Abstract
MicroRNAs (miRNAs) are important post-transcriptional regulators of nearly every biological process in the cell and play key roles in the pathogenesis of human disease. As a result, there are many drug discovery programs that focus on developing miRNA-based therapeutics. The most advanced of these programs targets the liver-expressed miRNA-122 using the locked nucleic acid (LNA)–modified antisense oligonucleotide miravirsen. Here, we describe the discovery of miravirsen, which is currently in phase 2 clinical trials for treatment of hepatitis C virus (HCV) infection.
The discovery of microRNAs: From worm genetics to human disease
The first miRNA gene, lin-4, was discovered in Caenorhabditis elegans by Victor Ambros and Gary Ruvkun in 1993 (Fig. 1), and shown to control developmental timing in the worm by binding to partially complementary sites in the 3′ untranslated region (UTR) of lin-14 mRNA (Lee et al., 1993; Wightman et al., 1993). For several years lin-4 was considered an oddity in worm genetics, until the discovery of a second, small regulatory RNA in the worm, called let-7, which was found to be highly conserved among animals, including humans (Pasquinelli et al., 2000; Reinhart et al., 2000). These seminal discoveries attracted the research laboratories of Victor Ambros, David Bartel, and Thomas Tuschl to search for similar RNAs in animals by small RNA cloning, which led to the discovery of numerous miRNAs in C. elegans, Drosophila embryos, and human HeLa cells (Fig. 1; Lagos-Quintana et al., 2001; Lau et al., 2001; Lee and Ambros, 2001). Soon after, the first studies linking miRNA dysregulation with human disease were published (Fig. 1). The human miRNA genes mir-15a and mir-16-1 were found to be deleted or down-regulated in the majority of B cell chronic lymphocytic leukemia (CLL) cases (Calin et al., 2002). Furthermore, the miR-17-92 cluster, which is amplified in many cancers, including B cell lymphomas, and miR-155, which is overexpressed in hematological malignancies, were reported as the first human oncogenic miRNAs (Eis et al., 2005; He et al., 2005; Costinean et al., 2006). Since then, 18,226 miRNAs have been annotated in animals, plants, and viruses, including 1,921 miRNAs encoded in the human genome (Kozomara and Griffiths-Jones, 2011). Indeed, miRNAs are predicted to repress a large fraction of all protein-coding genes and to participate in the regulation of almost every biological process in the cell (Kloosterman and Plasterk, 2006; Bushati and Cohen, 2007; Bartel, 2009; Friedman et al., 2009; Ambros, 2011). Moreover, recent work implies that miRNA dysregulation is frequently associated with the pathogenesis of human diseases (Gottwein and Cullen, 2008; Ventura and Jacks, 2009; Williams et al., 2009; Mendell and Olson, 2012), and has led to the discovery of several disease-implicated miRNAs that show promise as therapeutic targets (Stenvang et al., 2012; van Rooij et al., 2012).
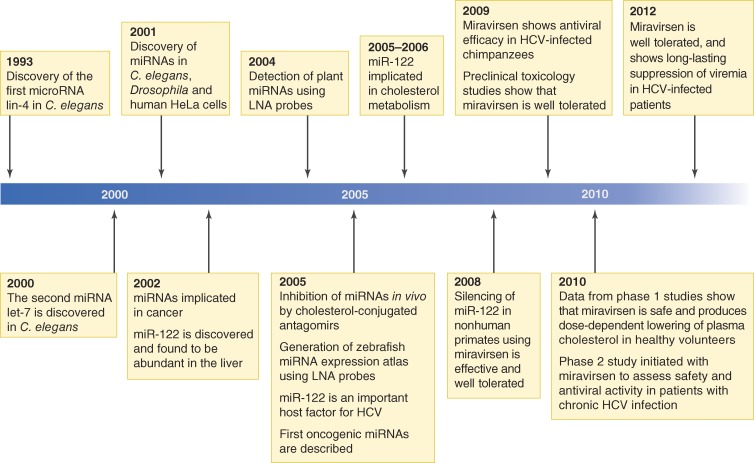
Figure 1.
Key findings in miRNA research and the discovery of the first miRNA-targeted drug, miravirsen. The timeline indicates the key events from the discovery of the first microRNA, lin-4, in C. elegans to clinical testing of miravirsen in HCV-infected patients.
