Abstract
Bone marrow-derived multipotent stromal cells (MSCs), also known as mesenchymal stem cells, have great promise due to their capacity for tri-lineage differentiation and immunosuppressive properties, which allows for their allogeneic use and ultimately may allow for treatment of many diseases. MSCs will require extensive expansion and passaging to obtain cells in sufficient numbers necessary for cell therapies. MSCs from many donors could potentially be used. Because of this, there is a need to understand the role of passaging and donor differences on differentiation capacity using quantitative approaches. Here, we evaluated MSCs from two donors (noted as PCBM1632 and PCBM1641 by the manufacturer) at tissue culture passages 3, 5, and 7. We used a colony forming unit (CFU) assay and limiting dilution to quantify clonogenicity and precursor frequency during adipogenesis, and quantitative real-time-polymerase chain reaction for adipogenic markers to evaluate changes on a gene expression level. Further, we observed changes in cell size, and we sorted small and large populations to evaluate size-related adipogenic potential. While the adipogenic precursor frequency of ∼1 in 76 cells remained similar through passages for cells from PCBM1641, we found a large decrease in the adipogenic potential of MSCs from PCBM1632, with 1 in 2035 cells being capable of differentiating into an adipocyte at passage 7. MSCs from both donors showed an increase in cell diameter with increasing passage, which correlates with a decrease in clonogenicity by CFU analysis. We also measured adipose lineage gene expression following induction of adipocyte differentiation. Expression of these genes decreased with passage number for MSCs from PCBM1632 and correlated with the decrease in adipogenic potential by passage 7. In contrast, MSCs from PCBM1641 showed increased expression of these genes with increasing passage. We have shown that several quantitative assays can detect differences in MSC differentiation capacity, clonogenicity, and cell size between donors and passages. These quantitative methods are useful to assess the quality of MSCs.
Introduction
Human multipotent stromal cells (hMSCs), often termed mesenchymal stem cells, represent a promising source of adult stem cells for regenerative medicine. There are nearly 200 clinical trials underway utilizing MSCs.1
MSCs are readily available from adult tissues and can be derived from fat,2–6 bone marrow,7–13 muscle,14–17 and other sources.18–20 MSCs have the potential to differentiate along several pathways including adipogenic,21–25 osteogenic,26–31 and chondrogenic lineages,32–36 provided they receive the appropriate environmental cues. Not only do MSCs have the capacity to differentiate, they also possess immunosuppressive capabilities,37–43 which allow for allogeneic uses. Because large amounts of MSCs can be made from healthy donors and MSCs can be used in allogeneic settings, they potentially can be used to treat a wide spectrum of diseases.
MSCs have proven to be easy to expand and differentiate in culture. MSCs are characterized by their adherent properties, expression of several surface antigens including CD73, CD105, and CD90, and tri-lineage differentiation44; however, investigators are continually trying to improve characterization due to MSC heterogeneity. Within a population of MSCs, variability in cell properties such as proliferation, morphology, differentiation capacity, and cell surface marker expression profiles has been widely observed.45 These “intra-population” MSC heterogeneities and their innate plasticity may arise due to the in vivo microenvironment or also due to long-term in vitro culture.46 It is this heterogeneous nature of MSCs that may allow them to effectively respond to a wide variety of cues in their local microenvironment to carry out a particular biological function.
As these cells are widely used for investigational clinical applications, it would be useful to develop new quantitative bioassays to measure donor variability and the effect of passaging. Such tools could help to determine the suitability of a particular population of MSCs in treating a particular disease. Further, these quantitative tools could be used to assess differences in parameters such as cell source (fat, bone marrow, and muscle), cell selection for enrichment, culture media, cell density, and the effects of different protocols for expansion of MSCs. Finally, these tools could enhance our understanding of MSC heterogeneity. As stated by Wagner and Ho,45 there is an urgent need for more precise cellular and molecular markers to define subsets of MSCs.
While qualitative and some quantitative approaches to assess MSCs from different donors currently exist, we are developing robust quantitative measurements that can detect changes as a result of passaging, donor differences, and differences in subpopulations of MSCs. The ability to undergo adipogenic differentiation is often determined by a qualitative assay, using the presence of Oil Red O lipid droplets after MSCs are exposed to adipogenic stimuli. Other quantitative techniques using pixel quantitation or alcohol extraction of the differentiated MSCs, followed by spectrophotometric determination of Oil Red O dye quantity has also been used.47,48 We wanted to develop a quantitative method that could reliably measure the frequency of adipogenic cells, on a per cell basis, in populations of MSCs from different donors and at different passages in tissue culture.
In this work, we report on the use of several quantitative bioassays including limiting dilution to detect differences in donors and passage number that may be related to MSC differentiation capacity and multipotency. The overall goal of this work is to develop and employ quantitative measurements that will improve characterization MSCs by determining the role of donor variability and passage number as it relates to the biological properties of MSCs.
Materials and Methods
Expansion of hMSCs
Human bone marrow-derived MSCs from two donors (PCBM1641 and PCBM1632) were purchased (All Cells) at passage 1. According to the manufacturer, the time in culture to passage 1 at 80% confluence was 14 and 15 days for PCBM1641 and PCBM1632, respectively. The donors were 23 (PCBM1641) and 24 (PCBM1632) years old. MSCs were plated at 60 cells/cm2 in T175 flasks (Cellstar), cultured in expansion medium containing α-MEM (Invitrogen) supplemented with 16.5% fetal bovine serum (FBS; JMBioscience), l-glutamine, and penstrep (Invitrogen), and cultured at 37°C and 5% CO2, as outlined by Wolfe et al.49 The same serum lot was used for all culture and experiments. At 80% confluence, MSCs were harvested using 0.25% Trypsin/EDTA (Invitrogen) and cryopreserved in freezing medium containing 5% dimethyl sulfoxide (Sigma-Aldrich), 30% FBS, 1% penicillin (100 units/mL), and streptomycin (100 μg/ml; Invitrogen) at 1×106 cells/ml at passages 3 (P3), 5 (P5), and 7 (P7). Passage number was the number of times the cells were trypsinized up until cryopreservation. For PCBM1641 the time to P3, P5, and P7 was 7, 8, and 8 days, respectively. For PCBM1632 the time to P3, P5, and P7 was 7, 9, and 17 days, respectively.
Upon harvest, an aliquot of MSCs from five to eight pooled T175 flasks was diluted 1:1 with trypan blue and counted using an automated cell counter (Nexcelom Cellometer). In addition to cell number, the Cellometer measures the pixel area of the cells, converts that measurement to a surface area, and calculates a cell diameter based on the assumption that the cells are circular within user-defined parameters. The frequency of cell sizes falling within a 1 μm diameter bin was calculated and graphed to determine the size distribution.
Proliferation kinetics
MSCs from both donors at P3, P5, and P7 were plated at 2000 cells/cm2 in 24-well plates (Corning). Following cell adhesion, cells were placed in an Incucyte Live Cell Imager (Essen Bioscience). Nine images per well were taken every 3 h. Percent confluence measurements were averaged for each well (n=9) and then at each passage (n=4), and plotted as a function of time to measure proliferation capacity.
Colony forming unit assay
For colony forming unit (CFU) assays, MSCs from both donors at P3, P5, and P7 were plated in 10 cm tissue culture dishes (BD Falcon) at 100 cells/dish. Plates were kept at 37°C and 5% CO2 for 14 days without changing the media. After 14 days, the media were removed, plates were washed with phosphate-buffered saline (PBS), and stained with 3% Crystal Violet in 100% methanol for 10 min at room temperature. Plates were rinsed and examined under an inverted microscope (Olympus IMT-2). The number of colonies >2 mm in diameter was recorded. The percent CFUs was calculated from the number of colonies counted.
Limiting dilution technique
MSCs were plated at 1000, 500, 250, 125, 63, and 32 cells/well at 48 wells/dilution in 96-well plates. After cells were allowed to adhere for 24 h, expansion media were removed and 100 μL/well adipogenic differentiation media (NH AdipoDiff; Miltenyi Biotec) were added (day 0). Differentiation media were supplemented every 3–5 days until day 21. At day 21, plates were removed from culture, fixed with 10% formalin, and stained with Oil Red O dye (Sigma).
