Abstract
Accurate determination of micrograph focuses is essential for averaging multiple images to reach high-resolution 3-D reconstructions in electron cryo-microscopy (cryo-EM). Current methods use iterative fitting of focus-dependent simulated power spectra to the power spectra of experimental images, with the fitting performed independently for different images. Here we have developed a novel graph theory based method in which the rotational average focus and individual angular sector focuses of all images are determined simultaneously in closed form using the least square solution of overdetermined linear equations. The new method was shown to be fast, accurate, and robust in tests with large datasets of experimental low dose cryo-EM images. Its integration with three classic power spectra fitting methods also allows cross validation of the results by these vastly different methods. The new integrated focus determination method will improve reliability of automated focus determination for large-scale data processing that is increasingly common in the cryo-EM field.
Keywords: cryo-EM, contrast transfer function, defocus, linear least square, graph theory
Introduction
Due to inherently low contrast in electron cryo-microscopy (cryo-EM) images of biological samples, it is common to increase contrast by imaging the samples with small under-focuses, typically a fraction of a micrometer to a few micrometers (Saad et al., 2001). However, the focus-dependent contrast transfer function (CTF) of the TEM instrument differentially modulates the images at different spatial frequencies (Erikson and Klug, 1970; Thon, 1971). It is essential to obtain accurate focus values of all images for subsequent correction of CTF modulations to coherently merge multiple images and to obtain high-resolution 3-D reconstructions.
Based on weak phase approximation (Erikson and Klug, 1970; Thon, 1971), image formation in a TEM instrument can be modeled in Fourier space as shown in equation 1:
| (1) |
in which s⃗ represents the spatial frequency, Fi (s⃗) represents the Fourier transform of the image, Fs (s⃗) represents the structural factor of the sample, Ctf (s⃗, f) represents the instrument contrast transfer function which is dependent of the focus (f ), and N(s⃗) represents the noise. Based on this image formation model, the corresponding power spectrum is:
| (2) |
in which s⃗ represents the spatial frequency, Ii (s⃗) represents the power spectra of the image, Is (s⃗) represents the structural factor of the sample.
Due to uncertainties in setting focuses or even with intentionally varying focuses, the exact focus values of the micrographs must be determined computationally after imaging. Current methods determine the focus value by iterative fitting of the simulated power spectra to the power spectra of experimental images by varying focus values to optimize the matching of the simulated and experimental power spectra (Equation 2). The fitting can be interactively performed using graphical user interface, for example, the EMAN ctfit program (Ludtke et al., 1999), or automated fitting methods (Huang et al., 2003; Mallick et al., 2005; Mindell and Grigorieff, 2003; Sander et al., 2003; Sorzano et al., 2007; Velazquez-Muriel et al., 2003; Yang et al., 2009). As can be seen in equation 2, the modulation of a micrograph depends only on its own CTF and in principal the focus values of different micrographs can be independently determined. Current fitting methods employ this independence to fit the focus value of different micrographs separately.
In recent years, graph theory has been increasingly used to represent data and relationships within the data as networks. Valuable information can be extracted from the data networks, for example, atomic coordinates from NMR measurements (Huang et al., 2006), protein functions from protein-protein interaction networks (Koyutürk et al., 2011), and accurate Web search results from Google PageRank link analysis (Brin and Page, 1998). Here we have developed a novel focus determination method (s2focus) by employing the radial scales of oscillations in power spectra of micrographs (i.e. Thon rings) at different focuses and the simple relationships between the scales and focuses. This new method determines the focus values of all micrographs simultaneously in closed-form by solving overdetermined linear equations constructed from the scale relationship graph. It is drastically different from all current methods that treat each micrograph and its focus as a separate optimization (i.e. search) problem.
Methods
Relationship between focus and scale of Thon rings in power spectra
The detailed Ctf (s, f) function based on weak phase approximation (Erikson and Klug, 1970; Thon, 1971) is shown in equation 3 and 4:
| (3) |
| (4) |
in which Q represents the amplitude contrast (Q ≪ 1), φ0 represents the phase term corresponding to amplitude contrast, λ represents the electron wavelength, Cs represents the spherical aberration coefficient of objective lens. It is obvious that the CTF function is a sine function that oscillates with its period determined by focus, spherical aberration, and wavelength (Jiang and Chiu, 2001). Since it is a sine function of s2 and s4, the oscillation becomes more frequent at larger s (i.e. at high resolutions). At sufficiently high resolution, the oscillation can be so frequent that a small change of focus can dramatically shift of the position of the CTF function. As a result, the oscillations are complicated position-dependent functions of the focus values. Current fitting methods aim to match power spectra computed using equations 2–4 to power spectra of experimental images by iteratively searching focus values.
By replacing s with s′ = s2 in equation 4, we can transform the formula to
| (5) |
in which the sine function includes both s′ and s′2 terms. Since the s range we are interested is small (0 < s < 0.25 for 4 Angstrom resolution target) for CTF fitting, the s′2 term can be effectively ignored as higher order perturbations. The CTF function can thus be simplified as
| (6) |
in which CTF function becomes simply a sine wave of s′ with its oscillation frequency linearly proportional to focus values. We will call this transformed power spectra s2 power spectra while the regular power spectra s1 power spectra. Two images with different focuses will have their s2 power spectra as sine waves of different frequency (i.e. Thon rings of different spacing). One sine wave can be scaled along the s2 axis to match another sine wave. The relative scale between the s2 power spectra of two images is simply the ratio of their focus values:
| (7) |
in which fi and fj represents the focus value of image i and j respectively, and si, j represents the amount of scaling to match s2 power spectra of image i to that of image j.