The first challenge: Developing the right tools
As the miRNA research field progressed, the first challenge was to develop and improve miRNA detection and functional analysis tools given the small size and sometimes low level of expression of different miRNAs. An important addition to the miRNA toolbox came from LNA (locked nucleic acid), a bicyclic high-affinity RNA analogue in which the ribose ring is chemically locked in an N-type (C3′-endo) conformation by the introduction of a 2′-O,4′-C methylene bridge (Koshkin et al., 1998). LNA-modified oligonucleotides possess high thermal stability when hybridized with their cognate mRNA target molecules (Koshkin et al., 1998; Braasch and Corey, 2001; Kurreck et al., 2002), and therefore appeared ideally suited for targeting of small RNAs, such as miRNAs. Thus, Sakari Kauppinen, who had been working with the development of LNA research tools since 2001, initiated a collaboration with the laboratories of Zoltán Havelda and József Burgyán to assess the utility of LNA probes in miRNA detection in plants. Indeed, this research team first reported the improved sensitivity and high specificity of LNA-modified oligonucleotide probes in detecting different plant miRNAs by small RNA Northern blot analysis (Válóczi et al., 2004) and in situ hybridization (Fig. 1; Válóczi et al., 2006). Encouraged by these findings, Kauppinen collaborated with Ronald Plasterk, then at the Hubrecht Institute, who applied LNA probes in the detection of zebrafish miRNAs. In 2005, the Plasterk group reported the spatial and temporal expression patterns of 115 conserved vertebrate miRNAs that were determined by whole-mount in situ hybridizations in zebrafish embryos with LNA-modified DNA probes similar to those used in miRNA detection in plants (Fig. 1; Wienholds et al., 2005). In a follow-up study, Kloosterman et al. (2006) demonstrated the utility of LNA-based miRNA in situ hybridization in mouse embryos, whereas another report showed that LNA-modified antisense oligonucleotides, termed antimiRs, could be used to inhibit miRNA function in cultured cells (Ørom et al., 2006). From these studies, LNAs emerged as an important tool in miRNA research.
The second challenge: Finding the right target
microRNA-122: From zebrafish to therapeutic target in liver disease.
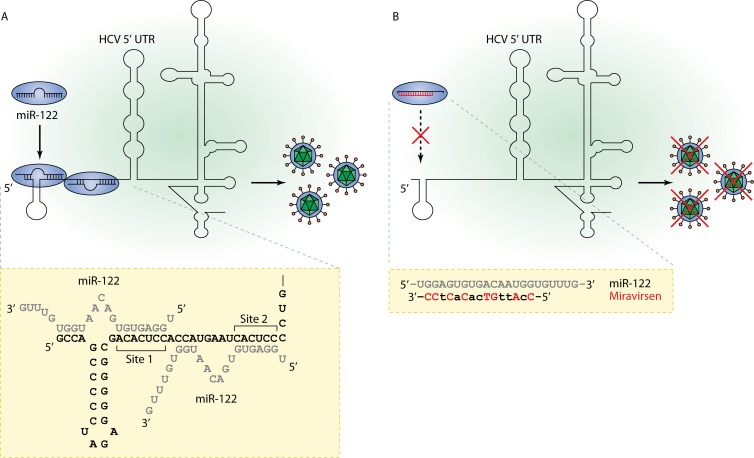
In 2005, several independent discoveries in the miRNA field led us to embark on the goal of developing the world’s first microRNA therapeutic (Fig. 1). First, emerging evidence indicated that miRNA dysregulation was associated with the pathogenesis of human disease. Second, as described in the previous section, the successful use of short LNA probes with high melting temperatures in miRNA in situ detection and in inhibiting miRNA function in cells suggested that the LNA chemistry would enable the design of high affinity antimiR oligonucleotides for pharmacological inhibition of disease-implicated miRNAs in vivo, provided that we could tackle the delivery issues. Third, cloning of tissue-specific miRNAs from mouse alongside the zebrafish miRNA expression atlas featured an animal miRNA named miR-122 (Lagos-Quintana et al., 2002; Wienholds et al., 2005) that caught our attention. Not only was miR-122 completely conserved in all vertebrates from zebrafish to man, but it also turned out to be highly abundant and expressed almost exclusively in the liver. These characteristics made miR-122 especially attractive for therapeutic targeting because lessons learned from three decades of research on phosphorothioate (PS)-modified antisense oligonucleotides had shown that they accumulated efficiently in the liver (Crooke, 2007). This work suggested that antimiR oligonucleotides with PS backbone modifications could similarly be delivered to the liver without additional conjugation or formulation chemistries. Finally, in a seminal study published in 2005, Peter Sarnow and his team reported that the liver-expressed miR-122 interacts with the HCV RNA by binding to two miR-122 seed sites located in the 5′ UTR of the virus genome, leading to up-regulation of the viral RNA in infected liver cells (Fig. 2 A; Jopling et al., 2005). This implied that miR-122 was an important host factor for the virus and, hence, a potential therapeutic target for treatment of HCV infection, a leading cause of liver disease with over 170 million infected individuals worldwide. The idea of developing a miR-122–based antiviral therapy was especially appealing, due to the many limitations and side effects of the current standard of care HCV therapy.
