Phosphocaveolin-1 regulates a positive feedback loop that responds to mechanical stress to induce caveola biogenesis by relieving Egr1 transcriptional inhibition of caveolin-1 and cavin-1.
Abstract
Caveolin-1 (Cav1) is an essential component of caveolae whose Src kinase-dependent phosphorylation on tyrosine 14 (Y14) is associated with regulation of focal adhesion dynamics. However, the relationship between these disparate functions remains to be elucidated. Caveola biogenesis requires expression of both Cav1 and cavin-1, but Cav1Y14 phosphorylation is dispensable. In this paper, we show that Cav1 tyrosine phosphorylation induces caveola biogenesis via actin-dependent mechanotransduction and inactivation of the Egr1 (early growth response-1) transcription factor, relieving inhibition of endogenous Cav1 and cavin-1 genes. Cav1 phosphorylation reduces Egr1 binding to Cav1 and cavin-1 promoters and stimulates their activity. In MDA-231 breast carcinoma cells that express elevated levels of Cav1 and caveolae, Egr1 regulated Cav1, and cavin-1 promoter activity was dependent on actin, Cav1, Src, and Rho-associated kinase as well as downstream protein kinase C (PKC) signaling. pCav1 is therefore a mechanotransducer that acts via PKC to relieve Egr1 transcriptional inhibition of Cav1 and cavin-1, defining a novel feedback regulatory loop to regulate caveola biogenesis.
Introduction
Caveolin-1 (Cav1) was originally identified as a component of caveolae and a major tyrosine-phosphorylated substrate of Src kinase (Glenney and Soppet, 1992; Kurzchalia et al., 1992; Rothberg et al., 1992). Caveola formation is critically dependent on expression of Cav1 as well as cavin-1/PTRF (polymerase I and transcript release factor), which along with cavin-2, -3, and -4 forms a protein family whose interaction with Cav1 impacts Cav1 stability and caveola biogenesis, morphology, and dynamics (Hill et al., 2008; Liu and Pilch, 2008; Bastiani et al., 2009; Hansen et al., 2009; McMahon et al., 2009). Tyrosine phosphorylation of Cav1 on Y14 is associated with Rous sarcoma virus transformation (Glenney, 1989; Li et al., 1996) and also occurs in response to growth factor stimulation and integrin-mediated mechanotransduction (Mastick et al., 1995; Kim et al., 2000; Lee et al., 2000; Fielding et al., 2004; Radel and Rizzo, 2005). pCav1 promotes integrin and focal adhesion dynamics, activates RhoA GTPase, and stimulates cell migration (del Pozo et al., 2005; Grande-García et al., 2007; Goetz et al., 2008; Joshi et al., 2008). pCav1 also mediates raft-dependent internalization of signaling complexes upon cell detachment as well as lipid order and organization within focal adhesions (del Pozo et al., 2005; Gaus et al., 2006). Tyrosine phosphorylation of Cav1 is required for EGF induction of caveola formation and E-cadherin endocytosis at epithelial junctions (Orlichenko et al., 2006, 2009). However, whether and how tyrosine phosphorylation of Cav1 impacts on caveola formation and function remain unknown.
Egr1 is a zinc finger protein and transcription factor induced by multiple cellular stimuli, including oxidative or mechanical stress (Nose and Ohba, 1996; Stula et al., 2000; Wu et al., 2010). The GC-rich consensus sequences that function as promoter-binding elements for Egr1 overlap with binding sites for the SP1 transcription factor. Competition of Egr1 with SP1 for these promoter binding sites can silence or decrease target gene expression (Silverman et al., 1997; Raychowdhury et al., 2002; Kubosaki et al., 2009). Egr1 is the substrate of multiple kinases, including PKC, and when phosphorylated fails to bind target promoter DNA (Huang and Adamson, 1995; Jain et al., 1996; Huang et al., 1998). We show here that pCav1 is a mechanotransducer that induces transcriptional activation via phosphorylation of the master regulator Egr1 and of the Cav1 and cavin-1 genes, thereby inducing caveola formation. This therefore represents the first mechanistic link between Cav1 induction of caveolae and focal adhesion regulation by pCav1. Together with the role of caveolae as a plasma membrane reservoir to buffer membrane surges upon mechanical cell stress (Sinha et al., 2011), this study defines Cav1 as a critical mechanotransducer that regulates the cellular response to mechanical stress.
Results and discussion
Cav1 Y14 phosphorylation induces caveola biogenesis through expression of Cav1 and cavin-1
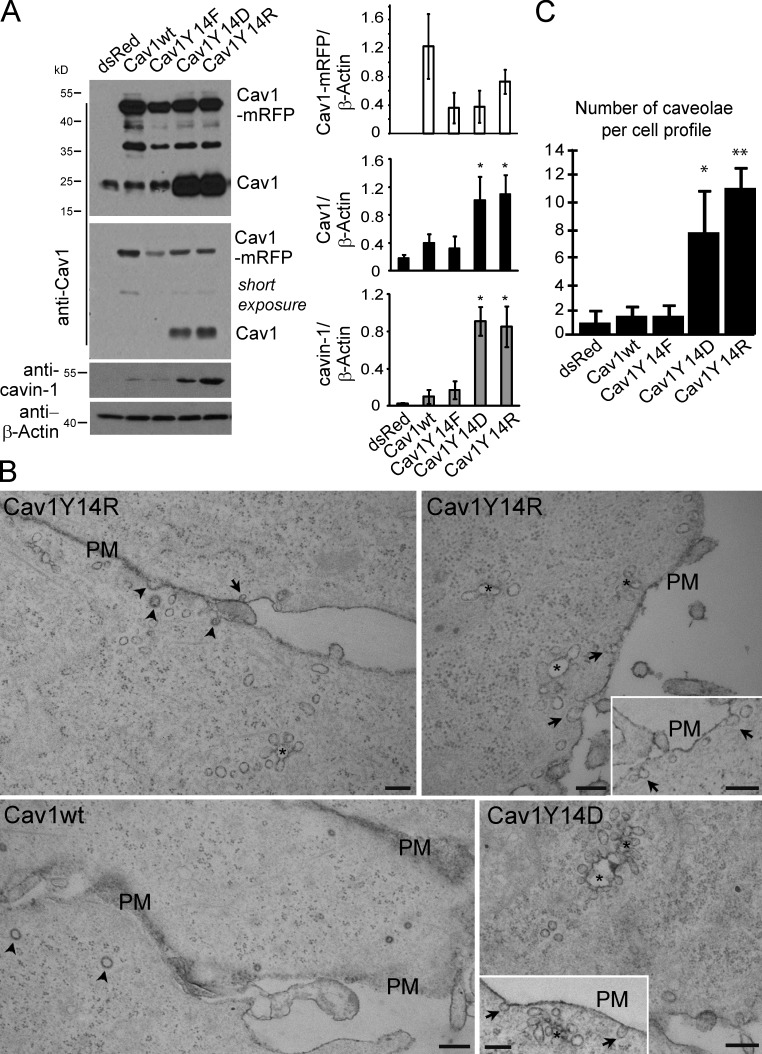
The MDA-435 cancer cell line expresses low levels of Cav1 and few caveolae (Kojic et al., 2007). Stable transfection of MDA-435 cells with wild-type, dominant-negative Y14F, phosphomimetic Y14D, or positively charged Y14R C-terminal monomeric RFP (mRFP)–tagged Cav1 (Goetz et al., 2008; Joshi et al., 2008) results in stable expression of the exogenous, higher molecular weight Cav1-mRFP constructs (Fig. 1 A). Expression of wild-type Cav1-mRFP is elevated relative to endogenous Cav1 as well as to the Cav1 Y14 mutants and comparable with endogenous Cav1 levels in caveolae-expressing MDA-231 breast carcinoma cells (Fig. S1). Interestingly, stable expression of Cav1Y14D and Y14R in MDA-435 cells results in elevated levels of endogenous Cav1 and cavin-1. Consistently, Cav1Y14D- and Y14R-expressing MDA-435 cells presented numerous caveolae and caveolae clusters by EM (Fig. 1, B and C). Surprisingly, expression of Cav1wt does not induce a significant increase in caveola density (Fig. 1, B and C). However, in Cav1−/− mouse embryo fibroblasts (MEFs), in which exogenous Cav1 induces caveola formation (Kirkham et al., 2008), all the Y14 Cav1-myc-mRFP constructs induce caveola formation (Fig. S2 A). Furthermore, interaction between Cav1-mRFP and cavin-1–GFP, measured using fluorescence lifetime measurement (FLIM), is equivalent for all Y14 mutants (Fig. S2 B). This suggests that Cav1 Y14 phosphorylation impacts neither on Cav1 interaction with cavin-1 nor on the ability of Cav1 to induce caveolae. In Cav1wt-expressing MDA-435 cells, Cav1 levels either remain below the threshold for caveola formation, or endogenous cavin-1 levels are insufficient. Induction of caveolae in the Cav1Y14D and Y14R MDA-435 transfectants may be related to elevated expression of endogenous Cav1 and cavin-1.