Computation of s2 power spectra
The s2 power spectra are computed through the following steps. The standard 2-D s1 power spectra of the micrograph are first computed using the same approach as in existing CTF fitting methods by incoherently averaging the power spectra of individual particles in a micrograph (Supplementary Fig. 1a and 2a) (Saad et al., 2001; Yang et al., 2009). Since the power spectra include contributions not only from CTF but also from sample structural factors and background noises (equation 2), values in power spectra usually span large ranges with low-resolution values orders larger than high-resolution values (Supplementary Fig. 2a). To minimize the overall slope from low to high resolutions and to make the CTF oscillations more dominant, several filtering steps were applied. The 2-D s1 power spectra are first log transformed by replacing the amplitudes (I) with their log function (lnI) (Supplementary Fig. 1b and 2b). The log-transformed s1 power spectra are then high pass filtered (Gaussian filter with sigma set to 1/10 of Nyquist frequency) (Supplementary Fig. 1c and 2c). The resulted s1 power spectra are now essentially free of overall slope. The s1 power spectra are then further filtered using a Gaussian low pass filter (sigma set to 2/5 of Nyquist frequency) to remove some noises (Supplementary Fig. 1d and 2d). The Thon ring oscillation amplitudes gradually decrease toward high-resolution regions due to instrument damping envelope functions. To concentrate on the resolution regions with oscillations, the filtered s1 power spectra are then truncated at user specified resolution (6 Å in Fig. 1a, 2a, Supplementary Fig. 1e and 2e). Finally the truncated s1 power spectra are transformed by rescaling nonlinearly along the radial direction so that the radius is proportional to s2 instead of s to obtain the s2 power spectra (Fig. 1b and Supplementary Fig. 1f). The nonlinear rescaling effectively compresses small radii regions to make the Thon rings oscillate more frequently but dilates larger radii regions to make the Thon rings oscillate less frequently. This nonlinear rescaling results in uniform oscillation rate for Thon rings from small to large radii in the s2 power spectra. The entire 2-D s2 power spectra are then rotationally averaged to obtain 1-D s2 power spectra (Fig. 1b and Supplementary Fig. 2f). When astigmatism is considered, the 2-D s2 power spectra (Fig. 6b) can be divided into multiple angular sectors and then a 1-D s2 power spectra can be obtained for each of the sectors by limiting the rotational averaging within each sector (Fig. 6c).
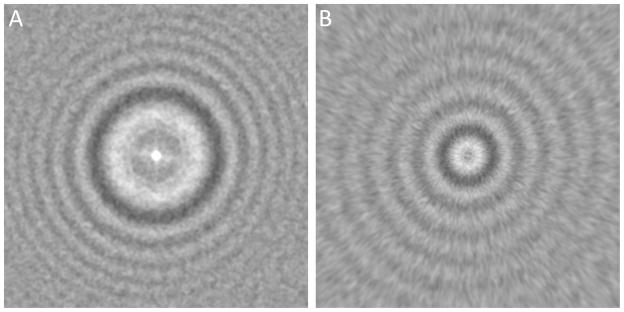
Figure 1. Comparison of 2-D regular s1 power spectra and s2 power spectra.
A. regular s1 power spectra of a micrograph of bacteriophage T7 MLDII with 93 particles and at 1.06 μm under-focus; B. s2 power spectra generated from A. Both power spectra were truncated to 6 Å resolution at the edges.
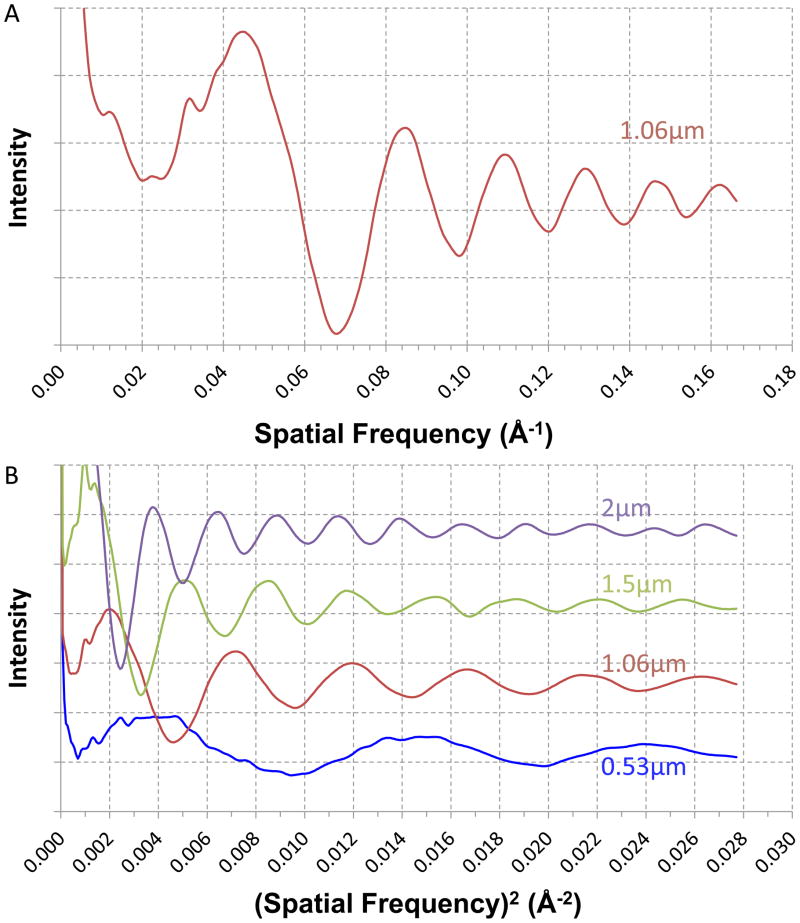
Figure 2. Comparison of 1-D regular s1 power spectra and s2 power spectra.
A. rotational average of s1 power spectra shown in Figure 1A; B. 1-D s2 power spectra of micrographs at different focuses. Shown are spectra from bacteriophage T7 MLDII images at 0.53, 1.06, 1.5, 2 μm under-focus respectively. The 1.06 μm under-focus curve is rotational average of s2 power spectra shown in Figure 1B. The curves were shifted along y-axis to avoid excessive overlapping. All power spectra curves were truncated at 6 Å resolution.
