KIF3A is important for the intracellular transport of HIV-containing compartments and HIV release from infected macrophages.
Abstract
Macrophages are long-lived target cells for HIV infection and are considered viral reservoirs. HIV assembly in macrophages occurs in virus-containing compartments (VCCs) in which virions accumulate and are stored. The regulation of the trafficking and release of these VCCs remains unknown. Using high resolution light and electron microscopy of HIV-1–infected primary human macrophages, we show that the spatial distribution of VCCs depended on the microtubule network and that VCC-limiting membrane was closely associated with KIF3A+ microtubules. Silencing KIF3A strongly decreased virus release from HIV-1–infected macrophages, leading to VCC accumulation intracellularly. Time-lapse microscopy further suggested that VCCs and associated KIF3A move together along microtubules. Importantly, KIF3A does not play a role in HIV release from T cells that do not possess VCCs. These results reveal that HIV-1 requires the molecular motor KIF3 to complete its cycle in primary macrophages. Targeting this step may lead to novel strategies to eliminate this viral reservoir.
Introduction
HIV-1 infects two main cellular targets: CD4+ T lymphocytes and macrophages, which can both contribute to the formation of reservoirs (Centlivre et al., 2011). In contrast to T cells, which usually die rapidly upon infection, HIV-1–infected macrophages survive for weeks or months both in vitro and in vivo. Newly formed virions accumulate intracellularly and remain infectious for extended periods of time (Sharova et al., 2005). HIV-1–infected macrophages are therefore considered as one of the main viral reservoirs. Whether they could fuel relapse after the arrest of highly active anti-retroviral therapy remains to be established (Igarashi et al., 2001; Zhu et al., 2002; Alexaki et al., 2008; Centlivre et al., 2011).
In T lymphocytes, assembly, budding, and fission of HIV take place at the plasma membrane. In contrast, in infected macrophages, profiles of viral budding are observed at the limiting membrane of intracellular virus-containing compartments (VCCs; Gendelman et al., 1988; Orenstein et al., 1988), indicating that they represent sites of viral assembly (Orenstein et al., 1988; Raposo et al., 2002; Pelchen-Matthews et al., 2003; Jouve et al., 2007). The origin of these internal VCCs is not yet clear, but it is proposed that they represent specialized domains of the plasma membrane sequestered intracellularly (Jouvenet et al., 2006; Deneka et al., 2007; Welsch et al., 2007).
Gag, the viral component that orchestrates viral assembly, is synthesized in the cytosol as a Pr55Gag precursor. When the viral particles are formed, the viral protease gets activated and cleaves the precursor into essentially four polypeptides, the matrix (MA), the capsid (CA or p24), the nucleocapsid (NC), and the late domain (p6). Studies suggest that Pr55Gag coordinates the recruitment of different host and viral proteins necessary for its transport to the assembly site and for the formation of new viral particles (Marsh et al., 2009). In T cells, the microtubule cytoskeleton appears to be involved in virus cell-to-cell transfer but not in viral particle release from T cells to the extracellular medium (Jolly et al., 2007). However, this conclusion relies on drug treatments and the molecular mechanisms involved remain to be identified. The contribution of microtubules to viral replication in macrophages remains unknown.
Microtubules are part of the cytoskeleton and provide structural support for the cytosolic transport of small protein complexes as well as vesicles and organelles. Kinesins and dyneins are families of molecular motors that literally walk on microtubules. Most members of the large kinesin family transport their cargo from the microtubule-organizing center located near the nucleus toward the plus (+) end of microtubules at the periphery (Hirokawa et al., 2009).
Most of the current knowledge on HIV cell biology comes from well-established, reliable, and easy-to-manipulate cell systems. In contrast, cell biology studies performed on primary human macrophages infected by HIV remain scarce. The development of advanced microscopy techniques and new cell biology technologies allowed us to perform the present study under more physiological although still challenging conditions.
In this study, we investigated the role of the microtubule network in the transport of the VCC and the release of the virus from HIV-1–infected primary human macrophages. We report that the kinesin KIF3A drives the intracellular transport of VCCs along microtubules allowing viral release. The role of KIF3A is restricted to VCC transport in macrophages, as KIF3A depletion does not modify the production of HIV-1 by T cells that do not form VCCs. Thus, KIF3A plays a pivotal role in the HIV-1 cycle in primary human macrophages.
Results
Microtubule-dependent distribution of VCCs
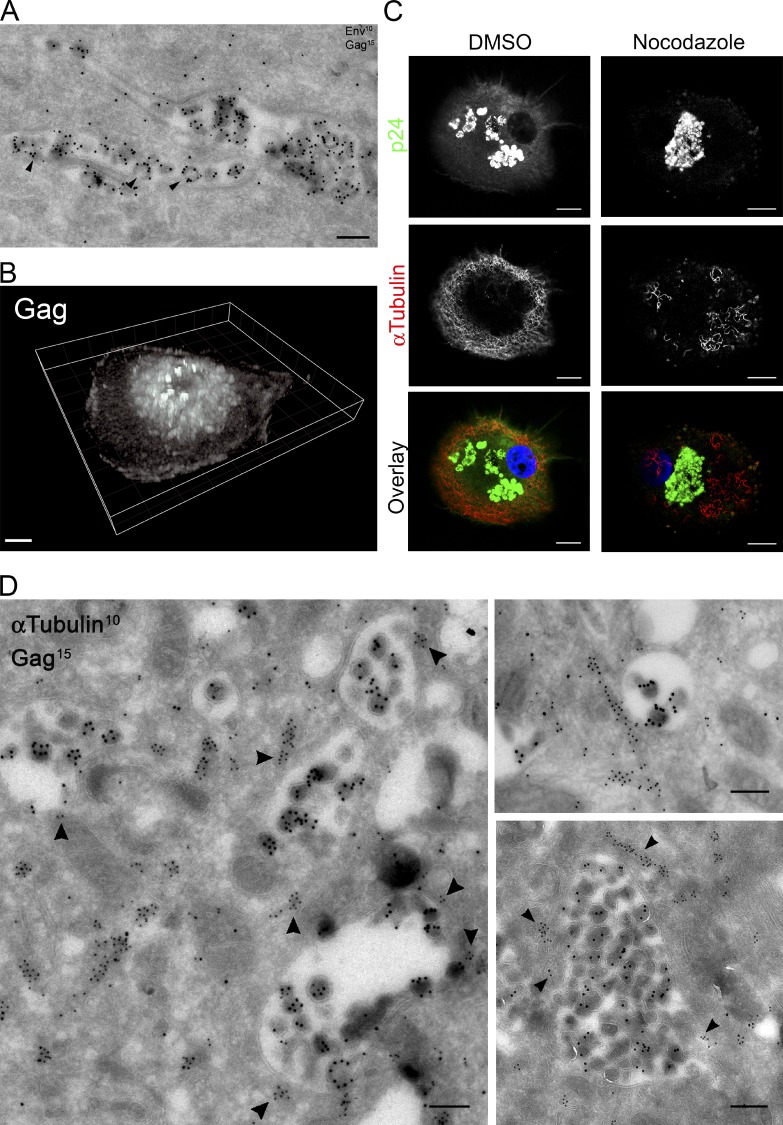
We first studied the role of the microtubule cytoskeleton in the intracellular transport and spatial organization of the VCCs. Analysis of HIV-1–infected macrophages by immunoelectron microscopy (immuno-EM) revealed VCCs with viral budding profiles typical of viral assembly at their limiting membrane (Fig. 1 A, arrowheads). Examination by confocal microscopy of HIV-1–infected macrophages revealed that large Gag+ compartments were present and frequently scattered in the central area of the cells (Fig. 1, B and C; and Video 1). Of note, the spatial distribution of these compartments was rather heterogeneous among the infected cells. Treatment of HIV-1–infected macrophages with nocodazole for 1 h disrupted most of the microtubule network concentrating all Gag+ compartments in the perinuclear area (Fig. 1 C). Time-lapse imaging also allowed visualization of the nocodazole-induced centripetal movement of the VCCs within a given macrophage (Fig. S1 A). Quantification of the effect of the nocodazole treatment on the spatial distribution of the Gag+ compartments by automated image analysis (see details in Fig. S1 B) fully confirmed this observation. Upon nocodazole washout, we observed that within 15 min the Gag+ compartments once again became spatially dispersed (Fig. S1 C), supporting the idea that the positioning of Gag+ compartments requires an intact microtubule network.
Figure 1.
