SUMMARY
In T cells the adapter Bam32 is coupled to Erk activation downstream of the TCR by an unknown mechanism. We characterized in Jurkat cells and primary T lymphocytes, a pathway dependent on Bam32-PLC-γ1-Pak1 complexes, in which Pak1 kinase activates Raf-1 and Mek-1, both upstream of Erk. In the Bam32-PLC-γ1-Pak1 complex, catalytically inactive PLC-γ1 is used as a scaffold linking Bam32 to Pak1. PLC-γ1(C-SH2) directly binds S141 of Bam32, preventing LAT-mediated activation of Ras by PLC-γ1. The Bam32-PLC-γ1 interaction enhances the binding of the SH3 domain of the phospholipase with Pak1. The PLC-γ1(SH3)-Pak1 interaction activates Pak1 independently of the small GTPases Rac1/Cdc42, previously described as being the only activators of Pak1 in T cells. Direct binding of the SH3 domain of PLC-γ1 to Pak1 dissociates inactive Pak1 homodimers, a mechanism required for Pak1 activation. We have thus uncovered a LAT/Ras-independent, Bam32-nucleated pathway that activates Erk signaling in T cells.
Keywords: T cell signaling, Bam32, PLC-γ1, Pak1, Erk
INTRODUCTION
T cell stimulation via the T cell antigen receptor (TCR) results in very robust Ras activation (Downward et al., 1990). Critical to activation of the Ras/MAPK pathway is phosphorylation of the adapter LAT. Proteins recruited to phosphorylated LAT activate the Ras/MAPK pathway by at least two different cascades (Sommers et al., 2004): (1) PLC-γ1 recruitment to LAT is required for its activation in which PIP2 is hydrolyzed into DAG and IP3. DAG activates several signaling proteins including RasGRP, a GEF that activates Ras (Dower et al., 2000). (2) LAT binds the complex of the adapter Grb2 and the Ras GEF SOS. Recent evidence suggests that SOS and RasGRP pathways are interconnected and are both needed for efficient Ras activation in T cells (Roose et al., 2007). The Ras/Raf/Mek/Erk pathway is often drawn in a linear manner such that Ras activates Raf, which in turn activates Mek, which in turn activates Erk. While this classical way of thinking about the pathway is correct, other mechanisms for Erk activation, some independent of Ras, are possible.
The adapter protein B cell adapter molecule of 32 kDa (Bam32) (Marshall et al., 2000) is potentially coupled to Erk activation in a different manner (Han et al., 2003; Sommers et al., 2008). Bam32 contains a SH2 domain, one potential Y phosphorylation site (Y139), and a PH domain. The Bam32 SH2 domain interacts with PLC-γ2, the B cell homologue of PLC-γ1 (Marshall et al., 2000). In B cells the effect of Bam32 on PLC-γ2 activation is controversial (Allam and Marshall, 2005). The putative MAP4K HPK1 is the second known Bam32 partner (Han et al., 2003). How Bam32 and HPK1 are connected is still unknown. Importantly the activation of Erk via Bam32 is also unexplained.
This effect of Bam32 on Erk in B cells led us to investigate the role of Bam32 in TCR-induced Erk activation in T cells. We published that Bam32 is also expressed in mouse CD4+ T cells (Sommers et al., 2008) and that it is required for optimal MAPK activation in these cells (Sommers et al., 2008). Bam32 deficiency also has a significant negative impact on TCR-induced cytokine production, proliferation, and spreading. The mechanism by which Bam32 activates Erk signaling in T cells has been investigated in the present study and we found that an interaction between Bam32 and Pak1, a well-known Erk activator that is related to HPK1, controls Erk activation in this setting.
The six serine/threonine kinases of the Pak family are involved in multiple cellular processes, including MAPK signaling, cytoskeletal reorganization, apoptotic signaling, cell migration, and growth factor-induced neurite outgrowth. Accumulating evidence implicates Pak kinases in oncogenic growth (Dummler et al., 2009). All Paks are characterized by a regulatory domain and a highly conserved kinase domain. The regulatory domain consists of a GTPase-binding domain (PBD) and several proline-rich regions, which serve as docking sites for SH3 domain-containing proteins. Pak1–3 also possess an autoinhibitory domain (PID) overlapping with the PBD. In resting cells Pak1 is a trans-inhibited homodimer in which the PID of one molecule binds to and inhibits the kinase domain of the other. Binding of activated Rac/Cdc42 to the PBD dissociates the dimer and activates Pak1 molecules by releasing the PID-mediated inhibition leading to autophosphorylation of T423 in the activation loop (Parrini et al., 2002). In T cells, different mechanisms of Pak1 activation have been described (Bubeck Wardenburg et al., 1998; Ku et al., 2001; Yablonski et al., 1998a; Yablonski et al., 1998b) and different pools of Pak1 might be differentially regulated. In T cells, Pak1 has only been shown to play a role in cytoskeletal rearrangement. Its role in MAPK activation has not been previously investigated.
In the present study we demonstrate the existence of a Bam32-PLC-γ1-Pak1 complex both in Jurkat T cells and in primary CD4+ cells. This complex works in a cooperative manner to activate Erk after TCR engagement via activation of the kinase Pak1, whose direct targets are Raf-1 S338 and Mek-1 S298 (Frost et al., 1997; Zang et al., 2002), two kinases that are upstream of Erk. Direct binding of the PLC-γ1 C terminal-SH2 domain to Bam32 requires S141 of Bam32, and the PLC-γ1 SH3 domain binds a proline-rich domain of Pak1. Interestingly we demonstrated that the role of Bam32 in Erk activation is independent of its PH and SH2 domains and residue Y139, which are necessary for other previously described Bam32 functions (Allam and Marshall, 2005). The phospholipase function of PLC-γ1 is not required for Bam32-mediated Erk activation; instead, PLC-γ1 has a scaffolding role in this setting. The trimolecular complex Bam32-PLC-γ1-Pak1 seems to be sufficient by itself to increase Pak1 signaling, as we demonstrated its independence from LAT, SLP-76, and Rac1/Cdc42, molecules previously shown to regulate Pak1 activity in T cells. The proposed mechanism of Pak1 activation is the dissociation of trans-inhibited dimers into active Pak1 monomers by binding of the SH3 domain of PLC-γ1, an interaction that is favored by the expression of Bam32. Thus, in this study, we describe a LAT-independent, Bam32-nucleated pathway that, complementing previously described LAT-dependent mechanisms, positively regulates Erk signaling in T cells.
RESULTS
Existence of a Bam32-Pak1 complex that activates Raf-1, Mek-1 and Erk
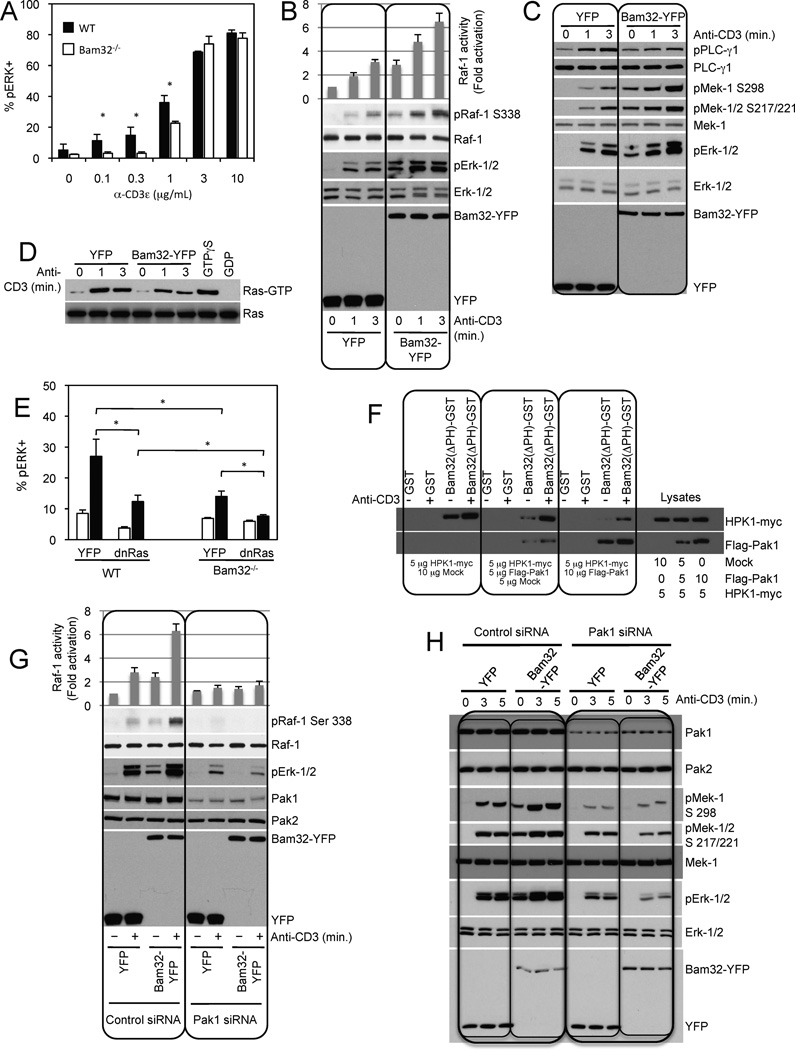
We have previously demonstrated by western-blotting that CD4+ T cells from Bam32 KO mice display less Erk phosphorylation than those from wild type mice following plate-bound anti-CD3ε stimulation (Sommers et al, 2008). In the current study, we stimulated CD4+ T cells with soluble anti-CD3ε mAb and measured phosphorylated Erk (pErk) by flow cytometry. Figure 1A shows that the percentage of T cells positive for pErk was considerably lower in Bam32 KO than in WT mice, especially at low doses of anti-CD3ε mAb. At high doses of stimulatory mAb, pErk reached the same levels in Bam32 KO and WT mice, thus demonstrating that Bam32 KO T cells were healthy. Comparing Bam32 KO and WT mice, we observed a greater difference in pErk when T cells were activated with soluble stimulation than when they were activated via plate-bound stimulation and concluded that a Bam32-coupled pathway is involved in a portion of Erk activation in T cells.
Figure 1. Existence of a Bam32-Pak1 complex that activates Mek-1 and Erk.
(A) Quantification of pErk-1/2 staining in WT and Bam32−/− CD4+ T cells stimulated with αCD3ε for 2 min. The average and SD from 3 independent experiments are shown. * p<0.05.
(B) JE6.1 cells transfected with YFP or Bam32-YFP were stimulated with αCD3. Unless otherwise indicated, αCD3 stimulations were at 1 µg/ml for 3 min. Raf-1 activity was measured with a kinase assay. Averages and standard deviations are representative of two independent experiments. Representative WBs are shown (n=2).
(C) Transfections and stimulation as in (A). Representative WBs are shown (n=4).
(D) Transfections and stimulation as in (A). Ras-GTP pull down was carried out using Raf-RBD-GST incubated with lysates. GTPγS- and GDP-loaded lysates were used as controls. Total Ras and active Ras were detected by WB. A representative WB is shown (n=2).
(E) Quantification of pErk-1/2 staining in YFP-gated WT CD4+ T cells and Bam32−/− CD4+ T cells transfected with YFP +/− dnRasG15A and stimulated with αCD3ε (1 µg/ml, 2 min., black bars). The average and SD from 3 independent experiments are shown. * p<0.05.