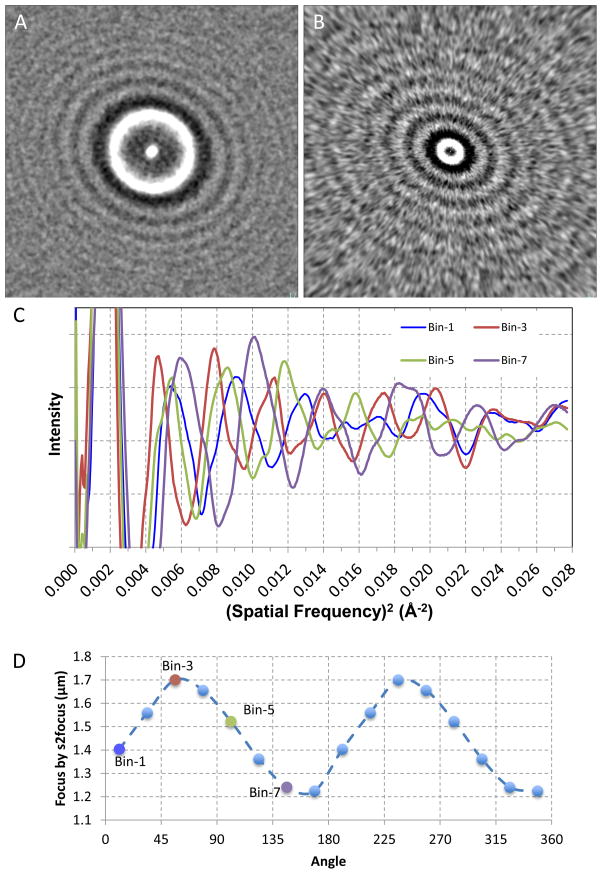
Figure 6. Determination of astigmatism.
A and B, regular s1 power spectrum (A) and s2 power spectrum (B) of an astigmatic image of bacterial phage T7 virion with 58 particles and at 1.42 μm under-focus; C. 1-D s2 power spectra of 4 out of 16 angular sectors; D. Plot of focus values for the angular sectors determined by s2 focus method.
Relative scale determination
The relative scales of the 1-D s2 power spectra of micrographs at different focuses are then determined by a simple search for the best scaling factor to bring the 1-D s2 power spectra of one image to match that of another image. As can be seen in Fig. 2b, the very low-resolution region of the s2 power spectra is still dominated by the sample structural factors. In general these regions (for example, Fourier origin to 30 Å or equivalently 0 to 0.0011 Å−2) are excluded in the calculation of matching scores (negative normalized correlation coefficients). The scale-score plots for several micrograph pairs (Supplementary Fig. 3) show that correct relative scales can be determined from their 1-D s2 power spectra. From these plots it is evident that matching of the oscillations dominates the matching scores despite the influences of residual structural factors and envelope functions that do not follow the linear relationship between focus and oscillation frequency.
Graph representation of image relationships
If we represent each 1-D s2 power spectrum as a node and the relative scale of the s2 power spectra as the edge between the corresponding nodes, a graph describing the relative scale of s2 power spectra of different micrographs can be constructed. Note that we use “image” and “micrograph” in this work interchangeably. One can link all pair of nodes to construct a complete graph or just link a subset of the nodes for a partially connected graph. Care must be taken to avoid isolated nodes by connecting a node to at least another node and to make sure that every node can directly or indirectly reach every other node. For N images, there will be N nodes and edges in the graph.
Determination of focuses as a linear least square problem
From the above graph representation, we can derive a closed-form solution to simultaneously determine the focus values of all images. To rearrange and expand equation 7 to include the focus values of all images, we can obtain
| (8) |
for each edge in the graph. Stacking the corresponding equations of all edges in the graph, we will obtain M equations relating the focus values of all images (Equation 9).
| (9) |
This set of equations corresponds exactly to a standard linear system Ax = b with x as the focus values, b = 0, and A as the matrix of relative scales of the s2 power spectra. The solution to this type of linear system is known as the least square solution if there are more equations than the unknowns. In our case, we will easily satisfy this requirement by connecting a node to more than one node for a graph of N ≥ 3 nodes. We will call this new focus determination method s2focus method.
Bootstrap the least square solution
However, the solution for equation 9 is all zero since this is the trivial solution when b = 0. In fact, equation 9 has infinite number of solutions because any solution multiplied by a constant (including 0) is also a solution. An additional constraint or equation is needed to break the constant factor degeneracy and to allow unique solutions at the correct absolute scale (i.e. the actual focus values).
If the focus value of an image is known, it can be used to bootstrap the solution into correct scale and to break the above constant factor degeneracy problem. The known focus value fi can be added to equation 9 as an additional row in the following form:
| (10) |
Though an arbitrary number of such equations can be added, one additional equation is sufficient. Now the composite of equations 9 and 10 is a truly overdetermined linear equation system with a unique solution. It can be solved by either the regular least square method or by the total least square method. While the regular least square solution considers errors only in observations (i.e. b in Ax = b ), the total least square solution considers errors in both sampling positions and observations (i.e. A and b in Ax = b ) (Wikipedia, 2012). Since our A matrix consists of the relative scales of s2 power spectra and can potentially have some errors, the total least square method should, in theory, provide a more robust solution. Our experimental results confirm this and thus we have chosen to use total least square solution in this work. We used the svd method in the scipy software (Jones et al., 2012) to solve the total least square problem (Wikipedia, 2012). To make the method even more robust against outliers in the data, we further extended the solution to iterative weighted total least square solution by dampening the weights of rows with large errors.
There are several ways to provide the required bootstrap. First, one can simply pre-determine the focus value of one image using any existing method. Second, one can fit a sine wave to the s2 power spectra and determine its focus (Supplementary Fig. 4). In both cases, the known focus value can be used as shown in equation 10. However, any error for this focus value will be carried over as multiplicative errors to all other focus values. Third, one can first set the least square solution to an arbitrary scale (for example, mean focus → 1) as shown in equation 11:
| (11) |
The solved focus values can then be adjusted by setting their mean value to match the mean focus value of another set of focus values independently determined using the traditional power spectra fitting method, for example, our earlier method (Yang et al., 2009). All three approaches were implemented. We prefer the last approach as the two sets of focus values determined independently by these drastically different approaches can also serve as cross validation for improved reliability of the solutions (see Results section, Fig. 7, Supplementary Figs. 5 and 6).