Figure 1.
Cav1 Y14 phosphomimetic induces expression of endogenous Cav1 and cavin-1 and caveolae. (A) Cell lysates of stably transfected MDA-435 cell lines expressing dsRed, RFP-tagged wild-type Cav1 (Cav1wt), or mutant RFP-tagged Cav1Y14F, Y14D, or 14R were Western blotted for Cav1, showing expression of higher molecular weight RFP-tagged Cav1 and endogenous Cav1, as well as with antibodies to cavin-1 and β-actin. A shorter exposure of the Cav1 blot shows dramatic increase in endogenous Cav1 and cavin-1 in Cav1Y14D and Y14R MDA-435 cell lines. Densitometric quantification of exogenous Cav1-mRFP, endogenous Cav1, and cavin-1 relative to β-actin are shown (*, P < 0.05). (B) Representative electron micrographs of stable Cav1wt-, Y14D-, and Y14R-expressing MDA-435 cells. Cav1Y14D and Y14R cells express increased numbers of single caveolae (arrows indicate typical caveolae connected to the cell surface) and surface-connected (ruthenium red positive) caveola clusters (asterisks show connections out of the plane of the section). Arrowheads indicate clathrin-coated pits. Insets show additional images of caveolae in Cav1Y14D- and Cav1Y14R-transfected cells. PM, plasma membrane. Bars, 200 nm. (C) The number of caveolae per cell profile was quantified for stable MDA-435 transfectants (*, P < 0.01; **, P < 0.001). Error bars show means ± SEM.
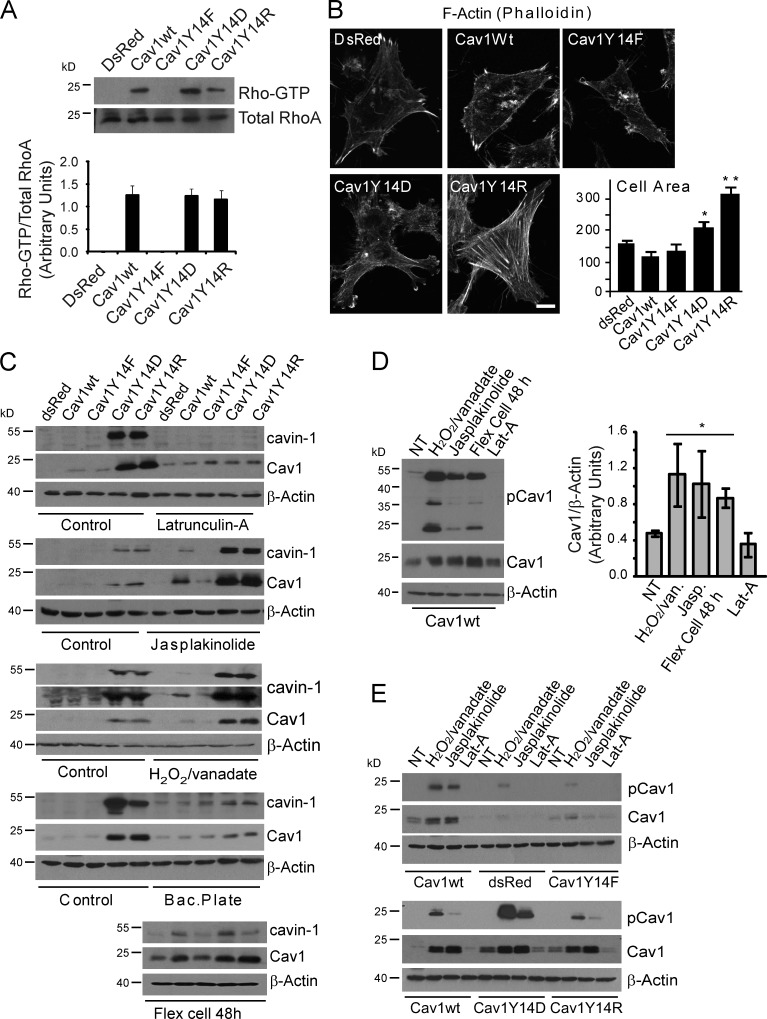
Both Cav1Y14D and Y14R functionally mimic tyrosine-phosphorylated Cav1, activating RhoA GTPase and stabilizing FAK-GFP in focal adhesions (Goetz et al., 2008; Joshi et al., 2008). RhoA GTP levels are elevated in stable Cav1wt, Y14D, and Y14R MDA-435 transfectants (Fig. 2 A). RhoA activation and recruitment of its effector Rho-associated kinase (ROCK) leads to focal adhesion maturation and actin stress fiber formation (Jaffe and Hall, 2005); however, only the Cav1Y14D and Y14R MDA-435 transfectants present increased actin stress fibers and cell spreading (Fig. 2 B). Similar effects of both negatively charged Y14D and positively charged Y14R Cav1 mutants suggest that pCav1 is not acting via SH2 domain recruitment of Csk, Grb2, or other effectors (Lee et al., 2000; Cao et al., 2002) but rather through conformational changes associated with altered actin cytoskeleton organization (Goetz et al., 2008).
Figure 2.
pCav1 impacts actin stress fiber formation and cell spreading. (A) Activated RhoA-GTP was assessed by rhotekin pull-down in MDA-435 stable transfectants expressing dsRed, Cav1wt, or Y14 mutants. (B) MDA-435 stable transfectants were fluorescently labeled with Alexa Fluor 488–phalloidin, and cell spreading was determined by scan array 48 h after plating (*, P < 0.05; **, P < 0.001). Bar, 10 µm. (C) Effect of actin depolymerization with LatA, actin stabilization with jasplakinolide and H2O2/vanadate, prevention of cell spreading by plating cells on bacterial plates (Bac.Plate), and mechanical cell stretching (36 cycles of 30-min stretched and relaxed cycles on Bioflex 6-well plates) on endogenous Cav1, cavin-1, and β-actin levels was determined by Western blotting in MDA-435 Cav1 transfectants. (D) Cav1wt cells were not treated (NT) or treated with H2O2/vanadate, jasplakinolide (Jasp.), mechanical cell stretching, or LatA and Western blotted for pCav1, Cav1, and β-actin. Densitometric quantification is presented as a bar graph (*, P < 0.05). (E) MDA-435 stable transfectants were treated with H2O2/vanadate, jasplakinolide, or LatA and Western blotted for pCav1, Cav1, and β-actin. Error bars show means ± SEM.