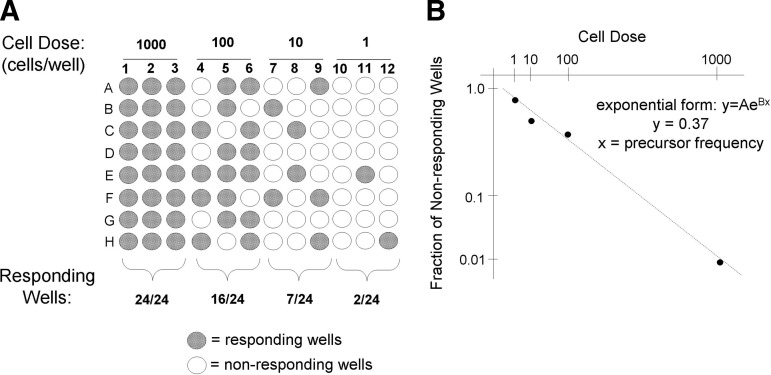
All wells were scanned visually by eye. Wells containing at least one differentiated cell (stained by Oil Red O) were scored positive. Precursor frequencies were determined by plotting the fraction of nonresponding wells versus cell dilution on a semi-logarithmic plot. For calculating the precursor frequency, we plotted a value of 0.0001 for the lowest cell dose where all wells responded. In these instances, data for higher cell doses were not plotted. This approach was used to employ only data that generate a linear dose response. The inverse of the cell dose corresponding to 37% of the nonresponding wells is the precursor frequency.50 Figure 1 shows an example of how data from this technique can be used to determine the frequency of adipogenic precursors in a batch of MSCs.
FIG. 1.
Illustration of limiting dilution methodology to calculate precursor frequency. (A) Cells are plated after serial dilution (cell dose), and are subjected to a stimulus, that is, differentiation medium. The response at each dosage is quantified and the number of responding wells is determined. (B) The number of nonresponding wells at each dilution is plotted on a semi-log plot as a function of cell dose, and points are best fit to a trend line. Using the exponential form of the equation, the precursor frequency (x) can be calculated assuming y=0.37.
Flow cytometry
MSCs were thawed, cultured to 80% confluence, and then incubated with 2.4G2 antibody (ATCC) at 4°C for 30 min to block nonspecific binding. Next, fluorochrome-labeled antibodies (phycoerythrin [PE]; allophycocyanin [APC]; and fluorescein thiocyanate [FITC]) were added to aliquots of MSCs. Positive markers for MSCs included: anti-CD29-APC, anti-CD44-APC, anti-CD90-FITC, anti-CD73-PE (Becton Dickinson), anti-CD166-FITC (US Biological), anti-CD105-PE (Beckman Coulter), and anti-STRO-1-Alexa647 (Biolegend). Negative markers included: anti-CD45-PE-Cy7, anti-CD34-PE, anti-CD14-ECD, anti-CD79α-PE-Cy5, anti-CD117-APC (Beckman Coulter), and anti-HLA-DR-FITC (BD Pharmingen). Data were acquired using either the FACSCalibur or FACS Canto II (BD Biosciences) flow cytometer and data analysis used FlowJo Analysis Software.
Cell sorting
PCBM1641 P7 cells were thawed, expanded, and sterilely sorted into populations of small and large cells based on forward scatter (FSC) properties using the FACS Aria (BD Biosciences). FSC correlates with cell size, so separate sorting gates were applied to approximately the upper (large cells) and lower (small cells) thirds of an FSC histogram plot. For example, for one experiment with PCBM1641 P7 cells, this corresponded to a gate from 45 to 105 FSC units (small cells) and from 170 to 258 FSC units (large cells). Cells in the middle third were not analyzed. Sorted populations were then counted and plated for limiting dilution analysis as described above.
RNA isolation and quantitative real-time polymerase chain reaction
MSCs were seeded at 3.75×105 cells per 10 cm dish, and cultured in adipogenic medium for 3, 7, 14, and 21 days. At harvest, cells were rinsed with PBS and lysed with RLT Buffer (Qiagen) for RNA isolation. For controls, undifferentiated MSCs were thawed at appropriate passages, cultured in expansion medium until 80% confluence, and then harvested for RNA preparation. RNA was made using QIAShredder and RNeasy columns following manufacturer's instructions (Qiagen). Total RNA quantity was determined using a NanoDrop (Thermo Scientific) and purity was determined using a Bioanalyzer (Agilent).
cDNA was synthesized using the Quantiscript Reverse Transcription Kit according to manufacturer's instructions (Qiagen) at 42°C for 15 min, followed by inactivation at 95°C for 3 min. Quantitative polymerase chain reaction (qPCR) analysis was performed using the QuantiTect SYBR Green PCR Kit (Qiagen) per manufacturer's instructions. 15 ng RNA-equivalents of cDNA was added to each reaction. The primer sets were based on genes previously reported to be involved in adipogenesis (Table 1). PCR was performed in an Applied Biosystems 7900: 95°C for 15 min; 40 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 45 s per cycle. Final extension was carried out at 72°C for 7 min. Melting curve analysis ruled out nonspecific amplification. All conditions were repeated in triplicate with 25 μL reaction volume per well.
Table 1.
Forward and Reverse Primer Sequences for Adipocyte-Specific Genes Used in Quantitative Polymerase Chain Reaction
| Gene name | Forward sequence (5′–3′) | Reverse sequence (5′–3′) | Melting temperature (°C) | Amplicon size (bp) |
|---|---|---|---|---|
| Adipsin | ACGACCTCCTGCTGCTAC | AGGAGTGGATGACTTCATTG | 55 | 493 |
| Adiponectin | ATCTCCTCCTCACTTCCATT | ATGTCTCCCTTAGGACCAAT | 55 | 304 |
| PPARg | CCTGAAACTTCAAGAGTACCA | TCATGAGGCTTATTGTAGAGC | 55 | 96 |
| C/EBPa | GAAGTCGGTGGACAAGAAC | CATTGTCACTGGTCAGCTC | 55 | 140 |
| LPL | AGACACAGCTGAGGACACTT | GCACCCAACTCTCATACATT | 55 | 137 |
| FABP4 | ACTGCAGATGACAGGAAAGT | CACCACCAGTTTATCATCCT | 55 | 126 |
| ACSS2 | GCAGGGTAAACTGAAAGAGA | GTGACGTAGGAATGACCAGT | 55 | 315 |
Statistical analysis
Statistical analyses were carried out using Microsoft Excel. Quantitative real-time PCR (qRT-PCR) experiments were completed in triplicate and analyzed using a two-tailed paired t-test. Cell size data used an unpaired t-test. Data are expressed as mean ± standard deviation in qRT-PCR data and ± standard error of the mean in cell size data. For limiting dilution, we used two-tailed and unpaired t-tests. p-Values <0.05 were considered significant.
Results
CFU assay and proliferation potential
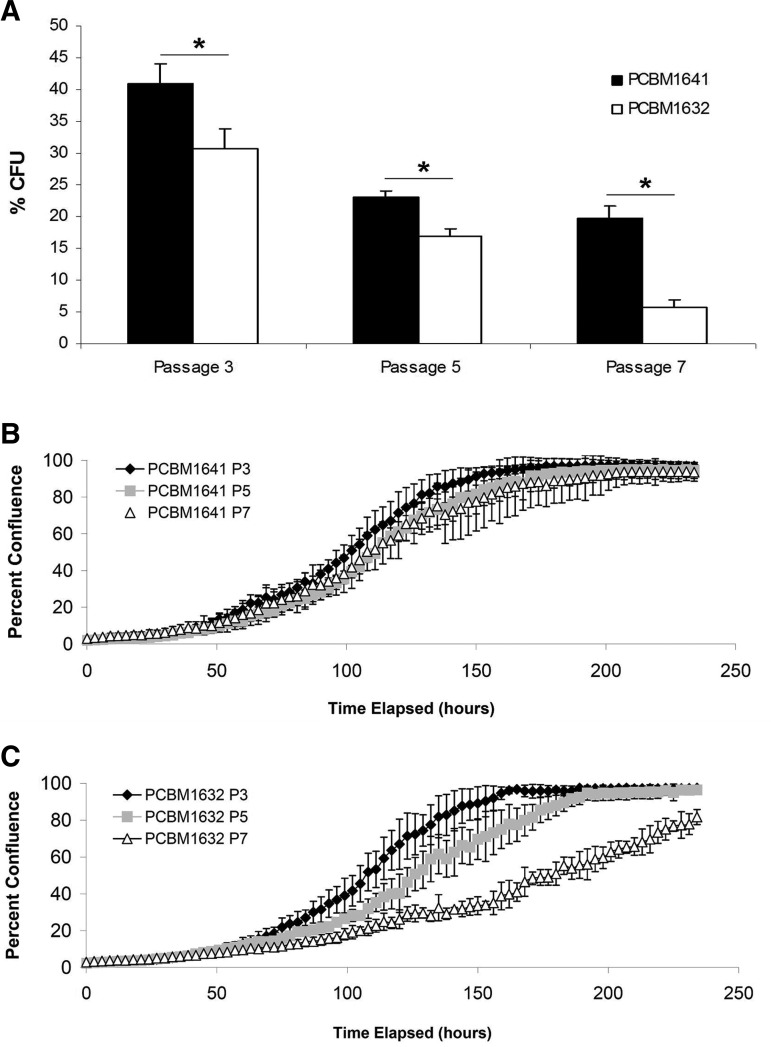
CFU assays are useful to measure the clonogenicity of a heterogeneous population of MSCs.9,51 We used this assay to detect differences in MSCs from different donors and passages. MSCs from both donors demonstrated a decrease in % CFUs with increasing passage (Fig. 2A). However, PCBM1641 was consistently higher at all passages compared with PCBM1632. Percent CFUs ranged from 41%, 23%, and 19.7% at P3, P5, and P7 respectively in PCBM1641, while they were less in PCBM1632 at 30.7%, 17%, and 5.7% at P3, P5, and P7 respectively. Data were statistically significant between P3, P5, and P7 and between both donors (p≤0.014).