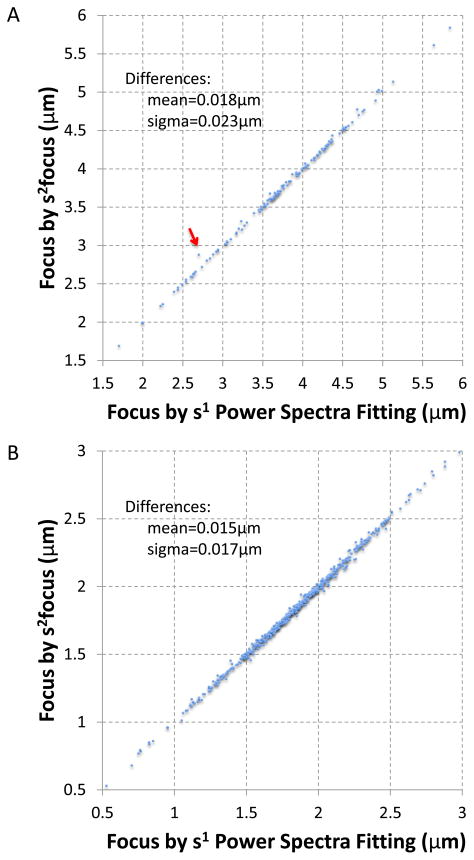
Figure 7. Cross validations of s2 focus and our earlier power spectra fitting s1psfit method.
A. 141 image dataset of Sindbis virus. The red arrow points to the image with largest focus value difference between these two methods; B. 644 image dataset of bacterial phage T7 MLDII capsid. The axes ranges in both A and B were limited to focus on the focus range of most images. Full range plots were shown in Supplementary Figs. 5a and 6a respectively.
Determination of Astigmatism
The s2 focus method can be easily extended to determine the focus values at different directions of an image and thus the astigmatisms of images. In addition to the rotational average s2 power spectra, we can also compute the s2 power spectra of angular sectors and use them to construct a graph including both the rotational average and angular sectors. The whole graph can then be used to solve the least square solution of focus values for the angular sectors and the rotational averages of all images simultaneously. The angular distribution of focus values is then used to describe the image astigmatism using a cosine function of azimuthal angles (Huang et al., 2003; Yang et al., 2009).
Focus determination by direct s2 power spectra fitting
As discussed in earlier sections, the Thon ring oscillation rate in a s2 power spectrum is proportional to the focus value (Equation 6). This simple relationship between focus and Thon ring oscillation rate can be employed to determine the focus value by fitting sine waves of different oscillation frequencies to the s2 power spectrum. A simple 1-D search of focus values will identify the focus value that gives rise to a best matching sine wave (Supplementary Fig. 4). We will call this direct s2 power spectra fitting method s2psfit method. To contrast with this new s2psfit method, we will refer to our earlier automated s1 power spectra fitting method (Yang et al., 2009) as s1psfit method in this publication.
Implementation
We have integrated the new s2focus method, the new s2psfit method, and our earlier s1psfit method into a single python program (fitctf2.py) for easy usage and for cross-validating the fitting results of all three methods. This program is parallelized using the multiprocessing module to use all CPU cores to speed up computation. EMAN2 library (Tang et al., 2007) was used for image IO, and to compute and transform the power spectra. To facilitate the testing of CTFFIND3 method (Mindell and Grigorieff, 2003), we also implemented a python script ctffind.py to run the ctffind3.exe binary in parallel for large number of micrographs and to format the fitting results for more convenient comparison with the results of s2focus, s2psfit and s1psfit methods. The fitctf2.py program examines the focus values by these three (or four if CTFFIND3 results are also included) methods and alerts the user with the list of micrographs with large inconsistencies (for example, > 0.1 μm) in order of decreasing level of inconsistency. Though currently tested only on Linux systems, both Python scripts should work on all platforms that the dependent software (python, EMAN2 and CTFFIND3) are available. Both python scripts, together with test data and usage examples, will be freely downloadable via our web site (http://jiang.bio.purdue.edu).
Test datasets
Three experimental datasets were used to test the performance of our new s2focus method and for the cross-validation of results from the s2 focus method and our earlier s1psfit method, our new s2psfit method, and the CTFFIND3 method. The bacteriophage T7 virion dataset (360 micrographs) and MLDII capsid dataset (644 micrographs) were acquired using a FEI Titan Krios cryo-TEM (300kV, FEG gun) sampled at 1.1 Å/pixel. The Sindbis virus dataset (141 micrographs) was acquired using a Philips CM200 cryo-TEM (200kV, FEG gun) sampled at 1.62 Å/pixel. The focus values of all these micrographs were originally determined by s1psfit method and subsequently verified using the EMAN ctfit graphic program (Ludtke et al., 1999). These verified focus values were used as ground truth in performance tests for the s2 focus, s2psfit and CTFFIND3 methods.
Results
Generation of s2 power spectra
Figure 1 compares 2-D s2 power spectra (Fig. 1b) with 2-D regular s1 power spectra (Fig. 1a). Since the power spectra includes the sample structural factor of which the magnitude can be several orders different from low to high resolution (i.e. center to edge) (Supplementary Figs. 1a and 2a), it is important to minimize such difference. Here we performed log-transform of the original power spectra as in our earlier fitting method (Yang et al., 2009) (Supplementary Figs. 1b and 2b). The remaining intensity gradient can be further removed by high pass filter (Supplementary Figs. 1c and 2c). By now the remaining intensity variations in the transformed power spectra were dominated by the CTF oscillations as seen in Fig. 1a and Supplementary Figs. 1c and 2c). After further denoising by low pass filter (Supplementary Figs. 1d and 2d) and truncation to exclude noises at outer radii (Supplementary Figs. 1e and 2e), a final step of s → s2 transform produced s2 power spectra in which the oscillations have apparently uniform periods from center to edge (2-D) (Fig. 1b and Supplementary Fig. 1f) or from left to right (1-D) (Fig. 2b and Supplementary Fig. 2f) instead of increasingly frequent oscillations in regular power spectra (Fig. 1a, Fig. 2a, Supplementary Figs. 1e and 2e).