(F) JE6.1 cells were transfected with 5 µg of HPK1-myc cDNA alone or + Flag-Pak1 cDNA at 5 or 10 µg. Cells ± αCD3 were lysed. Extracts were incubated with GST or Bam32(ΔPH)-GST proteins. Bound proteins were analyzed by WB. Input was 10%. Shown is a representative WB (n=2).
(G) JE6.1 cells were transfected with Pak1 siRNAs or control siRNAs. After 24h, cells were transfected with YFP or Bam32-YFP cDNAs. After an additional 20h, cells ± αCD3 were analyzed by WB. A representative WB is shown (n=2). Raf-1 activity was measured with a kinase assay. Averages and standard deviations are representative of 2 independent experiments.
(H) Transfections and stimulation as in (G). Representative WBs are shown (n=2).
Exogenous Bam32-YFP expression in Jurkat T cells increased Erk activation (Fig. 1B/1C), which correlated with changes in two of the core kinase components of the Erk cascade, Raf-1 (Fig. 1B) and Mek-1 (Fig. 1C). Raf-1 catalytic activity was elevated and increased pRaf-1 on the activating S338 site was observed (Fig. 1B); Similarly we detected an increase of pMek-1 on both the activation loop phosphorylation sites S217/S221 and S298 (Fig. 1C). These effects of Bam32 expression on Raf-1 and Mek-1 were observed when cells were stimulated by activation of the TCR using αCD3 stimulation and they were also observed under basal conditions. Therefore we evaluated the TCR-dependence of the Bam32 pathway by overexpressing Bam32-YFP in Jurkat T cells deficient for ZAP-70 kinase (P116 cells). ZAP-70 is recruited to the TCR after TCR engagement and then it phosphorylates LAT and other substrates. Bam32 fails to increase Erk activation in P116 cells, whereas it augments pErk in P116 + ZAP-70, thus demonstrating that the Bam32 pathway depends upon TCR activation (Fig. S1A).
In contrast to the Bam32-induced increase in pErk, cells expressing Bam32 showed reduced pPLC-γ1 (Fig. 1C) and reduced levels of Ras-GTP (Fig. 1D). The most critical GEF for Ras in T cells is RasGRP (Roose et al., 2007), which is activated by DAG produced by PLC-γ1. Thus these results suggest that the Bam32 pathway might bypass the activation of Ras to activate Erk. To test this hypothesis we overexpressed a dominant negative mutant of Ras (dnRasG15A) in CD4+ T cells from Bam32 KO and WT mice and measured levels of pErk by flow cytometry (Fig. 1E). Demonstrating the importance of Ras in the Erk pathway, dnRasG15A greatly decreased pErk levels in WT cells. However Erk activity was not totally inhibited. Interestingly dnRasG15A also diminished Erk activation in Bam32 KO CD4+ T cells, demonstrating that the Ras pathway and the Bam32 pathway do not overlap. The combined absence of Ras-dependent and Bam32-dependent pathways resulted in nearly complete loss of TCR-induced Erk activity.
We have thus uncovered a functional link between Bam32 and the Erk cascade. Next we probed for a physical interaction between Bam32 and components of the Erk cascade. Pak1 was of interest for two reasons. (1) Raf-1 S338 and Mek-1 S298 are known targets of Pak1 (Frost et al., 1997; Zang et al., 2002). (2) Pak1 is related to HPK1, which was previously reported to bind Bam32 in B cells. Thus we tested whether Bam32 and Pak1 interact in T cells and, if so, whether Pak1 competes with HPK1 for Bam32. Jurkat cells were transfected with a constant amount of HPK1-myc and increasing amounts of Flag-Pak1 (Fig. 1F). A Bam32(ΔPH)-GST chimeric molecule was used as an affinity reagent. We confirmed the interaction between Bam32(ΔPH) and HPK1, now in T cells, and we demonstrated an interaction between Bam32(ΔPH) and Pak1. Furthermore, HPK1 overexpression increased the level of Bam32(ΔPH)-HPK1 complexes but limited the amount of Bam32(ΔPH)-Pak1 complexes (Fig. 1F). The reciprocal experiment was performed (Fig. S1B). Jurkat T cells were transfected with a constant amount of HPK1-myc and increasing quantities of Flag-Pak1. With more Flag-Pak1 expression, more Bam32(ΔPH)-Pak1 complexes were found at the expense of Bam32(ΔPH)-HPK1 complexes. In conclusion, Bam32-Pak1 and Bam32-HPK1 complexes are distinct entities and the two kinases compete for Bam32 binding.
We next investigated whether Pak1 is required for the effects of Bam32 on Erk phosphorylation. Jurkat cells were transfected with Pak1 siRNAs and Bam32-YFP or YFP. Pak1 expression was reduced by 90%. Pak2 levels were examined as specificity and loading controls (Fig. 1G/1H). Bam32 increased both the pS338 and the kinase activity of Raf-1 in a Pak1-dependent manner (Fig. 1G). The basal activity and pS338 state of Raf-1 was enhanced in the presence of Bam32 and was further increased with TCR activation. Moreover, the effects of Bam32 on Raf-1 were no longer observed when Pak1 was depleted. Bam32-YFP overexpression increased Mek-1 phosphorylation on S217/S221 (activation loop) and also on S298 (Pak1 site) (Figure 1H). Erk phosphorylation was also augmented. This increase in pErk was not only lost after Pak1 knockdown (KD) but was actually replaced by clear inhibition (Figure 1G/1H). In contrast Pak2 KD minimally impacted the effect of Bam32 on pMek-1/pErk (Fig. S1C). Therefore we showed that Bam32 works through Raf-1 to mediate the phosphorylation of the Mek-1 activation loop sites. Additionally, we demonstrated that Bam32 modulates signaling through the Erk cascade in two ways: (1) the Pak1-dependent phosphorylation and activation of Raf-1 and (2) the Pak1-dependent phosphorylation of Mek-1 on S298. Therefore Bam32 stimulates Raf-1, Mek-1 and Erk activity via Pak1 and acts in a dominant negative fashion on Erk cascade activation in the absence of Pak1. Our subsequent experiments focus on Mek-1 and Erk as readouts in this system.
Bam32 interacts with Pak1 via PLC-γ1 and Bam32-PLC-γ1-Pak1 complexes activate Mek-1 and Erk
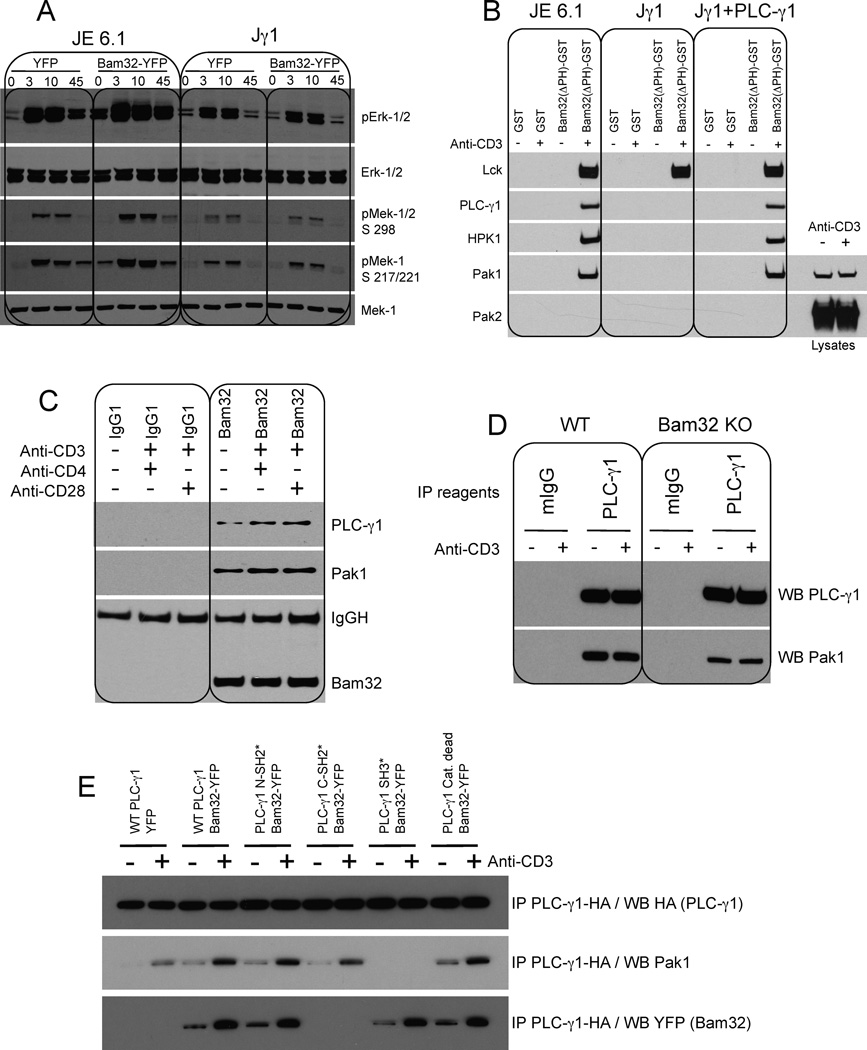
The Bam32-Pak1 interaction in Jurkat cells could be direct or indirect. Because Pak2 KD does not modify the effect of Bam32 on Erk cascade signaling, and because the main differences between Pak1 and Pak2 are in their proline-rich domains, we considered that an SH3-containing molecule binding Pak1 might serve as an intermediate between Bam32 and Pak1. As previously shown (Fig. 1C), Bam32-YFP overexpression diminished PLC-γ1 activation. Interestingly PLC-γ1 contains one SH3 domain, and we considered whether it might be the binding intermediate. We studied the effect of Bam32-mediated Erk activation in Jurkat T cells deficient for PLC-γ1 (Jγ1 cells). Bam32-YFP increased Mek-1 and Erk activation in WT Jurkat cells, but its effect was lost in Jγ1, suggesting that PLC-γ1 could be the link between Bam32 and Pak1 (Fig. 2A). To confirm this hypothesis a pulldown was performed in Jγ1, Jγ1 cells reconstituted with PLC-γ1 and in WT Jurkat cells (JE6.1). Fig. 2B shows a Bam32-Lck interaction (further characterized in Fig. S3B). The presence of this complex was used as a marker of cell activation. The pulldown experiment demonstrates that Bam32(ΔPH)-HPK1 and Bam32(ΔPH)-Pak1 interactions are dependent on PLC-γ1, as neither kinase co-precipitated with Bam32 in Jγ1 cells. In contrast to Pak1, Pak2 did not bind Bam32 confirming the previous KD experiment. A similar pulldown was performed with JE6.1 lysates immunodepleted of PLC-γ1. Bam32(ΔPH) bound Pak1 only when PLC-γ1 was present (Fig. S2 A3). We also performed a pulldown using (SH3)PLC-γ1-GST. We demonstrated that (SH3)PLC-γ1 basally bound Pak1. The interaction was slightly enhanced by αCD3 stimulation (Fig. S2 A3). Importantly the lysates immunodepleted for PLC-γ1 showed a loss of Pak1 of approximately 20% demonstrating that 20% of total Pak1 molecules are associated with PLC-γ1 (Fig. S2 A2). This result suggests an important role for the PLC-γ1/Pak1 pathway in T cells.
Figure 2. Bam32 interacts with Pak1 via PLC-γ1 and Bam32-PLC-γ1-Pak1 cooperative complexes increase Mek-1 and Erk phosphorylation.
