myh2, Myh4, and Myh7. Mstn treatment led to 4 times upregulation of slow Myh7 expression and 87% reduction of fast Myh4 expression (Fig. 3D). The expression of Type IIA fast oxidative myosin (Myh2) was also moderately reduced by recombinant Mstn treatment (Fig. 3D). These results suggest that Mstn not only enhances the expression of slow myosin heavy chain gene, but also inhibits the expression of fast myosin heavy chain genes.
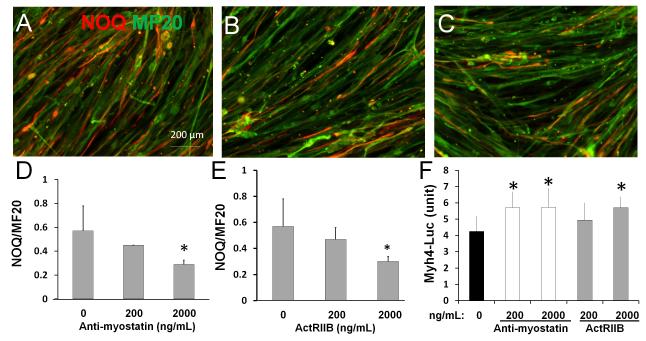
Blockage of Mstn signaling inhibits slow but promotes fast myosin heavy chain expression
We next asked if Mstn acts through canonical activin receptors to regulate myosin heavy chain expression. To this end, we used an Mstn antibody (Anti-myo) and a soluble activin receptor type IIB (ActRIIB) to block the Mstn signal transduction [24,25]. We treated primary myoblasts from wildtype mice with Anti-myo and ActRIIB while they were in differentiation medium. After 6 days of differentiation, immunostaining was performed to detect slow myosin heavy chain and total myosin heavy chain expression using NOQ and MF20 antibodies, respectively (Fig. 4A-C). Analysis of NOQ and MF20 signal areas with ImageJ software indicates that Mstn antibody dose-dependently reduces the expression of slow myosin heavy chain expression (Fig. 4D). Soluble ActRIIB similarly inhibited slow myosin heavy chain expression in nascent myotubes (Fig. 4E). Thus, inhibition of Mstn signaling pathway reduces formation of slow type myosin heavy chain during myogenic differentiation.
Figure 4. Blockage of myostatin signaling inhibits slow myosin expression and increases fast myosin expression.
(A-C) Primary myoblasts isolated from limb muscles of 2-month-old wildtype mice were induced to differentiate for 6 days in the presence of Mstn antibody, soluble Activin receptor type IIB (ActIIB) and vehicle control. Nascent myotubes were fixed and stained with NOQ (red) and MF20 (green) antibodies to label the slow and pan- myosin heavy chain proteins. (A-C) NOQ and MF20 staining on newly differentiated myotubes without treatment (A, vehicle control); with 2000 ng/ml myostatin antibody (B); and with 2000 ng/ml ActIIB (C). (D) The ratios of NOQ/MF20 signals measured with Image J in myotubes treated with Mstn antibody at the concentration shown. (E) The ratios of NOQ/MF20 signals measured with Image J in myotubes treated with soluble ActRIIB at the concentration shown. (F) Primary myoblasts from adult wildtype mice were transfected with the fast fiber (Myh4) specific luciferase reporter construct and pRL-TK plasmid expressing Renilla luciferase. Cells were then induced to differentiate and treated with Mstn antibody or ActRIIB for 5 days at the concentrations shown. The Y-axis shows the ratio of Myh4 luciferase and Renilla luciferase. Error bars represent SEM (N = 3). *p <0.05 (student’s T test).
We finally examined if Mstn antagonism also upregulates fast Myh4 gene expression in addition to suppressing slow Myh7. We transfected a Myh4-luciferase reporter plasmid into primary myoblasts, and then treated the cells with Mstn antibody or soluble ActRIIB for 5 days in differentiation medium. Both Mstn antibody and ActRIIB treatment increased the luciferase reporter activity (Fig 4F). These results indicate that Mstn antagonism enhance the transcription of the fast Myh4 gene. Together, the Mstn ligand and receptor blockage experiments provide strong evidence that Mstn acting through activin receptors to enhance slow myosin heavy expression and inhibit fast myosin heavy chain expression.
