Abstract
Herein we describe the utility of chaperone probes and a bead-based signal enhancement strategy for the analysis of full length messenger RNA transcripts using arrays of silicon photonic microring resonators. Changes in the local refractive index near microring sensors associated with biomolecular binding events are transduced as a shift in the resonant wavelength supported by the cavity, enabling the sensitive analysis of numerous analytes of interest. We employ the sensing platform for both the direct and bead-enhanced detection of three different mRNA transcripts, achieving a dynamic range spanning over four orders of magnitude, and demonstrating expression profiling capabilities in total RNA extracts from the HL-60 cell line. Small, dual-use DNA chaperone molecules were developed and found to both enhance the binding kinetics of mRNA transcripts by disrupting complex secondary structure and serve as sequence-specific linkers for subsequent bead amplification. Importantly, this approach does not require amplification of the mRNA transcript, thereby allowing for simplified analyses that do not require expensive enzymatic reagents or temperature ramping capabilities associated with RT-PCR-based methods.
Keywords: mRNA, sub-micron beads, silicon photonics, microring resonator, HL-60, biosensing
INTRODUCTION
The concept of personalized medical diagnostics is based upon the premise that panels of biomarkers can predict disease progression or otherwise determine an individual patient’s most effective treatment regimen.1–4 Advances in laboratory-based, high throughput measurement technologies have greatly advanced our understanding of the molecular basis of disease at a systems level, revealing a myriad of potential biomarkers at the level of DNA, RNA, proteins, and metabolites.5–7 through the interrogation of numerous classes of biomarkers, thus analysis techniques which are amenable to studying multiple classes of biomarkers have a distinct advantage over those limited to a specific class. Given this, recent biosensor efforts have been aimed at creating platforms that are scalable, cost effective, and flexible towards the detection of multiple marker classes, as the increase in measured biological information content should translate into more molecularly insightful diagnostic capabilities.
Due to their key position at the intersection between the genome and proteome, messenger RNA (mRNA) transcripts are particularly informative biomarkers in research and medical diagnostic applications.8 mRNA abundance has been used extensively to study disease classification,9,10 gene function,11 regulatory interactions,12 and therapeutic efficacy,13–16 among other applications. However, the naturally low abundance of mRNA, as well as challenges associated with their size and extensive secondary structure, present complications to many hybridization-based detection methodologies.17
Since the original extension of polymerase chain reaction (PCR) to RNA in the form of reverse transcription polymerase chain reaction (RT-PCR),18 numerous transduction methods have been developed to identify and quantitate the resultant amplicons including microarrays,19 fluorescent reporters,20 and sequencing.21 More recently other optical22,23 and electrochemical24–27 techniques have been successfully applied to the analysis of in vitro transcribed mRNA and mRNA isolated from cell lines. While these approaches to mRNA analysis provide robust analytical capabilities and high sensitivity, further improvements, particularly in multiplexing capability, time-to-result, and potential amenability to the simultaneous analysis of multiple classes of biomarkers, would be of potential value to the clinical diagnostic community.
Silicon photonic microring resonators have emerged as a promising technology for biomolecular analysis. These sensors are sensitive to binding-induced changes in the local refractive index and, after functionalization with appropriate capture agents, have been used to quantitate a range of biomolecular analytes including DNA,28 miRNA,29,30 proteins,31–40 bacteria,41 and whole virus particles.42 In this work, we apply this technology to the detection of three full length mRNA transcripts—c-myc, β-actin, and IL-8. These mRNAs serve as potentially useful diagnostic or theragnostic markers based on their involvement and abnormal expression in numerous oncological processes.43–46 mRNA analysis was performed in a multiplexed format to profile expression changes in HL-60 cells upon differentiation. Importantly, we utilized an signal enhancement approach combining short DNA chaperones and sub-micron beads that dramatically increased the assay sensitivity by removing the secondary structure of the mRNA and also increasing the refractive index change associated with the each target strand bound to the sensor. The end result is a quantitative, multiplexed, and PCR-free assay to analyze full length mRNA with a limit of detection of 512 amol.
EXPERIMENTAL
Materials
PBS buffer was reconstituted with deionized water from Dulbecco’s Phosphate Buffered Saline packets purchased from Sigma-Aldrich (St. Louis, MO), and the pH adjusted to 7.4(PBS-7.4) or 6.0 (PBS-6). A PBS + 0.5% Tween buffer (PBST) was created by the addition of Tween-20 to PBS-7.4. A high stringency hybridization buffer consisting of 30% formamide, 0.2% sodium dodecyl sulfate, 4X saline-sodium phosphate-EDTA buffer (SSPE, USB Corp.), and 3X Denhardt’s Solution (Invitrogen) was used for all hybridization steps. Bio-Streptavidin Plus 114 nm magnetic beads were obtained from Ademtech and buffer exchanged in PBST in 3 kDA MWCO Vivaspin columns (Sartorius) prior to use to remove free streptavidin from solution. Starting Block was obtained from Thermo-Scientific and used to prevent nonspecific fouling of streptavidin-coated beads. The silane 3-N-((6-(N′-Isopropylidene-hydrazino))nicotinamide)propyltriethyoxysilane (HyNic Silane) and succinimidyl-4-formylbenzamide (S-4FB) were purchased from Solulink. All other reagents were purchased from Fisher, unless otherwise noted, and used as received.