(A) JE6.1 and Jγ1 cells transfected with YFP or Bam32-YFP cDNAs were sorted for equal YFP fluorescence intensity. Cells ± αCD3 were analyzed by WB. A representative WB is shown (n=4).
(B) JE6.1, Jγ1 and Jγ1 cells expressing PLC-γ1 ± αCD3 were lysed. Extracts (1 mg total protein) were incubated with GST or Bam32(ΔPH)-GST proteins. Bound proteins were analyzed by WB. For Pak1 and Pak2 analysis WCL from resting and OKT3-stimulated Jγ1 cells were loaded to confirm Pak2 expression in Jγ1. A representative WB is shown (n=2).
(C) Human CD4+ T cells were stimulated with αCD3 (5 µg/ml) + αCD4 (10 µg/ml) or αCD28 (10 µg/ml). Lysates were subjected to a Bam32 or control IP. IPs were analyzed by WB. A representative WB is shown (n=2).
(D) CD4+ T cells from WT or Bam32−/− mice were stimulated with αCD3ε (1 µg/ml, 2 min.). Lysates were subjected to a PLC-γ1 or control IP. IPs were analyzed by WB. A representative WB is shown (n=4).
(E) JE6.1 cells were transfected with YFP or WT Bam32-YFP + WT PLC-γ1-HA or mutants of PLC-γ1-HA (10 µg each). Lysates from cells ± αCD3 stimulation were subjected to an HA IP to pull down PLC-γ1. A representative WB is shown (n=3).
Next we looked for endogenous Bam32-PLC-γ1-Pak1 complexes in human primary CD4+ T cells. Bam32-PLC-γ1-Pak1 complexes were found in resting CD4+ T cells, and cell stimulation moderately increased their levels (Fig. 2C). The constitutive binding may explain the Bam32-mediated increase of Erk activation observed prior to Jurkat cell stimulation. The Pak1 molecules bound to the Bam32-PLC-γ1 complexes are phosphorylated indicating that the kinases in the trimolecular complexes were active (Fig. S2B). We also examined the cooperative nature of the Bam32-PLC-γ1-Pak1 complex. Cells expressing increasing amounts of Bam32-YFP were stimulated by αCD3, PLC-γ1 was immunoprecipitated and bound Pak1 was studied. Bam32-YFP greatly enhanced the binding between PLC-γ1 and Pak1 (Fig. S2C). In a more physiological context, we verified that the PLC-γ1-Pak1 interaction was compromised in CD4+ T cells from Bam32 KO mice. Fig. 2D demonstrates that in the absence of Bam32, the PLC-γ1 molecules bound Pak1 kinase less efficiently than in normal CD4+ T cells.
To further characterize Bam32-PLC-γ1-Pak1 interactions we mapped the relevant binding domains. WT PLC-γ1 and various mutants (N-SH2*, C-SH2*, SH3* and catalytically dead enzyme) were cotransfected with Bam32-YFP and complexes were studied by immunoprecipitation (IP). PLC-γ1 C-SH2 domain bound Bam32, whereas the PLC-γ1 SH3 domain bound Pak1 (Fig. 2E). The catalytically dead enzyme bound Bam32 and Pak1 normally. In Fig. S2D, we demonstrated that these two interactions (Bam32-PLC-γ1 and PLC-γ1-Pak1) were both direct using far western-blotting. (C-SH2)PLC-γ1-GST directly bound WT Bam32-YFP whereas (N-SH2)PLC-γ1 did not. (N-SH2)PLC-γ1-GST did bind LAT extracted from activated cells, as a positive control. We also demonstrated that (SH3)PLC-γ1 directly bound Pak1. GST fusions of the SH2 domain and SH3 domain of the PI-3K subunit p85, used as specificity controls, bound neither Bam32 nor Pak1, respectively.
Other SH3 domain-containing molecules bind the proline-rich domains of Pak1. Grb2 (Puto et al., 2003), Nck (Bokoch et al., 1996), and the Rac1/Cdc42 GEF PIX (Manser et al., 1998), all can bind Pak1. We asked whether these other Pak1 partners were present in the Bam32-PLC-γ1-Pak1 complex (Fig. S2 E–G). To increase the likelihood of detecting interaction with Pak1 in the Bam32-PLC-γ1-Pak1 complex, we overexpressed each member of the complex (Bam32-YFP, PLC-γ1-HA, and Flag-Pak1). Because Grb2 (25 kDa) and Nck (47 kDa) comigrate with IgG light and heavy chains, we overexpressed them with tags (Grb2-YFP and Nck1-CFP) for better detection after IP. The results show that neither Nck1-CFP nor endogenous βPIX was detected in the Bam32-PLC-γ1-Pak1 complex. Only small amounts of Grb2-YFP were recruited to the complex in activated cells (Fig. S2 E, F, G).
Relative importance of Bam32 Y139 and S141 for the formation of the Bam32-PLC-γ1-Pak1 complex and for αCD3-induced Erk activation
Bam32 pY139 is crucial for Rac1 and Jnk activation in B cells (Allam et al., 2004). We investigated whether Bam32 is also Y phosphorylated in T cells and how such phosphorylation might impact Erk activation. Jurkat cells overexpressing Bam32-YFP were stimulated with the αTCR mAb C305 at very high concentrations and Bam32-YFP and LAT were immunoprecipitated to study phosphorylation kinetics (Fig. S3A). Strikingly the pY kinetics of these two proteins were very different. pLAT was rapid and transient. In contrast Bam32 pY began 15 min after TCR stimulation and lasted at least 45 min. We also studied the kinetics of the Bam32-Lck interaction, which did not depend on PLC-γ1, to determine if this interaction followed the time-course of Bam32 pY. Lck co-IP with Bam32-YFP was first detected 15 min after TCR stimulation (Fig. S3B). Note that the interactions between Bam32, PLC-γ1, and Pak1 were almost completely disrupted 15 min after TCR stimulation. The Bam32-Lck interaction peaked at 25 min and was disrupted 40 min post stimulation. Fig. S3A and S3B show that Bam32 pY and Bam32-Lck interaction follow similar kinetics, suggesting that Lck might be the kinase responsible for pY139 in T cells.
However given the late time course of Bam32 pY and the fact that Bam32-mediated Erk activation was observed in resting cells prior to any stimulation, we postulated that phosphorylation at the Bam32 Y139 residue would not be involved in the Erk pathway. As other phosphorylation pathways might be significant in Bam32-mediated Erk activation, we sought to identify additional potential phosphoresidues. One such residue, Bam32 S141 is a known site of phosphorylation on murine Bam32 identified in a mass spectral analysis (Cao et al., 2007).
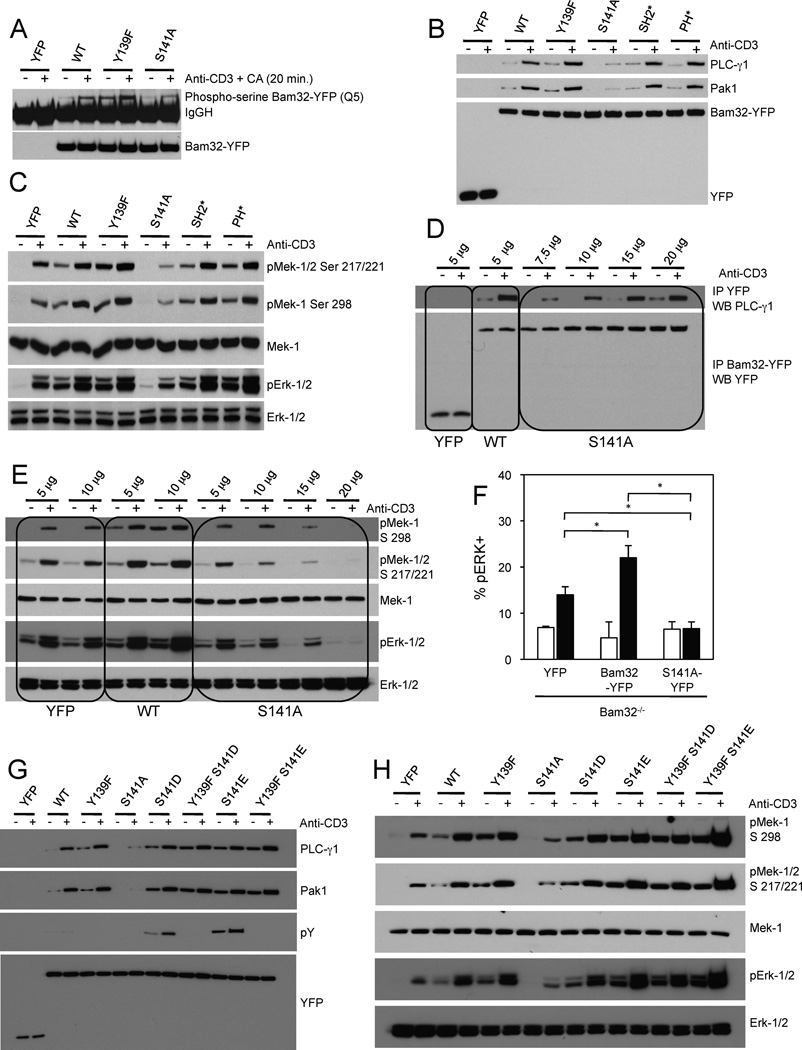
S141 is very well conserved among species though some species have T in this position, a conservative substitution (Fig. S3C). To better understand the roles of both Bam32 Y139 and S141 we introduced substitutions at these two residues. The corresponding Bam32-YFP mutants, Y139F and S141A, were overexpressed in Jurkat cells and were stimulated with αCD3 in combination with Calyculin A, a S/T phosphatase inhibitor (Fig. 3A). Bam32-YFP WT and mutants were immunoprecipitated and their phosphoserine status was analyzed by WB with an anti-pS mAb. As a specificity control, no pS was observed in cells transfected only with YFP. Interestingly WT Bam32-YFP was basally phosphorylated on S and the phosphorylation was greatly enhanced after cell stimulation. The Bam32 mutant S141A showed less than 50% phosphorylation than WT suggesting that S141 was the main acceptor for S phosphorylation. In addition, the mutant Y139F was more S-phosphorylated than WT suggesting that pY139 might negatively regulate pS of the nearby residue S141.
Figure 3. The relative importance of Bam32 Y139 and S141 for formation of the Bam32-PLC-γ1-Pak1 complex and αCD3-induced Erk activation.
(A) JE6.1 cells transfected with the indicated constructs (10 µg) were sorted for same YFP fluorescence intensity. Cells were treated or not with αCD3 + Calyculin A (CA) at 50 nM for 20 min. Lysates were subjected to a YFP IP. A representative WB is shown (n=3).
(B) Jurkat E6.1 cells were transfected with YFP, WT or mutants of Bam32-YFP cDNAs (all were 10 µg except mutant S141A, which was 12.5 µg). Lysates from cells ± αCD3 were subjected to a YFP IP. A representative WB is shown (n=3).
(C) Lysates (prepared in B) were studied by WB. A representative WB is shown (n=3).
(D) JE6.1 cells transfected with the indicated constructs ± αCD3 were lysed. Lysates were subjected to a YFP IP. A representative WB is shown (n=2).
(E) Transfection and stimulation as in (D). WCL were studied by WB. A representative WB is shown (n=2).