Figure 2.
A model for interaction of miR-122 with the 5′ UTR of the HCV RNA and miravirsen’s mechanism of action. (A) The liver-expressed miR-122 is an important host factor for HCV and interacts with the 5′ untranslated region (UTR) of the virus genome by binding to two miR-122 seed sites in association with Ago2. By forming an oligomeric miR-122-HCV complex, miR-122 protects the 5′ UTR from nucleolytic degradation, thereby promoting viral RNA stability and propagation (Jopling et al., 2005; Machlin et al., 2011; Shimakami et al., 2012). (B) Miravirsen is a high-affinity 15-mer LNA-modified antimiR oligonucleotide (red uppercase, LNA modifications; lowercase, DNA) that acts by sequestering mature miR-122, leading to inhibition of miR-122 function and thereby suppression of HCV.
The third challenge: Identifying a promising lead compound
Our first goal was to obtain a fast proof-of-concept for targeting of miR-122 using an LNA-modified antimiR oligonucleotide, and, thus, we chose a molecular design similar to that used in LNA probes for miRNA Northern blots and miRNA in situ hybridization, and adapted it for in vivo use by substituting the phosphodiester linkages with PS linkages. While this work was in progress in our laboratory, two studies reported on pharmacological inhibition of miR-122 in the mouse liver. Krützfeldt et al. (2005) described for the first time effective miR-122 silencing in mice using 3′ cholesterol-conjugated, 2′-O-Me oligonucleotides, termed antagomirs, after three intravenous doses of 80 mg/kg antagomir-122, whereas Esau et al. (2006) used an unconjugated 2′-O-methoxyethyl–modified PS oligonucleotide and achieved inhibition of miR-122 in mice after four weeks of treatment by twice weekly intraperitoneal doses ranging from 12.5 to 75 mg/kg/dose. Notably, both studies implicated miR-122 as a modulator of hepatic lipid metabolism and reported a 30–40% decrease in plasma cholesterol levels in the treated mice, which thereby provided a highly useful biomarker for assessing antimiR-122 activity in vivo. Our strategy for inhibiting miR-122 in the mouse liver turned out to be successful: a 16-mer LNA-modified antimiR oligonucleotide reproduced the low cholesterol phenotype upon systemic delivery in mice (Elmén et al., 2008a).
In parallel with this work, we were exploring different LNA designs to improve the antimiR drug potency. To this end, we synthesized a small library of LNA-modified antimiRs, in which we incorporated several features that we considered as important in the design of potent miRNA inhibitors (Elmén et al., 2008b). First, the use of LNA enabled us to synthesize short oligonucleotides with high melting temperatures, and we therefore decided to truncate the antimiR oligonucleotides from a fully complementary format to designs comprising 13-mer to 16-mer antimiRs with a high melting temperature of at least 60°C. Second, the antimiRs were all designed to target the 5′ end of miR-122 due to the importance of the miRNA seed region (nucleotides 2–8) in miRNA target recognition (Krek et al., 2005; Lewis et al., 2005). Third, we excluded the nucleotide complementary to the first nucleotide in the miRNA sequence because structural studies had indicated that it was hidden in a separate binding pocket in the Argonaute protein (Ma et al., 2005: Parker et al., 2005). Fourth, to prevent possible slicing of the antimiR by the RISC complex, we introduced LNA modifications at the site of passenger strand cleavage (Matranga et al., 2005; Leuschner et al., 2006). Finally, to enhance the pharmacokinetic properties and improve nuclease resistance, all antimiR compounds were synthesized with a complete PS backbone and preferentially with LNA modifications at the 5′ and 3′ end of the antimiR oligonucleotides (Elmén et al., 2008b).