The distribution of VCCs is dependent on the integrity of the microtubule network. (A) Ultrathin cryosections of macrophages infected with HIV-1 NLAD8 for 15 d were prepared and labeled with anti-Env antibodies and protein A coupled to gold particles of 10-nm diameter (PAG10) and with anti-Pr55Gag antibodies and PAG15. Arrowheads point to viral particles in the process of budding. Bar, 200 nm. (B) 3D reconstruction of macrophages infected with HIV-1 NLAD8 for 7 d and stained for Gag (see Video 1). (C) Confocal micrographs (one plane) of HIV-1–infected macrophages exposed to DMSO or nocodazole (10 µM) for 1 h. Cells were fixed and stained with antibodies specific for the indicated proteins. Bar, 5 µm. These experiments have been repeated three times. (D) Ultrathin cryosections of macrophages infected with HIV-1 NLAD8 for 15 d were double-immunogold labeled for Pr55Gag with PAG15 and for α-tubulin with PAG10. Three representative profiles are presented. Tubulin staining was present at the limiting membrane of VCCs (see arrowheads). Bars, 200 nm.
At the ultrastructural level, microtubules showing specific α-tubulin staining were present in the vicinity of compartments containing Gag+ particles but were mostly absent from the viral particles (Fig. 1 D). Strikingly, 85% of the VCCs had at least one detectable microtubule at less than 100 nm from their limiting membrane (Fig. 1 D, arrowheads; n = 32). The mean number of microtubules associated per compartment was 3 ± 2. Taken together, these results suggested that VCCs were closely surrounded by microtubules that play a critical role in their spatial distribution.
HIV release by macrophages is dependent on the kinesin KIF3A
Cargo transport along microtubules often involves kinesins, making them ideal candidate proteins to be involved in VCC movement. Two genome-wide RNAi screens have identified two kinesin chains, KIF3A and KIF3C, as host proteins involved in the HIV-1 cycle in cell lines (Brass et al., 2008; Zhou et al., 2008). In the kinesin-2 family, KIF3A can form heterodimers with KIF3C or KIF3B, whereas KIF3B and KIF3C cannot associate (Muresan et al., 1998; Hirokawa, 2000). In addition, our screening by RNAi of the human kinesins potentially involved in the HIV-1 cycle in primary macrophages also identified KIF3A (unpublished data).
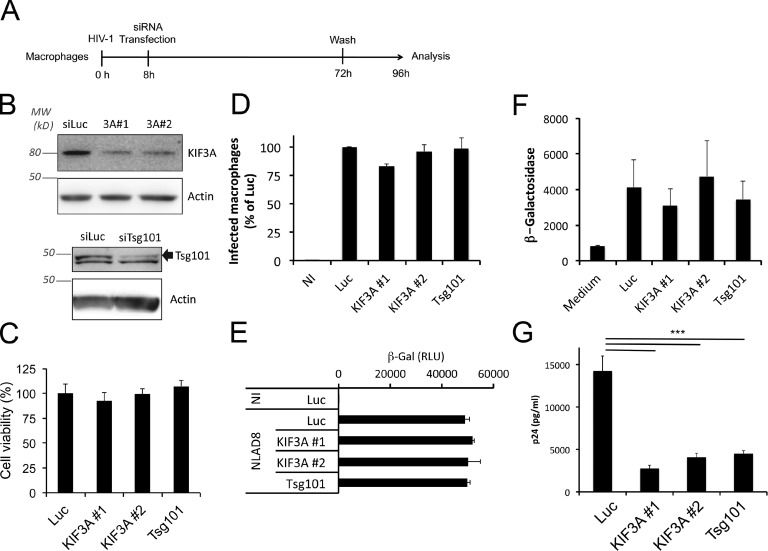
We therefore focused on the potential role of KIF3A on HIV-1 secretion by macrophages. Transfection of two different siRNA specific for KIF3A was performed in primary infected macrophages following the timeline presented in Fig. 2 A. SiRNA specific for luciferase and Tsg101 were included as negative and positive controls, respectively. Tsg101 is a member of the endosomal sorting complex required for transport (ESCRT) that is recruited together with Alix by the p6 late domain of Gag to allow HIV-1 budding and fission (Garrus et al., 2001; Strack et al., 2003). Efficiency of KIF3A depletions ranged from 55 to 95% as determined by immunoblot analysis (Fig. 2 B). Cell viability and percentage of infected macrophages were similar in the different cell populations upon viral infection and siRNA transfection (Fig. 2, C and D). The effects of KIF3A depletion were tested in the HeLa-derived TZM-bl cell line that reports on early viral protein expression (tat), as it carries a tat-sensitive promoter driving the expression of β-galactosidase, which activity reflects the rate of infection. We thus silenced KIF3A expression in TZM-bl before infection with HIV-1. β-Galactosidase activity remained similar, suggesting that viral entry and early steps of infection were not affected by KIF3A silencing (Fig. 2 E). We also used TZM-bl to test the infectious capacity of the viral particles produced by the KIF3A-depleted macrophages. For this purpose, similar amounts of viral particles (normalized by p24 content) produced by macrophages depleted or not of KIF3A were used to infect TZM-bl. The absence of significant difference in β-galactosidase activity (Fig. 2 F) suggested that viral particles still produced by KIF3A-depleted macrophages had no alteration in their infective capacity. Interestingly, KIF3A depletion by RNAi in HIV-1–infected macrophages led to a strong reduction (up to 75%) of the amount of p24 secreted in the supernatant, which reflects the secretion of viral particles (Fig. 2 G). The extent of the inhibition was similar to that obtained when the ESCRT protein Tsg101, which is crucial for viral budding (Garrus et al., 2001), was knocked down. We conclude that silencing of KIF3A inhibits the release of HIV-1 particles by infected macrophages.
Figure 2.
KIF3A is required for viral production by macrophages. (A) Schematic representation of the experimental design. (B) Immunoblot analysis of KIF3A and Tsg101 expression in macrophages transfected with the indicated siRNA. After 4 d, both siRNA specific for KIF3A significantly reduced its level of expression in primary macrophages. (C) Macrophages were infected by HIV-1 and transfected with siRNA as described in A. Cell viability was measured at d 4 p.i. using the CellTiter Glo kit and normalized to the control (siLuc). (D) Primary macrophages were infected with HIV Gag-iGFP ΔEnv pseudotyped with VSV-G and transfected with siRNA as described in A. This virus has a single cycle in macrophages and does not induce syncitia formation. Percentages of GFP+ macrophages were estimated by flow cytometry at d 4 p.i. (E) Infectivity of the virions produced by the macrophages subjected to the indicated siRNA was evaluated using the same amount of p24 (2 ng; see Materials and methods). (F) KIF3A depletion does not affect the early steps of infection. The reporter cell line TZM-bl was transfected with the indicated siRNA. 2 d later, cells were infected with HIV-1. Cells were washed 8 h later, reincubated for an additional 16 h, and assayed for β-gal activity, whose expression is driven by a Tat-sensitive promoter. (G) Dosage of p24 Gag in the 24-h culture supernatants harvested as indicated in A. *** indicates that the difference between the two histogram bars is statistically significant (P < 0.001). Experiments from D–F have been repeated at least two times and from B, C, and G at least three times in quadruplicate.
The role of KIF3A in HIV-1 release is cell type specific
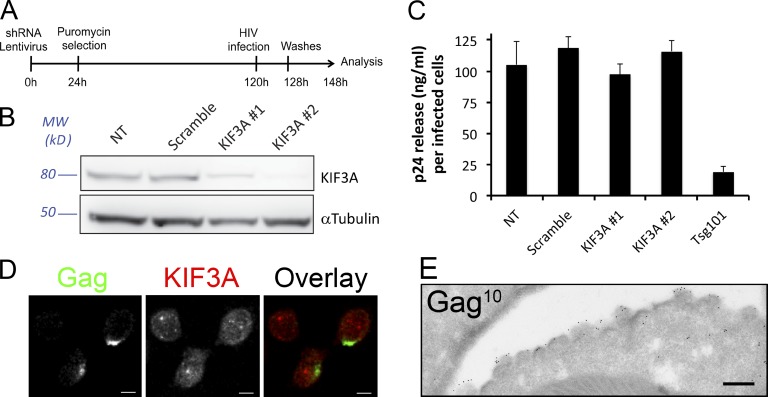
To determine whether the effect of KIF3A in the HIV-1 cycle was specific to macrophages, the role of KIF3A was assessed in the Jurkat T cell line. To achieve efficient silencing in these cells, we used shRNA delivered by lentiviral vectors (Fig. 3, A and B). KIF3A depletion had no impact on p24 release by Jurkat cells, whereas Tsg101 depletion led to an efficient reduction (80%) of p24 release (Fig. 3 C). In addition, staining of HIV-1–infected Jurkat T cells (not depicted) and primary CD4+ T cells (Fig. 3 D) revealed that Gag and KIF3A had completely different intracellular distributions. Furthermore, ultrastructural analysis of HIV-1–infected Jurkat T cells confirmed that viral assembly only takes place at the plasma membrane where zones of intense budding can be seen (Fig. 3 E). These results indicate that KIF3A does not play a role in the HIV-1 cycle in T cells, suggesting that KIF3A is required when viral assembly takes place in internal compartments but is dispensable when it occurs at the plasma membrane.