FIG. 2.
Colony forming unit (CFU) frequency can differ between cell lines and with passaging. This corresponds with a decrease in proliferation potential and an increase in cell passage. (A) Percent CFUs were determined by dividing the number of colonies detected by the number of cells plated for cells from PCBM1641 (black) and PCBM1632 (white) at P3, P5, and P7 as described in Materials and Methods. Error bars represent standard deviation; n=3. Asterisk indicates significant difference (p<0.014) between both donors at that passage, marked with a horizontal bar. Significance was also seen within both donors at P3 versus P5, P5 versus P7, and P3 versus P7 (p<0.038). (B, C) Percent confluence as measured by the Incucyte Live Cell Imager for PCBM 1641 (B) and PCBM1632 (C) as a function of time. Error bars represent standard deviation; n=4. P, passage.
MSCs from both donors vary in their proliferative capacity between both donors and with passage. Time to 50% confluence for PCBM1641 was 4.3, 4.6, and 4.8 days at P3, P5, and P7 respectively, while PCBM1632 reached 50% confluence at 4.5, 5.2, and 7.5 days respectively (Figs. 2B, C).
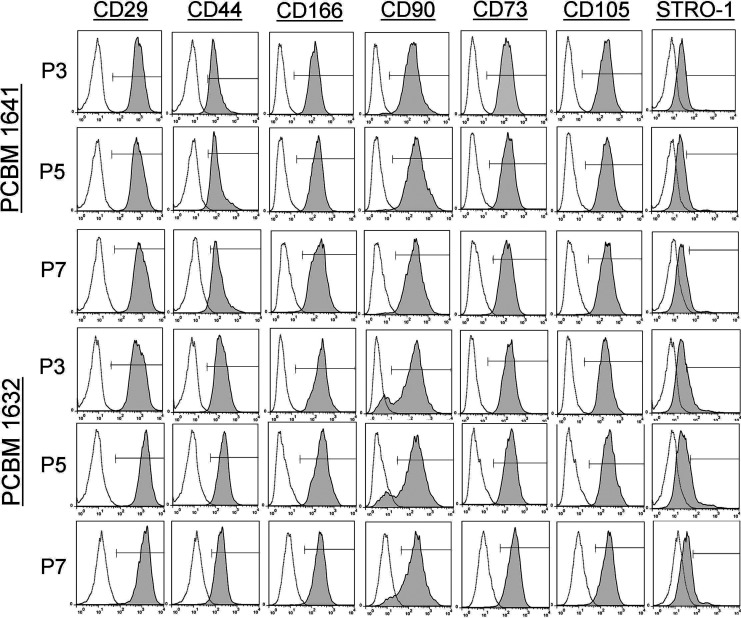
MSC cell surface marker expression
Cells were analyzed via flow cytometry to determine any differences related to passaging and donor variability (Fig. 3, Table 2). Cells from both donors and all passages were at least 90% positive for CD29, CD44, CD166, CD90, CD73, and CD105. As expected, MSCs from PCBM1632 and PCBM1641 were both <5% positive for antibodies to CD45, CD34, CD14, CD79α, CD117, and HLA-DR at all passages (data not shown).
FIG. 3.
MSC surface marker analysis reveals limited change during cell passaging. Single color cell surface marker analysis of cells from PCBM1641 and PCBM1632 at P3, P5, and P7 with antibodies specific for CD29, CD44, CD166, CD90, CD73, CD105, and STRO-1. Unstained controls are shown as the white histogram; shaded histograms represent samples stained with surface maker antibodies. The marker in each panel shows the gates used to identify percent positive cells. MSC, multipotent stromal cells.
Table 2.
MSC Cell Surface Marker Analysis
| CD29 | CD44 | CD166 | CD90 | CD73 | CD105 | STRO-1 | ||
|---|---|---|---|---|---|---|---|---|
| PCBM1641 | P3 | 99.9 | 94.3 | 99.8 | 98.9 | 99.8 | 99.9 | 10.8 |
| P5 | 99.9 | 96.4 | 99.7 | 98.7 | 99.7 | 99.7 | 12.3 | |
| P7 | 99.7 | 91.1 | 98.6 | 97.9 | 98.7 | 99.1 | 9.7 | |
| PCBM1632 | P3 | 99.9 | 99.4 | 99.9 | 99.4 | 99.9 | 99.9 | 27.2 |
| P5 | 99.9 | 99.3 | 99.5 | 99.7 | 99.5 | 99.5 | 17.1 | |
| P7 | 99.5 | 95.5 | 98.7 | 97.7 | 96.6 | 96.6 | 12.6 |
The top row shows the antibodies used for multipotent stromal cell surface marker analysis by flow cytometry. The cell line designation and passage are shown in the first two columns on the left. Numbers show the percent positive cells for each of the indicated antibodies. MSC, multipotent stromal cells.
In contrast to the other MSC cell surface markers, STRO-1 expression exhibited changes in levels of expression for one of the two MSC lines. In PCBM1641, STRO-1 expression was maintained at 9.7%–12.3% between the three passages. In contrast, PCBM1632 revealed higher STRO-1 expression at P3 (27.2% positive), but this decreased to 17.1% and 12.6% at P5 and P7 respectively.
Analysis of adipogenic precursor frequency by limiting dilution
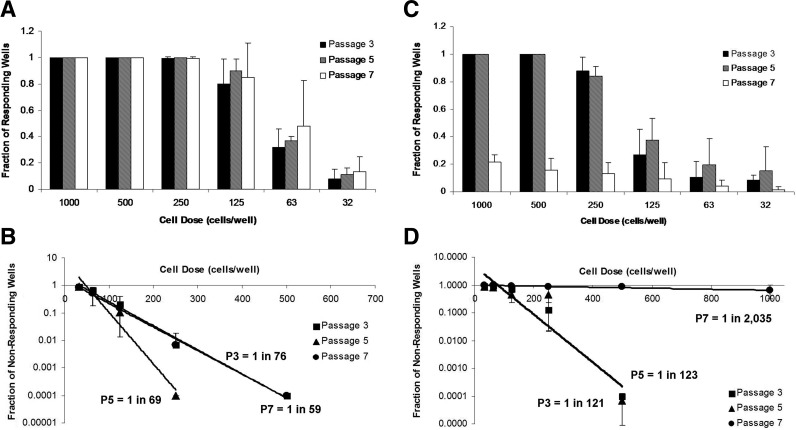
Limiting dilution has traditionally been used in immunology to determine the frequency of cells that have a particular function in a mixed population.50,52–54 We applied this method to stem cell differentiation to quantify the adipocyte precursor frequency in MSC populations. PCBM1641 responded similarly within each cell dose at P3, P5, and P7, while the fraction of responding wells at P7 in PCBM1632 drops greatly at all cell dilutions (Fig. 4A, C). As noted in the methods, we plotted a value of 0.0001 for the lowest cell dose where all wells responded. For example, in Figure 4A, P3 cell doses >500 cells per well were not plotted in Figure 4B.
FIG. 4.
Adipogenic precursor frequency can differ between cell lines and with cell passaging. Limiting dilution was conducted as described in Materials and Methods. The fraction of responding wells is represented as a function of dose of PCBM1641 (A) or PCBM1632 (C) at P3, P5, and P7. (B and D) show plots of the fraction of nonresponding wells as a function of cell dose on a semi-log plot for PCBM1641 (B) or PCBM1632 (D) at P3, P5, and P7. The trend lines were used to calculate the frequency of adipogenic precursors. Error bars represent standard deviation; n=3. R2 values were >0.96 for PCBM1641 and >0.85 for PCBM1632 at all passages.
The adipocyte precursor frequency for PCBM1641 was maintained at 1 in 76, 1 in 69, and 1 in 59 cells at P3, P5, and P7 respectively (Fig. 4C, D). PCBM1632 precursor frequencies were similar at P3 and P5 at 1 in 121 and 1 in 123 respectively. However, the precursor frequency for PCBM1632 dropped drastically by P7, where only 1 in 2035 cells were capable of differentiating into an adipocyte (p<0.0002, P7 compared with P3 and P5).