Sensor Arrays, Read-Out Mechanism, and Instrumentation
Sensor chips and read-out instrumentation were obtained from Genalyte, Inc (San Diego, CA), and have been described previously.40,47 Sensor chips were fabricated at a silicon foundry on 8″ silicon-on-insulator wafers using deep UV photolithography and dry etch methods, spin-coated with a fluoropolymer cladding layer, and diced into individual 6 × 6 mm chips, each having an array of 32 individually addressable microrings. The fluoropolymer cladding is selectively removed from 24 of the rings leaving these exposed to the solution and responsive to binding events. The eight occluded rings serve as control elements for subtracting thermal drift. Chips were fitted with a laser etched Mylar gasket, which defined flow chambers when sandwiched with a Teflon lid, and loaded into the read-out instrumentation. All experiments were performed at a flow rate of 20 μL/min unless noted otherwise.
As reported previously,47 resonant wavelengths for each microring were determined by coupling a tunable laser source (centered at 1550 nm) into an adjacent linear waveguide via on-chip grating couplers. The laser output was then swept through and appropriate spectral window and the light intensity in the linear waveguide past the microring was used to determine the resonance wavelength. This process was then serially repeated for each ring in the array and the resultant shifts in resonance as a function of time are recorded.
The resonance condition is given by:
where m equals a nonzero integer, λ is the wavelength of propagating light, r is the microring radius, and neff is the effective refractive index of the local microring environment. Therefore, the binding of higher refractive index biomolecules and accompanying displacement of water results in a resonance shift to longer wavelengths—a positive shift that is listed in units of Δ picometers (Δpm).
Nucleic Acid Sequences
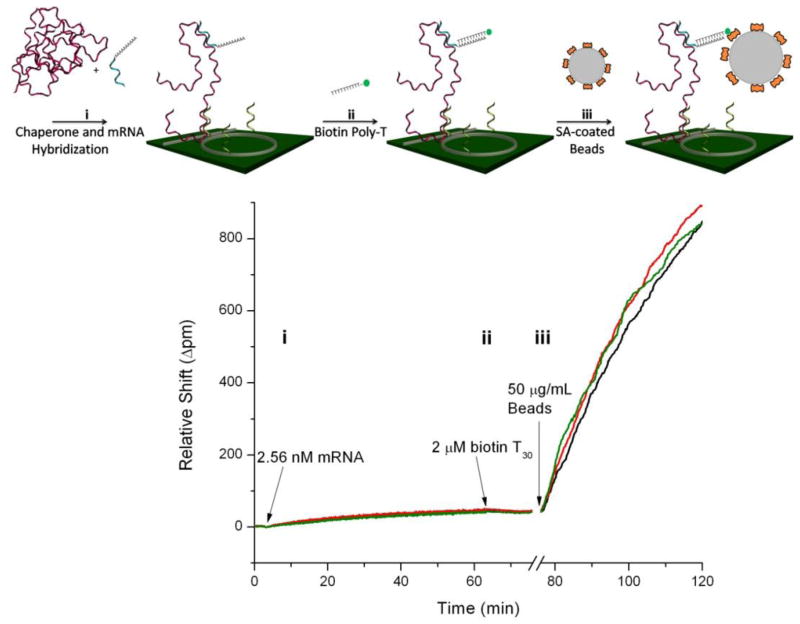
All synthetic nucleic acid sequences were obtained from Integrated DNA Technologies (Coralville, IA). Three kinds of synthetic oligonucleotides were used in this work. Single stranded DNA (ssDNA) capture probes, 5′ amine terminated for covalent surface immobilization, were designed to target specific mRNA target regions having minimal secondary structure so that hybridization would link the molecule to the biosensor surface. To enable conjugation to the sensor surface, capture probes were reacted with a 10-fold molar excess of S-4FB in a 1:1 solution of DMSO:H2O for two hours, followed by buffer exchange in 3kDa MWCO Vivaspin columns (Sartorius) to remove unreacted S-4FB. DNA chaperones were designed with two functional regions: the first complementary to regions immediately adjacent to capture probe binding epitopes to disrupt target mRNA secondary structure, and a second polyA region to serve as a linker for subsequent bead recognition. Thirdly, poly(T) linkers with a biotin moiety were employed to link the chaperone-primed, surface immobilized mRNA targets with streptavidin coated beads. For clarity, this sequential molecular linkage is illustrated in Figure 3a. All DNA was resuspended in PBST and buffer exchanged prior to use.
Figure 3.
(a) Schematic of the mRNA assay, in which target mRNA is first annealed and hybridized to short ‘chaperone’ DNA molecules prior to hybridization to surface immobilized DNA capture probes (i). Following the binding of biotinylated T30 linker strands (ii) and a blocking step, streptavidin coated beads are introduced and bind to the biotin activated mRNA (iii), resulting in significant signal enhancement. (b) Response curves for three microring resonators illustrate each step of the previous schematic, demonstrating the significant signal enhancement capabilities conferred by bead based amplification. Note the protein blocking step at ~76 minutes, as well as the bulk refractive index shift due to changing buffers between direct and bead-based detection, is omitted for clarity.
mRNA was synthesized via in vitro transcription using a Promega T7 RiboMAX Express Large Scale RNA Production System and Origene TrueClone cDNA clones following the manufacturers recommended procedures. A Qiagen RNeasy MinElute cleanup kit was used for subsequent mRNA purification and final mRNA quality was assessed via 1% agarose gels. The sequences of all synthetic nucleic acids used in this work are listed in the Supplementary Info Table #1–3; primary mRNA sequences can be found online at the National Center for Biotechnology Information48.