Figure 3.
KIF3A is dispensable for HIV-1 production by infected T cells. (A) Schematic representation of the experimental design. (B) Immunoblot analysis of KIF3A and α-tubulin expression in Jurkat T cells expressing the indicated shRNA. After 3 d, all shRNA specific for KIF3A reduced its level of expression down to less than 20% in Jurkat T cells. (C) Dosage of p24 Gag in 20-h culture supernatants of NL4-3 ΔEnv-infected Jurkat T cells harvested as indicated in A. Values presented have been corrected to the number of HIV-infected cells present in each sample, as calculated using CellTiter Glo (for cell viability) and anti-Gag staining analyzed by flow cytometry (for percentage of infection). (D) Confocal micrographs of primary T cells infected with VSV-G–pseudotyped HIV-1 NL4-3 for 24 h and stained for the indicated markers. Note that Gag distribution is highly polarized at the plasma membrane, whereas KIF3A distribution is totally different. Bars, 5 µm. (E) Ultrathin cryosection of Jurkat T cells infected with HIV-1 NL4-3 and immunogold labeled for Pr55Gag with PAG10. Bar, 200 nm.
The kinesin KIF3A associates with microtubules at the limiting membrane of VCCs
Focusing on KIF3A intracellular distribution in HIV-infected macrophages, and using the tetraspanin CD81 that is considered as a specific VCC marker (Deneka et al., 2007), we observed that KIF3A was associated with CD81+Gag+VCCs (Fig. S2 A). In contrast, the distribution of KIF15, a plus end–directed kinesin-12 member, appeared completely different from that of Gag (Fig. S2 B).
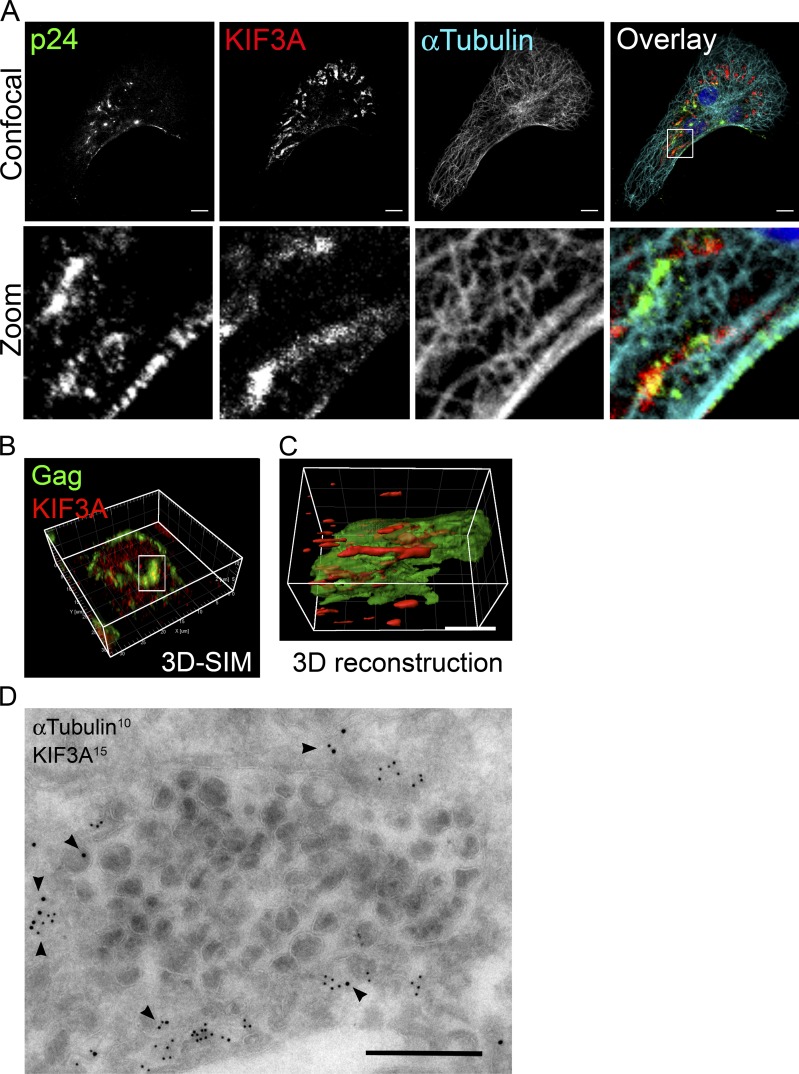
We also observed that while Gag+ compartments were heterogeneous in size and often rather large (up to several µm in diameter), KIF3A staining tended to localize to one or several restricted zones at the periphery of the Gag+ compartments together with tubulin (Fig. 4 A). 3D reconstructions from confocal images further showed association of patches of KIF3A staining with the edge of Gag+ compartments (Fig. S3 A). The Gag+KIF3A+ compartments were also positive for Env (Fig. S3 B), suggesting that they contained viral particles, and thus were bona fide VCCs.
Figure 4.
The VCCs are associated with tubulin and the kinesin KIF3A. (A) Confocal micrographs of HIV-1–infected macrophages. Cells were fixed and stained with antibodies specific for the indicated proteins. A z-projection of a stack of images is presented on the top row, and the bottom row shows one confocal plane of the magnification of the boxed area. Bar, 10 µm. (B) Structured illumination microscopy of HIV-1–infected macrophages stained for p24 and KIF3A. (C) 3D reconstruction with half opacity of the region boxed in B. Bar, 3 µm. (D) Immuno-EM of ultrathin cryosection of macrophages infected with HIV-1 NLAD8 for 7 d labeled for the indicated proteins. Arrowheads point to KIF3A staining associated with α-tubulin at the limiting membrane of a VCC. Bar, 500 nm. Experiments have been repeated at least twice.
Similar samples were examined by structured illumination microscopy (SIM), which provides a higher resolution (Schermelleh et al., 2010). 3D reconstructions from SIM images showed that KIF3A was localized along the VCC and further suggested that KIF3A was aligned on longitudinal structures that, in all likelihood, were microtubules (Fig. 4, B and C). Finally, immuno-EM unambiguously established that compartments containing viral particles had, at their limiting membrane, microtubules that stained positive for KIF3A (Fig. 4 D). Nocodazole exposure of HIV-1–infected macrophages led to the redistribution of Gag+ compartments together with KIF3A toward one pole of the nucleus (Fig. S3 C). Washout of nocodazole allowed regrowth of microtubules and spreading of the Gag+KIF3A+ compartments (not depicted). Thus, analysis of HIV-1–infected macrophages by confocal microscopy, SIM, and immuno-EM all indicated that KIF3A is associated with microtubules that are in contact with the limiting membrane of the VCC.
KIF3A silencing leads to intracellular accumulation of VCCs
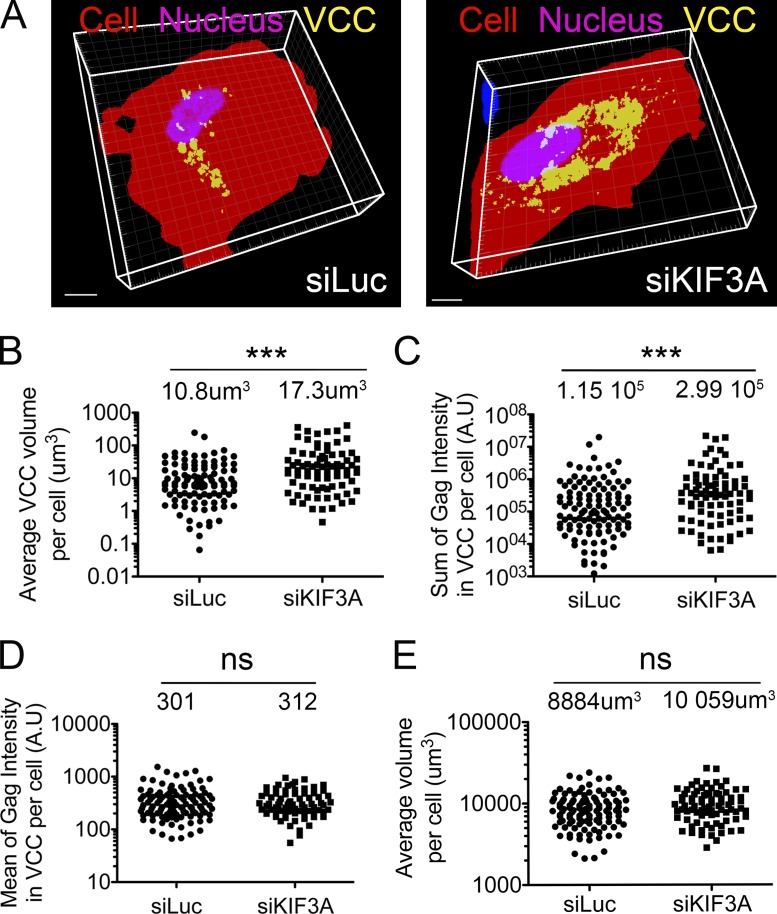
We reasoned that if KIF3A knockdown prevents transport of VCC and thus release of the virions contained therein, then VCCs should accumulate intracellularly. Thus, we compared HIV-1–infected macrophages from eight different donors treated with siRNA specific for KIF3A or for luciferase (control) and stained for Gag by immunofluorescence. Large sets of images were collected for both cell populations by confocal microscopy. 3D reconstruction of Gag+ compartments (Fig. 5 A), image segmentation, and quantification allowed the extraction of multiple parameters related to these compartments. The average volume of Gag+ compartments per cell as well as the sum of Gag intensity in VCC per cell were significantly increased upon KIF3A depletion (Fig. 5, B and C). Similarly, the ratio of the volume of Gag+ compartment/cytosolic volume per cell also increased in KIF3A knockdown cells (not depicted). In contrast, Gag mean intensity within individual VCCs remained the same (Fig. 5 D), suggesting that the density of Gag was similar and thus that the viral assembly process itself was not affected by the depletion. Finally, the cell area (not depicted) and the total cell volume (Fig. 5 E) were not statistically different between both cell populations.
