Abstract
Stuttering is a common but poorly understood speech disorder. Consistent evidence for the involvement of genetic factors in stuttering has motivated studies aimed at identifying causative genetic variants that could shed light on the underlying molecular and cellular deficits in this disorder. Such studies have begun to identify causative genes. The purpose of this review is to summarize the gene discoveries to date, and to cover the subsequent functional studies that are beginning to provide insights into how these gene mutations might cause stuttering. Surprisingly, the first variant genes to be associated with stuttering are those encoding the lysosomal targeting system, GNPTAB, GNPTG, and NAGPA. Although mutations in NAGPA have not been associated with a disorder in humans, mutations in GNPTAB and GNPTG cause mucolipidosis types II and III, which are rare autosomal recessive lysosomal storage disorders, associated with pathology of bone, connective tissue, liver, spleen, and brain. Analysis of mutations in these genes has so far identified predominantly missense mutations in stuttering, in contrast to the truncating and other mutations that result in very low GNPTAB/G enzyme activity and are historically associated with mucolipidosis. Genetic evidence for the role of lysosomal targeting mutations in stuttering has now been buttressed by biochemical studies of the mutant enzymes found in this disorder. While data on the GlcNAc phosphotransferase encoded by GNPTAB/G remains limited and only suggestive, a study of the enzyme encoded by NAGPA has shown that the mutations found in stuttering reduce the overall cellular activity of this enzyme by about half, and that they result in deficits in intracellular processing and trafficking that lead to a reduced cellular half life. How these deficits result in the presumed speech-specific neuropathology associated with stuttering is not yet known. However these findings have opened several new lines of inquiry, including studies in mice carrying human stuttering mutations, that represent promising approaches to this disorder.
1. Genetic studies of stuttering
1.1 Genetic factors in stuttering
Stuttering is a disorder affecting the flow of speech, characterized by repetitions or prolongations of syllables and words, and by interruptions in the flow of speech known as blocks. Stuttering typically arises in young children ages three to five, where it affects up to five percent of children in this age group. The majority of affected children (75–80%) recover, either spontaneously or with the aid of speech therapy. Nevertheless, in some the disorder persists, leading to a frequency of stuttering in the general population of approximately one percent. Our studies are focused on persistent developmental stuttering, which occurs in populations and language groups worldwide, and despite being recognized and well described since ancient times, little is known about its causes [1]. However substantial evidence has accumulated that genetic factors play an important role in stuttering. Twin studies have repeatedly demonstrated a substantial genetic component to this disorder, with heritability estimates ranging as high as 0.8 [2,3,4,5,6,7]. Adoption studies have also been performed, and while the statistical power of these studies has been limited, no support has been obtained for a substantial role of environment in this disorder [8,9]. While stuttering displays clear familial aggregation [10,11], segregation analyses of the disorder in families have produced mixed results [12,13], and Mendelian transmission is frequently not observed.
Nevertheless, based on the strong overall evidence for genetic factors in stuttering, a number of genetic linkage studies have been undertaken. The results of these early linkage studies, which analyzed relatively small families from outbred populations, were disappointing. Few convincing linkage scores were obtained, and linkage loci often were not replicated across different studies [14,15,16]. This led to an alternative strategy for linkage studies, using large, highly consanguineous families with many affected members. Several such studies have employed consanguineous Pakistani kindreds and have produced strong evidence for linkage. A study of 44 such families identified clear linkage on chromosome 12q that reached genome-wide significance using multiple non-parametric analysis methods [17]. More recently, a study of a single Pakistani family identified linkage on chromosome 3q under a model of autosomal recessive transmission, and generated a maximum LOD score of 4.23 [18]. An additional recent study of a single Pakistani family generated a LOD score of 4.42 on chromosome 16q, again under an autosomal recessive model of inheritance [19]. In both of the latter studies, linkage scores were greatly reduced under other modes of inheritance. Beyond Pakistan, a linkage study has been performed in a large non-consanguineous family from Cameroon that contains many affected individuals. In this family, no single chromosomal locus generated significant linkage scores in the full pedigree. However analysis of different combinations of sub-pedigrees generated significant evidence for linkage on chromosomes 2p, 3p, 3q, and 14 under a recessive mode of inheritance [20]. This suggested that, like other studies in non-consanguineous families, no single highly penetrant gene variant was responsible for the many cases of stuttering observed in this family. Rather gene variants at multiple loci, perhaps brought together by assortative mating, seem to be causative in this family. Taken together, the recent linkage studies of stuttering indicate that highly significant linkage scores can be obtained, that evidence supporting autosomal recessive transmission is frequently observed, and that substantial locus heterogeneity exists for this disorder.