Inhibition of actin polymerization with latrunculin A (LatA) prevents, whereas actin stabilization with jasplakinolide enhances, induction of Cav1 and cavin-1 in Cav1Y14D- and Y14R-transfected MDA-435 cell lines (Fig. 2 C). Jasplakinolide treatment also induces Cav1/cavin-1 expression in Cav1wt-transfected cells but not control or Cav1Y14F-transfected cells as does stimulation of Cav1 phosphorylation by oxidative stress using hydrogen peroxide and sodium orthovanadate (H2O2/vanadate; Volonté et al., 2001). Prevention of cell spreading on bacterial plates inhibits induction of endogenous Cav1 and cavin-1 in Cav1Y14D and Y14R cells, whereas mechanical stretching (repeated [36×] 30-min stretched and relaxed cycles) on a FlexCell device induces Cav1 and cavin-1 in Cav1wt-transfected cells (Fig. 2 C). Tyrosine phosphorylation of both endogenous Cav1 and Cav1wt-RFP are induced by H2O2/vanadate, jasplakinolide, and mechanical stretching, whereas LatA eliminates the detectable phosphorylation of Cav1wt-RFP (Fig. 2 D). H2O2/vanadate and jasplakinolide also stimulate pCav1 expression in Cav1Y14D and Y14R, but not in dsRed or Cav1Y14F, stable MDA-435 transfectants (Fig. 2 E). Tyrosine phosphorylation of Cav1 is therefore a mechanotransducer that regulates expression of Cav1 and cavin-1 and thereby caveolae.
pCav1 regulation of Cav1 and cavin-1 is mediated by the Egr1 transcription factor
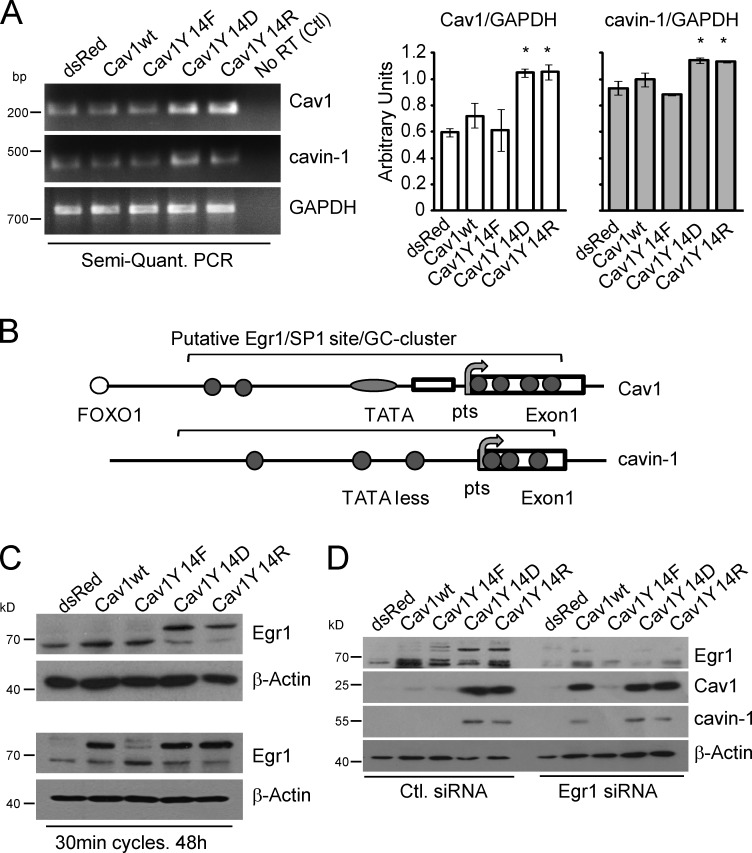
Semiquantitative RT-PCR shows the significant elevation of Cav1 and, to a lesser extent, cavin-1 mRNA in Cav1Y14D- and Y14R-transfected MDA-435 cell lines (Fig. 3 A), suggesting that their expression is regulated at the transcriptional level. Egr1 is a zinc finger protein and transcription factor that binds promoter regions overlapping consensus Sp1 sites (Shingu and Bornstein, 1994; Kubosaki et al., 2009). RhoA activation, actin stress fiber formation, and mechanical stretching increase expression of Egr1 (Nose and Ohba, 1996; Chen et al., 2010; Wu et al., 2010). Promoter analysis identifies several putative Egr1 binding sites within GC-rich clusters overlapping SP1 binding sites in the 1,000-bp promoter region upstream of the start codon and the initial 200 bp of the first exon of both the Cav1 and cavin-1 promoters (Fig. 3 B).
Figure 3.
pCav1 transcriptional regulation of Cav1 and cavin-1 is mediated by Egr1. (A) Semiquantitative (Semi-Quant.) RT-PCR of Cav1 and cavin-1 transcript levels in stable MDA-435 Cav1 transfectants. (right) Bar graphs show normalized (against glyceraldehyde 3-phosphate dehydrogenase [GAPDH]) values for Cav1 and cavin-1 PCR products (*, P < 0.05). Error bars show means ± SEM. (B) Schematic diagrams of Cav1 and cavin-1 promoter analysis show several putative Egr1 binding sites (gray circles) within an ∼1-kb area (between −1,000 and 200) comprising the 1,000 bp upstream from the start codon and the first exon of the Cav1 and cavin-1 promoters. Egr1 sites overlap with SP1 binding sites and reside within GC-rich clusters. (C) Cell lysates of stable MDA-435 Cav1 transfectants were Western blotted for Egr1 and β-actin. Cav1Y14D- and Y14R-expressing cells present reduced levels of active nonphosphorylated (bottom band) and increased levels of the inactive phosphorylated (top band) Egr1. (bottom) Mechanical stretching (30-min stretched and relaxed cycles over 48 h) induces phosphorylation of Egr1 in Cav1wt- but not control dsRed- or dominant-negative Cav1Y14F-expressing MDA-435 stable cell lines. (D) Egr1 siRNA knockdown induces Cav1 and cavin-1 in Cav1wt- but not control dsRed- or dominant-negative Cav1Y14F-expressing MDA-435 stable cells. Ctl, control.
Egr1 is a substrate of PKC and casein kinase II and, when phosphorylated, fails to avidly bind to DNA (Huang and Adamson, 1995; Jain et al., 1996; Huang et al., 1998; Srivastava et al., 1998). The dramatic shift in Egr1 mobility in Cav1Y14D- and Y14R-expressing cells is indicative of its phosphorylation (Fig. 3 C; Cao et al., 1990; Day et al., 1990). dsRed-, Cav1wt-, and Y14F-expressing MDA-435 cell lines express the 60-kD nonphosphorylated Egr1 protein, whereas both Cav1Y14D and Y14R transfectants have significantly higher levels of inactive phosphorylated Egr1 (∼80 kD). Egr1 phosphorylation is induced in Cav1wt- but not dsRed- or Cav1Y14F-expressing cells by mechanical stretching cycles that increase endogenous Cav1 and cavin-1 levels (Fig. 3 C). siRNA knockdown of Egr1 increases levels of endogenous Cav1 and cavin-1 but only in Cav1wt-expressing line (Fig. 3 D). No effect is observed in dsRed or Cav1Y14F stable MDA-435 cell lines, suggesting that the mechanotransduced Egr1 suppressive effect is dependent on Cav1Y14 phosphorylation. Our data suggest that mechanical stress increases Egr1 phosphorylation and inhibition of its suppressive activity. As a suppressor of both Cav1 and cavin-1 transcription, Egr1 is therefore a critical and novel regulator of caveola biogenesis.