Cell size as an indicator of multipotency
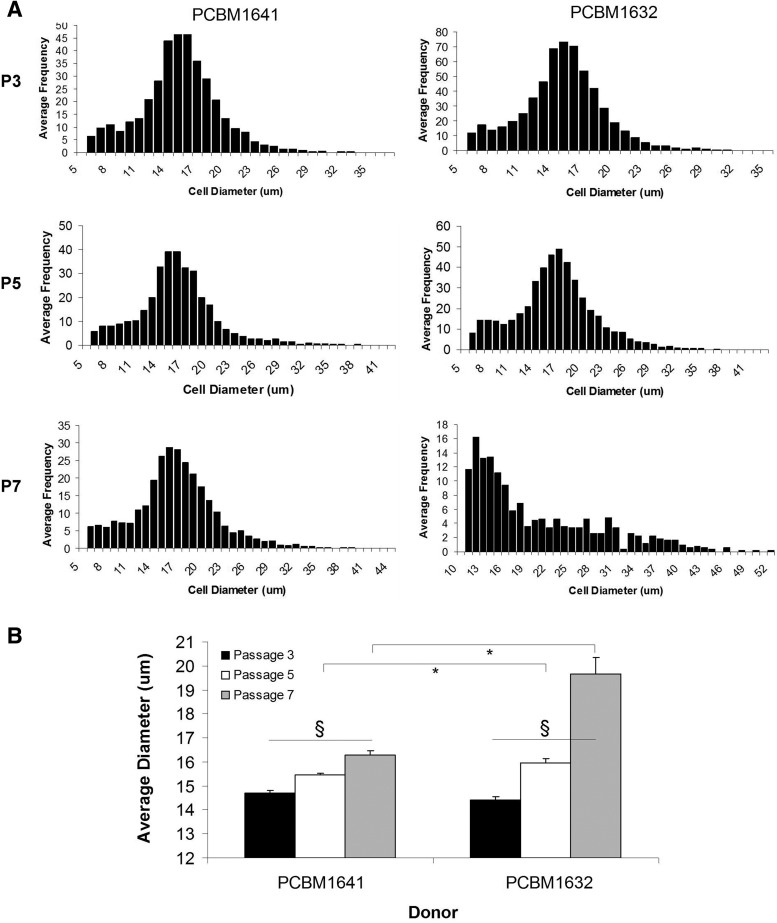
We observed that cell size increases with increasing passage for both PCBM1632 and PCBM1641 (Fig. 5A, B). The average diameter of PCBM1632 was significantly larger than PCBM1641 by P7 (Fig. 5B, p≤0.024). Further, the distribution of cell diameter can change with increasing passage, as seen in PCBM1632, P7 cells (Fig. 5A).
FIG. 5.
MSC sizes can differ between cell lines and with cell passaging. Cell sizes were measured as described in Materials and Methods. (A) Bars represent size bins for pooled individual cells of PCBM1641 or PCBM1632 at P3, P5, and P7. (B) Average size of cells from PCBM1641 (black) and PCBM1632 (white) as a function of passage. Sample sizes were the following: PCBM1641: P3 (n=13), P5 (n=16), and P7 (n=15). PCBM1632: P3 (n=13), P5 (n=13), and P7 (n=5). Error bars represent standard error of the mean. Asterisk indicates significant difference (p<0.019) between donors at that passage, marked with a horizontal bar. Significance was also seen within both donors at P3 versus P5, P5 versus P7, and P3 versus P7 (p≤0.004), as indicated by §.
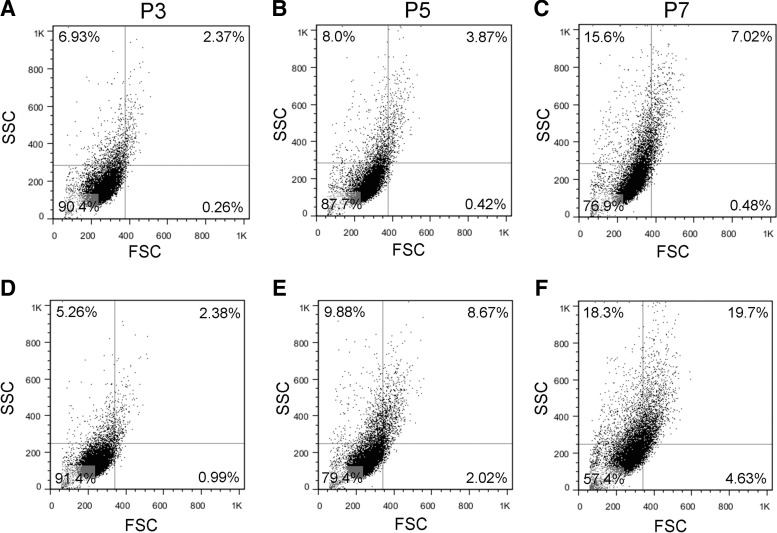
Changes in cell size between MSC lines and passage was further demonstrated by evaluating forward scatter (FSC) and side scatter (SSC) using a flow cytometer (Fig. 6). FSC correlates with cell size and SSC with granularity. The percent positive cells with high FSC and SSC (i.e., “large cells”) in the upper right (UR) quadrant increases with increasing passage for both cell lines, while cells with low FSC and SSC (i.e., “small cells”) in the lower left quadrant decrease with increasing passage, indicating a shift toward an increase in cell size and granularity with increasing passage. PCBM1641 and PCBM1632 at P3 displayed similarities in the percent positive in the UR quadrant (2.37% and 2.38% respectively). PCBM1641 increased to 3.87% and 7.02% at P5 and P7 respectively, while PCBM1632 increased to 8.67% and 19.7% in the UR quadrant at P5 and P7 respectively.
FIG. 6.
Flow cytometry analysis reveals changes in size in cells from PCBM1641 and PCBM1632 at P3, P5, and P7. Quadrant gating was applied to dot plots of forward scatter (FSC) versus side scatter (SSC) for cells from the indicated cells and passages. Percent of cells in each quadrant are shown for PCBM1641 (A–C) and PCBM1632 (D–F) at P3, P5, and P7.
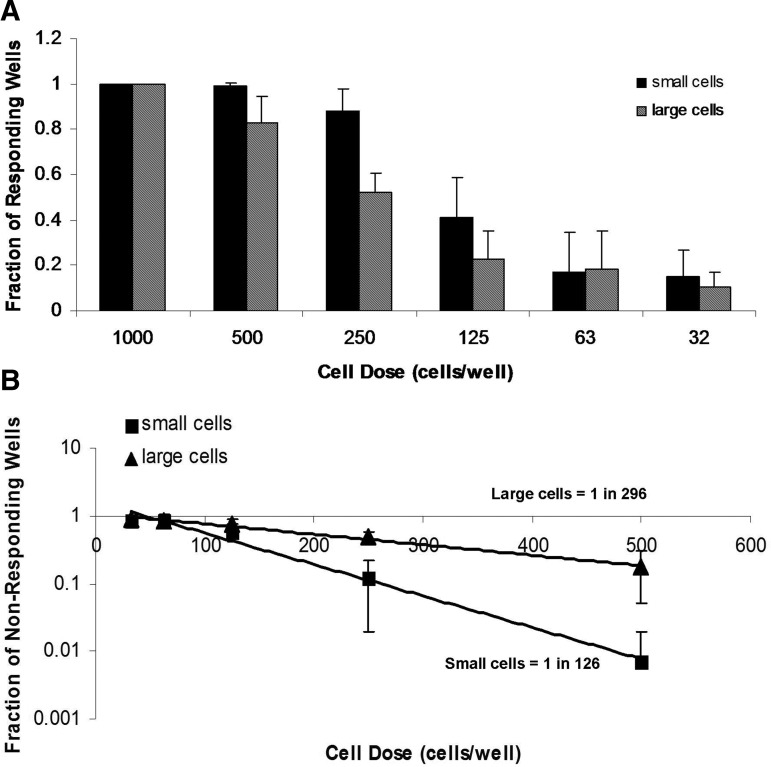
Because there were detectable changes in the FSC profile between passages, we utilized this property to sort small and large cells. We then applied the adipogenic precursor limiting dilution method to the sorted populations to determine whether cell size affects differentiation. PCBM1641 cells at P7 were sorted into high FSC (“large” cells) and low FSC populations (“small” cells). The average cell diameter post-sort was 14.6 and 19.3 μm for the small and large populations respectively. The number of cells that were capable of differentiating into an adipocyte in the small population was 1 in 126 cells, while in the large population it was 1 in 296 cells (Fig. 7), which was statistically significant (p≤0.024).