1.2 The identification of causative genes for stuttering
One linkage locus has led to the identification of a causative gene for stuttering. An analysis of the genomic region identified on chromosome 12 q [17] initially identified a mis-sense mutation in the GNPTAB gene in 28 affected members of one large Pakistani stuttering family. This mutation substituted a lysine residue for the highly conserved glutamic acid found at amino acid position 1200 in the protein product of this gene. The same mutation was also found in the available affected members of three other Pakistani stuttering families (a total of 3 individuals), as well as in nine unrelated stuttering individuals in the Pakistani and Indian populations [21]. A detailed analysis of the chromosomes containing this mutation revealed that it occurs in a conserved haplotype, indicating that this represents a founder mutation, with a likely single origin rather than arising by multiple mutational events at this position in the gene. Analysis of the existing variation in the chromosomal region surrounding this mutation showed that the conserved haplotype was quite short (approximately 7 kb), suggesting that this mutation is relatively old, having originated perhaps 14,000 years ago [22]. A number of other mutations in the GNPTAB gene were subsequently identified in unrelated individuals who stutter (123 Pakistani, 270 North American subjects) but not in normal controls (96 Pakistani, 276 North American subjects), and several of these mutations were observed in multiple unrelated individuals. Therefore there may be a limited spectrum of mutations in this gene, several of which are repeatedly observed, that are associated with stuttering.
1.3 Enzymology of the lysosomal targeting system
The GNPTAB gene encodes the α and β subunits of the enzyme GlcNAc-phosphotransferase (EC 2.7.8.17), which is involved in the generation of the mannose 6-phosphate (Man-6-P) targeting signal that directs a diverse group of acid hydrolases to the lysosome [23]. These lysosomal hydrolases comprise ~60 enzymes capable of metabolizing a great variety of biologic materials, breaking down these macromolecules to their component parts that can then be re-used. The acid hydrolases are synthesized in the endoplasmic reticulum (ER) where they acquire Asn-linked high mannose glycans and undergo folding, followed by transport to the Golgi complex where Man-6-P tags are attached. These tags allow binding to Man-6-P receptors in the late Golgi followed by transport to the lysosomes.
GlcNAc-phosphotransferase contains the product of another gene designated GNPTG, which encodes the γ subunit of the GNPTAB/G enzyme that enhances the generation of the Man-6-P recognition marker on a subset of the acid hydrolases [24]. Thus GNPTG became a biological candidate gene for stuttering. Sequencing of this gene in unrelated stuttering cases and controls identified a number of different mis-sense mutations associated with stuttering, none of which appeared in controls.
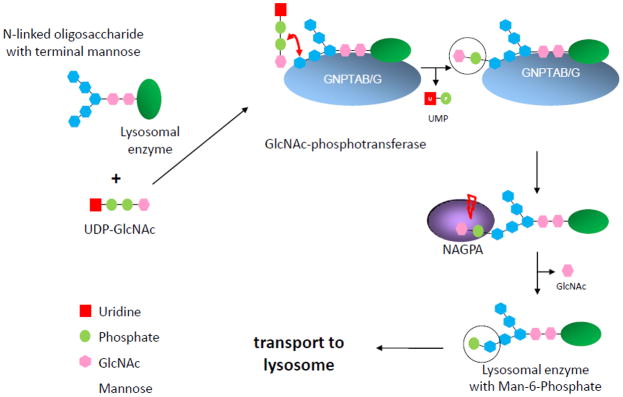
The Man-6-P signal is synthesized by a two-step process (Figure 1). First, GlcNAc-phosphotransferase covalently links GlcNAc-1-P to specific mannose residues of the Asn-linked high mannose glycans present on the acid hydrolases. In the second step, the GlcNAc is removed, leaving the exposed Man-6-P targeting signal. This second step is performed by the enzyme GlcNAc-1-phosphodiester-N-acetylglucosaminidase (NAGPA, EC 3.1.4.45, also known as the uncovering enzyme (UCE)). This suggested that the NAGPA gene might also be a source of mutations that are associated with stuttering. Evaluation of this gene identified a number of mutations in stuttering cases that were not found in controls. Overall, a mutation in GNPTAB, GNPTG or NAGPA was found in approximately 9% of unrelated individuals with familial stuttering [21]. This indicated that mutations that affect the lysosomal targeting pathway can account for a small but significant portion of stuttering.
Fig. 1.
The lysosomal enzyme-targeting pathway. In a first step, GNPTAB/G transfers GlcNAc-1-phosphate from UDP-GlcNAc onto the terminal mannose of N-linked oligosaccharides. In a second step, the GlcNAc residue is cleaved off by NAGPA, thereby uncovering the mannose-6-phosphate targeting signal.
