Abstract
Background
EF-hand proteins can be activated by the binding of various heavy metals other than calcium, and such complexes can disturb the calcium-signaling pathway and cause toxicity and disease causing state. So far, no comprehensive study has been done to understand different heavy metals binding to calcium signaling proteins.
Results
In this work, the flexibility of the EF-hand motifs are examined by crystallographic and thermodynamic studies of binding of Pb2+, Ba2+ and Sr2+ to Calcium Binding Protein-1 from Entamoeba histolytica (EhCaBP1). The structures of the EhCaBP1- heavy metal complexes are found to be overall similar, nevertheless specific differences in metal coordination, and small differences in the coordination distances between the metal and the ligands in the metal binding loop. The largest such distances occur for the Ba2+- EhCaBP1 complex, where two bariums are bound with partial occupancy at the EF2 motif. Thermodynamic studies confirm that EhCaBP1 has five binding sites for Ba2+ compared to four binding sites for the other metals. These structures and thermodynamic studies reveal that the EF-hand motifs can accommodate several heavy atoms with similar binding affinities. The binding of Ca2+ to the 1st, 2nd and 4th sites and the binding of Ba2+ to the 1st, 2nd, 4th and 5th sites are both enthalpically and entropically driven, whereas the binding of Sr2+ to the 1st, 2nd and 4th sites are simply enthalpy driven, interestingly in agreement with ITC data, Sr2+ do not coordinate with water in this structure. For all the metals, binding to the 3rd site is only entropy driven.
Conclusion
Energetically, Ca2+ is preferred in three sites, while in one site Ba2+ has better binding energy. The Sr2+-coordination in the EF hand motifs is similar to that of the native Ca2+ bound structure, except for the lack of water coordination. Sr2+ coordination seems to be a pre-formed in nature since all seven coordinating atoms are from the protein itself, which also correlates with entropy contributions in Sr2+ binding. These findings improve our understanding of metal association with calcium binding proteins and of metal induced conformational changes.
Keywords: Calcium sensor, Calcium binding protein, Coordination geometry, EF-hand motifs, Anthropogenic toxicant, Domain swapped manner, Anomalous signal
Background
The presence of toxic heavy metals in the environment is of particular concern since they can accumulate in the body and cause significant health problems even at low concentrations [1,2]. Although the coordination geometries of many proteins are well suited for binding essential metal ions, they can also often interact with a number of heavy metals. Pb2+ is the most studied and prominent such toxicant: it displaces both Ca2+ and Zn2+ in proteins, affects neurotransmitter release and neuronal growth, and causes anemia, kidney damage, hypertension as well as male infertility [3-5] At the molecular level, Pb2+ has been shown to specifically target voltage-gated calcium channels [6], skeletal muscle troponin C (TnC) [7] and can also activate calmodulin (CaM) at low concentration [8].
Cationic lead is able to substitute for Ca2+ in the regulation of CaM’s function [9,10]. The activity levels of many target proteins are indeed affected by Pb-CaM as they are by Ca-CaM [11], leading to abnormal responses. Other metal ions such as Mg2+, Ba2+, Sr2+, Hg2+, Cd2+ and most lanthanides also show affinity with natural and engineered calcium binding proteins (CaBPs) including CaM [12-15]. However, the maximum activation decreases in the order Pb2+ > Ca2+ > Sr2+ > Ba2+ > Cd2+ as demonstrated previously in the EF-hand motif of D-galactose binding protein [16]. Moreover, the activity of downstream CaM-mediated functions depends on the concentration of the metal ion.
Structural and binding affinity investigations of calcium binding proteins aimed at understanding the flexibility and plasticity of EF-hand motifs, as well as understanding the structures of their interactions with various heavy metal ions, are very limited. Among the EF-hand containing proteins, the structure of CaM has been determined in complex with couple of heavy metals: one case with Pb2+[17,18] and another with partly bound Ba2+, in which only EF-hand motif 2 was bound with the ion [17].
So far, detailed comparative binding studies of different heavy atoms to any single CaBP, including CaM, have not been carried out. Here we have carried out such studies in detail with EhCaBP1, which is a calcium binding protein from Entamoeba histolytica. EhCaBP1 has four EF-hand motifs, each of them able to bind one calcium ion similar to that of calmodulin. The crystal structure of EhCaBP1 has been reported previously at 2.4 Å resolution by our group, where only the N-terminal half was traced and the C-terminal domain was found to be missing due to the presence of the flexible linker region between these two domains [19]. The overall structure is trimeric in which EF-hand 1 of one molecule interacts with EF-hand 2 of the other molecule in a domain swapped manner to form an assembled domain similar to that of the calmodulin N-terminal domain (Figure 1). The trimeric and monomeric forms of EhCaBP1 are in equilibrium with each other depending on the solvent conditions [20].
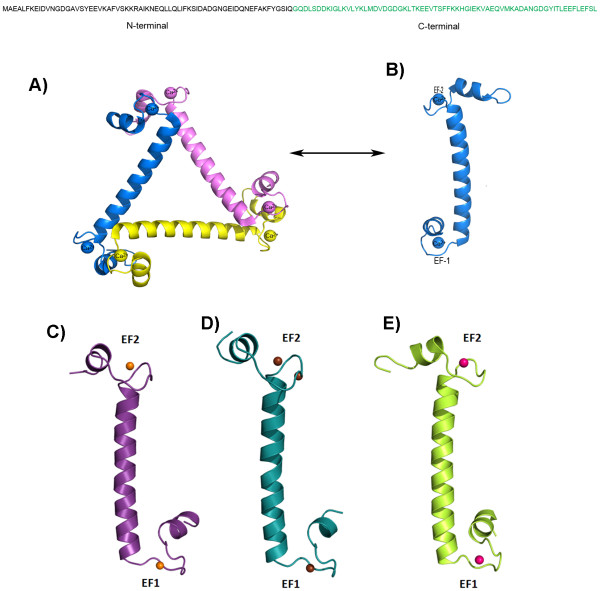
Figure 1.
Crystal structure of EhCaBP1. The sequence on the top shown with the N and C-terminal domains. In the structure only the N-terminal domain was traced since the C-terminal domain is disordered. A) The three molecules of the N-terminal domain come together to form a trimer. B) Each monomer is an extended structure with two EF hand motifs separated by a long helix. This calcium bound structure was reported earlier by Kumar et al., 2007. C-E) The heavy atom complex structures. C) Sr2+ complex, D) Ba2+ complex and E) Pb2+ complex of EhCaBP1 have overall similar structures to the native Ca2+ bound structure.