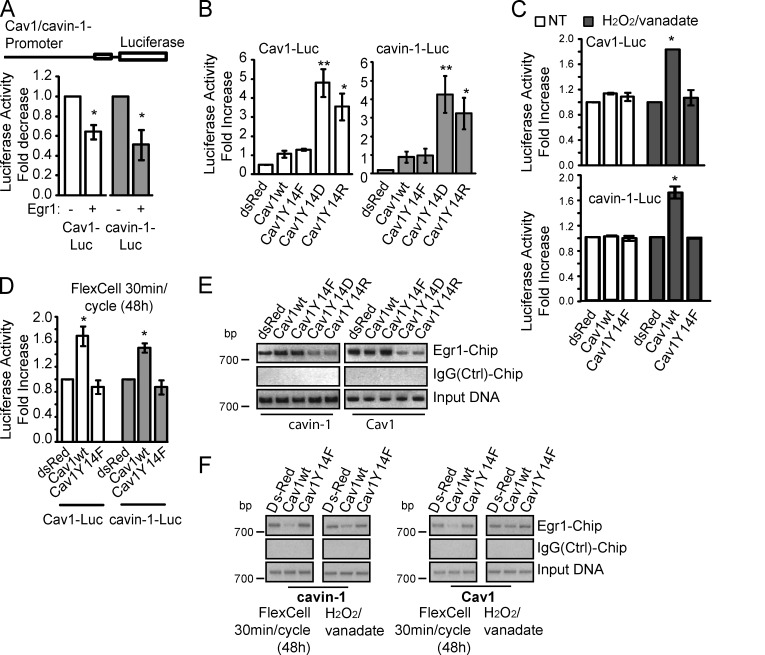
Cotransfection of Cav1 and cavin-1 promoter luciferase reporter plasmids with an Egr1-expressing plasmid reduces the activity of both promoters (Fig. 4 A). Furthermore, Cav1 and cavin-1 promoter activity is selectively elevated in Cav1Y14D- and Y14R-expressing MDA-435 cell lines, matching the increased endogenous Cav1 and cavin-1 protein expression in these cells (Fig. 4 B). Consistently, oxidative stress (H2O2/vanadate) and mechanical stretching selectively induce Cav1 and cavin-1 promoter activity in Cav1wt- but not dsRed control- or Cav1Y14F-expressing MDA-435 cells (Fig. 4, C and D). Egr1 regulation of Cav1 and cavin-1 promoter activity is therefore pCav1 dependent.
Figure 4.
Egr1 suppression of Cav1 and cavin-1 promoters is pCav1 dependent. (A) MDA-435 cells were transiently transfected with control pRL, Cav1-luciferase (Luc), or cavin-1–luciferase promoter constructs with or without pcDNA3-Egr1 and subjected to luciferase reporter assay. (B) Stable MDA-435 Cav1 transfectants were transfected with Cav1-luciferase and cavin-1–luciferase reporter constructs and subjected to luciferase reporter assay. (C) Cav1-luciferase and cavin-1–luciferase promoter activity in nontreated (NT) or H2O2/vanadate-stimulated MDA-435 Cav1 transfectants. (D) FlexCell mechanical stretching induces Cav1 and cavin-1 promoter reporter activity in MDA-435 Cav1 transfectants. (E) CHIP assays of stable Cav1 MDA-435 transfectants showed markedly reduced promoter occupancy by Egr1 in Cav1Y14D- and Y14R-expressing stables. Negative control IgG immunoprecipitation is shown. (F) H2O2/vanadate or FlexCell mechanical stretching reduce Egr1 occupation of Cav1 and cavin-1 promoter sites in Cav1wt relative to dsRed- and Cav1Y14F-transfected MDA-435 cell lines (*, P < 0.05; **, P < 0.001). Ctrl, control. Error bars show means ± SEM.
Occupancy of the endogenous Cav1 or cavin-1 promoters by Egr1 was determined using chromatin immunoprecipitation (CHIP) assays. In Cav1Y14D and Y14R MDA-435 cell lines, Egr1 binding to the ∼1-kb promoter region of both the Cav1 and cavin-1 genes is reduced relative to dsRed, Cav1wt, and Y14F lines (Fig. 4 E). Equivalent input DNA for all the lines and the lack of a PCR product in the IgG pull-down indicate the specificity of Egr1 binding. FlexCell mechanical stretching and H2O2/vanadate both selectively reduce Egr1 interaction with the promoter regions of the Cav1 and cavin-1 genes in Cav1wt-expressing cells (Fig. 4 F). These results suggest a feedback mechanism whereby pCav1 actively employs the Egr1 transcription factor to autoregulate its own expression as well as that of cavin-1.
Endogenous Cav1 regulates Egr1 promoter regulation in MDA-231 breast cancer cells
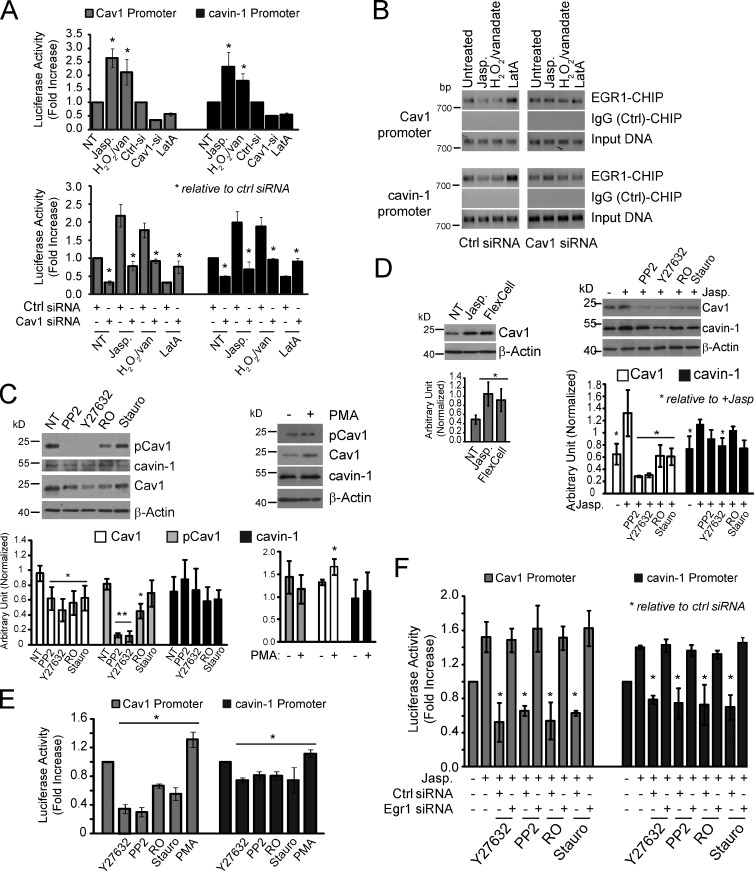
In the metastatic MDA-231 breast carcinoma cell line that expresses elevated Cav1, pCav1, and caveolae (Kojic et al., 2007; Joshi et al., 2008), Cav1 and cavin-1 promoter activity is enhanced by actin stabilization with jasplakinolide or by H2O2/vanadate and reduced by Cav1 knockdown and LatA treatment (Fig. 5 A). The ability of jasplakinolide, H2O2/vanadate, and LatA to modulate Cav1 and cavin-1 promoter activity is Cav1 dependent as promoter activity is restored to control levels in the presence of Cav1 siRNA. CHIP assays were performed on the same set of cells transfected with control or Cav1-specific siRNAs (Fig. 5 B). Treatment with H2O2/vanadate or jasplakinolide reduces physical occupancy by Egr1 of the Cav1 and cavin-1 promoters, whereas LatA treatment enhances Egr1 promoter occupancy. However, these effects on Egr1 promoter occupancy are not observed upon siRNA knockdown of Cav1 in MDA-231 cells. Endogenous Cav1 is therefore required for Egr1 regulation of Cav1 and cavin-1 promoter activity in these metastatic breast cancer cells.
Figure 5.