(F) Quantification of pErk-1/2 staining in YFP-gated Bam32−/− CD4+ T cells transfected with YFP, Bam32-YFP, or Bam32 S141A-YFP and stimulated with αCD3ε (1 µg/ml, 2 min.). The average and SD from 3 independent experiments are shown. * p<0.05.
(G) Jurkat E6.1 cells transfected with the indicated constructs (10 µg) were sorted for equivalent YFP expression. Lysates from cells ± αCD3 were subjected to a YFP IP. A representative WB is shown (n=2).
(H) Lysates prepared in G were analyzed by WB. A representative WB is shown (n=2).
Next, the mutants Y139F and S141A were studied for their ability to bind PLC-γ1 and Pak1 by co-IP. In addition to these two mutants, the Bam32-YFP mutant SH2* and the mutant PH* were also included (Fig. 3B). Because the transfection efficiency of each construct was different, we transfected different amounts of the plasmids to guarantee equal levels of protein expression (Fig. S3D). Neither a functional Bam32 SH2 domain nor PH domain was required for the formation of the Bam32-PLC-γ1-Pak1 complex. These two mutants increased Mek-1/Erk activation with the same magnitude as WT Bam32-YFP (Fig. 3C). As expected, PLC-γ1 and Pak1 bound the mutant with Y139F. Interestingly, almost no Bam32/PLC-γ1/Pak1 complex was immunoprecipitated when Bam32-YFP S141 was mutated to A indicating that this site is crucial for complex formation (Fig. 3B). The Y139F mutant increased Mek and Erk phosphorylation to the same extent as WT Bam32 whereas the mutant S141A partially decreased their activation (Fig. 3C). Weak binding between mutant S141A and PLC-γ1/Pak1 was detectable (Fig. 3B). Indeed when the mutant S141A was overexpressed 3.5 times more than WT, the amount of bound PLC-γ1 was approximately 80% of the amount bound to WT (Fig. 3D). The effect of the Bam32(S141A)/PLC-γ1/Pak1 complex on Mek-1/Erk activation was then studied (Fig. 3E–F). As more S141A mutant was expressed, decreased Mek-1/Erk activation was observed. Furthermore expression of WT Bam32-YFP favored PLC-γ1-Pak1 interaction, whereas the S141A mutant disrupted it (Fig. S3E).
The dominant negative effect of the S141A mutant was also studied in CD4+ T cells from Bam32 KO mice (Fig. 3F). WT Bam32-YFP and S141A Bam32-YFP were expressed in Bam32 KO CD4+ T cells and pErk was measured by flow cytometry. As expected WT Bam32-YFP increased the proportion of pErk-positive CD4+ T cells, whereas the S141A Bam32-YFP mutant, even at low levels of expression, inhibited pErk. The effect of Bam32 S141A mutation was further studied in Jurkat cells (Fig. S3F). WT Bam32-YFP and S141A mutant were expressed at two concentrations in Jurkat cells and the interaction between LAT and PLC-γ1 was studied by IP/WB. WT Bam32, which strongly binds PLC-γ1, disrupts LAT-PLC-γ1 interactions in a dose-dependent manner. Interestingly the S141A mutant, which weakly binds PLC-γ1, also disrupts LAT-PLC-γ1 interactions in a dose-dependent fashion, but to a lesser degree than WT Bam32 (Fig. S3F). Combined together, these results suggest that the binding of the S141A mutant to PLC-γ1 sequesters PLC-γ1 from LAT (Fig. S3F) and Pak1 (Fig. S3E). This Bam32 mutant disrupts both the LAT-PLC-γ1-DAG-RasGRP pathway and the PLC-γ1-Pak1 pathway. As a result, expression of the S141A mutant inhibits the Erk pathway by inhibiting, at the same time, two concurrent pathways both involving PLC-γ1. The dominant negative effect of the mutant S141A was also evaluated on pPak1. The mutant S141A inhibited Pak1 pSer199/204 (Fig. S3G).
Next we mutated S141 to amino acids that would be predicted to have phosphomimetic properties. We observed that the pY of mutants S141D and S141E was considerably higher in resting cells and was further enhanced after αCD3 stimulation (Fig. 3G). Having previously ruled out the contribution of Bam32 Y139 in the Erk pathway, we created double mutants (Y139F plus S141 phosphomimetic mutants) to further study the role of pS141 independent of pY139. Double mutants (Y139F/S141D and Y139F/S141E) were not Y phosphorylated demonstrating that Y139 is the only Y phosphorylation site on Bam32 in T cells. Both single and double mutants bound more PLC-γ1 and Pak1 than WT Bam32. A phosphomimetic at position 141 increased both Mek-1 and Erk activation whether Y139 was present or not (Fig. 3H).
The Bam32 pathway regulates Erk activation independently of the canonical LAT pathway
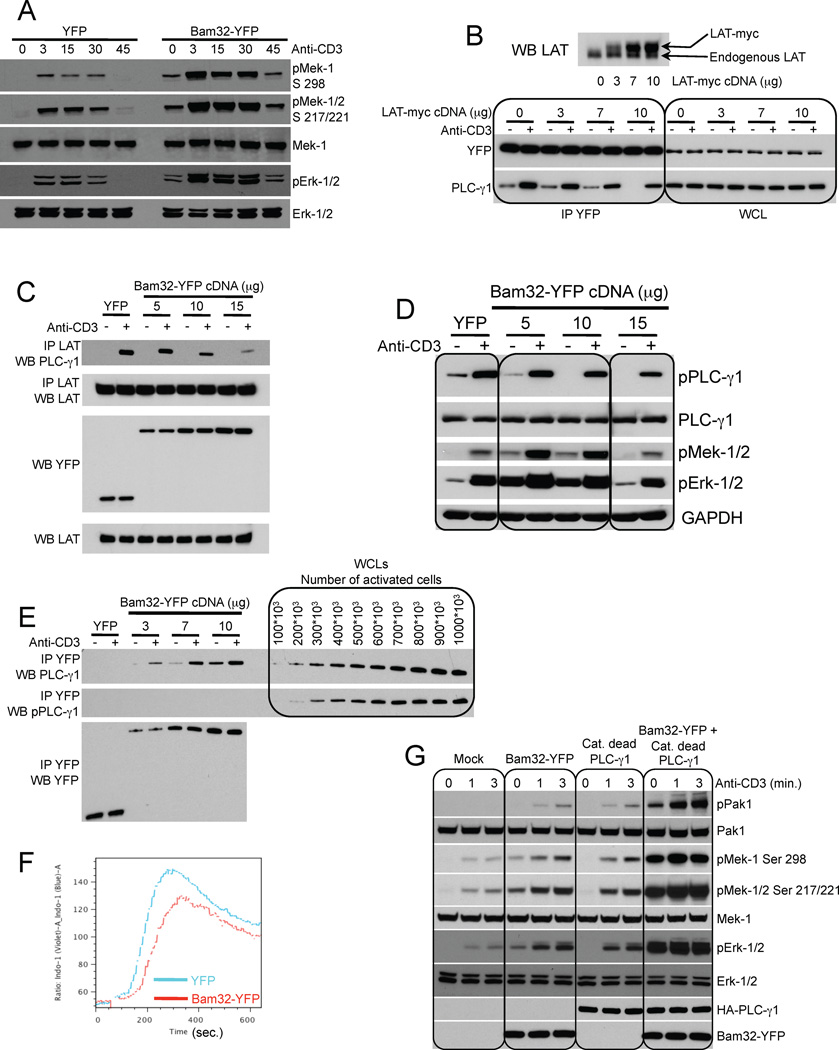
Extensive analysis has shown that PLC-γ1 enzymatic activity depends on its binding to LAT and SLP-76 (Balagopalan et al., 2010). However Pak1 regulation through an interaction with LAT or SLP-76 is controversial and has been questioned (Bubeck Wardenburg et al., 1998; Phee et al., 2005). We investigated whether Pak1 in complex with PLC-γ1 and Bam32 was regulated by LAT (Fig. 4) and/or SLP-76 (Fig. 5). Bam32 overexpression in Jurkat T cells deficient for LAT (JCam2.5) resulted in a robust increase in Mek and Erk activation demonstrating that the Bam32-PLC-γ1-Pak1 pathway is independent of LAT (Fig. 4A). Next we investigated the relationship of LAT-PLC-γ1 and Bam32-PLC-γ1. Increasing amounts of LAT-myc cDNA were cotransfected with constant amounts of Bam32-YFP cDNA. The amount of PLC-γ1 bound to Bam32-YFP was studied by co-IP. The more LAT-myc was overexpressed, the less PLC-γ1 bound Bam32-YFP before or after activation (Fig. 4B). Next increasing amounts of LAT-YFP were overexpressed and the impact of LAT-YFP on the interaction between Bam32 and PLC-γ1 was studied by GST affinity purification. Again, the more LAT-YFP was overexpressed, the less PLC-γ1 was available for binding to Bam32 (Fig. S4A, B). To confirm that LAT and Bam32 compete for PLC-γ1, we also performed the reciprocal experiment. Bam32-YFP was overexpressed in increasing amounts and the fraction of PLC-γ1 bound to LAT was studied by co-IP. The more Bam32-YFP was expressed, the less LAT bound PLC-γ1 (Fig. 4C). Together these results demonstrate a competition between Bam32 and LAT for PLC-γ1. Furthermore pPLC-γ1 decreased proportionally as the amount of Bam32-YFP expressed increased while Mek and Erk activation showed a biphasic response (Fig. 4D). Low and intermediate levels of Bam32-YFP (5 and 10 µg) increased Mek-1/Erk signaling whereas a higher dose did not (15 µg).
Figure 4. The Bam32 pathway regulates Erk activation independently of the canonical LAT pathway.
(A) JCam 2.5 cells stably transfected with either YFP or Bam32-YFP cDNAs were sorted for the same YFP intensity. Cells were stimulated or not with αCD3. WCL were analyzed by WB. A representative WB is shown (n=2).
(B) LAT-myc cDNA was overexpressed in JE6.1 cells stably expressing Bam32-YFP. Lysates from cells ± αCD3 were subjected to a YFP IP. WCL were run to monitor the input. Upper panel is a LAT WB showing the increase of LAT-myc in addition to endogenous LAT. A representative WB is shown (n=2).
(C, D) JE6.1 cells were transfected with YFP or increasing amounts of WT Bam32-YFP. Cells ± αCD3 were lysed. Lysates were split in half. Half of the lysates were subjected to a LAT IP (panel C) and the other half was used to make WCL (panel D). Samples were analyzed by WB. Representative WBs are shown (n=2).
(E) JE6.1 cells were transfected with YFP (7 µg) or increasing amounts of WT Bam32-YFP cDNAs. Cells ± strong αCD3 (10 µg/ml) were lysed. Lysates were subjected to a Bam32-YFP IP. The amount of PLC-γ1 bound to Bam32-YFP and its phosphorylation state were studied by WB. In parallel, JE6.1 cells were stimulated under the same conditions and lysed. Increasing amounts of this WCL were loaded on the same gel. A representative WB is shown (n=2).
(F) JE6.1 cells were transfected with YFP or WT Bam32-YFP cDNAs (10 µg). Cells loaded with Indo-1 AM were stimulated with αCD3 and cytosolic Ca2+ concentration was assayed by flow cytometry. Data are representative of 4 independent experiments.
(G) JE6.1 cells were transfected with WT Bam32-YFP, catalytically dead PLC-γ1 or both (7.5 µg). Cells were stimulated or not with αCD3 (500 ng/ml, 3 min.). WCL were analyzed by WB. A representative WB is shown (n=2).