Figure 5.
KIF3A depletion results in intracellular accumulation of VCCs. (A) Examples of 3D reconstructions obtained after thresholding of image stacks acquired by confocal microscopy of HIV-1–infected macrophages transfected with the indicated siRNA and stained for Gag. (B–E) Effects of KIF3A depletion on the indicated parameters within HIV-1–infected macrophages. Reconstructions as seen in A were segmented and quantified using Imaris software. KIF3A knockdown was estimated in parallel by immunoblot in the eight donors used for these experiments. The average efficiency of silencing was 72.9%, ranging from 55 to 95%. n.s., not statistically significant.
We also noticed that the secretion of MIP-1β, also known as CCL4, which is produced by macrophages upon HIV-1 infection (Fig. S4 A), was not affected by KIF3A depletion (Fig. S4 B). Because MIP-1β possesses a signal peptide and thus probably follows the classical secretory pathway, these results indicate that KIF3A depletion does not affect signal peptide–dependent secretion. We conclude that in the absence of KIF3A, VCCs accumulate intracellularly, explaining the observed reduction of p24 release.
Dynamics of KIF3A and VCC transport in live macrophages
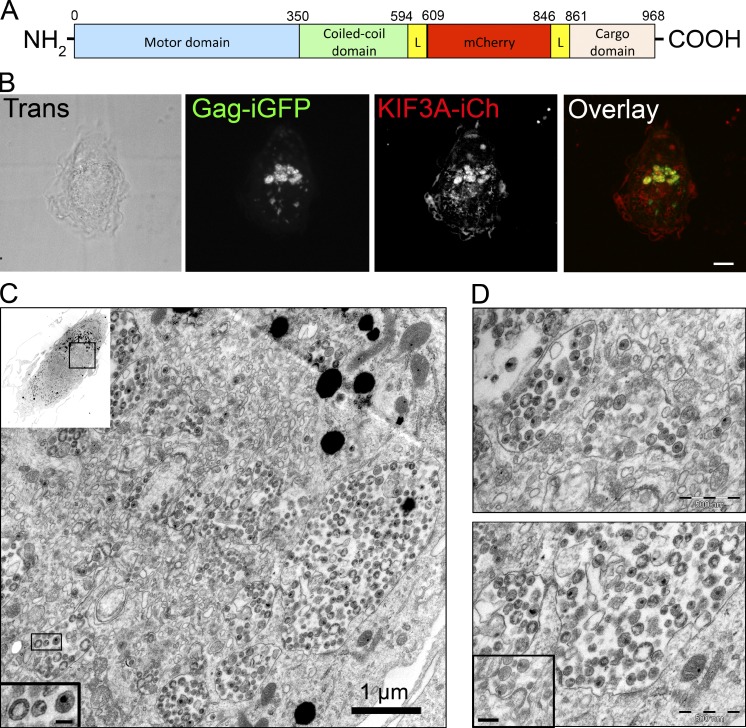
To analyze the dynamics of VCCs and KIF3A in live macrophages, we used two fusion proteins to simultaneously follow Gag and KIF3A. First, we derived HIV Gag-iGFP, a replication-competent and macrophage-tropic version of an internally GFP-tagged HIV-1 (Hübner et al., 2007). It has been shown that HIV Gag-iGFP shares essential features with wild-type HIV-1 and is suitable for kinetic studies in macrophages (Koppensteiner et al., 2012). Second, we generated a construct in which the fluorescent protein mCherry was inserted between the stalk and the cargo domains of KIF3A, flanked by two spacers to provide flexibility (Fig. 6 A). Of note, kinesins are often tagged by replacing their cargo domain by XFPs (Cai et al., 2009), but this could have prevented KIF3A to associate with VCCs. Our KIF3A-internal mCherry construct (KIF3A-iCh) was correctly expressed as seen by immunoblot and immunofluorescence in 293T cells (Fig. S5, A and B). Moreover, KIF3A-iCh, but not KIF15, was coimmunoprecipitated together with KIF3B-GFP using GFP-TRAP beads on cell lysates of HeLa cells expressing both fusion proteins, i.e., KIF3B-GFP and KIF3A-iCh (Fig. S5 C). In contrast, no KIF3A protein was detected in GFP immunoprecipitates from HeLa cells expressing GFP and KIF3A-iCh. Therefore, our data suggest that insertion of Cherry into KIF3A does not affect its capacity to associate with its known partners like KIF3B. In noninfected cells, the KIF3A-iCh fusion protein was observed to move along microtubules that we identified by coexpression of EGFP–α-tubulin (Fig. S5 D). From the analysis of time-lapse recording by spinning disk microscopy (Video 2), we estimated the average speed of KIF3A-iCh+ structures at 0.59 µm/s ± 0.65 (n = 643), and the average length of displacement at 5.3 µm ± 4.6, as expected for kinesin-2 movements (Loubéry et al., 2008; Cai et al., 2009).
Figure 6.
KIF3A-iCh is recruited to active VCCs. (A) Schematic representation of the KIF3A-iCh construct. (B and C) Correlative light and electron microscopy. (B) Primary macrophages grown on coverslips with coordinates were coinfected with HIV-1 Gag-iGFP and KIF3A-iCherry lentiviral vector, fixed, and imaged by spinning disk microscopy at d 7 p.i. Bar, 5 µm. (C) The same coverslips were then embedded in Epon and processed for EM. An overview of the macrophage imaged in B is presented in the top left inset. Example of mature and immature virions are presented in the bottom left inset. Bar, 100 nm. The VCCs are in the top third of the cell. (D) Two magnifications of the VCC-rich region, with a viral budding profile shown in the inset (bar, 200 nm).
To establish whether KIF3A-iCh associated with VCCs like the wild-type protein, we performed correlative light and electron microscopy. Primary macrophages were coinfected with HIV Gag-iGFP virus and KIF3A-iCh lentiviral vector. After 7 d of infection, macrophages were fixed on coverslips containing etched grids with coordinates and imaged by spinning disk microscopy. Co-expression of both fluorescent proteins was visualized in large intracellular compartments (Fig. 6 B). Samples were imbedded and sectioned for electron microscopy (EM). The coordinates of the cell on the grid allowed us to find the very same cell previously observed by fluorescence microscopy and perform correlative EM. As seen in Figs. 6 (B and C) and S5 E, the same features were observed both in photonic and electron microscopy, confirming that the very same cell was analyzed. At low magnification, EM revealed the position of the VCCs within the cell (Fig. 6 C). At high magnification, we observed that compartments corresponding to Gag-iGFP+ KIF3A-iCh+ contained numerous viral particles (Fig. 6, C and D). These compartments were active in HIV assembly, as budding profiles were visible at their limiting membrane (Fig. 6 D, bottom, inset). Both immature (electron dense at the periphery and electron lucent at the core) and mature (electron dense at the core) viral particles were present in VCCs (Fig. 6 C, inset). These results establish that the Gag-iGFP+ KIF3A-iCh+ compartments, observed in infected macrophages, are indeed VCCs where assembly and storage of viral particles take place.
