Summary
Cells constantly adjust their metabolism in response to environmental conditions, yet major mechanisms underlying survival remain poorly understood. We discover a post-transcriptional mechanism that integrates starvation response with GTP homeostasis to allow survival, enacted by the nucleotide (p)ppGpp, a key player in bacterial stress response and persistence. We reveal that (p)ppGpp activates global metabolic changes upon starvation, allowing survival by regulating GTP. Combining metabolomics with biochemical demonstrations, we find that (p)ppGpp directly inhibits the activities of multiple GTP biosynthesis enzymes. This inhibition results in robust and rapid GTP regulation in Bacillus subtilis, which we demonstrate is essential to maintaining GTP levels within a range that supports viability even in the absence of starvation. Correspondingly, without (p)ppGpp, gross GTP dysregulation occurs, revealing a vital housekeeping function of (p)ppGpp; in fact, loss of (p)ppGpp results in death from rising GTP, a severe and previously unknown consequence of GTP dysfunction.
Introduction
To adapt to environmental constraints such as nutrient availability, organisms alter their transcriptome, proteome, and metabolome to allocate and conserve resources (Buescher et. al., 2012; Scott et al., 2010). The ubiquitous nature of environmental challenge and the dire consequences to organisms incapable of robust response necessitate the study of key factors underlying global alterations and supporting the stability of metabolism.
GTP, an important player in metabolism, is required for multiple cellular processes. Reduction of GTP levels lowers transcription of rRNA (Krasny and Gourse, 2004), triggers sporulation in B. subtilis (Lopez et al., 1981), and slows growth of yeast (Iglesias-Gato et al., 2011). GTP dysfunction may also have consequences in metazoans: Increased capacity for GTP biosynthesis is a long-suspected feature of malignancy (Jackson et al., 1975), and mutations in IMP dehydrogenase, a GTP biosynthesis enzyme, are linked to hereditary retinal disease (Bowne et al., 2002; Kennan et al., 2002). Regulation of GTP levels by homeostatic mechanisms has previously been demonstrated (Lehninger et al., 2000; Ebbole and Zalkin, 1989), but it is unknown whether these mechanisms are sufficient to control GTP production robustly.
The starvation-inducible nucleotide guanosine (penta)tetraphosphate ((p)ppGpp) exists in bacteria, plants, and possibly metazoans and is crucial for bacterial fitness, persistence, virulence, and development (Dalebroux and Swanson, 2012; Potrykus and Cashel, 2008; Sun et al., 2010). (p)ppGpp is produced from GTP/GDP and ATP, and its production during amino acid starvation accompanies a decrease in cellular GTP levels (Lopez et al., 1981; Gallant et al., 1971). In Escherichia coli, (p)ppGpp enables resistance to starvation by binding to RNA polymerase (RNAP), in synergy with the transcription factor DksA (Paul et al., 2004), directly altering transcription of many genes, including rRNA and amino acid biosynthesis genes (Barker et al., 2001; Murphy and Cashel, 2003). This global transcriptional response to starvation is the best-characterized physiological role of (p)ppGpp (Durfee et al., 2008; Traxler et al., 2008). However, the mechanism in E. coli does not explain the action(s) of (p)ppGpp in other bacteria, in which (p)ppGpp does not affect RNAP directly (Krasny and Gourse, 2004; Vrentas et al., 2008). Although (p)ppGpp has multiple targets other than RNAP, their regulation by (p)ppGpp has not been shown to be critical for survival (Dalebroux and Swanson, 2012), and it is unclear what may be the direct critical target(s) of (p)ppGpp in organisms other than E. coli.
Here we characterize the direct regulation of cellular GTP levels by (p)ppGpp and show that this regulation is essential for survival during starvation and perturbations to GTP homeostasis. We found that (p)ppGpp elicits global metabolic changes in B. subtilis upon starvation, dramatically reducing GTP levels. By quantitatively comparing metabolome and transcriptome results, we identified two GTP biosynthesis enzymes, Gmk and HprT, as major post-transcriptional targets of (p)ppGpp whose activities are strongly inhibited by (p)ppGpp in vitro. This regulation of GTP levels is sufficient for resisting starvation; suppressor mutations that reduce GTP synthesis restore viability, revealing different critical (p)ppGpp targets in B. subtilis versus E. coli. This regulation is also required for general GTP homeostasis: GTP levels can rise uncontrollably in (p)ppGpp-deficient cells, even in the absence of starvation, leading to cell death.
We conclude that (p)ppGpp plays a crucial role in B. subtilis to regulate GTP homeostasis in response to extrinsic stress and intrinsic cell status, thus preventing death-by-GTP and preserving metabolic stability. This implies an important and multifunctional role for (p)ppGpp as a global player in the metabolome, and regulation of GTP levels by (p)ppGpp may be a common strategy employed by many bacteria and beyond.
Results
Starvation Induces Profound, Predominately (p)ppGpp-Dependent Metabolic Changes
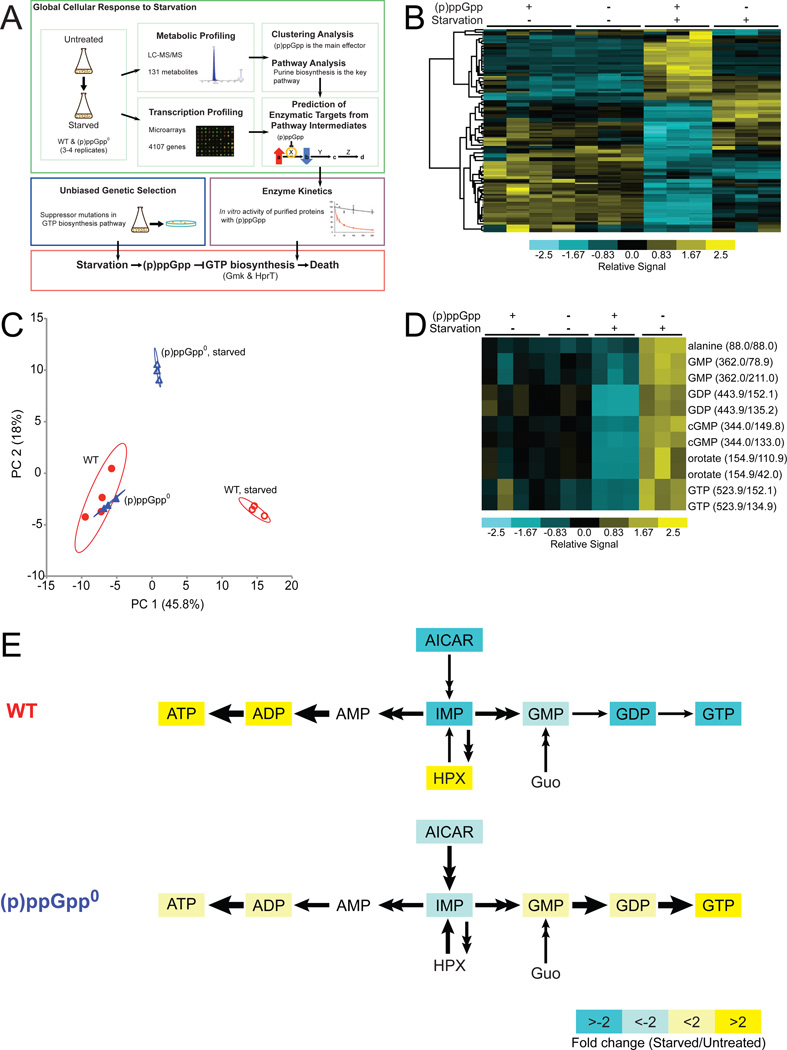
To understand how cellular metabolism globally responds to environmental stressors, we extracted metabolites from exponentially growing and amino acid-starved B. subtilis cells and quantified 131 metabolites with liquid chromatography-tandem mass spectrometry (LC-MS/MS) (Tu et al., 2007) (Figure 1A). We identified 96 abundant species with high quality values (the remaining 35 were low-abundance species or could not be unambiguously quantified) (Table S1). These 96 metabolites exhibited quantitative consistency among biological replicates yet displayed profound changes upon amino acid starvation, with half (48/96) altered significantly during starvation (1.5–15 fold change) (Figure 1B, Figure S1, and Table S2).
Figure 1. Metabolic Profiling of Wild-type and (p)ppGpp0 Cells upon Amino Acid Starvation.
(A) Discovery of critical direct targets of (p)ppGpp during starvation in B. subtilis. For metabolic profiling, wild-type (WT) and (p)ppGpp0 cells were starved and metabolites were quantified by LC-MS/MS. Changes in metabolite signatures were analyzed to reveal (p)ppGpp as the major effector of the starvation response and to identify purine biosynthesis as significantly altered. To identify targets of (p)ppGpp, the pathway was examined for steps in which levels of a substrate increased while levels of the product decreased. Transcriptional profiling indicated that the identified targets were regulated post-transcriptionally. Predicted targets were assayed for enzymatic inhibition by (p)ppGpp in vitro. In parallel, a genetic screen was performed in (p)ppGpp0 cells, which identified mutations that lower GTP biosynthesis. These results revealed that the crucial function of (p)ppGpp during starvation is to directly inhibit the activities of GTP biosynthesis enzymes.
(B) Clustering analysis of metabolite profiles obtained by LC-MS/MS. Rows correspond to 55 metabolites that display significant changes in WT ((p)ppGpp +) or (p)ppGpp0 ((p)ppGpp −) cells in the presence (+) or absence (−) of starvation. Columns correspond to samples labeled above the heat map; ≥ 3 experimental replicates are shown. Samples and metabolites were hierarchically clustered using Cluster and plotted with TreeView. Metabolite levels are normalized to total ion current (TIC) and are indicated by color: high levels are yellow and low levels are blue. An expanded, annotated view is presented in Figure S1.