Biochemical Modification of the Silicon Photonic Microring Resonator Surface
Prior to chemical modification, sensor chips were first immersed in a piranha solution (3:1 H2SO4:30% H2O2) for 30 seconds to clean the surface (Caution! Piranha solutions are extremely dangerous and react explosively with trace amounts of organics). Subsequently a 1 mg/mL solution of HyNic Silane in ethanol was applied to the surface for 20 minutes to activate the surface towards S-4FB modified DNA capture probes. Following a 7 minute sonication rinse in ethanol, chips were dried under a stream of N2 and manually spotted in a spatially controlled manner with ~10 μM solution of 4FB modified DNA capture probes in PBS and incubated overnight to covalently modify the surface. Immediately prior to an experiment, chips were sonicated in 8 M Urea for 7 minutes and rinsed in deionized water to remove physisorbed DNA capture probes.
mRNA Analysis and Nanoparticle Amplification
In vitro transcribed mRNA was first incubated with a 5-fold excess of polyadenylated DNA chaperones in hybridization buffer at 95 °C for 3 minutes, followed by 30 minutes at room temperature. The 200 μL mRNA sample was then introduced to the sensor chip and recirculated for 60 minutes and the binding response was monitored as a shift in microring resonance wavelength. Following a 5 minute PBST rinse, a 2 μM biotinylated T30 linker solution in PBST was hybridized to the polyA sequence on the surface immobilized chaperone-mRNA complex in preparation for binding of streptavidin coated beads. After surface blocking for 10 minutes in Starting Block to prevent non-specific bead binding, a 50 μg/mL bead solution in PBST was introduced at 10 μL/min and the binding response arising from the streptavidin-biotin interaction was monitored. Importantly, beads were buffer exchanged twice immediately prior to use to remove free streptavidin from solution, as this can out-compete bead immobilized streptavidin for surface binding sites due to its more rapid diffusion rate.
Data Analysis
All data was analyzed in Origin Pro 8. Direct mRNA hybridization was quantitated by recording the resonance shift after 60 minutes; the bead-amplified signal was recorded at 45 minutes. Both of these times reflect pseudo-equilibrium points and were chosen with respect to overall assay time and quantitation considerations. Control rings functionalized with non-complementary DNA were subtracted from direct mRNA hybridization signals to account for temperature fluctuations during the experiment. Calibration data was fit to a logistic function,
where A1 is the initial value limit, A2 is the final value limit, c is the center of the fit, and p is the power of the fit.
Cell Culture and Total RNA Analysis
The promyelocytic HL-60 cell line was purchased from ATCC (CCL-240) and propagated in accordance with ATCC recommended protocols. Cells were cultured in RPMI media + 10% heat inactivated FBS + Penicillin and Streptomycin and passaged every 2–3 days to maintain the cell density between 0.13–1×107 cells/mL. Cells were differentiated by the addition of DMSO to a final concentration of 1.3%, and subsequently incubated for 7 days in these conditions prior to RNA extraction. Total RNA extracted with Qiagen RNeasy Mini kits was assessed for purity and quantity using a Thermo-Fisher Nanodrop UV-Vis spectrometer, and stored at −80 C until further use. Total RNA samples were analyzed identically to in vitro mRNA studies, excepting the addition of total RNA extracts instead of in vitro transcribed mRNA.
RESULTS and DISCUSSION
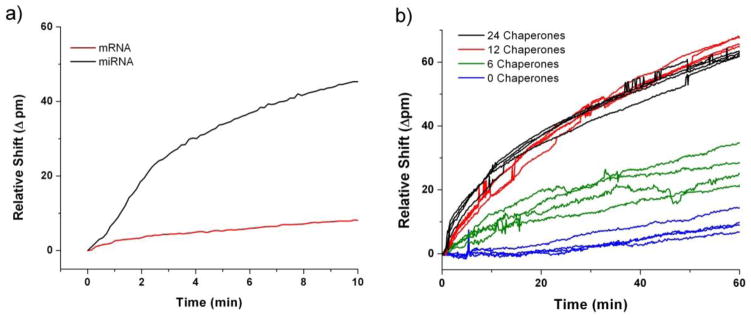
Full length mRNA transcripts average 2200 ribonucleotides in length and can pose several significant challenges with respect to their analysis when compared to smaller fragments (ex. miRNA). Specifically, full length mRNA transcripts often feature extensive secondary structure that can hinder hybridization to surface immobilized capture probes. That is to say that targeted binding regions of the molecule can be effectively screened from interacting with surface bound capture probes by ‘spectator’ regions of the transcript.49 Diffusion of mRNA is also considerably slower than for smaller RNA molecules, necessitating longer assay times to achieve comparable sensor response.50 Finally, the larger size of the mRNA molecule increases the probability that its orientation, with respect to a given surface immobilized capture probe, will not be optimal for hybridization. Combined, these effects conspire to reduce mRNA detection efficiency and kinetics. Figure 1a illustrates this point by comparing the hybridization binding curves of a 22 nt miRNA to a full-length, 1820 nt mRNA at the identical concentration. As shown, the mRNA hybridization response is dramatically slower even though the concentration of target is identical to that of the miRNA.
Figure 1.
(a) Real-time shifts in microring resonances accompanying target hybridization highlight that despite over an 80-fold increase in size, full length mRNA elicits a much lower sensor response than smaller miRNA molecules based on steric inhibition and diffusion limitations. (b) Small DNA chaperones were pre-hybridized to a 2.56 nM concentration of myc mRNA before being flowed across an array of ssDNA-presenting microring resonators. The chaperone/mRNA mixture was introduced to the sensor array at t=0 minutes, and the resultant responses demonstrate the enhanced rate of target binding in the presence of chaperones.