In this study, we investigate the specificity and flexibility of EF-hand motifs of EhCaBP1 in detail, using three heavy atoms: Pb2+, Ba2+ and Sr2+. We have successfully replaced the calcium ions in EhCaBP1 with each of these heavy atoms and have determined their structures. The association profile of Ca2+, Sr2+ and Ba2+ to EhCaBP1 has been determined by isothermal titration calorimetry (ITC) at physiological pH. Sr2+ and Ba2+ have similar binding affinities but a little lower binding energy (ΔG) compared to Ca2+ for the first, third, and fourth sites, while for the second site, Sr2+ and Ba2+ have better binding affinities. The heavy metal complex structures are similar to the native structure, with minor differences in the EF-hand motifs. There are small differences in the coordination distances between the metals and the ligands; the distances are largest for EhCaBP1-Ba. These results help explain the physiological basis for toxicity of these heavy metals by providing structural and biochemical insights into how these metal ions can effectively substitute for Ca2+ in a molecule that is central to several regulatory processes.
Results
Lead complex
In calmodulin (CaM), Pb2+ has been reported to exhibit higher affinity towards the calcium binding site than Ca2+ itself, and induce the protein into a constitutively active state [8,11]. The calcium ions were indeed replaced by lead ions, in both EF hands, when EhCaBP1 was co-crystallized with Pb2+, as indicated by the anomalous signal (Table 1 and see below), peaks in difference fourier maps, and improved crystallographic refinement upon including the heavy metal. As is found in the native, Ca2+-bound EhCaBP1 structure, the EF-hand motifs are also connected by a long helix in the Pb2+-bound EhCaBP1 structure. Since Pb2+ has one extra lone pair of electrons in the outermost shell, in contrast to Ca2+, Pb2+ generally interacts with six ligands, in contrast to Ca2+, which interacts with seven ligands. In the complex structures, Pb2+ thus cannot interact with the additional water molecule that Ca2+ interacts with. In the EhCaBP1-Pb2+ complex structure, six oxygens from the both EF hand motifs are interacting with Pb2+ (Figure 2B) as expected. The average coordination distances between Pb2+ and its ligands are a little lower than between Ca2+ and its ligands in the EF1 motif, but they are almost the same in the EF2 motif. Note that the Ca2+ and Pb2+ coordination distances were shown to be the same in the Calmodulin complex structures [17].
Table 1.
Anomalous signal evaluation
| Data set | CaBP1-Ba | CaBP1-Pb |
|---|---|---|
| Wavelength (Å) |
1.54178 |
1.54178 |
| Anomalous f” |
9.025 |
8.934 |
| Anomalous signal |
0.1071 |
0.0622 |
| Bijvoet pairs |
4892 |
7281 |
| Lone Bijvoet mates |
187 |
24 |
|
Expected Anomalous signal | ||
|
AutoSol | ||
| No. of Heavy atom sites |
6 |
4 |
| FOM |
0.40 |
0.18 |
| CC |
0.67 |
0.41 |
| Residues built |
91 |
75 |
| R/Free_R (%) |
0.41/0.48 |
53.0/56.0 |
|
AutoBuild | ||
| Residues built (%) |
80 |
65 |
| Map CC |
0.72 |
0.64 |
| R/Free_R | 0.42/0.47 | 47.0/55.0 |
Figure 2.
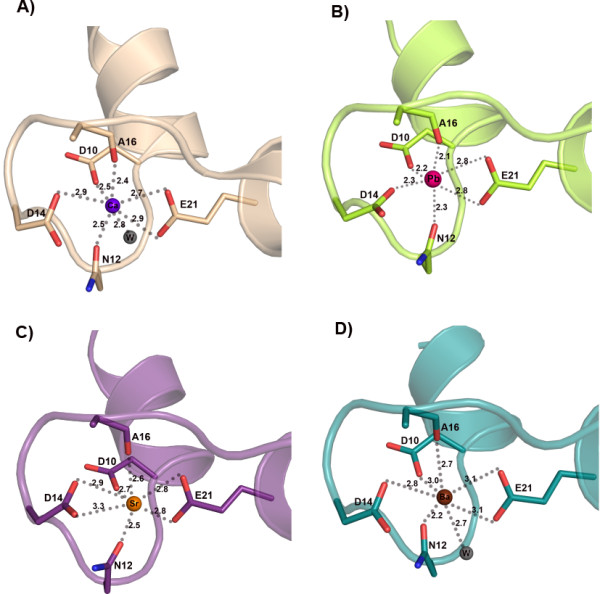
EF-hand motif 1 of EhCaBP1 showing the heavy-metal coordination. (A) Calcium coordination in the calcium binding loop-1 of EhCaBP1. Calcium coordinates with seven of its ligands including one water molecule to satisfy the coordination geometry. The coordination distances are shown. (B) Lead coordination in the calcium binding loop-1 of EhCaBP1. Lead co-ordinates with six ligands in the EF-hand motifs. The coordination distances between the Pb2+ and ligand oxygen atoms for EF-hand 1 motif range between 2.1 Ǻ and 2.8 Ǻ. (C) Strontium coordination in the calcium binding loop-1 of EhCaBP1. Strontium co-ordinates with seven ligands in the EF-hand motif. The coordination of Sr2+ is similar to that of Ca2+ coordination except for the absence of water molecule in the coordination. The 3rd aspartate residue is donating both of its oxygen ligands to satisfy the coordination geometry. The coordination distances between the ion and oxygen atom for the EF-hand 1 motif range between 2.1 Ǻ and 2.8 Ǻ. (D) Barium coordination in the calcium binding loop-1 of EhCaBP1. Barium coordinates with seven ligands including one water molecule to satisfy the coordination geometry. The coordination distances are shown. The coordination of Ba2+ is similar to that of Ca2+. The coordination distances between the ion and oxygen atom for EF-hand 1 motif range between 2.4 Ǻ and 2.9 Ǻ.
Strontium complex
The affinity of Sr2+ for CaM is lower than that of Ca2+ and Pb2+, but higher than that of Ba2+. Moreover, strontium and calcium have comparable cation sizes and hydration energies [10]. The overall structures of the Sr2+ bound and Ca2+ bound EhCaBP1 complexes are similar to each other, except for the metal coordination. A water molecule was not found to be coordinating Sr2+, as is observed in the Ca2+ bound as well as Ba2+ bound structures (Figure 2C). Both oxygens of the 3rd aspartate residue (residue 14) of the calcium-binding loop interact with Sr2+, compensating for the loss of water coordination. The average coordination distances with Sr2+ are a bit higher in the EF1 motif (by 0.1 Å) and almost same in the EF2 motif, compared to the average Ca2+-coordination distances.