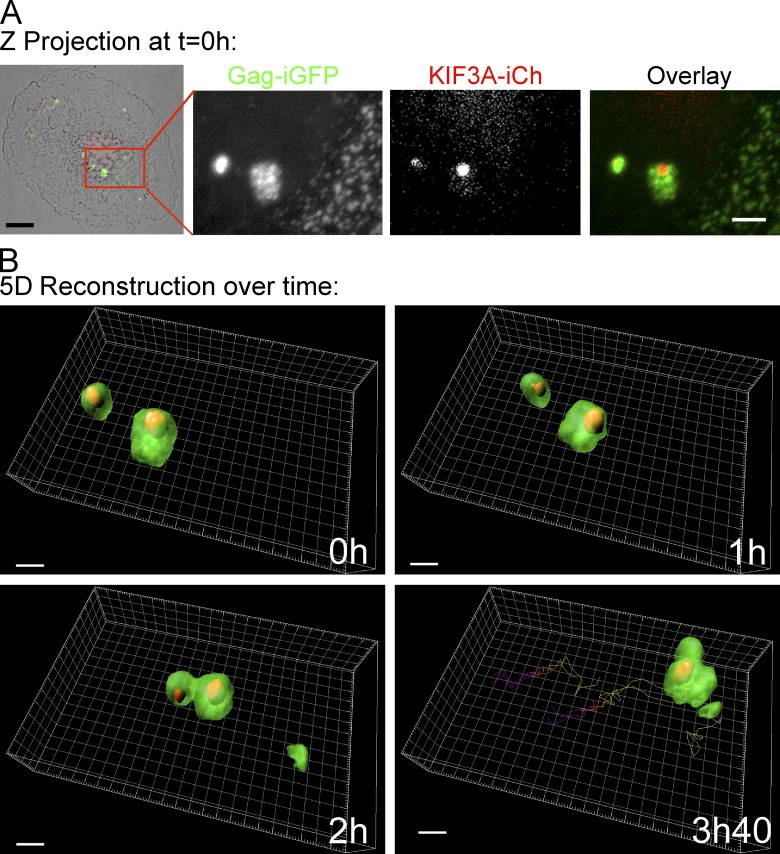
Time-lapse spinning-disk microscopy was finally performed on primary macrophages coinfected with HIV Gag-iGFP ΔEnv virus and KIF3A-iCh lentiviral vector for 5 d. Observation of a z-projection at the beginning of the video revealed that KIF3A-iCh was associated in a polarized manner with the periphery of Gag-iGFP+ compartments (Fig. 7), in agreement with our observations regarding endogenous KIF3A distribution in HIV-1–infected macrophages (Fig. 4). The 5D movies revealed that both fluorescent proteins remained associated over time and moved together in a directional manner (Fig. 7, A and B; see tracking and Videos 3 and 4). Thus, our results indicate that VCCs might be transported by KIF3A toward the periphery of macrophages, suggesting that KIF3A is required for their release into the extracellular milieu.
Figure 7.
Dynamics of internally tagged forms of KIF3A and Gag in primary macrophages. General view of a macrophage coinfected with HIV-1 Gag-iGFP ΔEnv and KIF3A-iCherry lentiviral vector and cultured for 5 d. The cell was imaged for 5 h by spinning disc microscopy. (A) Z-projection of the overlaid transmission, GFP and mCherry fluorescent intensities at time 0 (left, bar, 5 µm). The red square region is magnified for GFP and mCherry channels and the overlay of the two (panels 2–4; bar, 2 µm). Time 0, 1, 2, and 3 h 40 of a 3D reconstruction of the boxed area are presented with the tracking of the VCCs visible on the 3 h 40 panel (see Videos 3 and 4). Bar, 2 µm.
Discussion
In this study, we show the involvement of the microtubule network and the requirement for a processive motor, the kinesin KIF3A, for the late phases of the HIV-1 cycle in primary macrophages. Such motors of the kinesin-2 family drive their cargo, such as organelles, vesicles, or proteins from the microtubule-organizing center near the nucleus toward the cell periphery (Hirokawa et al., 2009). We observed that nocodazole exposure leads to the redistribution of the VCCs toward the nucleus. This may suggest that VCCs are exposed to opposite forces probably involving multiple molecular motors. A similar phenotype was reported about MIIC compartments from dendritic cells that collapse and merge upon nocodazole exposure (Vyas et al., 2007). Of note, members of the kinesin-2 family have been reported to stabilize microtubules and to have a shorter run length than kinesin-1 motors, suggesting that kinesin-2 motors may be well suited for bidirectional mobility (Verhey et al., 2011).
KIF3A has been reported to be involved in different cellular processes, such as intraflagellar transport within the primary cilia and the flagella (Scholey, 2008; Verhey and Hammond, 2009), axonal elongation and nodal flow (Hirokawa, 2000), and transport of late endosomes (Bananis et al., 2004; Brown et al., 2005; Loubéry et al., 2008). Within the kinesin-2 family, KIF3A is central as it can form heterodimers with KIF3B or KIF3C. In contrast, KIF3B or KIF3C cannot associate with each other due to the presence of similarly charged residues on both chains, which can associate with opposite charged residues present on KIF3A (Muresan et al., 1998; Chana et al., 2002; Chana et al., 2005). In addition, KAP3 (KIF-associated protein 3, also known as KIFAP3) is a large nonmotor protein that binds to the tail of KIF3 heterodimers. This protein containing many Armadillo repeats has been proposed to stabilize the association of the kinesin heterodimer (Doodhi et al., 2009) and to make the link between motor and cargo. Although expression of KIF3A and B is ubiquitous, to date KIF3C expression has been shown in neuronal and epithelial cells (Muresan et al., 1998). We have detected KIF3C expression in infected macrophages (unpublished data). However, the exact composition of the heterotrimeric KIF3 complex that is involved in VCC transport in HIV-1–infected macrophages remains to be determined.
Analysis of KIF3A distribution by immunofluorescence revealed some cytosolic staining, but concentration of KIF3A at the limiting membrane of VCCs or other organelles clearly occurs in HIV-1–infected macrophages (see Figs. 4, 6, 7, S2, and S3). In their extensive study, Muresan et al. (1998) established that KIF3C labeling in cells permeabilized with saponin before fixation revealed fine punctuated structures in neuronal cells. This labeling was sensitive to extraction with Triton X-100, suggesting that these structures may represent cargo vesicles.
KIF3A was previously shown to be required for virus-like particle release by HeLa cells expressing Gag-GFP (Azevedo et al., 2009) and to be involved in the transport of a viral protein from Kaposi’s sarcoma-associated herpes virus, named ORF45 (Sathish et al., 2009). In contrast, we show that KIF3A was dispensable for HIV-1 production by Jurkat T cells. Therefore, KIF3A appears as the first cellular protein specifically required in macrophages for HIV-1 late phase. It is tempting to link this result to the location of viral assembly: KIF3A might be required when viral assembly takes place in intracellular compartments, like in macrophages, and might be dispensable when it occurs at the plasma membrane, like in T cells. This hypothesis is supported by our proposed role for KIF3A driving the motion of the VCC toward the cell periphery. However, we cannot rule out that KIF3A can also play a role in T cells, but that its depletion is compensated by the activity of other kinesin(s). Our data further suggest that other cellular proteins than KIF3A might be involved in a macrophage-specific manner for the delivery of intracellular HIV-1 particles to the extracellular medium.
In the present study, we generated an internally tagged version of KIF3A for time-lapse microscopy. In contrast to most current fusions in which the cargo-binding domain is replaced by a fluorescent protein, in our construct, the fluorophore is inserted between two functional domains of the kinesin motor, flanked by flexible spacers. Appropriate insertion and spacing is crucial for proper folding and thus functioning of the protein. The KIF3A-iCherry fusion protein presents, like the endogenous protein, both diffused and punctuated patterns when transduced in macrophages (Fig. 6 B). Our approach represents a general strategy to efficiently tag such motors.
The 5D microscopy movies recorded on primary macrophages are striking, as they suggest that KIF3A is at the edge of VCCs during their motion. The term 5D microscopy (Roux et al., 2004) refers to three-dimensional fluorescence (3D) time-lapse (4D) imaging together with multispectral acquisition (5D). Given the way VCCs appear surrounded by microtubules and their relatively large size (micron range), cooperation between KIF3A and possibly other molecular motors at the limiting membrane of a given VCC probably aids its motion on microtubule filaments. Of note, different steps of the VCC life cycle may require different kinesins.
Our results are compatible with a model wherein the KIF3A complex drives VCCs toward the plasma membrane and may aid in the last step of the process, i.e., the release of virions into the extracellular space or to neighboring cells across virological synapses. Identification of KIF3A as a major player in the HIV cycle in primary macrophages opens new possibilities to specifically target this type of viral reservoir. The accumulation of intracellular stocks of infectious viral particles by macrophages represents a constant threat of potential escape from the control maintained by the immune system or by antiviral treatments. Targeting KIF3A activity may provide a new strategy for therapeutic intervention to be combined with existing therapies.