Transcriptional regulation of Cav1 and cavin-1 through endogenous Cav1 in metastatic MDA-231 breast cancer cells. (A, top) Metastatic MDA-231 breast cancer cells treated with jasplakinolide (Jasp.), H2O2/vanadate (H2O2/van), or LatA or transfected with control (Ctrl si) or Cav1 siRNAs were then transiently transfected with control pRL, Cav1-luciferase (Luc), or cavin-1–luciferase constructs for 48 h and subjected to luciferase assay. (bottom) Altered promoter activity caused by jasplakinolide, H2O2/vanadate, or LatA treatment is Cav1 dependent as it is not observed in Cav1 siRNA–transfected cells. (B) CHIP assays for Egr1 binding to Cav1 and cavin-1 promoters in MDA-231 cells were performed after H2O2/vanadate, jasplakinolide, or LatA treatment on control (Ctrl) or Cav1 siRNA–transfected cells. Negative control IgG immunoprecipitation is shown. (C) MDA-231 cells were not treated (NT) or treated with PP2, Y27632, R0, or staurosporine (Stauro; left) or with PMA (right) and then Western blotted for pCav1, Cav1, cavin-1, and β-actin (**, P < 0.001). (D) MDA-231 cells treated with jasplakinolide or mechanically stretched (left) or treated with jasplakinolide and either PP2, Y27632, R0, or staurosporine (right) were Western blotted for Cav1, cavin-1, and β-actin. (E) MDA-231 cells transiently transfected with Cav1-luciferase or cavin-1–luciferase for 48 h were subjected to luciferase assay after treatment with PP2, Y27632, R0, staurosporine (Stauro), or PMA. (F) MDA-231 cells were cotransfected with Cav1-luciferase or cavin-1–luciferase and either control (siCtl) or Egr1 (siEgr1) siRNA and treated with jasplakinolide and either PP2, Y27632, R0, or staurosporine before luciferase assay (*, P < 0.05). Error bars show means ± SEM.
To identify signaling pathways that mediate pCav1-Egr1 control of Cav1 and cavin-1 gene expression, we treated MDA-231 cells with the Src kinase inhibitor PP2, the ROCK inhibitor Y27632, and the PKC inhibitors R0 and staurosporine. In PP2- and Y27632-treated MDA-231 cells, pCav1 is no longer detected, as previously reported (Joshi et al., 2008), but pCav1 is still present in cells treated with the PKC inhibitors R0 and staurosporine. The PKC activator PMA does not affect pCav1 levels but does significantly increase Cav1 expression (Fig. 5 C). As observed for Cav1-expressing MDA-435 cells, both jasplakinolide treatment and cell stretching induce Cav1 expression in MDA-231 cells. All four kinase inhibitors reduce jasplakinolide induction of Cav1, although not cavin-1, levels (Fig. 5 D). This suggests that although PP2 and Y27632 prevent jasplakinolide induction of Cav1 by preventing Cav1 phosphorylation, PKC acts downstream of pCav1 to regulate actin-dependent induction of Cav1.
We then tested the effect of these drugs on Cav1 and cavin-1 promoter activity. PP2, Y27632, R0, and staurosporine reduce and PMA increases Cav1 and, to a lesser extent, cavin-1 promoter activity in untreated (Fig. 5 E) and jasplakinolide-treated (Fig. 5 F) MDA-231 cells. Src, ROCK, and PKC inhibition of jasplakinolide-induced Cav1 and cavin-1 promoter activity is not observed in Egr1 knockdown cells and therefore mediated by Egr1 (Fig. 5 F). Relative to Cav1, control of cavin-1 promoter activity and protein expression levels by pCav1 and Egr1 is reduced in MDA-231 cells (Fig. 5, C–E). Similarly, cavin-1 mRNA induction in Cav1Y14D and Y14R MDA-435 cells, while significant, is reduced in amplitude relative to Cav1 mRNA (Fig. 3 A). Differential impact on Cav1 and cavin-1 expression at both the mRNA and protein levels in MDA-231 cells may reflect cell-specific promoter regulation and protein stability. Cav1 gene expression is regulated by NF-κB and SP1 (Dasari et al., 2006; Volonte and Galbiati, 2011), and pCav1-PKC-Egr1 may be one of multiple mechanisms that regulate Cav1 and cavin-1 expression. Indeed, Cav1 and cavin-1 gene expression is selectively induced in Cav1wt- but not control dsRed- or dominant-negative Cav1Y14F-transfected MDA-435 cells by actin stabilization, mechanical stretch, or Egr1 knockdown (Figs. 2 C and 3 D). In the breast cancer cell models studied here, Egr1 transcriptional regulation of Cav1 and cavin-1 gene expression therefore requires pCav1 and is specific to Cav1-expressing cells.
Cav1 therefore represents a mechanosensor that transduces increased actin cytoskeleton–focal adhesion tension to regulate Cav1 and cavin-1 gene expression and thereby supply caveolae to buffer membrane tension in response to mechanical stress (Sinha et al., 2011). Endothelial Cav1 and caveolae are required for mechanotransduction in blood vessels (Yu et al., 2006). Mechanical and oxidative stress induce Cav1 phosphorylation (Volonté et al., 2001; Radel and Rizzo, 2005), including a role for p38 MAPK/Sp1 gene transcription (Dasari et al., 2006; Volonte and Galbiati, 2011). In cancer cells, tissue stiffness promotes malignancy through integrin-mediated focal adhesion and Rho/ROCK-generated contractility (Paszek et al., 2005). Together with pCav1 activation of RhoA and Rho/ROCK-dependent phosphorylation of Cav1 (Grande-García et al., 2007; Joshi et al., 2008), this suggests that pCav1 may be a central regulator of a positive feedback loop that responds to mechanical stress and thereby promotes Rho/ROCK-dependent tumor cell migration.
In conclusion, we have identified a pathway linking force-induced phosphorylation of Cav1 with transcriptional regulation of key components of caveolar biogenesis. Posttranslational modification of Cav1 (i.e., Y14 tyrosine phosphorylation) enables it to function as a reporter of cellular tension that feedbacks to regulate, via Egr1 transcriptional regulation of Cav1 and cavin-1, the plasma membrane abundance of an organelle critical to the cellular response to mechanical stress.
Materials and methods
Antibodies and reagents
Antibodies to Egr1 and RhoA were obtained from Santa Cruz Biotechnology, Inc., to β-actin were obtained from Sigma-Aldrich, to Cav1 and cavin-1 were obtained from BD, and to pCav1 were obtained from Cell Signaling Technology. Alexa Fluor 488–phalloidin was purchased from Molecular Probes. Jasplakinolide, LatA, R0, staurosporine, PMA, hydrogen peroxide (H2O2), and sodium orthovanadate (Na2VO4) were obtained from Sigma-Aldrich. PP2 and Y27632 were obtained from EMD Millipore. Egr1-targeting SMARTpool siRNA was purchased from Thermo Fisher Scientific. OneStep StopGlo was obtained from Promega.
Plasmids, luciferase, and CHIP assays
C-terminal–tagged myc/mRFP-tagged Cav1wt, Y14F, Y14D, and Y14R under control of the cytomegalovirus promoter in pcDNA3 plasmid and cavin-1–GFP in pEGFP-N1 were as previously described (Goetz et al., 2008; Hill et al., 2008; Joshi et al., 2008). Control pRL, Cav1-luciferase, and cavin-1–luciferase plasmids were purchased from SwitchGear, and their expression was assayed using a luciferase detection kit (Promega) and a luminometer (Lumat LB 9507; Berthold Technologies). CHIP assays were performed as previously described (Joshi et al., 2005). In brief, cells treated with 1% formaldehyde for 10 min at room temperature to cross-link protein–DNA complexes were harvested, washed three times with PBS, lysed in 1% SDS, 50 mM Tris-HCI, pH 8.1, and 1 mM EDTA, and sonicated twice (10 W for 15 s). Cell debris was pelleted at 4°C, and the supernatant was incubated with either 3 µg anti-Egr1 antibody or IgG in dilution buffer (0.01% SDS, 16.7 mM Tris-HCI, pH 8.1, 1.2 mM EDTA, 150 mM NaCl, and 1% Triton X-100) and rotated overnight at 4°C. Antibody DNA–protein complexes were pulled down with protein A–Sepharose beads, washed, and then decross-linked by incubating at 65°C for 4 h. DNA was ethanol precipitated, subjected to proteinase K treatment for 30 min at 37°C, and repurified before using for PCR. The following sets of primers were used to assess Egr1 binding to Cav1 or cavin-1 promoters: 5′-TGGAGAGGCAGATAGCAGAAGCGG-3′ and 5′-GATCTTCATTTCTTATTTCGAAGCG-3′; or 5′-TGCCCATCTTAATTCTACTCTGTTAGG-3′ and 5′-GACTTGATCAGCTCTTCTGAG-3′.