To illustrate the linear relationship between the focus and the oscillation frequency in s2 power spectra, the 1-D s2 power spectra of four cryo-EM images of varying focuses are shown in Figure 2b. It is clear that oscillations indeed become more frequent as focus increases. The oscillation frequency doubles as focus doubles. Since the oscillation frequency is inverse to the oscillation period, the equivalent interpretation of Figure 2b is that the oscillation period doubles if the focus is halved. The 1-D s2 power spectra of larger focus will need to be scaled up along the s2-axis (i.e. stretched) to match that of smaller focuses. Correct scaling factors can be determined by 1-D search as shown in Supplementary Fig. 3. These data confirmed the theoretical prediction of the relationship between s2 power spectra and focus that serves as the basis of the s2 focus method.
Performance tests
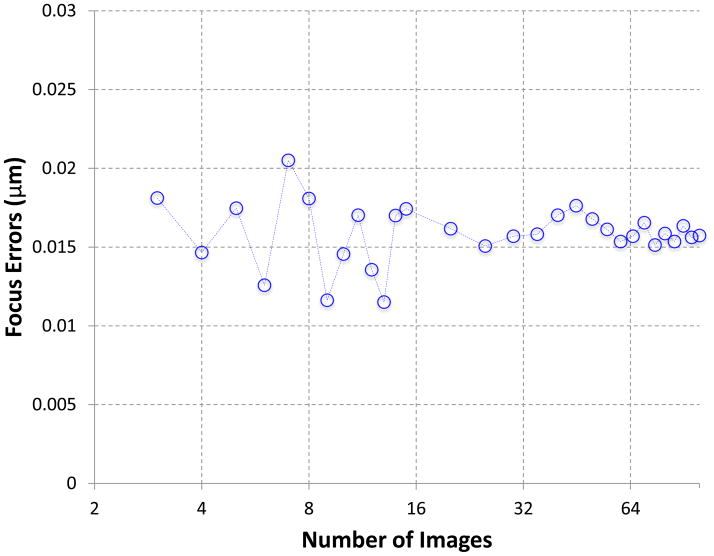
We first tested the s2 focus method on datasets consisting of different numbers of images. Theoretically, the s2 focus method requires N ≥ 3 as there must be at least as many edges as the number of nodes in the graph to provide a sufficient number of equations (i.e. number of edges) for the unknowns (i.e. number of nodes). From the full bacteriophage T7 MLDII capsid test dataset, we randomly selected different numbers of images (N=3 to 100), constructed a complete graph (i.e. the relative scale of s2 power spectra of all pair of images were identified), and solved the focus values. From the results (Figure 3), we can see that the method work reliably across all dataset sizes when N ≥ 3 with the mean “errors” from the reference values clustered in the range of 0.01 to 0.02 μm. The performance becomes more stable with a larger number of images. These results suggest that the s2 focus method works well for both small and large datasets.
Figure 3. Performance with varying number of images.
The focus values fitted using our earlier power spectra fitting s1psfit method (Yang et al., 2009) and then graphically verified were used as references to compute the “errors” of the focus values determined using the new s2 focus method. In these tests, complete graphs were constructed for the least square solutions. The plot shows the average of five runs with the varying number of images randomly selected from a 100-image dataset.
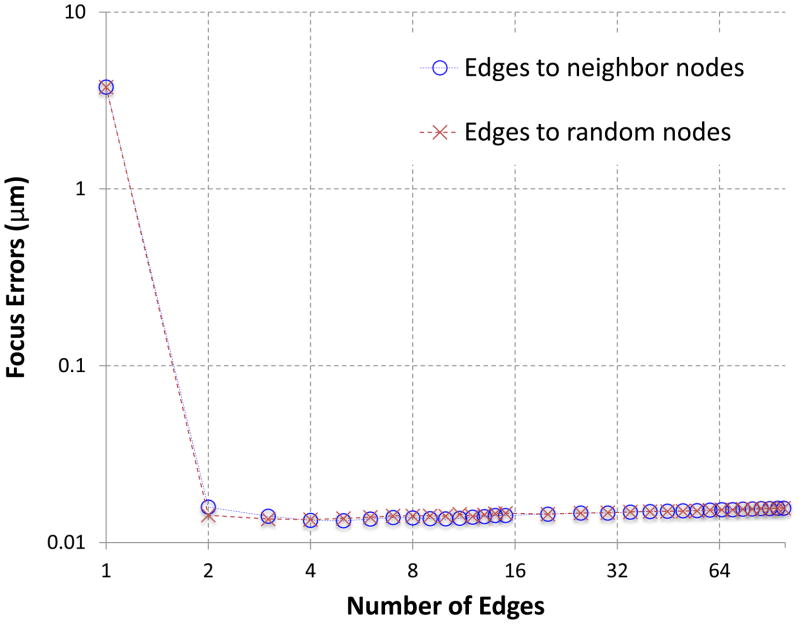
We then tested the s2 focus method with different levels of connectivity on the graph. Using the 100 image dataset, we gradually reduced the number of edges in the graph until there was only one edge for each node. Both random selection of the edges and patterned selection of the edges were tested. In the patterned selection, each node (i.e. image) was only connected to its neighbors in the node list that were in turn arranged according to the order being imaged. From the test results (Figure 4), we determine that the s2 focus method works reliably across a wide range of connectivity levels (from complete graph to 2-connected graphs) and only fails when there is only one edge for each node. The failure with the one-edge graph is expected since there is no longer the sufficient number of edges for any node to reach any other node (i.e. image) on the graph. Both random selection of edges and selection of neighbor edges give similar levels of performance.
Figure 4. Performance with varying levels of graph connectivity.
Both randomly connected edges and edges between neighboring nodes were tested. The same 100 images used in Figure 3 were used in these tests.