Materials and methods
Plasmids, antibodies, and reagents
The HIV Gag-iGFP T-tropic envelope construct was a gift from B. Chen (Mount Sinai School of Medicine, New York, NY; Hübner et al., 2007). In brief, HIV-1 molecular clone NL4-3 was modified by Chen’s group by standard overlap extension PCR methods to insert a polylinker between the MA and CA domains of Gag. Then, the enhanced fluorescent protein GFP (Takara Bio Inc.) was amplified by PCR and inserted into the polylinker region. The resulting clone expresses an 8–amino acid repeat, SQNYPIVQ, flanking either side of the GFP. We used this construct to derive the HIV-1 Gag-iGFP carrying an R5-tropic envelope with the V3-loop V92th014.12 (Koppensteiner et al., 2012). We also derived an env-defective version of the provirus, which does not express Env or Vpr and is referred to as HIV Gag-iGFP ΔEnv. pENTR KIF3A (GenBank/EMBL/DDBJ accession no. NM_007054.1) has been purchase from GeneCopoeia. To construct KIF3A-iCherry, the cDNA for the mCherry fluorescent protein, flanked by linkers (amino acid sequence GGGGSGGGGSGGGGS), has been inserted between the stalk and the cargo domain region of KIF3A (Fig. 7 A, see site of insertion). The resulting cDNA KIF3A-iCherry has been cloned with Gateway technology (Invitrogen) into pDEST DH1, a modified version of pCDH1 from System Bioscience in which the MCS has been replaced by a gateway cassette.
Goat polyclonal antibodies anti-p24, mouse monoclonal antibodies anti–human CD81 (clone TS81), KIF15 and Tsg101, and rabbit antibodies anti–human KIF3A (ab11259) were purchased from Abcam. Rat antibodies specific for α-tubulin (AbD Serotec) as well as antibodies specific for Lamp1 (BD) and for actin (MP Biomedicals) were used according to the manufacturers’ recommendations. Rabbit antibodies specific for mCherry were kindly provided by A. Marjou (Institut Curie, Paris, France). The following reagents were obtained through the NIH AIDS Research and Reference Reagent Program, Division of AIDS (National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, MD): HIV-1 gp120 monoclonal antibody (2G12, from H. Katinger [Institute of Applied Microbiology, Vienna, Austria]), and rabbit polyclonal antibodies specific for p17 as well as p55 (NIH AIDS reagents 4811, from P. Spearman [Emory University, Atlanta, GA]). Secondary antibodies conjugated with Alexa Fluor 488 or 647 (Invitrogen), or Cy3 or Cy5 (Jackson ImmunoResearch Laboratories) were used according to the manufacturers’ instructions. Nocodazole (Sigma-Aldrich) was dissolved in DMSO at a 10-mM stock concentration. M-CSF (EMD Millipore) was dissolved in ultrapure water at a 25-µM stock.
Primary cells and cell lines
PBMCs were purified from blood from healthy donors and monocytes were isolated by positive selection using CD14+ microbeads from Miltenyi Biotec. Monocytes were differentiated for 7 d to macrophages by culture on nontreated culture plastic (Thermo Fisher Scientific) in RPMI 1640 supplemented with 10% FCS (Gibco) and M-CSF (25 µg/ml final). Ghost, HEK293T, and TZM-Bl cells were cultured in DME Glutamax. Both mediums were supplemented with 10% FCS (Gibco).
CD4+ primary T cells were purified by negative selection of PBMC using the CD4+ T Cell Isolation kit II (Miltenyi Biotec). CD4+ cells were cultured in RPMI 1640 supplemented with 10% FCS (Gibco), 5 µg/ml PHA-L, and 20 U/ml IL-2 for 2 d, then resuspended in RPMI 10% FCS containing only IL-2. Cells were used for experiments on the fourth day after purification.
Jurkat T cells clone 20 were kindly provided by O. Schwartz (Pasteur Institute, Paris, France) and were maintained in RPMI 1640 supplemented with 10% FCS (Gibco).
Virus preparations and infections
The following virus strains have been produced and used in the present study: NL4-3 AD8 (NLAD8, R5-tropic), NL4-3 ΔEnv, HIV Gag-iGFP (R5-tropic, expressing GFP between MA and CA domains of Gag), and HIV Gag-iGFP ΔEnv (not expressing Env or Vpr). Viruses were produced by transfection of the corresponding proviral cDNA in 293T cells (ATCC CRL-11268) by transfection with polyethylenimine. The plasmid pVSV-G (BD) was eventually used for pseudotyping. Supernatants were harvested 48–72 h after transfection, and ultracentrifuged at 100,000 g for 90 min. Pellets were resuspended in 2% BSA in PBS. As described previously (Mörner et al., 1999), virus preparations were titrated by infecting the Ghost reporter cell line and infectivity was measured 24 h after infection by measuring the percentage of GFP-positive cells by flow cytometry.
Lentiviral vectors encoding either KIF3A-iCherry or the different shRNA (MISSION; Sigma-Aldrich) were produced by cotransfection of the VSV-G plasmid and the packaging psPax2 plasmid in 293T cells.
NLAD8-infected macrophage samples were prepared for immunoelectron microscopy as described previously (Jouve et al., 2007). In brief, macrophages were fixed with 4% paraformaldehyde in 0.1 M phosphate buffer, pH 7.4. Ultrathin cryosections were collected with a mixture of 2.3 M sucrose/methylcellulose. Single- or double-immunogold labeling on ultrathin cryosections was performed using protein A–gold conjugates (PAG) of indicated sizes. Macrophages were infected at a multiplicity of infection of 2 and kept in culture for the indicated times.
Viral production and infectivity assays
Measure of HIV p24 was performed using the Innotest HIV Ag mAb Screening kit (InGen). Cell viability was evaluated using CellTiter Glo (Promega).
For infectivity assays, virion-containing supernatants were normalized for equal amounts of p24 and infectious titers were determined using TZM-bl indicator cells as described previously (Martin-Serrano et al., 2001). The TZM-bl cell line was generated from HeLa cells expressing CCR5 and CD4 by introducing separate integrated copies of the luciferase and β-galactosidase genes under control of the HIV-1 promoter. In brief, the volume of supernatant equivalent to 2 ng of p24 was incubated with 10,000 TZM-bl cells for 24 h. Then, luminescent detection of β-galactosidase activity in the cell lysates was performed using the Gal-Screen kit (Applied Biosystems).
Immunoblotting
Cells were lysed in 0.5% NP-40 lysis buffer + 2 mM PMSF for 30 min on ice. Cell lysates were loaded on 4–12% Bis-Tris acrylamide gels (NuPAGE system; Invitrogen), transferred to a PVDF membrane, blocked, and immunoblotted in PBS 0.1% Tween 20 and 5% milk.
RNA interference
The various siRNA and shRNA used in the present study are indicated in Tables S1 and S2. Macrophages were transfected using Interferin (Polyplus). In brief, siRNA (100 nM final) were diluted in 250 µl of OptiMEM (Gibco) mixed with 5 µl of Interferin and left at room temperature for 15 min. Complexes were added drop-wise onto the macrophage culture. Cells were assayed 3 d later (Fig. 2 A). In all the experiments where macrophages were silenced for KIF3A expression (Figs. 2, 3, and 5), we checked for the efficiency of the knockdown by immunoblotting. Chemiluminescence acquisition was performed using an imaging system (LAS-3000; Fujifilm) allowing estimation of the band intensities. Silencing efficiency expressed as percentage of KIF3A intensity divided by the tubulin intensity and normalized to control ranged from 55 to 95% with a 72.9% average.
Immunostaining and microscope image acquisition
Cells were either fixed in methanol at −20°C for 5 s and incubated in 0.2% BSA in PBS (Figs. 3 D; 4 [A–C]; S1 [B and C]; S2 [A and B]; and S5 B) or fixed in 4% PFA for 15 min and permeablized in 0.05% saponin, 0.2% BSA in PBS (Figs. 1 [B and C]; and 5 A). Antibody staining and washes were performed in the same buffers, before mounting coverslips in DAPI Fluoromount G (SouthernBiotech). Samples were then imaged on a Ti inverted microscope (Nikon) fitted with a confocal A1R system, using a 60x oil immersion objective, NA 1.4 (Figs. 1 [B and C]; 3 D; 4 A; 5 A; S1 [B and C]; S2 [A and B]; S3 [A–C]; and S5 B). Confocal images were collected as snapshot or 3D stacks with a focal step size from 0.25 to 0.5 µm. Three-dimensional structured illumination microscopy (3D-SIM) was performed on an inverted Microscope (Eclipse Ti; Nikon) with a MCL Piezo stage and a 100x oil immersion objective, NA 1.49. Stacks were acquired every 0.1 µm (Fig. 4, B and C).The confocal and SIM microscopes were running with NIS Elements (Nikon) and were used for imaging of fixed samples at room temperature.