FIG. 7.
MSCs of different sizes have different adipogenic precursor frequencies. Cell size thresholds were based on separate FSC sorting gates that were applied to the upper (large cells) and lower (small cells) thirds of an FSC histogram plot. The middle third was not analyzed. (A) The fraction of responding wells was plotted as a function of cell dose in sorted small and large cells from PCBM1641 following limiting dilution analysis. (B) A plot of nonresponding wells as a function of cell dose. The trend line was used to calculate precursor frequency in populations of small and large cells. Error bars represent standard deviation; n=3 for both cell populations. R2 values were 0.99 for small cells and 0.99 for large cells.
Adipogenic potential as measured by qRT-PCR analysis
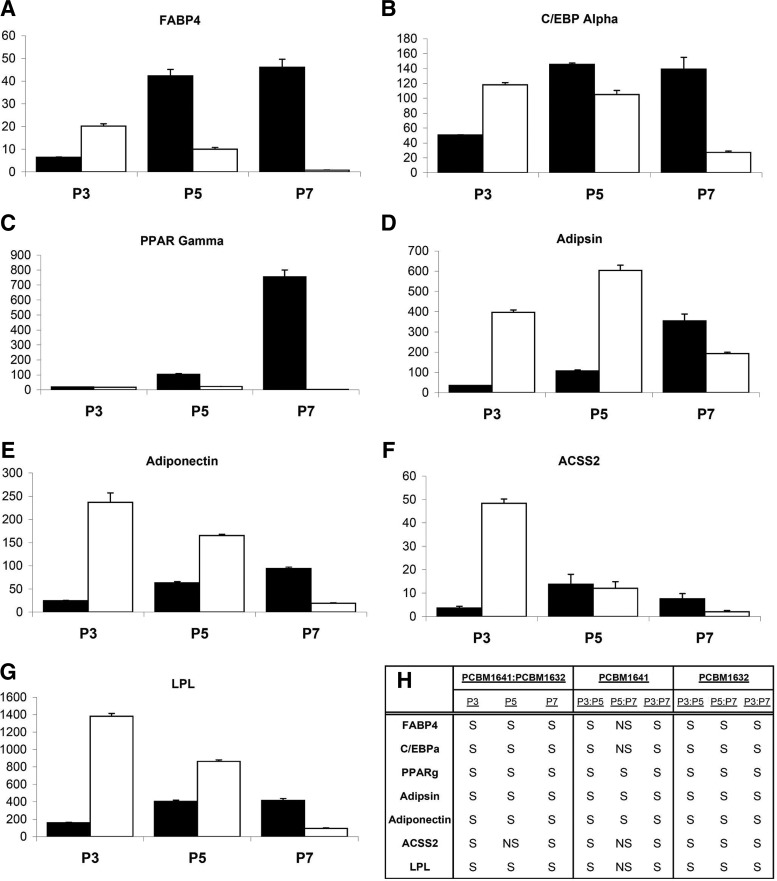
To further examine differences between MSCs from different donors and passages on the ability to differentiate toward an adipocyte lineage, we evaluated gene expression levels for seven genes known to be associated with adipogenesis.
MSCs undergo terminal differentiation into a mature adipocyte following growth arrest, and expression of mRNAs including lipoprotein lipase (LPL), which is involved in lipogenesis.21 This is followed by activation of transcriptional factors including CCAAT/enhancer binding protein alpha (C/EBPα) and peroxisome proliferator-activated receptor gamma (PPARγ).55 Other markers of adipogenesis include adipsin, a protease involved in glucose transport stimulation; adiponectin, a protein in involved in insulin sensitivity regulation;56 and acyl-CoA synthetase short-chain family member 2 (ACSS2), which is involved in lipid synthesis.21 PCBM1641 and PCBM1632 at P3, P5, and P7 were differentiated toward an adipogenic lineage, and gene expression was analyzed at days 3, 7, 14, and 21 during differentiation. Relative fold change in gene expression was calculated taking into account the efficiency of each primer set used. Ct values for undifferentiated control samples measuring LPL and adiponectin were undetectable and therefore were assigned a Ct value of 40 to allow for a fold change calculation. Because the same amount of cDNA was used in all reactions for all primer sets, the data were not normalized to an internal control.57 In general, PCBM1641 revealed an increase in the relative fold change in gene expression with increasing passage, while PCBM1632 decreased in fold change with increasing passage. Day 21 data are shown in Figure 8. PCBM1641 is significantly different from PCBM1632 (p≤0.021) within each passage comparison for all genes and all passages, with the exception of ACSS2 at P5 (Fig. 8F). The greatest differences in fold change between donors were seen for PPARγ (Fig. 8C) and FABP4 (Fig. 8A) at P7, where PCBM1641 is 295- and 58-fold greater in fold change than PCBM1632 respectively. The greatest decrease in fold change from P3 to P7 was seen for PCBM1632 in FABP4 (Fig. 8A) and ACSS2 (Fig. 8F) expression, where a 25- and 23-fold decrease in expression was detected respectively. A 40-fold increase in fold change from P3 to P7 was seen in PPARγ (Fig. 8C) for PCBM1641, the largest fold increase detected between passages. Day 7 and 14 analyses show similar patterns but less expression relative to day 21 (data not shown). Ct values at day 3 were undetectable and therefore fold changes could not be calculated.
FIG. 8.
Differentiation induces differences in adipocyte-specific gene expression levels that vary between cell lines and passages. The relative fold change in RNA expression for (A) fatty acid binding protein 4 (FABP4), (B) CCAAT/enhancer binding protein alpha (C/EBPα), (C) peroxisome proliferator-activated receptor gamma (PPARγ), (D) Adipsin, (E) Adiponectin, (F) acyl-CoA synthetase short-chain family member 2 (ACSS2), and (G) lipoprotein lipase (LPL) was measured for PCBM1641 and PCBM1632 cells from P3, P5, and P7 21 days after induction of adipose-lineage differentiation. Black and white bars represent PCBM1641 and PCBM1632 respectively. The y-axis indicates fold change in gene expression, and is represented in thousands in FABP4, Adiponectin, and LPL. Error bars represent standard deviation; n=3 for all samples. (H) Statistical analysis summary of data represented in A–G. Analysis completed between donors and between P3, P5, and P7; S indicates significance at p<0.05, NS indicates nonsignificance at p>0.05.
Discussion
Because of the heterogeneous nature of MSCs, the establishment and application of quantitative bioassays that detect differences between MSCs from different donors and passages would be of great value. Such quantitative bioassays could be used to compare biological properties of MSCs from different donors and to evaluate the effect of different tissue culture conditions and duration. These quantitative bioassays could provide information that would potentially aid in identification of MSC product characteristics predictive of clinical outcome. Accordingly, we developed a novel quantitative assay of adipose differentiation capacity and employed several other existing assays to study MSCs from two different donors at three passages and to assess whether or not any of these assays can detect quantitative differences between MSCs from different donors and passages.
Flow cytometry is a cell-by-cell quantitative assay to assess cell surface marker patterns and can be used to determine levels of cell surface antigen expression. As outlined in a white paper by the International Society for Cell Therapy,44 MSCs are characterized as being adherent to plastic, capable of tri-lineage differentiation, and >95% positive for CD73, CD90, and CD105 by flow cytometry. In addition to these markers, we included other MSC markers in our flow cytometry analyses, namely CD29, CD44, CD166, and STRO-1. MSCs from two different donors and three different passages maintained expression of all three positive CD markers mentioned in the ISCT paper above 95%. Expression of these positive markers did not change or correlate with changes observed using other bioassays conducted with the two MSC lines.
In contrast, STRO-1 revealed differences between cells from different donors and passages. Interestingly, while PCBM1641 maintained similar expression of STRO-1 in all three passages, PCBM1632 demonstrated higher expression at P3 and decreased in percent positive cells with increasing passage. These differences correlated with differences seen with the limiting dilution assay. Adipogenic precursor frequencies calculated for PCBM1641 remained fairly consistent between passages, while PCBM1632 demonstrated an ∼20-fold decrease in precursor frequency by P7. This observation shows that precursor frequency can be donor- and passage-dependent and that STRO-1 expression pattern may correlate with adipogenic capacity. Others have also shown that STRO-1 is potentially a useful marker of MSC quality because STRO-1+ cells were shown to give rise to adipocytes, smooth muscle, and fibroblasts, and shown by Gronthos et al. to give rise to cells of the osteogenic lineage.58 Also, STRO-1 expression decreases with increasing passage in rat bone marrow-derived MSCs.59 Investigators continue to use STRO-1 as a marker of a clonogenic population of MSCs,60–62 and a recent article suggests that STRO-1 may be implicated in cardiovascular paracrine activity in vitro, as evidenced by migration, endothelial tube formation assays, and cytokine expression.63
A traditional measure of clonogenicity in MSCs is a CFU assay, which will quantify the number of single cells capable of proliferating sufficiently to form a detectable colony. Our observations for both donors are consistent with and extend other findings in that both decrease in % CFUs with increasing passage, as demonstrated by other investigators.9 While both donors demonstrated similar proliferation potential at P3 and P5, proliferative capacity in PCBM1632 decreased noticeably at P7. Although the same number of cells were used to initiate the CFU assay, it is possible that the dramatically decreased CFU capability of PCBM1632 by P7 was at least partially the result of its decreased proliferation rate. These findings correlate with our observations that cell size and morphological changes increase at higher passages. Two distinct cell morphologies have been observed by others at low passages; small spindle-shaped cells that are termed “rapidly self-renewing” cells and more mature, large, and flattened cells that replicate more slowly.51,64 These investigators have also documented that an increase in the percentage of self-renewing cells correlates with CFUs. Cell size may therefore serve well as a parameter that correlates well with stemness of MSCs.