These findings were surprising because mutations in the GNPTAB and GNPTG genes had been previously well known to be the underlying cause of the lysosomal storage disorders know as mucolipidoses types II and III. Mutations in GNPTAB are causative in mucolipidosis types II (MLII, also known as I-cell disease, MIM #252500) and IIIA (MLIIIA, also known as pseudo-Hurler polydystrophy, MIM #252600) [23,25], while mutations in the GNPTG are associated with mucolipidosis type IIIC (MLIIIC, variant pseudo-Hurler polydystrophy, MIM #252605) [26,27]. These three diseases are rare autosomal recessive disorders characterized primarily by abnormalities of bone, connective tissue, liver, heart, and brain. MLII is typically diagnosed at birth and has a severe course, leading to death within the first decade of life. MLIIIA and MLIIIC are less serious diseases with a widely varying course, but they are also ultimately fatal, often in young adulthood [28]. Mutations in NAGPA had not been previously associated with any human disorder. This has been puzzling, as mutations in this gene might be expected to produce mucolipidosis. We hypothesize that the most common manifestation of mutations in NAGPA is persistent developmental stuttering.
Our findings immediately raised the question of whether our stuttering subjects with mutations in these three genes displayed symptoms of MLII or MLIII. A detailed evaluation of a limited number of such subjects (two subjects heterozygous for the p.Ala455Ser mutation in GNPTAB, one homozygous for the p.Arg328Cys mutation in NAGPA, and one heterozygous for the p.Phe513SerfsX113 mutation in NAGPA) at the National Institutes of Health Clinical Center failed to reveal any such symptoms, thus the mutations we identified appeared to be associated with persistent developmental stuttering in the absence of any other medical symptoms. This led to the question of why these mutations, some of which occurred in homozygous form in some stuttering individuals, failed to generate frank MLII or MLIII. Although we currently do not have an answer to this question, we have some suggestive observations. First, mutations associated with MLII are frequently truncating mutations that eliminate or drastically reduce the activity of the GNPTAB/G enzyme [23,25,26,29], while with one exception (discussed further below), the mutations associated with stuttering are mis-sense mutations [21]. It is possible that mutations that cause mucolipidosis cause a more severe deficit in enzyme function than those associated with stuttering. Second, MLII and MLIII are highly penetrant autosomal recessive disorders, associated with homozygosity for mutations in GNPTAB or GNPTG. Most of the individuals in our stuttering sample are heterozygous, carrying a single mutant copy. This observation has led to the question of why parents of children with MLII or MLIII, who are obligate carriers of mutations in GNPTAB or GNPTG do not appear to stutter or display other speech pathologies [30]. Again, the answer to this question is unknown. However, it is possible that truncating mutations associated with mucolipidosis may not be transcribed or if they are, the transcripts are subject to nonsense mediated decay [31]. This would effectively eliminate transcripts from the mutant allele, and leave carriers with transcripts only from the normal allele, although perhaps at reduced levels. Genes containing mis-sense mutations, on the other hand, are likely to be transcribed and translated, and the resulting protein incorporated into the GlcNAC-phosphotransferase enzyme, which exists as an 2α, 2β, 2γ heterohexamer. This could result in the production of normal amounts of an enzyme composed of a mixture of wild type and mis-sense mutant subunits that displays reduced activity.
We mentioned above that all mutations associated with stuttering to date have been mis-sense mutations with one exception. That exception is a 16 bp deletion of NAGPA, resulting in p.Phe513SerfsX113 in the encoded protein. In this mutant protein, the normal phenylalanine at amino acid position 513 is replaced by serine, and the last 2 amino acids and stop codon are missing, leaving an open reading frame that results in the addition of 113 nonsense amino acids until the occurrence of a stop codon. It is of interest that this mutation was found in a 67 year old woman of European descent, who was one of the individuals who underwent a detailed medical examination at the NIH Clinical Center. This individual displayed exceptionally severe lifelong stuttering and no evidence of mucolipidosis.