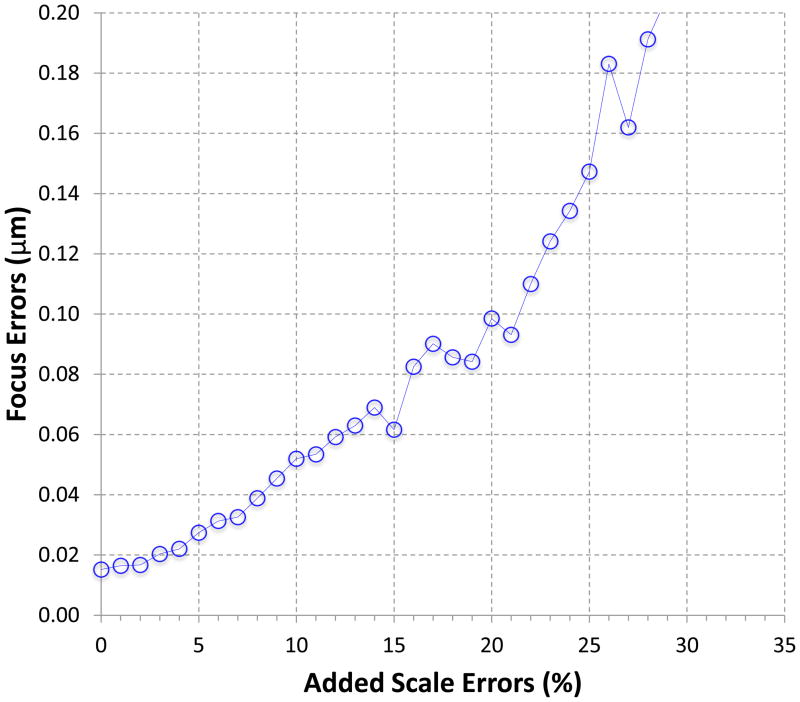
The above tests have shown that the s2 focus method works reliably with experimental data. Since correct solution of focus values relies on correctly identified relative scale values in s2focus method, these results reaffirm that the relative scales of the s2 power spectra can be reliably determined as shown in Supplementary Fig. 3. To further test the robustness of s2focus method against errors in the relative scales, we intentionally added synthetic errors to the identified relative scales between s2 power spectra of images at different focuses. Random scale errors in Gaussian distributions of zero mean and different sigma values were added. The results shown in Figure 5 indicate that the s2 focus method can determine accurate focus values despite the increased level of errors in the relative scale values. Using 0.06 μm as the cutoff (i.e. about one particle diameter), the s2 focus method can tolerate additional 12% errors in the scale values.
Figure 5. Performance with increased “errors” in the relative scale values.
Random synthetic errors were added to the determined scales. The same 100 images for Figure 3 and 4 and 10 edges for each node were used in these tests. The plot shows the average of five runs.
Determination of astigmatism
While high-resolution cryo-EM imaging requires careful instrument alignment that in general can reduce astigmatism to minimal levels, astigmatic images are occasionally found in production datasets. Here we tested the s2 focus method on astigmatic images. As seen from the elliptic Thon rings in regular s1 power spectrum (Fig. 6a) and the corresponding s2 power spectrum (Fig. 6b), this image has significant astigmatism. The Thon rings in s2 power spectra appear to be significantly more elliptic. The oscillations in the 1-D s2 power spectra along different directions are offset from each other (Fig. 6c). By using these 1-D s2 power spectra of different angular directions together with the rotational average 1-D s2 power spectra, s2 focus can simultaneously determine direction-specific focus values together with average focus values in a single least square linear solution. The angular distribution of focus values (Fig. 6d) clearly follows a sinusoidal pattern with a period of 180 degrees, characteristic of 2-fold astigmatism for TEM. By fitting the angular distribution of these focus values to a cosine function (Yang et al., 2009), the direction of smallest focus was determined at 155 degrees, consistent with the most elongated Thon ring direction in Fig. 6a and 6b.
Cross validation with large experimental datasets
From the above performance tests, we found that the s2 focus method works reliably even with limited number of edges per node in the graph. This allows the method to scale to large number of images as it reduces the computational complexity of the method from O(N2) for complete graph to sparsely connected graph with O(aN) in which N is the number of images and a is a small constant factor (a ≪ N ). We thus tested the s2 focus method with two full experimental datasets with 141 and 644 micrographs respectively. In these tests, we only connected every node to other 10 randomly selected nodes. As can be seen in the results (Figure 7), the determined focus values for both datasets are very consistent with the reference focus values based on graphically verified results of our earlier s1psfit method (Yang et al., 2009). The average difference is at about 0.02 μm, which is significantly smaller than the diameter (~0.06 μm) of both virus particles. Since several near atomic resolution (3–4 Å) 3-D reconstructions have been reported for viruses of comparable or larger sizes without correction of focus variations within the same virus particle (Grigorieff and Harrison, 2011; Jiang et al., 2008; Zhang et al., 2010), we assume that such small fitting errors as shown in our tests are insignificant.
As these two focus determination methods are drastically different, consistent results serve as excellent cross validation for both methods when the ground truth is unknown. In the test results with the Sindbis dataset, we also found an outlier with largest focus value difference between these two methods (2.87 μm by s2 focus vs. 2.7 μm by s1psfit) (Fig 7a). Subsequent examination using a graphic program (EMAN ctfit) verified that 2.87 μm by the new s2 focus method is more accurate. It is worth pointing out that the inaccurate 2.7 μm reference focus value by s1psfit method should have already been corrected by the graphic verification step prior to this test as it is part of our standard procedure in image processing. The failure to detect and correct such inaccuracy reflects the occasional omissions associated with repetitive human user operations when processing large data.