We and others have demonstrated the utility in using cell size measurements as a correlate of MSC clonogenicity. We have also shown that we can detect differences in cell size and cell size distribution between donors and passages. MSCs from both donors show an increase in average diameter with increasing passage. Interestingly by P7, the size distribution and average size of PCBM1632 is significantly larger than PCBM1641, which correlates with the sharp decrease in CFUs and adipogenic precursor frequency in PCBM1632 P7 MSCs. Also, as shown by Larson et al., we demonstrated that MSC size distribution could be detected by flow cytometry using FSC and SSC analysis.65 We extended these results by showing that we could correlate cell size with differentiation capacity in MSCs. We observed higher adipocyte precursor frequencies in the sorted small population when compared with the large. Consistent with earlier publications,10,51,64 this data suggest that small cells, which are also more frequent in low passage populations, demonstrate greater multipotentiality and differentiation capacity than larger cells. Further, cell sorting for the small population may be useful to enrich for subpopulations with more stem cell characteristics than that of the original MSC population. Limiting dilution revealed not only a qualitative difference between the small and large population, but also shows these differences can be quantified. MSC subpopulations can be purified and studied separately using limiting dilution, which will provide quantitative measurements to better understand the role of cell size parameters in differentiation and stemness. Future refinements to define donor-specific cell size thresholds for differentiation will provide utility in establishing predictive models for differentiation capacity in MSCs.
One plausible explanation for the dramatically decreased adipogenic potential of PCBM1632 at P7 is that the rate of proliferation has decreased. However, all adipogenesis experiments were initiated at the same cell densities, and we did not observe any differences in the number of cells in wells at the end of the experiment. Also, others have shown that induction of adipogenesis halts proliferation of bone marrow-derived MSCs.66
We showed that different populations of MSCs can respond differently to adipogenic stimulation, and then used qRT-PCR to determine whether expression of certain adipose lineage-specific genes correlated with our quantitative measures of cellular differentiation capacity. Gene expression data during adipogenesis from PCBM1641 and PCBM1632 demonstrated marked differences in trends between cell lines and passages. When MSCs were differentiated to adipocytes at P3, P5, and P7, genes related to adipogenesis are downregulated with increasing passage in PCBM1632. This is closely correlated with the decrease in PCBM1632 precursor frequency during adipogenesis, the decrease in % CFUs, and the decreasing expression of STRO-1. In contrast, PCBM1641 shows a different pattern of upregulated adipogenic gene expression with increasing passage. The reason for this remains unclear; however, this highlights the effect of donor variability on adipogenesis.
While the age of both MSC donors was similar, cells from the PCBM1641 donor may have had more multipotent stem cells at the start of in vitro culture. The propensity for PCBM1641 to continue to maintain its differentiation capacity at P7 underscores this idea. Taken together, these inherent differences in MSCs from donors of similar age highlight the potential for other donor-related factors in MSC biological variability, which may play a role in their clinical usefulness or performance in various model systems. This re-emphasizes the need to determine potential biomarkers that are predictive of donor multipotentiality and stemness. Microarray analyses completed by other laboratories have determined early biomarkers such as SRY sex determining region Y-box 11 (Sox11), which may correlate with MSC clonogenicity. Larson et al. determined that Sox11 was downregulated with increasing passage. Subsequent knockdown studies showed Sox11 decreased proliferation and osteogenic potential of MSCs.65 Our data also show that expression of genes associated with late steps of adipogenesis are not necessarily predictive of differentiation capacity in MSCs. PCBM1632 was consistently higher than PCBM1641 in adipogenic gene expression at P3; however by P7, PCBM1641 demonstrated significantly higher fold changes in gene expression of all markers of adipogenesis. Microarray analysis will be useful in the future to detect early biomarkers that correlate with gene expression patterns during differentiation and passaging.
Quantitative assessments of the biological characteristics of MSCs will allow for the detection of donor and passage-specific differences and may be of great utility in the discovery of molecular markers that correlate with differentiation and proliferation capacity of MSCs. We showed that limiting dilution is capable of determining a precursor frequency that provides a quantitative measure of the adipogenic differentiation capacity in populations of MSCs. We have demonstrated that differentiation capacity varies between MSCs from different donors and passages, and that mean cell size increases with cell passage. Also, cell size distribution can change dramatically (PCBM1632, Fig. 5). Future work will focus more on cell size and assessing differentiation in sorted small and large cells, and validation of our limiting dilution approach using other quantitative methods such as automated microscopy, while also quantifying additional differentiation pathways and different donors.
Acknowledgments
This project was supported in part by Jessica Lo Surdo's appointment to the Research Participation Program at CBER administered by the Oak Ridge Institute for Science and Education through US DOE and US FDA. This work was also supported in part by the FDA Modernizing Science grant program, a BARDA grant, and Division of Cell and Gene Therapies. The authors would like to acknowledge Drs. Cheng-Hong Wei and Jeffry Smith for reviewing this article and Howard Mostowski for his assistance in sorting MSC samples. Research support came from FDA's Office of the Chief Scientist, BARDA, and operating funds from the Division of Cellular and Gene Therapies.
Authors' Contribution
Jessica Lo Surdo and Steve Bauer planned experiments and wrote the article. Ms. Lo Surdo performed experiments and analyzed data for this article.
Disclosure Statement
No competing financial interests exist.