Cell lines, cell stretching, and drug treatment
Stable MDA-435 cell lines expressing dsRed, Cav1wt-mRFP, Cav1Y14F-mRFP, Cav1Y14R-mRFP, or Cav1Y14D-mRFP were generated as described previously (Joshi et al., 2008). In brief, 48 h after Effectene transfection, cells were selected with 400 µg/ml G418 for 2 wk, and resistant cells were FACS sorted. Cells were maintained in DME medium containing 10% FBS, penicillin/streptomycin, and 50 µg/ml geneticin (Gibco; Invitrogen). All cell lines were passaged at least twice after recovery from frozen stocks before initiating experiments and maintained in culture for a maximum of 8–10 passages to minimize phenotypic drift. For cyclic stretch experiments, cells were seeded and grown on untreated silicone elastomer-bottomed plates (Bioflex) overnight before subjecting them to cyclic stretch in a strain unit (FX-4000T Flexercell; FlexCell). Cells were exposed to an equiaxial stretch of 5% at a frequency of 1 Hz after a cycle of 30-min stretching and 30-min relaxation for a total of 36 repeats. Cells were treated with 3 mM hydrogen peroxide–1 mM sodium vanadate for 45–60 min to induce oxidative stress and with 150 nM LatA for 30 min to depolymerize actin microfilaments and jasplakinolide (MDA-231, 100 nm; MDA-435, 150 nm) for 2 h to stabilize the actin cytoskeleton (Sun et al., 2009; Shankar et al., 2010). Cells transiently transfected with reporter constructs were treated 36–42 h after transfection. Where indicated, cells were treated with 5 µM PP2, 10 µM Y27632, 4 µM R0, 100 nM staurosporine, or 150 nM PMA for 2 h.
EM
Stable MDA-435 cell lines or transiently transfected Cav1−/− immortalized MEFs expressing the indicated constructs were fixed in the presence of ruthenium red to highlight the cell surface (Hill et al., 2008) and processed by conventional methods for Epon embedding. Cav1−/− cells were sectioned parallel to the substratum, and images were captured using a transmission electron microscope (1011; JEOL) equipped with a cooled charge-coupled device camera (Morada; Olympus) and with AnalySIS software (Olympus) at a magnification of 15,000–20,000×. For quantification of caveolae density in MDA-435 cell lines, sections were cut perpendicular to the cell surface (vertical sections) to ensure random sampling of the entire plasma membrane. Complete cells were identified by EM, and the number of ruthenium red–labeled 55–70-nm uncoated circular profiles (i.e., caveolae) was counted per cell profile. Data were generated from analysis of ≥45 cell profiles per line (from three independent cultures of each line).
FLIM, immunofluorescence, cell spreading, Western blots, RhoA-GTP assay, and semiquantitative RT-PCR
FLIM of Cav1−/− MEFs transfected with cavin-1–GFP (donor) and mRFP-Cav1wt and Y14 mutants (acceptor) was performed using a lifetime fluorescence imaging attachment (Lambert Instruments) on an inverted microscope (IX71; Olympus) as previously described (Hill et al., 2008). Cells were excited using a sinusoidally modulated 3-W 470-nm light-emitting diode at 80 MHz under epi-illumination and imaged with a 60×, NA 1.45 oil objective using an appropriate GFP filter set. The phase and modulation were determined from a set of 12 phase settings, and pixel resolution of two lifetimes in the frequency domain was performed using the polar plot method as implemented in the manufacturer’s software.
To label F-actin, cells were fixed with 3% paraformaldehyde and permeabilized with 0.1% Triton X-100 before incubation with Alexa Fluor 568–phalloidin. Coverslips were mounted in Airvol (Air Products, Inc.), and images were collected with the 100× Plan Apochromat objective, NA 1.35, of a confocal microscope (FV1000; Olympus). To quantify cell area, cells labeled with Alexa Fluor 568–phalloidin and Hoechst on 96-well plates were scanned using the 10× objective of an automated fluorescence microscope (ArrayScan; Cellomics, Inc.), and cell area was measured on five fields/well using the Morphology Explorer BioApplication software (Cellomics, Inc.) module as previously described (Shankar et al., 2010). Western blots and semiquantitative RT-PCR were as previously described (Joshi et al., 2008). Antibody dilutions for Western blots were as follows: 1:1,000 for pCav1, Cav1, and cavin-1, 1:500 for Egr1, 1:10,000 for β-actin, and 1:3,000 for HRP anti–mouse or anti–rabbit. RhoA-GTP pull-downs were performed with rhotekin beads according to the supplier’s protocol (EMD Millipore) and probed by Western blotting for RhoA (active and total; 1:1,000 antibody dilution) as previously described (Joshi et al., 2008). Transcript levels were determined by PCR (25 cycles) from total cDNA using primer sets for Cav1, 5′-TCTCTACACCGTTCCCATCC-3′ and 5′-TCCCTTCTGGTTCTGCAATC-3′; cavin-1, 5′-CAAGGCCACCGAGATGGTGGA-3′ and 5′-GAAGTTCGGAAGGAGCGAGGA-3′; and glyceraldehyde 3-phosphate dehydrogenase, 5′-GGTCGGAGTCAACGGATTTGGTCG-3′ and 5′-CCTCCGACGCCTGCTTCACCAC-3′.
Online supplemental material
Fig. S1 shows Cav1 expression by Western blotting in the MDA-435 Cav1 Y14 transfectants relative to endogenous Cav1 in MDA-231 cells. Fig. S2 shows that Cav1−/− MEF expression of mRFP-tagged Cav1wt, Y14F, Y14D, or Y14R induces caveolae by EM and their equivalent interaction with cavin-1–GFP by FLIM analysis. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201207089/DC1.
Supplementary Material
Acknowledgments
We thank Dr. Alex Smith for providing access to the FlexCell device and Charles Ferguson and Nicole Schieber for assistance with EM.
This work was supported by a grant from the Canadian Institutes for Health Research (MOP-43938 to I.R. Nabi) and by a grant and fellowship from the National Health and Medical Research Council of Australia (511005 and 569452 to R.G. Parton).