NIS Elements, MetaMorph (Molecular Devices), and ImageJ (National Institutes of Health, Bethesda MD) softwares were used for image processing. Figs. 1 B, 4 [B and C], 5, 7 B, and S3 A were 3D reconstructed using Imaris software x64 7.0 (Bitplane), based on fluorescence intensity of each channel. Background subtraction (Gaussian filter), thresholding, and isosurfaces were performed on Figs. 4 C, 5, 7 B, and S3 A, based on individual channel fluorescence intensity.
Live imaging was performed on a Ti inverted microscope (Nikon) fitted with a video-rate confocal system consisting of a spinning disk confocal head (Yokogawa Corporation of America). Cells were seeded in video chamber (MatTek Corporation) in cell culture medium. Videos were acquired at 37°C in a humid atmosphere with 5% CO2. Figs. 6 B, 7 [A and B], and S5 D were imaged using a 100x oil immersion objective, NA 1.4, with an HQ2 camera (Photometrics) and the microscope was driven by MetaMorph software.
Correlative light-electron microscopy
Purified monocytes were plated with M-CSF on CELLocate coverslips (Eppendorf) that etched grids with coordinates, allowing the cell of interest to be found through all of the steps of the procedure. After 7 d, macrophages were coinfected with HIV-1 Gag-iGFP and KIF3A-iCherry lentiviral vector. At d 7 after infection, cells were fixed in 2.5% glutaraldehyde in 0.2 M cacodylate buffer and imaged with a spinning disk confocal microscope. Then, coverslips were embedded in Epon and processed for electron microscopy (Jouve et al., 2007).
Image quantification
Quantification in Fig. 5 was performed as follows: macrophages from eight donors were infected with NLAD8 and transfected with either siLuc or siKIF3A as described in Fig. 2. Confocal planes were acquired for more than 100 cells per condition. Cells were 3D reconstructed and individually analyzed in a blind manner using Imaris x64 7.0 software. The low Gag fluorescence intensity was used to define the cell edges with a 1-µm smoothing (Fig. 5 A, red) without background subtraction. Higher Gag fluorescence intensity corresponding to the VCCs was detected and smoothed at 0.1 µm (Fig. 5 A, yellow). Cells with a volume under 1,000 µm3 were not taken into account as well as VCCs under 0.050 µm3. Automated statistical analysis was run to measure for each condition the average VCC volume per cell (Fig. 5 B), the sum of Gag intensity into VCCs per cell (Fig. 5 C), the mean of Gag intensity into VCCs per cell (Fig. 5 D), and the average volume of a cell (Fig. 5 E).
Spreading index in Fig. S1 B was evaluated as follows: single confocal planes of infected cells were acquired. Using ImageJ, vesicles were thresholded based on fluorescence intensity of the Gag staining and counted for each cell using the “analyze particles” tool with exclusion of small objects. The mask generated by the threshold was extended twice using the “dilate” tool of ImageJ and the number of vesicles was counted again. The spreading index corresponds to the number of vesicles counted after dilation divided by the number of vesicles counted before dilation.
Online supplemental material
Fig. S1 shows that the distribution of Gag+ compartments depends on the integrity of the microtubule network. Fig. S2 shows that KIF3A but not KIF15 is associated with the VCCs. Fig. S3 shows that the kinesin KIF3A is associated with the VCC. Fig. S4 shows that KIF3A depletion does not affect MIP-1β secretion by HIV-infected macrophages. Fig. S5 shows characterization of the KIF3A-iCherry construct. Video 1 shows an animation of a 3D reconstruction of an HIV-1–infected macrophage stained for Gag presented in Fig. 1 B. Video 2 shows dynamic of KIF3A-iCh and GFP-tubulin in live RPE1 cells presented in Fig. S5 D. Videos 3 and 4 show dynamics of KIF3A-iCherry and HIV-1 Gag-iGFP in primary macrophages presented in Fig. 7, A and B. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201201144/DC1.
Supplementary Material
Acknowledgments
We thank M. Linder for the KIF3B-GFP construct. The authors greatly acknowledge the Nikon Imaging Center at Institut Curie-CNRS as well as S. Amigorena, F. Perez, N. Manel, and A.-M. Lennon-Dumenil for discussions. We thank S. Watson, C. Banning, T. Jones, M. Touzot, and O. Lantz for help. We thank the BioPhenics platform of the translational department of the Insitut Curie for its help in automated image acquisition and analysis.
This work was supported by grants from Agence Nationale de Recherche contre le SIDA (ANRS) and Ensemble contre le SIDA (Sidaction) to P. Benaroch and fellowships to R. Gaudin (ANRS), E. Le Bouder (Sidaction), S. Bèrre (Curie Institute), and B.C. de Alencar (Fondation pour la Recherche Médicale).
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- HIV
- human immunodeficiency virus
- PAG
- protein A coupled to gold particles
- PBMC
- peripheral blood mononuclear cell
- SIM
- structured illumination microscopy
- VCC
- virus-containing compartment
References
- Alexaki A., Liu Y., Wigdahl B. 2008. Cellular reservoirs of HIV-1 and their role in viral persistence. Curr. HIV Res. 6:388–400 10.2174/157016208785861195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azevedo C., Burton A., Ruiz-Mateos E., Marsh M., Saiardi A. 2009. Inositol pyrophosphate mediated pyrophosphorylation of AP3B1 regulates HIV-1 Gag release. Proc. Natl. Acad. Sci. USA. 106:21161–21166 10.1073/pnas.0909176106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bananis E., Nath S., Gordon K., Satir P., Stockert R.J., Murray J.W., Wolkoff A.W. 2004. Microtubule-dependent movement of late endocytic vesicles in vitro: requirements for Dynein and Kinesin. Mol. Biol. Cell. 15:3688–3697 10.1091/mbc.E04-04-0278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brass A.L., Dykxhoorn D.M., Benita Y., Yan N., Engelman A., Xavier R.J., Lieberman J., Elledge S.J. 2008. Identification of host proteins required for HIV infection through a functional genomic screen. Science. 319:921–926 10.1126/science.1152725 [DOI] [PubMed] [Google Scholar]
- Brown C.L., Maier K.C., Stauber T., Ginkel L.M., Wordeman L., Vernos I., Schroer T.A. 2005. Kinesin-2 is a motor for late endosomes and lysosomes. Traffic. 6:1114–1124 10.1111/j.1600-0854.2005.00347.x [DOI] [PubMed] [Google Scholar]
- Cai D., McEwen D.P., Martens J.R., Meyhofer E., Verhey K.J. 2009. Single molecule imaging reveals differences in microtubule track selection between Kinesin motors. PLoS Biol. 7:e1000216 10.1371/journal.pbio.1000216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centlivre M., Legrand N., Steingrover R., van der Sluis R., Grijsen M.L., Bakker M., Jurriaans S., Berkhout B., Paxton W.A., Prins J.M., Pollakis G. 2011. Altered dynamics and differential infection profiles of lymphoid and myeloid cell subsets during acute and chronic HIV-1 infection. J. Leukoc. Biol. 89:785–795 10.1189/jlb.0410231 [DOI] [PubMed] [Google Scholar]
- Chana M., Tripet B.P., Mant C.T., Hodges R.S. 2002. The role of unstructured highly charged regions on the stability and specificity of dimerization of two-stranded alpha-helical coiled-coils: analysis of the neck-hinge region of the kinesin-like motor protein Kif3A. J. Struct. Biol. 137:206–219 10.1006/jsbi.2002.4446 [DOI] [PubMed] [Google Scholar]
- Chana M.S., Tripet B.P., Mant C.T., Hodges R. 2005. Stability and specificity of heterodimer formation for the coiled-coil neck regions of the motor proteins Kif3A and Kif3B: the role of unstructured oppositely charged regions. J. Pept. Res. 65:209–220 10.1111/j.1399-3011.2005.00210.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deneka M., Pelchen-Matthews A., Byland R., Ruiz-Mateos E., Marsh M. 2007. In macrophages, HIV-1 assembles into an intracellular plasma membrane domain containing the tetraspanins CD81, CD9, and CD53. J. Cell Biol. 177:329–341 10.1083/jcb.200609050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doodhi H., Ghosal D., Krishnamurthy M., Jana S.C., Shamala D., Bhaduri A., Sowdhamini R., Ray K. 2009. KAP, the accessory subunit of kinesin-2, binds the predicted coiled-coil stalk of the motor subunits. Biochemistry. 48:2248–2260 10.1021/bi8018338 [DOI] [PubMed] [Google Scholar]