Previous studies have also reported that loss of Mstn expression is associated with a decrease in the slow-twitch myofibers [12,13]. However, it is has been unclear how and when Mstn works to affect myofiber type composition. It has been suggested that Mstn regulates fiber-type composition by regulating myocyte enhancer factor 2 (MEF2) and MyoD gene expression [10]. In our study, we show for the first time that mstn is differentially expressed in different populations of myogenic progenitors (myoblasts) to specify their developmental fate to become either fast or slow myofibers. Our results provide novel insights into the molecular regulation of skeletal myofiber type specification. Such information will lead to strategies that target myostatin signaling to alter the ratio of fast and slow myofibers, which is a critical determinant of athletic motor performance [37]. Specific combinations of fast and slow muscles also affect the quality and taste attributes of meat products in animal production industry [38].
Highlights.
Myostatin (Mstn) is a TGFβ family master regulator of muscle mass
Mstn is differentially expressed by progenitors in fast and slow muscles
Mstn promotes slow but inhibits fast myosin heavy chain expression
Mstn specifies fast/slow fiber types during myogenic differentiation
These results establish a new role of Mstn in myogenesis
Acknowledgement
We thank members of Kuang lab for technical assistance and Jun Wu for mouse colony support. The project is partially supported by funding from MDA, NIH and USDA to SK. The authors declare no conflict of interests.
Abbreviations
- ActRIIB
activin receptor type IIB
- EDL
Extensor digitorum longus
- Mstn
Myostatin
- Myh
Myosin heavy chain
- SOL
soleus
- TGFβ
Transforming growth factor β
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].McPherron AC, Lawler AM, Lee SJ. Regulation of skeletal muscle mass in mice by a new TGF-beta superfamily member. Nature. 1997;387:83–90. doi: 10.1038/387083a0. [DOI] [PubMed] [Google Scholar]
- [2].Hill JJ, Qiu Y, Hewick RM, Wolfman NM. Regulation of myostatin in vivo by growth and differentiation factor-associated serum protein-1: a novel protein with protease inhibitor and follistatin domains. Mol Endocrinol. 2003;17:1144–1154. doi: 10.1210/me.2002-0366. [DOI] [PubMed] [Google Scholar]
- [3].Hill JJ, Davies MV, Pearson AA, Wang JH, Hewick RM, Wolfman NM, Qiu Y. The myostatin propeptide and the follistatin-related gene are inhibitory binding proteins of myostatin in normal serum. J Biol Chem. 2002;277:40735–40741. doi: 10.1074/jbc.M206379200. [DOI] [PubMed] [Google Scholar]
- [4].Zimmers TA, Davies MV, Koniaris LG, Haynes P, Esquela AF, Tomkinson KN, McPherron AC, Wolfman NM, Lee SJ. Induction of cachexia in mice by systemically administered myostatin. Science. 2002;296:1486–1488. doi: 10.1126/science.1069525. [DOI] [PubMed] [Google Scholar]
- [5].Massague J. TGF-beta signal transduction. Annu Rev Biochem. 1998;67:753–791. doi: 10.1146/annurev.biochem.67.1.753. [DOI] [PubMed] [Google Scholar]
- [6].Shi Y, Massague J. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell. 2003;113:685–700. doi: 10.1016/s0092-8674(03)00432-x. [DOI] [PubMed] [Google Scholar]
- [7].Kollias HD, McDermott JC. Transforming growth factor-beta and myostatin signaling in skeletal muscle. J Appl Physiol. 2008;104:579–587. doi: 10.1152/japplphysiol.01091.2007. [DOI] [PubMed] [Google Scholar]
- [8].Lee SJ. Regulation of muscle mass by myostatin. Annu Rev Cell Dev Biol. 2004;20:61–86. doi: 10.1146/annurev.cellbio.20.012103.135836. [DOI] [PubMed] [Google Scholar]
- [9].Schiaffino S, Reggiani C. Fiber Types in Mammalian Skeletal Muscles. Physiological Reviews. 2011;91:1447–1531. doi: 10.1152/physrev.00031.2010. [DOI] [PubMed] [Google Scholar]
- [10].Hennebry A, Berry C, Siriett V, O’Callaghan P, Chau L, Watson T, Sharma M, Kambadur R. Myostatin regulates fiber-type composition of skeletal muscle by regulating MEF2 and MyoD gene expression. Am J Physiol Cell Physiol. 2009;296:C525–534. doi: 10.1152/ajpcell.00259.2007. [DOI] [PubMed] [Google Scholar]