Barium complex
In CaM, Ba2+ has low binding affinity for EF-hand motifs compared to Ca2+[10] For EhCaBP1 also, Ba2+ has lower binding energy compared to Ca2+, even though the difference in binding energy is not very high, therefore it cannot replace the calcium at low concentrations. Hence, the EhCaBP1 was denatured and refolded in Ba2+ containing buffer as described above.
The overall structure of the Ba2+-EhCaBP1 complex, including the coordination geometry, is similar to that seen in the native structure, except for a couple of features (Figure 1) The coordination distances in the EF1 motif are on average 0.1 Å longer in Ba2+-EhCaBP1 than in the Ca2+-bound structure. This difference suggests that the EF-hand motif is flexible enough to accommodate metal ions of varied size (Figure 2D).
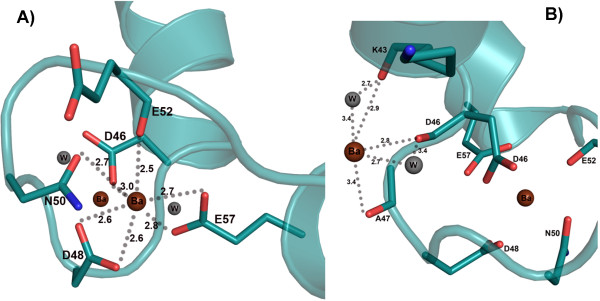
Moreover, a significant difference occurs in the EF2 motif, where two barium sites are observed. One barium is located at the same site as calcium The extra barium is in the close vicinity of EF-hand 2 loop, with partial occupancy and coordinated by residues K43, D46 and D48 (Figure 3). Both barium ions in the EF2 motif seem to be partially occupied, according to analyses of temperature factors and difference Fourier peaks. When assigned full occupancies, these bariums in the EF2 hand refined with high temperature factors compared to the barium ion bound in the EF1 hand loop as well as compared to other atoms in the EF2 hand. After reducing the occupancy to 0.5, the temperature factors were comparable to those of the barium atom in the EF1 motif. These two sites were confirmed by anomalous map and difference Fourier map up to 5σ before fitting barium in the electron density map. Before placing the heavy atoms, the fully occupied barium ion site had difference Fourier electron density at about 10σ and partially occupied barium ions has about 5σ level, clearly indicating the occupancy level. Similar binding pattern was also observed in EF-hand 2 of barium-soaked CaM crystal structure, but no barium was found to be bound to its other EF hand motifs [17].
Figure 3.
Unusual binding of barium to the EF-hand 2 of EhCaBP1. Barium has dual, partial occupancy to the EF hand 2 motif. (A) Barium coordinates with seven ligands from the protein to satisfy the coordination geometry. The coordination distances are shown. (B) Another barium ion in the close vicinity of EF-hand 2 motif is coordinated with the backbone oxygen atoms of K43, D46 and D48 residues of the protein and with two water molecules. The coordination distances between the barium ion and oxygen atoms range between 2.7 Ǻ and 3.4 Ǻ.
Anomalous signal analysis and verification of heavy atom binding
The barium complex shows a good anomalous signal of 0.1071, the signal for the lead complex is 0.0622, and the strontium complex data exhibits no anomalous signal at the respective wavelengths for which the data sets were collected. The calculated anomalous signals of the three data sets, the number of heavy atom binding sites in the proteins, as well as the f’ values of the heavy atoms all follow a similar trend. Since the Ba2+ is bound at three sites per molecule (one ion with full occupancy and two ions with partial occupancy) in the Ba+2-EhCaBP1 structure, its data showed the highest anomalous signal.
Thermodynamics of EhCaBP1-Ca/Sr/Ba complex formation
The ITC data indicate that EhCaBP1 has four sequential binding sites for Ca2+ and Sr2+, whereas the protein has five sequential binding sites for Ba2+ (Figure 4), consistent with the trends observed in the crystal structures described above. (Note that throughout the manuscript, “sites 1, 2, 3, 4 and 5” only correspond to the order of occupied sites during the fitting of ITC data by sequential mode. These ITC identifiers of binding sites do not necessarily correspond to the names of the EF hand motifs depicted in the crystal structures).
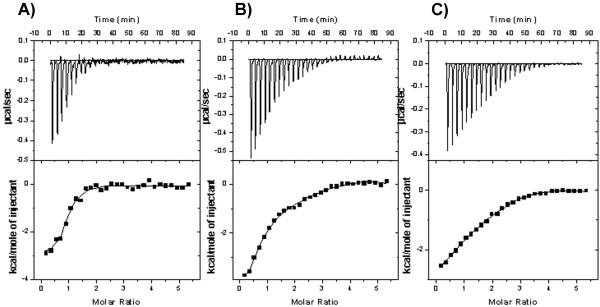
Figure 4.
ITC profile of the EhCaBP1-Ca (A), EhCaBP1-Sr (B), and EhCaBP1-Ba (C) associations at 25°C. Upper panel– raw data for sequential 10 μL injections of 0.5 mM metals into 0.02 mM EhCaBP1 solution; Lower panel–the data points obtained by integration of the peaks in the upper panel, and plotted against the molar ratio (metal: protein). The squares are the experimental data and the solid line represents the best fit obtained from the data fitting by sequential mode. Further details are given in the materials and methods section.
It is well known that metals induce large conformational changes in calmodulin-like proteins, including EhCaBP1 [21]. Hence it is very difficult to conclude whether the obtained thermodynamic parameters are associated only with binding a metal ion or with synchronous conformational changes. But the accurate determination of the thermodynamic parameters of protein-ligand interaction incorporated with ligand-induced conformational alterations, different contributions in the change of free energy, enthalpy and entropy are helpful in understanding ligand binding and conformational changes in protein structure. The binding of ligand to protein would be expected to be enthalpically-driven in the absence of any additional conformational changes in the protein. Replacing water molecules with ligand will, however, influence the entropy.