We further tested the performance of two additional fitting methods, the new s2psfit method developed in this work and the CTFFIND3 method (Mindell and Grigorieff, 2003), on these two datasets (Supplementary Figs. 5 and 6). We found that both s2psfit and CTFFIND3 could determine correct focus values for majority of the images while only failed for small number of images for both datasets. The failure rates for the Sindbis dataset are modestly low (~2%) for both methods. However, the failure rate for CTFFIND3 on the larger T7 MLDII capsid dataset is significantly higher (~11%), a failure rate larger than the reported 5% maximal rate (Mindell and Grigorieff, 2003). In contrast, other than small (<0.1μm) differences, s2focus method did not fail for these two test datasets. Patterns can be recognized in the failures by both s2psfit and CTFFIND3. Further investigations will be needed to understand the source(s) of these failure patterns. However, a more interesting observation for these failures is that the failures by different methods occurred for different subset of the datasets. The uncorrelated failures are consistent with the fact that these methods are based on different principals and their errors should be susceptible to different factors. In our integrated method we take advantage of the uncorrelated failures to examine the consistency of the focus values determined by these different methods and alert the users with the list of micrographs with inconsistent results.
Discussion
We have developed a novel focus determination method s2 focus for cryo-EM images using graph theory and total least square solution of linear systems. It can determine both the average focus and the anisotropic focus distribution of astigmatic images. Systematic tests have found that this method works accurately and robustly with datasets of a variety of sizes. Its good performance with sparsely connected graphs allows it to scale to large experimental datasets. It takes only seconds to a few minutes on a desktop computer to complete the two large experimental datasets with hundreds of micrographs (Supplementary Table I).
This s2 focus method is the first method that employs the relationships among different images to simultaneously determine the focuses of all images. In contrast, current methods work on individual images and the fitting process is independent for different images. In the s2 focus approach, determination of astigmatism and average focus values are also unified in a single least square solution that are solved simultaneously. This is in stark contrast to existing methods in which the focus values of angular sectors are determined separately from the rotational average focus (Huang et al., 2003; Yang et al., 2009) or the Thon rings are explicitly located using image feature detection methods to approximate the ellipticity and then astigmatism (Mallick et al., 2005). It is thus appropriate to classify our new s2 focus method as the only global method while current methods are all local methods. In addition, the new s2 focus method is a closed-form method that utilizes the robust total least square solution of overdetermined linear systems. In contrast, all existing methods formulate focus determination as an iterative non-linear search/optimization problem.
During TEM instrument alignment, an essential task is to minimize objective lens astigmatism by visually monitoring the apparent ellipticity of Thon rings of image power spectra. It is thus beneficial to enhance ellipticity of the Thon rings to help further reduce the residual astigmatism. As shown in Figure 6, Thon rings of astigmatic images are apparently more elliptic in s2 power spectra (Fig. 6b) than in regular s1 power spectra (Fig. 6a). This observation suggests that s2 power spectra can be potentially used to facilitate visual detection and further minimization of residual astigmatism during instrument alignment and to improve cryo-EM image quality.
It is worth pointing out that mean focus and astigmatism of the entire micrograph are only an approximation to the true focus and astigmatism for particles scattered across the micrograph. The intentional tilt of the specimen to alleviate preferred particle orientation (Frank and Radermacher, 1992) or the local tilt due to cryo-crinking of the sample grid (Booy and Pawley, 1993) will result in planar distribution of the focus values. The positioning of particles at different Z-heights in the embedding vitreous ice will result in more randomly varying focuses. The systematic variations of focuses caused by tilt can be determined using local fitting with optional planar constraints (Mindell and Grigorieff, 2003; van Heel et al., 2000). Though not currently implemented, it will be straightforward to extend the s2focus method to also deal with the tilt by dividing the entire micrograph into multiple blocks and treating each block as a different micrograph. In our practical image processing strategy, we instead choose to include further focus refinement in the iterative image alignment and 3-D reconstruction process (Chen et al., 2011). Such focus refinement requires pre-determined orientation and center parameters as prior conditions but it can use both the amplitude and phase information to achieve higher accuracy and higher resolution 3-D reconstruction (Chen et al., 2011). In contrast, all Thon-ring based focus determination methods only use amplitude information although no pre-determined particle orientation and center parameters are needed.
Though our earlier iterative fitting method s1psfit is a high quality method as shown in (Yang et al., 2009) and in the tests of this work, we still always use a graphic program (EMAN ctfit) to interactively verify the fitting results of all images. This is due to the lack of a reliable indicator for the correctness of the determined focuses and the lack of reliable self-detection of rare failures as shown in Figure 7a. Tests with two additional fitting methods, our new s2psfit method and the CTFFIND3 method, found that both methods also fail with small number of micrographs (Supplementary Figs. 5 and 6). It is probably unrealistic to reach 100% accuracy and reliability for any single method. In this work, we have found that the rare inaccuracies and failures in the determined focus values can be conveniently detected by comparing the results of different methods. A very valuable advantage arisen from this work is that we now have multiple methods (s2focus, s2psfit, s1psfit, and optionally CTFFIND3) integrated in a single program for reliable automated focus determination of experimental images. Since these methods are based on drastically different principals, the occasional failures by each of the methods are uncorrelated. The consistency or discrepancy of the results of these methods can thus provide rigorous cross-validation for the results of individual methods. In our opinion, availability of cross-validation method is essential for quality assurance of all tasks (Henderson et al., 2012; Read et al., 2011) though it is often missing in many systems including cryo-EM 3-D reconstructions (Henderson et al., 2012). Here we demonstrated a reliable cross validation method for focus determination, an essential early step in high-resolution cryo-EM image processing and 3-D reconstruction. Coupled with its scalability to a large number of images, the focus determination method with reliable internal cross-validations developed in this work will allow more robust automated image processing of increasingly more common large datasets for high-resolution 3-D reconstructions.