References
- 1.http://www.clinicaltrials.gov/ [Oct.2011 ]. http://www.clinicaltrials.gov/
- 2.Banas A. Teratani T. Yamamoto Y. Tokuhara M. Takeshita F. Quinn G. Okochi H. Ochiya T. Adipose tissue-derived mesenchymal stem cells as a source of human hepatocytes. Hepatology. 2007;46:219. doi: 10.1002/hep.21704. [DOI] [PubMed] [Google Scholar]
- 3.Brayfield C. Marra K. Rubin J.P. Adipose stem cells for soft tissue regeneration. Handchir Mikrochir Plast Chir. 2010;42:124. doi: 10.1055/s-0030-1248269. [DOI] [PubMed] [Google Scholar]
- 4.Cherubino M. Marra K.G. Adipose-derived stem cells for soft tissue reconstruction. Regen Med. 2009;4:109. doi: 10.2217/17460751.4.1.109. [DOI] [PubMed] [Google Scholar]
- 5.Kokai L.E. Rubin J.P. Marra K.G. The potential of adipose-derived adult stem cells as a source of neuronal progenitor cells. Plast Reconstr Surg. 2005;116:1453. doi: 10.1097/01.prs.0000182570.62814.e3. [DOI] [PubMed] [Google Scholar]
- 6.Zuk P.A. Zhu M. Mizuno H. Huang J. Futrell J.W. Katz A.J. Benhaim P. Lorenz H.P. Hedrick M.H. Multilineage cells from human adipose tissue: implications for cell-based therapies. Tissue Eng. 2001;7:211. doi: 10.1089/107632701300062859. [DOI] [PubMed] [Google Scholar]
- 7.Bosnakovski D. Mizuno M. Kim G. Takagi S. Okumura M. Fujinaga T. Isolation and multilineage differentiation of bovine bone marrow mesenchymal stem cells. Cell Tissue Res. 2005;319:243. doi: 10.1007/s00441-004-1012-5. [DOI] [PubMed] [Google Scholar]
- 8.Delorme B. Charbord P. Culture and characterization of human bone marrow mesenchymal stem cells. Methods Mol Med. 2007;140:67. doi: 10.1007/978-1-59745-443-8_4. [DOI] [PubMed] [Google Scholar]
- 9.Digirolamo C.M. Stokes D. Colter D. Phinney D.G. Class R. Prockop D.J. Propagation and senescence of human marrow stromal cells in culture: a simple colony-forming assay identifies samples with the greatest potential to propagate and differentiate. Br J Haematol. 1999;107:275. doi: 10.1046/j.1365-2141.1999.01715.x. [DOI] [PubMed] [Google Scholar]
- 10.Prockop D.J. Sekiya I. Colter D.C. Isolation and characterization of rapidly self-renewing stem cells from cultures of human marrow stromal cells. Cytotherapy. 2001;3:393. doi: 10.1080/146532401753277229. [DOI] [PubMed] [Google Scholar]
- 11.Spees J.L. Whitney M.J. Sullivan D.E. Lasky J.A. Laboy M. Ylostalo J. Prockop D.J. Bone marrow progenitor cells contribute to repair and remodeling of the lung and heart in a rat model of progressive pulmonary hypertension. FASEB J. 2008;22:1226. doi: 10.1096/fj.07-8076com. [DOI] [PubMed] [Google Scholar]
- 12.Friedenstein A.J. Gorskaja J.F. Kulagina N.N. Fibroblast precursors in normal and irradiated mouse hematopoietic organs. Exp Hematol. 1976;4:267. [PubMed] [Google Scholar]
- 13.Gregory C.A. Prockop D.J. Spees J.L. Non-hematopoietic bone marrow stem cells: molecular control of expansion and differentiation. Exp Cell Res. 2005;306:330. doi: 10.1016/j.yexcr.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 14.Deasy B.M. Jankowski R.J. Huard J. Muscle-derived stem cells: characterization and potential for cell-mediated therapy. Blood Cells Mol Dis. 2001;27:924. doi: 10.1006/bcmd.2001.0463. [DOI] [PubMed] [Google Scholar]
- 15.Jankowski R.J. Deasy B.M. Huard J. Muscle-derived stem cells. Gene Ther. 2002;9:642. doi: 10.1038/sj.gt.3301719. [DOI] [PubMed] [Google Scholar]
- 16.Huard J. Regenerative medicine based on muscle stem cells. J Musculoskelet Neuronal Interact. 2008;8:337. [PubMed] [Google Scholar]
- 17.Bellayr I.H. Gharaibeh B. Huard J. Li Y. Skeletal muscle-derived stem cells differentiate into hepatocyte-like cells and aid in liver regeneration. Int J Clin Exp Pathol. 2010;3:681. [PMC free article] [PubMed] [Google Scholar]
- 18.Castrechini N.M. Murthi P. Gude N.M. Erwich J.J. Gronthos S. Zannettino A. Brennecke S.P. Kalionis B. Mesenchymal stem cells in human placental chorionic villi reside in a vascular Niche. Placenta. 2010;31:203. doi: 10.1016/j.placenta.2009.12.006. [DOI] [PubMed] [Google Scholar]
- 19.Park S.B. Seo M.S. Kang J.G. Chae J.S. Kang K.S. Isolation and characterization of equine amniotic fluid-derived multipotent stem cells. Cytotherapy. 2010;13:341. doi: 10.3109/14653249.2010.520312. [DOI] [PubMed] [Google Scholar]
- 20.Yoon B.S. Moon J.H. Jun E.K. Kim J. Maeng I. Kim J.S. Lee J.H. Baik C.S. Kim A. Cho K.S. Lee H.H. Whang K.Y. You S. Secretory profiles and wound healing effects of human amniotic fluid-derived mesenchymal stem cells. Stem Cells Dev. 2009;19:887. doi: 10.1089/scd.2009.0138. [DOI] [PubMed] [Google Scholar]
- 21.Sekiya I. Larson B.L. Vuoristo J.T. Cui J.G. Prockop D.J. Adipogenic differentiation of human adult stem cells from bone marrow stroma (MSCs) J Bone Miner Res. 2004;19:256. doi: 10.1359/JBMR.0301220. [DOI] [PubMed] [Google Scholar]
- 22.Hung S.C. Chang C.F. Ma H.L. Chen T.H. Low-Tone Ho L. Gene expression profiles of early adipogenesis in human mesenchymal stem cells. Gene. 2004;340:141. doi: 10.1016/j.gene.2004.06.028. [DOI] [PubMed] [Google Scholar]
- 23.Janderova L. McNeil M. Murrell A.N. Mynatt R.L. Smith S.R. Human mesenchymal stem cells as an in vitro model for human adipogenesis. Obes Res. 2003;11:65. doi: 10.1038/oby.2003.11. [DOI] [PubMed] [Google Scholar]
- 24.Nakamura T. Shiojima S. Hirai Y. Iwama T. Tsuruzoe N. Hirasawa A. Katsuma S. Tsujimoto G. Temporal gene expression changes during adipogenesis in human mesenchymal stem cells. Biochem Biophys Res Commun. 2003;303:306. doi: 10.1016/s0006-291x(03)00325-5. [DOI] [PubMed] [Google Scholar]
- 25.Post S. Abdallah B.M. Bentzon J.F. Kassem M. Demonstration of the presence of independent pre-osteoblastic and pre-adipocytic cell populations in bone marrow-derived mesenchymal stem cells. Bone. 2008;43:32. doi: 10.1016/j.bone.2008.03.011. [DOI] [PubMed] [Google Scholar]
- 26.Augello A. De Bari C. The regulation of differentiation in mesenchymal stem cells. Hum Gene Ther. 2010;21:1226. doi: 10.1089/hum.2010.173. [DOI] [PubMed] [Google Scholar]
- 27.Briggs T. Treiser M.D. Holmes P.F. Kohn J. Moghe P.V. Arinzeh T.L. Osteogenic differentiation of human mesenchymal stem cells on poly(ethylene glycol)-variant biomaterials. J Biomed Mater Res A. 2009;91:975. doi: 10.1002/jbm.a.32310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen D.F. Li H. Zhou J.H. Xie Y. Li Y.W. Du S.H. Zhang Y. Huang J. Xu M. Osteogenesis characteristics of cultured rat mesenchymal stem cells under bone induction condition. J Chin Integr Med. 2004;2:375. doi: 10.3736/jcim20040519. [DOI] [PubMed] [Google Scholar]
- 29.de Girolamo L. Sartori M.F. Albisetti W. Brini A.T. Osteogenic differentiation of human adipose-derived stem cells: comparison of two different inductive media. J Tissue Eng Regen Med. 2007;1:154. doi: 10.1002/term.12. [DOI] [PubMed] [Google Scholar]
- 30.Donzelli E. Salvade A. Mimo P. Vigano M. Morrone M. Papagna R. Carini F. Zaopo A. Miloso M. Baldoni M. Tredici G. Mesenchymal stem cells cultured on a collagen scaffold: In vitro osteogenic differentiation. Arch Oral Biol. 2007;52:64. doi: 10.1016/j.archoralbio.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 31.Long M.W. Osteogenesis and bone-marrow-derived cells. Blood Cells Mol Dis. 2001;27:677. doi: 10.1006/bcmd.2001.0431. [DOI] [PubMed] [Google Scholar]
- 32.Arufe M.C. De la Fuente A. Fuentes I. de Toro F.J. Blanco F.J. Chondrogenic potential of subpopulations of cells expressing mesenchymal stem cell markers derived from human synovial membranes. J Cell Biochem. 2010;111:834. doi: 10.1002/jcb.22768. [DOI] [PubMed] [Google Scholar]
- 33.Kim H.J. Lee J.H. Im G.I. Chondrogenesis using mesenchymal stem cells and PCL scaffolds. J Biomed Mater Res A. 2009;92:659. doi: 10.1002/jbm.a.32414. [DOI] [PubMed] [Google Scholar]