- [11].Allen DL, Loh AS. Posttranscriptional mechanisms involving microRNA-27a and b contribute to fast-specific and glucocorticoid-mediated myostatin expression in skeletal muscle. Am J Physiol Cell Physiol. 2011;300:C124–137. doi: 10.1152/ajpcell.00142.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Stavaux D, Art T, McEntee K, Reznick M, Lekeux P. Muscle fibre type and size, and muscle capillary density in young double-muscled blue Belgian cattle. Zentralbl Veterinarmed A. 1994;41:229–236. doi: 10.1111/j.1439-0442.1994.tb00089.x. [DOI] [PubMed] [Google Scholar]
- [13].Girgenrath S, Song K, Whittemore LA. Loss of myostatin expression alters fiber-type distribution and expression of myosin heavy chain isoforms in slow- and fast-type skeletal muscle. Muscle Nerve. 2005;31:34–40. doi: 10.1002/mus.20175. [DOI] [PubMed] [Google Scholar]
- [14].McCroskery S, Thomas M, Maxwell L, Sharma M, Kambadur R. Myostatin negatively regulates satellite cell activation and self-renewal. J Cell Biol. 2003;162:1135–1147. doi: 10.1083/jcb.200207056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].McFarlane C, Hennebry A, Thomas M, Plummer E, Ling N, Sharma M, Kambadur R. Myostatin signals through Pax7 to regulate satellite cell self-renewal. Experimental Cell Research. 2008;314:317–329. doi: 10.1016/j.yexcr.2007.09.012. [DOI] [PubMed] [Google Scholar]
- [16].McFarland DC, Velleman SG, Pesall JE, Liu C. Effect of myostatin on turkey myogenic satellite cells and embryonic myoblasts. Comp Biochem Physiol A Mol Integr Physiol. 2006;144:501–508. doi: 10.1016/j.cbpa.2006.04.020. [DOI] [PubMed] [Google Scholar]
- [17].Gilson H, Schakman O, Kalista S, Lause P, Tsuchida K, Thissen JP. Follistatin induces muscle hypertrophy through satellite cell proliferation and inhibition of both myostatin and activin. American Journal of Physiology-Endocrinology and Metabolism. 2009;297:E157–E164. doi: 10.1152/ajpendo.00193.2009. [DOI] [PubMed] [Google Scholar]
- [18].Yamada M, Tatsumi R, Yamanouchi K, Hosoyama T, Shiratsuchi S, Sato A, Mizunoya W, Ikeuchi Y, Furuse M, Allen RE. High concentrations of HGF inhibit skeletal muscle satellite cell proliferation in vitro by inducing expression of myostatin: a possible mechanism for reestablishing satellite cell quiescence in vivo. American Journal of Physiology-Cell Physiology. 2010;298:C465–C476. doi: 10.1152/ajpcell.00449.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Huang Z, Chen X, Yu B, He J, Chen D. MicroRNA-27a promotes myoblast proliferation by targeting myostatin. Biochem Biophys Res Commun. 2012;423:265–269. doi: 10.1016/j.bbrc.2012.05.106. [DOI] [PubMed] [Google Scholar]
- [20].Garikipati DK, Rodgers BD. Myostatin stimulates myosatellite cell differentiation in a novel model system: evidence for gene subfunctionalization. Am J Physiol Regul Integr Comp Physiol. 2012;302:R1059–1066. doi: 10.1152/ajpregu.00523.2011. [DOI] [PubMed] [Google Scholar]
- [21].Amthor H, Otto A, Vulin A, Rochat A, Dumonceaux J, Garcia L, Mouisel E, Hourde C, Macharia R, Friedrichs M, Relaix F, Zammit PS, Matsakas A, Patel K, Partridge T. Muscle hypertrophy driven by myostatin blockade does not require stem/precursor-cell activity. Proc Natl Acad Sci U S A. 2009;106:7479–7484. doi: 10.1073/pnas.0811129106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Rachagani S, Cheng Y, Reecy JM. Myostatin genotype regulates muscle-specific miRNA expression in mouse pectoralis muscle. BMC Res Notes. 2010;3:297. doi: 10.1186/1756-0500-3-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Waddell JN, Zhang PJ, Wen YF, Gupta SK, Yevtodiyenko A, Schmidt JV, Bidwell CA, Kumar A, Kuang SH. Dlk1 Is Necessary for Proper Skeletal Muscle Development and Regeneration. Plos One. 2010;5 doi: 10.1371/journal.pone.0015055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Kim YS, Bobbili NK, Paek KS, Jin HJ. Production of a monoclonal anti-myostatin antibody and the effects of in ovo administration of the antibody on posthatch broiler growth and muscle mass. Poult Sci. 2006;85:1062–1071. doi: 10.1093/ps/85.6.1062. [DOI] [PubMed] [Google Scholar]