Next, to establish a sensitive assay that would allow us to quickly screen our antimiR inhibitor library in cultured cells, we cloned a perfect match miR-122 target site into the 3′ UTR of the Renilla luciferase reporter gene. Using this miR-122 luciferase reporter assay as a workhorse, we found that inhibition of miR-122 function in cultured human Huh-7 hepatoma cells by the different LNA oligonucleotides was affinity dependent. Our screen identified a 15-mer LNA-modified antimiR, named SPC3649, with a high melting temperature of 80°C, which mediated robust inhibition of miR-122 function in the liver cells when transfected at low nanomolar concentrations (Elmén et al., 2008b). Moreover, in close collaboration with the Sarnow laboratory at Stanford University, we found that SPC3649 was also the most potent inhibitor of HCV RNA accumulation in Huh-7 cells harboring the HCV-N replicon (Fig. 2 B; Elmén et al., 2008b). Finally, SPC3649 showed markedly improved efficiency in antagonizing miR-122 in mice compared with animals that were treated with either cholesterol-conjugated antagomir-122 or with other unconjugated antimiR oligonucleotides (Krützfeldt et al., 2005; Esau et al., 2006; Elmén et al., 2008b). These experiments identified SPC3649 (later named miravirsen) as the lead candidate for the world’s first miRNA-targeted drug.
The fourth challenge: Testing in nonhuman primates
An important step in the development of miravirsen was to ask whether it could mediate pharmacological inhibition of miR-122 in nonhuman primates. Collaborating with Matthew Lawrence and his team at RxGen, we performed a pharmacology study in African green monkeys at the research facilities on St. Kitts in the West Indies. Indeed, miravirsen’s efficacy in mice translated very well to primates, as shown by monitoring plasma cholesterol levels in monkeys over time after administration of PBS-formulated miravirsen at doses ranging from 1 to 10 mg/kg with three intravenous injections over five days (Fig. 1). This treatment resulted in efficient sequestration of miR-122, leading to dose-dependent and long-lasting decrease of total plasma cholesterol that returned to baseline over a three-month period (Elmén et al., 2008b). We also found miravirsen to be well tolerated without any acute or subchronic toxicity in the monkeys. Indeed, these data showed for the first time that efficient antagonism of miR-122 can be achieved in nonhuman primates by simple delivery of unconjugated LNA-modified antimiR oligonucleotides. Moreover, the safety profile in this study as well as those in subsequent preclinical toxicology studies in cynomolgus monkeys (Hildebrandt-Eriksen et al., 2012) were found to be very favorable. Highly encouraged by these findings, we were thus prepared to take the next steps toward the clinic.
Miravirsen shows efficacy in HCV-infected chimpanzees.
Because miravirsen was a first-in-class inhibitor of a miRNA, we wanted to obtain proof-of-concept in an animal model of HCV before advancing it to clinical trials. Together with Robert Lanford at the Southwest Foundation of Biomedical Research we performed an efficacy study in the only validated animal model for HCV: chronically HCV-infected chimpanzee (Lanford et al., 2010). In this study, two chimpanzees were administered with 12 weekly intravenous doses of 5 mg/kg and two with 12 doses of 1 mg/kg of miravirsen. A significant decline of the HCV viral titer was detected in the sera of the two chimpanzees in the 5 mg/kg miravirsen dose group with a maximum decrease of 2.6 orders of magnitude two weeks after last miravirsen dose, with no evidence of side effects in the treated animals. Importantly, no viral rebound was detected during the 12-wk treatment phase, which was consistent with the lack of adaptive mutations in the two miR-122 seed sites of the HCV 5′ UTR, suggesting that miravirsen had a high barrier to viral resistance. These findings are in stark contrast to what has been observed with direct acting antivirals, where resistance and viral breakthrough in the worst cases develop already after 6–7 days of treatment (Cooper et al., 2009). In addition, the fact that the two critical miR-122 binding sites are completely conserved in all HCV genotypes implied that the antiviral effect of miravirsen on HCV would be genotype independent, which has recently been confirmed by another study (Li et al., 2011).