- 34.Mueller M.B. Fischer M. Zellner J. Berner A. Dienstknecht T. Prantl L. Kujat R. Nerlich M. Tuan R.S. Angele P. Hypertrophy in mesenchymal stem cell chondrogenesis: effect of TGF-beta isoforms and chondrogenic conditioning. Cells Tissues Organs. 2010;192:158. doi: 10.1159/000313399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ronziere M.C. Perrier E. Mallein-Gerin F. Freyria A.M. Chondrogenic potential of bone marrow- and adipose tissue-derived adult human mesenchymal stem cells. Biomed Mater Eng. 2010;20:145. doi: 10.3233/BME-2010-0626. [DOI] [PubMed] [Google Scholar]
- 36.Worster A.A. Brower-Toland B.D. Fortier L.A. Bent S.J. Williams J. Nixon A.J. Chondrocytic differentiation of mesenchymal stem cells sequentially exposed to transforming growth factor-beta1 in monolayer and insulin-like growth factor-I in a three-dimensional matrix. J Orthop Res. 2001;19:738. doi: 10.1016/S0736-0266(00)00054-1. [DOI] [PubMed] [Google Scholar]
- 37.Aggarwal S. Pittenger M.F. Human mesenchymal stem cells modulate allogeneic immune cell responses. Blood. 2005;105:1815. doi: 10.1182/blood-2004-04-1559. [DOI] [PubMed] [Google Scholar]
- 38.Le Blanc K. Mesenchymal stromal cells: Tissue repair and immune modulation. Cytotherapy. 2006;8:559. doi: 10.1080/14653240601045399. [DOI] [PubMed] [Google Scholar]
- 39.Le Blanc K. Tammik C. Rosendahl K. Zetterberg E. Ringden O. HLA expression and immunologic properties of differentiated and undifferentiated mesenchymal stem cells. Exp Hematol. 2003;31:890. doi: 10.1016/s0301-472x(03)00110-3. [DOI] [PubMed] [Google Scholar]
- 40.Le Blanc K. Tammik L. Sundberg B. Haynesworth S.E. Ringden O. Mesenchymal stem cells inhibit and stimulate mixed lymphocyte cultures and mitogenic responses independently of the major histocompatibility complex. Scand J Immunol. 2003;57:11. doi: 10.1046/j.1365-3083.2003.01176.x. [DOI] [PubMed] [Google Scholar]
- 41.Ramasamy R. Tong C.K. Seow H.F. Vidyadaran S. Dazzi F. The immunosuppressive effects of human bone marrow-derived mesenchymal stem cells target T cell proliferation but not its effector function. Cell Immunol. 2008;251:131. doi: 10.1016/j.cellimm.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 42.Siegel G. Schafer R. Dazzi F. The immunosuppressive properties of mesenchymal stem cells. Transplantation. 2009;87:S45. doi: 10.1097/TP.0b013e3181a285b0. [DOI] [PubMed] [Google Scholar]
- 43.Uccelli A. Moretta L. Pistoia V. Immunoregulatory function of mesenchymal stem cells. Eur J Immunol. 2006;36:2566. doi: 10.1002/eji.200636416. [DOI] [PubMed] [Google Scholar]
- 44.Dominici M. Le Blanc K. Mueller I. Slaper-Cortenbach I. Marini F. Krause D. Deans R. Keating A. Prockop D. Horwitz E. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy. 2006;8:315. doi: 10.1080/14653240600855905. [DOI] [PubMed] [Google Scholar]
- 45.Wagner W. Ho A.D. Mesenchymal stem cell preparations—comparing apples and oranges. Stem Cell Rev. 2007;3:239. doi: 10.1007/s12015-007-9001-1. [DOI] [PubMed] [Google Scholar]
- 46.Pevsner-Fischer M. Levin S. Zipori D. The origins of mesenchymal stromal cell heterogeneity. Stem Cell Rev. 2011;7:560. doi: 10.1007/s12015-011-9229-7. [DOI] [PubMed] [Google Scholar]
- 47.Taipaleenmaki H. Abdallah B.M. AlDahmash A. Saamanen A.M. Kassem M. Wnt signalling mediates the cross-talk between bone marrow derived pre-adipocytic and pre-osteoblastic cell populations. Exp Cell Res. 2011;317:745. doi: 10.1016/j.yexcr.2010.12.015. [DOI] [PubMed] [Google Scholar]
- 48.Dexheimer V. Mueller S. Braatz F. Richter W. Reduced reactivation from dormancy but maintained lineage choice of human mesenchymal stem cells with donor age. PLoS One. 2011;6:e22980. doi: 10.1371/journal.pone.0022980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wolfe M. Pochampally R. Swaney W. Reger R.L. Isolation and culture of bone marrow-derived human multipotent stromal cells (hMSCs) Methods Mol Biol. 2008;449:3. doi: 10.1007/978-1-60327-169-1_1. [DOI] [PubMed] [Google Scholar]
- 50.Carneiro J. Duarte L. Padovan E. Limiting dilution analysis of antigen-specific T cells. Methods Mol Biol. 2009;514:95. doi: 10.1007/978-1-60327-527-9_7. [DOI] [PubMed] [Google Scholar]
- 51.Colter D.C. Class R. DiGirolamo C.M. Prockop D.J. Rapid expansion of recycling stem cells in cultures of plastic-adherent cells from human bone marrow. Proc Natl Acad Sci U S A. 2000;97:3213. doi: 10.1073/pnas.070034097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Frisan T. Levitsky V. Masucci M. Limiting dilution assay. Methods Mol Biol. 2001;174:213. doi: 10.1385/1-59259-227-9:213. [DOI] [PubMed] [Google Scholar]
- 53.Strijbosch L.W. Buurman W.A. Does R.J. Zinken P.H. Groenewegen G. Limiting dilution assays. Experimental design and statistical analysis. J Immunol Methods. 1987;97:133. doi: 10.1016/0022-1759(87)90115-3. [DOI] [PubMed] [Google Scholar]
- 54.Taswell C. Limiting dilution assays for the determination of immunocompetent cell frequencies. I. Data analysis. J Immunol. 1981;126:1614. [PubMed] [Google Scholar]
- 55.Rosen E.D. Walkey C.J. Puigserver P. Spiegelman B.M. Transcriptional regulation of adipogenesis. Genes Dev. 2000;14:1293. [PubMed] [Google Scholar]
- 56.Odrowaz-Sypniewska G. Markers of pro-inflammatory and pro-thrombotic state in the diagnosis of metabolic syndrome. Adv Med Sci. 2007;52:246. [PubMed] [Google Scholar]
- 57.Livak K.J. Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 58.Gronthos S. Graves S.E. Ohta S. Simmons P.J. The STRO-1+ fraction of adult human bone marrow contains the osteogenic precursors. Blood. 1994;84:4164. [PubMed] [Google Scholar]
- 59.Harting M. Jimenez F. Pati S. Baumgartner J. Cox C., Jr. Immunophenotype characterization of rat mesenchymal stromal cells. Cytotherapy. 2008;10:243. doi: 10.1080/14653240801950000. [DOI] [PubMed] [Google Scholar]
- 60.Nasef A. Zhang Y.Z. Mazurier C. Bouchet S. Bensidhoum M. Francois S. Gorin N.C. Lopez M. Thierry D. Fouillard L. Chapel A. Selected Stro-1-enriched bone marrow stromal cells display a major suppressive effect on lymphocyte proliferation. Int J Lab Hematol. 2009;31:9. doi: 10.1111/j.1751-553X.2007.00997.x. [DOI] [PubMed] [Google Scholar]
- 61.Gronthos S. McCarty R. Mrozik K. Fitter S. Paton S. Menicanin D. Itescu S. Bartold P.M. Xian C. Zannettino A.C. Heat shock protein-90 beta is expressed at the surface of multipotential mesenchymal precursor cells: generation of a novel monoclonal antibody, STRO-4, with specificity for mesenchymal precursor cells from human and ovine tissues. Stem Cells Dev. 2009;18:1253. doi: 10.1089/scd.2008.0400. [DOI] [PubMed] [Google Scholar]
- 62.Bensidhoum M. Chapel A. Francois S. Demarquay C. Mazurier C. Fouillard L. Bouchet S. Bertho J.M. Gourmelon P. Aigueperse J. Charbord P. Gorin N.C. Thierry D. Lopez M. Homing of in vitro expanded Stro-1- or Stro-1+ human mesenchymal stem cells into the NOD/SCID mouse and their role in supporting human CD34 cell engraftment. Blood. 2004;103:3313. doi: 10.1182/blood-2003-04-1121. [DOI] [PubMed] [Google Scholar]
- 63.Psaltis P.J. Paton S. See F. Arthur A. Martin S. Itescu S. Worthley S.G. Gronthos S. Zannettino A.C. Enrichment for STRO-1 expression enhances the cardiovascular paracrine activity of human bone marrow-derived mesenchymal cell populations. J Cell Physiol. 2010;223:530. doi: 10.1002/jcp.22081. [DOI] [PubMed] [Google Scholar]
- 64.Colter D.C. Sekiya I. Prockop D.J. Identification of a subpopulation of rapidly self-renewing and multipotential adult stem cells in colonies of human marrow stromal cells. Proc Natl Acad Sci U S A. 2001;98:7841. doi: 10.1073/pnas.141221698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Larson B.L. Ylostalo J. Lee R.H. Gregory C. Prockop D.J. Sox11 is expressed in early progenitor human multipotent stromal cells and decreases with extensive expansion of the cells. Tissue Eng Part A. 2010;16:3385. doi: 10.1089/ten.tea.2010.0085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Qian S.W. Li X. Zhang Y.Y. Huang H.Y. Liu Y. Sun X. Tang Q.Q. Characterization of adipocyte differentiation from human mesenchymal stem cells in bone marrow. BMC Dev Biol. 2010;10:47. doi: 10.1186/1471-213X-10-47. [DOI] [PMC free article] [PubMed] [Google Scholar]