2. Biochemical studies of mutant enzymes found in stuttering
While the occurrence of mutations in the GNPTAB, GNPTG, and NAGPA genes made a strong case for the role of these mutations in stuttering, functional evidence for a resulting deficit in the encoded enzymes was initially lacking. Fortunately, because these enzymes had been identified and well studied in the context of the human disease mucolipidosis, a number of important tools for the study of these enzymes, including in vitro and cell-based assays, have been previously developed [32,33]. Large reductions in the activity of GlcNAc-phosphotransferase are often observed in individuals with this disease. When synthetic α-methyl mannoside was used for the enzyme substrate, fibroblasts from ML II individuals typically display 0–1% of normal activity, and those from ML IIIA individuals display 0.8–14% of normal activity, although normal activity was shown in ML IIIC patients [26]. In an assay using the lysosomal enzyme uteroferrin as a substrate, ML IIIC patients showed less than 10 % of normal activity [27]. Because no human disorder had previously been associated with mutations in NAGPA, data on clinical-pathological correlations for such mutations were unavailable. However, a significant amount was known about the structure, synthesis, and maturation of this enzyme [34,35], and assays for activity of this enzyme had been developed.
Skin biopsies from two affected individuals, one carrying the p.Arg328Cys mutation and another carrying the p.F513SfsX113 mutation in NAGPA were obtained, and fibroblast cell lines were subsequently cultured from these individuals. Biochemical assays were performed to measure the uncovering enzyme activity in these mutant cell lines using synthetic [3H]GlcNAc-α-Me-P-Mannoside as a substrate. The fibroblast cell line harboring the p.Arg328Cys mutation in UCE showed 54% of wild-type activity (p < 0.01), while the fibroblasts carrying the p.F513SfsX113 mutation showed 40% of wild-type activity (p < 0.001). In addition to measurements on synthetic substrates, the activities of these mutant enzymes on oligosaccharides that reside on lysosomal enzymes were also measured. When measured as the ability to generate Man-6-P monoester from diester, the activities of mutant UCE’s were reduced; 37% (p = 0.01) in the both p.Arg328Cys and p.F513SfsX113 mutants compared to 51% in the wild-type [36]. The partial loss of uncovering activities in these two mutant NAPGA products may explain why these mutation carriers did not show any of the phenotypes associated with mucolipidosis.
Cell-based studies were used to better understand the intracellular effects conferred by mutations found in stuttering. It is known that newly synthesized UCE is initially localized in the ER where it acquires high mannose type glycans. It then moves to the trans-Golgi network where further glycosylation takes place to generate complex type N-linked oligosaccharides on this protein. Analysis of HeLa cells transfected with cDNAs encoding wt and the three UCE mutants revealed that the mutants were retained in the ER and subjected to ER-associated degradation, resulting in a reduction in their half-lives (t1/2). Thus the t1/2 values of the p.Arg328Cys, p.His84Gln and p.F513SfsX113 proteins were 7hr, 6hr and < 30min respectively, compared to a t1/2 of 14hr for the wild-type UCE. Additional analysis of the UCE carrying the p.F513SfsX113 mutation showed that the 113 amino acid nonsense sequence generated by this frameshift contains a VWLL sequence that causes rapid degradation via the proteasomal system [36].
The conclusion from these studies is that mutations in the NAGPA gene found in stuttering individuals disrupt proper folding of their protein products and is followed by rapid degradation in the ER, which results in decreases in the uncovering activities present in these cells. Thus these biochemical studies provided additional support for role of mutant UCE in stuttering. However, it is still unclear why reduced uncovering activities lead to stuttering in the absence of other pathology.
For the analysis of mutations found in the GNPTAB gene, studies to date have been limited to a single fibroblast line from a stuttering individual, who carries the p.Ala455Ser mutation. Analysis of the activity of this mutant GNPTAB in the fibroblasts showed a decrease in the synthesis of the Man-6-P signal of about 50%. Although this limited data is only suggestive, it is in general agreement with the hypothesis that modest deficits in enzymes in the lysosomal targeting pathway, substantially less severe than the deficits observed in mucolipidosis patients, are associated with non-syndromic persistent developmental stuttering. However further detailed studies of the folding, trafficking, and enzymatic activity of mutant GNPTAB proteins are needed to make conclusions about the cellular pathology associated with stuttering.
Individuals with mucolipidosis present with deficits in speech production, particularly deficits in expressive speech including complete absence of speech in ML II and stuttering in ML III [25,29,37]. Since ML II individuals typically display developmental delay and mental retardation, their speech deficits have often been thought of as secondary to more general neurological impairments. However there are anecdotal reports of ML II children who display age-normal behavior in some aspects [38]. This gives rise to the hypothesis that the speech deficits in such children are primary results of lesions in GNPTAB/G, and that perhaps such children are viewed as more developmentally delayed than they are because they generally do not gain the ability to speak.