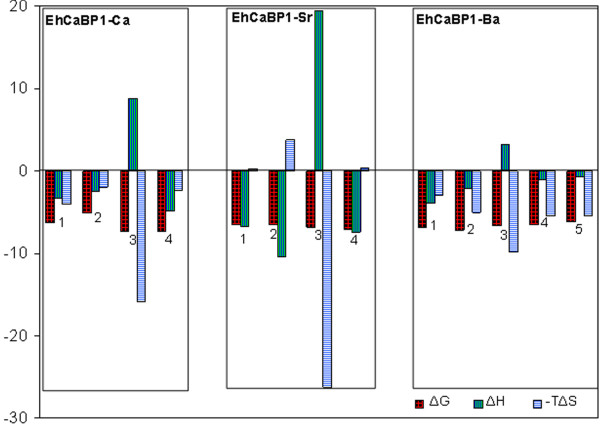
The change in free energy (ΔG) of the binding of metal to EhCaBP1 is negative for every case (Table 2), revealing that binding occurs spontaneously. Except for site 2 of Ca2+, it is observed that although the binding enthalpies and entropies of Ca2+, Sr2+ and Ba2+ to EhCaBP1 interaction differ a lot, their binding affinities (ΔG) remain effectively the same because changes in the binding enthalpy are compensated by changes in the binding entropy (Figure 5, Table 2). Consequently, enthalpically more favorable binding essentially results in greater entropic constraint, and thus in more unfavorable entropy. It is possible that a large increase in entropy derived from changes in water structure drives the entropically contributed interaction in the first site of Ca2+ as well as sites 2, 4 and 5 of Ba2+.
Table 2.
Thermodynamic parameters of metal binding to EhCaBP1in 20 mM MOPS buffer pH 7.4 at 25°C obtained and calculated from ITC results
| Metal | Site* | Ka(M-1) | ΔH(kcal.mol-1) | ΔS(Cal.mol-1.K-1) | ΔG(kcal.mol-1) | Dominating forces (inferred) |
|---|---|---|---|---|---|---|
| Ca2+ |
1 |
2.56E5 ± 9.1E4 |
−3.33 ± 0.30 |
13.6 |
−7.38 |
H-bonding/hydrophobic interactions |
| |
2 |
1.60E3 ± 9.0E2 |
−2.42 ± 0.28 |
6.54 |
−4.37 |
H-bonding/hydrophobic interactions |
| |
3 |
2.42E5 ± 8.3E4 |
8.80 ± 0.36 |
53.2 |
−7.34 |
Hydrophobic interactions |
| |
4 |
1.97E5 ± 8.6E4 |
−4.85 ± 0.94 |
7.94 |
−7.22 |
H-bonding/hydrophobic interactions |
| Sr2+ |
1 |
5.57E4 ± 9.6E3 |
−6.74 ± 0.51 |
−0.90 |
−6.47 |
H-bonding /conformational change |
| |
2 |
6.44E4 ± 7.0E3 |
−10.35 ± 1.98 |
−12.7 |
−6.56 |
H-bonding /conformational change |
| |
3 |
1.06E5 ± 1.5E4 |
19.42 ± 2.71 |
88.1 |
−6.83 |
Hydrophobic interactions |
| |
4 |
1.49E5 ± 2.1E4 |
−7.44 ± 1.31 |
−1.28 |
−7.06 |
H-bonding /conformational change |
| Ba2+ |
1 |
1.02E5 ± 1.5E4 |
−3.84 ± 0.17 |
10.0 |
−6.82 |
H-bonding/hydrophobic interactions |
| |
2 |
1.88E5 ± 2.0E4 |
−2.15 ± 0.39 |
16.9 |
−7.18 |
H-bonding/hydrophobic interactions |
| |
3 |
7.15E4 ± 6.1E3 |
3.23 ± 1.02 |
33.1 |
−6.62 |
Hydrophobic interactions |
| |
4 |
6.25E4 ± 9.3E3 |
−1.09 ± 2.14 |
18.3 |
−6.55 |
H-bonding/hydrophobic interactions |
| 5 | 3.16E4 ± 1.9E3 | −0.69 ± 0.23 | 18.3 | −6.14 | H-bonding/hydrophobic interactions |
*Sites 1, 2, 3, 4 and 5 only correspond to the order of occupied sites during the non-linear fitting of ITC data by sequential mode. These are not exchange digits for the loop of EF-I, EF-II etc.
Figure 5.
Thermodynamic signatures for Ca2+, Sr2+and Ba2+associations to EhCaBP1. The corresponding binding sites are in Roman capitals.
The most favorable binding reactions are both enthalpy and entropy driven due to specific hydrogen-bond formation and van der Waals forces as well as hydrophobic interactions [22]. Hence, EhCaBP1-metal associations with Ca2+ and Ba2+ except their site 3 are favorable reactions. In site 3, for every metal (Ca2+, Sr2+ and Ba2+), the interaction is highly entropically favored (despite with positive enthalpy) (Figure 5). These parameters for site 3 may be explained by an opening of the lobe and exposure of hydrophobic pockets, which are target sites in signal transduction. The transformation from the apo to metal bound form of a protein, through rearrangement of electrostatic bonds where metals and charged amino acids interact with each other through electrostatic forces, is a determining factor for the protein’s plasticity. Therefore, the degree to which electrostatic interactions (side chains) stabilize the protein may be determined in its flexible region.
The interaction of the “extra” Ba2+ with the K43/D46/A47 oxygens in EF-2 as observed in the crystal structure (Figure 3B) is directed by a combination of both the coordination and electrostatic forces. This interaction could be the 5th site according to ITC studies. In general, if ΔH ≈ 0, ΔS >0, and the opposite charges are less than 3.5 Å apart, the interaction is mainly governed by electrostatic force; Such is the case for the binding of Ba2+ to the 5th site, where ΔH, ΔS and the distances are −0.7 kCal.mol-1, 18.3 Cal.mol-1.K-1 and 2–3 Å respectively.
The binding of Sr2+ to site 1, site 4, and especially site 2 are purely enthalpy driven (Table 2). A high binding enthalpy may also be used for the prediction that the binding of Sr2+ to the loop displaces the bridging water molecule. Hence, structural alterations at the binding site due to the binding event may contribute to this enthalpy. The largest –TΔS value for the binding of Sr2+ is to site 2 and should correspond to the opening of the binding pocket. The results for site 1 and 4 may be due to the binding to the heterogeneous native state structure, which may be ensembles of minimum energy conformations. Further, like other metals, only the binding of Sr2+ to site 3 is purely entropic (Table 2, Figure 5). Hence, enthalpic contributions dominate the association of Sr2+ with EhCaBP1.