To address this challenge, several methods were investigated to enhance the sensor response from primary mRNA binding events. In all cases, mRNA targets were annealed at 95° C for 3 minutes prior to other treatment or detection methodologies to denature secondary structure. To improve the accessibility of capture agents to complementary epitopes on the mRNA target, small DNA helper strands, or ‘chaperones’, were introduced to unravel portions of the secondary structure and enhance binding kinetics. These 20-nucleotide long chaperones were designed to hybridize starting immediately adjacent to the mRNA epitope targeted by capture agents, and extending with each successive chaperone away from the binding epitope. This creates a localized duplex region with reduced secondary structure, and thus hopefully improving target accessibility. A total of 6, 12, or 24 unique chaperone sequences were designed to be adjacent to the three regions targeted by the myc DNA capture probes. These chaperone sequences were incubated all together with the full length mRNA and their resultant hybridization therefore resulted in 20, 40, or 80 base pair double stranded regions flanking both sides of the binding epitope, effectively linearizing the regions required for binding to the surface immobilized capture probes.
Figure 1b shows that an enhanced response is observed with increasing chaperone number up to 12 chaperones added simultaneously, at which point additional chaperones have a minimal effect on binding kinetics. Given the close surface proximity and 3′ to 5′ orientation of the final surface immobilized mRNA, additional experiments were performed containing chaperones on only the 3′ or 5′ side of the epitope to confirm that the formation of a rigid, double stranded region immediately adjacent to the surface did not produce unfavorable steric conditions for mRNA binding. No difference in sensor response was observed for either configuration (data not shown). Consequently, all analyses were run using 12 mRNA-specific chaperones targeting both flanking regions directly adjacent to mRNA epitope targeted by the capture probe.
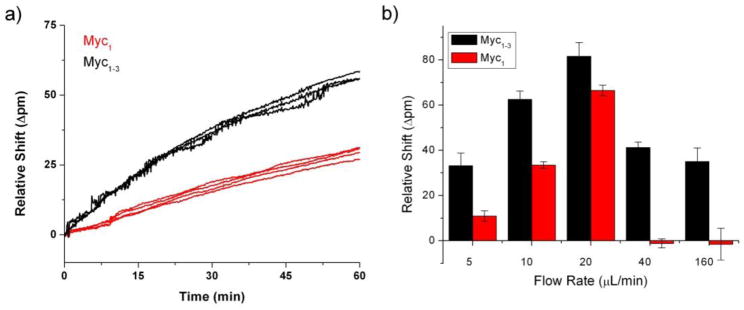
To improve the likelihood that an encounter with the surface would result in hybridization, we increased the number of capture probes targeting each mRNA in hopes of enhancing the probability of a hybridization event occurring upon every target-surface interaction. Capture probes were selected to target unique identifier regions of the primary sequence as determined by BLAST searches to successfully eliminate cross-reactivity amongst other mRNA species, as shown in Figure S-1. Additionally, regions which showed conserved regions of minimal secondary structure based on Mfold calculations were preferentially targeted.51 As shown in Figure 2a for myc mRNA, using three ssDNA capture probes (myc1–3), as opposed to only one (myc1), enhanced the signal by a factor of two after 60 minutes as compared to a single capture agent.
Figure 2.
(a) Increasing the number of DNA capture agents from one to three probes significantly enhanced the kinetics of mRNA capture. Shown here is 2.56 nM myc mRNA hybridizing to either myc1 capture probe or an equimolar mixture of three myc capture probes (myc1–3). (b) Flow rates ranging from 5 to 160 μL/min were tested and the net response was quantitated at 60 minutes. The signals increased for both the single and multi-capture probe sensors up until the maximum response at 20 μL/min. Further increases in flow rate led to the complete loss of hybridization for the single probe-functionalized rings while the multi-probe rings retained binding capabilities. Note the non-linear x-axis.
This finding was expected given the higher probability of accessing and hybridizing to the target region using multiple capture agents. Additionally, the possibility exists for multiple binding interactions with a single target, resulting in an increase in binding strength over time as initial capture with a single capture probe can transition to a multivalent interaction. A concomitant reduction in the overall desorption constant would also enhance signal response. Given these improvements, three capture agents were used for all further experimentation.
Further support for multivalent interactions was found as we evaluated the optimal flow rate for mRNA hybridization experiments on both myc1 and myc1–3 functionalized sensors. As shown in Figure 2b, enhanced target binding was observed up to flow rates of 20 μL/min for both single and multiple capture probe-modified sensors. However, at higher flow rates the hybridization response, which was recorded at a defined time point of 60 minutes of hybridization, was negligible for the myc1 sensors, while the myc1–3 microrings still showed significant hybridization response. In general, faster flow rates are useful in biosensor systems, as mass transport is less of a limitation under this scenario, up until the point that the target dwell time competes with the period needed to form a stable interaction with the capture agent. In this case, the reduced rate of target desorption achieved by multivalent surface interactions help to preserve a significant hybridization response. Nonetheless, the hybridization response was still found to be maximized at 20 μL/min and this flow rate was used subsequently for all detection experiments.
Another challenge associated with mRNA analysis is the naturally low abundance of many transcripts, which can be over 1000 times less abundant than miRNAs, for example. This necessitates further sensitivity enhancements to enable comprehensive mRNA expression profiling in relevant samples and biological systems. As no further binding efficiency optimization could circumvent the limitations of direct, label-free detection, we decided to pursue signal-enhancement methodology by modifying the DNA chaperones to accommodate subsequent recognition by a large refractive index label (~114 nm magnetic bead, Ademtech). Importantly, this approach does not require PCR, thereby eliminating the need for enzymatic reagents, variable temperature control, and avoiding potential sequence biases during amplification. Finally, the lack of reliance upon PCR leaves this approach potentially amenable to multi-class biomarker analysis (RNA and proteins).