The fifth challenge: Advancing miravirsen from bench to bedside
The miR-122 inhibitor miravirsen is the first and to date only miRNA-targeted drug to have entered human clinical trials. Two phase 1 safety studies have been performed in healthy male volunteers: (1) a single ascending dose study with doses up to 12 mg/kg, and (2) a multiple ascending dose study with up to five doses of 5 mg/kg. Data from these studies demonstrated dose-dependent pharmacology as measured by lowering of plasma cholesterol, and showed that miravirsen was well tolerated with no dose-limiting toxicities (Hildebrandt-Eriksen et al., 2009). In 2010, Santaris Pharma initiated a phase 2a study to assess the safety and antiviral activity of miravirsen in treatment-naive HCV patients (Janssen et al., 2011). In this study, 36 patients with chronic HCV genotype 1 infection were enrolled and randomized to three cohorts with nine miravirsen-treated and three placebo-treated subjects in each cohort. Treatment with miravirsen at doses of 3, 5, or 7 mg/kg as a total of 5 weekly subcutaneous injections over 29 days provided dose-dependent and long-lasting antiviral activity with a mean decrease of 2–3 logs from baseline HCV RNA levels (log10 IU/ml). Furthermore, HCV RNA became undetectable in four out of nine patients in the high dose cohort (Janssen et al., 2011). The reported adverse events were infrequent and mostly mild, such as headache or coryza, and no serious adverse events or clinically significant changes in safety tests, vital signs, or electrocardiograms were observed (Janssen et al., 2011). Taken together, these data indicate that a four-week miravirsen monotherapy provides long-lasting suppression of viremia, has a high barrier to viral resistance, and is well tolerated in patients with chronic HCV infection.
So what began with the seminal discovery of the founding member of microRNAs, lin-4, in the worm just two decades ago, has now translated to clinical testing of the world’s first miRNA-targeted drug in HCV-infected patients, thereby setting the stage for improved anti-HCV therapy. We believe that a large part of the success and the remarkable speed by which miravirsen has advanced from bench to bedside is due to the many highly fruitful and agile collaborations between researchers in academia and the biotech industry with a shared vision of translating an exciting scientific discovery into a new therapy to improve human health. Thus, we hope that this story can serve as an inspiration and example for the benefit of both science and patients and look forward to seeing the next microRNA-based drug enter clinical development.
Acknowledgments
We would like to thank our colleagues at Santaris Pharma and all our collaborators who have contributed to the miravirsen endeavor over the years.
The miravirsen project has been supported by grants 006-2005-1 and 044-2007-1 from the Danish National Advanced Technology Foundation (to S. Kauppinen). Illustrations were provided by Neil Smith, www.neilsmithillustration.co.uk.