(C) PCA analyzes correlations among the metabolite profiles in WT and (p)ppGpp0 samples and identifies independent factors (principal components) that explain the variation between samples. Principal components 1 (PC1) and 2 (PC2) comprise nearly 64% of the total variation. The 95% confidence interval is circled around each sample set.
(D) A cluster is magnified from a complete sub-branch of 1B. Metabolites were quantified by the levels of their daughter ions (1–2 daughter ions), with parent/daughter ion masses indicated.
(E) Starvation-induced changes in the purine biosynthesis pathway. Metabolites with significant changes (q-value < 1% for both daughter ions) in WT and (p)ppGpp0 cells are colored: yellow and blue indicate increased and decreased levels, respectively; white indicates no significant change. HPX: hypoxanthine, Guo: guanosine.
We then examined the dependence of these metabolic changes on a single regulator, (p)ppGpp, which is rapidly induced to high concentration during amino acid starvation (Potrykus and Cashel, 2008). We created (p)ppGpp-deficient cells (termed (p)ppGpp0 (Potrykus and Cashel, 2008)) by deleting the three genes encoding (p)ppGpp synthetases: RelA (Wendrich and Marahiel, 1997), YjbM, and YwaC (Nanamiya et al., 2008; Srivatsan et al., 2008). Compared to wild-type cells, we observed significantly attenuated or opposite metabolic responses in (p)ppGpp0 cells upon starvation (Figure 1B, Figure S1, and Table S2). Among the 48 metabolites that changed significantly in wild-type cells, only 11 changed independently of (p)ppGpp, and most of these changes were mild (Table S2).
As a complementary approach, we performed principal component analysis (PCA) to separate our samples by metabolic features (Figure 1C). We verified that profiles from the same strain and treatment are located near each other in the PCA plot, demonstrating that the experiments are reproducible. Untreated wild-type and (p)ppGpp0 samples are located in overlapping regions, indicating that they have similar metabolic profiles. In contrast, profiles of starved wild-type cells are located in a distinct cluster along the first principal component axis (PC 1), separate from those of starved (p)ppGpp0 cells and untreated cells. This accounts for the largest difference among metabolic profiles and thus supports that (p)ppGpp drives the metabolic response to starvation.
To identify metabolic pathways affected by (p)ppGpp during starvation, we performed pathway analysis using MetaboAnalyst (Xia and Wishart, 2011). The highest-ranking pathway (p-value = 1.01 × 10−5) differentially altered was purine biosynthesis (Table S3). We observed differential changes between wild-type and (p)ppGpp0 cells in the pathways leading to production of ATP and GTP from IMP (Figures 1D and 1E). GMP, GDP, and GTP levels were reduced in starved wild-type cells but elevated in starved (p)ppGpp0 cells. In contrast, ADP and ATP were elevated in starved wild-type cells, but the increase was attenuated in (p)ppGpp0 cells.
(p)ppGpp Post-transcriptionally Blocks Two Steps in GTP Biosynthesis
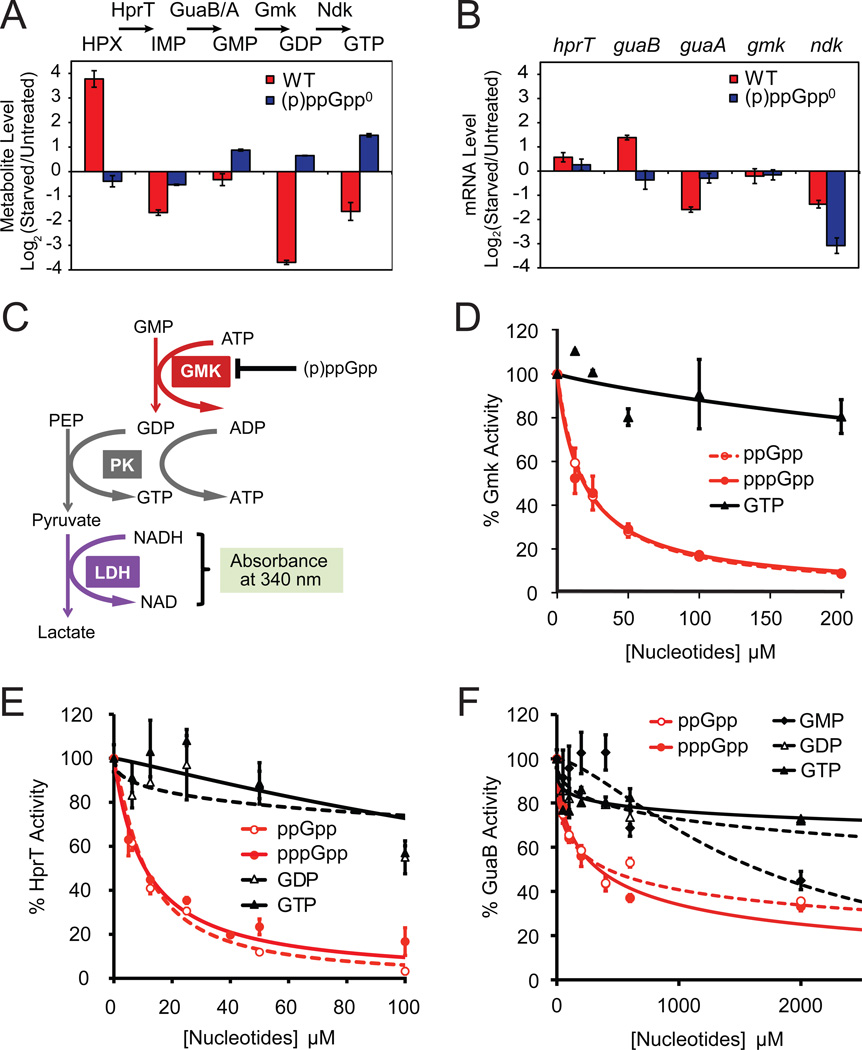
Our metabolic profiling supports previous observations that starvation-induced reduction of GTP levels correlates with (p)ppGpp induction (Lopez et al., 1981; Krasny and Gourse, 2004). It also shows (p)ppGpp-dependent reduction of multiple intermediates in the GTP biosynthesis pathway. To identify key targets of (p)ppGpp, we quantitatively analyzed our results, focusing on adjacent intermediates in the GTP biosynthesis pathway (Figure 2A) and reasoning that (p)ppGpp inhibition of a reaction would result in substrate accumulation and decreased product levels (Figure 1A). We observed such changes at the transitions from GMP to GDP (~10 fold decrease) and hypoxanthine to IMP (~40 fold decrease) upon starvation in wild-type cells but not in (p)ppGpp0 cells (Figure 2A). This suggests two enzymes—guanylate kinase (Gmk, which is proposed to convert GMP to GDP) and HprT (which converts hypoxanthine to IMP and guanine to GMP)—as in vivo targets of (p)ppGpp.
Figure 2. (p)ppGpp Directly Inhibits Multiple GTP Biosynthesis Enzymes.
(A) Quantification of LC-MS/MS data suggests that upon starvation, conversions from hypoxanthine (HPX) to IMP (by HprT) and from GMP to GDP (by Gmk) are blocked in WT but not (p)ppGpp0 cells. Log2 ratios of metabolite levels between starved and untreated samples are plotted. Red: WT; blue: (p)ppGpp0 cells. One daughter ion is used for calculation; nearly identical results were obtained from the second daughter ion (not shown). Error bars = standard error (n ≥ 3) for this and all subsequent figures.
(B) Microarray-based profiling shows that mRNA levels of hprT and gmk are not down-regulated by starvation. Log2 ratios of mRNA levels between starved and untreated samples are plotted. Red: WT; blue: (p)ppGpp0 cells.
(C) Schematic of the in vitro three-step coupled assay for Gmk enzymatic activity. PK: pyruvate kinase, LDH: lactate dehydrogenase, PEP: Phospho(enol)pyruvate.
(D) (p)ppGpp potently inhibits the enzymatic activity of Gmk in vitro. Gmk activity was measured with 10 nM Gmk, 4 mM ATP, 1.5 mM PEP, 150 µM NADH, 2U PK, 2.64U LDH, 50 µM GMP. The relative activity (V0 with inhibitors / without inhibitors) is plotted as a function of concentration of the indicated nucleotide inhibitors. V0 is obtained as in Figure S2C.
(E) (p)ppGpp potently inhibits B. subtilis HprT activity in vitro. HprT activity was measured with 20 nM HprT, 50 µM guanine and 1 mM PRPP, and plotted as a function of concentration of the indicated inhibitors. V0 is obtained as in Figure S2D.
(F) (p)ppGpp and GMP modestly inhibit GuaB activity in vitro. GuaB activity was measured with 50 nM GuaB, 400 µM IMP, and 2.5 mM NAD, and plotted as a function of concentration of the indicated inhibitors. V0 is obtained as in Figure S2E.
To determine whether (p)ppGpp down-regulates Gmk and HprT via transcriptional versus post-transcriptional mechanisms, we examined transcript levels using microarrays. gmk and hprT mRNAs were not reduced following starvation (Figure 2B), indicating that (p)ppGpp-mediated regulation of Gmk and HprT occurs post-transcriptionally.