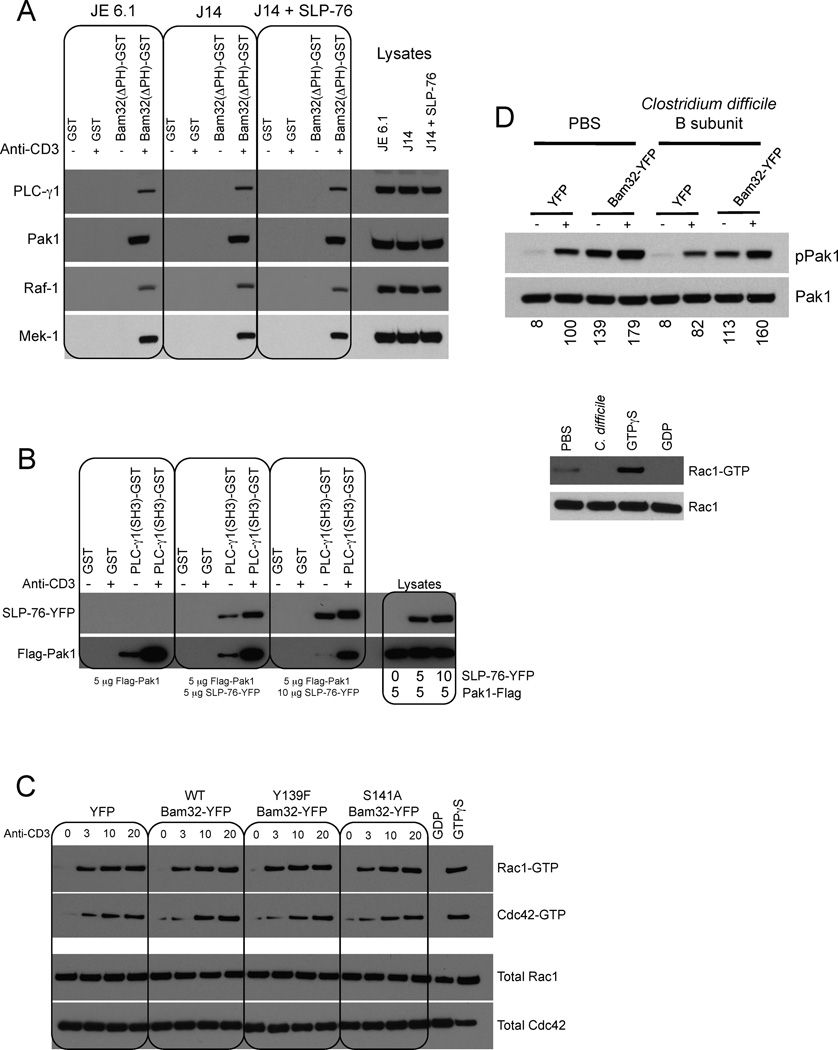
Figure 5. Pak1 in the Bam32-PLC-γ1-Pak1 complex is not regulated by SLP-76 nor by Rac1.
(A) JE6.1, J14 and J14 cells expressing SLP-76-YFP ± αCD3 were lysed. Extracts were incubated with GST or Bam32(ΔPH)-GST proteins. Bound proteins were subjected to WB. The interaction of Bam32 with PLC-γ1 and Pak1 was studied using 1 mg lysate. Interaction between Bam32 and Raf-1 and Mek-1 was studied using 2 mg extracts. A representative WB is shown (n=2).
(B) JE6.1 cells were transfected with 5 µg of Flag-Pak1 cDNA ± SLP-76-YFP cDNA at 5 or 10 µg. Cells ± αCD3 were lysed. Extracts were incubated with GST or (SH3)PLC-γ1-GST proteins. Bound proteins were analyzed by WB. A representative WB is shown (n=2).
(C) JE6.1 cells transfected with the indicated constructs (10 µg) were sorted for equal YFP fluorescence intensity. Cells ± αCD3 were lysed. Rac1-GTP and Cdc42-GTP pulldown assays were carried out using Pak1-CRIB-GST. GTPγS- and GDP-loaded lysates were used as controls. Total Rac1 and Cdc42, and active Rac1 and Cdc42, were detected by WB. A representative WB is shown (n=2).
(D) JE6.1 cells transfected with YFP or Bam32-YFP cDNAs (10 µg) and pretreated or not with C. difficile toxin B (100 ng/ml, 1h) were stimulated with αCD3. WCL were analyzed by WB. Densitometric analysis was done using ImageJ and results are expressed as percentages of “stimulated YFP cells” condition. Efficiency of the toxin treatment was verified with a Rac1-GTP pulldown assay. A representative WB is shown (n=2).
Next we interrogated the phosphorylation state of the PLC-γ1 molecules bound to Bam32-YFP. Increasing amounts of Bam32-YFP cDNA were transfected and cells were stimulated with saturating doses of αCD3 (Fig. 4E). For comparison, increasing amounts of cells stimulated under the same conditions were loaded onto the same gel. As expected, the more Bam32-YFP was overexpressed, the more PLC-γ1 associated with it. The fraction of PLC-γ1 bound to Bam32-YFP was not phosphorylated (Fig. 4E). Cells overexpressing Bam32-YFP fluxed less Ca2+ than control cells in response to αCD3 stimulation (Fig. 4F) indicating that Bam32 inhibits PLC-γ1 activity. We also tested whether a catalytically dead PLC-γ1 could synergize with Bam32 to activate Pak1 and downstream Mek-1 and Erk (Fig. 4G). To inhibit PLC-γ1 lipase activity, H335, a critical residue in the split catalytic domain X, was mutated to F. Cells were stimulated with a low dose of αCD3 for short time periods because activated Pak1 is unstable and undergoes rapid degradation. Bam32-YFP and inactive PLC-γ1-HA synergized resulting in enhanced pPak1. The extra band above Pak1 represents super-activated Pak1 with a mobility shift (Strochlic et al., 2010). Downstream of Pak1, Mek-1 and Erk were also strongly activated by the coexpression of Bam32-YFP and catalytically inactive PLC-γ1. To summarize this set of experiments, we demonstrated that Bam32 diverts PLC-γ1 away from LAT. PLC-γ1 in the Bam32-PLC-γ1-Pak1 complex is inactive, but has scaffolding ability and activates Pak1. To further validate this model, we asked whether Pak1 overexpression could mimic the effect of Bam32 on LAT-PLC-γ1 interaction and PLC-γ1 activity. Increasing Pak1 partially disrupted the LAT-PLC-γ1 complex (Fig. S4C). As a result, pPLC-γ1 (Fig. S4D) and Ca2+ flux (Fig. S4E) were reduced.
Pak1 in the Bam32-PLC-γ1-Pak1 complex is not regulated by SLP-76
The potential for SLP-76 regulation of the Pak1 present in Bam32-PLC-γ1-Pak1 complexes is of importance as some models postulate regulation of total Pak1 by SLP-76 (Bubeck Wardenburg et al., 1998). Therefore we tested whether the interactions between Bam32, Pak1 and the components of the Erk cascade downstream of Pak1 depended on SLP-76 (Fig. 5A). We performed a pulldown assay in Jurkat cells deficient for SLP-76 (J14 cells). PLC-γ1 and Pak1 interacted with Bam32 whether SLP-76 was expressed or not (Fig. 5A). Downstream of Pak1, both Raf-1 and Mek-1 were associated with Bam32 independently of SLP-76 expression. A pulldown assay was performed in cells transfected with Pak1 siRNAs. The interaction between Bam32(ΔPH) and Mek-1 is mediated by Pak1 (Fig. S5A). Indeed, the interaction was lost in cells in which Pak1 expression was reduced to 8% after Pak1 siRNA treatment. The HPK1-Bam32 interaction was not affected by Pak1 reduction.
We then questioned whether Pak1 would compete with SLP-76 for PLC-γ1 binding in view of the observation that the interaction between Pak1 and PLC-γ1 requires an intact PLC-γ1 SH3 domain as does the interaction of PLC-γ1 with a proline-rich domain in SLP-76 (Yablonski et al., 2001). Increasing amounts of SLP-76-YFP cDNA were cotransfected with a constant amount of Flag-Pak1 and a pulldown was performed using (SH3)PLC-γ1-GST (Fig. 5B). With increased SLP-76-YFP overexpression, greater SLP-76-YFP-PLC-γ1 binding was observed, whereas binding between PLC-γ1 and Pak1 was reduced. Reciprocally, the more Flag-Pak1 was overexpressed, the less the SH3 domain of PLC-γ1 was able to bind SLP-76-YFP (Fig. S5). From these results, we conclude that Pak1 and SLP-76 compete for the SH3 domain of PLC-γ1. This result demonstrates that Bam32-PLC-γ1-Pak1 and LAT-SLP-76-PLC-γ1 complexes are distinct.
The Bam32-PLC-γ1-Pak1 complex is not regulated by Rac1
The interplay between Pak1 in the Bam32-PLC-γ1-Pak1 complex and Rac1/Cdc42 was studied, as Bam32 activates Rac1 in B cells (Allam et al., 2004). Notably in B cells Bam32-mediated Rac1 activation requires both the Bam32 Y139 and the Bam32 SH2 domain. We observed in T cells that the Bam32 Y139F mutant increased pPak1 (Fig. S3D). This correlates with the fact that in T cells, neither the mutation of Bam32 Y139 nor the mutation of its SH2 domain had a negative impact on Bam32-mediated Erk activation (Fig. 3C). These results strongly suggest that it is unlikely that Bam32 activates Pak1 via Rac1. This hypothesis was confirmed in Fig. 5C as we did not detect an increase in Rac1-GTP after Bam32 overexpression in T cells. We then used Clostridium difficile toxin B, a Rac1 and Cdc42 inhibitor, to study Pak1 activation in Jurkat cells. Treatment efficiency was verified with a Rac-1-GTP pulldown experiment. Cells expressing only the YFP control and treated with the toxin exhibited only 18% less phospho-Pak1 than non-treated YFP cells (Fig. 5D) suggesting that the conventional pathway, in which Rac1 and Cdc42 are the main Pak1 activators, is not preponderant in T cells. Importantly the impact of Bam32-YFP on pPak1 at 3 min. was almost the same whether the cells were treated with the toxin or not. Indeed Bam32-YFP overexpression induced a 177% increase of pPak1 in non-treated cells (YFP cells compared to Bam32-YFP cells), whereas the increase due to Bam32-YFP was 193% in toxin-treated Jurkat cells. Thus Pak1 molecules in the Bam32-PLC-γ1-Pak1 complex are not regulated by Rac1/Cdc42.