Discussion
We have successfully replaced the calcium of EhCaBP1 with the heavy metal ions Pb2+, Ba2+ and Sr2+, crystallized the complexes, and determined their structures. The difference Fourier electron density and anomalous signal confirms the presence of these heavy metal ions in place of calcium at the calcium binding loops in the respective crystal structures. The overall conformation and metal-coordination geometry of these complexes are quite similar to those of Ca2+-bound EhCaBP1, except for some relatively minor differences (Figure 1). This overall similarity provides a structural rationale for the ability of EhCaBP1 complexed with Pb2+, Sr2+ or Ba2+ to bind and to activate some of the biological targets of Ca2+- EhCaBP1.
Lead has been reported several times to be the most effective heavy metal for replacing calcium in the CaM activation cascade [8,9]. No significant structural rearrangements occur upon replacement of calcium with lead, and the coordination geometry is similar [18]. As a result of lead’s high binding affinity, it is tightly bound to the EF hand motif even at relatively low concentrations, unlike Ca2+, thus providing a structural reason for the reported constitutively active state of Pb2+ -CaM [10,11]. It is probable that lead has similar effects on EhCaBP1. Since lead has a relatively high binding affinity for the calcium binding site and can replace calcium very easily in EhCaBP1 as well. It may very well also result in the aberrant activation of this (EhCaBP1) protein’s downstream signaling pathways.
Although the complexes of EhCaBP1 with the various metal ions are similar to one another, the binding of Sr2+ and Ba2+ to the protein show a couple of special features. It appears that the EhCaBP1-Sr association is of a pre-formed nature since all seven coordinating atoms are from the protein itself (Figure 2); in contrast, in EhCaBP1-Ca and EhCaBP1-Ba, only six coordinating atoms are supplied by the protein and the remaining one is from water. Hence, among the three heavy metal ions, it can be concluded that Sr2+ shows the most specific interactions with EF-hand motifs. In the crystal structures of EhCaBP1, for each of the EF-I and EF-II binding sites for Ca2+, and for the EF-I binding site for Ba2+, the metal coordinates one water molecule (Figure 2). In the EF2 motif, Ba2+ binds with partial occupancy to two adjacent binding sites, and, in one of these sites, coordinates with several water molecules (Figure 3). The ITC data of the association of EhCaBP1 with Ca2+ and Ba2+ show a positive change in entropy in all of the sites (Table 2). In contrast, in EhCaBP1-Sr, where both the EF-I and EF-II binding sites are devoid of water, the change in entropy is positive for only one site (site 3) and negative for the others (site 1, 2 and 4). This result suggests that if there is ligated water in an EhCaBP1-metal complex, the change in entropy will be positive. Therefore, in EhCaBP1-Sr, the only site with positive entropy of binding, i.e. site 3, must be involved in opening of hydrophobic pocket as discussed above (Figure 5). These findings also imply that the unusual thermodynamic signatures in EhCaBP1-Sr (Table 2) are just a result of the absence of any ligated water molecule.
It has been already reported that in the different signal transducers, calmodulin (CaM) and troponin C (TnC), calcium-induced conformational alterations resulting in exposure of hydrophobic target sites are mainly due to motions of secondary structural units [23]. The gross structural morphology, secondary structural contents, and target binding of EhCaBP1 are related to those of vertebrate CaM [24]. According to this, upon binding a metal (apo → holo), the molecule changes conformation from a closed to an open state conformation, with many H-bonds from waters to side chains and backbone being rearranged, as is observed in CaM and also opening of hydrophobic core. Upon metal interaction in the first site, the most compact loop is formed by the rearrangement of van der Waals and ion pair contacts. Consequently the energetic barriers for the formation of another binding loop will be lowered, and the loop will properly position binding residues. The same pattern could then be sequentially transmitted in a cooperative manner to the next binding site.
Conclusion
Here, we have reported for the first time, the structural analysis of any calmodulin like protein, EhCaBP1 in complex with strontium. The Sr2+ -coordination in the EF hand motifs is similar to that of the native Ca2+ bound structure, except for the lack of water coordination. Sr2+ coordination seems to be a pre-formed in nature since all seven coordinating atoms are from the protein itself, which also correlates with entropy contributions in Sr2+ binding, in contrast, in EhCaBP1-Ca and EhCaBP1-Ba, only six coordinating atoms are supplied by the protein and the remaining one is from water. Our work has also provided the first successful example of replacing Ca2+ with Ba2+ in the EF1 motif of a calcium binding protein. Previously such information was restricted to the EF2 motif in CaM [17], and here as in our results for EhCaBP1 there are two bariums ions bound to EF2 motif. This is first time, any calcium binding protein structure is reported with four different heavy atoms, albeight with low resolution.
The crystallographic and ITC results clearly indicate that EhCaBP1 is also capable of binding and being activated by metal ions other than Ca2+, providing a better understanding of the activation mechanisms of EhCaBP1 by different heavy metals, and also give better insight into the flexibility of the calcium-binding loops of EhCaBP1. These results can be also extended to all EF hand containing proteins like Calmodulin. These results help us to explain the physiological basis for toxicity of these heavy metals by providing structural and biochemical insights into how these metal ions can effectively substitute for Ca2+ in several signaling pathways which are central to several regulatory processes.
Methods
Preparation of EhCaBP1-Pb complex, crystallization and data collection
EhCaBP1 was over-expressed and purified as previously described [19] and concentrated to 20 to 25 mg/ml. Since Pb2+ can easily replace Ca2+ from the calcium-binding loops of CaM, [10,11,18] Pb(NO3)2 at an initial concentration of 1 M was directly added to the purified EhCaBP1 to bring the final concentration of Pb2+ to 5 mM. Upon addition of Pb2+, the solution became turbid and protein precipitated. The solution was partially clarified upon addition of 1 M sodium acetate pH 4.0 to a final concentration of 100 mM. The EhCaBP1-Pb complex was centrifuged at 12,000 rpm to remove any precipitate, and the supernatant was used for crystallization.
Hanging-drop vapor-diffusion crystallization trials were carried out under conditions similar to that used for crystallization of the native protein [19]. Crystals suitable for diffraction were obtained by mixing 5 μl of EhCaBP1-Pb complex and 2 μl of reservoir solution containing 60–65% MPD in 50 mM sodium acetate pH 3.6. Rod shaped crystals (250 × 250 × 150 μm) appeared after 7–10 days of equilibration (Figure 1A).