Footnotes
Abbreviations used in this paper:
- HCV
- hepatitis C virus
- LNA
- locked nucleic acid
- miRNA
- microRNA
- PS
- phosphorothioate
- UTR
- untranslated region
References
- Ambros V. 2011. MicroRNAs and developmental timing. Curr. Opin. Genet. Dev. 21:511–517 10.1016/j.gde.2011.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel D.P. 2009. MicroRNAs: target recognition and regulatory functions. Cell. 136:215–233 10.1016/j.cell.2009.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braasch D.A., Corey D.R. 2001. Locked nucleic acid (LNA): fine-tuning the recognition of DNA and RNA. Chem. Biol. 8:1–7 10.1016/S1074-5521(00)00058-2 [DOI] [PubMed] [Google Scholar]
- Bushati N., Cohen S.M. 2007. microRNA functions. Annu. Rev. Cell Dev. Biol. 23:175–205 10.1146/annurev.cellbio.23.090506.123406 [DOI] [PubMed] [Google Scholar]
- Calin G.A., Dumitru C.D., Shimizu M., Bichi R., Zupo S., Noch E., Aldler H., Rattan S., Keating M., Rai K., et al. 2002. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. USA. 99:15524–15529 10.1073/pnas.242606799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper C., Lawitz E.J., Ghali P., Rodriguez-Torres M., Anderson F.H., Lee S.S., Bédard J., Chauret N., Thibert R., Boivin I., et al. 2009. Evaluation of VCH-759 monotherapy in hepatitis C infection. J. Hepatol. 51:39–46 10.1016/j.jhep.2009.03.015 [DOI] [PubMed] [Google Scholar]
- Costinean S., Zanesi N., Pekarsky Y., Tili E., Volinia S., Heerema N., Croce C.M. 2006. Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in E(mu)-miR155 transgenic mice. Proc. Natl. Acad. Sci. USA. 103:7024–7029 10.1073/pnas.0602266103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooke S.T. 2007. Antisense Drug Technology. 2nd ed. CRC Press, Boca Raton, FL: 848 pp [Google Scholar]
- Eis P.S., Tam W., Sun L., Chadburn A., Li Z., Gomez M.F., Lund E., Dahlberg J.E. 2005. Accumulation of miR-155 and BIC RNA in human B cell lymphomas. Proc. Natl. Acad. Sci. USA. 102:3627–3632 10.1073/pnas.0500613102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmén J., Lindow M., Silahtaroglu A.N., Bak M., Christensen M., Lind-Thomsen A., Hedtjärn M., Hansen J.B., Hansen H.F., Straarup E.M., et al. 2008a. Antagonism of microRNA-122 in mice by systemically administered LNA-antimiR leads to up-regulation of a large set of predicted target mRNAs in the liver. Nucleic Acids Res. 36:1153–1162 10.1093/nar/gkm1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmén J., Lindow M., Schütz S., Lawrence M., Petri A., Obad S., Lindholm M., Hedtjärn M., Hansen H.F., Berger U., et al. 2008b. LNA-mediated microRNA silencing in non-human primates. Nature. 452:896–899 10.1038/nature06783 [DOI] [PubMed] [Google Scholar]
- Esau C., Davis S., Murray S.F., Yu X.X., Pandey S.K., Pear M., Watts L., Booten S.L., Graham M., McKay R., et al. 2006. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 3:87–98 10.1016/j.cmet.2006.01.005 [DOI] [PubMed] [Google Scholar]
- Friedman R.C., Farh K.K.-H., Burge C.B., Bartel D.P. 2009. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 19:92–105 10.1101/gr.082701.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottwein E., Cullen B.R. 2008. Viral and cellular microRNAs as determinants of viral pathogenesis and immunity. Cell Host Microbe. 3:375–387 10.1016/j.chom.2008.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L., Thomson J.M., Hemann M.T., Hernando-Monge E., Mu D., Goodson S., Powers S., Cordon-Cardo C., Lowe S.W., Hannon G.J., Hammond S.M. 2005. A microRNA polycistron as a potential human oncogene. Nature. 435:828–833 10.1038/nature03552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrandt-Eriksen E.S., Bagger Y.Z., Knudsen T.B. 2009. A unique therapy for HCV inhibits microRNA-122 in humans and results in HCV RNA suppression in chronically infected chimpanzees: results from primate and first-in-human studies. Hepatology. 50:12A [Google Scholar]
- Hildebrandt-Eriksen E.S., Aarup V., Persson R., Hansen H.F., Munk M.E., Ørum H. 2012. A locked nucleic acid oligonucleotide targeting microRNA 122 is well-tolerated in cynomolgus monkeys. Nucleic Acid Ther. 22:152–161 [DOI] [PubMed] [Google Scholar]