3. Pathophysiology
3.1 Clinical effects of mutations associated with stuttering
While at least some of the mutations associated with stuttering result in enzymes with reduced activity, it is not yet clear how this reduction in enzyme activity leads to stuttering. In general, neurologic deficits are seen in many lysosomal storage disorders. However other very specific pathologies are often observed, where some organs, or cell types within organs, are severely compromised and others largely spared, despite the fact that lysosomal enzymes are expressed widely throughout the body [28]. We have hypothesized that in stuttering, the neurological deficits conferred by these mutations are manifest in a set of neurons that are uniquely dedicated to speech production, and that these neurons are particularly sensitive to relatively modest impairments in enzyme activity. Our suggestion that these neurons are uniquely dedicated to speech is based on the fact that clinical examination of stuttering individuals carrying these mutations has failed to identify any other neurological abnormalities. However, it should be noted that the clinical evaluations performed to date have looked at standard measures in clinical neurology, and not utilized more subtle or specialized laboratory-based tests of neurologic function, such as motor learning tests. Thus the conclusion that our subjects are completely neurologically normal other than their stuttering needs further confirmation.
3.2 Elucidation of neuropathology
The results demonstrating deficits in the lysosomal targeting pathway in stuttering have provided a surprising new insight into the underlying molecular causes of this disorder. How these molecular deficits give rise to the disorder at the cellular and organ-system level remains unknown, however, and the elucidation of these pathophysiological mechanisms is of great interest. It is difficult to perform these studies in human subjects due to the several factors, including the fact that the pathology of interest likely resides within the brain, which presents many obstacles to study at the molecular level in living humans. Alternatively, the mouse provides a powerful mammalian model for human disease studies. Mouse models have been constructed that carry targeted inactivation of each of the three genes encoding the lysosomal targeting pathway, GNPTAB, GNPTG, and NAGPA [39,40,41]. While a number of the features similar to human mucolipidosis are present in GNPTAB and GNPTG knock out lines, inactivation of the NAGPA gene initially leads to no obvious phenotypic abnormalities, although elevated plasma levels of a number of lysosomal hydrolases suggest the presence of a physiological deficit in this line [41]. Subsequent observations have suggested that older NAGPA knock-out animals may develop neurologic abnormalities [42], which could be consistent with neurologic deficits in humans produced by the UCE mutations observed in stuttering. Because the mutations we’ve indentified in stuttering subjects do not result in detectable symptoms of mucolipidosis, creation of an appropriate mouse model will require generation of knock-in lines that carry human stuttering mutations, in addition to the existing knock-out lines. Such knock-in lines may reveal specific neuropathology that could be very helpful for understanding the human neuropathology that accompanies these cases of stuttering, regardless of any effect of such targeted mutations on mouse vocalization.
The overall suitability of the mouse as a model for human speech disorders is less certain. Mice display rich vocalizations, many of which are ultrasonic and remain incompletely understood [43,44,45]. However, mouse models of human verbal dyspraxia created by alterations in the FOXP2 gene have demonstrated altered vocalization [46,47,48], which suggests that a mouse model of stuttering may be possible.
4. Genetic insights into a broader fraction of stuttering
Mutations in the GNPTAB, GNPTG, and NAGPA genes have been found in less than 10% of unrelated stutterers who have a family history of the disorder [21]. However, the recent linkage studies discussed in Section 1.1 above have given rise to optimism that additional genes, which could explain a larger fraction of stuttering, can be identified [18,19]. At the present time the causative genes that reside at these loci remain unidentified. It is not possible to know if mutations in these genes give rise to cellular metabolic deficits that could provide further insights into the pathophysiology of persistent stuttering, or what fraction of familial stuttering can be accounted for by mutations in such genes. Nevertheless it is clear that genetic approaches to this disorder can be fruitful, and are likely to continue to be so.
Highlights.
-
1
Using consanguineous families, linkage studies are rapidly identifying new loci for stuttering
-
2
Studies to date have found mutations in GNPTAB, GNPTG, and NAGPA associated with stuttering.
-
3
These genes encode the lysosomal enzyme targeting pathway, defective in mucolipidosis II and III.
-
4
Stuttering individuals carrying mutations in these genes showed no symptoms of mucolipidosis.
-
6
Biochemical studies of these mutants show deficits in intracellular trafficking and reduced half life.
-
7
Vocalization in knock-out and knock-in mouse lines with these mutations is being evaluated.