- Garrus J.E., von Schwedler U.K., Pornillos O.W., Morham S.G., Zavitz K.H., Wang H.E., Wettstein D.A., Stray K.M., Côté M., Rich R.L., et al. 2001. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell. 107:55–65 10.1016/S0092-8674(01)00506-2 [DOI] [PubMed] [Google Scholar]
- Gendelman H.E., Orenstein J.M., Martin M.A., Ferrua C., Mitra R., Phipps T., Wahl L.A., Lane H.C., Fauci A.S., Burke D.S., et al. 1988. Efficient isolation and propagation of human immunodeficiency virus on recombinant colony-stimulating factor 1-treated monocytes. J. Exp. Med. 167:1428–1441 10.1084/jem.167.4.1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirokawa N. 2000. Stirring up development with the heterotrimeric kinesin KIF3. Traffic. 1:29–34 10.1034/j.1600-0854.2000.010105.x [DOI] [PubMed] [Google Scholar]
- Hirokawa N., Noda Y., Tanaka Y., Niwa S. 2009. Kinesin superfamily motor proteins and intracellular transport. Nat. Rev. Mol. Cell Biol. 10:682–696 10.1038/nrm2774 [DOI] [PubMed] [Google Scholar]
- Hübner W., Chen P., Del Portillo A., Liu Y., Gordon R.E., Chen B.K. 2007. Sequence of human immunodeficiency virus type 1 (HIV-1) Gag localization and oligomerization monitored with live confocal imaging of a replication-competent, fluorescently tagged HIV-1. J. Virol. 81:12596–12607 10.1128/JVI.01088-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igarashi T., Brown C.R., Endo Y., Buckler-White A., Plishka R., Bischofberger N., Hirsch V., Martin M.A. 2001. Macrophage are the principal reservoir and sustain high virus loads in rhesus macaques after the depletion of CD4+ T cells by a highly pathogenic simian immunodeficiency virus/HIV type 1 chimera (SHIV): Implications for HIV-1 infections of humans. Proc. Natl. Acad. Sci. USA. 98:658–663 10.1073/pnas.98.2.658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolly C., Mitar I., Sattentau Q.J. 2007. Requirement for an intact T-cell actin and tubulin cytoskeleton for efficient assembly and spread of human immunodeficiency virus type 1. J. Virol. 81:5547–5560 10.1128/JVI.01469-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouve M., Sol-Foulon N., Watson S., Schwartz O., Benaroch P. 2007. HIV-1 buds and accumulates in “nonacidic” endosomes of macrophages. Cell Host Microbe. 2:85–95 10.1016/j.chom.2007.06.011 [DOI] [PubMed] [Google Scholar]
- Jouvenet N., Neil S.J., Bess C., Johnson M.C., Virgen C.A., Simon S.M., Bieniasz P.D. 2006. Plasma membrane is the site of productive HIV-1 particle assembly. PLoS Biol. 4:e435 10.1371/journal.pbio.0040435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koppensteiner H., Banning C., Schneider C., Hohenberg H., Schindler M. 2012. Macrophage internal HIV-1 is protected from neutralizing antibodies. J. Virol. 86:2826–2836 10.1128/JVI.05915-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loubéry S., Wilhelm C., Hurbain I., Neveu S., Louvard D., Coudrier E. 2008. Different microtubule motors move early and late endocytic compartments. Traffic. 9:492–509 10.1111/j.1600-0854.2008.00704.x [DOI] [PubMed] [Google Scholar]
- Marsh M., Theusner K., Pelchen-Matthews A. 2009. HIV assembly and budding in macrophages. Biochem. Soc. Trans. 37:185–189 10.1042/BST0370185 [DOI] [PubMed] [Google Scholar]
- Martin-Serrano J., Zang T., Bieniasz P.D. 2001. HIV-1 and Ebola virus encode small peptide motifs that recruit Tsg101 to sites of particle assembly to facilitate egress. Nat. Med. 7:1313–1319 10.1038/nm1201-1313 [DOI] [PubMed] [Google Scholar]
- Mörner A., Björndal A., Albert J., Kewalramani V.N., Littman D.R., Inoue R., Thorstensson R., Fenyö E.M., Björling E. 1999. Primary human immunodeficiency virus type 2 (HIV-2) isolates, like HIV-1 isolates, frequently use CCR5 but show promiscuity in coreceptor usage. J. Virol. 73:2343–2349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muresan V., Abramson T., Lyass A., Winter D., Porro E., Hong F., Chamberlin N.L., Schnapp B.J. 1998. KIF3C and KIF3A form a novel neuronal heteromeric kinesin that associates with membrane vesicles. Mol. Biol. Cell. 9:637–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orenstein J.M., Meltzer M.S., Phipps T., Gendelman H.E. 1988. Cytoplasmic assembly and accumulation of human immunodeficiency virus types 1 and 2 in recombinant human colony-stimulating factor-1-treated human monocytes: an ultrastructural study. J. Virol. 62:2578–2586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelchen-Matthews A., Kramer B., Marsh M. 2003. Infectious HIV-1 assembles in late endosomes in primary macrophages. J. Cell Biol. 162:443–455 10.1083/jcb.200304008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raposo G., Moore M., Innes D., Leijendekker R., Leigh-Brown A., Benaroch P., Geuze H. 2002. Human macrophages accumulate HIV-1 particles in MHC II compartments. Traffic. 3:718–729 10.1034/j.1600-0854.2002.31004.x [DOI] [PubMed] [Google Scholar]
- Roux P., Münter S., Frischknecht F., Herbomel P., Shorte S.L. 2004. Focusing light on infection in four dimensions. Cell. Microbiol. 6:333–343 10.1111/j.1462-5822.2004.00374.x [DOI] [PubMed] [Google Scholar]
- Sathish N., Zhu F.X., Yuan Y. 2009. Kaposi’s sarcoma-associated herpesvirus ORF45 interacts with kinesin-2 transporting viral capsid-tegument complexes along microtubules. PLoS Pathog. 5:e1000332 10.1371/journal.ppat.1000332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schermelleh L., Heintzmann R., Leonhardt H. 2010. A guide to super-resolution fluorescence microscopy. J. Cell Biol. 190:165–175 10.1083/jcb.201002018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholey J.M. 2008. Intraflagellar transport motors in cilia: moving along the cell’s antenna. J. Cell Biol. 180:23–29 10.1083/jcb.200709133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharova N., Swingler C., Sharkey M., Stevenson M. 2005. Macrophages archive HIV-1 virions for dissemination in trans. EMBO J. 24:2481–2489 10.1038/sj.emboj.7600707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strack B., Calistri A., Craig S., Popova E., Göttlinger H.G. 2003. AIP1/ALIX is a binding partner for HIV-1 p6 and EIAV p9 functioning in virus budding. Cell. 114:689–699 10.1016/S0092-8674(03)00653-6 [DOI] [PubMed] [Google Scholar]
- Verhey K.J., Hammond J.W. 2009. Traffic control: regulation of kinesin motors. Nat. Rev. Mol. Cell Biol. 10:765–777 10.1038/nrm2782 [DOI] [PubMed] [Google Scholar]
- Verhey K.J., Kaul N., Soppina V. 2011. Kinesin assembly and movement in cells. Annu Rev Biophys. 40:267–288 10.1146/annurev-biophys-042910-155310 [DOI] [PubMed] [Google Scholar]
- Vyas J.M., Kim Y.M., Artavanis-Tsakonas K., Love J.C., Van der Veen A.G., Ploegh H.L. 2007. Tubulation of class II MHC compartments is microtubule dependent and involves multiple endolysosomal membrane proteins in primary dendritic cells. J. Immunol. 178:7199–7210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsch S., Keppler O.T., Habermann A., Allespach I., Krijnse-Locker J., Kräusslich H.G. 2007. HIV-1 buds predominantly at the plasma membrane of primary human macrophages. PLoS Pathog. 3:e36 10.1371/journal.ppat.0030036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H., Xu M., Huang Q., Gates A.T., Zhang X.D., Castle J.C., Stec E., Ferrer M., Strulovici B., Hazuda D.J., Espeseth A.S. 2008. Genome-scale RNAi screen for host factors required for HIV replication. Cell Host Microbe. 4:495–504 10.1016/j.chom.2008.10.004 [DOI] [PubMed] [Google Scholar]
- Zhu T., Muthui D., Holte S., Nickle D., Feng F., Brodie S., Hwangbo Y., Mullins J.I., Corey L. 2002. Evidence for human immunodeficiency virus type 1 replication in vivo in CD14(+) monocytes and its potential role as a source of virus in patients on highly active antiretroviral therapy. J. Virol. 76:707–716 10.1128/JVI.76.2.707-716.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.