X-ray diffraction experiments were done at 100 K with EhCaBP1-Pb crystals mounted on cryoloops in mother liquor and flash frozen in liquid nitrogen. These crystals diffracted to a resolution of 3.0 Å with an in-house Bruker Microstar rotating anode generator and a MAR345 image plate detector (Advanced Instrumentation Research Facility, JNU). The data sets were indexed, processed and scaled using the program Automar [25]. The crystals belong to the space group P63 (Table 3) with two molecules per asymmetric unit, similar to that of the native structure [19] (Figure 1).
Table 3.
Crystallographic data- statistics
| Data set | EhCaBP1-Pb | EhCaBP1-Sr | EhCaBP1-Ba |
|---|---|---|---|
|
Crystallographic Data | |||
| X-ray Source |
MicroStar |
Elettra |
MicroStar |
| Wavelength (Å) |
1.5418 |
1.0 |
1.5418 |
| Space group |
P63 |
P63 |
P63 |
| Unit-cell parameters (Å) |
a = 95.264, |
a = 95.429, |
a = 95.210, |
| b = 95.264, |
b = 95.429, |
b = 95.210, |
|
| c = 64.597 |
c = 63.973 |
c = 62.986 |
|
| Resolution range (Å) |
28.08-3.0 |
30-3.0 |
50-3.2 |
| Rsym (%) |
4.66 (36.4) |
6.80 (45.4) |
6.70 (62.8) |
| Completeness (%) |
99.8 (100) |
94.9 (96.8) |
99.6 (98.7) |
| Observations (N) |
24659 |
26059 |
28321 |
| Unique observations (N) |
6763 |
6442 |
5485 |
| Redundancy |
8.8 (8.1) |
4.0 (4.0) |
5.2 (4.4) |
| Average I/s (I) |
15.2 (1.9) |
18.81 (2.8) |
15.4 (1.8) |
| Crystal mosaicity (º) |
0.5 |
0.5 |
1.0 |
|
Refinement | |||
| Resolution (Å) |
28.08-3.0 |
30-3.0 |
50-3.2 |
| R-factor (%) |
24.24 |
25.23 |
24.70 |
| Free R-factor (%) |
28.67 |
27.99 |
31.40 |
| Mean B-factor(Å2) |
71.80 |
95.18 |
122.09 |
| Atoms (N) Protein/HA/ACT/water |
996/4/2/45 |
1000/4/0/27 |
966/6/0/21 |
|
RMS deviations | |||
| Bonds (Å) |
0.009 |
0.008 |
0.010 |
| Bond angles (°) |
1.7 |
1.2 |
1.3 |
| Cross validated error | 0.47 | 0.50 | 0.49 |
HA = Heavy Atom, ACT = Acetate.
Values in parentheses are for the highest resolution shell.
Free R-factors were calculated with a subset of randomly selected reflections (10.7% for EhCaBP1-Pb, 9.4% for EhCaBP1-Sr and 4.4% for EhCaBP1-Ba complex).
Preparation of EhCaBP1-Ba complex, crystallization and data collection
High concentrations of Ba2+ is required for the activation of CaM compared to Pb2+[26] Since Ba2+ has lower binding affinity for EF-hand motifs than does Ca2+, it cannot simply replace Ca2+. To replace all of the bound calcium ions from the calcium binding loops, EhCaBP1 was unfolded and calcium ions were removed by treatment with 8 M urea and 5 mM EGTA, The protein was dialyzed against 10 mM barium chloride in Tris buffer (pH 7.5) several times to remove urea and EGTA and then the protein was refolded in presence of excess barium ions. The EhCaBP1-Ba complex was then concentrated to 20–25 mg/ml, and crystallized by the hanging-drop vapor-diffusion method using conditions similar to that used for the native protein. Crystals suitable for diffraction were obtained by mixing 5 μl of EhCaBP1-Ba complex and 5 μl of the reservoir solution (60–65% MPD, 50 mM sodium acetate pH 3.8). Rod shaped crystals with oiling appeared after 15–20 days of equilibration.
The X-ray diffraction experiments were done at 100 K with EhCaBP1-Ba crystals mounted on cryoloops in mother liquor and flash frozen in liquid nitrogen stream. These crystals diffracted to a resolution of 3.2 Å using an in-house rotating anode generator (Advanced Instrumentation Research Facility, JNU). The data sets were indexed, processed and scaled using Automar [25]. They belong to the space group P63 (Table 3) with two molecules per asymmetric unit, similar to that of the native structure [19].
Preparation of EhCaBP1-Sr complex, crystallization and data collection
The EhCaBP1-Sr complex was prepared and crystallized following protocols similar to those used for the EhCaBP1-Ba complex. Crystals suitable for diffraction were obtained by mixing 3 μl of the EhCaBP1-Sr complex and 3 μl of the reservoir solution (60–65% MPD, 50 mM sodium acetate pH 3.6). Hexagonal crystals appeared after 15–20 days of equilibration. The X-ray diffraction experiments were done at 100 K with EhCaBP1-Sr crystals mounted on cryoloops in mother liquor and flash frozen in liquid nitrogen stream. These crystals diffracted to a resolution of 3.0 Å at beamline BL 5.2 R, XRD1, Elettra Synchrotron Source, Trieste, Italy. The data sets were indexed, processed and scaled with HKL2000 program [27]. They, too, belong to space group P63 with two molecules per asymmetric unit.
Structure determination
All three complex structures were solved by molecular replacement with the program Phaser [28] using the unliganded native structure of EhCaBP1 (2NXQ) as the search model. Iterative model building by the COOT graphics package [29] combined with Phenix.refine and Refmac5 for refinement was carried out to determine the EhCaBP1-Ba and EhCaBP1-Pb complex structures. CNS refinement with conjugate-gradient minimization and bulk solvent correction [30] was used for both the EhCaBP1-Sr and complex, whose structures are more ordered than the EhCaBP1-Ba complex structure (Table 3).