Acknowledgments
We thank Stuart Kornfeld, Thomas Friedman, and Allen Braun for valuable comments on the manuscript. This work was supported by the NIH/NIDCD intramural grant Z01-000046-12 and by the Stuttering Foundation of America.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bloodstein O, Ratner N. A handbook of stuttering. 6. Thomson Delmar Learning; Clifton Park, NY: 2008. [Google Scholar]
- 2.Andrews G, Morris-Yates A, Howie P, Martin N. Genetic factors in stuttering confirmed. Arch Gen Psychiatry. 1991;48:1034–1035. doi: 10.1001/archpsyc.1991.01810350074012. [DOI] [PubMed] [Google Scholar]
- 3.Howie P. Concordance for stuttering in monozygotic and dizygotic twin pairs. J Speech Hear Res. 1981;24:317–321. doi: 10.1044/jshr.2403.317. [DOI] [PubMed] [Google Scholar]
- 4.Ooki Genetic and environmental influences on stuttering and tics in Japanese twin children. Twin Res Hum Genet. 2005;8:69–75. doi: 10.1375/1832427053435409. [DOI] [PubMed] [Google Scholar]
- 5.Felsenfeld S, Kirk K, Zhu G, Statham D, Neale M, Martin N. A study of the genetic and environmental etiology of stuttering in a selected twin sample. Behav Genet. 2000;30:359–66. doi: 10.1023/a:1002765620208. [DOI] [PubMed] [Google Scholar]
- 6.Dworzynski K, Remington A, Rijsdijk P, Howell F, Plomin R. Genetic etiology in cases of recovered and persistent stuttering in an unselected, longitudinal sample of young twins. Am J Speech Lang Pathol. 2007;16:169–78. doi: 10.1044/1058-0360(2007/021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fagnani C, Fibiger S, Skythe A, Hjelmborg J. Heritability and environmental effects for self-reported periods with stuttering: a twin study from Denmark. Logoped Phoniatr Vocol. 2011;36:114–20. doi: 10.3109/14015439.2010.534503. [DOI] [PubMed] [Google Scholar]
- 8.Bloodstein O. Stuttering in families of adopted stutterers. J Speech Hear Disord. 1961;26:395–396. doi: 10.1044/jshd.2604.395. [DOI] [PubMed] [Google Scholar]
- 9.Felsenfeld S, Plomin R. Epidemiological and offspring analyses of developmental speech disorders using data from the Colorado Adoption Project. J Speech Lang Hear Res. 1997;40:778–791. doi: 10.1044/jslhr.4004.778. [DOI] [PubMed] [Google Scholar]
- 10.Seider R, Gladstein K, Kidd K. Recovery and persistence of stuttering among relatives of stutterers. J Speech Lang Hear Res. 1983;31:62–71. doi: 10.1044/jshd.4804.402. [DOI] [PubMed] [Google Scholar]
- 11.Kidd K, Kidd J, Records M. The possible causes of the sex ratio in stuttering and its implications. J Fluency Disord. 1978;3:13–23. [Google Scholar]
- 12.Cox N, Kramer P, Kidd K. Segregation analysis of stuttering. Genetic Epidemiology. 1984;1:245–253. doi: 10.1002/gepi.1370010304. [DOI] [PubMed] [Google Scholar]
- 13.Viswantath N, Lee R, Chakraborty H. Evidence for a major gene influence on persistent developmental stuttering. Human Biology. 2004;76:401–412. doi: 10.1353/hub.2004.0050. [DOI] [PubMed] [Google Scholar]
- 14.Shugart Y, Mundorff J, Kilshaw J, Doheny K, Doan B, Wanyee J, Green E, Drayna D. Results of a genome-side linkage scan for stuttering. Am J Med Genet A. 2004;124A:133–5. doi: 10.1002/ajmg.a.20347. [DOI] [PubMed] [Google Scholar]