(p)ppGpp Directly and Potently Inhibits the Enzymatic Activities of Gmk and HprT
We next examined whether (p)ppGpp directly inhibits the enzymatic activities of Gmk and HprT. In B. subtilis, Gmk is an essential enzyme whose putative function is to convert GMP to GDP (Kobayashi et al., 2003). We purified B. subtilis Gmk (Figure S2A) and used a coupled enzymatic assay to verify that Gmk converts GMP to GDP (Figure 2C and Figure S2C). We found that pppGpp and ppGpp, but not GTP, potently inhibit Gmk activity, achieving 50% inhibition at ~20 µM (p)ppGpp (Figure 2D and Table S4). This inhibition is specific to Gmk and not attributable to inhibition of the coupling enzymes (Figure S2B). We conclude that (p)ppGpp is a specific, direct, and potent inhibitor of Gmk enzymatic activity.
HprT, which converts both guanine to GMP and hypoxanthine to IMP, was previously suggested as a potential target of (p)ppGpp during amino acid starvation in B. subtilis (Beaman et al., 1983). To test whether (p)ppGpp inhibits HprT in vitro, we purified B. subtilis HprT (Figure S2A) and performed kinetic assays (Figure S2D). We found that pppGpp and ppGpp are potent inhibitors of HprT, achieving 50% inhibition at ~11 µM (Figure 2E and Table S4).
Inhibition of GuaB Plays a Minor Role in Starvation Response
GuaB (IMP dehydrogenase) converts IMP to XMP and is a proposed target of (p)ppGpp (Gallant et al., 1971; Lopez et al., 1981). To test whether (p)ppGpp inhibits B. subtilis GuaB in vitro, we purified GuaB (Figure S2A) and performed kinetic assays (Figure S2E). GMP moderately inhibits GuaB activity (Figure 2F), similar to results obtained in E. coli (Gallant et al., 1971). However, pppGpp and ppGpp only moderately inhibit GuaB activity, and 50% inhibition of GuaB activity requires relatively high levels of pppGpp and ppGpp (~0.3–0.5 mM, respectively) (Table S4).
Thus, ~10–20 µM (p)ppGpp significantly inhibits both HprT and Gmk activity, while even at 2 mM (p)ppGpp, more than 30% of GuaB activity remains (Figure 2F). In relevant context, as the in vivo concentration of (p)ppGpp increases up to 1–2 mM during amino acid starvation, it should be sufficient to strongly inhibit Gmk and HprT activity, thus lowering GTP pools in response to starvation; however, inhibition of GuaB activity by (p)ppGpp is likely a minor contributor. Correspondingly, our metabolomic data did not show a major block after IMP (Figure 2A), and overexpressing guaB did not increase GTP levels during amino acid starvation (Figure S2F).
(p)ppGpp is Indispensable for GTP Homeostasis, via Negative Feedback Control
While high levels of (p)ppGpp upon starvation strongly inhibit GTP synthesis, basal levels of (p)ppGpp in cells during normal growth (~10–20 µM) are comparable to the in vitro (p)ppGpp concentrations at which ~50% of activities of HprT and Gmk are inhibited. Therefore, we hypothesized that (p)ppGpp might regulate GTP levels even in the absence of starvation.
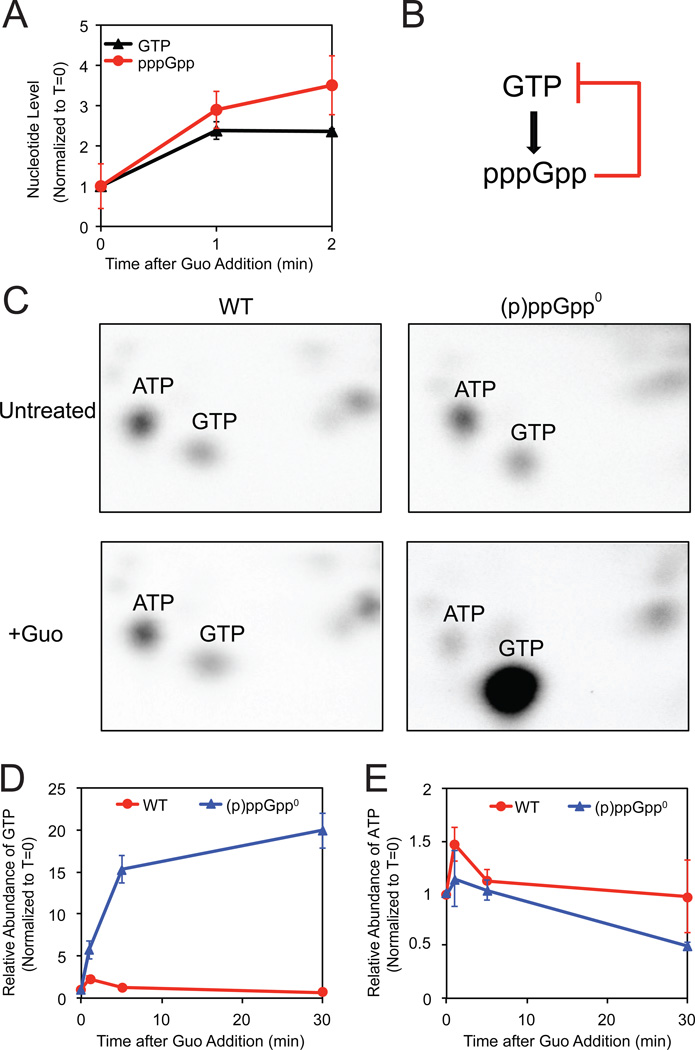
Interestingly, we found that pppGpp, produced from GTP, is in fact moderately induced by increased GTP levels in the absence of starvation. To increase GTP levels transiently, we added guanosine, which is converted to GTP via the salvage pathway (Figure 1E), and we measured levels of GTP and (p)ppGpp by thin layer chromatography (TLC). Following guanosine addition, pppGpp levels rise concomitantly with GTP levels (Figure 3A). Although the pppGpp level is much lower than that induced during amino acid starvation, it should be sufficient to inhibit GTP biosynthesis via HprT and Gmk inhibition, based on the in vitro potency of (p)ppGpp-dependent regulation (Figures 2D and 2E).
Figure 3. pppGpp Mediates Feedback Control of GTP Levels.
(A) pppGpp levels increase concomitantly with increased GTP levels in WT cells following guanosine treatment (Guo). 32P-labeled WT cells were treated with 1 mM Guo. At the indicated times after treatment, nucleotides were extracted, measured by TLC, and normalized to T=0.
(B) Schematic of a negative feedback loop of GTP synthesis with pppGpp.
(C) Two-dimensional TLC of 32P-labeled nucleotides with ATP and GTP indicated. WT (left panels) and (p)ppGpp0 (right panels) cells before (upper panels) and 20 minutes after addition of 1 mM Guo (lower panels) are shown.
(D–E) Quantification of GTP (D) and ATP (E) levels by LC-MS/MS of WT (red) and (p)ppGpp0 (blue) cells in a time course after treatment with 1 mM Guo, normalized to T=0.
Therefore, we proposed that pppGpp might be globally involved in GTP homeostasis via a negative feedback mechanism, as it is induced by increased GTP and subsequently inhibits GTP synthesis (Figure 3B). We thus examined GTP homeostasis in (p)ppGpp0 cells: Surprisingly, we observed complete dysregulation of cellular GTP levels. Correspondingly, two-dimensional TLC (to visualize nucleotides in 32P-labeled cell extracts sampled 20 minutes after guanosine addition) revealed no significant changes in GTP levels with respect to other nucleotides in wild-type cells (Figure 3C, left), indicative of tight feedback control of GTP levels. However, in the absence of (p)ppGpp, GTP levels strikingly rose to become the most dominant spot (Figure 3C, right).
We confirmed this result by quantifying label-free GTP levels with targeted LC-MS/MS. In wild-type cells, GTP levels initially rise following guanosine addition but quickly re-equilibrate within 5 minutes (Figure 3D), likely due to negative feedback regulation by increased levels of pppGpp. In (p)ppGpp0 cells, in contrast, GTP levels continuously increase up to ~20 fold (Figure 3D), while ATP levels are not dramatically altered (Figure 3E). Examination of the metabolites along the GTP salvage pathway in guanosine-treated cells verified that a major block occurs before GMP formation (Figure S3), supporting our in vitro results showing that (p)ppGpp inhibits HprT activity (Figure 2E).
In summary, we found that previously characterized homeostatic mechanisms are insufficient to protect GTP levels from perturbations in B. subtilis, and we demonstrate that pppGpp not only facilitates but is indispensable for maintaining GTP homeostasis via negative feedback control.