Supplementary Material
Acknowledgments
We would like to thank Dr. Philips Sewer for the T7 phage sample, Dr. Richard Kuhn for the Sindbis virus sample, and Mr. Samir Parmar for his assistance in preparation of the manuscript. The cryo-EM images were taken in the Purdue Biological Electron Microscopy Facility. This research is funded by grants from NIH (S10RR023011, P01AI055672, R01AI072035).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Booy FP, Pawley JB. Cryo-Crinkling - What Happens to Carbon-Films on Copper Grids at Low-Temperature. Ultramicroscopy. 1993;48:273–280. doi: 10.1016/0304-3991(93)90101-3. [DOI] [PubMed] [Google Scholar]
- Brin S, Page L. The anatomy of a large-scale hypertextual Web search engine. Computer networks and ISDN systems. 1998;30:107–117. [Google Scholar]
- Chen DH, Baker ML, Hryc CF, DiMaio F, Jakana J, et al. Structural basis for scaffolding-mediated assembly and maturation of a dsDNA virus. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:1355–1360. doi: 10.1073/pnas.1015739108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erikson HP, Klug A. The Fourier transform of an electron micrograph: effects of defocussing and aberrations, and implications for the use of underfocus contrast enhancement. Berichte der Bunsen-Gesellschaft. 1970;74:1129–1137. [Google Scholar]
- Frank J, Radermacher M. Three-dimensional reconstruction of single particles negatively stained or in vitreous ice. Ultramicroscopy. 1992;46:241–262. doi: 10.1016/0304-3991(92)90018-f. [DOI] [PubMed] [Google Scholar]
- Grigorieff N, Harrison SC. Near-atomic resolution reconstructions of icosahedral viruses from electron cryo-microscopy. Current opinion in structural biology. 2011;21:265–273. doi: 10.1016/j.sbi.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson R, Sali A, Baker ML, Carragher B, Devkota B, et al. Outcome of the first electron microscopy validation task force meeting. Structure. 2012;20:205–214. doi: 10.1016/j.str.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Tejero R, Powers R. A Topology-Constrained Distance Network Algorithm for Protein Structure Determination From NOESY Data. PROTEINS: Structure, Function, and Bioinformatics. 2006;62:587–603. doi: 10.1002/prot.20820. [DOI] [PubMed] [Google Scholar]
- Huang Z, Baldwin PR, Mullapudi S, Penczek PA. Automated determination of parameters describing power spectra of micrograph images in electron microscopy. Journal of structural biology. 2003;144:79–94. doi: 10.1016/j.jsb.2003.10.011. [DOI] [PubMed] [Google Scholar]
- Jiang W, Chiu W. Web-based Simulation for Contrast Transfer Function and Envelope Functions. Microscopy and microanalysis: the official journal of Microscopy Society of America, Microbeam Analysis Society, Microscopical Society of Canada. 2001;7:329–334. doi: 10.1017.S1431927601010315. [DOI] [PubMed] [Google Scholar]
- Jiang W, Baker ML, Jakana J, Weigele PR, King J, et al. Backbone structure of the infectious epsilon15 virus capsid revealed by electron cryomicroscopy. Nature. 2008;451:1130–1134. doi: 10.1038/nature06665. [DOI] [PubMed] [Google Scholar]
- Jones E, Oliphant T, Peterson P, et al. SciPy: Open Source Scientific Tools for Python. 2012 http://www.scipy.org.
- Koyutürk M, Subramaniam S, Grama A. Functional Coherence of Molecular Networks in Bioinformatics. Springer Verlag; 2011. [DOI] [PubMed] [Google Scholar]
- Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. Journal of structural biology. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- Mallick SP, Carragher B, Potter CS, Kriegman DJ. ACE: automated CTF estimation. Ultramicroscopy. 2005;104:8–29. doi: 10.1016/j.ultramic.2005.02.004. [DOI] [PubMed] [Google Scholar]
- Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. Journal of structural biology. 2003;142:334–347. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- Read RJ, Adams PD, Arendall WB, 3, Brunger AT, Emsley P, et al. A new generation of crystallographic validation tools for the protein data bank. Structure. 2011;19:1395–1412. doi: 10.1016/j.str.2011.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saad A, Ludtke SJ, Jakana J, Rixon FJ, Tsuruta H, et al. Fourier amplitude decay of electron cryomicroscopic images of single particles and effects on structure determination. Journal of structural biology. 2001;133:32–42. doi: 10.1006/jsbi.2001.4330. [DOI] [PubMed] [Google Scholar]
- Sander B, Golas MM, Stark H. Automatic CTF correction for single particles based upon multivariate statistical analysis of individual power spectra. Journal of structural biology. 2003;142:392–401. doi: 10.1016/s1047-8477(03)00072-8. [DOI] [PubMed] [Google Scholar]
- Sorzano CO, Jonic S, Nunez-Ramirez R, Boisset N, Carazo JM. Fast, robust, and accurate determination of transmission electron microscopy contrast transfer function. Journal of structural biology. 2007;160:249–262. doi: 10.1016/j.jsb.2007.08.013. [DOI] [PubMed] [Google Scholar]
- Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, et al. EMAN2: an extensible image processing suite for electron microscopy. Journal of structural biology. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- Thon F. Phase contrast electron microscopy. Academic Press; New York: 1971. [Google Scholar]
- van Heel M, Gowen B, Matadeen R, Orlova EV, Finn R, et al. Single-particle electron cryo-microscopy: towards atomic resolution. Quarterly reviews of biophysics. 2000;33:307–369. doi: 10.1017/s0033583500003644. [DOI] [PubMed] [Google Scholar]
- Velazquez-Muriel JA, Sorzano CO, Fernandez JJ, Carazo JM. A method for estimating the CTF in electron microscopy based on ARMA models and parameter adjustment. Ultramicroscopy. 2003;96:17–35. doi: 10.1016/S0304-3991(02)00377-7. [DOI] [PubMed] [Google Scholar]
- Wikipedia. Wikipedia: Total Least Squares. 2012 http://en.wikipedia.org/wiki/Total_least_squares.
- Yang C, Jiang W, Chen DH, Adiga U, Ng EG, et al. Estimating contrast transfer function and associated parameters by constrained non-linear optimization. Journal of microscopy. 2009;233:391–403. doi: 10.1111/j.1365-2818.2009.03137.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Jin L, Fang Q, Hui WH, Zhou ZH. 3.3 A cryo-EM structure of a nonenveloped virus reveals a priming mechanism for cell entry. Cell. 2010;141:472–482. doi: 10.1016/j.cell.2010.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.