- Janssen H.L., Reesink H.W., Zeuzem S., Lawitz E., Rodriguez-Torres M., Chen A., Davis C., King B., Levin A.A., Hodges M.R. 2011. A randomized, double-blind, placebo (plb) controlled safety and anti-viral proof of concept study of miravirsen (MIR), an oligonucleotide targeting miR-122, in treatment naïve patients with genotype 1 (gt1) chronic HCV infection. Hepatology. 54:1430A [Google Scholar]
- Jopling C.L., Yi M., Lancaster A.M., Lemon S.M., Sarnow P. 2005. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 309:1577–1581 10.1126/science.1113329 [DOI] [PubMed] [Google Scholar]
- Kloosterman W.P., Plasterk R.H.A. 2006. The diverse functions of microRNAs in animal development and disease. Dev. Cell. 11:441–450 10.1016/j.devcel.2006.09.009 [DOI] [PubMed] [Google Scholar]
- Kloosterman W.P., Wienholds E., de Bruijn E., Kauppinen S., Plasterk R.H.A. 2006. In situ detection of miRNAs in animal embryos using LNA-modified oligonucleotide probes. Nat. Methods. 3:27–29 10.1038/nmeth843 [DOI] [PubMed] [Google Scholar]
- Koshkin A.A., Singh S.K., Nielsen P., Rajwanshi V.K., Kumar R., Meldgaard M., Olsen C.E., Wengel J. 1998. LNA (Locked Nucleic Acids): Synthesis of the adenine, cytosine, guanine, 5-methylcytosine, thymine and uracil bicyclonucleoside monomers, oligomerisation, and unprecedented nucleic acid recognition. Tetrahedron. 54:3607–3630 10.1016/S0040-4020(98)00094-5 [DOI] [Google Scholar]
- Kozomara A., Griffiths-Jones S. 2011. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 39(Database issue):D152–D157 10.1093/nar/gkq1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krek A., Grün D., Poy M.N., Wolf R., Rosenberg L., Epstein E.J., MacMenamin P., da Piedade I., Gunsalus K.C., Stoffel M., Rajewsky N. 2005. Combinatorial microRNA target predictions. Nat. Genet. 37:495–500 10.1038/ng1536 [DOI] [PubMed] [Google Scholar]
- Krützfeldt J., Rajewsky N., Braich R., Rajeev K.G., Tuschl T., Manoharan M., Stoffel M. 2005. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 438:685–689 10.1038/nature04303 [DOI] [PubMed] [Google Scholar]
- Kurreck J., Wyszko E., Gillen C., Erdmann V.A. 2002. Design of antisense oligonucleotides stabilized by locked nucleic acids. Nucleic Acids Res. 30:1911–1918 10.1093/nar/30.9.1911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagos-Quintana M., Rauhut R., Lendeckel W., Tuschl T. 2001. Identification of novel genes coding for small expressed RNAs. Science. 294:853–858 10.1126/science.1064921 [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M., Rauhut R., Yalcin A., Meyer J., Lendeckel W., Tuschl T. 2002. Identification of tissue-specific microRNAs from mouse. Curr. Biol. 12:735–739 10.1016/S0960-9822(02)00809-6 [DOI] [PubMed] [Google Scholar]
- Lanford R.E., Hildebrandt-Eriksen E.S., Petri A., Persson R., Lindow M., Munk M.E., Kauppinen S., Ørum H. 2010. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science. 327:198–201 10.1126/science.1178178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau N.C., Lim L.P., Weinstein E.G., Bartel D.P. 2001. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 294:858–862 10.1126/science.1065062 [DOI] [PubMed] [Google Scholar]
- Lee R.C., Ambros V. 2001. An extensive class of small RNAs in Caenorhabditis elegans. Science. 294:862–864 10.1126/science.1065329 [DOI] [PubMed] [Google Scholar]
- Lee R.C., Feinbaum R.L., Ambros V. 1993. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 75:843–854 10.1016/0092-8674(93)90529-Y [DOI] [PubMed] [Google Scholar]
- Leuschner P.J.F., Ameres S.L., Kueng S., Martinez J. 2006. Cleavage of the siRNA passenger strand during RISC assembly in human cells. EMBO Rep. 7:314–320 10.1038/sj.embor.7400637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis B.P., Burge C.B., Bartel D.P. 2005. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 120:15–20 10.1016/j.cell.2004.12.035 [DOI] [PubMed] [Google Scholar]