A Genetic Screen Reveals that (p)ppGpp Targets GTP Biosynthesis, not RNAP
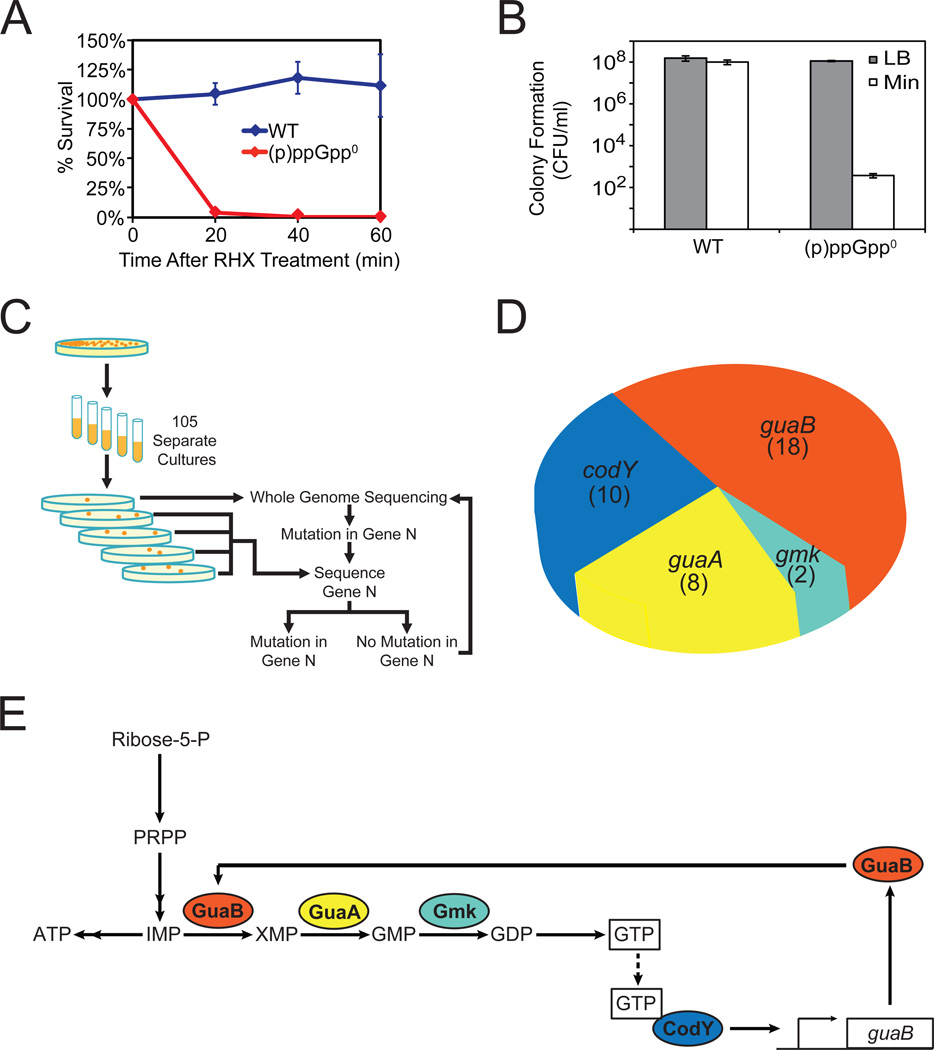
We found that (p)ppGpp not only elicits changes in metabolites but also is required for B. subtilis cells to survive nutrient stress. Within 20 minutes of amino acid starvation, only ~3% of (p)ppGpp0 cells survive, compared to ~100% of wild-type cells (Figure 4A). In addition, similar to E. coli results, loss of (p)ppGpp renders B. subtilis cells unable to form colonies on minimal medium without amino acid supplementation (Figure 4B).
Figure 4. (p)ppGpp0 Phenotype and a Genetic Selection for Suppressor Mutations.
(A) (p)ppGpp0 cells do not survive sudden starvation. WT and (p)ppGpp0 cells were treated with 0.5 mg/ml RHX for the indicated time and plated on LB. Percent survival was calculated by counting the number of cells able to form colonies the next day, normalized to T=0.
(B) (p)ppGpp0 cells fail to form colonies on minimal medium plates. WT and (p)ppGpp0 cells were plated on LB and minimal medium plates, and colonies were counted the next day. CFU/ml: colony forming unit (CFU) per ml of culture, normalized by OD600.
(C) Schematic of genetic selection for (p)ppGpp0 suppressor mutants that can form colonies on minimal medium. Suppressors were selected on minimal medium plates. Whole genome sequencing was conducted on one suppressor to identify a mutation in one gene, followed by targeted sequencing of this gene in all other suppressors. An additional suppressor was subsequently identified by whole-genome sequencing. This process was repeated iteratively.
(D) Distributions of (p)ppGpp0 suppressor mutations in the indicated genes. The numbers indicate independently isolated suppressors with mutations in each gene. Details are listed in Table S5.
(E) Schematic of the pathways affected by the (p)ppGpp0 suppressor mutations. GuaB, GuaA, and Gmk are enzymes in the GTP biosynthesis pathway. CodY is a pleiotropic transcription factor that is activated by GTP and activates transcription of guaB. The colors of the gene products match that of the genes in Figure 4D.
To understand how (p)ppGpp exerts its protective role during nutrient stress, we performed an unbiased genetic selection for mutations that allow (p)ppGpp0 cells to form colonies on minimal medium. We inoculated (p)ppGpp0 cells in separate liquid cultures and plated them on minimal medium plates. We obtained a single colony from each plate (Figure 4C) and examined these mutants by first sequencing the genes encoding the β and β' subunits of RNAP, rpoB, and rpoC, as a similar screen performed in E. coli found mutations in rpoB and rpoC (Xiao et al., 1991; Murphy and Cashel, 2003). Interestingly, among the 105 suppressors we isolated, none contained mutations in rpoB or rpoC, suggesting that the physiologically critical targets of (p)ppGpp in B. subtilis differ from those in E. coli.
Combining Illumina whole-genome sequencing with gene-targeted DNA sequencing (Figure 4C), we identified mutations in 37 suppressors (Figure 4D and Table S5). Most mutations are located in genes along the de novo GTP biosynthesis pathway—guaA, guaB and gmk (Figure 4E)—and several have mutations in the −10 and −35 canonical promoter sequences, presumably resulting in reduced transcription (Table S5). This suggests that partial loss-of-function mutations in GTP biosynthesis genes rescue colony formation. Correspondingly, we placed the endogenous guaB locus under the control of an IPTG-inducible promoter and observed that depletion of guaB, upon removal of IPTG, also completely rescued colony formation (Figure S4A).
In addition, a number of suppressor mutations map to codY, which encodes a GTP-regulated transcription factor with numerous targets (Molle et al., 2003). The majority of codY mutants contained frame-shift mutations (Table S5), and deletion of codY partially rescued colony formation (Figure S4B and Figure 5A). Although CodY is not directly involved in GTP biosynthesis, it activates transcription of guaB (Figure 4E) (Molle et al., 2003), and loss-of-function mutations in codY could decrease GTP levels.
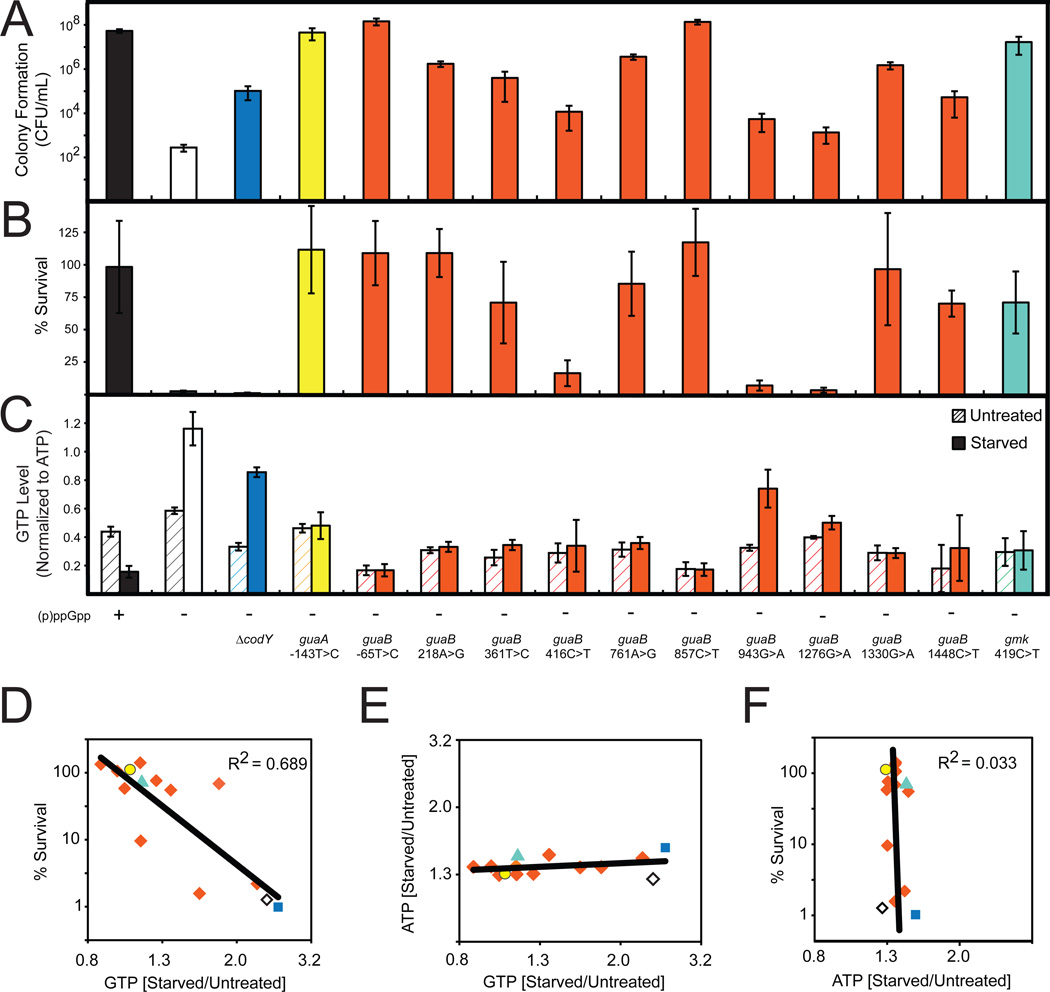
Figure 5. Decreased GTP Levels Allow Survival of Amino Acid Starvation.