Footnotes
Abbreviations used in this paper:
- Cav1
- caveolin-1
- CHIP
- chromatin immunoprecipitation
- FLIM
- fluorescence lifetime measurement
- LatA
- latrunculin A
- MEF
- mouse embryo fibroblast
- mRFP
- monomeric RFP
- ROCK
- Rho-associated kinase
References
- Bastiani M., Liu L., Hill M.M., Jedrychowski M.P., Nixon S.J., Lo H.P., Abankwa D., Luetterforst R., Fernandez-Rojo M., Breen M.R., et al. 2009. MURC/Cavin-4 and cavin family members form tissue-specific caveolar complexes. J. Cell Biol. 185:1259–1273 10.1083/jcb.200903053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H., Courchesne W.E., Mastick C.C. 2002. A phosphotyrosine-dependent protein interaction screen reveals a role for phosphorylation of caveolin-1 on tyrosine 14: recruitment of C-terminal Src kinase. J. Biol. Chem. 277:8771–8774 10.1074/jbc.C100661200 [DOI] [PubMed] [Google Scholar]
- Cao X.M., Koski R.A., Gashler A., McKiernan M., Morris C.F., Gaffney R., Hay R.V., Sukhatme V.P. 1990. Identification and characterization of the Egr-1 gene product, a DNA-binding zinc finger protein induced by differentiation and growth signals. Mol. Cell. Biol. 10:1931–1939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Wang S., Zhou Y., Wu X., Entin I., Epstein J., Yaccoby S., Xiong W., Barlogie B., Shaughnessy J.D., Jr, Zhan F. 2010. Identification of early growth response protein 1 (EGR-1) as a novel target for JUN-induced apoptosis in multiple myeloma. Blood. 115:61–70 10.1182/blood-2009-03-210526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasari A., Bartholomew J.N., Volonte D., Galbiati F. 2006. Oxidative stress induces premature senescence by stimulating caveolin-1 gene transcription through p38 mitogen-activated protein kinase/Sp1-mediated activation of two GC-rich promoter elements. Cancer Res. 66:10805–10814 10.1158/0008-5472.CAN-06-1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day M.L., Fahrner T.J., Aykent S., Milbrandt J. 1990. The zinc finger protein NGFI-A exists in both nuclear and cytoplasmic forms in nerve growth factor-stimulated PC12 cells. J. Biol. Chem. 265:15253–15260 [PubMed] [Google Scholar]
- del Pozo M.A., Balasubramanian N., Alderson N.B., Kiosses W.B., Grande-García A., Anderson R.G., Schwartz M.A. 2005. Phospho-caveolin-1 mediates integrin-regulated membrane domain internalization. Nat. Cell Biol. 7:901–908 10.1038/ncb1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fielding P.E., Chau P., Liu D., Spencer T.A., Fielding C.J. 2004. Mechanism of platelet-derived growth factor-dependent caveolin-1 phosphorylation: relationship to sterol binding and the role of serine-80. Biochemistry. 43:2578–2586 10.1021/bi035442c [DOI] [PubMed] [Google Scholar]
- Gaus K., Le Lay S., Balasubramanian N., Schwartz M.A. 2006. Integrin-mediated adhesion regulates membrane order. J. Cell Biol. 174:725–734 10.1083/jcb.200603034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glenney J.R., Jr 1989. Tyrosine phosphorylation of a 22-kDa protein is correlated with transformation by Rous sarcoma virus. J. Biol. Chem. 264:20163–20166 [PubMed] [Google Scholar]
- Glenney J.R., Jr, Soppet D. 1992. Sequence and expression of caveolin, a protein component of caveolae plasma membrane domains phosphorylated on tyrosine in Rous sarcoma virus-transformed fibroblasts. Proc. Natl. Acad. Sci. USA. 89:10517–10521 10.1073/pnas.89.21.10517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetz J.G., Joshi B., Lajoie P., Strugnell S.S., Scudamore T., Kojic L.D., Nabi I.R. 2008. Concerted regulation of focal adhesion dynamics by galectin-3 and tyrosine-phosphorylated caveolin-1. J. Cell Biol. 180:1261–1275 10.1083/jcb.200709019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grande-García A., Echarri A., de Rooij J., Alderson N.B., Waterman-Storer C.M., Valdivielso J.M., del Pozo M.A. 2007. Caveolin-1 regulates cell polarization and directional migration through Src kinase and Rho GTPases. J. Cell Biol. 177:683–694 10.1083/jcb.200701006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen C.G., Bright N.A., Howard G., Nichols B.J. 2009. SDPR induces membrane curvature and functions in the formation of caveolae. Nat. Cell Biol. 11:807–814 10.1038/ncb1887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill M.M., Bastiani M., Luetterforst R., Kirkham M., Kirkham A., Nixon S.J., Walser P., Abankwa D., Oorschot V.M., Martin S., et al. 2008. PTRF-Cavin, a conserved cytoplasmic protein required for caveola formation and function. Cell. 132:113–124 10.1016/j.cell.2007.11.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang R.P., Adamson E.D. 1995. A biological role for Egr-1 in cell survival following ultra-violet irradiation. Oncogene. 10:467–475 [PubMed] [Google Scholar]
- Huang R.P., Fan Y., deBelle I., Ni Z., Matheny W., Adamson E.D. 1998. Egr-1 inhibits apoptosis during the UV response: correlation of cell survival with Egr-1 phosphorylation. Cell Death Differ. 5:96–106 10.1038/sj.cdd.4400322 [DOI] [PubMed] [Google Scholar]
- Jaffe A.B., Hall A. 2005. Rho GTPases: biochemistry and biology. Annu. Rev. Cell Dev. Biol. 21:247–269 10.1146/annurev.cellbio.21.020604.150721 [DOI] [PubMed] [Google Scholar]
- Jain N., Mahendran R., Philp R., Guy G.R., Tan Y.H., Cao X. 1996. Casein kinase II associates with Egr-1 and acts as a negative modulator of its DNA binding and transcription activities in NIH 3T3 cells. J. Biol. Chem. 271:13530–13536 10.1074/jbc.271.23.13530 [DOI] [PubMed] [Google Scholar]
- Joshi B., Ordonez-Ercan D., Dasgupta P., Chellappan S. 2005. Induction of human metallothionein 1G promoter by VEGF and heavy metals: differential involvement of E2F and metal transcription factors. Oncogene. 24:2204–2217 10.1038/sj.onc.1208206 [DOI] [PubMed] [Google Scholar]
- Joshi B., Strugnell S.S., Goetz J.G., Kojic L.D., Cox M.E., Griffith O.L., Chan S.K., Jones S.J., Leung S.P., Masoudi H., et al. 2008. Phosphorylated caveolin-1 regulates Rho/ROCK-dependent focal adhesion dynamics and tumor cell migration and invasion. Cancer Res. 68:8210–8220 10.1158/0008-5472.CAN-08-0343 [DOI] [PubMed] [Google Scholar]
- Kim Y.N., Wiepz G.J., Guadarrama A.G., Bertics P.J. 2000. Epidermal growth factor-stimulated tyrosine phosphorylation of caveolin-1. Enhanced caveolin-1 tyrosine phosphorylation following aberrant epidermal growth factor receptor status. J. Biol. Chem. 275:7481–7491 10.1074/jbc.275.11.7481 [DOI] [PubMed] [Google Scholar]
- Kirkham M., Nixon S.J., Howes M.T., Abi-Rached L., Wakeham D.E., Hanzal-Bayer M., Ferguson C., Hill M.M., Fernandez-Rojo M., Brown D.A., et al. 2008. Evolutionary analysis and molecular dissection of caveola biogenesis. J. Cell Sci. 121:2075–2086 10.1242/jcs.024588 [DOI] [PubMed] [Google Scholar]
- Kojic L.D., Joshi B., Lajoie P., Le P.U., Cox M.E., Turbin D.A., Wiseman S.M., Nabi I.R. 2007. Raft-dependent endocytosis of autocrine motility factor is phosphatidylinositol 3-kinase-dependent in breast carcinoma cells. J. Biol. Chem. 282:29305–29313 10.1074/jbc.M704069200 [DOI] [PubMed] [Google Scholar]