- 15.Wittke-Thompson J, Ambrose N, Yairi E, et al. Genetic studies of stuttering in a founder population. J Fluency Disord. 2007;32:33–50. doi: 10.1016/j.jfludis.2006.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Suresh R, Ambrose N, Roe C, et al. New complexities in the genetics of stuttering: significant sex-specific linkage signals. Am J Hum Genet. 2006;78:554–63. doi: 10.1086/501370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Riaz N, Steinberg S, Ahmad J, Pluzhnikov A, Riazuddin S, Cox N, Drayna D. Genomewide Significant Linkage to Stuttering on Chromosome 12. Am J Hum Genet. 2005;76:647–651. doi: 10.1086/429226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Raza M, Riazuddin S, Drayna D. Identification of an autosomal recessive stuttering locus on chromosome 3q13.2–3q13.33. Hum Genet. 2010;128:461–463. doi: 10.1007/s00439-010-0871-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Raza M, Amjad R, Riazuddin S, Drayna D. Studies in a consanguineous family reveal a novel locus for stuttering on chromosome 16q. Hum Genet. 2012:311–313. doi: 10.1007/s00439-011-1134-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Raza M, Gertz E, Mundorff J, Lukong J, Kuster J, Schäffer A, Drayna D. Linkage analysis of a large African family segregating stuttering suggests polygenic inheritance and assortative mating. doi: 10.1007/s00439-012-1252-5. Manuscript submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kang C, Riazuddin S, Mundorff J, Krasnewich D, Friedman P, Mullikin J, Drayna D. Mutations in the lysosomal enzyme-targeting pathway and persistent stuttering. New Engl J Med. 2010;362:677–685. doi: 10.1056/NEJMoa0902630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fedyna A, Drayna D, Kang C. Characterization of a mutation commonly associated with persistent stuttering: evidence for a founder mutation. J Hum Genet. 2011;56:80–82. doi: 10.1038/jhg.2010.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tiede S, Storch S, Lübke T, Henrissat B, Bargal R, Raas-Rothschild A, Braulke T. Mucolipidosis II is caused by mutations in GNPTA encoding the alpha/beta GlcNAc-1-phosphotransferase. Nat Med. 2005;11:1109–1112. doi: 10.1038/nm1305. [DOI] [PubMed] [Google Scholar]
- 24.Qian Y, Lee I, Lee W, Qian M, Kudo M, Canfield W, Lobel P, Kornfeld S. Functions of the alpha, beta, and gamma subunits of UDP-GlcNAc:lysosomal enzyme N-acetylglucosamine-1-phosphotransferase. J Biol Chem. 2010;285:3360–3370. doi: 10.1074/jbc.M109.068650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Otomo T, Muramatsu T, Yorifuji T, Okuyama T, Nakabayashi H, Fukao T, et al. Mucolipidosis II and III alpha/beta: mutation analysis of 40 Japanese patients showed genotype phenotype correlation. J Hum Genet. 2009;54:145–51. doi: 10.1038/jhg.2009.3. [DOI] [PubMed] [Google Scholar]
- 26.Kudo M, Brem M, Canfield W. Mucolipidosis II (I-cell disease) and mucolipidosis IIIA (classical pseudo-Hurler polydystrophy) are caused by mutations in the GlcNAc-phosphotransferase alpha/beta - subunits precursor gene. Am J Hum Genet. 2006;78:451–63. doi: 10.1086/500849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raas-Rothschild A, Cormier-Daire V, Bao M, Genin E, Salomon R, Brewer K, et al. Molecular basis of variant pseudo-Hurler polydystrophy (mucolipidosis IIIC) J Clin Invest. 2000;105:673–81. doi: 10.1172/JCI5826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kornfeld S, Sly W. The metabolic and molecular bases of inherited disease. 8. Vol. 3. New York: McGraw-Hill; 2010. I-cell disease and pseudo-Hurler polydystrophy: disorders of lysosomal enzyme phosphorylation and localization; pp. 3469–82. [Google Scholar]
- 29.Tappino B, Chuzhanova N, Regis S, Dardis A, Corsolini F, Tonoli E, Beccari T, Rosano C, Mucha J, Blanco M, Szlago M, Di Rocco M, Cooper D, Filocamo M. Molecular characterization of 22 novel UDP-N-acetylglucosamine-1-phosphate transferase alpha- and beta-subunit (GNPTAB) gene mutations causing mucolipidosis types IIalpha/beta and IIIalpha/beta in 46 patients. Hum Mutat. 2009;30:E956–73. doi: 10.1002/humu.21099. [DOI] [PubMed] [Google Scholar]
- 30.Leroy J. Personnal communication. 2011.