(A) Colony formation of WT ((p)ppGpp +), (p)ppGpp0 ((p)ppGpp −) and the indicated suppressor mutants on minimal medium. For this and subsequent panels, the colors denote the different genes that have mutations in the (p)ppGpp0 suppressors and are coded as in Figure 4D.
(B) Percent survival of WT, (p)ppGpp0, and the indicated suppressor mutants upon amino acid starvation. Cells were treated with 0.5 mg/ml RHX for 40 minutes and then plated on LB. Percent survival was calculated as in Figure 4A.
(C) GTP levels of WT, (p)ppGpp0, and the indicated suppressor mutants before and after treatment with 0.5 mg/ml RHX for 10 minutes were quantified by TLC and normalized to initial levels of ATP.
(D) GTP levels are correlated with survival of amino acid starvation. Starvation-induced changes of GTP levels in the indicated suppressor mutants were obtained by TLC and plotted against % survival. Averages from three independent experiments are plotted for each suppressor allele. To assess correlation, the coefficient of determination (R2 value) was calculated. Strains are color coded as in Figure 4D. ◇: (p)ppGpp0; ■: codY, ○: guaA, ◆: guaB, and ▲: gmk suppressor alleles.
(E) Starvation-induced changes of GTP levels of the indicated suppressor mutants are plotted against changes of ATP levels, similar to Figure 5D.
(F) Starvation-induced changes of ATP levels of the indicated suppressor mutants were obtained by TLC and plotted against % survival, similar to Figure 5D.
(p)ppGpp Regulation of GTP Biosynthesis is Essential for Survival During Starvation
Next, we tested whether these suppressors could survive sudden amino acid starvation. Interestingly, suppressor mutations that best rescue colony formation on minimal medium can prevent cell death completely upon starvation in liquid culture (Figures 5A and 5B). On the other hand, deletion of codY, which does not completely rescue colony formation on minimal medium, did not prevent cell death.
We confirmed that GTP levels in both untreated and starved suppressor mutants are decreased in comparison to (p)ppGpp0 cells (Figure 5C), suggesting that the mutants resist amino acid starvation due to lowered GTP levels. We also noticed that stronger suppressors had lower GTP levels and survived starvation in liquid culture, while weaker suppressors had higher GTP levels and did not survive. GTP levels upon amino acid starvation negatively correlate with the ability to survive starvation (Figure 5D) and to form colonies on minimal medium (Figure S5A). Although changes in ATP and GTP levels are inversely coupled during starvation in wild-type cells (Figure 1E), GTP but not ATP levels varied greatly from one suppressor allele to another (Figure 5E). There was also no significant correlation between ATP levels and resistance to starvation (Figure 5F and Figure S5B). Our results indicate that GTP levels or GTP/ATP ratios, but not ATP levels, correlate with the ability to withstand amino acid limitation.
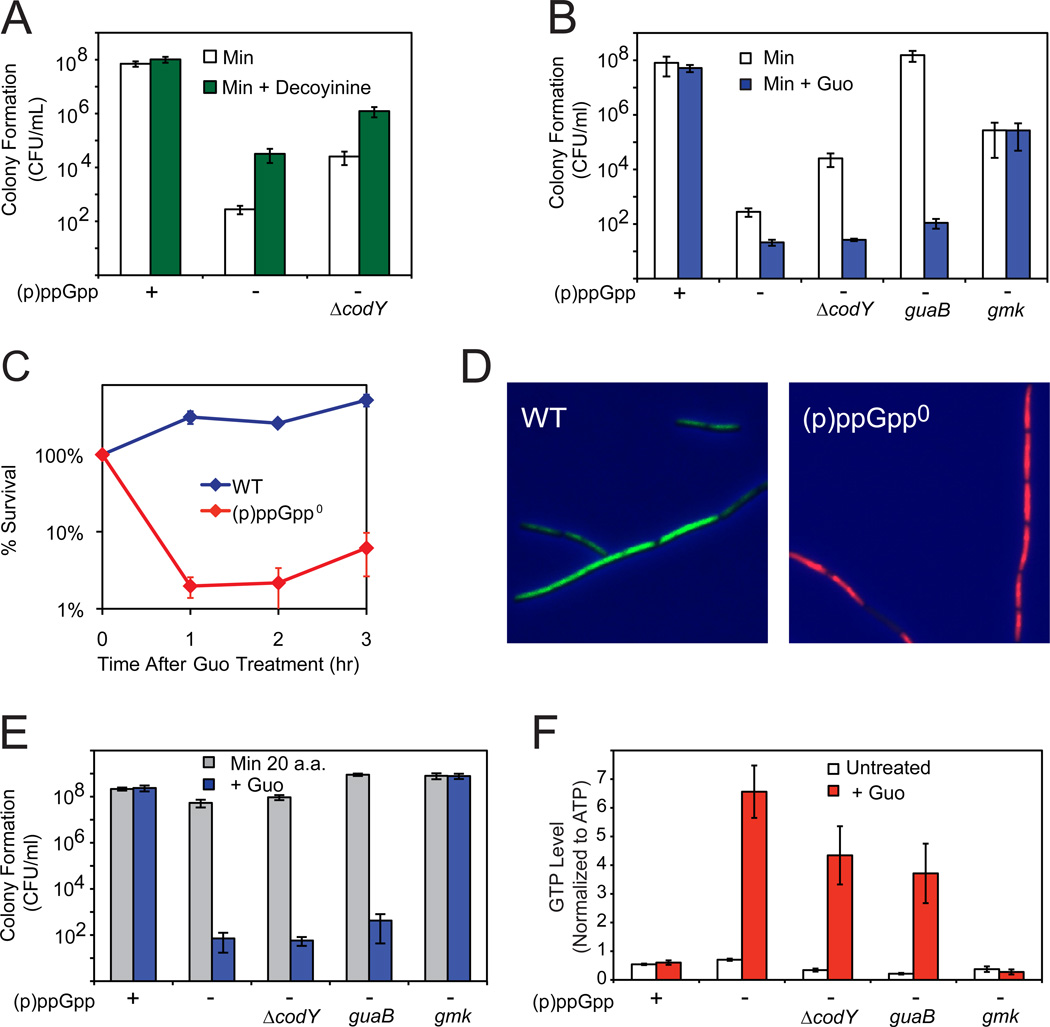
To test the causal relationship between GTP levels and resistance to amino acid limitation, we treated cells with the GMP synthetase (GuaA) inhibitor decoyinine (Lopez et al., 1981) to inhibit GTP biosynthesis and found that it increased the ability of (p)ppGpp0 and ΔcodY (p)ppGpp0 cells to form colonies on minimal medium (Figure 6A). Conversely, increasing GTP levels by guanosine addition abolished the ability of the suppressors to form colonies on minimal medium (Figure 6B), demonstrating that lowering GTP levels enhances (and increasing GTP levels diminishes) resistance to amino acid limitation.
Figure 6. High Levels of GTP Kill (p)ppGpp0 Cells Independent of Starvation.
(A) Decreasing GTP levels by decoyinine (50 µg/ml) enhances colony formation of (p)ppGpp0 and ΔcodY (p)ppGpp0 cells on minimal medium.
(B) Increasing GTP levels by guanosine (0.1 mM Guo) decreases colony formation of (p)ppGpp0 suppressors on minimal medium. WT, (p)ppGpp0, (p)ppGpp0 ΔcodY, guaB-65T>C (guaB), and gmk419C>T (gmk) mutants were plated on medium with or without 0.1 mM Guo.
(C) (p)ppGpp0 cells do not survive Guo addition. WT and (p)ppGpp0 cells were grown in liquid medium with casamino acids, treated with 1 mM Guo, and then plated on LB plates at the indicated times after treatment.
(D) Exponential phase cultures 30 minutes after treatment with 1 mM Guo are stained with the fluorescent dyes SYTO9 (green) and propidium iodide (red) to label live and dead cells, respectively.
(E) A mutation in gmk, but not codY or guaB, enables (p)ppGpp0 cells to form colonies in the presence of Guo. WT, (p)ppGpp0, (p)ppGpp0 ΔcodY, (p)ppGpp0 guaB-65T>C (guaB), and (p)ppGpp0 gmk419C>T (gmk) mutants were plated on medium supplemented with 20 amino acids, with or without 0.1 mM Guo.
(F) A mutation in gmk, but not codY and guaB, prevents high levels of GTP in (p)ppGpp0 cells. GTP levels for untreated or Guo-treated (1 mM, 20 minutes) samples were determined by TLC for the same strains as 6E.
Regulation of GTP Homeostasis Dictates Survival
Finally, we found that loss of (p)ppGpp-mediated GTP homeostasis drastically reduces cell viability even in the absence of starvation. Addition of guanosine to (p)ppGpp0 cells, thereby increasing GTP levels (Figures 3C and 3D), kills ~99% of cells within an hour (Figure 6C; confirmed using a Live/Dead test, Figure 6D). (p)ppGpp0 cells also fail to form colonies on plates with guanosine even in the presence of all 20 amino acids (Figure 6E).
We found that the sensitivity of (p)ppGpp0 cells to guanosine is attributable to high levels of GTP (or potentially GDP), as suppressors affecting different steps of the GTP biosynthesis pathway show differential resistance to guanosine addition (Figures 6E and 6F). Upon guanosine addition, suppressors that inhibit only the de novo GTP biosynthesis pathway (guaB, codY) have high levels of GTP (Figure 6F) and cannot form colonies even when all amino acids are present (Figure 6E). In contrast, a suppressor with a gmk mutation, which blocks both the de novo and salvage pathways prior to GDP formation, does not have high levels of GTP upon guanosine addition (Figure 6F) and can form colonies (Figures 6B and 6E). Our results demonstrate that high GTP (or GDP) levels, but not their precursors, are toxic to cells and that this effect is independent of amino acid availability.