Calcium binding sites showed significant density peaks in the difference fourier maps of all the three heavy atom complexed structures compared to calcium bound structure. The peak in the barium bound difference map was strongest, at 6σ, Lead at 4σ, whereas that of the strontium bound structure was at a 3σ level, following the trend of the molecular weights of heavy atoms bound compared to calcium. The best R-factors were not achieved until the replacement of Ca2+ with the respective heavy atoms. The final models refined well, fitting most of the electron density (for the N-terminal domain), and yielded crystallographic Rfactor and Rfree values that are within the range of average values for structures refined at the given resolutions [31]. The 2Fo-Fc electron density maps for the heavy atoms were so strong that the density was even seen at 10σ for Barium, 6σ for lead and 7σ for strontium ( Additional file 1: Figure S1). The temperature factors for Lead were very high compared to temperature factor from Wilson plot. The occupancy for lead was adjusted to 0.5 to get comparable temperature factors. Generally, the protein was urea denatured followed by refolding in presence of the respective heavy metals containing buffers except for lead. Lead has more affinity than calcium, supposed to occupy the calcium-binding sites more easily. This may be the reason why the lead ions have half occupancies. Despite acceptable refinement statistics, electron density for the C-terminal half of the molecule was absent, as occurs in the native structure. In the final stages of refinement, water molecules were added manually where the Fo-Fc electron density exceeded 3.0 σ and when justified by hydrogen bonds. Only a few water molecules met these criteria.
Protein data bank accession codes
Coordinates and structure factors have been deposited in PDB with accession code 3PX1 for EhCaBP1-Sr, 3QJK for EhCaBP1-Pb and 3ULG for EhCaBP1-Ba complexes.
Anomalous signal analysis
To confirm the presence of heavy atoms at the EF-hand motifs, the anomalous signal was checked. Barium and lead atoms have f” of about 9 electrons at Cu-Kα wavelength of 1.54 Å, where as strontium has F” of 1.845 electrons at 1.2 Å. The anomalous signal was calculated using phenix.xtriage [32] for all data sets (Table 1).
Isothermal titration calorimetry (ITC)
All solutions were demetallized by a Chelex-100 resin (BioRad). The protein concentration was determined by using ε = 5120 M-1 cm-1 at 280 nm [33]. The calorimetric measurements were carried out using a titration calorimeter from Microcal (Northampton, MA) at 25°C. The solutions of EhCaBP1 and metals were prepared in the same buffer. The 0.02 mM EhCaBP1 solution in the 1.44 ml sample cell was titrated with 0.5 mM calcium chloride (Ca2+), strontium chloride (Sr2+) and barium chloride (Ba2+) dissolved in the same buffer using a 288 μl automatic rotating syringe stirring at 307 rpm. Titration experiments consisted of 28 injections of 10 μl each of duration 20s with 2 s filter period and 180 s spacing between each injection. The analog input range was +/− 1.25 V and the reference power was set at 20 μcal s-1. The heat associated with each injection was observed, as a peak that corresponded to the power required to keep the sample and reference cells at identical temperatures. Control experiments were performed by titrating metals into the same buffer to obtain the heats of ligand dilution. The net enthalpy for each EhCaBP1-metal association was determined by subtraction of the component heats of dilution from each injection heat pulse. Integration with respect to time of the heats produced per serial injection of metal yielded the corresponding binding isotherm. The binding isotherms were fitted for a model of sequential binding sites by using Origin 7.0 provided with the MicroCal instrument (Table 2). The titration with Pb2+ could not be done, as the lead salts were insoluble at physiological pH 7.4. The details of curve fitting and determination of number of binding sites for each heavy atom are described in Additional file 1.
Abbreviations
TnC: Troponin C; CaM: Calmodulin; MPD: 2-methyl-2, 4-pentanediol; EhCaBP1: Entamoeba histolytica calcium binding protein 1; PDB: Protein data bank.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
SK and SK purified the protein, crystallized and determined the structures with different heavy metals, analyzed and submitted the coordinates. EA and RHK did ITC experiments, analyzed the data and presented. SG planned, conceptualized, written the manuscript, raised the funds, also discussed and helped SK and SK in every step, where ever needed. All authors read and approved the final manuscript.
Supplementary Material
Electron density for heavy metals and Details of ITC experiments.
Contributor Information
Shivesh Kumar, Email: 2shivesh@gmail.com.
Ejaz Ahmad, Email: ejazaaa@gmail.com.
Sanjeev Kumar, Email: sanjeev.b23@gmail.com.
Rizwan Hasan Khan, Email: rizwanhkhan@hotmail.com.
Samudrala Gourinath, Email: samudralag@yahoo.com.
Acknowledgement
Authors thank DBT-IYBA and DST, Govt. of India for financial support. Authors would like to thank the staff members of Elettra, Trieste for beam time and assistance during data collection and Indo-Italian POC in science & tech, DST, India & MAE, Italy for financial support for data collection. SK thanks UGC for fellowship. We thank Dr. Jerry Brown, Brandeis University for proof reading and editing the manuscript.
References
- Cline HT, Witte S, Jones KW. Low lead levels stunt neuronal growth in a reversible manner. Proc Natl Acad Sci U S A. 1996;93:9915–9920. doi: 10.1073/pnas.93.18.9915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lidsky TI, Schneider JS. Lead neurotoxicity in children: basic mechanisms and clinical correlates. Brain. 2003;126:5–19. doi: 10.1093/brain/awg014. [DOI] [PubMed] [Google Scholar]
- Apostoli P, Bellini A, Porru S, Bisanti L. The effect of lead on male fertility: a time to pregnancy (TTP) study. Am J Ind Med. 2000;38:310–315. doi: 10.1002/1097-0274(200009)38:3<310::AID-AJIM10>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Hernandez-Ochoa I, Garcia-Vargas G, Lopez-Carrill L, Rubio-Andrade M, Moran-Martinez J, Cebrian ME, Quintanilla-Vega B. Low lead environmental exposure alters semen quality and sperm chromatin condensation in northern Mexico. Reprod Toxicol. 2005;20:221–228. doi: 10.1016/j.reprotox.2005.01.007. [DOI] [PubMed] [Google Scholar]