PLC-γ1 dissociates inactive Pak1 dimers into monomers allowing their subsequent autophosphorylation
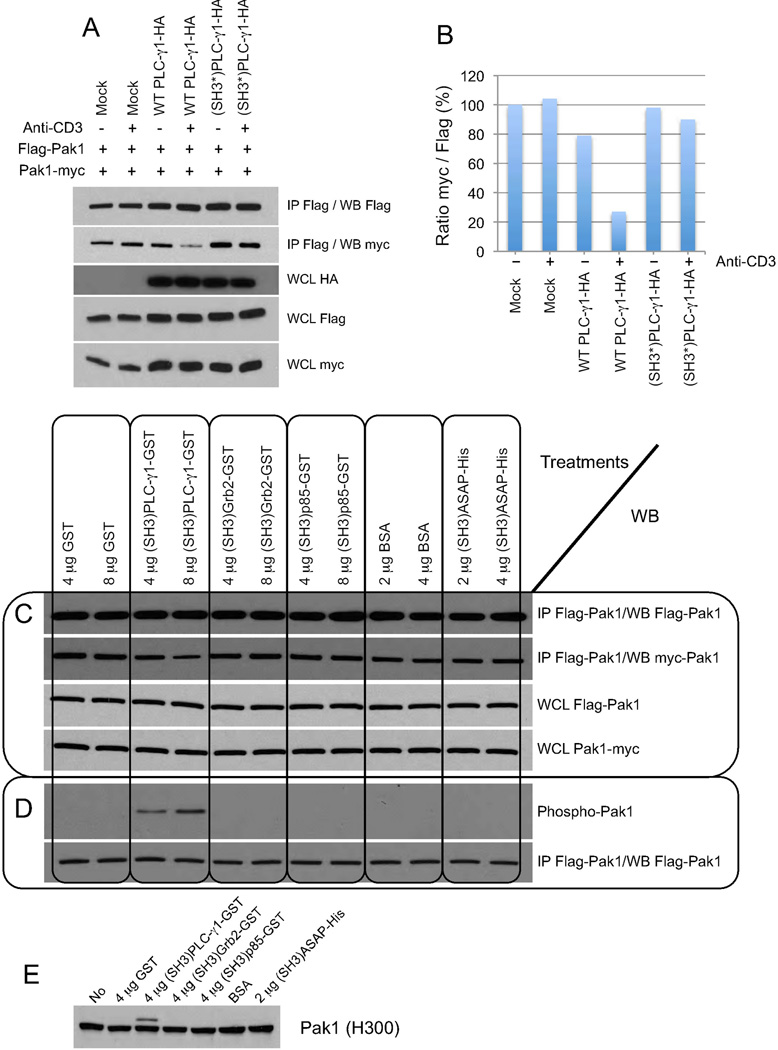
Pak1 forms homodimers and dimerized Pak1 adopts a trans-inhibited conformation in which the inhibitory portion of one Pak1 in the dimer binds and inhibits the catalytic domain of the other. The consensus model for Pak1 activation is that interaction with an active GTPase results in a change in conformation and activation of both Pak1 molecules (Parrini et al., 2002). Because we previously showed that Pak1 in the Bam32-PLC-γ1-Pak1 complex is not regulated by small GTPases, we asked whether PLC-γ1 instead of a GTPase could induce the dissociation of Pak1 dimers. In Fig. 6A, Jurkat cells were cotransfected with Flag-Pak1 and Pak1-myc in combination with WT HA-PLC-γ1 or (SH3*)-HA-PLC-γ1, which cannot bind Pak1 (Fig. 6A). We confirmed the dimerization of Pak1 by co-IP. We also observed that PLC-γ1-HA approximately doubles the expression of Pak1 (tagged with either Flag or myc) possibly via a previously described mechanism involving enhanced transcription (Fig. S6 A, B) (Huang et al., 1995) and that it substantially disrupts the interaction between the two tagged Pak1. Indeed despite an increased level of Pak1-Flag directly precipitated in cells overexpressing PLC-γ1-HA, less Pak1-myc was detected in the co-IP (see Fig. 6A and quantification in Fig. 6B). As expected (SH3*)PLC-γ1-HA, which cannot bind Pak1, had no effect on Pak1 dimerization. αCD3 stimulation had only a modest effect on Pak1 dissociation likely because we had to use a very low amount of OKT3 (300 ng/ml) to avoid Pak1 degradation, an event that follows its activation (Weisz Hubsman et al., 2007). To conclude, we demonstrated that PLC-γ1 disrupts Pak1 dimers in vivo.
Figure 6. PLC-γ1 binding converts inactive Pak1 dimers into monomers allowing their subsequent autophosphorylation.
(A) JE6.1 cells were transfected with WT PLC-γ1-HA or (SH3*)PLC-γ1-HA cDNAs (5 µg each) + the mixture Flag-Pak1 (5 µg) + Pak1-myc (5 µg). Cells ± αCD3 (500 ng/ml, 2 min.) were lysed. Lysates were subjected to Flag IP. The presence of Flag-Pak1 and bound Pak1-myc was studied by WB. A representative WB is shown (n=2).
(B) Densitometric analysis of the WB in (A).
(C) JE6.1 cells were transfected with Flag-Pak1 + Pak1-myc (5 µg each) and lysates were subjected to Flag IP. IPs were incubated with (SH3)PLC-γ1-GST or other SH3 domains from various proteins. Presence of Flag-Pak1 and bound Pak1-myc was studied by WB. WCL were analyzed by WB. A representative WB is shown (n=2).
(D) JE6.1 cells were transfected with Flag-Pak1 (10 µg) and lysates were subjected to Flag IP. IPs were washed with cold kinase assay buffer. Buffer containing (SH3)PLC-γ1-GST protein or other SH3 domains from various proteins were exchanged against cold kinase assay buffer. Flag-Pak1 IPs were incubated with (SH3)PLC-γ1-GST protein or the other SH3 domains in cold kinase assay buffer containing 2X protease inhibitors for 1h at RT. Samples were studied by WB. A representative WB is shown (n=2).
(E) JE6.1 cells were lysed in 1% Triton X-100 containing 2X protease inhibitors. WCL were incubated with (SH3)PLC-γ1-GST or various SH3 domains for 10 min. at 37°C. Aliquots were subjected to SDS-PAGE and the membrane was blotted with αPak1 Ab (H300, Santa-Cruz). A representative WB is shown (n=4).
Next we confirmed the Pak1 dissociation by PLC-γ1 using an in vitro assay (Fig. 6C). Flag-Pak1 and Pak1-myc cDNAs were cotransfected and Flag-Pak1 immunoprecipitates were incubated with increasing amounts of either (SH3)PLC-γ1-GST or other SH3 domains from various proteins. Residual Pak1-myc bound to Flag-Pak1 was detected by blotting. The direct binding of (SH3)PLC-γ1-GST to Pak1 specifically disrupted Pak1 dimers in a dose-dependent manner (for densitometric analysis see Fig. 6C). The other SH3 domains tested had no effect on Pak1 dimerization. Next we investigated the phosphorylation of Pak1 monomers. Flag-Pak1 immunoprecipitates were incubated with either (SH3)PLC-γ1-GST or other SH3 domains in cold kinase assay buffer (Fig. 6D). The direct binding of (SH3)PLC-γ1-GST to Pak1 specifically induced Pak1 autophosphorylation on S199/204. Pak1 autophosphorylation was proportional to the amount of PLC-γ1 incubated with Pak1 immunoprecipitates. We also incubated Triton X-100 lysates with either (SH3)PLC-γ1-GST or other SH3 domains and we observed that (SH3)PLC-γ1-GST specifically induced a molecular weight shift of Pak1 molecules generating a species referred to as super-activated Pak1 (Fig. 6E). Jurkat lysates were then incubated with (SH3)PLC-γ1-GST to activate Pak1 in vitro (Fig. S6D), and the GST fusion protein was precipitated using glutathione-coated beads. The ratio of shifted-Pak1/unshifted Pak1 is greater in the WCL than in the pull-down. It has previously been shown that Nck and PIX bind polyphosphorylated Pak1 with lower affinity than nonphosphorylated Pak1 (Zhao et al., 2000).
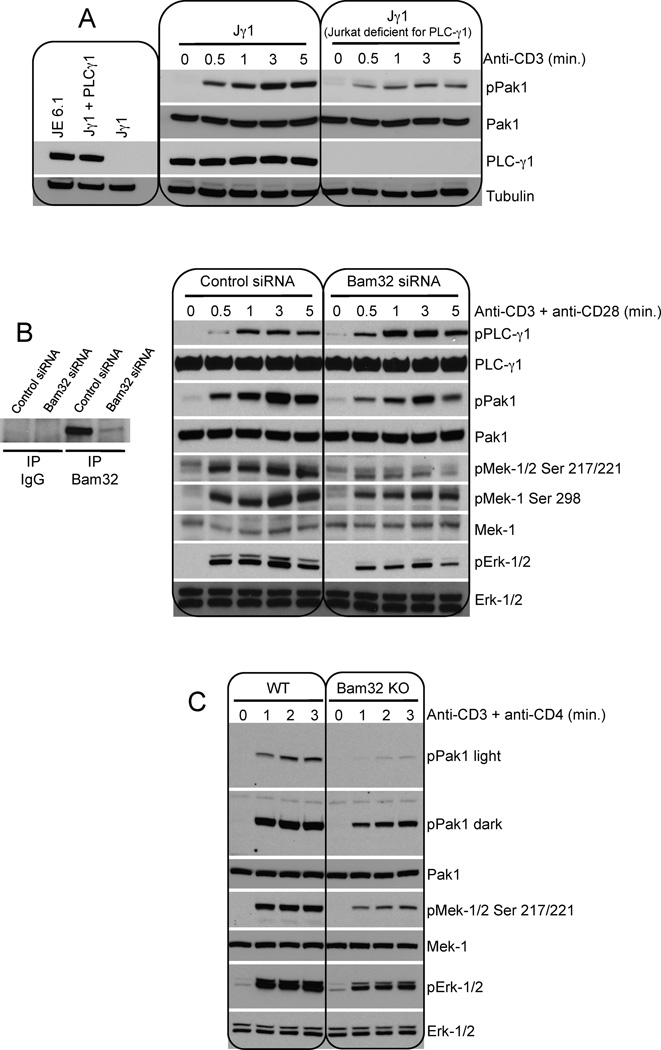
The importance of PLC-γ1 for pPak1 was confirmed by the following experiment. Jurkat cells deficient for PLC-γ1 (Jγ1 cells) compared to Jγ1 cells reexpressing PLC-γ1 show a great reduction in Pak1 activation (Fig. 7A). Densitometric analysis demonstrated that pPak1 was down 3-fold in the absence of PLC-γ1.
Figure 7. Bam32 deficiency in human and mouse CD4+ T cells considerably reduces the activation of Pak1 and downstream Mek and Erk.
(A) Jγ1 (Jurkat deficient for PLC-γ1) and Jγ1 reconstituted with PLC-γ1 were stimulated with αCD3. WCL were analyzed by WB. A representative WB is shown (n=3).
(B) Human CD4+ T cells were transfected with Bam32 siRNAs or control siRNAs and were maintained in culture media without IL-2. CD4+ T cells were stimulated with αCD3 (5 µg/ml) + αCD28 (10 µg/ml). WCL were analyzed by WB. A representative WB is shown (n=2).
(C) CD4+ T cells from WT or Bam32−/− mice were stimulated with αCD3ε (1 µg/ml, 2 min.). WCL were analyzed by WB. A representative WB is shown (n=2).