Discussion
In this study, we profiled metabolic changes in B. subtilis cells and found that amino acid starvation significantly alters half of the representative metabolites. The starvation-inducible nucleotide (p)ppGpp, which strongly inhibits GTP biosynthesis, actively mediates most of these changes. We showed that (p)ppGpp reduces GTP levels during starvation by directly inhibiting the activities of two enzymes, Gmk and HprT, at critical steps in GTP biosynthesis. This (p)ppGpp-mediated regulation also prevents GTP from surging to high levels even in the absence of starvation, revealing a central mechanism of GTP homeostasis—regulation that protects cells from starvation and death-by-GTP. While it is unclear how high levels of GTP result in cell death, the regulation of GTP by (p)ppGpp is crucial for survival under multiple conditions and may be conserved in other species.
Critical Targets of (p)ppGpp Differ Between E. coli and B. subtilis
p)ppGpp has multiple previously identified targets, including E. coli RNAP (Barker et al., 2001), translation factor IF2 (Milon et al., 2006), lysine decarboxylase (Kanjee et al., 2011), GTPase Obg (Buglino et al., 2002), exopolyphosphatase (Kuroda et al., 1997), and primase (Wang et al., 2007). However, the only target previously known to have a major physiological impact is E. coli RNAP (Barker et al., 2001). Here we identify direct (p)ppGpp targets with major impact in B. subtilis: the GTP biosynthesis enzymes Gmk and HprT. Although GuaB and HprT are regulated by (p)ppGpp in E. coli, inhibition requires higher levels of (p)ppGpp (Hochstadt-Ozer and Cashel, 1972). In contrast, we show potent inhibition of Gmk and HprT in B. subtilis (IC50 ~20 µM and ~10 µM, respectively), which demonstrates the major biological relevance of these targets, allowing protection of GTP levels independently of starvation even at barely detectable (p)ppGpp levels.
We note that (p)ppGpp regulation of Gmk, HprT, and GuaB may not be exclusive; our data suggest additional layers of regulation could exist upstream of IMP in de novo purine biosynthesis (Figure 1E). Thus, (p)ppGpp may affect multiple components of purine biosynthesis both directly or via transcriptional control, adjusting GTP levels robustly in fluctuating environmental conditions.
Importantly, we demonstrate that regulation of GTP biosynthesis enzymes by (p)ppGpp is critical for B. subtilis viability. In E. coli, genetic selections identified mutations in RNAP (β and β’ subunits) that enable (p)ppGpp0 cells to grow on minimal medium (Murphy and Cashel, 2003), indicating that the crucial function of (p)ppGpp is to regulate RNAP. In performing a similar genetic selection in B. subtilis, we found no suppressors with mutations in RNAP. Instead, all mutations lead to decreased GTP levels. Decreasing GTP levels by decoyinine addition also suppresses the (p)ppGpp0 phenotype, and increasing GTP levels by guanosine addition abolishes the ability of the identified suppressors to form colonies on minimal medium (Figures 6A and 6B). Thus, (p)ppGpp enables B. subtilis cells to form colonies on minimal medium by reducing GTP levels.
Our results indicate that bacteria use (p)ppGpp in different ways to survive starvation. In one strategy, (p)ppGpp regulates transcription by directly interacting with RNAP (in synergy with DksA) in bacteria such as E. coli. Our work suggests that a second strategy, direct regulation of GTP levels by (p)ppGpp, may be applicable to other bacteria such as B. subtilis, allowing (p)ppGpp to regulate transcription indirectly via GTP levels and to mediate protective effects beyond transcription. In T. thermophilus, (p)ppGpp does not affect transcription directly (Vrentas et al., 2008) despite forming a complex with RNAP in a high-resolution structural study (Artsimovitch et al., 2004). Like B. subtilis, T. thermophilus lacks DksA and GTP levels decrease upon starvation (Kasai et al., 2006); thus, it will be interesting to test whether T. thermophilus uses (p)ppGpp-mediated regulation of GTP synthesis for starvation response.
From Stress Response to Homeostasis: A Master Regulator of GTP
Interestingly, our results show that (p)ppGpp-mediated regulation of GTP biosynthesis is a critical event not only during starvation but also upon other perturbations to GTP homeostasis. GTP homeostasis is tightly controlled, and multiple well-established mechanisms were thought to be sufficient to maintain GTP homeostasis (Lehninger et al., 2000). These include negative feedback by IMP/AMP/GMP to inhibit the de novo synthesis pathway and mechanisms that maintain balance between GTP and ATP levels. Unexpectedly, we found that GTP homeostasis is abolished in (p)ppGpp0 cells, with GTP levels rising uncontrollably to ~10 mM or higher (Figure 3C and 3D). This dysregulation indicates that (p)ppGpp not only contributes to but is a master regulator of GTP homeostasis.
Several features of this direct enzymatic feedback inhibition advance our understanding of GTP homeostasis. First, most known mechanisms regulate the de novo pathway; (p)ppGpp blocks both de novo and salvage GTP biosynthesis, globally regulating GTP production from all sources. Second, transcriptional feedback mechanisms that regulate GTP biosynthesis genes (Ebbole and Zalkin, 1989; Belitsky and Sonenshein, 2011) may not prevent accumulation of GTP as rapidly as direct enzymatic inhibition by (p)ppGpp. Third, previously known mechanisms of GTP homeostasis involve only precursors and intermediates of purine biosynthesis; pppGpp is an off-pathway product synthesized from GTP to provide a negative feedback loop (Figure 3B). Because increases in GTP produce pppGpp to buffer GTP levels against fluctuations, the role of (p)ppGpp in maintaining GTP homeostasis may lie beyond protection from imbalanced external guanosine and may constitute a fundamental aspect of GTP control. Inhibition of GTP biosynthesis appears to allow two major functions of (p)ppGpp as a master regulator of GTP homeostasis: (1) at lower (p)ppGpp levels, buffering GTP against fluctuations and (2) at higher levels, modulating GTP levels to stabilize metabolism in response to external stressors (Figure 7).
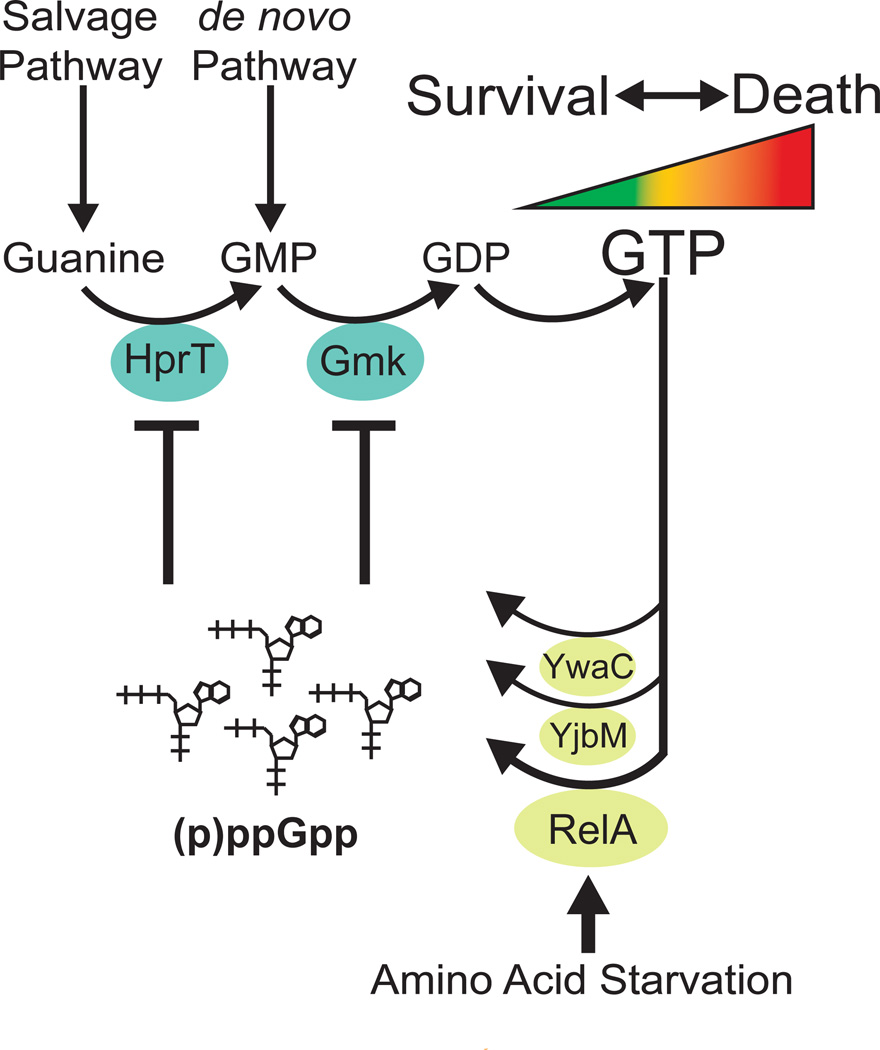
Figure 7. (p)ppGpp governs starvation-sensing negative feedback control of GTP levels via direct enzymatic inhibition, thus preventing death.