- Li Y.P., Gottwein J.M., Scheel T.K., Jensen T.B., Bukh J. 2011. MicroRNA-122 antagonism against hepatitis C virus genotypes 1-6 and reduced efficacy by host RNA insertion or mutations in the HCV 5′ UTR. Proc. Natl. Acad. Sci. USA. 108:4991–4996 10.1073/pnas.1016606108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J.-B., Yuan Y.-R., Meister G., Pei Y., Tuschl T., Patel D.J. 2005. Structural basis for 5′-end-specific recognition of guide RNA by the A. fulgidus Piwi protein. Nature. 434:666–670 10.1038/nature03514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machlin E.S., Sarnow P., Sagan S.M. 2011. Masking the 5’ terminal nucleotides of the hepatitis C virus genome by an unconventional microRNA-target RNA complex. Proc. Natl. Acad. Sci. USA. 108:3193–3198 10.1073/pnas.1012464108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matranga C., Tomari Y., Shin C., Bartel D.P., Zamore P.D. 2005. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell. 123:607–620 10.1016/j.cell.2005.08.044 [DOI] [PubMed] [Google Scholar]
- Mendell J.T., Olson E.N. 2012. MicroRNAs in stress signaling and human disease. Cell. 148:1172–1187 10.1016/j.cell.2012.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ørom U.A., Kauppinen S., Lund A.H. 2006. LNA-modified oligonucleotides mediate specific inhibition of microRNA function. Gene. 372:137–141 10.1016/j.gene.2005.12.031 [DOI] [PubMed] [Google Scholar]
- Parker J.S., Roe S.M., Barford D. 2005. Structural insights into mRNA recognition from a PIWI domain-siRNA guide complex. Nature. 434:663–666 10.1038/nature03462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquinelli A.E., Reinhart B.J., Slack F., Martindale M.Q., Kuroda M.I., Maller B., Hayward D.C., Ball E.E., Degnan B., Müller P., et al. 2000. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 408:86–89 10.1038/35040556 [DOI] [PubMed] [Google Scholar]
- Reinhart B.J., Slack F.J., Basson M., Pasquinelli A.E., Bettinger J.C., Rougvie A.E., Horvitz H.R., Ruvkun G. 2000. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 403:901–906 10.1038/35002607 [DOI] [PubMed] [Google Scholar]
- Shimakami T., Yamane D., Welsch C., Hensley L., Jangra R.K., Lemon S.M. 2012. Base pairing between hepatitis C virus RNA and microRNA 122 3′ of its seed sequence is essential for genome stabilization and production of infectious virus. J. Virol. 86:7372–7383 10.1128/JVI.00513-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenvang J., Petri A., Lindow M., Obad S., Kauppinen S. 2012. Inhibition of microRNA function by antimiR oligonucleotides. Silence. 3:1 10.1186/1758-907X-3-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Válóczi A., Hornyik C., Varga N., Burgyán J., Kauppinen S., Havelda Z. 2004. Sensitive and specific detection of microRNAs by northern blot analysis using LNA-modified oligonucleotide probes. Nucleic Acids Res. 32:e175 10.1093/nar/gnh171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Válóczi A., Várallyay E., Kauppinen S., Burgyán J., Havelda Z. 2006. Spatio-temporal accumulation of microRNAs is highly coordinated in developing plant tissues. Plant J. 47:140–151 10.1111/j.1365-313X.2006.02766.x [DOI] [PubMed] [Google Scholar]
- van Rooij E., Purcell A.L., Levin A.A. 2012. Developing microRNA therapeutics. Circ. Res. 110:496–507 10.1161/CIRCRESAHA.111.247916 [DOI] [PubMed] [Google Scholar]
- Ventura A., Jacks T. 2009. MicroRNAs and cancer: short RNAs go a long way. Cell. 136:586–591 10.1016/j.cell.2009.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wienholds E., Kloosterman W.P., Miska E., Alvarez-Saavedra E., Berezikov E., de Bruijn E., Horvitz H.R., Kauppinen S., Plasterk R.H.A. 2005. MicroRNA expression in zebrafish embryonic development. Science. 309:310–311 10.1126/science.1114519 [DOI] [PubMed] [Google Scholar]
- Wightman B., Ha I., Ruvkun G. 1993. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 75:855–862 10.1016/0092-8674(93)90530-4 [DOI] [PubMed] [Google Scholar]
- Williams A.H., Liu N., van Rooij E., Olson E.N. 2009. MicroRNA control of muscle development and disease. Curr. Opin. Cell Biol. 21:461–469 10.1016/j.ceb.2009.01.029 [DOI] [PMC free article] [PubMed] [Google Scholar]