Based upon the previously optimized conditions for disrupting mRNA secondary structure and improving surface hybridization, we developed an assay to detect full length mRNAs from both buffer and isolated total RNA from cell lysate. This multi-step assay is schematically illustrated in Figure 3a, with representative sensor responses in Figure 3b.
An initial thermal denaturation step is performed to allow polyadenylated DNA chaperones to hybridize to the messenger RNA target, thus reducing secondary structure, while also incorporating recognition motifs for subsequent bead binding. The mRNA sample is then flowed across a sensor array presenting microring sensors previously modified with ssDNA capture probes and hybridization is monitored as a function of time as a shift in the resonant wavelength of the microring resonators. To further increase the signal resulting from mRNA hybridization, a biotinylated T30 linker and protein blocking solution are sequentially introduced, followed by exposure to streptavidin-coated beads. Bead binding greatly increases the refractive index change near the sensor surface and therefore offers a further method of signal enhancement.
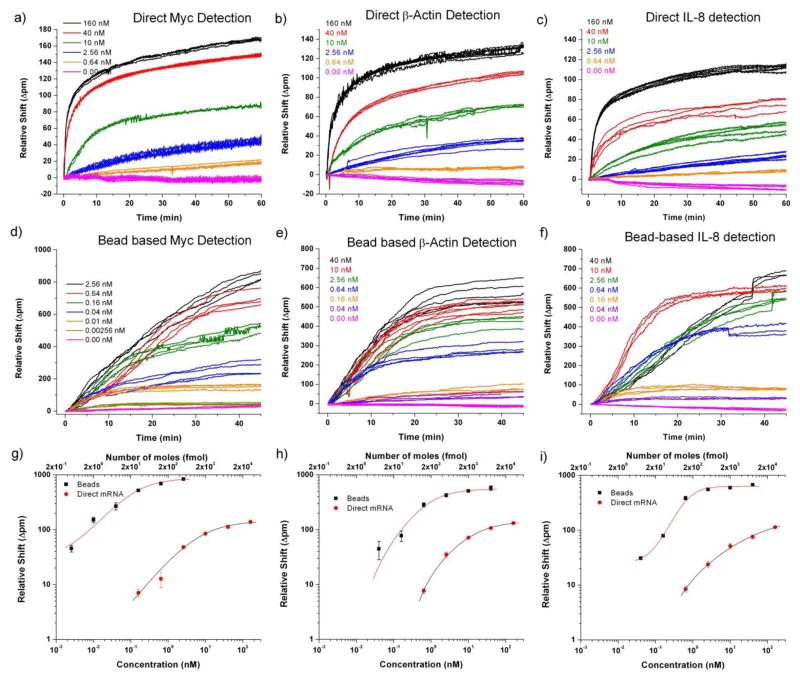
Using the chaperone-assisted detection methodology described above, we were able to quantitate three mRNA targets over a 2- to 3-order of magnitude concentration range and achieve a 32 fmol detection limit. Pursuing the bead-based amplification strategy detailed in the latter part of Figure 3, we demonstrated a 16- to 64-fold increase in sensitivity over direct, label-free detection with the total dynamic range exceeding 4 orders of magnitude. Figure 4 summarizes the quantitative capabilities afforded by this joint methodology utilizing both direct and bead-enhanced detection. All assays were performed followed the procedures described previously in the Experimental section. A single mRNA concentration was assayed on each chip, with chips subsequently regenerated and re-functionalized for the next measurement. Consequently calibration plots were made across multiple chips, demonstrating high reproducibility and minimal chip-to-chip variability.
Figure 4.
The concentration-dependent sensor response for direct (a–c) and bead-enhanced (d–f) mRNA detection. In each trace sample or beads was injected at t=0 minutes. Direct hybridization responses (a–c) were quantitated at 60 minutes and bead-enhanced responses (d–f) at 45 minutes. These pseudo-equilibrium time points were as a balance between minimizing total assay time while still maintaining quantitation capabilities. Calibration plots (g–i) demonstrate the full 3–4 order of magnitude dynamic range achieved combining direct and bead-based detection.
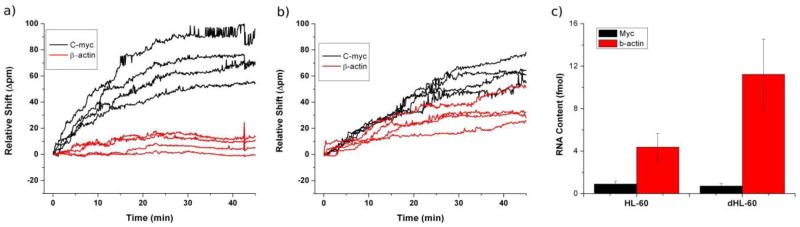
To demonstrate the amenability of this assay to a more challenging matrix, we performed expression profiling of the three mRNA transcripts in total RNA extracted from HL-60 cells using the same procedures optimized for studies in buffer. Following standard protocols, we differentiated HL-60 cells with 1.3% DMSO and harvested total RNA after 7 days, in tandem with RNA extraction from undifferentiated cells.52 Twenty micrograms of total RNA was analyzed on a sensor chip functionalized towards all three mRNAs of interest. As shown in Figure 5, myc expression decrease upon differentiation by a very small amount while β-actin abundance increased by a factor of ~3 accompanying visual changes in the cellular morphology associated with the differentiation process. Literature reports corroborate these changes in the expression levels of both myc and β-actin mRNA upon differentiation,53,54 albeit in a qualitative manner. We attribute any remaining discrepancies in expression levels not to the measurement process, but rather to the incomplete cellular differentiation achieved in this study (See Supplementary Figure 3 and Table 4 for Flow Cytometry data).