(p)ppGpp, produced from GTP(GDP) and ATP, directly inhibits the GTP biosynthesis enzymes HprT and Gmk to rapidly block de novo and salvage GTP biosynthesis. (p)ppGpp is induced (1) to low levels by increased GTP levels, to provide negative feedback control of GTP; and (2) to high levels by amino acid starvation, via activation of the main (p)ppGpp synthetase RelA, to decrease GTP levels. The regulation of GTP by (p)ppGpp is important for cell viability as high levels of GTP result in cell death.
Discovering a Severe Metabolic Consequence: Death-by-GTP
GTP is involved in numerous cellular processes, but unlike ATP, a well-known indicator of metabolic states, less is known about the cellular effects of GTP concentrations. By removing (p)ppGpp-dependent GTP homeostatic control, we uncovered a drastic consequence of GTP dysregulation—massive and rapid cell death. This death is independent of starvation, accompanied by excess GTP, and rescued by decreased GTP synthesis; therefore, we deem it death-by-GTP.
The mechanisms underlying death-by-GTP remain unknown. Elevated GTP levels might compete with ATP and poison ATP-utilizing proteins required for viability. They might also lead to deleterious alterations of the transcriptome and may directly up-regulate transcription of rRNA (Krasny and Gourse, 2004), leading to over-investment in translation machinery at the expense of other essential products. GTP is the precursor of dGTP, and dysregulation of GTP may perturb DNA replication due to dGTP-pool overexpansion, as suggested in yeast (Breton et al., 2008). Dysregulation of GTP may also alter other GTP-dependent processes such as translation, secretion, ribosome biogenesis, signaling, or cell division, resulting in loss of viability. While future work will be necessary to determine the mechanisms of death-by-GTP, it is clear that a vital role of (p)ppGpp in B. subtilis is to prevent this death.
Maintenance of GTP levels across species is critical to fitness, and GTP dysregulation has relevance to malignancy and genetic disease. (p)ppGpp is a key player in bacterial stress response and pathogenesis. Our discovery in the broadly applicable organism B. subtilis–that GTP modulation involves direct regulation of enzymatic activities by (p)ppGpp—expands the (p)ppGpp paradigm established by decades of research in E. coli and demonstrates that previously characterized homeostatic mechanisms are insufficient to protect GTP levels from perturbations. Many stressors induce (p)ppGpp; it is feasible that (p)ppGpp regulates GTP levels during a multitude of conditions. Thus, (p)ppGpp plays a crucial role: regulating GTP homeostasis in response to extrinsic stress and intrinsic cell status, preserving metabolic stability. Future experiments should test whether (p)ppGpp modulation of GTP levels is a general strategy used by organisms across the domains of life.
Experimental Procedures
Strains, Growth Conditions, and Protein Assays
All strains and plasmids used are listed in Table S6. Construction of strains and plasmid, protein purification and enzymatic assays are described in Supplemental Material. Unless otherwise indicated, all cells were grown in S7 defined medium (Vasantha and Freese, 1980); MOPS was used at 50 mM rather than 100 mM, supplemented with 0.1% glutamate, 1% glucose, and 0.5% casamino acids at 37°C with shaking. For metabolic profiling and measurement of nucleotide levels, phosphate was 0.5 mM instead of 5 mM.
Metabolite Extraction and LC-MS/MS Quantification
The procedure for metabolite extraction was adapted from published protocols (Kiefer et al., 2008). Wild-type and (p)ppGpp0 cells were grown to an OD600 ~0.4–0.5 and treated with 0.5 mg/ml arginine hydroxamate (RHX), an analog of arginine, to induce amino acid starvation for 10 minutes. 10 ml samples were collected and washed on two GHP membrane filters by vacuum filtration within 30 sec (0.45 µm; 5 ml per filter). Filters were immediately transferred to boiling water for 15 minutes to extract metabolites. Extracts were centrifuged to remove cell debris, frozen in liquid nitrogen, and vacuum dried. Metabolites were quantified by LC-MS/MS as described in Supplemental Materials.
Suppressor Selection and Identification
To isolate suppressor mutants, single colonies of the strain JDW755 were inoculated in individual tubes. Cells were grown either to exponential or stationary phase, collected by centrifugation, washed thrice with Spizizen’s minimal salts (Spizizen, 1958), and selected on minimal plates supplemented with tryptophan and methionine. One colony on each plate was selected.
To identify suppressor mutations, whole genome sequencing was first performed for one suppressor strain with the Illumina platform as described (Srivatsan et al., 2008). This sequence was compared to that of the parental strain to reveal a single difference in codY. To verify the suppression phenotype, the mutation was delivered to the parental strain as described in Supplemental Experimental Procedures. Next, Sanger sequencing of all other suppressors was performed specifically at the codY locus to identify additional codY mutants. Finally, whole genome sequencing was applied to an additional suppressor strain with a wild-type codY locus to identify mutations in other genes.
Microscopic Evaluation of Cell Death
The Live/Dead BacLight Viability Kit (Molecular Probes) was applied, in which live and dead cells are labeled with SYTO9 and propidium iodide, respectively. Cells were fixed on pads of 1% agarose in Spizizen's salts and visualized on a Zeiss Axiovert 200 microscope, with a 100× phase contrast objective. Images were captured on a Hamamatsu Digital CCD camera.
Measurement of Intracellular Nucleotides by TLC
Measurement of nucleotides was performed as described (Wang et al., 2007). For two-dimensional TLC, 1.75 M morpholine, 0.1 M boric acid, 1.4 M HCl (pH 8.7) was used for the first dimension solvent, and 3 M (NH4)2SO4 with 2% disodium EDTA (pH 5.5) was used for the second dimension solvent (Lee et al., 1983).
Supplementary Material
Highlights.
(p)ppGpp allows survival of amino acid starvation by reducing GTP levels
(p)ppGpp directly and potently inhibits multiple GTP biosynthesis enzymes
(p)ppGpp is a key component of GTP homeostasis
In the absence of (p)ppGpp, high GTP levels lead to cell death
Acknowledgements
We thank S. Stibitz, J. Berger, R. Britton and M. Cashel for reagents; E. White for help on the manuscript; G. Allen, B. Bochner, S. Brinsmade, M. Cashel, R. Gourse, C. Herman, S. Rosenberg, W. Ross, L. Sonenshein, J. Wilson, and the Wang Lab for discussions and comments. JDW is supported by NIGMS R01GM084003 and Welch Grant Q-1698. BPT is supported by NIGMS R01GM094314 and Welch Grant I-1697. ANB was supported by a fellowship from GCC (T90 DA022885-05).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession Numbers
Microarray data are available in the NCBI GEO database under accession number GSE39758 (http://www.ncbi.nlm.nih.gov/geo/).
References
- Artsimovitch I, Patlan V, Sekine S, Vassylyeva MN, Hosaka T, Ochi K, Yokoyama S, Vassylyev DG. Structural basis for transcription regulation by alarmone ppGpp. Cell. 2004;117:299–310. doi: 10.1016/s0092-8674(04)00401-5. [DOI] [PubMed] [Google Scholar]
- Barker MM, Gaal T, Josaitis CA, Gourse RL. Mechanism of regulation of transcription initiation by ppGpp. I. Effects of ppGpp on transcription initiation in vivo and in vitro. J Mol Biol. 2001;305:673–688. doi: 10.1006/jmbi.2000.4327. [DOI] [PubMed] [Google Scholar]
- Beaman TC, Hitchins AD, Ochi K, Vasantha N, Endo T, Freese E. Specificity and control of uptake of purines and other compounds in Bacillus subtilis. J Bacteriol. 1983;156:1107–1117. doi: 10.1128/jb.156.3.1107-1117.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belitsky BR, Sonenshein AL. CodY-Mediated Regulation of Guanosine Uptake in Bacillus subtilis. J Bacteriol. 2011;193:6276–6287. doi: 10.1128/JB.05899-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowne SJ, Sullivan LS, Blanton SH, Cepko CL, Blackshaw S, Birch DG, Hughbanks-Wheaton D, Heckenlively JR, Daiger SP. Mutations in the inosine monophosphate dehydrogenase 1 gene (IMPDH1) cause the RP10 form of autosomal dominant retinitis pigmentosa. Hum Mol Genet. 2002;11:559–568. doi: 10.1093/hmg/11.5.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breton A, Pinson B, Coulpier F, Giraud MF, Dautant A, Daignan-Fornier B. Lethal accumulation of guanylic nucleotides in Saccharomyces cerevisiae HPT1-deregulated mutants. Genetics. 2008;178:815–824. doi: 10.1534/genetics.107.083295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buescher JM, Liebermeister, et al. Global network reorganization during dynamic adaptations of Bacillus subtilis metabolism. Science. 2012;335:1099–1103. doi: 10.1126/science.1206871. [DOI] [PubMed] [Google Scholar]