- Moore MR, Goldberg A, Yeung-Laiwah AA. Lead effects on the heme biosynthetic pathway. Relationship to toxicity. Ann N Y Acad Sci. 1987;514:191–203. doi: 10.1111/j.1749-6632.1987.tb48774.x. [DOI] [PubMed] [Google Scholar]
- Atchison WD. Effects of toxic environmental contaminants on voltage-gated calcium channel function: from past to present. J Bioenerg Biomembr. 2003;35:507–532. doi: 10.1023/b:jobb.0000008023.11211.13. [DOI] [PubMed] [Google Scholar]
- Chao SH, Bu CH, Cheung WY. Activation of troponin C by Cd2+ and Pb2+ Arch Toxicol. 1990;64:490–496. doi: 10.1007/BF01977632. [DOI] [PubMed] [Google Scholar]
- Aramini JM, Hiraoki T, Grace MR, Swaddle TW, Chiancone E, Vogel HJ. NMR and stopped-flow studies of metal ion binding to alpha-lactalbumins. Biochim Biophys Acta. 1996;1293:72–82. doi: 10.1016/0167-4838(95)00223-5. [DOI] [PubMed] [Google Scholar]
- Fullmer CS, Edelstein S, Wasserman RH. Lead-binding properties of intestinal calcium-binding proteins. J Biol Chem. 1985;260:6816–6819. [PubMed] [Google Scholar]
- Chao SH, Bu CH, Cheung WY. Stimulation of myosin light-chain kinase by Cd2+ and Pb2+ Arch Toxicol. 1995;69:197–203. doi: 10.1007/s002040050158. [DOI] [PubMed] [Google Scholar]
- Habermann E, Crowell K, Janicki P. Lead and other metals can substitute for Ca2+ in calmodulin. Arch Toxicol. 1983;54:61–70. doi: 10.1007/BF00277816. [DOI] [PubMed] [Google Scholar]
- Forsen S, Thulin E, Lilja H. 113Cd NMR in the study of calcium binding proteins: troponin C. FEBS Lett. 1979;104:123–126. doi: 10.1016/0014-5793(79)81097-2. [DOI] [PubMed] [Google Scholar]
- Pidcock E, Moore GR. Structural characteristics of protein binding sites for calcium and lanthanide ions. J Biol Inorg Chem. 2001;6:479–489. doi: 10.1007/s007750100214. [DOI] [PubMed] [Google Scholar]
- Wang CL, Leavis PC, Gergely J. Kinetic studies show that Ca2+ and Tb3+ have different binding preferences toward the four Ca2 + −binding sites of calmodulin. Biochemistry. 1984;23:6410–6415. doi: 10.1021/bi00321a020. [DOI] [PubMed] [Google Scholar]
- Yang W, Wilkins AL, Ye Y, Liu ZR, Li SY, Urbauer JL, Hellinga HW, Kearney A, van der Merwe PA, Yang JJ. Design of a calcium-binding protein with desired structure in a cell adhesion molecule. J Am Chem Soc. 2005;127:2085–2093. doi: 10.1021/ja0431307. [DOI] [PubMed] [Google Scholar]
- Vyas MN, Jacobson BL, Quiocho FA. The calcium-binding site in the galactose chemoreceptor protein. Crystallographic and metal-binding studies. J Biol Chem. 1989;264:20817–20821. [PubMed] [Google Scholar]
- Kursula P, Majava V. A structural insight into lead neurotoxicity and calmodulin activation by heavy metals. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2007;63:653–656. doi: 10.1107/S1744309107034525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson MA, Brunger AT. Domain flexibility in the 1.75 A resolution structure of Pb2+−calmodulin. Acta Crystallogr D: Biol Crystallogr. 2003;59:1782–1792. doi: 10.1107/S0907444903016846. [DOI] [PubMed] [Google Scholar]
- Kumar S, Padhan N, Alam N, Gourinath S. Crystal structure of calcium binding protein-1 from Entamoeba histolytica: a novel arrangment of EF hand motifs. Proteins. 2007;68:990–998. doi: 10.1002/prot.21455. [DOI] [PubMed] [Google Scholar]
- Kumar S, Ahmad E, Mansuri MS, Kumar S, Jain R, Khan RH, Gourinath S. Crystal structure and trimer-monomer transition of N-terminal domain of EhCaBP1 from Entamoeba histolytica. Biophys J. 2010;98:2933–2942. doi: 10.1016/j.bpj.2010.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopal B, Swaminathan CP, Bhattacharya S, Bhattacharya A, Murthy MR, Surolia A. Thermodynamics of metal ion binding and denaturation of a calcium binding protein from Entamoeba histolytica. Biochemistry. 1997;36:10910–10916. doi: 10.1021/bi9702546. [DOI] [PubMed] [Google Scholar]
- Velazquez-Campoy A, Todd MJ, Freire E. HIV-1 protease inhibitors: enthalpic versus entropic optimization of the binding affinity. Biochemistry. 2000;39:2201–2207. doi: 10.1021/bi992399d. [DOI] [PubMed] [Google Scholar]
- Strynadka NC, Cherney M, Sielecki AR, Li MX, Smillie LB, James MN. Structural details of a calcium-induced molecular switch: X-ray crystallographic analysis of the calcium-saturated N-terminal domain of troponin C at 1.75 A resolution. J Mol Biol. 1997;27:238–255. doi: 10.1006/jmbi.1997.1257. [DOI] [PubMed] [Google Scholar]
- Jain R, Kumar S, Gourinath S, Bhattacharya S, Bhattacharya A. N- and C-terminal domains of the calcium binding protein EhCaBP1 of the parasite Entamoeba histolytica display distinct functions. PLoS One. 2009;4:e5269. doi: 10.1371/journal.pone.0005269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartels KS, Klein C. The AUTOMAR Manual, v.1.4. Norderstedt, Germany: MAR Research GmbH; 2003. [Google Scholar]
- Yamazaki J, Urushidani T, Nagao T. Barium activates rat cerebellar nitric oxide synthase. Jpn J Pharmacol. 1996;70:351–354. doi: 10.1254/jjp.70.351. [DOI] [PubMed] [Google Scholar]
- Otwinowski O, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Meth Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Storoni LC, McCoy AJ, Read RJ. Likelihood-enhanced fast rotation functions. Acta Crystallogr D: Biol Crystallogr. 2004;60:432–438. doi: 10.1107/S0907444903028956. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D: Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D: Biol Crystallogr. 1998;54:905–921. doi: 10.1107/S0907444998003254. [DOI] [PubMed] [Google Scholar]
- Kleywegt GJ, Brunger AT. Checking your imagination: applications of the free R value. Structure. 1996;4:897–904. doi: 10.1016/S0969-2126(96)00097-4. [DOI] [PubMed] [Google Scholar]
- Zwart PH. Anomalous signal indicators in protein crystallography. Acta Crystallogr D: Biol Crystallogr. 2005;61:1437–1448. doi: 10.1107/S0907444905023589. [DOI] [PubMed] [Google Scholar]
- Gopal B, Krishna Rao JV, Thomas CJ, Bhattacharya A, Bhattacharya S, Murthy MR, Surolia A. Induction of a spectroscopically defined transition by guanidinium hydrochloride on a recombinant calcium binding protein from Entamoeba histolytica. FEBS Lett. 1998;441:71–76. doi: 10.1016/S0014-5793(98)01513-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Electron density for heavy metals and Details of ITC experiments.