Figure 5.
Microring resonators addressed with 20 μg of total RNA harvested from either (a) proliferating or (b) differentiated HL-60 cells show differing responses reflective of the molecular changes accompanying the phenotypic change. (c) Using the concentration calibrations to quantitate these responses, we observed the expected increase in β-actin production upon cell differentiation, and a marginal decrease in myc expression upon differentiation. This residual myc response is explained by the incomplete differentiation of HL-60 cells.
We were unable to accurately quantitate IL-8 mRNA transcripts due to their lower abundance, demonstrating the need for further sensitivity increases in future work. Further improvements in assay performance are still required before this assay is competitive with conventional PCR-based assays in terms of sensitivity; however we feel that the insights communicated herein may be important to related surface hybridization-based detection methods such as surface plasmon resonance or microcantilevers, and establish this platform’s potential for full-length mRNA detection. Additionally, numerous enzymatic signal amplification methodologies could be readily modified to augment the methods developed in this work, including horseradish peroxidase55, duplex specific nuclease56, and RNA-assisted Klenow enzyme57.
In this work we have shown the quantitation of whole mRNA in a multiplexed format, achieving a dynamic range over four orders of magnitude and detecting as little as 512 amol of mRNA. This demonstration is significant because it enables analysis of whole mRNA transcripts, without requiring either fragmentation of mRNA or PCR. The lack of reliance upon PCR, or any other class-specific detection strategy, seems particularly compelling as the microring resonator platform has been previously applied to the separate detection of DNA, miRNA, and proteins. Although admittedly a long term vision still requiring many advances in terms of enhanced sensitivity and sample handling, we feel that the realization of an integrated platform for simultaneously profiling biomarkers from multiple, distinct classes of biomolecules is a compelling aspiration.
CONCLUSION
mRNAs represent an important class of potential biomarkers for understanding disease onset and progression, but their analysis using surface hybridization-based biosensors can be complicated by the size and secondary structure of the full length transcript. In this work we have developed methodologies to enable multiplexed mRNA profiling using arrays of silicon photonic microring resonators, and demonstrated mRNA expression profiling capabilities in total RNA samples. Importantly, we found that the addition of short, secondary structure-disrupting chaperones and the use of multiple, co-localized capture probes significantly improved the hybridization kinetics of full length mRNA molecules. Furthermore, we showed that these chaperones can be designed so that a subsequent bead recognition step significantly enhances the observed resonance shift allows quantitation over a broad dynamic range. Future efforts will focus on further improving methodological measurement sensitivity, enabling the analysis of lower abundance transcripts and simultaneously reducing the amount of sample input required. These capabilities, in conjunction with previous demonstrations of the detection of DNA, miRNA, and protein biomarkers suggest that chip-integrated, multi-class biomarker analysis might eventually be achievable using this scalable analytical platform.
Supplementary Material
Acknowledgments
The authors gratefully acknowledge financial support from the National Institutes of Health (NIH) Director’s New Innovator Award Program, part of the NIH Roadmap for Medical Research, through grant number 1-DP2-OD002190-01 and. JTK was supported by a National Science Foundation Graduate Research Fellowship. This material is based upon work supported by the National Science Foundation Graduate Research Fellowship under Grant No. DGE 07-15088-FLW. RCB was a Beckman Fellow of the Center for Advanced Study at the University of Illinois at Urbana-Champaign and is a research fellow of the Alfred P. Sloan Foundation.
References
- 1.Yu X, Schneiderhan-Marra N, Joos TO. Clin Chem. 2010;56:376. doi: 10.1373/clinchem.2009.137158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Robin X, Turck N, Hainard A, Lisacek F, Sanchez JC, Muller M. Expert Rev Proteomics. 2009;6:675. doi: 10.1586/epr.09.83. [DOI] [PubMed] [Google Scholar]
- 3.Ginsburg GS, Haga SB. Expert Rev Mol Diagn. 2006;6:179. doi: 10.1586/14737159.6.2.179. [DOI] [PubMed] [Google Scholar]