- Buglino J, Shen V, Hakimian P, Lima CD. Structural and biochemical analysis of the Obg GTP binding protein. Structure. 2002;10:1581–1592. doi: 10.1016/s0969-2126(02)00882-1. [DOI] [PubMed] [Google Scholar]
- Dalebroux ZD, Swanson MS. ppGpp: magic beyond RNA polymerase. Nat Rev Microbiol. 2012;10:203–212. doi: 10.1038/nrmicro2720. [DOI] [PubMed] [Google Scholar]
- Durfee T, Hansen AM, Zhi H, Blattner FR, Jin DJ. Transcription profiling of the stringent response in Escherichia coli. J Bacteriol. 2008;190:1084–1096. doi: 10.1128/JB.01092-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebbole DJ, Zalkin H. Bacillus subtilis pur operon expression and regulation. J Bacteriol. 1989;171:2136–2141. doi: 10.1128/jb.171.4.2136-2141.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallant J, Irr J, Cashel M. The mechanism of amino acid control of guanylate and adenylate biosynthesis. J Biol Chem. 1971;246:5812–5816. [PubMed] [Google Scholar]
- Hochstadt-Ozer J, Cashel M. The regulation of purine utilization in bacteria. V. Inhibition of purine phosphoribosyltransferase activities and purine uptake in isolated membrane vesicles by guanosine tetraphosphate. J Biol Chem. 1972;247:7067–7072. [PubMed] [Google Scholar]
- Iglesias-Gato D, Martin-Marcos P, Santos MA, Hinnebusch AG, Tamame M. Guanine nucleotide pool imbalance impairs multiple steps of protein synthesis and disrupts GCN4 translational control in Saccharomyces cerevisiae. Genetics. 2011;187:105–122. doi: 10.1534/genetics.110.122135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson RC, Weber G, Morris HP. IMP dehydrogenase, an enzyme linked with proliferation and malignancy. Nature. 1975;256:331–333. doi: 10.1038/256331a0. [DOI] [PubMed] [Google Scholar]
- Kanjee U, Gutsche I, Alexopoulos E, Zhao B, El Bakkouri M, Thibault G, Liu K, Ramachandran S, Snider J, Pai EF, Houry WA. Linkage between the bacterial acid stress and stringent responses: the structure of the inducible lysine decarboxylase. EMBO J. 2011;30:931–944. doi: 10.1038/emboj.2011.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasai K, Nishizawa T, Takahashi K, Hosaka T, Aoki H, Ochi K. Physiological analysis of the stringent response elicited in an extreme thermophilic bacterium, Thermus thermophilus. J Bacteriol. 2006;188:7111–7122. doi: 10.1128/JB.00574-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennan A, Aherne A, Palfi A, Humphries M, McKee A, Stitt A, Simpson DA, Demtroder K, Orntoft T, Ayuso C, Kenna PF, Farrar GJ, Humphries P. Identification of an IMPDH1 mutation in autosomal dominant retinitis pigmentosa (RP10) revealed following comparative microarray analysis of transcripts derived from retinas of wildtype and Rho(−/−) mice. Hum Mol Genet. 2002;11:547–557. doi: 10.1093/hmg/11.5.547. [DOI] [PubMed] [Google Scholar]
- Kiefer P, Portais JC, Vorholt JA. Quantitative metabolome analysis using liquid chromatography-high-resolution mass spectrometry. Anal Biochem. 2008;382:94–100. doi: 10.1016/j.ab.2008.07.010. [DOI] [PubMed] [Google Scholar]
- Kobayashi K, Ehrlich SD, et al. Essential Bacillus subtilis genes. Proc Natl Acad Sci U S A. 2003;100:4678–4683. doi: 10.1073/pnas.0730515100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasny L, Gourse RL. An alternative strategy for bacterial ribosome synthesis: Bacillus subtilis rRNA transcription regulation. EMBO J. 2004;23:4473–4483. doi: 10.1038/sj.emboj.7600423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda A, Murphy H, Cashel M, Kornberg A. Guanosine tetra- and pentaphosphate promote accumulation of inorganic polyphosphate in Escherichia coli. J Biol Chem. 1997;272:21240–21243. doi: 10.1074/jbc.272.34.21240. [DOI] [PubMed] [Google Scholar]
- Lee PC, Bochner BR, Ames BN. Diadenosine 5',5"'-P1,P4-tetraphosphate and related adenylylated nucleotides in Salmonella typhimurium. J Biol Chem. 1983;258:6827–6834. [PubMed] [Google Scholar]
- Lehninger AL, Nelson DL, Cox MM. Principles of Biochemistry. N.Y.: Worth Publisher Inc.; 2000. [Google Scholar]
- Lopez JM, Dromerick A, Freese E. Response of guanosine 5'-triphosphate concentration to nutritional changes and its significance for Bacillus subtilis sporulation. J Bacteriol. 1981;146:605–613. doi: 10.1128/jb.146.2.605-613.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milon P, Tischenko E, Tomsic J, Caserta E, Folkers G, La Teana A, Rodnina MV, Pon CL, Boelens R, Gualerzi CO. The nucleotide-binding site of bacterial translation initiation factor 2 (IF2) as a metabolic sensor. Proc Natl Acad Sci U S A. 2006;103:13962–13967. doi: 10.1073/pnas.0606384103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molle V, Nakaura Y, Shivers RP, Yamaguchi H, Losick R, Fujita Y, Sonenshein AL. Additional targets of the Bacillus subtilis global regulator CodY identified by chromatin immunoprecipitation and genome-wide transcript analysis. J Bacteriol. 2003;185:1911–1922. doi: 10.1128/JB.185.6.1911-1922.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy H, Cashel M. Isolation of RNA polymerase suppressors of a (p)ppGpp deficiency. Methods Enzymol. 2003;371:596–601. doi: 10.1016/S0076-6879(03)71044-1. [DOI] [PubMed] [Google Scholar]
- Nanamiya H, Kasai K, Nozawa A, Yun CS, Narisawa T, Murakami K, Natori Y, Kawamura F, Tozawa Y. Identification and functional analysis of novel (p)ppGpp synthetase genes in Bacillus subtilis. Mol Microbiol. 2008;67:291–304. doi: 10.1111/j.1365-2958.2007.06018.x. [DOI] [PubMed] [Google Scholar]
- Paul BJ, Barker MM, Ross W, Schneider DA, Webb C, Foster JW, Gourse RL. DksA: a critical component of the transcription initiation machinery that potentiates the regulation of rRNA promoters by ppGpp and the initiating NTP. Cell. 2004;118:311–322. doi: 10.1016/j.cell.2004.07.009. [DOI] [PubMed] [Google Scholar]
- Potrykus K, Cashel M. (p)ppGpp: still magical? Annu Rev Microbiol. 2008;62:35–51. doi: 10.1146/annurev.micro.62.081307.162903. [DOI] [PubMed] [Google Scholar]
- Scott M, Gunderson CW, Mateescu EM, Zhang Z, Hwa T. Interdependence of cell growth and gene expression: origins and consequences. Science. 2010;330:1099–1102. doi: 10.1126/science.1192588. [DOI] [PubMed] [Google Scholar]
- Spizizen J. TRANSFORMATION OF BIOCHEMICALLY DEFICIENT STRAINS OF BACILLUS SUBTILIS BY DEOXYRIBONUCLEATE. Proc Natl Acad Sci U S A. 1958;44:1072–1078. doi: 10.1073/pnas.44.10.1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivatsan A, Han Y, Peng J, Tehranchi AK, Gibbs R, Wang JD, Chen R. High-precision, whole-genome sequencing of laboratory strains facilitates genetic studies. PLoS Genet. 2008;4:e1000139. doi: 10.1371/journal.pgen.1000139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun D, Lee G, Lee JH, Kim HY, Rhee HW, Park SY, Kim KJ, Kim Y, Kim BY, Hong JI, Park C, Choy HE, Kim JH, Jeon YH, Chung J. A metazoan ortholog of SpoT hydrolyzes ppGpp and functions in starvation responses. Nat Struct Mol Biol. 2010;17:1188–1194. doi: 10.1038/nsmb.1906. [DOI] [PubMed] [Google Scholar]
- Traxler MF, Summers SM, Nguyen HT, Zacharia VM, Hightower GA, Smith JT, Conway T. The global, ppGpp-mediated stringent response to amino acid starvation in Escherichia coli. Mol Microbiol. 2008;68:1128–1148. doi: 10.1111/j.1365-2958.2008.06229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu BP, Mohler RE, Liu JC, Dombek KM, Young ET, Synovec RE, McKnight SL. Cyclic changes in metabolic state during the life of a yeast cell. Proc Natl Acad Sci U S A. 2007;104:16886–16891. doi: 10.1073/pnas.0708365104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasantha N, Freese E. Enzyme changes during Bacillus subtilis sporulation caused by deprivation of guanine nucleotides. J Bacteriol. 1980;144:1119–1125. doi: 10.1128/jb.144.3.1119-1125.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrentas CE, Gaal T, Berkmen MB, Rutherford ST, Haugen SP, Vassylyev DG, Ross W, Gourse RL. Still looking for the magic spot: the crystallographically defined binding site for ppGpp on RNA polymerase is unlikely to be responsible for rRNA transcription regulation. J Mol Biol. 2008;377:551–564. doi: 10.1016/j.jmb.2008.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JD, Sanders GM, Grossman AD. Nutritional control of elongation of DNA replication by (p)ppGpp. Cell. 2007;128:865–875. doi: 10.1016/j.cell.2006.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendrich TM, Marahiel MA. Cloning and characterization of a relA/spoT homologue from Bacillus subtilis. Mol Microbiol. 1997;26:65–79. doi: 10.1046/j.1365-2958.1997.5511919.x. [DOI] [PubMed] [Google Scholar]
- Xia J, Wishart DS. Metabolomic data processing, analysis, and interpretation using MetaboAnalyst. Chapter 14. Curr Protoc Bioinformatics. 2011 doi: 10.1002/0471250953.bi1410s34. Unit 14.10. [DOI] [PubMed] [Google Scholar]
- Xiao H, Kalman M, Ikehara K, Zemel S, Glaser G, Cashel M. Residual guanosine 3',5'-bispyrophosphate synthetic activity of relA null mutants can be eliminated by spoT null mutations. J Biol Chem. 1991;266:5980–5990. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.