Bam32 deficiency in human and mouse CD4+ T cells reduces the activation of Pak1 and downstream Mek and Erk
To confirm that Bam32 plays a critical role in the activation of Pak1 and downstream Mek and Erk in normal, non-transformed cells, we used siRNAs to knockdown Bam32 in human CD4+ T cells and we studied mouse CD4+ T cells from Bam32 KO mice. Human CD4+ T cells were purified and endogenous Bam32 was targeted by siRNAs. Bam32 expression was reduced to 11% after siRNA treatment. Bam32 depletion greatly enhanced pPLC-γ1 after αCD3/CD28 co-stimulation confirming that Bam32 inhibited the activation of the phospholipase. As expected Bam32 depletion also considerably reduced Pak1 activation. Downstream of Pak1, Mek-1 and Erk activation were also greatly reduced in T cells transfected with Bam32 siRNAs (Fig. 7B). Thus in these cells as well, Bam32 expression determines the balance between two competing pathways as mediated by levels of two different protein complexes, LAT-PLC-γ1 versus PLC-γ1-Pak1. Finally mouse CD4+ T cells from Bam32 KO mice compared to WT cells displayed a significant reduction in Pak1 activation. Downstream Mek and Erk activation were also significantly impaired in CD4+ T cells from Bam32 KO mice (Fig. 7C).
DISCUSSION
In this study we characterized the Bam32-PLC-γ1-Pak1 pathway that is, in part, responsible for Raf-1, Mek and Erk activation downstream of the TCR (see Fig. S7 for model). PLC- γ1 serves as a catalytically inactive scaffold and as such is distinct from the classical LAT-PLC-γ1-DAG-RasGRP-Ras-Erk pathway in which PLC-γ1 is catalytically active. Additionally Pak1 activation by PLC-γ1 is different from the other published mechanisms that activate Pak1 in T cells as it is Rac1/Cdc42-independent. Interestingly we observed that Bam32-PLC-γ1-Pak1 complexes compete with Bam32-PLC-γ1-HPK1 complexes. Pak1 is related to HPK1, which is, in contrast to Pak1, a negative regulator of Erk activation (Shui et al., 2007). The competition between these two complexes could fine tune Erk activation. Erk activation by the Bam32-PLC-γ1-Pak1 pathway is Ras-independent.
We do not wish to underestimate the considerable work that has shown the importance of Ras in Erk signaling in T cells as it explains to a great extent how a robust Erk signal can be generated. Instead, our work reveals additional complexity to the current pathway for Erk activation in T cells. We believe that our study of the Bam32-PLC-γ1-Pak1 complex supports the existence of an additional Erk pathway needed to generate Erk activation even in the presence of Ras desensitization. It is known that Erk activation leads to a negative feedback loop for Ras activation by at least two distinct mechanisms. (1) The activation of the Ras-Erk pathway results in the phosphorylation of SOS and subsequent dissociation of SOS from Grb2 (Langlois et al., 1995). (2) The Ras-Erk pathway results in phosphorylation of LAT at T155, and T155 phosphorylation decreases the affinity of PLC-γ1 for LAT (Matsuda et al., 2004). As a consequence, it attenuates the signaling events downstream of PLC-γ1, including the activation of Erk itself by inhibiting the PLC-γ1-DAG-RasGRP pathway. An additional Ras-independent Erk pathway would help to provide a more robust signal to Erk in this setting. The significance of the additional Bam32-mediated Ras-independent pathway can be inferred by examination of Erk responses in Bam32 KO T cells (Han et al., 2003; Sommers et al., 2008) and in our experiments in which Bam32 or Pak1 deficiency depresses Erk activity in human and mouse T cells (Fig. 1A, 1G, 1H, 7B, 7C).
We demonstrated that the interaction in the Bam32-PLC-γ1-Pak1 complex between Bam32 S141 and the PLC-γ1 C-SH2 domain is direct using far-western blotting. In addition to pY-interactions, pS/T- and E-dependent binding to SH2 domains in several other molecules have been reported. Bcr contains two S/T- and E-rich regions that bind the c-Abl SH2 domain (Pendergast et al., 1991) (E mimicking a pS/T motif). The specific binding of the S/T kinase p130PITSLRE to the SH2 domain of Blk is mediated by a p130PITSLRE pS and E-rich cluster of amino acids. Some SH2 domains bind neither pY nor pS/T or E but nonphosphorylated peptides. For example, the C-SH2 domain of PLC-γ1 binds non-phosphorylated Itk (Min et al., 2009). Therefore although our description of a SH2 domain-pS interaction is unusual, it is not unprecedented.
Interestingly expression of Bam32 activated Raf-1, Mek-1 and Erk prior to cell stimulation and αCD3 treatment increased the effect of Bam32. In accordance with this observation, Bam32 exhibits low baseline levels of phosphorylated S141 and TCR stimulation greatly enhanced Bam32 pS. Bam32 S141 is crucial for the integrity of the Bam32-PLC-γ1-Pak1 complex and for the activation of Erk. Mutation of the phosphorylatable Y139 of Bam32 does not disrupt the Bam32-PLC-γ1 interaction nor impair Bam32-induced Erk activation. However pY139 and pS141 are interdependent, as our results suggest that pS141 increases pY139 whereas pY139 decreases pS141. This negative feedback loop could represent a fine-tuning of MAPK activation by Bam32. Bam32 with a phosphomimetic at S141 bound PLC-γ1 better than WT Bam32 and also increased Erk activity.
We showed that pPLC-γ1 decreased proportionally with Bam32 expression and that Mek and Erk activation is biphasic. The MAPKs are strongly activated when Bam32 is expressed at low or intermediate levels and the increase is lost with higher doses of Bam32. At this point, this biphasic response can be interpreted in at least two different ways: (1) as Bam32 amounts increase relative to PLC-γ1 and Pak1, the Bam32-PLC-γ1-Pak1 trimolecular complex may be titrated away in favor of bimolecular complexes. This is consistent with the model that a scaffolding protein, if present in excess of its constituent binding partners, impairs signaling by assembling non-productive complexes (Scott and Pawson, 2009); (2) the increase in Pak1 activation is proportional to the amount of Bam32 but at some point the PLC-γ1-Pak1 pathway likely cannot compensate for the strong decrease in MAPK activation from the PLC-γ1-DAG pathway.
The Bam32-PLC-γ1-Pak1 pathway and the coincident LAT-PLC-γ1-DAG-RasGRP pathway are competing pathways both utilizing PLC-γ1. In addition to its role in Pak1 dissociation/activation, PLC-γ1, because of its PH domain, might localize Bam32-PLC-γ1-Pak1 complexes to membranes enriched in PIP2. Because the PLC-γ1 in these complexes is catalytically inactive, it cannot hydrolyze PIP2. Thus the accumulation of PIP2, an essential coactivator for Pak1, would result in stronger Pak1 activation (Strochlic et al., 2010).
Two separate pathways that activate Pak1 in T cells have been described. The first one involves the adapters LAT, SLP-76, and Nck, which synergistically activate Vav1, a GEF for Rac1/Cdc42 (Bubeck Wardenburg et al., 1998). We observed that Pak1 and SLP-76 compete for binding to PLC-γ1. Thus SLP-76 is unlikely to regulate Pak1 in the Bam32-PLC-γ1-Pak1 pathway, a conclusion supported by the presence of this complex in cells lacking SLP-76. The second pathway uses the adapter GIT and PIX, another GEF for Rac1 and Cdc42 (Phee et al., 2005). These two pathways may regulate different subsets of Pak1. Those studies were focused on Pak1 but by extrapolation the two pathways can be applied to Pak2. Thus the Bam32-PLC-γ1 pathway is unique for two reasons, first it specifically activates Pak1 and not Pak2 and second the activation is PIX-, Nck-, and Rac1/Cdc42-independent.
We demonstrated that the SH3 domain of PLC-γ1 disrupts Pak1 dimers in vivo and in vitro and allows their subsequent autophosphorylation on S199/204. Phosphorylation on S in the regulatory portion, such as S199/204, may block Pak1 reassociation. It is noteworthy that pS199 decreases by 6-fold the affinity of Pak1 for PIX (Mott et al., 2005) suggesting why the Pak1 molecules in the Bam32-PLC-γ1-Pak1 complex do not bind PIX. We thus propose that PLC-γ1 acts in place of the small GTPases and its binding dirupts Pak1 dimers to activate Pak1.
The importance of the pathway we have described for Erk activation in T cells is supported by numerous observations: (1) Erk activation is enhanced by Bam32 overexpression and diminished in cells deficient for Bam32, (2) Erk activation is diminished in PLC-γ1- or Pak1-deficient cells, (3) almost 20% of Pak1 is associated with PLC-γ1 in resting Jurkat cells, (4) disruption of the Bam32-PLC-γ1-Pak1 complex, using Bam32 mutants, diminished Erk activation, and (5) Bam32-mediated Erk activation is important for TCR-induced cytokine production and proliferation of normal T cells, and for the progression of disease in a murine lymphoproliferation model (Miyaji et al., 2009). Studies exploring the TCR regulation of this pathway are underway. Additionally, the discovery of the Bam32-PLC-γ1-Pak1-Erk pathway highlights the potential for finding novel strategies to limit Pak1 oncogenicity (Dummler et al., 2009).
EXPERIMENTAL PROCEDURES
An overview of experimental procedures is provided below. For additional details see Supplemental Experimental Procedures.
Cell lines, cell culture, plasmids, siRNA reagents, transfection and cell stimulation
Most of the experiments were carried out using WT Jurkat E6.1 cells or Jurkat variants described elsewhere. Cells were cultured under standard conditions in RPMI 1640. Plasmids were described elsewhere or were purchased from GeneCopoeia. Protein expression was studied 20h post-transfection. siRNA reagents were from Dharmacon. Protein KD was studied 48h post-transfection. Cells were transfected with the Lonza system. More details are available in the Supplemental Experimental Procedures. Except when specified differently, Jurkat cells were stimulated with αCD3 mAb OKT3 for 3 min. at 2 µg/ml.
Human CD4+ T cell isolation and preparation and utilization of murine CD4+ T cells
Human PBMCs were isolated from buffy coats by centrifugation on a Histopaque gradient. CD4+ T cells were isolated by negative selection using an EasySep CD4+ T cell enrichment cocktail. CD4+ T cells stimulated with PHA and IL-2 were starved for 48h prior to utilization. More details are available in the Supplemental Experimental Procedures.
Ca2+ flux measurement
Ca2+ flux measurement was made using Indo-1-AM; cells were analyzed by flow cytometry. More details are in the Supplemental Experimental Procedures.
Mutagenesis
To generate Bam32 mutants we used the QuickChange II site directed mutagenesis kit (Agilent Stratagene, 200524-5). Mutants were verified by sequencing. Primers are available in the Supplemental Experimental Procedures.
Expression and purification of GST fusion proteins
Expression and purification of Bam32(ΔPH)-GST fusion protein and GST alone were achieved according to the basic protocol 1 (Harper and Speicher, 2008).
GST pulldown assays
GST pulldown assays were carried out as described in Nature Methods, vol. 1, no. 3, December 2004.
Detection of active Ras, Rac1, and Cdc42
Detection of active Ras, Rac1, and Cdc42 was achieved using kits from Thermo Scientific (respectively cat. numbers 16117, 16118, and 16119)
Immunoprecipitation, SDS-PAGE and WB
Briefly antibodies were incubated with Protein A/G-Agarose beads. Cells were lysed with 1% Brij 97/0.5% octylglucoside. Pre-cleared lysates were incubated with the beads coated with antibodies. Immunoprecipitates were analyzed by SDS-PAGE and WB. A detailed protocol is available in the Supplemental Experimental Procedures.
Kinase assays and phospho-Erk measurement via flow cytometry
Raf-1 and Pak1 kinase assay protocols and the quantification of pErk by flow cytometry are detailed in the Supplemental Experimental Procedures.
Supplementary Material
Highlights.