- 4.McKee AE, Silver PA. Cell Res. 2007;17:581. doi: 10.1038/cr.2007.54. [DOI] [PubMed] [Google Scholar]
- 5.Ideker T, Galitski T, Hood L. Annu Rev Genom Hum G. 2001;2:343. doi: 10.1146/annurev.genom.2.1.343. [DOI] [PubMed] [Google Scholar]
- 6.Weston AD, Hood L. J Proteome Res. 2004;3:179. doi: 10.1021/pr0499693. [DOI] [PubMed] [Google Scholar]
- 7.Kitano H. Science. 2002;295:1662. doi: 10.1126/science.1069492. [DOI] [PubMed] [Google Scholar]
- 8.Sunde RA. J Nutr Biochem. 2010;21:665. doi: 10.1016/j.jnutbio.2009.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chung CH, Bernard PS, Perou CM. Nat Genet. 2002;32:533. doi: 10.1038/ng1038. [DOI] [PubMed] [Google Scholar]
- 10.Ramaswamy S, Tamayo P, Rifkin R, Mukherjee S, Yeang CH, Angelo M, Ladd C, Reich M, Latulippe E, Mesirov JP, Poggio T, Gerald W, Loda M, Lander ES, Golub TR. P Natl Acad Sci USA. 2001;98:15149. doi: 10.1073/pnas.211566398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giaever G, Chu AM, Ni L, Connelly C, Riles L, Veronneau S, Dow S, Lucau-Danila A, Anderson K, Andre B, Arkin AP, Astromoff A, El-Bakkoury M, Bangham R, Benito R, Brachat S, Campanaro S, Curtiss M, Davis K, Deutschbauer A, Entian KD, Flaherty P, Foury F, Garfinkel DJ, Gerstein M, Gotte D, Guldener U, Hegemann JH, Hempel S, Herman Z, Jaramillo DF, Kelly DE, Kelly SL, Kotter P, LaBonte D, Lamb DC, Lan N, Liang H, Liao H, Liu L, Luo C, Lussier M, Mao R, Menard P, Ooi SL, Revuelta JL, Roberts CJ, Rose M, Ross-Macdonald P, Scherens B, Schimmack G, Shafer B, Shoemaker DD, Sookhai-Mahadeo S, Storms RK, Strathern JN, Valle G, Voet M, Volckaert G, Wang CY, Ward TR, Wilhelmy J, Winzeler EA, Yang Y, Yen G, Youngman E, Yu K, Bussey H, Boeke JD, Snyder M, Philippsen P, Davis RW, Johnston M. Nature. 2002;418:387. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- 12.Gardner TS, di Bernardo D, Lorenz D, Collins JJ. Science. 2003;301:102. doi: 10.1126/science.1081900. [DOI] [PubMed] [Google Scholar]
- 13.Bollag G, Hirth P, Tsai J, Zhang J, Ibrahim PN, Cho H, Spevak W, Zhang C, Zhang Y, Habets G, Burton EA, Wong B, Tsang G, West BL, Powell B, Shellooe R, Marimuthu A, Nguyen H, Zhang KY, Artis DR, Schlessinger J, Su F, Higgins B, Iyer R, D’Andrea K, Koehler A, Stumm M, Lin PS, Lee RJ, Grippo J, Puzanov I, Kim KB, Ribas A, McArthur GA, Sosman JA, Chapman PB, Flaherty KT, Xu X, Nathanson KL, Nolop K. Nature. 2010;467:596. doi: 10.1038/nature09454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Druker BJ, Lydon NB. J Clin Invest. 2000;105:3. doi: 10.1172/JCI9083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mdivani N, Li H, Akhalaia M, Gegia M, Goginashvili L, Kernodle DS, Khechinashvili G, Tang YW. Clin Chem. 2009;55:1694. doi: 10.1373/clinchem.2009.124396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hudis CA. N Engl J Med. 2007;357:39. doi: 10.1056/NEJMra043186. [DOI] [PubMed] [Google Scholar]
- 17.Carrascosa LG, Gomez-Montes S, Avino A, Nadal A, Pla M, Eritja R, Lechuga LM. Nucleic Acids Research. 2012:40. doi: 10.1093/nar/gkr1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saiki RK, Gelfand DH, Stoffel S, Scharf SJ, Higuchi R, Horn GT, Mullis KB, Erlich HA. Science. 1988;239:487. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- 19.Lockhart DJ, Winzeler EA. Nature. 2000;405:827. doi: 10.1038/35015701. [DOI] [PubMed] [Google Scholar]
- 20.Nolan T, Hands RE, Bustin SA. Nat Protoc. 2006;1:1559. doi: 10.1038/nprot.2006.236. [DOI] [PubMed] [Google Scholar]
- 21.Velculescu VE, Zhang L, Vogelstein B, Kinzler KW. Science. 1995;270:484. doi: 10.1126/science.270.5235.484. [DOI] [PubMed] [Google Scholar]
- 22.Takahashi H, Matsumoto A, Sugiyama S, Kobori T. Anal Biochem. 2010;401:242. doi: 10.1016/j.ab.2010.03.011. [DOI] [PubMed] [Google Scholar]
- 23.Jin-Lee H, Goodrich TT, Corn RM. Anal Chem. 2001;73:5525. doi: 10.1021/ac010762s. [DOI] [PubMed] [Google Scholar]
- 24.Lee AC, Dai Z, Chen B, Wu H, Wang J, Zhang A, Zhang L, Lim TM, Lin Y. Anal Chem. 2008;80:9402. doi: 10.1021/ac801263r. [DOI] [PubMed] [Google Scholar]
- 25.Wu MS, Qian GS, Xu JJ, Chen HY. Anal Chem. 2012;84:5407. doi: 10.1021/ac3009912. [DOI] [PubMed] [Google Scholar]
- 26.Fang Z, Kelley SO. Anal Chem. 2009;81:612. doi: 10.1021/ac801890f. [DOI] [PubMed] [Google Scholar]
- 27.Chen X, Roy S, Peng Y, Gao Z. Anal Chem. 2010;82:5958. doi: 10.1021/ac1003135. [DOI] [PubMed] [Google Scholar]