- [25].Kim YS, Kim KH, Kim CJ. Production of bioactive extracellular domain of pig and chicken activin type IIB receptors in Pichiapastoris. Process Biochemistry. 2012;47:139–146. [Google Scholar]
- [26].Shi H, Scheffler JM, Pleitner JM, Zeng C, Park S, Hannon KM, Grant AL, Gerrard DE. Modulation of skeletal muscle fiber type by mitogen-activated protein kinase signaling. The FASEB Journal. 2008;22:2990–3000. doi: 10.1096/fj.07-097600. [DOI] [PubMed] [Google Scholar]
- [27].Larsson L, Edstrom L. Effects of Age on Enzyme-Histochemical Fiber Spectra and Contractile Properties of Fast-Twitch and Slow-Twitch Skeletal-Muscles in the Rat. Journal of the Neurological Sciences. 1986;76:69–89. doi: 10.1016/0022-510x(86)90143-7. [DOI] [PubMed] [Google Scholar]
- [28].Kalhovde JM, Jerkovic R, Sefland I, Cordonnier C, Calabria E, Schiaffino S, Lomo T. ‘Fast’ and ‘slow’ muscle fibres in hindlimb muscles of adult rats regenerate from intrinsically different satellite cells. Journal of Physiology-London. 2005;562:847–857. doi: 10.1113/jphysiol.2004.073684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Negredo P, Rivero JLL, Gonzalez B, Ramon-Cueto A, Manso R. Slow- and fast-twitch rat hind limb skeletal muscle phenotypes 8 months after spinal cord transection and olfactory ensheathing glia transplantation. Journal of Physiology-London. 2008;586:2593–2610. doi: 10.1113/jphysiol.2007.149120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Manzano R, Toivonen JM, Calvo AC, Miana-Mena FJ, Zaragoza P, Munoz MJ, Montarras D, Osta R. Sex, Fiber-Type, and Age Dependent In Vitro Proliferation Of Mouse Muscle Satellite Cells. Journal of Cellular Biochemistry. 2011;112:2825–2836. doi: 10.1002/jcb.23197. [DOI] [PubMed] [Google Scholar]
- [31].Liu WY, Liu YQ, Lai XS, Kuang SH. Intramuscular adipose is derived from a non-Pax3 lineage and required for efficient regeneration of skeletal muscles. Developmental Biology. 2012;361:27–38. doi: 10.1016/j.ydbio.2011.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Carlson CJ, Booth FW, Gordon SE. Skeletal muscle myostatin mRNA expression is fiber-type specific and increases dining hindlimb unloading. American Journal of Physiology-Regulatory Integrative and Comparative Physiology. 1999;277:R601–R606. doi: 10.1152/ajpregu.1999.277.2.r601. [DOI] [PubMed] [Google Scholar]
- [33].Chanaud CM, Pratt CA, Loeb GE. Functionally complex muscles of the cat hindlimb. V. The roles of histochemical fiber-type regionalization and mechanical heterogeneity in differential muscle activation. Exp Brain Res. 1991;85:300–313. doi: 10.1007/BF00229408. [DOI] [PubMed] [Google Scholar]
- [34].Wang LC, Kernell D. Recovery of type I fiber regionalization in gastrocnemius medialis of the rat after reinnervation along original and foreign paths, with and without muscle rotation. Neuroscience. 2002;114:629–640. doi: 10.1016/s0306-4522(02)00315-9. [DOI] [PubMed] [Google Scholar]
- [35].Dahiya S, Bhatnagar S, Hindi SM, Jiang C, Paul PK, Kuang S, Kumar A. Elevated levels of active matrix metalloproteinase-9 cause hypertrophy in skeletal muscle of normal and dystrophin-deficient mdx mice. Hum Mol Genet. 2011;20:4345–4359. doi: 10.1093/hmg/ddr362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Matsakas A, Otto A, Elashry MI, Brown SC, Patel K. Altered Primary and Secondary Myogenesis in the Myostatin-Null Mouse. Rejuvenation Research. 2010;13:717–727. doi: 10.1089/rej.2010.1065. [DOI] [PubMed] [Google Scholar]
- [37].Edstrom L, Grimby L. Effect of exercise on the motor unit. Muscle Nerve. 1986;9:104–126. doi: 10.1002/mus.880090203. [DOI] [PubMed] [Google Scholar]
- [38].Hwang YH, Kim GD, Jeong JY, Hur SJ, Joo ST. The relationship between muscle fiber characteristics and meat quality traits of highly marbled Hanwoo (Korean native cattle) steers. Meat Science. 2010;86:456–461. doi: 10.1016/j.meatsci.2010.05.034. [DOI] [PubMed] [Google Scholar]