- 31.Chang Y, Imam J, Wilkinson M. The nonsense-mediated decay RNA surveillance pathway. Annu Rev Biochem. 2007;76:51–74. doi: 10.1146/annurev.biochem.76.050106.093909. [DOI] [PubMed] [Google Scholar]
- 32.Reitman M, Kornfeld S. Lysosomal enzyme targeting. NAcetylglucosaminylphosphotransferase selectively phosphorylates native lysosomal enzymes. J Biol Chem. 1981;256:11977–11980. [PubMed] [Google Scholar]
- 33.Varki A, Kornfeld S. Purification and characterization of rat liver alpha-N-acetylglucosaminyl phosphodiesterase. J Biol Chem. 1981;256:9937–9943. [PubMed] [Google Scholar]
- 34.Wei Y, Yen T, Cai J, Trent J, Pierce W, Young W. Structural features of the lysosomal hydrolase mannose 6-phosphate uncovering enzyme. Glycoconj J. 2005;(1–2):13–19. doi: 10.1007/s10719-005-0846-8. [DOI] [PubMed] [Google Scholar]
- 35.Do H, Lee W, Ghosh P, Hollowell T, Canfield W, Kornfeld S. Human mannose 6-phosphate-uncovering enzyme is synthesized as a proenzyme that is activated by the endoprotease furin. J Biol Chem. 2002;277:29737–29744. doi: 10.1074/jbc.M202369200. [DOI] [PubMed] [Google Scholar]
- 36.Lee W, Kang C, Drayna D, Kornfeld S. Analysis of mannose 6-phosphate uncovering enzymes associated with persistent stuttering. J Biol Chem. 2011;286:39786–39793. doi: 10.1074/jbc.M111.295899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smuts I, Potgieter D, Hendrikus van der Westhuizenc F. Combined Tarsal and Carpal Tunnel Syndrome in Mucolipidosis Type III A Case Study and Review. Ann N Y Acad Sci. 2009;1151:77–84. doi: 10.1111/j.1749-6632.2008.03451.x. [DOI] [PubMed] [Google Scholar]
- 38.Leroy J, Cathey S, Friez M. Mucolipidosis II. In: Pagon R, Bird T, Dolan C, Stephens K, editors. GeneReviews. Seattle: Univ. Wash; 2010. http://www.ncbi.nlm.nih.gov/books/NBK1828/ [Google Scholar]
- 39.Gelfman C, Vogel P, Issa T, Turner C, Lee W, Kornfeld S, Rice D. Mice lacking alpha/beta subunits of GlcNAc-1-phosphotransferase exhibit growth retardation, retinal degeneration, and secretory cell lesions. Invest Ophthalmol Vis Sci. 2007;48:5221–8. doi: 10.1167/iovs.07-0452. [DOI] [PubMed] [Google Scholar]
- 40.Vogel P, Payne B, Read R, Lee W, Gelfman C, Kornfeld S. Comparative pathology of murine mucolipidosis types II and IIIC. Vet Pathol. 2009;46:313–324. doi: 10.1354/vp.46-2-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boonen M, Vogel P, Platt K, Dahms N, Kornfeld S. Mice lacking mannose 6-phosphate uncovering enzyme activity have a milder phenotype than mice deficient for N-acetylglucosamine-1-phosphotransferase activity. Molec Biol Cell. 2009;20:4381–4389. doi: 10.1091/mbc.E09-05-0398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kornfeld S. Personnal communication. 2012.
- 43.Holy T, Guo Z. Ultrasonic Songs of Male Mice. PLoS Biology. 2005;3:2177–2186. doi: 10.1371/journal.pbio.0030386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Scattoni M, Crawley J, Ricceri L. Ultrasonic vocalizations: A tool for behavioural phenotyping of mouse models of neurodevelopmental disorders. Neurosci Biobehav Rev. 2009;33:508–515. doi: 10.1016/j.neubiorev.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grimsley J, Monaghan J, Wenstrup J. Development of social vocalizations in mice. PLoS one. 2011;6:1–15. doi: 10.1371/journal.pone.0017460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fujita E, Tanabe Y, Shiota A, Ueda M, Suwa K, Momoi M, Momoi T. Ultrasonic vocalization impairment of Foxp2 (R552) knockin mice related to speech-language disorder and abnormality of Purkinje cells. Proc Natl Acad Sci USA. 2008;105:3117–3122. doi: 10.1073/pnas.0712298105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shu W, Cho J, Jiang Y, Zhang M, Weisz D, Elder G, Schmeidler J, De Gasperi R, Sosa M, Rabidou D, Santucci A, Perl D, Morrisey E, Buxbaum J. Altered ultrasonic vocalization in mice with a disruption in the Foxp2 gene. Proc Natl Acad Sci USA. 2005;102:9643–9648. doi: 10.1073/pnas.0503739102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Enard W, Gehre S, Hammerschmidt K, Holter S, Blass T, Somel M, Bruckner M, Schreiweis C, Winter C, Sohr R, Becker L, Wiebe V, Nickel B, Giger T, Muller U, Groszer M, Adler T, Aguilar A, Bolle I, Calzada-Wack J, Dalke C, Ehrhardt N, Favor J, Fuchs H, Gailus-Durner V, Hans W, Holzlwimmer G, Javaheri A, Kalaydjiev S, Busch D, Graw J, Ivandic B, Klingerspor M, Klopstock T, Ollert M, Quintanilla-Martinez L, Schulz H, Wold E, Wurst W, Zimmer A, Fisher S, Morgenstren R, Arendt T, de Angelis M, Fischer J, Schwarz S, Paabo S. A humanized version of Foxp2 affects cortico-basal ganglia circuits in mice. Cell. 2009;137:961–971. doi: 10.1016/j.cell.2009.03.041. [DOI] [PubMed] [Google Scholar]