- 28.Qavi AJ, Mysz TM, Bailey RC. Anal Chem. 2011;83:6827. doi: 10.1021/ac201659p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qavi AJ, Bailey RC. Angew Chem Int Edit. 2010;49:4608. doi: 10.1002/anie.201001712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qavi AJ, Kindt JT, Gleeson MA, Bailey RC. Anal Chem. 2011;83:5949. doi: 10.1021/ac201340s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu DX, Densmore A, Delage A, Waldron P, McKinnon R, Janz S, Lapointe J, Lopinski G, Mischki T, Post E, Cheben P, Schmid JH. Opt Express. 2008;16:15137. doi: 10.1364/oe.16.015137. [DOI] [PubMed] [Google Scholar]
- 32.Claes T, Molera JG, De Vos K, Schacht E, Baets R, Bienstman P. IEEE Photon J. 2009;1:197. [Google Scholar]
- 33.De Vos K, Girones J, Claes T, De Koninck Y, Popelka S, Schacht E, Baets R, Bienstman P. IEEE Photon J. 2009;1:225. [Google Scholar]
- 34.Xu DX, Vachon M, Densmore A, Ma R, Delâge A, Janz S, Lapointe J, Li Y, Lopinski G, Zhang D, Liu QY, Cheben P, Schmid JH. Opt Lett. 2010;35:2771. doi: 10.1364/OL.35.002771. [DOI] [PubMed] [Google Scholar]
- 35.Kirk JT, Fridley GE, Chamberlain JW, Christensen ED, Hochberg M, Ratner DM. Lab on a Chip. 2011;11:1372. doi: 10.1039/c0lc00313a. [DOI] [PubMed] [Google Scholar]
- 36.Luchansky MS, Washburn AL, Martin TA, Iqbal M, Gunn LC, Bailey RC. Biosens Bioelectron. 2010;26:1283. doi: 10.1016/j.bios.2010.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Washburn AL, Gomez J, Bailey RC. Anal Chem. 2011;83:3572. doi: 10.1021/ac200317z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Luchansky MS, Bailey RC. J Am Chem Soc. 2011;133:20500. doi: 10.1021/ja2087618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Washburn AL, Luchansky MS, Bowman AL, Bailey RC. Anal Chem. 2010;82:69. doi: 10.1021/ac902451b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Washburn AL, Gunn LC, Bailey RC. Anal Chem. 2009;81:9499. doi: 10.1021/ac902006p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ramachandran A, Wang S, Clarke J, Ja SJ, Goad D, Wald L, Flood EM, Knobbe E, Hryniewicz JV, Chu ST, Gill D, Chen W, King O, Little BE. Biosensors & Bioelectronics. 2008;23:939. doi: 10.1016/j.bios.2007.09.007. [DOI] [PubMed] [Google Scholar]
- 42.McClellan MS, Domier LL, Bailey RC. Biosensors & Bioelectronics. 2012;31:388. doi: 10.1016/j.bios.2011.10.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bonello TT, Stehn JR, Gunning PW. Future Med Chem. 2009;1:1311. doi: 10.4155/fmc.09.99. [DOI] [PubMed] [Google Scholar]
- 44.Gearhart J, Pashos EE, Prasad MK. N Engl J Med. 2007;357:1469. doi: 10.1056/NEJMp078126. [DOI] [PubMed] [Google Scholar]
- 45.Shahzad A, Knapp M, Lang I, Kohler G. Int Arch Med. 2010;3:11. doi: 10.1186/1755-7682-3-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pelengaris S, Khan M, Evan GI. Cell. 2002;109:321. doi: 10.1016/s0092-8674(02)00738-9. [DOI] [PubMed] [Google Scholar]
- 47.Iqbal M, Gleeson MA, Spaugh B, Tybor F, Gunn WG, Hochberg M, Baehr-Jones T, Bailey RC, Gunn LC. IEEE J Sel Top Quant Electron. 2010;16:654. [Google Scholar]
- 48.Sayers EW, Barrett T, Benson DA, Bolton E, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Federhen S, Feolo M, Fingerman IM, Geer LY, Helmberg W, Kapustin Y, Krasnov S, Landsman D, Lipman DJ, Lu ZY, Madden TL, Madej T, Maglott DR, Marchler-Bauer A, Miller V, Karsch-Mizrachi I, Ostell J, Panchenko A, Phan L, Pruitt KD, Schuler GD, Sequeira E, Sherry ST, Shumway M, Sirotkin K, Slotta D, Souvorov A, Starchenko G, Tatusova TA, Wagner L, Wang YL, Wilbur WJ, Yaschenko E, Ye J. Nucleic Acids Research. 2012;40:D13. doi: 10.1093/nar/gkr1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wei T, Pearson MN, Armstrong K, Blohm D, Liu J. Mol Biosyst. 2012;8:1325. doi: 10.1039/c2mb05300d. [DOI] [PubMed] [Google Scholar]
- 50.Squires TM, Messinger RJ, Manalis SR. Nat Biotechnol. 2008;26:417. doi: 10.1038/nbt1388. [DOI] [PubMed] [Google Scholar]
- 51.Zuker M. Nucleic Acids Res. 2003;31:3406. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Collins SJ, Ruscetti FW, Gallagher RE, Gallo RC. Proc Natl Acad Sci U S A. 1978;75:2458. doi: 10.1073/pnas.75.5.2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Colbert DA, Fontana JA, Bode U, Deisseroth AB. Cancer Res. 1983;43:229. [PubMed] [Google Scholar]
- 54.Collins SJ. Blood. 1987;70:1233. 22. [PubMed] [Google Scholar]
- 55.van de Corput MP, Dirks RW, van Gijlswijk RP, van Binnendijk E, Hattinger CM, de Paus RA, Landegent JE, Raap AK. J Histochem Cytochem. 1998;46:1249. doi: 10.1177/002215549804601105. [DOI] [PubMed] [Google Scholar]
- 56.Shagin DA, Rebrikov DV, Kozhemyako VB, Altshuler IM, Shcheglov AS, Zhulidov PA, Bogdanova EA, Staroverov DB, Rasskazov VA, Lukyanov S. Genome Res. 2002;12:1935. doi: 10.1101/gr.547002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nelson PT, Baldwin DA, Scearce LM, Oberholtzer JC, Tobias JW, Mourelatos Z. Nat Methods. 2004;1:155. doi: 10.1038/nmeth717. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.