-
●
Bam32-mediated Erk activation is LAT/SLP-76- and Ras-independent
-
●
Bam32-PLC-γ1-Pak1 complexes activate Raf-1, Mek-1 and Erk downstream of Pak1
-
●
Bam32 S141 is crucial for Bam32-PLC-γ1-Pak1 complex formation and for Erk activation
-
●
Independently of Rho GTPases, PLC-γ1(SH3) dissociates/activates Pak1 dimers
ACKNOWLEDGEMENTS
We thank B. Rellahan and K. DeBell for PLC-γ1 constructs, B. Taylor for cell sorting, C. Regan and C. Hatheway for technical help. This research was supported by the Intramural Research Program of the NIH, NCI, CCR.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTAL INFORMATION
Supplemental Information includes 7 Fig., Supplemental Experimental Procedures with 2 tables, and Supplemental References.
REFERENCES
- Allam A, Marshall AJ. Role of the adaptor proteins Bam32, TAPP1 and TAPP2 in lymphocyte activation. Immunol Lett. 2005;97:7–17. doi: 10.1016/j.imlet.2004.09.019. [DOI] [PubMed] [Google Scholar]
- Allam A, Niiro H, Clark EA, Marshall AJ. The adaptor protein Bam32 regulates Rac1 activation and actin remodeling through a phosphorylation-dependent mechanism. J Biol Chem. 2004;279:39775–39782. doi: 10.1074/jbc.M403367200. [DOI] [PubMed] [Google Scholar]
- Balagopalan L, Coussens NP, Sherman E, Samelson LE, Sommers CL. The LAT story: a tale of cooperativity, coordination, and choreography. Cold Spring Harb Perspect Biol. 2010;2 doi: 10.1101/cshperspect.a005512. a005512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokoch GM, Wang Y, Bohl BP, Sells MA, Quilliam LA, Knaus UG. Interaction of the Nck adapter protein with p21-activated kinase (PAK1) J Biol Chem. 1996;271:25746–25749. doi: 10.1074/jbc.271.42.25746. [DOI] [PubMed] [Google Scholar]
- Bubeck Wardenburg J, Pappu R, Bu JY, Mayer B, Chernoff J, Straus D, Chan AC. Regulation of PAK activation and the T cell cytoskeleton by the linker protein SLP-76. Immunity. 1998;9:607–616. doi: 10.1016/s1074-7613(00)80658-5. [DOI] [PubMed] [Google Scholar]
- Cao L, Yu K, Banh C, Nguyen V, Ritz A, Raphael BJ, Kawakami Y, Kawakami T, Salomon AR. Quantitative time-resolved phosphoproteomic analysis of mast cell signaling. J Immunol. 2007;179:5864–5876. doi: 10.4049/jimmunol.179.9.5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dower NA, Stang SL, Bottorff DA, Ebinu JO, Dickie P, Ostergaard HL, Stone JC. RasGRP is essential for mouse thymocyte differentiation and TCR signaling. Nat Immunol. 2000;1:317–321. doi: 10.1038/79766. [DOI] [PubMed] [Google Scholar]
- Downward J, Graves JD, Warne PH, Rayter S, Cantrell DA. Stimulation of p21ras upon T-cell activation. Nature. 1990;346:719–723. doi: 10.1038/346719a0. [DOI] [PubMed] [Google Scholar]
- Dummler B, Ohshiro K, Kumar R, Field J. Pak protein kinases and their role in cancer. Cancer Metastasis Rev. 2009;28:51–63. doi: 10.1007/s10555-008-9168-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost JA, Steen H, Shapiro P, Lewis T, Ahn N, Shaw PE, Cobb MH. Cross-cascade activation of ERKs and ternary complex factors by Rho family proteins. EMBO J. 1997;16:6426–6438. doi: 10.1093/emboj/16.21.6426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han A, Saijo K, Mecklenbrauker I, Tarakhovsky A, Nussenzweig MC. Bam32 links the B cell receptor to ERK and JNK and mediates B cell proliferation but not survival. Immunity. 2003;19:621–632. doi: 10.1016/s1074-7613(03)00275-9. [DOI] [PubMed] [Google Scholar]
- Harper S, Speicher DW. Expression and purification of GST fusion proteins. Curr Protoc Protein Sci. 2008;Chapter 6(Unit 6):6. doi: 10.1002/0471140864.ps0606s52. [DOI] [PubMed] [Google Scholar]
- Huang PS, Davis L, Huber H, Goodhart PJ, Wegrzyn RE, Oliff A, Heimbrook DC. An SH3 domain is required for the mitogenic activity of microinjected phospholipase C-gamma 1. FEBS Lett. 1995;358:287–292. doi: 10.1016/0014-5793(94)01453-8. [DOI] [PubMed] [Google Scholar]
- Ku GM, Yablonski D, Manser E, Lim L, Weiss A. A PAK1-PIX-PKL complex is activated by the T-cell receptor independent of Nck, Slp-76 and LAT. EMBO J. 2001;20:457–465. doi: 10.1093/emboj/20.3.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langlois WJ, Sasaoka T, Saltiel AR, Olefsky JM. Negative feedback regulation and desensitization of insulin- and epidermal growth factor-stimulated p21ras activation. J Biol Chem. 1995;270:25320–25323. doi: 10.1074/jbc.270.43.25320. [DOI] [PubMed] [Google Scholar]
- Manser E, Loo TH, Koh CG, Zhao ZS, Chen XQ, Tan L, Tan I, Leung T, Lim L. PAK kinases are directly coupled to the PIX family of nucleotide exchange factors. Mol Cell. 1998;1:183–192. doi: 10.1016/s1097-2765(00)80019-2. [DOI] [PubMed] [Google Scholar]
- Marshall AJ, Niiro H, Lerner CG, Yun TJ, Thomas S, Disteche CM, Clark EA. A novel B lymphocyte-associated adaptor protein, Bam32, regulates antigen receptor signaling downstream of phosphatidylinositol 3-kinase. J Exp Med. 2000;191:1319–1332. doi: 10.1084/jem.191.8.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda S, Miwa Y, Hirata Y, Minowa A, Tanaka J, Nishida E, Koyasu S. Negative feedback loop in T-cell activation through MAPK-catalyzed threonine phosphorylation of LAT. EMBO J. 2004;23:2577–2585. doi: 10.1038/sj.emboj.7600268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min L, Joseph RE, Fulton DB, Andreotti AH. Itk tyrosine kinase substrate docking is mediated by a nonclassical SH2 domain surface of PLCgamma1. Proc Natl Acad Sci U S A. 2009;106:21143–21148. doi: 10.1073/pnas.0911309106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyaji M, Kortum RL, Surana R, Li W, Woolard KD, Simpson RM, Samelson LE, Sommers CL. Genetic evidence for the role of Erk activation in a lymphoproliferative disease of mice. Proc Natl Acad Sci U S A. 2009;106:14502–14507. doi: 10.1073/pnas.0903894106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mott HR, Nietlispach D, Evetts KA, Owen D. Structural analysis of the SH3 domain of beta-PIX and its interaction with alpha-p21 activated kinase (PAK) Biochemistry. 2005;44:10977–10983. doi: 10.1021/bi050374a. [DOI] [PubMed] [Google Scholar]
- Parrini MC, Lei M, Harrison SC, Mayer BJ. Pak1 kinase homodimers are autoinhibited in trans and dissociated upon activation by Cdc42 and Rac1. Mol Cell. 2002;9:73–83. doi: 10.1016/s1097-2765(01)00428-2. [DOI] [PubMed] [Google Scholar]
- Pendergast AM, Muller AJ, Havlik MH, Maru Y, Witte ON. BCR sequences essential for transformation by the BCR-ABL oncogene bind to the ABL SH2 regulatory domain in a non-phosphotyrosine-dependent manner. Cell. 1991;66:161–171. doi: 10.1016/0092-8674(91)90148-r. [DOI] [PubMed] [Google Scholar]
- Phee H, Abraham RT, Weiss A. Dynamic recruitment of PAK1 to the immunological synapse is mediated by PIX independently of SLP-76 and Vav1. Nat Immunol. 2005;6:608–617. doi: 10.1038/ni1199. [DOI] [PubMed] [Google Scholar]
- Puto LA, Pestonjamasp K, King CC, Bokoch GM. p21-activated kinase 1 (PAK1) interacts with the Grb2 adapter protein to couple to growth factor signaling. J Biol Chem. 2003;278:9388–9393. doi: 10.1074/jbc.M208414200. [DOI] [PubMed] [Google Scholar]
- Roose JP, Mollenauer M, Ho M, Kurosaki T, Weiss A. Unusual interplay of two types of Ras activators, RasGRP and SOS, establishes sensitive and robust Ras activation in lymphocytes. Mol Cell Biol. 2007;27:2732–2745. doi: 10.1128/MCB.01882-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott JD, Pawson T. Cell signaling in space and time: where proteins come together and when they're apart. Science. 2009;326:1220–1224. doi: 10.1126/science.1175668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommers CL, Gurson JM, Surana R, Barda-Saad M, Lee J, Kishor A, Li W, Gasser AJ, Barr VA, Miyaji M, et al. Bam32: a novel mediator of Erk activation in T cells. Int Immunol. 2008;20:811–818. doi: 10.1093/intimm/dxn039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommers CL, Samelson LE, Love PE. LAT: a T lymphocyte adapter protein that couples the antigen receptor to downstream signaling pathways. Bioessays. 2004;26:61–67. doi: 10.1002/bies.10384. [DOI] [PubMed] [Google Scholar]
- Strochlic TI, Viaud J, Rennefahrt UE, Anastassiadis T, Peterson JR. Phosphoinositides are essential coactivators for p21-activated kinase 1. Mol Cell. 2010;40:493–500. doi: 10.1016/j.molcel.2010.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisz Hubsman M, Volinsky N, Manser E, Yablonski D, Aronheim A. Autophosphorylation-dependent degradation of Pak1, triggered by the Rho-family GTPase, Chp. Biochem J. 2007;404:487–497. doi: 10.1042/BJ20061696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yablonski D, Kadlecek T, Weiss A. Identification of a phospholipase C-gamma1 (PLC-gamma1) SH3 domain-binding site in SLP-76 required for T-cell receptor-mediated activation of PLC-gamma1 and NFAT. Mol Cell Biol. 2001;21:4208–4218. doi: 10.1128/MCB.21.13.4208-4218.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yablonski D, Kane LP, Qian D, Weiss A. A Nck-Pak1 signaling module is required for T-cell receptor-mediated activation of NFAT, but not of JNK. EMBO J. 1998a;17:5647–5657. doi: 10.1093/emboj/17.19.5647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yablonski D, Kuhne MR, Kadlecek T, Weiss A. Uncoupling of nonreceptor tyrosine kinases from PLC-gamma1 in an SLP-76-deficient T cell. Science. 1998b;281:413–416. doi: 10.1126/science.281.5375.413. [DOI] [PubMed] [Google Scholar]
- Zang M, Hayne C, Luo Z. Interaction between active Pak1 and Raf-1 is necessary for phosphorylation and activation of Raf-1. J Biol Chem. 2002;277:4395–4405. doi: 10.1074/jbc.M110000200. [DOI] [PubMed] [Google Scholar]
- Zhao ZS, Manser E, Lim L. Interaction between PAK and nck: a template for Nck targets and role of PAK autophosphorylation. Mol Cell Biol. 2000;20:3906–3917. doi: 10.1128/mcb.20.11.3906-3917.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.