- Kubosaki A., Tomaru Y., Tagami M., Arner E., Miura H., Suzuki T., Suzuki M., Suzuki H., Hayashizaki Y. 2009. Genome-wide investigation of in vivo EGR-1 binding sites in monocytic differentiation. Genome Biol. 10:R41 10.1186/gb-2009-10-4-r41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurzchalia T.V., Dupree P., Parton R.G., Kellner R., Virta H., Lehnert M., Simons K. 1992. VIP21, a 21-kD membrane protein is an integral component of trans-Golgi-network-derived transport vesicles. J. Cell Biol. 118:1003–1014 10.1083/jcb.118.5.1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H., Volonte D., Galbiati F., Iyengar P., Lublin D.M., Bregman D.B., Wilson M.T., Campos-Gonzalez R., Bouzahzah B., Pestell R.G., et al. 2000. Constitutive and growth factor-regulated phosphorylation of caveolin-1 occurs at the same site (Tyr-14) in vivo: identification of a c-Src/Cav-1/Grb7 signaling cassette. Mol. Endocrinol. 14:1750–1775 10.1210/me.14.11.1750 [DOI] [PubMed] [Google Scholar]
- Li S., Seitz R., Lisanti M.P. 1996. Phosphorylation of caveolin by src tyrosine kinases. The alpha-isoform of caveolin is selectively phosphorylated by v-Src in vivo. J. Biol. Chem. 271:3863–3868 10.1074/jbc.271.7.3863 [DOI] [PubMed] [Google Scholar]
- Liu L., Pilch P.F. 2008. A critical role of cavin (polymerase I and transcript release factor) in caveolae formation and organization. J. Biol. Chem. 283:4314–4322 10.1074/jbc.M707890200 [DOI] [PubMed] [Google Scholar]
- Mastick C.C., Brady M.J., Saltiel A.R. 1995. Insulin stimulates the tyrosine phosphorylation of caveolin. J. Cell Biol. 129:1523–1531 10.1083/jcb.129.6.1523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon K.-A., Zajicek H., Li W.-P., Peyton M.J., Minna J.D., Hernandez V.J., Luby-Phelps K., Anderson R.G.W. 2009. SRBC/cavin-3 is a caveolin adapter protein that regulates caveolae function. EMBO J. 28:1001–1015 10.1038/emboj.2009.46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nose K., Ohba M. 1996. Functional activation of the egr-1 (early growth response-1) gene by hydrogen peroxide. Biochem. J. 316:381–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlichenko L., Huang B., Krueger E., McNiven M.A. 2006. Epithelial growth factor-induced phosphorylation of caveolin 1 at tyrosine 14 stimulates caveolae formation in epithelial cells. J. Biol. Chem. 281:4570–4579 10.1074/jbc.M512088200 [DOI] [PubMed] [Google Scholar]
- Orlichenko L., Weller S.G., Cao H., Krueger E.W., Awoniyi M., Beznoussenko G., Buccione R., McNiven M.A. 2009. Caveolae mediate growth factor-induced disassembly of adherens junctions to support tumor cell dissociation. Mol. Biol. Cell. 20:4140–4152 10.1091/mbc.E08-10-1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paszek M.J., Zahir N., Johnson K.R., Lakins J.N., Rozenberg G.I., Gefen A., Reinhart-King C.A., Margulies S.S., Dembo M., Boettiger D., et al. 2005. Tensional homeostasis and the malignant phenotype. Cancer Cell. 8:241–254 10.1016/j.ccr.2005.08.010 [DOI] [PubMed] [Google Scholar]
- Radel C., Rizzo V. 2005. Integrin mechanotransduction stimulates caveolin-1 phosphorylation and recruitment of Csk to mediate actin reorganization. Am. J. Physiol. Heart Circ. Physiol. 288:H936–H945 10.1152/ajpheart.00519.2004 [DOI] [PubMed] [Google Scholar]
- Raychowdhury R., Schäfer G., Fleming J., Rosewicz S., Wiedenmann B., Wang T.C., Höcker M. 2002. Interaction of early growth response protein 1 (Egr-1), specificity protein 1 (Sp1), and cyclic adenosine 3′5′-monophosphate response element binding protein (CREB) at a proximal response element is critical for gastrin-dependent activation of the chromogranin A promoter. Mol. Endocrinol. 16:2802–2818 10.1210/me.2001-0292 [DOI] [PubMed] [Google Scholar]
- Rothberg K.G., Heuser J.E., Donzell W.C., Ying Y.-S., Glenney J.R., Anderson R.G.W. 1992. Caveolin, a protein component of caveolae membrane coats. Cell. 68:673–682 10.1016/0092-8674(92)90143-Z [DOI] [PubMed] [Google Scholar]
- Shankar J., Messenberg A., Chan J., Underhill T.M., Foster L.J., Nabi I.R. 2010. Pseudopodial actin dynamics control epithelial-mesenchymal transition in metastatic cancer cells. Cancer Res. 70:3780–3790 10.1158/0008-5472.CAN-09-4439 [DOI] [PubMed] [Google Scholar]
- Shingu T., Bornstein P. 1994. Overlapping Egr-1 and Sp1 sites function in the regulation of transcription of the mouse thrombospondin 1 gene. J. Biol. Chem. 269:32551–32557 [PubMed] [Google Scholar]
- Silverman E.S., Khachigian L.M., Lindner V., Williams A.J., Collins T. 1997. Inducible PDGF A-chain transcription in smooth muscle cells is mediated by Egr-1 displacement of Sp1 and Sp3. Am. J. Physiol. 273:H1415–H1426 [DOI] [PubMed] [Google Scholar]
- Sinha B., Köster D., Ruez R., Gonnord P., Bastiani M., Abankwa D., Stan R.V., Butler-Browne G., Vedie B., Johannes L., et al. 2011. Cells respond to mechanical stress by rapid disassembly of caveolae. Cell. 144:402–413 10.1016/j.cell.2010.12.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava S., Weitzmann M.N., Kimble R.B., Rizzo M., Zahner M., Milbrandt J., Ross F.P., Pacifici R. 1998. Estrogen blocks M-CSF gene expression and osteoclast formation by regulating phosphorylation of Egr-1 and its interaction with Sp-1. J. Clin. Invest. 102:1850–1859 10.1172/JCI4561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stula M., Orzechowski H.D., Gschwend S., Vetter R., von Harsdorf R., Dietz R., Paul M. 2000. Influence of sustained mechanical stress on Egr-1 mRNA expression in cultured human endothelial cells. Mol. Cell. Biochem. 210:101–108 10.1023/A:1007126218740 [DOI] [PubMed] [Google Scholar]
- Sun Y., Hu G., Zhang X., Minshall R.D. 2009. Phosphorylation of caveolin-1 regulates oxidant-induced pulmonary vascular permeability via paracellular and transcellular pathways. Circ. Res. 105:676–685 10.1161/CIRCRESAHA.109.201673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volonte D., Galbiati F. 2011. Polymerase I and transcript release factor (PTRF)/cavin-1 is a novel regulator of stress-induced premature senescence. J. Biol. Chem. 286:28657–28661 10.1074/jbc.C111.235119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volonté D., Galbiati F., Pestell R.G., Lisanti M.P. 2001. Cellular stress induces the tyrosine phosphorylation of caveolin-1 (Tyr(14)) via activation of p38 mitogen-activated protein kinase and c-Src kinase. Evidence for caveolae, the actin cytoskeleton, and focal adhesions as mechanical sensors of osmotic stress. J. Biol. Chem. 276:8094–8103 10.1074/jbc.M009245200 [DOI] [PubMed] [Google Scholar]
- Wu X., Cheng J., Li P., Yang M., Qiu S., Liu P., Du J. 2010. Mechano-sensitive transcriptional factor Egr-1 regulates insulin-like growth factor-1 receptor expression and contributes to neointima formation in vein grafts. Arterioscler. Thromb. Vasc. Biol. 30:471–476 10.1161/ATVBAHA.109.184259 [DOI] [PubMed] [Google Scholar]
- Yu J., Bergaya S., Murata T., Alp I.F., Bauer M.P., Lin M.I., Drab M., Kurzchalia T.V., Stan R.V., Sessa W.C. 2006. Direct evidence for the role of caveolin-1 and caveolae in mechanotransduction and remodeling of blood vessels. J. Clin. Invest. 116:1284–1291 10.1172/JCI27100 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.